Abstract
Exposure to human pathogenic viruses in recreational waters has been shown to cause disease outbreaks. In the context of Article 14 of the revised European Bathing Waters Directive 2006/7/EC (rBWD, CEU, 2006) a Europe-wide surveillance study was carried out to determine the frequency of occurrence of two human enteric viruses in recreational waters. Adenoviruses were selected based on their near-universal shedding and environmental survival, and noroviruses (NoV) selected as being the most prevalent gastroenteritis agent worldwide. Concentration of marine and freshwater samples was done by adsorption/elution followed by molecular detection by (RT)-PCR. Out of 1410 samples, 553 (39.2%) were positive for one or more of the target viruses. Adenoviruses, detected in 36.4% of samples, were more prevalent than noroviruses (9.4%), with 3.5% GI and 6.2% GII, some samples being positive for both GI and GII. Of 513 human adenovirus-positive samples, 63 (12.3%) were also norovirus-positive, whereas 69 (7.7%) norovirus-positive samples were adenovirus-negative. More freshwater samples than marine water samples were virus-positive. Out of a small selection of samples tested for adenovirus infectivity, approximately one-quarter were positive. Sixty percent of 132 nested-PCR adenovirus-positive samples analysed by quantitative PCR gave a mean value of over 3000 genome copies per L of water. The simultaneous detection of infectious adenovirus and of adenovirus and NoV by (RT)PCR suggests that the presence of infectious viruses in recreational waters may constitute a public health risk upon exposure. These studies support the case for considering adenoviruses as an indicator of bathing water quality.
Keywords: Adenoviruses, Noroviruses, Bathing water, River water, Sea water, Recreational water, Water quality
1. Introduction
Enteric viruses have frequently been implicated in recreational water-related gastro-intestinal (G.I.) disease (Sinclair et al., 2009). Studies in Europe and the US suggest that most infections contracted as a result of swimming, canoeing or other recreational use of sewage-polluted water may be viral in nature (e.g. Medema et al., 1995, Gray et al., 1997). Enteric viruses may cause asymptomatic or mild infections in humans, but these faecal-orally transmitted viruses may also cause more serious disease, such as hepatitis and meningitis, especially in vulnerable groups, e.g. young children (Nwachuku and Gerba, 2006). Enteric viruses are recognized as agents that can cause large outbreaks throughout the world with thousands of cases (Sarguna et al., 2007, Bucardo et al., 2007, Iijima et al., 2008, Zhang et al., 2009). Novel emerging viruses such as SARS coronavirus, human parechovirus and zoonotic influenza viruses also appear to be excreted in faeces but the evidence for enteric transmission is not always clear (Ding et al., 2004). Transmission routes for enteric viruses may be diverse such as person–person, food- or waterborne associated with insufficient hygiene and sanitation (Koopmans et al., 2002, Wyn-Jones and Sellwood, 2001). Disease outbreaks associated with enteric viruses, such as noroviruses and astroviruses, in bathing water have been described (Hauri et al., 2005, Maunula et al., 2004). However, bathing water-related outbreaks may be easily missed due to either unidentified source or unidentified agent, or both.
Enteric viruses in water may originate from discharges of raw or treated sewage, run-off of animal manure or directly from humans or animals. Viruses commonly associated with waterborne disease include the human adenoviruses (HAdVs), noroviruses (NoVs), hepatitis A and E viruses (HAV, HEV), parvoviruses, enteroviruses, and rotaviruses (RVs). In addition, sewage, especially from slaughterhouses, may contain (for example) animal adenoviruses, sapoviruses, and HEV (Hundesa et al., 2006), which may be zoonotic. Viruses originating from (un)treated sewage can contaminate bathing water after discharge into surface waters (in)directly used for recreational water activities. All are capable of infection by ingestion. Some multiply in the intestine and may cause diarrhoea and/or vomiting, while some are associated with tissues (e.g. the liver) other than the gut. The viruses responsible for waterborne infections are not usually identified at the time of a disease outbreak following recreational water activity, and robust associations between the simultaneous presence of virus in faeces of affected individuals and in the water are only occasionally demonstrated (e.g. Hoebe et al., 2004). The epidemiological picture of disease associated with recreational use of water is therefore far from complete, and measures to limit enteric disease after exposure to recreational water are based on water quality parameters built on the detection of faecal bacterial indicator organisms (FIOs). However, it has been shown that water conforming to bacterial standards may contain high levels of human enteric viruses and that FIOs often fail to predict the risk for waterborne pathogens including enteric viruses (Gerba et al., 1979, Lipp et al., 2001). Further, several studies have shown that levels of indicator bacteria do not correlate with those of viruses, particularly when faecal indicator concentrations are low (Contreras-Coll et al., 2002). Viruses are known to be more resistant to environmental degradation than bacteria (Vasl et al., 1981, Thurston-Enriquez et al., 2003, Rzezutka and Cook, 2004, de Roda Husman et al., 2009). Together with the understanding that G.I. illness may be due to viruses rather than bacteria, this provides a case for using a viral indicator of human faecal pollution rather than to rely exclusively on bacterial parameters.
Bathing water quality in the European Union (EU) has been regulated since 1976 by the Bathing Water Directive (76/160/EEC). In 2006 this was revised (rBWD, CEU, 2006) by including enterococci (and, in fresh waters, Escherichia coli) as the principal microbial determinants which placed the microbiological parameters on a firmer scientific footing (Kay et al., 1994, Kay et al., 2004: Wiedenmann et al., 2006, WHO, 2003) and allowed classification of bathing waters to be undertaken with more confidence. When tested at sufficient frequency E. coli may be a useful indicator of faecal pollution and therefore of the probability of waterborne disease. However, in the EU Directive the frequency is only about once in two weeks and testing takes two days.
The earlier Directive included an enterovirus parameter which stipulated that 95% of 10-L water samples taken during the bathing season should contain no (zero p.f.u.) enteroviruses. This was based on early work (described by Farrah and Bitton, 1990) which suggested that, for poliovirus, Coxsackie A and Coxsackie B viruses, between one and twenty virus infectious units might be sufficient to cause infection. The pathogenesis of enterovirus infections is now better understood, and this belief is considered unsound in determining water quality. Further, although important pathogens in many contexts, the presence of enteroviruses in water does not necessarily correlate with the presence of pathogens such as hepatitis A virus (Dubrou et al., 1991, Pina et al., 1998). The enterovirus parameter was removed during the revision of the 1976 Directive.
Concentrations of some viruses in surface waters can be determined by cell culture monolayer plaque assays, but the technique is not applicable to most viruses of prime interest. Furthermore, cell culture is expensive and time-consuming, and detection of viruses is now done mainly by molecular methods such as reverse transcription RT-PCR or nucleic acid sequence-based amplification (NASBA) which amplify RNA/DNA. Although mainly described as end-point assays, amplification products of both techniques can be detected by real-time methods. The major advantages of real-time detection are hands-on time and the ability to quantify amplification products, which is very important in being able to estimate the public health risks of low levels of enteric viruses in bathing water.
A viral indicator may be better suited to indicate the risk of human pathogenic viruses in bathing waters. However, cell culture-based methods for viral detection are costly, require specialised skills and equipment, and have too long a turnaround time. For this reason, the EU Framework 6 Project VIROBATHE was done to devise a robust, rapid and cost-efficient method for routine compliance monitoring of enteric viruses in recreational waters. Part of the work involved Europe-wide surveillance of recreational waters to determine the frequency of target virus occurrence and, to a limited extent, serotypes and quantities. So that the virus levels could be seen in the context of compliance-related water quality, the work also included determination of FIO levels to provide general water quality data. The viruses selected as targets were adenoviruses and noroviruses. The former are shed by many individuals (often without showing symptoms), they are more environmentally robust than enteroviruses (Enriquez et al., 1995, Thurston-Enriquez et al., 2003), they have been found in surveys of polluted waters (e.g. Pina et al., 1998, Laverick et al., 2004, Lee et al., 2004, Miagostovich et al., 2008), and have been associated with outbreaks of disease in swimming pools (e.g. Papapetropolou and Vantarakis, 1995, Harley et al., 2001) and other recreational waters (Sinclair et al., 2009). Being DNA viruses, their detection by PCR does not have the problems associated with the genetic variation seen with RNA viruses. They are also more likely to be detected in recreational water samples (e.g. Pina et al., 1998, Miagostovich et al., 2008), especially if sensitive nucleic acid detection methods are used, and they may therefore provide the best indicator of viral faecal pollution. Noroviruses are the most important cause of acute viral gastroenteritis in people of all age groups and many waterborne outbreaks have been reported. Sinclair et al. (2009) reviewed 55 recreational water-related G.I. disease outbreaks of which 25 (46%) were reported as caused by noroviruses.
The study reported here was performed to demonstrate that a common concentration protocol could be used across recreational waters in widely diverse geographical areas, that viruses concentrated by this protocol could be detected by a rapid molecular method, that it was possible to enumerate viruses and to investigate whether there was a range of sero/genotypes of the target viruses present across the locations studied.
