Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) is an inefficient inducer of interferon (IFN) response. It expresses various proteins that effectively circumvent IFN production at different levels via distinct mechanisms. Through the construction of recombinant IBV expressing proteins 8a, 8b and 8ab encoded by SARS-CoV ORF8, we demonstrate that expression of 8b and 8ab enables the corresponding recombinant viruses to partially overcome the inhibitory actions of IFN activation to achieve higher replication efficiencies in cells. We also found that proteins 8b and 8ab could physically interact with IRF3. Overexpression of 8b and 8ab resulted in the reduction of poly (I:C)-induced IRF3 dimerization and inhibition of the IFN-β signaling pathway. This counteracting effect was partially mediated by protein 8b/8ab-induced degradation of IRF3 in a ubiquitin-proteasome-dependent manner. Taken together, we propose that SARS-CoV may exploit the unique functions of proteins 8b and 8ab as novel mechanisms to overcome the effect of IFN response during virus infection.
Keywords: IFN antagonist, IRF3, SARS-CoV, ORF8, Ubiquitin-dependent degradation
Highlights
-
•
Recombinant IBV expressing SARS-CoV protein 8b or 8ab replicates better than wild type in cells pre-treated with poly(I:C).
-
•
8b interacts with the IAD domain of IRF3.
-
•
Overexpression of 8b or 8ab reduces poly(I:C)-induced IRF3 dimerization and interferon induction.
-
•
8b and 8ab induce degradation of IRF3 in a ubiquitin-proteasome-dependent manner.
-
•
8b and 8ab suppress interferon response induced by constitutively active IRF3.
1. Introduction
When challenged by a viral infection, the host mounts an immediate innate immune response, leading to the production of type I interferons (IFN-α and IFN-β) and the expression of hundreds of downstream IFN-stimulated genes (ISGs) (Stetson and Medzhitov, 2006, Suhara et al., 2002). Central to the induction of type I IFN is interferon regulatory factor 3 (IRF3) (Hiscott, 2007, Taniguchi et al., 2001). The initial step of the signaling cascade leading to IRF3 activation is recognition of specific viral pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs) in two major pathways. These include cell surface toll-like receptors (TLRs), such as TLR3, TLR7, TLR8 and TLR9, which sense viral components (Kumagai et al., 2008, Kumar et al., 2009a, Kumar et al., 2009b), and cytosolic RNA helicases such as retinoic acid-inducible gene I (RIG-I) and/or melanoma differentiation-associated gene 5 (MDA5), which detect viral RNA (Loo et al., 2008, Onomoto et al., 2007). Upon binding to their ligands, PRRs recruit adaptor proteins to set off a series of signaling cascades to phosphorylate and dimerize IRF3 (Fitzgerald et al., 2003). The activated IRF3 homodimer then translocates to the nucleus, switching on IFN synthesis (Lin et al., 1998; Suhara et al., 2002; Thanos and Maniatis, 1995). Many viruses have also evolved strategies to counteract the IFN action, including mechanisms that allow virus to evade recognition by the immune surveillance system, so as to inhibit IFN induction by hijacking molecules involved in IFN activation pathways or by inhibiting downstream signal transduction (Goodbourn et al., 2000, Versteeg et al., 2007, Weber et al., 2003).
Severe acute respiratory syndrome coronavirus (SARS-CoV) was the etiological agent of the SARS epidemic in 2003 (Guan et al., 2003, Marra et al., 2003). Apart from four typical structural proteins, nucleocapsid (N), envelope (E), membrane (M) and spike (S) proteins, and approximately 16 non-structural proteins (Nsp1-16) involved in viral replication, SARS-CoV encodes an exceptionally high number of accessory proteins that bear little resemblance to accessory genes of other coronaviruses (Liu et al., 2014, Narayanan et al., 2008b). Similar to other coronaviruses, SARS-CoV is an inefficient inducer of IFN-β response in cell culture system (Spiegel et al., 2005) and is sensitive to the antiviral state induced by IFNs (Spiegel et al., 2004, Zheng et al., 2004). Its genome may therefore encode proteins that allow this virus to effectively circumvent IFN production in order to overcome limitations imposed by the IFN action.
Together with a few structural proteins and Nsps, many coronavirus accessory proteins could suppress IFN production by targeting different aspects of the IFN signaling cascade (Lim et al., 2016, Liu et al., 2014, Zhong et al., 2012). For instance, SARS-CoV Papain-like protease (PLpro) attenuates IFN synthesis by abrogating IRF3 phosphorylation and nuclear translocation by physically interacting with IRF3 (Devaraj et al., 2007). IFN induction mediated by a constitutively active IRF3 is also inhibited by the de-ubiquitination activity of SARS-CoV PLpro (Matthews et al., 2014). On the other hand, Nsp1 of PEDV does not interfere IRF3 phosphorylation and nuclear translocation, but interrupts the enhanceosome assembly of IRF3 and CREB-binding protein (CBP) by promoting proteasomal degradation of CBP (Zhang et al., 2016). Targeting further upstream, SARS-CoV M protein prevents IRF3 phosphorylation by inhibiting the assembly of TBK1/IKK complex (Siu et al., 2009), whereas MERS-CoV M protein interacts with TRAF3 and disrupts TRAF3-TBK1 association, leading to reduced IRF3 phosphorylation (Lui et al., 2016). Likewise, the accessory protein ORF4b of MERS-CoV has been shown to specifically bind to TBK1 and IKKε, thereby inhibiting IRF3 phosphorylation and IFN-β production (Yang et al., 2015).
Signaling molecules downstream of IFN synthesis are also targets of SARS-CoV proteins. For instance, SARS-CoV Nsp1 inhibits STAT1-mediated transcription of ISGs by inhibiting its phosphorylation (Wathelet et al., 2007), apart from inducing degradation of a wide range of host mRNAs (Kamitani et al., 2006, Narayanan et al., 2008a). SARS-CoV ORF6 blocks STAT1 nuclear translocation by trapping the nuclear import factors in the endoplasmic reticulum and Golgi apparatus (Frieman et al., 2007). Among other IFN antagonists identified are nucleocapsid (N) protein of SARS-CoV and PEDV (Kopecky-Bromberg et al., 2007, Ding et al., 2014), accessory protein 4a of MERS-CoV (Siu et al., 2014), and PLpro domain 2 (PLP2) of MHV-A59 (Wang et al., 2011). IFN antagonism mediated by SARS-CoV N protein seemed to target a very early step of RNA recognition (Lu et al., 2011). Overexpression of SARS-CoV accessory protein 3a was found to down-regulate type I IFN receptor by promoting its ubiquitination and subsequent degradation via the lysosomal pathway (Minakshi et al., 2009).
Although both 8b and 8ab are encoded by SARS-CoV ORF8, they are expressed under distinct conditions. 8ab is expressed as a single protein encoded by the single continuous ORF (ORF8ab) found in SARS-CoV isolated from animals and early stage human isolates. In contrast, as a consequence of a 29-nt deletion that results in two separate overlapping ORFs (ORF8a ⁄ ORF8b), most human isolates obtained at the middle to later phase of the epidemic encode instead 8a and 8b as two distinct proteins (Guan et al., 2003, Oostra et al., 2007). ORF8 is presumably acquired from SARS-related coronavirus from greater horseshoe bats through recombination (Lau et al., 2015), and the 29-nt deletion may be an evolutionary adaptation for enhancing viral pathogenesis in the human host. Proteins 8a, 8b and 8ab may possess different biochemical properties and possibly cellular functions (Law et al., 2006, Le et al., 2007). Protein 8ab was shown to be a glycosylated ER resident protein which can activate ATF6 to modulate the unfolded protein response (Sung et al., 2009). Protein 8a was found to enhance viral replication and induce cell death (C.Y. Chen et al., 2007), while 8b induces DNA synthesis and down-regulates SARS-CoV E protein via a proteasome-independent pathway (Keng et al., 2006). SARS-CoV 8b and 8ab were also shown to bind to both mono- and poly-ubiquitin when expressed in cell culture (Keng et al., 2006). Whether these ubiquitin-binding properties allow them to interact with host cell proteins remains unknown. Interestingly, when an in vivo attenuated recombinant SARS-CoV lacking the full-length E gene is passaged in mice, the ORF8 sequence mutates to encode a PDZ-binding motif in protein 8a, and the virus regained virulence (Jimenez-Guardeño et al., 2015).
In this study, we show proteins 8b and 8ab as novel IFN antagonists. Evidence presented supports the direct interaction between these two proteins and IRF3. The two proteins were also found to partially suppress IFN induction by limiting IRF3 activation and/or promoting the proteasome-mediated degradation of IRF3.