2. Materials and methods
2.1. Survey design
Each of the 15 Surveillance Laboratories located in nine countries selected up to two sites for study which were sampled during the EU Bathing Season 2006, and samples were concentrated and analysed for the target viruses by molecular means. FIOs and various physico-chemical parameters were also determined. Data were sent to the co-ordinating Laboratory at the University of Aberystwyth for collation.
2.2. Sampling sites
Each laboratory selected up to two sites (main site and second site) for the study (Table 1 and Fig. 1 ). The principal criterion for a site being chosen was its current use for recreational water activity; sites were not chosen on the basis of being EU-designated bathing waters, nor because they had a history of pollution in the area, though several sites were known to be impacted by sewage effluent. A minimum of 80 10-L water samples from the main site was taken and up to 20 additional samples were taken in the event of (e.g.) heavy rain or when investigators considered that there was some other occurrence which may have resulted in deterioration of water quality. The second site could also be used if the main site yielded negative data in the first stages of sampling, or for taking the 20 additional samples following the 80 minimum to be taken at the main site. Thus, each laboratory could focus on one site (100 samples) or divide surveillance between the main site (80 samples) and the second site (20 samples). In practice both approaches were used, so in total, 24 sites were sampled. Sites were sampled at approximately weekly intervals from the end of May to the beginning of November 2006, which included the Bathing Season in all Member States. On each sampling occasion, four 10-L samples (a ‘tetrad’), plus one additional sample for positive Quality Control (QC) purposes, were collected from each site. One 250 mL sample for bacterial faecal indicators was also taken. In total each laboratory processed and analysed at least 100 water samples for virus detection and 25 samples for bacterial enumeration.
Table 1.
Location of sampling sites.
| Site* | Country | Location | Site name | Water type |
|---|---|---|---|---|
| 1 | Cyprus | Larnaca | Larnaca Marina | Marine |
| 2 | France | Nancy | Tomblaine | Fresh |
| 3 | Germany | Baden-Württemberg | Neckar River | Fresh |
| 4 | Germany | Baden-Württemberg | Kirchentellinsfurt Lake | Fresh |
| 5 | Germany | Bavaria | Amper Grasslfing | Fresh |
| 6 | Germany | Berlin | Wannsee | Fresh |
| 7 | Germany | Berlin | Landwehrkanal | Fresh |
| 8 | Italy | Pisa | San Rossore | Marine |
| 9 | Italy | Pisa | Bocca d’Arno | Marine |
| 10 | Italy | Castel Gandolfo | Castel Gandolfo Lake | Fresh |
| 11 | Italy | Ardea (Rome) | Fosso dell’Incastro | Marine |
| 12 | Italy | Pomezia (Rome) | Rio Torto | Marine |
| 13 | Netherlands | Durgerdam | Kinselmeer | Fresh |
| 14 | Netherlands | Leerdam | Linge | Fresh |
| 15 | Poland | Pulawy | VistulaRiver | Fresh |
| 16 | Portugal | Porto | Molhe South | Marine |
| 17 | Portugal | Porto | Molhe North | Marine |
| 18 | Spain | Barcelona | Gavà | Marine |
| 19 | Spain | Barcelona | Gavà | Marine |
| 20 | UK | York | Naburn Lock | Fresh |
| 21 | UK | Devon | Axmouth Harbour | Marine |
| 22 | UK | Devon | River Kenn | Marine |
| 23 | UK | Kew (London) | River Thames | Fresh |
| 24 | UK | Reading | River Thames | Fresh |
*See also Fig. 1 for site locations.
Fig. 1.
Location of sampling sites.
2.3. Sample processing
Many methods for the concentration and detection of enteric viruses in water samples have been described (Wyn-Jones and Sellwood, 2001). For virological water quality to be assessed on a comparable basis, a single method common to all laboratories was needed for each water type (fresh or coastal/transitional) analysed during the surveillance programme. Prior to the surveillance stage several different methods were evaluated (see Section 3.1) using HAdV2 and NoV GII.4, and the best in terms of virus recovery and capital/recurrent costs was selected. The HAdV2 was obtained from the UK Health Protection Agency (HPA) National Collection of Pathogenic Viruses (NCPV), where the virus genome was authenticated. Virus was grown and assayed by plaque assay in A549 cultures and distributed by the HPA Environmental Virology Unit to other laboratories. Norovirus GII.4 was identified in a faecal sample from an outbreak in a care home and the identity confirmed by sequencing. End-point dilution assay by RT-PCR gave a titre of 10−9. The suspension was distributed at 10−3 which provided sufficient virus for evaluation and quality control purposes for all laboratories throughout the project. Process characterisation was done by four experienced laboratories concentrating replicate samples of water spiked with HAdV2 and analysing the concentrates for recovered virus.
2.3.1. Concentration of freshwater samples by glass wool filtration
For freshwater samples a modification of the glass wool method of Vilaginès et al. (1993) was used. The glass wool filter was made by compressing 10 g glass wool (type 725; Rantigny, Saint-Gobain, France) into a 30 cm by 3 cm polystyrene column to obtain a filter height of 6–8 cm. The filter was washed by gravity with 50 mL volumes of (in order) 1 M HCl, tap water, and 1 M NaOH, followed by tap water until the filtrate pH was neutral. Water samples (10-L) were conditioned with 1 M or 0.1 M HCl to pH 3.5 to enhance binding of the viruses to the filter and passed through the filter at a rate not exceeding 1.5 L min−1. When all the sample had passed through the filter the virus was eluted from the glass wool by slow (20–30 min) passage of 200 mL 3% (w/v) beef extract at pH 9.5 in 0.05 M glycine buffer through the filter. The eluate was flocculated by the addition of 1 M and 0.1 M HCl until the pH reached 3.5. The resultant protein floc, containing virus, was deposited by centrifugation at 7500× g for 30 min, dissolved to a final volume of 10 mL phosphate buffered saline (PBS) and stored at −20 °C pending further analysis.
2.3.2. Concentration of marine water samples by nitrocellulose membrane filtration
Coastal/transitional water samples were processed by filtration through nitrocellulose membranes, elution and organic flocculation (Wyn-Jones et al., 2000). The sample, at pH 3.5, was passed through a 142 mm diameter glass fibre pre-filter and a nitrocellulose membrane in a Sartorius filter holder at a maximum rate of 1.5 L min−1. The filtrate was run to waste and the virus was then eluted from the membrane by slow passage (10 min) of 200 ml skimmed milk solution (0.1% in 0.05 M glycine buffer). The eluate was flocculated by reducing its pH to 4.5 with M HCl and centrifuging as above.
2.4. Extraction of nucleic acids from sample concentrates
Nucleic acid (NA) was extracted from 5 mL volumes of sample concentrate using the NucliSens® miniMAG™ system (Biomérieux, France) according to manufacturer’s instructions, with slight modifications comprising centrifugation at 1500× g for 2 min after addition of the silica suspension to reduce the chance of cross-contamination. The final 100 μL NA extract was centrifuged at 13,000× g for 1 min to pellet any remaining traces of silica which could inhibit downstream (RT)PCR reactions, the supernatant was transferred to a clean microfuge tube and was stored at −80 °C if not used immediately.
2.5. Human adenovirus PCR
For the detection of human adenovirus in the water samples the nested-PCR based on the method of Allard et al. (2001) was employed, using primers Hex1deg and Hex2deg for the first round of amplification and primers nehex3deg and nehex4deg for the second round. Additionally, an internal amplification control (IAC, see below) was incorporated in the assay, and a carryover contamination prevention system utilising uracil-N-glycosylase (UNG) in the first round PCR and dUTP (replacing dTTP) in both PCRs. The reaction incorporated a hot-start polymerase (Platinum® Taq DNA polymerase, Life Technologies Inc.).
The target amplicon sizes were 301 bp in the first round and 171 bp in the second round. The first round reaction conditions were as follows: 10 μL DNA, 1X Platinum® Taq buffer, 1.5 mM Mg++, 250 μM dNTPs, 0.5 μM primer Hex1deg, 0.5 μM primer Hex2deg, 1U Platinum® Taq (Life Technologies Inc.), and 1 U HK-UNG (Epicentre®, Madison, Wisconsin). Five μL IAC were added in the first round. Adenovirus DNA (20 ng μL−1), and ultrapure water were included as positive and negative reaction control, respectively. After 10 min at 50 °C (UNG) and 10 min at 95 °C (activation of Taq polymerase), cycling conditions included 45 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, followed by a final extension of 72 °C for 5 min. The second round reaction conditions were: 1X Platinum® Taq buffer, 1.5 mM Mg++, 100 μM dNTPs, 0.5 μM primer nehex3deg, 0.5 μM primer nehex4deg, and 1U Platinum® Taq. Two μL from the first round reaction were used as target. The thermocycling conditions were 94 °C for 3 min, then 45 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, followed by a final extension of 72 °C for 5 min. The amplicons were electrophoresed in a 2% agarose gel stained with 10 ng mL−1 ethidium bromide or equivalent nucleic acid staining methods such as SYBR-Gold, and subsequently visualised by UV transillumination.