2. Materials and methods
2.1. Cell culture
African green monkey kidney Cos-7 and Vero cells, human non-small cell lung carcinoma H1299 cells and human hepatocellular carcinoma Huh7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Hyclone) and 1% penicillin/streptomycin DMEM (Invitrogen) and maintained at 37 °C with 5% CO2. To inhibit the proteasome activity, MG132 (Sigma) at a final concentration of 10 µM was added to cells 2 h prior to harvest.
2.2. Virus growth curve and plaque assay
To compare the growth kinetics of various recombinant virus strains, Vero cells were infected with the respective recombinant IBV strains at a multiplicity of infection (MOI) of 0.5, and harvested at a 4 h interval within 20 h post-infection for virus titration through plaque assay.
A monolayer of Vero cells seeded on 6-well plates a day prior to infection was infected with 200 µl of 10-fold serially diluted virus stock. After 1 h of incubation at 37 °C with regular shaking to ensure even distribution of the virus, cells were washed with PBS and cultured in 3 ml of DMEM containing 1% carboxymethyl cellulose (CMC) for 3 days. The cells were then fixed with 4% paraformaldehyde and stained with 0.1% toluidine. The number of plaques was counted in duplicates and the virus titer was calculated as plaque-forming unit (Pfu) per ml.
2.3. Plasmid Construction
The pKTO-Flag-8b and pKTO-Flag-8ab plasmids were previously described (Le et al., 2007). The coding sequences of 8b or 8ab were also subcloned to the pXJ40-Flag vector, which contains the CMV promoter for expression in mammalian cell lines. For the construction of the pXJ40-Myc-IRF3 plasmid, human IRF3 gene was amplified from cDNA of H1299 cells using the forward primer 5′-AACGCCTCGACGGAACCCCAAAGCCACGGAT-3′and the reverse primer 5′-GCCGGTACCTTATTGGTTGAGGTGGTGGGG-3′ prior to ligation into pXJ40-My plasmid at XhoI and KpnI restriction sites. Truncated mutants were then constructed based on the pXJ40-Myc-IRF3 construct using the following primers for PCR amplification: for IRF3 (1−133), Forward 5′-CCGCTCGAGCGGATGATGGGAACCCCAAAGCCACG–3′and Reverse 5′–GGGGTACCCCTCAAGAAGTACTGCCTCCACCAT–3′; for IRF3 (399-379), Forward 5′-CCGCTCGAGCGGATGGATACCCAGGAAGACATTCT-3′ and Reverse 5′-GGGGTACCCCTCATCCAGGCAGCGTCCTGTCTC-3′; for IRF3 (241−427), Forward 5′-CCGCTCGAGCGGATGTGGCCAGTCACACTGCCAGA-3′ and Reverse 5′-GGGGTACCCCTCAGCTCTCCCCAGGGCCCT-3′; for IRF3 (134−427): Forward 5′-CCGCTCGAGCGGATGGATACCCAGGAAGACATTCT-3′ and Reverse 5′-GGGGTACCCCTCAGCTCTCCCCAGGGCCCT-3′. Constitutively active mutant pXJ40-Myc-IRF3-5D was generated by performing sequential site-directed mutagenesis PCR (QuikChange II site-directed mutagenesis kit; Stratagene) to replace amino acids at positions 396, 398, 402, 404, and 405 with the phosphomimetic aspartic acid.
2.4. Western blot analysis
Cells were lysed in RIPA buffer in the presence of protease inhibitors (Roche Diagnostics) and phosphatase inhibitors (Pierce). Protein lysates were separated by electrophoresis in 8% SDS polyacrylamide gels and transferred to nitrocellulose membrane (Amersham Biosciences) via wet transfer (Bio-rad). The membranes were blocked overnight at 4 °C with 10% non-fat milk in PBST before probing with specific primary antibodies, followed by horse-radish peroxidase (HRP)-conjugated anti-mouse, anti-rabbit or anti-goat IgG secondary antibodies (DAKO), respectively. The following commercial primary antibodies were used: β-tubulin (Sigma), IRF3 (Santa Cruz Biotechnology), pIRF3(398) (Cell Signaling) and β-actin (Santa Cruz Biotechnology). Polyclonal antibodies against IBV N were raised in rabbits by this laboratory (Li et al., 2005).
2.5. Co-immunoprecipitation
Monolayer cells grown overnight in 6-well plates (Nunc) were infected with recombinant vaccinia virus encoding the bacteriophage T7 RNA polymerase before transfection of plasmids using Effectene reagent (Qiagen), as previously described (Liu and Inglis, 1991, Liu and Inglis, 1992). Briefly, at 20 h post-transfection, cells were harvested with 500 µl of RIPA buffer in the presence of protease (Roche Diagnostics) and phosphatase inhibitors (Pierce). Lysates were centrifuged at 13,000×g at 4 °C for 15 min, and the supernatant obtained was immunoprecipitated directly with antibody-conjugated agarose beads for 2 h or with appropriate antibodies followed by incubation with protein A agarose beads (Sigma) for another 2 h at room temperature. The immunoprecipitated proteins were separated on SDS-PAGE and analyzed by western blot using appropriate antibodies. The HRP-conjugated anti-Myc and anti-Flag antibodies were purchased from Sigma, while antibodies against IgG, ISG56, β-actin and full-length IRF3 were from Santa Cruz Biotechnology.
2.6. Native PAGE analysis of IRF3
Cells were lysed in buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40) containing protease inhibitors and phosphatase inhibitors for 30 min at 4 °C. Proteins were then separated by electrophoresis in 8% non-denaturing polyacrylamide gels, with 1% sodium deoxycholate (Sigma) in the cathode buffer. IRF3 monomers and dimers were detected by Western blot analysis using polyclonal antibodies against full-length IRF3 (Santa Cruz Biotechnology).
2.7. IFN-β reporter assays
Huh7 cells seeded on a 12-well plate were transfected with a total of 4 μg of the appropriate plasmids using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. pIFN-β-Luc and pRL-TK were purchased from Promega. 10 µg of poly (I:C) complexed with 10 µl Lipofectamine 2000 was then introduced into the cells 20 h later. Cells were lysed 20 h post treatment in passive lysis buffer (Promega) and an aliquot of the lysates was measured for firefly and Renilla luciferase activities according to the manufacturer's instruction (Promega).
2.8. In vitro assembly and transcription of full-length cDNA clones
Construction of recombinant IBVs (rIBVs) was carried out essentially as previously described (Fang et al., 2007, Le et al., 2007, Tan et al., 2006). To generate the 8b mutant (8bm) containing lysine to arginine mutations at all three positions, standard PCR site-directed mutagenesis was performed using the construct containing ORF8 insertion as a template. The genotypes of rIBVs were validated by sequencing.
2.9. RNA Isolation and Quantitative RT-PCR
Total RNA was isolated using the Trizol reagent (Invitrogen) as described by manufacturer's protocol. Three µg total RNA was reversed transcribed (Roche). The relative abundance of IFN-β, ISG15, ISG56 and RANTES mRNAs in treated samples with respect to their mock treated counterparts was determined by real-time quantitative RT-PCR using the SYBR Green method (Roche). Briefly, a 20 µl PCR reaction containing cDNA template, the respective primers and LightCycler Fast Start SYBR Green I DNA Mastermix (Roche) was prepared and subjected to a qPCR program using the LightCycler (Roche). PCR cycling conditions comprised of an initial denaturation step at 94 °C for 10 min followed by an amplification program for 40 cycles of 30 s at 95 °C, 10 s at 55 °C, and 20 s at 72 °C with fluorescence acquisition at the end of each extension. The relative expression of each gene is calculated using the comparative ΔΔC T method, using the mock treated sample as calibrator and housekeeping gene GAPDH as internal control.
The following primer pairs were used: for GAPDH Forward 5′-GACAACTTTGGTATCTTGGAA-3′ and Reverse 5′-CCAGGAAATGAGCTTGACA-3′; for ISG56 Forward 5′-TCTCAG AGGAGCCTGGCTAAG-3′ and Reverse 5′-CCACACTGTATTTGGTGTCTAGG-3′; for ISG15 Forward 5′-TGGTGGACAAATCGCACGAA-3′ and Reverse 5′-CAGGCGCAGATTCATGAAC-3′; for RANTES Forward 5′-GGCACGCCTCGCTGTCATCCTCA-3′ Reverse 5′-CTTGATGTGGGCACGGGGCAGTG-3′; and for IFN-β Forward 5′-CTCTCCTGTTGTGCTTCTCCAC-3′ and Reverse 5′-TAGTCTCATTCCAGCCAGTGCT-3′.
3. Results
3.1. Characterization of recombinant IBV expressing SARS-CoV proteins 8b, 8ab and 8a/b
In a previous study, we reported the construction of two recombinant IBV (rIBV8b and rIBV8ab) expressing SARS-CoV proteins 8b and 8ab, respectively (Le et al., 2007). To assess the role of SARS-CoV 8b and 8ab in modulating viral replication, the growth properties and kinetics of the recombinant viruses were characterized and compared to wild type IBV (wtIBV). A new recombinant IBV expressing the 8a and 8b in separate ORFs (rIBV8a/b) was also constructed as a control. Vero cells, known to lack the expression of type I IFNs, were infected with wild type and recombinant IBV at an MOI of ~0.5 and harvested at every 4 h over a time course of 20 h for plaque assay to determine virus titers. Consistent with our previous report on the impediment of virus replication by 8b expression, recombinant viruses expressing 8b, 8ab and 8a/b replicated at a slightly slower rate, compared to wild type virus during the first 12 h of infection ( Fig. 1a & b). At 16 h post-infection, however, the three recombinant IBVs were able to attain titers comparable to that of wild type virus (Fig. 1b). Expression of protein 8b or 8ab could not be detected in cells infected with rIBV8a/b using Western blot analysis (data not shown), further supporting our previous observation that 8b is not expressed from this construct (Le et al., 2007). Taken together, these results demonstrate that the inclusion of 8b and 8ab does not render detectable enhancement effects on the replication and growth of IBV in culture cells. The slightly slower growth rates observed for the recombinant viruses at early time points of the infection cycle may be due to the introduction of extra sequences into the IBV genome. This is consistent with our previous observations that such genetic manipulations may alter the replication of IBV in cells (Le et al., 2007, Shen et al., 2009). As expression of proteins 8a, 8b and 8ab did not render growth advantages to IBV in normal cultured cells, these proteins may not have direct functions in viral replication, especially in a heterogeneous genome context.
Fig. 1.
Growth kinetics for recombinant viruses. a. Titration by plaque assay. Representative images showed that comparable numbers of plaques were formed by the respective recombinant viruses at 16 h post-infection. b. Growth curve of wild type and recombinant viruses over a time course 20 h. Plaque assays were performed with cell lysates infected with the respective recombinant viruses at an MOI of 0.5 and harvested every 4 h post-infection over a time course of 20 h. 10-fold serially diluted virus stock was then used for plaque assay to determine virus titer. Error bar shows standard deviation from 3 independent experiments.
3.2. Expression of 8b and 8ab confers growth and replication advantages to rIBV8b and rIBV8ab over wtIBV and rIBV8a/b in the presence of poly (I:C)
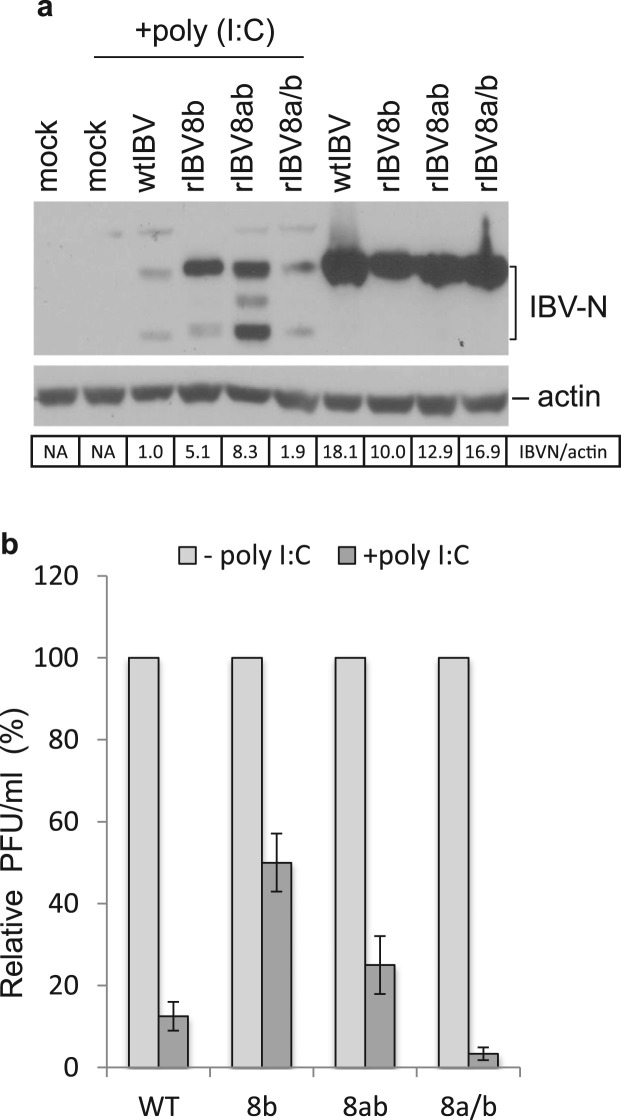
The ability to subvert the host innate immune response is a critical factor for establishing effective virus replication. To determine if 8b and 8ab may have a role in counteracting the action of IFN, one of the most common and potent host anti-viral defense mechanisms, we examined the relative replication efficacy of rIBV8b, rIBV8ab and rIBV8a/b in the presence of poly (I:C). For this purpose, H1299 cells were infected with wild type or the recombinant IBVs for 8 h prior to mock or poly (I:C) transfection. Cells were then further incubated for 10 h before lysates were harvested for analysis of viral protein expression ( Fig. 2a). In agreement with our growth kinetics studies earlier, the recombinant viruses replicated to similar levels by 18 h (Fig. 2a) in the absence of poly (I:C) treatment although the wild type virus was observed to replicate slightly faster as indicated by a higher abundance of IBV N protein (Fig. 2a).
Fig. 2.
Recombinant IBVs expressing 8b and 8ab replicate more efficiently in the presence of IFN induction by poly (I:C) treatment. a. Western blot analysis of IBV N protein expression in cells infected with wild type and recombinant IBVs in the presence and absence of IFN induction. Cells were infected with wtIBV, rIBV8b, rIBv8ab or rIBV8a/b for 8 h before transfection with poly (I:C). Whole cell lysate collected at 10 h post-poly (I:C) treatment was analyzed for IBV-N. Actin was probed as a loading control. The N protein expression efficiency was calculated as the band intensity of the protein after being normalized to the band intensity of actin, with the N protein expression from cells infected with wtIBV after poly (I:C) treatment as 1. b. Relative titers of wild type and recombinant IBV released from infected cells in the presence or absence of IFN induction. The number of infectious particles released in the supernatants was quantified using plaque assay in triplicates, and the average number of Pfu for each treatment was calculated. The relative amount of virus produced after poly (I:C) treatment was expressed as a percentage of their respective control cells not treated with poly (I:C). Error bars showed standard deviation from 3 independent experiments. Error bars showed standard deviation from 3 independent experiments.
Not surprisingly, replication of wild type and the three recombinant viruses were severely suppressed in cells stimulated by poly (I:C) (Fig. 2a), reflecting the sensitivity of coronavirus to interferon intervention. Compared to wild type and rIBV8a/b, however, rIBV8b and rIBV8ab were observed to replicate significantly better and express higher levels of N protein in cells stimulated by poly (I:C) (Fig. 2a). Using plaque assay, viral titers attained in infected cells exposed to poly (I:C) treatment were compared to their respective mock-treated counterparts. While poly (I:C)-treatment reduced the virus titers of wtIBV (from 1.1×106 to 2.75×105) and rIBV8a/b (from 1×106 to 4×104) drastically by 87.5% and 96%, respectively, titers of rIBV8b (from 8.2×105 to 4.1×105) and rIBV8ab (from 7.9×105 to 1.98×105) were reduced by a more modest 50% and 75%, respectively (Fig. 2b). These results suggest that proteins 8b and 8ab may have a functional role in modulating the IFN pathway. As expression of protein 8a (from rIBV8a/b) did not show a similar effect, we hypothesized that this effect may be unique to the 8b region of the two proteins.