2.6. Norovirus RT-PCR
To detect norovirus, the nested RT-PCR based on the method of Vennema et al. (2002) was used, and comprised amplification of norovirus RNA-dependent RNA polymerase (RdRp) gene sequences by RT-PCR followed by a semi-nested PCR for each genogroup (G). Depending on the laboratory, contamination carryover prevention was also incorporated utilising uracil-N-glycosylase (UNG) in the PCR. The target amplicon sizes were 327 bp in the RT-PCR, 188 bp in the GI nested PCR, and 237 bp in the GII nested PCR.
Reverse transcription PCR conditions were as follows: 1X OneStep buffer (Qiagen, UK), 400 μM each dNTP, 1X OneStep enzyme mix (Qiagen, UK), 0.5 μM primer JV12Y, 0.5 μM primer JV13i, and 50U RNasin (RNasin®Plus, Promega, UK), 1U Platinum® Taq (Life Technologies Inc.). Five μL IAC were added in the first round. A 10 μL sample of nucleic acid was used as target. The thermocycling conditions were 50 °C for 30 min, 95 °C for 15 min, then 40 cycles of 94 °C for 1 min, 37 °C for 1 min and 72 °C for 1 min, followed by a final extension of 72 °C for 10 min. The second round PCR conditions were as follows: 1X Platinum® Taq buffer, 2.0 mM Mg++, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 400 μM dUTP, 0.4 μM primer JV12Y, 0.4 μM primer Ni-R, 1U HK-UNG and 1U Platinum® Taq. One μL from the first round reaction was used as target. The thermocycling conditions were 50 °C for 10 min, 95 °C for 10 min, 96 °C for 3 min then 40 cycles of 95 °C for 1 min, 40 °C for 1 min and 72 °C for 1 min, followed by a final extension of 72 °C for 10 min. The amplicons were electrophoresed in a 2% agarose gel stained with 10 ng mL−1 ethidium bromide or equivalent nucleic acid staining methods such as SYBR-Gold, and subsequently visualised by UV transillumination.
2.7. Internal amplification controls (IACs)
The need to guard against false negative results required the use of a novel IAC in each PCR. For adenovirus IACs, oligonucleotides were constructed which contained the adenovirus primer sequences used in each round flanking primer sequences for amplification of invA sequences from Salmonella enterica (Malorny et al., 2003, Malorny et al., 2004). The amplicon was cloned into a plasmid (pGem T-Easy vector) by Yorkshire Bioscience Ltd. (York, UK). The resulting pADENOIAC plasmid was linearised at the unique PstI site downstream of the adenovirus IAC insert region. Yorkshire Bioscience supplied pADENOIAC in 100 μL volumes containing 1 mg mL−1 plasmid DNA in 10 mM Tris–HCl, 1 mM EDTA buffer pH 8.0. The IAC amplicon sizes were 384 bp in the first round and 337 bp in the second round.
For the norovirus IAC the RNA was synthesized by the addition of complementary sequences of the first round primers JV12Y and JV13i to part of the β-globin gene, resulting in a PCR product of 369 base pairs. In this same construct sequences complementary to the GI nested-primer Ni-R and to the GII nested-primer GI were included. The construct was subsequently cloned downstream of a T7 RNA-polymerase promoter. The RNA IAC was prepared by Yorkshire Bioscience Ltd. (York, UK) using plasmid pnJV IAC which was linearised with Sal1 restriction endonucleases and purified. The resulting RNA was transcribed using the T7 RNA polymerase transcription system. Template DNA was removed from the preparation during incubation with RNase-free DNase. The RNA was purified by LiCl precipitation followed by multiple phenol/chloroform extractions. The preparation was concentrated to 1.0 μg μL−1 by precipitation with ethanol and dissolving in a minimal volume of MilliQ/18.2 MΩ quality water. Both types of IAC were prepared in single batches by Yorkshire Bioscience, and checked and distributed to all participants by one of the participant laboratories (FERA).
Amplification products of the IAC with the GI specific primers produced a PCR product of 228 base pairs, GII-specific amplification resulted in a PCR product of 277 base pairs. The working concentration of each IAC (in 10 mM Tris–HCl, 1 mM EDTA buffer pH 8.0, plus 500 ng mL−1 bovine serum albumin) was empirically determined as the dilution which consistently (triplicate determinations) gave a positive signal. Aliquots were stored at −20 °C (adenovirus IAC) or −70 °C (norovirus IAC).
In a correctly functioning reaction, an IAC signal should always be produced in the absence of a target signal (high amounts of target can out-compete amplification of the IAC, but then the target signal itself shows that the reaction has worked). In this study, when an (RT)PCR of a sample produced neither IAC nor target signal, the presence of inhibitory substances derived from the water sample was assumed. Consequently, the nucleic extract was diluted ten-fold until the appearance of an IAC or target signal revealed that no inhibition was occurring.
2.8. Infectivity determination
At least 10 adenovirus-positive (by nested-PCR) samples from each Laboratory were tested for virus infectivity by integrated cell culture-PCR (ICC-PCR, Reynolds et al., 2001, Greening et al., 2002). If any of the four test samples in a tetrad was positive by human adenovirus nested-PCR then the sample concentrate which had given the strongest PCR band was tested for infectious adenovirus by inoculation of cell cultures and nested-PCR analysis of the cultures after zero and five days’ incubation. No infectivity assay was performed if the adenovirus nested-PCR on all four concentrates was negative. At least two 25 cm2 flasks, each containing a monolayer of confluent A549 cells (European Collection of Cell Culture, ECACC, UK) were inoculated with 1 mL of sample concentrate. At least one flask was incubated for five days (T = 5). One flask was analysed without incubation (T = 0), to guard against detection of seed virus. One negative control with cell culture medium only was set up. Following incubation, flasks in the first set (T = 5) were frozen and thawed three times and the separated supernatant analysed by the adenovirus nested PCR. A positive nested-PCR signal after five days, coupled with a negative reaction after zero days (confirming that inoculum was not being detected) was taken as evidence of virus multiplication, and hence of infectivity.
2.9. QPCR assay for the detection of HAdV DNA
Virus nucleic acid in at least 10 samples which were positive for adenovirus by nested-PCR from each Laboratory was quantified by real-time qPCR. The nucleic acid extracts from these samples were diluted as was found necessary to observe a signal in the PCR (see Section 2.7). Assays were done in 25-μL reaction mixtures each containing 10 μL of nucleic acid extract and 15 μL of TaqMan® Universal PCR Master Mix (Applied Biosystems) containing 0.9 μM of each primer (AdF and AdR) and 0.22 5 μM of fluorogenic probe (AdP1) as previously described (Hernroth et al., 2002).
Following activation of the uracil-N-glycosylase (2 min, 50 °C) and activation of the AmpliTaq Gold for 10 min at 95 °C, 45 cycles (15 s at 95 °C and 1 min at 60 °C) were performed.
A pBR322 plasmid containing the HAdV 41 hexon sequence kindly donated by Dr. Annika Allard from the University of Umeå, Sweden, was used to construct a standard containing 101–107 copies of DNA in the 10 μL added to the PCR reaction. Each dilution of standard DNA suspensions was run in triplicate. Ten μL of undiluted and a ten-fold dilution of the DNA suspensions obtained from water samples were run in duplicate. In all QPCRs the amount of DNA was defined as the mean of the data obtained. A non-template control and a non-amplification control were added to each run.
2.10. Sequence analysis
The amplicons obtained after nested-PCR assays of HAdV or NoV were purified using the QIAquick PCR purification kit (QIAGEN, Inc.). Purified DNA was directly sequenced with the ABI PRISM™ Dye Terminator Cycle Sequencing Ready Reaction kit version 3.1 with Ampli Taq® DNA polymerase FS (Applied Biosystems) following the manufacturer’s instructions. The conditions for the 25-cycle sequencing amplification were: denaturing at 96 °C for 10 s, annealing for 5 s at 50 °C and extension at 60 °C for 4 min. The nested primers were used for sequencing at a concentration of 0.05 μM.
The results were checked using the ABI PRISM 377 automated sequencer (Perkin–Elmer, Applied Biosystems). The sequences were compared with the GenBank and the EMBL (European Molecular Biology Library) using the basic BLAST program of the NCBI (The National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/BLAST/). Alignments of the sequences were carried out using the ClustalW program of the EBI (European Bioinformatics Institute of the EMBL, http://www.ebi.ac.uk/clustalw/).
2.11. Faecal indicator organisms
Detection of E. coli and intestinal enterococci was done according to ISO 9308-3 and ISO 7899-1 using Microtiter plates. One laboratory enumerated bacteria by colony-forming units (cfu).
2.12. Quality assurance – robustness of the concentration and detection methods
The robustness of the methods was calculated using the results obtained from the analysis of quality control samples. Nine laboratories participated in the trial of the methods for analysis of fresh waters and six laboratories participated in the trial of the methods for analysis of marine samples. Test samples comprised 1 mL aliquots of adenovirus Type 2 (containing 200 pfu of virus), and norovirus GII.4 which were added by the participants to their own water samples. A batch of adenovirus Type 2 and a batch of norovirus GII.4 were prepared, distributed into single-use ampoules and sent to each participant. On each sampling occasion 1 mL of the adenovirus Type 2 and 1 mL of the norovirus-positive control material were added to a separate 10-L quality control sample of the recreational water being tested. Negative samples were prepared from a mixture of de-ionised and tap water, or artificial seawater. Each participant analysed at least 25 sets of quality control samples.