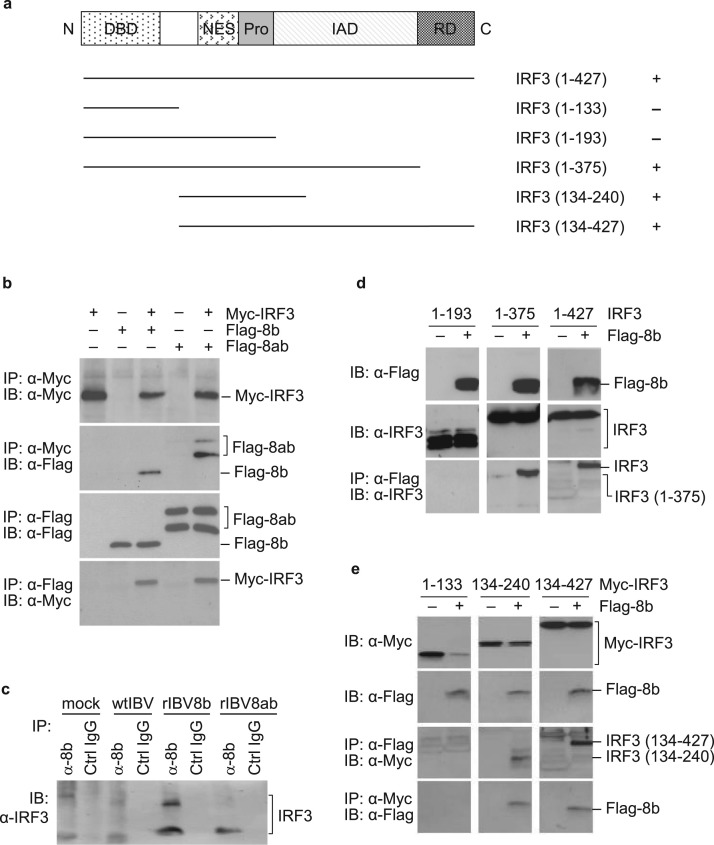
3.3. Proteins 8b and 8ab interact physically with IRF3
In view of the central role of IRF3 in regulating IFN activation during virus infection, 8b and 8ab with Flag epitope-tagged to their N-termini were co-expressed with Myc-tagged IRF3 ( Fig. 3a) in Cos-7 cells using the vaccinia/T7 expression system (Anderson et al., 1996, Lim and Liu, 2001) for co-immunoprecipitation assays to determine if there is any physical interaction between the proteins. IRF3 was precipitated from the cell lysates prepared from cells harvested at 20 h post-transfection using anti-Myc antibody-coated agarose beads, followed by Western blot analysis with antibodies against the Flag epitope. As shown in Fig. 3b, proteins 8b and 8ab were consistently pulled down with IRF3 in samples where they were co-expressed with IRF3. Similarly, when the experiment was repeated using anti-Flag coated agarose beads, IRF3 co-precipitation was detected with both proteins. These results demonstrate that IRF3 could physically interact with protein 8b and 8ab.
Fig. 3.
Interaction of proteins 8b and 8ab with IRF3. a. Schematic diagram showing the functional domains of IRF3 and various IRF3 deletion constructs used. The N-terminal DNA-binding domain (DBD), nuclear export signal (NES), proline-rich region (Pro), the IRF association domain (IAD) and the C-terminal repeat domain (RD) are shown. Fragments that immunoprecipitated with protein 8b are denoted with (+). b. Co-immunoprecipitation of Myc-tagged IRF3 with Flag-tagged 8b and 8ab, respectively. Cos7 cells were infected with the recombinant vaccinia/T7 virus at an MOI of approximately 5 per cell. After incubation for 1, cells were transfected with pMyc-IRF3, pFlag-8b, pMyc-IRF3+pFlag-8b, pFlag-8ab and pMyc-IRF3+pFlag-8ab, respectively. Cells were harvested at 18 h post-transfection, lysates prepared, and subjected to immunoprecipitation with either the Myc antibody-conjugated agarose beads (top two panels) or the Flag-antibody-conjugated beads (bottom two panels). The precipitates were separated on SDS-PAGE and analyzed by Western blot with either anti-Myc (top and bottom panels) or anti-Flag (2nd and 3rd panels) antibodies. c. Pull down of the endogenous IRF3 protein with protein 8b. H1299 cells were infected with wtIBV, rIBV8b and rIBV8ab, respectively. Cells were lysed 20 h post-infection and immunoprecipitated with either polyclonal antibodies raised against SARS-CoV 8b or control IgG antibodies. The precipitates were probed with antibodies against the full-length IRF3. d. Co-immunoprecipitation analysis of the Flag-tagged protein 8b co-expressed with the untagged IRF3 from 1–193, 1–375 and 1–427 (full-length). H1299 cells were infected with the recombinant vaccinia/T7 virus at an MOI of approximately 5 per cell. Immunoprecipitation was performed as described above. Total lysates and the precipitates were separated on SDS-PAGE and analyzed by Western blot with either anti-Myc or anti-Flag antibodies. e. Co-immunoprecipitation analysis of the Flag-tagged protein 8b co-expressed with the Myc-tagged IRF3 from 1–133, 134–240 and 134–427. H1299 cells were infected with the recombinant vaccinia/T7 virus at an MOI of approximately 5 per cell. Immunoprecipitation was performed as described above. Total lysates and the precipitates were separated on SDS-PAGE and analyzed by Western blot with either anti-Myc or anti-Flag antibodies.
To confirm that this interaction occurs in the context of coronavirus infection, total lysates prepared from cells infected with wtIBV, rIBV8b and rIBV8ab together with mock infected controls were also subjected to immunoprecipitation with either polyclonal antibodies raised against SARS-CoV 8b or IgG controls. Immunoprecipitated proteins were then analyzed for the presence of endogenous IRF3 using antisera against the protein. The detection of IRF3 in cells infected with rIBV8b and rIBV8ab, but not in mock- and wtIBV-infected cells (Fig. 3c) indicates that protein 8b and 8ab formed complexes with IRF3 during the virus replication process. It was noted that apart from the 55 kDa band corresponding to the endogenous monomeric IRF3, an additional band with the apparent molecular weight of approximately 85 kDa was detected (Fig. 3c) in cells infected by rIBV-8b. The identity of this band is not certain, but it may represent a modified form of IRF3. IRF3 is well documented to be heavily targeted for post-translational modifications such as phosphorylation, ubiquitination, SUMOylation, and neddylation (Bibeau-Poirier et al., 2006, Hiscott, 2007, Kubota et al., 2008, Ran et al., 2011).
Since proteins 8b and 8ab exhibited comparable efficiency in terms of pulling down IRF3, just the 8b region was then used for subsequent pull-down experiments to pinpoint the domains in IRF3 essential for the interaction. Four deletion constructs of IRF3 either with or without a Myc-tag at the N-termini were constructed (Fig. 3a) and co-expressed with the Flag-tagged 8b. Co-immunoprecipitation experiments showed that the N-terminal region covering the first 133 residues of IRF3 (Myc-IRF3(1-133)) failed to interact with protein 8b (Fig. 3e), suggesting that the N terminal DNA-binding domain (DBD) is dispensable for the interaction. Immunoprecipitation with fragments covering residues 134–240 and 134–427, respectively, resulted in effective pulldown of 8b (Fig. 3e), suggesting that the 134–240 region is likely to be important for the interaction. This region spans across several functional domains, including the nuclear export signal (NES), proline-rich region (Pro), and the first 47 residues of the IRF association domain (IAD). Interestingly, immunoprecipitation experiments repeated with just the N terminal fragment comprising of only the first 193 residues, thus excluding the 47 residues of IAD, abolished the binding (Fig. 3d), indicating that just the NES and Pro regions alone were insufficient for the binding. Consistently, the full-length IRF3 with the N-terminal Myc-tag and the N-terminal region covering the first 375 residues could be efficiently pulled down by protein 8b (Fig. 3d).
3.4. Proteins 8b and 8ab reduce poly (I:C)-induced IRF3 and IFN activation
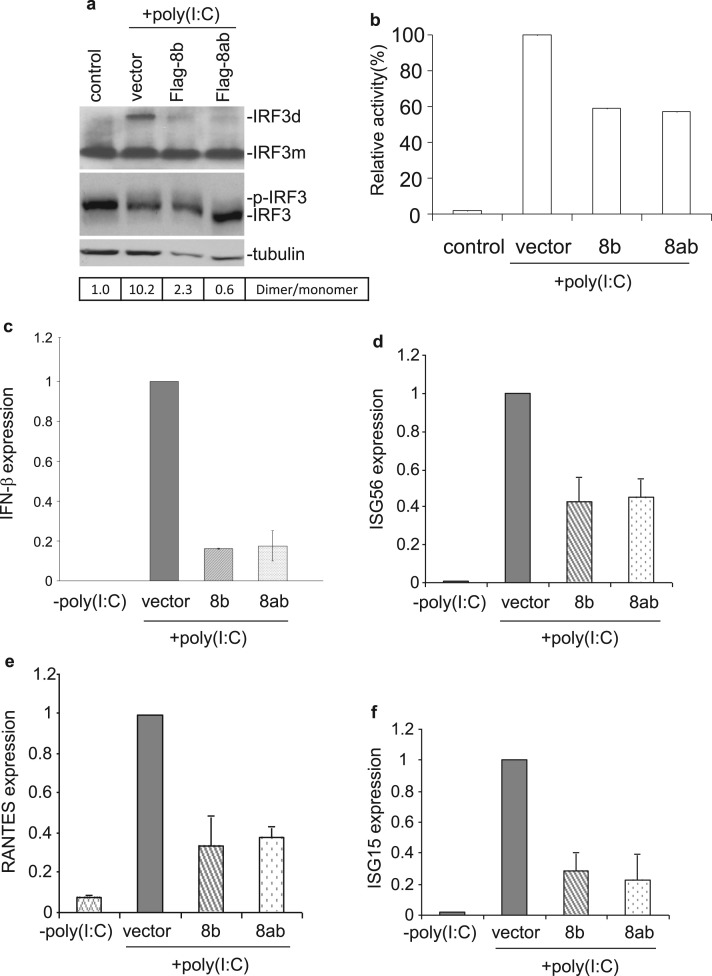
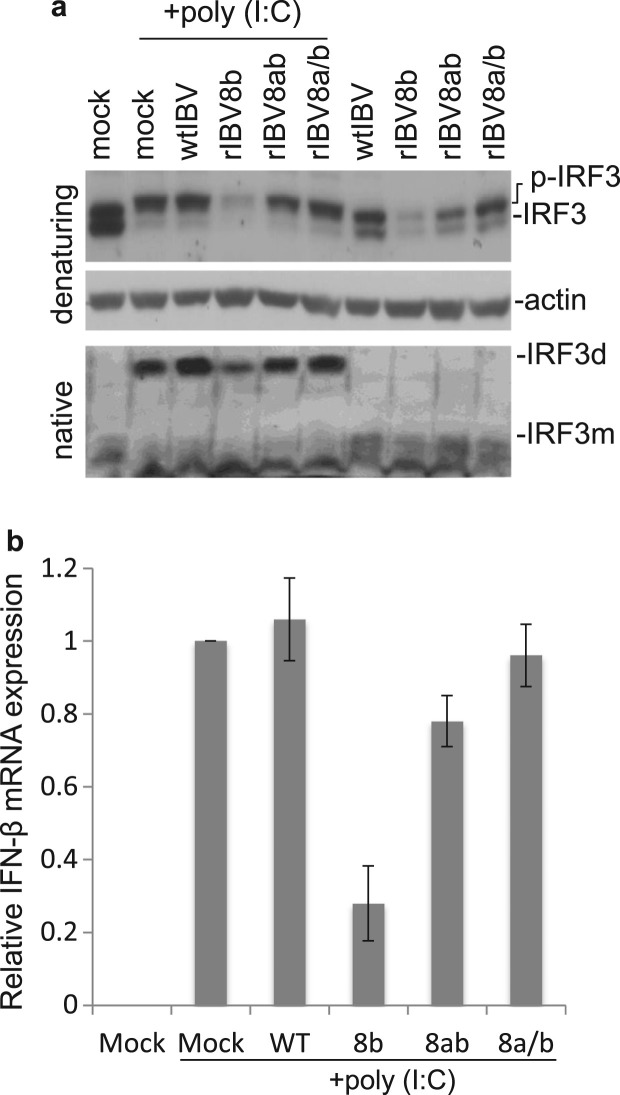
We next sought to test the effect of 8b and 8ab expression on IRF3 activation. Cells were transfected with either plasmid encoding Flag-8b, Flag-8ab or a corresponding control vector, before subjected to poly (I:C) treatment. Western blot analysis coupled with native PAGE revealed that the ectopic expression of proteins 8b and 8ab resulted in markedly decreased levels of poly (I:C)-induced IRF3 dimerization ( Fig. 4a). However, analysis of the same samples by SDS-PAGE showed no observable difference in the levels of hyper-phosphorylated IRF3 (p-IRF3) (appearing as more slowly migrating bands) in cells over-expressing proteins 8b and 8ab, compared to that in the control (Fig. 4a). It was also noted that the total amounts of IRF3 were approximately comparable in these transfected cells (Fig. 4a).
Fig. 4.
Suppression of poly (I:C)-induced IRF3 activation by protein 8b and 8ab. a. Suppression of IRF3 dimerization by proteins 8b and 8ab. Huh7 cells were transfected with 4 μg of empty vector, Flag-8b and Flag8ab, respectively, followed by stimulation with poly(I:C) for 20 h. Whole cell lysates were subjected to either native PAGE or SDS-PAGE and probed with anti-IRF3. Tubulin was included as a loading control. The ratio of dimeric IRF3 to monomeric IRF3 was calculated as the band intensity of monomer divided by the band intensity of dimer. b. Suppression of poly (I:C)-induced IFN-β promoter activity by proteins 8b and 8ab. Huh7 cells were transfected with control vector pcDNA3.1, pcDNA-8b and pcDNA-8ab, respectively, together with a luciferase reporter construct under the control of IFN-β promoter. At 24 h post-transfection, cells were then further transfected with poly (I:C). At 24 h post-stimulation, cells were lysed and measured for the firefly luciferase activity. pRL-TK was also co-transfected to serve as an internal control. Data were represented as mean of triplicates from 3 independent experiments. c-f. Huh7 cells were transfected with 4 μg of empty vector, Flag-8b and Flag8ab, respectively, followed by stimulation with poly(I:C) for 20 h. Total RNA was then extracted for quantitative real-time RT-PCR with specific primers for IFN−β (c), ISG56 (d), RANTES (e) and ISG15 (f). The expression of each gene was expressed relative to their respective control sample transfected with empty vector. Data were represented as mean of replicates from 2 independent experiments. GAPDH was used as internal control.
The concomitant decreased in IRF3 dimerization with 8b and 8ab expression lead us to examine the impact of such phenomenon on the activation of IFN-β and other known IRF3 downstream effectors. For the study on the effect on IFN-β promoter activity, a reporter construct expressing firefly luciferase driven by the IFN-β promoter was co-transfected with either 8b, 8ab or a control plasmid prior to poly (I:C) stimulation. At 16 h post poly (I:C) treatment, lysates were assayed for luciferase activity. Relative to cells transfected with control plasmid, poly (I:C)-induced IFN-β promoter activation in cells expressing 8b and 8ab were reduced to 59% and 57% respectively (Fig. 4b). This data mirrored the results obtained from the analysis of endogenous IFN-β transcript levels examined by quantitative real-time RT-PCR where relative levels of IFN-β mRNA in poly (I:C)-treated cells expressing 8b and 8ab were reduced to approximately 18–20% (Fig. 4c) of control. Similar to the effect on IFN-β expression, the mRNA levels of other IRF3 downstream target genes (Grandvaux et al., 2002), including ISG56, RANTES and ISG15, were significantly lower in cells over-expressing proteins 8b and 8ab than those of the control. The relative levels of ISG56 (Fig. 4d), RANTES (Fig. 4e) and ISG15 (Fig. 4f) were reduced to approximately 40–41, 35–40 and 25–30%, respectively, in poly (I:C)-treated cells transfected with 8b and 8ab, compared to those in cells transfected with the empty vector.
3.5. Induction of rapid degradation of IRF3 by SARS-CoV proteins 8b and 8ab
The negative modulation of 8b and 8ab expression on the activation of IRF3 was further verified from the analysis of IRF3 activation and stability in virus-infected cells shown in Fig. 2a. In the absence of poly (I:C) treatment, infection by wild type and all the three recombinant viruses did not induce a detectable level of IRF3 dimerization using native PAGE analysis ( Fig. 5a). While IRF3 dimerization was detected in all infected cells in the presence of poly (I:C) treatment (Fig. 5a), less poly (I:C)-induced IRF3 dimers could be detected in cells infected with rIBV-8b, compared to those infected by wtIBV (Fig. 5a). Reduction in IRF3 dimerization, although to a lesser extent, was also observed in cells infected with rIBV8ab (Fig. 5a). Interestingly, analysis of the same lysates by denaturing SDS-PAGE revealed that hyper-phosphorylated IRF3 was invariantly detected in the poly (I:C) treated cells regardless of whether they are mock infected, infected with wild type virus or the recombinant IBVs (Fig. 5a). These observations were in agreement with the data we described in Fig. 4a. However, we did observe that the overall level of IRF3 expression was significantly reduced in cells infected with rIBV8b, both in the presence and absence of poly (I:C) treatment (Fig. 5a). A more moderate reduction of IRF3 was also detected in cells infected with rIBV8ab (Fig. 5a).
Fig. 5.
Rapid degradation of IRF3 in cells infected with recombinant IBV expressing proteins 8b and 8ab. a. Western blot analysis of IRF3 in cells infected with wild type and recombinant IBV in the presence and absence of IFN induction. Cells were infected with wtIBV, rIBV8b, rIBv8ab and rIBV8a/b for 8 h before transfection with poly (I:C). Whole cell lysate collected 10 h post-poly (I:C) treatment was analyzed for IRF3 in denaturing SDS-PAGE. Actin was probed as a loading control. IRF3 dimerization in the same samples was detected by native PAGE. b. Relative expression of IFN-β mRNA in poly (I:C)-treated cells infected with recombinant viruses with respect to the wild type virus, was analyzed using quantitative real-time RT-PCR. Experiments were performed in triplicates. Data were represented as mean of replicates from 2 independent experiments. GAPDH was used as an internal control.