The raw data sent by each laboratory were statistically analysed according to the recommendations of Scotter et al. (2001) by the methods of Langton et al. (2002). The trial sensitivity was defined as the percentage of positive samples giving a correct positive signal, and trial specificity was defined as the percentage of negative samples giving a correct negative signal. Accordance (repeatability of qualitative data) was defined as the percentage chance of finding the same result, positive or negative, from two identical samples analysed in the same laboratory under predefined repeatability conditions, and concordance (reproducibility of qualitative data) was defined as the percentage chance of finding the same result, positive or negative, from two identical samples analysed in different laboratories under predefined repeatability conditions. These calculations take into account different replication in different laboratories by weighting results appropriately. The concordance odds ratio (COR) was the degree of inter-laboratory variation in the results, and expressed as the ratio between accordance and concordance percentages (Langton et al., 2002). The COR value may be interpreted as the likelihood of getting the same result from two identical samples, whether they are sent to the same laboratory or to two different laboratories. The closer the value is to 1.0, the higher is the likelihood of getting the same result. Confidence intervals for accordance, concordance and COR were calculated by the method of Davison and Hinckley (1997); each laboratory was considered representative of all laboratories in the “population” of laboratories, not just those participating in this analysis.
3. Results
The study surveillance period ran from the end of May until early November 2006. Nine participant Laboratories collected samples at both of their sampling sites, whereas six Laboratories took samples from only their main site. Thirteen fresh water sites and 11 marine sites were sampled (Table 1 and Fig. 1). A total of 1410 samples was taken of which 928 were from fresh water and 482 were from marine sites (Table 1).
3.1. Virus detection
Four experienced laboratories evaluated the concentration methods by processing replicate samples using different methods, and analysing the concentrates by (RT)PCR and, for HAdV2, monolayer plaque assay in A549 cultures. Concentration by three different methods gave mean recoveries of 49% and 37% of seeded HAdV2 from fresh and artificial sea water respectively, as measured by plaque assay. Across all evaluating laboratories, concentration of HAdV2 in spiked freshwater samples by glass wool with elution using beef extract gave a mean recovery of 57.1% (range 34.2%–78.2%), while concentration of virus in spiked artificial seawater samples with skimmed milk elution gave a mean recovery of 35.4% (range 22.5%–43.8%). The variation between laboratories’ performance made decisions on method choice based only on recovery values less than clearcut, which is why other factors such as cost were also taken into account.
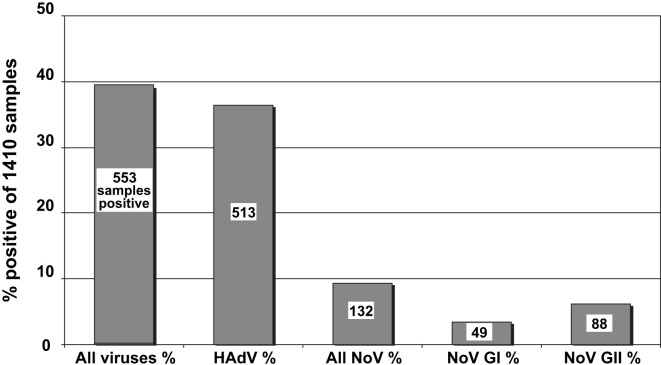
From the overall surveillance data 553 out of 1410 samples (39.2%) were positive for one or more of the target viruses (Fig. 2 ). This corresponded to 582 virus detections, some samples being positive for more than one kind of virus. Adenoviruses were detected more often than noroviruses, 513 (36.4%) samples being positive for one or more human adenovirus types, while 132 samples (9.4%) tested positive for one or both norovirus genogroups; these were divided between GI (49, 3.5% samples positive) and GII (88, 6.2%, Fig. 2). Five samples (two marine and three fresh water) were positive for both norovirus genogroups. Out of the 513 human adenovirus-positive samples, 63 (12.3%) were also positive for one or both NoV genogroups (33 out of 381 freshwater samples and 30 out of 132 marine samples). Just four samples (two fresh water and two marine), were positive for all three virus types. Interestingly, 69 samples (22 fresh water and 47 marine) were positive for one or both norovirus genogroups while testing negative for adenovirus.
Fig. 2.
Summary of virus detection in all water types.
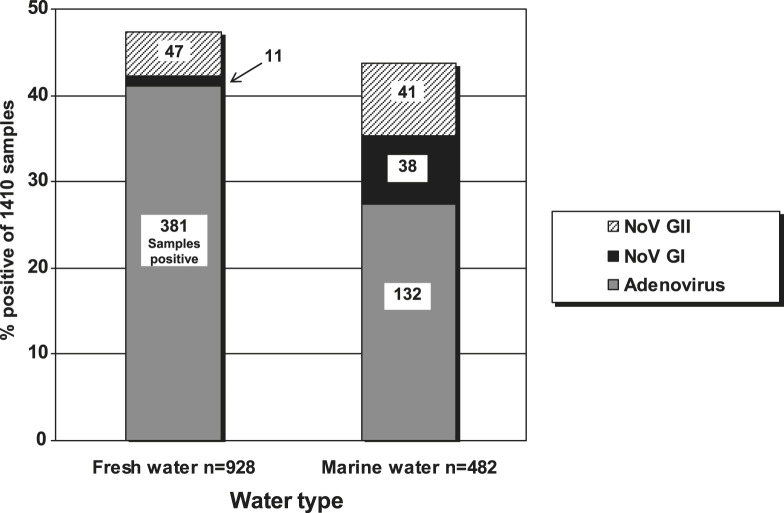
3.2. Water type
Freshwater sites showed a higher frequency of virus-positive samples than marine sites (Fig. 3 ). Adenoviruses were detected more often in fresh water (381 adenovirus-positive samples out of 928, 41.1%, Fig. 3) than in marine water (132 out of 482, 27.4%). Conversely, noroviruses (either GI or GII or both) were detected less often in freshwater samples (58 norovirus-positive samples out of 928, 6.3%) than in marine water (79 out of 482, 16.4%, Fig. 3). Further, in marine waters the detection rate of norovirus GI was almost as high as norovirus GII (7.9% compared with 8.5%), which differs from the clinical context where GI viruses are found much less frequently than GII types in patients from gastroenteritis outbreaks, even in surveys of unaffected individuals (e.g. Verhoef et al., 2009). However, these high GI detection rates were mainly due to just four sites having higher frequencies of NoV GI.
Fig. 3.
Adenovirus and norovirus detection in marine and fresh waters.
3.3. Variation according to site
Virus occurrence ranged widely between sites. Some Laboratories reported no viruses at all in any sample while others found many samples positive for at least one virus. Human adenoviruses were detected in all except two sites, one marine and one fresh water. Sites were chosen on the basis of their recreational use, and most were impacted by sewage effluent. Among the marine sites, 55% of samples from Pomezia, Rome, were positive for HAdV, while none was found at one of the Barcelona sites (though more samples were positive at the second site), and none was detected at Larnaca, Cyprus, where it is known no sewage is discharged. Among the freshwater sites, no HAdV was found at Kirchentellinsfurt Lake in Baden-Württemberg, while 80% of samples were HAdV-positive at Amper Grasslfing in Bavaria and 91% were positive at the site at Tomblaine, Nancy, a site well recognised for its anthropogenic effects as well as its recreational activities (mainly canoeing). With respect to noroviruses, five out of 11 marine water sites, and four out of 13 freshwater sites gave samples positive for GI noroviruses, the highest recovery from a marine site being 30% of samples positive at Pomezia (Rome), and that from a freshwater site being 10% of samples positive at Reading. Genogroup II noroviruses were detected at eight marine and eight freshwater sites, the highest frequencies being 16.3% positive samples at Ardea (Rome, marine), and 15% at Durgerdam (freshwater). Overall, the data showed that adenoviruses were present at more sites than noroviruses.