When the mRNA levels of IFN-β from these samples were analyzed, negligible IFN-β induction was observed in virus-infected cells in the absence of poly (I:C) stimulation (data not shown). Among the samples treated with poly (I:C), IFN-β induction in cells infected with rIBV8b and rIBV8ab infection was suppressed (25% and 75% relative to control wtIBV, respectively) (Fig. 5b). Taken together, these data suggest that the observed enhanced replication of rIBV8b and rIBV8ab in the presence of poly (I:C) may be due to the diminished IFN activation owing to the down-regulation of IRF3 levels coupled to reduced IRF3 dimer formation and IFN-β induction.
3.6. Degradation of IRF3 mediated by proteins 8b and 8ab is ubiquitin-dependent
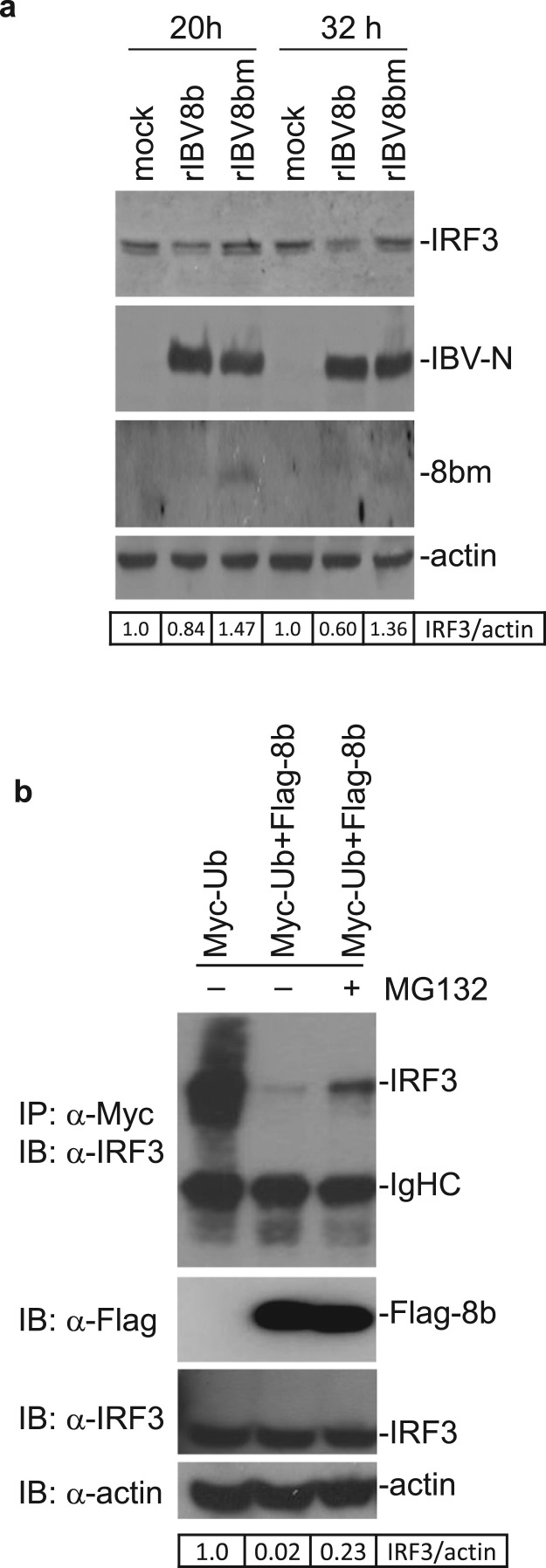
We had previously demonstrated that 8b binds to both poly- and mono-ubiquitin (Le et al., 2007). To address if the down-regulation of IRF3 in rIBV8b-infected cells is linked to its ubiquitin-binding activities, lysine knockout mutant of 8b, 8bm was generated by mutating all three lysine residues (K4, K26 and K81) in 8b to arginine. Mutation of these lysine residues enhanced the stability of the 8b protein. As shown in Fig. 6a, while the mutant 8bm protein was readily detected in rIBV8bm-infected cells at 20 and 32 h post-infection, protein 8b was not detected under the same conditions (Fig. 6a). This is consistent with previous reports of the instability of 8b protein and that protein 8b could only be detected in rIBV8b-infected cells in the presence of proteasome inhibitors (Le et al., 2007). IRF3 was down-regulated in cells infected with rIBV8b at both 20 and 32 h post-infection (Fig. 6a). Infection of cells with rIBV-8bm did not result in similar reductions in IRF3 protein levels, despite a similar replication efficiency of the two recombinant viruses (Fig. 6a).
Fig. 6.
Degradation of IRF3 by 8b in a ubiquitin/proteasome-dependent manner. a. Degradation of IRF3 by 8b but not by a lysine-knockout mutant 8b. A lysine knockout mutant of rIBV, rIBV8bm, was created by substituting the three lysine residues (K4, K26 and K81) with arginine. Cells were then infected up to 32 h with rIBV-8b and rIBV8bm, respectively, before analyzing for IRF3 expression. The same membrane was also probed with anti-IBV N, SARS-CoV protein 8b and actin antibodies. The relative amount of IRF3 was calculated as the band intensity of the protein divided by the band intensity of actin. b. Ubiquitin-dependent degradation of IRF3. Cells transfected with either Myc-tagged ubiquitin alone or co-transfected with Flag-tagged 8b, in the presence or absence of MG132, were subjected to immunoprecipitation with antibodies against Myc. Immunoprecipitates were then probed for pull-down of endogenous IRF3 with specific antibodies. The relative amount of IRF3 was calculated as the band intensity of the protein divided by the band intensity of actin.
As mentioned previously, IRF3 levels are regulated by ubiquitination during virus infection. However, some viruses exploit this mode of regulation by expressing viral proteins that promote untimely proteasomal degradation of IRF3 (Chen et al., 2007, Saira et al., 2009, Sen et al., 2009). To affirm the involvement of the ubiquitin-proteasome pathway in the down-regulation of IRF3 observed with 8b, IRF3 stability was studied in cells overexpressing protein 8b in the presence of ubiquitin. Over-expression of 8b together with ubiquitin resulted in a reduction of detectable levels of IRF3 compared to cells expressing ubiquitin alone (Fig. 6b). This degradation was partially suppressed in the presence of proteasome inhibitor MG132 (Fig. 6b). In a previous study, we found that proteasome-mediated rapid degradation of protein 8b could be much more efficiently inhibited by NLVS than did lactacystin (Le et al., 2007). As the supply of NLVS was discontinued, we chose to use MG132, a product shown similarly mild inhibitory effect as lactacystin in the suppression of proteasome-mediated degradation of protein 8b, in this study.
3.7. Suppression of IRF3-5D-induced interferon response by proteins 8b and 8ab
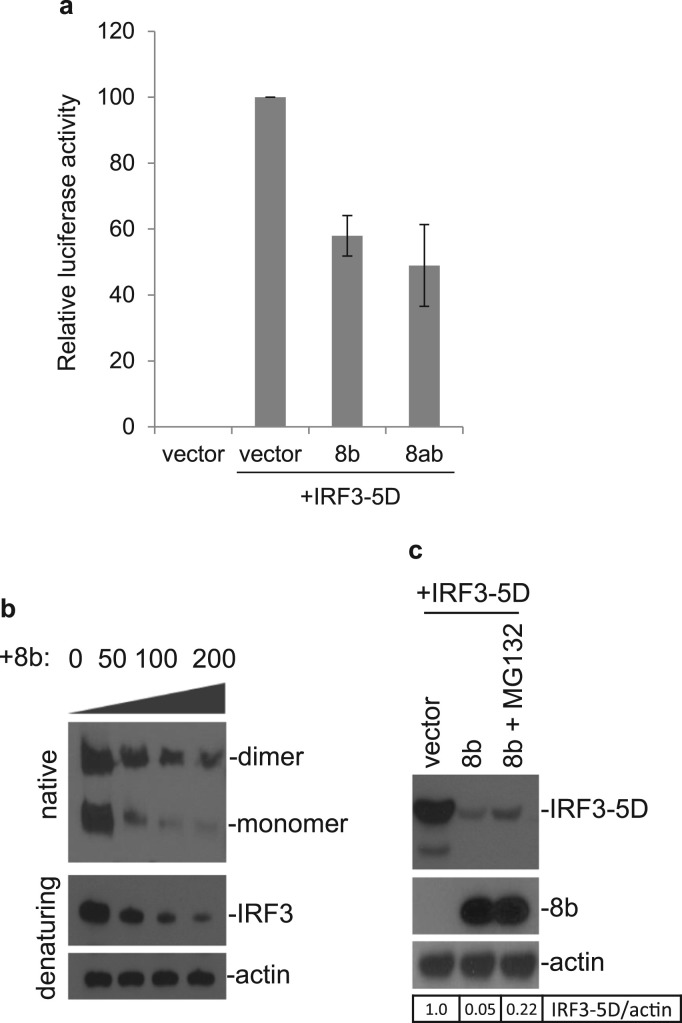
Finally, to address the observation of the negligible impact exerted by 8b and 8ab on IRF3 phosphorylation despite their capacity to suppress IRF3 dimer formation, we study the effect of 8b on activated IRF3. A construct expressing the phosphomimetic form of IRF3 (IRF3-5D) was constructed, which contains amino acid substitutions at positions 396, 398, 402, 404, and 405 by the phosphomimetic aspartic acid. IRF3-5D undergoes spontaneous dimerization leading to IFN-β induction (Grandvaux et al., 2002, Lin et al., 1999). Over-expression of proteins 8b and 8ab was able to reduce the IRF3-5D-induced IFN-β promoter activity to approximately 58% and 49% to that of control, respectively ( Fig. 7a). This suggests that 8b can act on IRF3 at step(s) that is downstream of its activation. When protein 8b is co-expressed with IRF3-5D, 8b reduces the expression of IRF3-5D in a dose-dependent manner (Fig. 7b). For reasons yet to be known, we observed that the expression of a monomeric form of IRF3-5D seemed to be more affected by the presence of 8b than its homodimeric counterpart. Similar to results presented earlier with endogenous IRF3, IRF3-5D levels was partially rescued with the addition of MG132, indicating the role of the ubiquitin-proteasome pathway (Fig. 7c).
Fig. 7.
Suppression of IRF3-5D-induced IFN-β expression by proteins 8b and 8ab. a. Control vector pcDNA3.1, pcDNA-8b or pcDNA-8ab together with a luciferase reporter construct under the control of the IFN- β promoter were transfected into Huh7 cells. At 24 h post-transfection, cells were transfected with pIRF3-5D and co-transfected with pRL-TK to serve as an internal control. At 24 h post-stimulation, cells were lysed and measured for the firefly luciferase activity. Data were represented as mean of triplicates from 3 independent experiments. b. Cells were co-transfected with 200 ng of pxj40-IRF3-5D and either 0, 50, 100, 200 ng of pKTO-8b. The total DNA transfected was made up to 400 ng using empty pKTO vector. Lysates harvested 20 h post-transfection were either subjected to native or SDS PAGE and probed with anti-IRF3 antibodies. Actin was also probed to serve as a loading control. c. IRF3-5D was co-transfected pKTO-8b. 20 h post transfection, co-transfected cells was either left untreated or treated with MG132 2 h prior to harvest. Lysates were then probed for IRF3, 8b and actin expression. The relative amount of IRF3-5D was calculated as the band intensity of the protein divided by the band intensity of actin.
4. Discussion
The SARS-CoV genome encodes an exceptionally high number of accessory genes that bear little resemblance to other known coronavirus accessory proteins. It is believed that while these unique proteins may not participate directly in viral replication, they possess biological functions that may enhance SARS-CoV pathogenesis in cells. By demonstrating that recombinant IBV expressing 8b or 8ab replicates more efficiently in the background of IFN activation, data presented in this study demonstrate that expression of SARS-CoV ORF8b and ORF8ab contributes positively to viral pathogenesis.
As the expression of 8b helped to overcome partially the potent inhibitory effect of IFN induction on coronavirus replication in cell culture, it suggests that 8b and 8ab are novel IFN antagonists. ORF8 failed to show up in the screen for interferon antagonist in a previous study (Kopecky-Bromberg et al., 2007), likely due to the incomplete inhibition of IFN activation exhibited by ORF8. Other reasons could include the low expression of 8b in their system owing to the inherent instability of the protein (Le et al., 2007), and/or that the antagonistic activity of 8b is not apparent in their NDV model because the IFN antagonistic activity of this protein may be specific to coronavirus infection due to the requirement of other viral proteins. However, this is quite unlikely because the inhibitory effect could also be observed with ectopic expression of 8b and 8ab in the absence of viral replication.
The other significant finding from this study is that 8b and 8ab directly physically interact with IRF3 and that 8b-IRF3 interaction involves part of the IAD domain that is responsible for the formation of IRF3 homodimers. Over-expression of 8b and 8ab appears to have a more profound effect on IRF3 dimerization than on its phosphorylation status. Although it is still unclear whether this is a consequence of assay insensitivity or that the inhibitory actions of 8b and 8ab bypass the step of phosphorylation and targets specifically on the event of IRF3 dimerization, we are inclined to believe that it is the latter owing to the ability of 8b to suppress IFN-β induced by constitutively active phosphomimetic IRF3-5D. While disruption to the dimerization event as a result of direct steric interference brought about by 8b interaction at the IAD domain is an attractive model, the exact mechanisms can only be ascertained through more in-depth studies such as structural analysis.
We previously reported that the 8b region of SARS-CoV proteins 8b and 8ab consists of domains that allow for ubiquitin binding, ubiquitination and glycosylation (Le et al., 2007). Based on these finding, we proposed that the 8b region may mediate the binding of 8b and 8ab to ubiquitinated cellular proteins, such as p53 and IκBα (Le et al., 2007). Here we showed that IRF3, another protein regulated by ubiquitination (Siu et al., 2009, Spiegel et al., 2004), interacts with proteins 8b and 8ab, suggesting that the ubiquitin-binding properties of 8b region could allow them to interact with multiple cellular proteins. It would be interesting to find out what other cellular targets bind to 8b and 8ab and whether they have a regulatory role during SARS-CoV infection. Furthermore, expression of 8b and 8ab appears to regulate the stability and function of IRF3 by promoting degradation of IRF3 in a ubiquitin/proteasome-dependent manner. Several viral proteins have been reported to cause proteasomal degradation of IRF3 (Chen et al., 2007, Saira et al., 2009, Sen et al., 2009). IRF3 degradation is typically triggered post-infection, when viral infection-induced IRF3 activation leads to the ubiquitination of the protein targeting it for proteasomal degradation (Higgs and Jefferies, 2008, Liu et al., 2005). This serves to regulate type I IFN production as excessive IFN is detrimental to the cells. In this study, we observed IRF3 degradation even in the absence of strong IRF3 activation in cells infected with rIBV8b and rIBV8ab, suggesting that the 8b- and 8ab-mediated IRF3 degradation is not the result of a typical negative feedback mechanism to bring the IFN level back to the physiological level at the end of viral infection as observed with other virus infection, but may be an active step undertaken by the virus to limit IRF3 activation during its course of replication.
At this stage, while we confirm the involvement of the proteasome, we do not know if other cellular factors are also recruited by 8b to mediate IRF3 degradation. Cellular factors, such as peptidyl-prolyl-isomerase Pin1 (Saitoh et al., 2006), Ro52/TRIM21 (Pin + Ro52) (Higgs et al., 2008) and E3 ubiquitin ligase RBCC protein interacting with PKC1 (RBCK1) (Zhang et al., 2008), were identified to participate in the negative regulation of IRF3 by targeting it for ubiquitination. Additional experiments such as mass spectrometry could perhaps help elucidate if 8b interaction with IRF3 also involves any of these reported proteins.
Considering the fact that 8b expression is unstable and that it is only expressed during the late stages of SARS-CoV infection, it seems counter-intuitive why the virus would express such a late stage IFN antagonist as 8b. This is especially true since SARS-CoV has a strong inhibitory effect on IFN induction plausibly owing to the expression of multiple viral proteins that antagonize the pathway in myriad ways during the earlier stages of infection. A possible explanation may come from a study carried out by Spiegel and coworkers who reported that while nuclear translocation of IRF3 remains unabated by SARS-CoV infection during the early stages of infection at 8 h post-infection, IRF3 activation is specifically blocked during later stages of SARS-CoV infection at 16 h post-infection (Spiegel et al., 2005). This coincides with the late expression of 8b during SARS-CoV infection (Keng et al., 2006). Hence, we hypothesize that the growth advantage conferred by the expression of 8b in recombinant IBV was due to the role of 8b in late-stage viral pathogenesis, when the expression of 8b aids in dampening the activation of IRF3 that may occur during the later phase of infection. Nevertheless, we do not rule out the possibility that 8b and 8ab may regulate IRF3 via mechanisms independent of its ubiquitin-binding activity as it has also been shown to down-regulate E protein via a ubiquitin-independent proteasomal pathway (Keng et al., 2006, Keng et al., 2011).