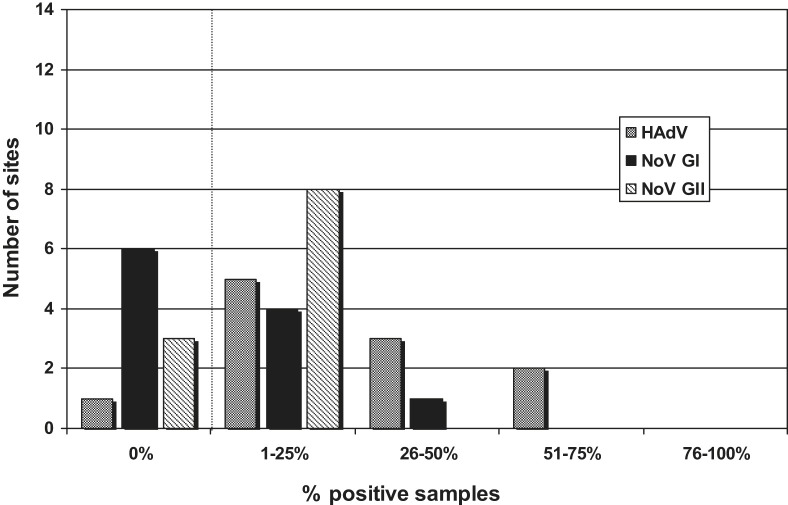
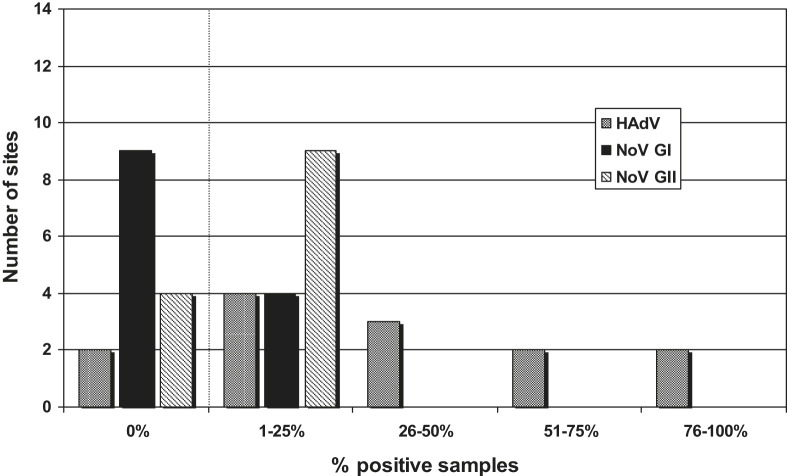
Some sites had more than 25% samples virus-positive in respect of both adenoviruses and noroviruses. To illustrate the distribution of sites relative to the frequency of virus detection, Fig. 4 (marine sites) and Fig. 5 (freshwater sites) show the frequencies of positive samples divided into five groups (0%, 1–25%, 26–50%, 51–75% and 76–100% positive samples) plotted against the number of sites in each group. Thus there was, for example, one of the 11 marine sites which reported no samples being HAdV-positive, five sites in which between 1% and 25% samples were HAdV-positive, three sites between 26% and 50% and two sites with between 51% and 75% HAdV-positive (Fig. 4). There were several sites where the adenovirus frequency was in the higher categories and two freshwater sites where over 76% samples were HAdV-positive.
Fig. 4.
Distribution of virus-positive sites – marine. Frequencies of positive samples divided into five groups (0%, 1–25%, 26–50%, 51–75% and 76–100% samples positive) plotted against the number of sites in each group.
Fig. 5.
Distribution of virus-positive sites – fresh water. Frequencies of positive samples divided into five groups (0%, 1–25%, 26–50%, 51–75% and 76–100% samples positive) plotted against the number of sites in each group.
Examination of the marine water norovirus GI data, when divided according to sites, shows that almost all the norovirus GI-positive samples (37/38) were found in four sites in Italy, the only other norovirus GI-positive marine water sample being found in one of the sites in Portugal. There was no evidence of outbreaks of norovirus-related disease in Italy in the areas local to the detection of GI virus in the environmental samples at the time when the samples were taken.
3.4. Virus infectivity by ICC-PCR
From each Laboratory, at least 10 samples that gave a strong HAdV-positive signal by nested PCR were analysed further by inoculation into cell culture and analysis of the supernatants by PCR. Fifty-one of 482 marine sample concentrates and 226 of 928 freshwater sample concentrates were tested. The results are shown in Table 2 . Twenty-four (47%) of the marine water samples were found to be positive by nested PCR following inoculation of A549 cell cultures and where uninoculated control cultures remained negative, and where cultures inoculated and sampled immediately after inoculation also remained negative. Forty-six (20%) freshwater samples were positive for infectious HAdV.
Table 2.
Adenovirus infectivity over all sampling sites.
| T = 0 | T = 5 | Number of samples | % of those tested | |
|---|---|---|---|---|
| Marine (51 tested) | –∗ | – | 15 | 29 |
| – | + | 24 | 47 | |
| + | – | 0 | 0 | |
| + | + | 12 | 24 | |
| Fresh (226 tested) | – | – | 169 | 75 |
| – | + | 46 | 20 | |
| + | – | 2 | 1 | |
| + | + | 9 | 4 |
∗ Nested-PCR test result on cell culture after zero (T = 0) or five (T = 5) days’ incubation.
3.5. QPCR assay for the detection of HAdV DNA
A total of 132 marine and freshwater samples which had previously tested HAdV-positive by nested-PCR were further analysed by the QPCR assay of Hernroth et al. (2002). Eighty (60.6%) samples were positive, with a mean value of 3260 genome copies (GC)/L of water. The percentage of positive samples was similar in both types of recreational water; 61.3% positive for fresh water with mean GC values of 558 GC/L versus 58.6% positive for marine waters with mean concentrations of 8810 GC/L.
3.6. Analysis of the sequence of the PCR products obtained
Fifty-three samples were further analysed to type the HAdV present. The most frequently detected HAdV serotypes were 12 (n = 4), 31 (n = 8), 40 (n = 4) and 41 (n = 22). Serotypes 1 and 19 were observed with lower frequency. Serotypes 1, 2, 3, 12, and 31 were observed after analysing 7 samples which had been cultured in A549 cells as part of the infectivity detections.
Nineteen samples were studied for determining NoV genotypes. Fifteen were confirmed as GII, with seven of them being GII.4. Four were GI, with one being GI.2. Over the last few years the most newly emerging NoV strains belong to GII.4 and show a global presence (Bull et al., 2006, Rowena et al., 2006).
3.7. Relationship of virus frequency to faecal indicators
Frequencies of virus-positive samples were compared with the threshold values for E. coli and intestinal enterococci defining “good” water quality in the rBWD. The levels specified in the Directive for E. coli are 500/100 mL (coastal/transitional waters) and 1000/100 mL (inland waters), and the corresponding values for intestinal enterococci are 200/100 mL (coastal/transitional waters) and 400/100 mL (inland waters). Matching E. coli and intestinal enterococci data were available for 193 adenovirus-positive samples of which 117 (60.6%) had E. coli concentrations below the thresholds for “good” water quality whilst 151 (78.2%) had intestinal enterococci concentrations below the “good” water quality thresholds. For norovirus, matching E. coli and intestinal enterococci data were available for 52 positive samples, and the E. coli concentration in 31 (59.6%) of these was below the rBWD thresholds for “good” water quality. For intestinal enterococci, 38 (73.1%) norovirus-positive samples had concentrations below the “good” water quality thresholds. These results demonstrate the presence of PCR-detected virus in samples that would be considered “clean”, and of low illness risk, in terms of their faecal indicator organism concentration.
3.8. Robustness of virus detection methods
The results of the robustness calculations of the virus/water detection methods are shown in Table 3 . With the adenovirus/freshwater method the trial sensitivity, or percentage of correctly identified positive samples, was 77.2%, and the concordance was lower than the accordance. A value of 1.0 lies just outside the COR 95% confidence intervals (CI), indicating that the method was not quite as reproducible as repeatable. The trial specificity, or percentage of correctly identified negative samples, was 96.1%, and 1.0 fell within the COR 95% CI, indicating that with identification of negative samples the method was as reproducible as it was repeatable. With the adenovirus/seawater method the trial sensitivity was 89.3%, and the concordance was lower than the accordance. Again, 1.0 lies just outside the COR 95% confidence intervals (CI). The trial specificity was 99.2%, and 1.0 fell within the COR 95% CI. With the norovirus/freshwater method the trial sensitivity was 91.4%, and the concordance was lower than the accordance, 1.0 lying just outside the COR 95% confidence intervals (CI). The trial specificity was 96.1%, and 1.0 fell within the COR 95% CI. With the norovirus/seawater method the trial sensitivity was 91.7%, and 1.0 fell within the COR 95% CI. The trial specificity was 92.6%, and 1.0 fell within the COR 95% CI.
Table 3.
Statistical evaluation of methods for virus detection from recreational waters.
| Method | Sample type | Sensitivity (%) | Specificity (%) | Accordance (%) | Concordance (%) | COR | |
|---|---|---|---|---|---|---|---|
| A | Adenovirus/freshwater | Positive | 77.2 (71.3–82.1)* | N/A | 73.9 (61.2–86.5) | 63.5 (50.9–81.7) | 1.63 (1.07–2.52) |
| Negative | N/A | 96.1 (92.8–98.0) | 93.0 (85.2–100) | 92.5 (84.8–100) | 1.08 (1.00–1.16) | ||
| B | Adenovirus/seawater | Positive | 89.3 (82.5–93.6) | N/A | 85.9 (68.9–94.9) | 79.6 (66.1–92.7) | 1.57 (1.01–2.29) |
| Negative | N/A | 99.2 (95.5–99.9) | 98.6 (97.4–100) | 98.3 (94.6–100) | 1.25 (0.97–1.44) | ||
| C | Norovirus/freshwater | Positive | 91.4 (87.1–94.3) | N/A | 86.2 (74.4–96.1) | 83.9 (71.9–95.7) | 1.2 (1.02–1.35) |
| Negative | N/A | 96.1 (92.8–98) | 92.9 (87–97.7) | 92.5 (86.8–97.5) | 1.06 (0.97–1.14) | ||
| D | Norovirus/seawater | Positive | 91.7 (85.5–95.5) | N/A | 85.3 (75.6–94.9) | 84.6 (75.3–94.9) | 1.05 (0.81–1.38) |
| Negative | N/A | 92.6 (86.5–96.0) | 88.0 (70.8–100) | 85.7 (70.1–100) | 1.22 (0.92–2.18) |
*Numbers in parentheses indicate lower and upper 95% confidence intervals.
4. Discussion
This study has shown clearly that it is possible to use relatively straightforward methods for the detection of two important enteric viruses in water samples across a range of geographical sites with varying degrees of pollution.
The common occurrence of adenoviruses (36.4% of samples tested) reflected the intermittent shedding of these viruses in the faeces by most adults. The difference in detection frequency may have been due to the greater dispersing and diluting power of the sea compared with that of the fresh waters. Alternatively, viruses may be less stable in marine waters due to the higher salt content, especially with higher temperatures (Hawley and Garver, 2008, Lo et al., 1976). The frequent detection of HAdVs by most laboratories reflected their known environmental robustness; though it was not possible to perform ICC-PCR on all the adenovirus-positive samples and thus show that all contained infectious viruses, it is known that adenoviruses can persist in an infectious state in various environments over long periods (Rzezutka and Cook, 2004). Charles et al. (2009) found a strong relation between PCR detection and infectivity of adenovirus Type 2 in groundwater over one year.
Although noroviruses are spread principally by person-to-person transmission, environmental spread is also important, for instance in outbreaks associated involving drinking water (e.g. Hewitt et al., 2007) and consumption of bivalve molluscs (Lees, 2000). In this study, the high frequency of NoV GI detection in two Laboratories suggests a higher level in the environment than was demonstrated by consideration of the rest of the data for this virus. Detection of GI noroviruses in the environment is not matched by their detection in clinical samples; GI NoV strains have been detected frequently in sewage, effluent, and surface waters (da Silva et al., 2007, Katayama et al., 2008, Myrmel et al., 2006), which contributes to the view that many norovirus infections are symptomless, with GI viruses being under-represented among those found in clinical cases. It is unclear whether this relates to our data as most of the GI isolates were found in only four sites. The frequency of GII norovirus detection (approximately 6%) was as expected. It is commonly accepted that norovirus-related disease shows a seasonal trend, with most outbreaks and sporadic cases occurring in winter. Whilst it would have been interesting to obtain a temporal distribution of environmental norovirus detection similar to that of Nordgren et al. (2009), this was not feasible in this study since it was specifically planned to be related to the EU bathing season, and in any case RT-PCR detection might not have provided resolution high enough to show temporal differences in norovirus levels. Further studies are planned using a norovirus QPCR to investigate this aspect.
The performance characteristics of the methods used for concentration and detection of HAdV and NoV in both fresh and marine water samples were determined. Recovery values of 49% (seeded fresh water) and 37% (seeded artificial sea water) were considered acceptable, though variations between laboratories prevented direct statistical comparisons of performance, and a modified method for marine water samples was developed during the project (Calgua et al., 2008). The percentage of correctly identified positive samples was around 90%, except for HAdV in freshwater, which showed a sensitivity of 77%, while the specificity of the methods was shown to be 93% or more. The sensitivity and specificity values compare well with those of some PCR-based methods for foodborne pathogen detection (Abdulmawjood et al., 2004, Malorny et al., 2004). The lower sensitivity value of the adenovirus/freshwater may be due to the fact that the HAdV concentration in the seeded sample was lower than the NoV concentration used. This may also explain the higher COR values for the HAdV-positive marine and freshwater samples. Furthermore, it should be noted that the samples used for the QC were not actually identical, whereas for the COR estimation this would be preferred. Each participant used the water from their own site(s), and this would differ from site to site and from week to week. River water, particularly, will contain varying levels of material that may reduce the effectiveness of the concentration method and/or inhibit the molecular assays. Notwithstanding this, the results demonstrate that the methods used are robust, although currently no criteria exist on lower limits of acceptability for robustness of methods for detection of viruses in water.
The theoretical limit of detection of the method reported here can be estimated. If an (RT)PCR signal was obtained from an undiluted nucleic acid extract, and the assumption is made that the assay could detect one target molecule, this signifies that there was one virus equivalent in 10 μL nucleic acid extract. There were thus 10 virus equivalents in 100 μL nucleic acid extract, and on the assumption that this extract was obtained from 5 mL concentrate with no loss of target nucleic acid, this implies that there were 20 virus particles in the 10 mL concentrate. Assuming that the concentrate was derived from the original sample with no loss of virus, the conclusion is that a signal from the neat extract indicates that there were at least 20 virus particles in the 10-L water sample. If the extract had to be diluted to 10−1, then there were 200 virus particles in the 10-L sample.
In selecting methods for concentration and detection of the target viruses practical and cost factors were considered in addition to recovery efficiency. Concentration by glass wool filtration is inexpensive requiring no specialised equipment beyond a centrifuge capable of 7000× g, and running costs are minimal. Membrane filtration is slightly more expensive, requiring a filtration stand, but again, running costs are low. Both approaches contrast with (for example) ultrafiltration (high costs of filtration units and pumps, or disposable cartridges) and ultracentrifugation, which is unlikely to be found in routine environmental virology laboratories. The time taken to process samples was also an important factor; using the selected methods it was possible to process up to eight samples in one day (including controls) following familiarisation with the method. For detection, cell culture was not considered for the surveillance stage, being too slow, expensive and requiring specialised facilities; the costs and labour time spent on molecular detection was as might be expected in any laboratory equipped for PCR and related techniques.
The amount of sewage discharged in the vicinity of many of the sites studied will affect the likelihood of human viruses being present in the water. Sewage input was not measured directly but the level of faecal indicators found reflects the contamination level. Viruses were found less often in sites where the sewage input was expected to be lower.
The influence of organic contaminants that occur naturally in water must not be underestimated. Reaction inhibition by substances in the sample is a well-known problem associated with analysis of environmental samples (e.g. da Silva et al., 2007), and was observed in this study. The use of the IACs in both NoV and HAdV PCRs was of significant benefit in guarding against false negative reactions. In the current study the norovirus RT-PCR suffered about 5.5% of reactions failing to give a conclusive result (4.4% of freshwater samples and 7.7% of marine samples). Samples were tested at a higher dilution (up to 10−3) to remove inhibition and achieve a positive IAC signal. Successive dilutions were done when a higher concentration failed to give a target signal or an IAC signal. Inhibition of the adenovirus PCR was much less problematic, with PCR reactions of 0.9% of freshwater samples and 5.6% of marine water samples being inhibited. Samples from one inland major river site (Kew Bridge, UK) had often to be diluted up to 10−3 and consequently unexpectedly low numbers of samples positive for adenovirus (23%) were recorded. Subsequent tests with bovine serum albumin (BSA) in the PCR reaction suggest that routine incorporation of this reagent in the reaction mix may reduce enzymatic inhibition.
Integrated cell culture-PCR provided a method of determining the infectivity of adenoviruses, which was particularly useful since naturally-occurring virus strains do not always grow in cell culture with the same rapidity nor with the same evidence of cellular destruction. The enteric Ad40 and Ad41 viruses cannot be grown in most cell culture systems that support the growth of adenoviruses from the other subgroups, A549, HeLa, primary human amnion and primary human embryo kidney cells (Tiemessen and Kidd, 1995). They have been shown to replicate in cell culture systems using Graham 293 cells, HEp-2 cells and HT-29 cells (Ko et al., 2003, Tiemessen and Kidd, 1995). Our data support these findings, because the presence of both Ad40 and Ad41 was shown by direct PCR, not in the cell culture-PCR assay using A549 cells. Direct inoculation of cell cultures followed by observation over an extended period would not provide a good indication of infectivity and would not be in the interests of providing a rapid test. The finding that about 20% of freshwater samples and about 47% of marine water samples contained infectious adenovirus supports laboratory observations (e.g. Thurston-Enriquez et al., 2003) that these agents are environmentally robust.
The FIO levels encountered in this project exhibited a wide range. Comparisons with FIO thresholds defined in the current European Directive bathing water standards (2006/7/EC) suggest that over 50% of samples that are relatively clean in terms of FIO concentrations and which exhibit “good” water quality, with a low associated illness risk, can be positive for adenovirus and norovirus. However, use of an adenovirus PCR, for example, as a means of determining recreational water quality would require the use of quantitative, rather than presence/absence detection. Quantitative PCRs for different types of environmental adenovirus are now available. Whether such a test would detect infectious virus may be addressed by, for example, detection of virus-specific mRNA, and also there is some evidence that in adenovirus preparations from which free DNA has been removed before analysis virus titres measured by infectivity and by QPCR are very similar (Gironès, personal communication). It would then be necessary to determine any association between adenovirus levels and health risk, and there is thus a need for further work before the viral parameters investigated here could be used in a regulatory framework prior to epidemiological investigation to provide an appropriate evidence-base for policy development.