Using an infectious clone system based on the Urbani strain of SARS-CoV, the 29-nt deletion is inserted to fuse ORF8a/b back into the single ORF8. Compared with the wild type control, this recombinant virus replicates similarly in both cell culture and in the murine model (Yount et al., 2005). Theoretically, only protein 8ab is produced in cells infected with this virus, whereas both proteins 8a and 8b are produced in the wild type control. Because both 8b and 8ab can antagonize IFN signaling by mediating IRF3 down-regulation, it is no surprise that the recombinant virus replicates similarly as the wild type control.
Deletion of accessory proteins 6, 7, 7b, 8a, 8b and 9b altogether in recombinant virus rSARS-CoV-Δ[6-9b] (Dediego et al., 2008) showed that the recombinant virus replicates as well as wild type control in both cell culture and transgenic mice expressing the SARS-CoV receptor human angiotensin converting enzyme-2 (hACE-2). Since IFN antagonist function is also possessed by other SARS-CoV proteins (such as nsp1, PLpro, M and N), it is possible that loss of functional 8b in rSARS-CoV-Δ[6-9b] is compensated, and thus the recombinant virus is not attenuated in vivo. Further studies using recombinant SARS-CoV with only 8b or 8ab deleted should be performed in cell culture and in appropriate animal models, to better characterize the detailed mechanisms of their involvement in modulating viral replication and pathogenesis.
Finally, Poly (I:C) is known to be able to induce both IFN and a subset of IFN-stimulated genes through activation of IRF3 in the absence of IFN. We believe that the observed antiviral effects of poly (I:C) on wild type and recombinant IBV in this study would be the combined action of poly (I:C)-induced IFN-stimulated genes either dependent upon or independent of IFN induction.
Acknowledgements
This work was partially supported by an Academic Research Fund (AcRF) Tier 2 grant (ACR47/14), Ministry of Education, Singapore and by Guangdong Province Key Laboratory of Microbial Signals and Disease Control grants MSDC-2017-05 and MSDC-2017-06, Guangdong, People's Republic of China.
References
- Anderson R.A., Liu D.X., Gompels U.A. Definition of a human herpesvirus-6 betaherpesvirus-specific domain in glycoprotein gH that governs interaction with glycoprotein gL: substitution of human cytomegalovirus glycoproteins permits group-specific complex formation. Virology. 1996;217:517–526. doi: 10.1006/viro.1996.0146. [DOI] [PubMed] [Google Scholar]
- Bibeau-Poirier A., Gravel S.-P., Clément J.-F., Rolland S., Rodier G., Coulombe P., Hiscott J., Grandvaux N., Meloche S., Servant M.J. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 2006;1950(177):5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Ping Y.H., Lee H.C., Chen K.H., Lee Y.M., Chan Y.J., Lien T.C., Jap T.S., Lin C.H., Kao L.S., Chen Y.M.A. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Rijnbrand R., Jangra R.K., Devaraj S.G., Qu L., Ma Y., Lemon S.M., Li K. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology. 2007;366:277–292. doi: 10.1016/j.virol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.-T.K., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Chen B., Tay F.P.L., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.-M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Didcock L., Randall R.E. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Grandvaux N., Servant M.J., tenOever B., Sen G.C., Balachandran S., Barber G.N., Lin R., Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., Luo S., Li P., Zhang L., Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Higgs R., Jefferies C.A. Targeting IRFs by ubiquitination: regulating antiviral responses. Biochem. Soc. Trans. 2008;36:453–458. doi: 10.1042/BST0360453. [DOI] [PubMed] [Google Scholar]
- Higgs R., Ní Gabhann J., Ben Larbi N., Breen E.P., Fitzgerald K.A., Jefferies C.A. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 2008;1950(181):1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castaño-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for thE Design of A Genetically Stable Vaccine. PLoS Pathog. 2015;11:e1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.-T., Akerström S., Leung C.S.-W., Poon L.L.M., Peiris J.S.M., Mirazimi A., Tan Y.-J. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. Microbes Infect. 2011;13:179–188. doi: 10.1016/j.micinf.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.-T., Choi Y.-W., Welkers M.R.A., Chan D.Z.L., Shen S., Gee Lim S., Hong W., Tan Y.-J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T., Matsuoka M., Chang T.-H., Tailor P., Sasaki T., Tashiro M., Kato A., Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Takeuchi O., Akira S. Pathogen recognition by innate receptors. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. Pathogen recognition in the innate immune response. Biochem. J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Feng Y., Chen H., Luk H.K.H., Yang W.-H., Li K.S.M., Zhang Y.-Z., Huang Y., Song Z.-Z., Chow W.-N., Fan R.Y.Y., Ahmed S.S., Yeung H.C., Lam C.S.F., Cai J.-P., Wong S.S.Y., Chan J.F.W., Yuen K.-Y., Zhang H.-L., Woo P.C.Y. Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.Y.P., Liu Y.-M., Geng H., Kwan K.H., Waye M.M.-Y., Ho Y.-Y. Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus. FEBS Lett. 2006;580:3643–3648. doi: 10.1016/j.febslet.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.M., Wong H.H., Tay F.P.L., Fang S., Keng C.-T., Tan Y.J., Liu D.X. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007;274:4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. The missing link in coronavirus assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016:4. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Pitha P.M., Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Mamane Y., Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C., Penninger J., Karin M. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 2005;5:941–952. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui P.-Y., Wong L.-Y.R., Fung C.-L., Siu K.-L., Yeung M.-L., Yuen K.-S., Chan C.-P., Woo P.C.-Y., Yuen K.-Y., Jin D.-Y. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microbes Infect. 2016;5:e39. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Matthews K., Schäfer A., Pham A., Frieman M. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol. J. 2014;11:209. doi: 10.1186/s12985-014-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K., Yoneyama M., Fujita T. Regulation of antiviral innate immune responses by RIG-I family of RNA helicases. Curr. Top. Microbiol. Immunol. 2007;316:193–205. doi: 10.1007/978-3-540-71329-6_10. [DOI] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A.M., Rottier P.J.M. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y., Liu T.-T., Zhou Q., Li S., Mao A.-P., Li Y., Liu L.-J., Cheng J.-K., Shu H.-B. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J. Mol. Cell Biol. 2011;3:283–292. doi: 10.1093/jmcb/mjr020. [DOI] [PubMed] [Google Scholar]
- Saira K., Zhou Y., Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) associates with interferon regulatory factor 7 and consequently inhibits beta interferon promoter activity. J. Virol. 2009;83:3977–3981. doi: 10.1128/JVI.02400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K.P., Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- Sen A., Feng N., Ettayebi K., Hardy M.E., Greenberg H.B. IRF3 inhibition by rotavirus NSP1 is host cell and virus strain dependent but independent of NSP1 proteasomal degradation. J. Virol. 2009;83:10322–10335. doi: 10.1128/JVI.01186-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Fang S.G., Chen B., Chen G., Tay F.P.L., Liu D.X. Towards construction of viral vectors based on avian coronavirus infectious bronchitis virus for gene delivery and vaccine development. J. Virol. Methods. 2009;160:48–56. doi: 10.1016/j.jviromet.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Kok K.-H., Ng M.-H.J., Poon V.K.M., Yuen K.-Y., Zheng B.-J., Jin D.-Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Yeung M.L., Kok K.-H., Yuen K.-S., Kew C., Lui P.-Y., Chan C.-P., Tse H., Woo P.C.Y., Yuen K.-Y., Jin D.-Y. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Mühlberger E., Haller O., Weber F. The antiviral effect of interferon-beta against SARS-coronavirus is not mediated by MxA protein. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004;30:211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Suhara W., Yoneyama M., Kitabayashi I., Fujita T. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 2002;277:22304–22313. doi: 10.1074/jbc.M200192200. [DOI] [PubMed] [Google Scholar]
- Sung S.C., Chao C.Y., Jeng K.S., Yang J.Y., Lai M. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Versteeg G.A., Bredenbeek P.J., van den Worm S.H.E., Spaan W.J.M. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS One. 2011;6:e17192. doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Kochs G., Haller O., Staeheli P. Viral evasion of the interferon system: old viruses, new tricks. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2003;23:209–213. doi: 10.1089/107999003765027410. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Tian Y., Wang R.-P., Gao D., Zhang Y., Diao F.-C., Chen D.-Y., Zhai Z.-H., Shu H.-B. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., He M.-L., Wong K.-L., Lum C.T., Poon L.L.M., Peng Y., Guan Y., Lin M.C.M., Kung H.-F. Potent inhibition of SARS-associated coronavirus (SCOV) infection and replication by type I interferons (IFN-alpha/beta) but not by type II interferon (IFN-gamma) J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2004;24:388–390. doi: 10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Tan Y.W., Liu D.X. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses. 2012;4:980–1010. doi: 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]