5. Conclusions
A comprehensive surveillance study of EU recreational waters was done through the 2006 bathing season. It may be concluded from the results that:
-
1.
Almost 40% of bathing water samples in Europe were virus-positive entailing a possible public health risk from bathing;
-
2.
Adenoviruses are more prevalent than noroviruses in both marine and fresh waters and appear to be a promising viral indicator for bathing water quality;
-
3.
A single concentration method can be used to concentrate adenoviruses and noroviruses in fresh water recreational samples and a further single method can be used for marine waters;
-
4.
Concentration and detection methods may be used effectively even in polluted waters;
-
5.
Though the majority of sites returned frequencies of 0–25% positive, some were so polluted that >50% of samples contained one or both target viruses;
-
6.
Adenoviruses remain infectious in the environment, and this may be true for other pathogenic viruses such as noroviruses.
6. The ‘Virobathe’ group
This work was performed by scientists and technicians from 16 Institutions across Europe. In addition to the Authors of this paper, those making significant contributions were as follows:
Dr. Silvia Bofill-Mash, Ms. Pilar Clementeh, Dr. Donia Domenicad, Ms. Alexandra Duarten, Dr. Inge Gräberl, Dr. Wafa Hollisterp, Ms. Stephanie Huberi, Dr. Marcello Iaconellim, Dr. Giuseppina La Rosam, Prof. Beata Cuvelierk, Ms. Leslie Orgorzalyf, Dr. Nicholas Pissarides, Dr. Gabrieli Rosannad, Ms. Elyne Salagnonc, Dr. Oliver Schneidere, Ms. Arieke Docters van Leeuwenj, Dr. Marco Veranib, and Mr. Steve Wildeg.
Acknowledgements
This work was funded by an EU contract number 513648 VIROBATHE, as part of the Sixth Framework Programme. The authors are grateful to Dr. Jan Vinjé for helpful comments on the manuscript.
References
- Abdulmawjood A., Bülte M., Roth S., Schönenbrücher H., Cook N., Heuvelink A.E., Hoorfar J. Development, validation, and standardization of polymerase chain reaction-based detection of E. coli O157. Journal of AOAC International. 2004;87(3):596–603. [PubMed] [Google Scholar]
- Allard A., Albinsson B., Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. Journal of Clinical Microbiology. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F., Karlsson B., Nordgren J., Paniagua M., González A., Amador J.J., Espinoza F., Svensson L. Mutated G4P[8] rotavirus associated with a nationwide outbreak of gastroenteritis in Nicaragua in 2005. Journal of Clinical Microbiology. 2007;45(3):990–997. doi: 10.1128/JCM.01992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R.A., Tu E.T., McIver C.J., Rawlinson W.D., White P.A. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. Journal of Clinical Microbiology. 2006;44(2):327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B., Mengewein A., Grunert A., Bofill-Mas S., Clemente-Casares P., Hundesa A., Wyn-Jones A.P., López-Pila J.M., Girones R. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. Journal of Virological Methods. 2008;153:79–83. doi: 10.1016/j.jviromet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- CEU, 2006. Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC.
- Charles K.J., Shore J., Sellwood J., Laverick M., Hart A., Pedley S. Assessment of the stability of human viruses and coliphage in groundwater by PCR and infectivity methods. Journal of Applied Microbiology. 2009;106:1827–1837. doi: 10.1111/j.1365-2672.2009.04150.x. [DOI] [PubMed] [Google Scholar]
- Contreras-Coll N., Lucena F., Mooijman K., Havelaar A., Pierz V., Boque M., Gawler A., Höller C., Lambiri M., Mirolo G., Moreno B., Niemi M., Sommer R., Valentin B., Wiedenmann A., Young V., Jofre J. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Research. 2002;36(20):4963–4974. doi: 10.1016/s0043-1354(02)00229-4. [DOI] [PubMed] [Google Scholar]
- da Silva A.K., le Saux J.-C., Parnaudeau S., Pommepuy M., Elimelech M., le Guyader F.S. Evaluation of removal of noroviruses during wastewater treatment using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Applied and Environmental Microbiology. 2007;73(24):7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A.C., Hinckley D.V. Cambridge University Press; Cambridge, UK: 1997. Bootstrap Methods and Their Application. [Google Scholar]
- de Roda Husman A.M., Lodder W.J., Rutjes S.A., Schijven J.F., Teunis P.F. Long-term inactivation study of three enteroviruses in artificial surface and groundwaters using PCR and cell culture. Applied and Environmental Microbiology. 2009;75(4):1050–1057. doi: 10.1128/AEM.01750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z., Geng J., Cai J., Han H., Li X., Kang W., Weng D., Liang P., Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. Journal of Pathology. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrou S., Kopecka H., Lopez-Pila J.M., Marechal J., Prevot J. Detection of hepatitis A virus and other enteroviruses in wastewater and surface water samples by gene probe assay. Water Science and Technology. 1991;24(2):267–272. [Google Scholar]
- Enriquez C.E., Hurst C.J., Gerba C.P. Survival of the enteric adenoviruses 40 and 41 in tap, sea and wastewater. Water Research. 1995;29:2548–2553. [Google Scholar]
- Farrah S.R., Bitton G. In: Jean-Marc Bollag, Stotzky Arthur Douglas, Peterson G., George H., editors. vol. 6. CRC Press; 1990. (Soil Biochemistry). [Google Scholar]
- Gerba C.P., Goyal S.M., LaBelle R.L., Cech I., Bodgan G.F. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. American Journal of Public Health. 1979;69:1116–1119. doi: 10.2105/ajph.69.11.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.J., Green J., Gallimore C., Lee J.V., Neal K., Brown D.W.G. Mixed genotype SRSV infections among a party of canoeists exposed to contaminated recreational water. Journal of Medical Virology. 1997;52:425–429. [PubMed] [Google Scholar]
- Greening G.E., Hewitt J., Lewis G.D. Evaluation of integrated cell culture-PCR (ICC-PCR) for virological analysis of environmental samples. Journal of Applied Microbiology. 2002;93(5):745–750. doi: 10.1046/j.1365-2672.2002.01741.x. [DOI] [PubMed] [Google Scholar]
- Harley D., Harrower B., Lyon M., Dick A. A primary school outbreak of pharyngoconjunctival fever caused by adenovirus type 3. Communicable Diseases Intelligence. 2001;25(1):9–12. doi: 10.33321/cdi.2001.25.2. [DOI] [PubMed] [Google Scholar]
- Hauri A.M., Schimmelpfennig M., Walter-Domes M., Letz A., Diedrich S., Lopez-Pila J., Schreier E. An outbreak of viral meningitis associated with a public swimming pond. Epidemiology and Infection. 2005;133(2):291–298. doi: 10.1017/s0950268804003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley L.M., Garver K.A. Stability of viral hemorrhagic septicemia virus (VHSV) in freshwater and seawater at various temperatures. Diseases of Aquatic Organisms. 2008;82:171–178. doi: 10.3354/dao01998. [DOI] [PubMed] [Google Scholar]
- Hernroth B.E., Conden-Hansson A.C., Rehnstam-Holm A.S., Girones R., Allard A.K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Applied and Environmental Microbiology. 2002;68(9):4523–4533. doi: 10.1128/AEM.68.9.4523-4533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J., Bell D., Simmons G.C., Rivera-Aban M., Wolf S., Greening G. Gastroenteritis outbreak caused by waterborne norovirus at a New Zealand ski resort. Applied and Environmental Microbiology. 2007;73:7853–7857. doi: 10.1128/AEM.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe C.J.P.A., Vennema H., de Roda Husman A.M., van Duynhoven Y.T.H.P. Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. Journal of Infectious Diseases. 2004;189:699–705. doi: 10.1086/381534. [DOI] [PubMed] [Google Scholar]
- Hundesa A., Maluquer de Motes C., Bofill-Mas S., Albinana-Gimenez N., Girones R. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Applied and Environmental Microbiology. 2006;72:7886–7893. doi: 10.1128/AEM.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Tanaka S., Ohishi H. Multiple outbreaks of gastroenteritis due to a single strain of genotype GII/4 norovirus in Kobe, Japan: risk factors for norovirus spread in health care settings. Japanese Journal of Infectious Diseases. 2008;61(5):419–422. [PubMed] [Google Scholar]
- Katayama H., Haramoto E., Oguma K., Yamashita H., Tajima A., Nakajima H., Ohgaki S. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Research. 2008;42:1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Kay D., Fleisher J.M., Salmon R.L., Jones F., Wyer M.D., Godfree A.F., Zelenauch-Jacquotte Z., Shore R. Predicting the likelihood of gastroenteritis from sea bathing – results from randomized exposure. The Lancet. 1994;344(8927):905–909. doi: 10.1016/s0140-6736(94)92267-5. [DOI] [PubMed] [Google Scholar]
- Kay D., Bartram J., Prüss A., Ashbolt N., Wyer M.D., Fleisher J.M., Fewtrell L., Rogers A., Rees G. Derivation of numerical values for the World Health Organization guidelines for recreational waters. Water Research. 2004;38(5):1296–1304. doi: 10.1016/j.watres.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Ko G., Cromeans T.L., Sobsey M.D. Detection of infectious adenovirus in cell culture by mRNA reverse transcriptase-PCR. Applied and Environmental Microbiology. 2003;69:7377–7384. doi: 10.1128/AEM.69.12.7377-7384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., von Bonsdorff C.H., Vinjé J., de Medici D., Monroe S. Foodborne viruses. FEMS Microbiology Reviews. 2002;26(2):187–205. doi: 10.1111/j.1574-6976.2002.tb00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton S.D., Chevennement R., Nagelkerke N., Lombard B. Analysing collaborative trials for qualitative microbiological methods: accordance and concordance. International Journal of Food Microbiology. 2002;79(3):175–181. doi: 10.1016/s0168-1605(02)00107-1. [DOI] [PubMed] [Google Scholar]
- Laverick M.A., Wyn-Jones A.P., Carter M.J. Quantitative RT-PCR for the enumeration of noroviruses (Norwalk-like viruses) in water and sewage. Letters in Applied Microbiology. 2004;39:127–136. doi: 10.1111/j.1472-765X.2004.01534.x. [DOI] [PubMed] [Google Scholar]
- Lee C., Lee S.H., Han E., Kim S.J. Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and enteroviruses in river water. Applied and Environmental Microbiology. 2004;70:6695–6705. doi: 10.1128/AEM.70.11.6695-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees D. Viruses and bivalve shellfish. International Journal of Food Microbiology. 2000;59(1–2):81–116. doi: 10.1016/s0168-1605(00)00248-8. [DOI] [PubMed] [Google Scholar]
- Lipp E.K., Farrah S.A., Rose J.B. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Marine Pollution Bulletin. 2001;42:286–329. doi: 10.1016/s0025-326x(00)00152-1. [DOI] [PubMed] [Google Scholar]
- Lo S., Gilbert J., Hetrick F. Stability of human enteroviruses in estuarine and marine waters. Applied and Environmental Microbiology. 1976;32:245–249. doi: 10.1128/aem.32.2.245-249.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorny B., Bunge C., Hoorfar J., Helmuth R. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Applied and Environmental Microbiology. 2003;69:290–296. doi: 10.1128/AEM.69.1.290-296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorny B., Cook N., D’Agostino M., de Medici D., Croci L., Abdulmawjood A., Fach P., Karpiskova R., Aymerich T., Kwaitek K., Kuchta T., Hoorfar J. Multicenter collaborative trial validation of a PCR-based method for detection of Salmonella in chicken and pig samples. Journal of AOAC International. 2004;87:861–866. [PubMed] [Google Scholar]
- Maunula L., Kalso S., von Bonsdorff C.-H., Pönkä A. Wading pool water contaminated with both noroviruses and astroviruses as the source of a gastroenteritis outbreak. Epidemiology and Infection. 2004;132(4):737–743. doi: 10.1017/s0950268804002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G.J., van Asperen I.A., Kokman-Houweling J.M., Nooitgedagt A., van de Laar M.J.W., Havelaar A.H. The relationship between health effects in triathletes and microbiological quality of freshwater. Water Science and Technology. 1995;31(5–6):19–26. [Google Scholar]
- Miagostovich M.P., Ferreira F.F., Guimarães F.R., Fumian T.M., Diniz-Mendes L., Luz S.L., Silva L.A., Leite J.P. Molecular detection and characterization of gastroenteritis viruses occurring naturally in the stream waters of Manaus, Central Amazonia, Brazil. Applied and Environmental Microbiology. 2008;74(2):375–382. doi: 10.1128/AEM.00944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrmel M., Berg E.M.M., Grinde B., Rimstad E. Enteric viruses in inlet and outlet samples from sewage treatment plants. Journal of Water and Health. 2006;4:197–209. [PubMed] [Google Scholar]
- Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P.-E. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Research. 2009;43:1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- Nwachuku N., Gerba C.P. Health risks of enteric viral infections in children. Reviews of Environmental Contamination and Toxicology. 2006;186:1–56. doi: 10.1007/0-387-32883-1_1. [DOI] [PubMed] [Google Scholar]
- Papapetropolou M., Vantarakis A.C. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. Journal of Infection. 1995;36:101–103. doi: 10.1016/s0163-4453(98)93414-4. [DOI] [PubMed] [Google Scholar]
- Pina S., Puig M., Lucena F., Jofre J., Girones R. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Applied and Environmental Microbiology. 1998;64:3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K.A., Gerba C.P., Abbaszadegan M., Pepper I.L. ICC-PCR detection of enteroviruses and hepatitis A virus in environmental samples. Canadian Journal of Microbiology. 2001;47:153–157. [PubMed] [Google Scholar]
- Rowena A., Bull Elise, Tu T.V., McIver Christopher J., Rawlinson William D., White Peter A. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. Journal of Clinical Microbiology. 2006;44(2):327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzezutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiology Reviews. 2004;28:441–453. doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Sarguna P., Rao A., Sudha Ramana K.N. Outbreak of acute viral hepatitis due to hepatitis E virus in Hyderabad. Indian Journal of Medical Microbiology. 2007;25(4):378–382. doi: 10.4103/0255-0857.37343. [DOI] [PubMed] [Google Scholar]
- Scotter S.L., Langton S., Lombard B., Lahellec C., Schulten S., Nagelkerke N., in’t Veld P.H., Rollier P. Validation of ISO method 11290 part 2. Enumeration of Listeria monocytogenes in foods. International Journal of Food Microbiology. 2001;70(1–2):121–129. doi: 10.1016/s0168-1605(01)00530-x. [DOI] [PubMed] [Google Scholar]
- Sinclair R.G., Jones E.L., Gerba C.P. Viruses in recreational water-borne disease outbreaks: a review. Journal of Applied Microbiology. 2009;107(6):1769–1780. doi: 10.1111/j.1365-2672.2009.04367.x. [DOI] [PubMed] [Google Scholar]
- Thurston-Enriquez J.A., Haas C.N., Jacangelo J., Gerba C.P. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Applied and Environmental Microbiology. 2003;69:577–582. doi: 10.1128/AEM.69.1.577-582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemessen C.T., Kidd A.H. The subgroup F adenoviruses. Journal of General Virology. 1995;76:481–497. doi: 10.1099/0022-1317-76-3-481. [DOI] [PubMed] [Google Scholar]
- Vasl R., Fattal B., Katzenelson E., Shuval H. Survival of enteroviruses and bacterial indicator organisms in the sea. In: Goddard M., Butler M., editors. Viruses and Wastewater Treatment. Pergamon Press; 1981. pp. 113–116. [Google Scholar]
- Vennema H., de Bruin E., Koopmans M. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. Journal of Clinical Virology. 2002;25(2):233–235. doi: 10.1016/s1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- Verhoef L.P., Kroneman A., van Duynhoven Y., Boshuizen H., van Pelt W., Koopmans M. Foodborne viruses in Europe network. Selection tool for foodborne norovirus outbreaks. Emerging Infectious Diseases. 2009;15(1):31–38. doi: 10.3201/eid1501.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaginès P., Sarrette B., Husson G., Vilaginès R. Glass wool for virus concentration at ambient water pH level. Water Science and Technology. 1993;27:299–306. [Google Scholar]
- WHO . World Health Organisation; Geneva: 2003. Guidelines for Safe Recreational Water Environments Volume 1: Coastal and Freshwaters. p. 219. [Google Scholar]
- Wiedenmann A., Krüger P., Dietz K., López-Pila J., Szewzyk R., Botzenhart K. A randomized controlled trial assessing infectious disease risks from bathing in fresh recreational waters in relation to the concentration of Escherichia coli, intestinal enterococci, Clostridium perfringens and somatic coliphages. Environmental Health Perspectives. 2006;8115:1–41. doi: 10.1289/ehp.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyn-Jones A.P., Sellwood J. Enteric viruses in the aquatic environment. Journal of Applied Microbiology. 2001;91:945–962. doi: 10.1046/j.1365-2672.2001.01470.x. [DOI] [PubMed] [Google Scholar]
- Wyn-Jones A.P., Pallin R., Dedoussis C., Shore J., Sellwood J. The detection of small round-structured viruses in water and environmental materials. Journal of Virological Methods. 2000;87:99–107. doi: 10.1016/s0166-0934(00)00157-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tan X.J., Wang H.Y., Yan D.M., Zhu S.L., Wang D.Y., Ji F., Wang X.J., Gao Y.J., Chen L., An H.Q., Li D.X., Wang S.W., Xu A.Q., Wang Z.J., Xu W.B. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. Journal of Clinical Virology. 2009;44(4):262–267. doi: 10.1016/j.jcv.2009.02.002. [DOI] [PubMed] [Google Scholar]