Abstract
Spike (S) glycoprotein on the viral envelope is the main determinant of infectivity. The S protein of coronavirus infectious bronchitis virus (IBV) contains 29 putative asparagine(N)-linked glycosylation sites. These post-translational modifications may assist in protein folding and play important roles in the functionality of S protein. In this study, we used bioinformatics tools to predict N-linked glycosylation sites and to analyze their distribution in IBV strains and variants. Among these sites, 8 sites were confirmed in the S protein extracted from partially purified virus particles by proteomics approaches. N-D and N-Q substitutions at 13 predicted sites were introduced into an infectious clone system. The impact on S protein-mediated cell-cell fusion, viral recovery and infectivity was assessed, leading to the identification of sites essential for the functions of IBV S protein. Further characterization of these and other uncharacterized sites may reveal novel aspects of N-linked glycosylation in coronavirus replication and pathogenesis.
Keywords: Coronavirus, Infectious bronchitis virus, Virus infectivity, Spike protein, N-linked glycosylation, Cell-cell fusion, Infectious cDNA, Clone
1. Introduction
Coronaviruses are positive stranded RNA viruses. A typical coronavirus consists of few basic structural components. These include the membrane (M), peplomer-like protein spike (S) and envelope (E) protein on the viral envelope, and the nucleocapsid (N) protein which wraps the genomic RNA inside the particles. Some coronaviruses encode an additional protein, the hemagglutinin-esterase (HE), a glycoprotein that forms smaller spikes on the exterior in addition to the S proteins.
Infectious bronchitis virus (IBV) is the coronavirus that plagues the domestic fowl Gallus gallus. Similar to other coronavirus S protein, IBV S protein is a type I glycoprotein and forms the peplomers on virion particles giving the crown-like appearance. The protein contains two glycopolypeptides S1 (90 kDa) and S2 (84 kDa) in equimolar proportions (Cavanagh, 1983) ( Fig. 1a). The S1 subunit is believed to form the globular head of the protein and contains a receptor binding domain (Kubo et al., 1994). The carboxy terminal S2 subunit, however, is conserved among all coronavirus spikes and forms a stalk-like structure that is embedded in the membrane (Masters, 2006). Overall this gives the spike protein a teardrop shaped structure (Masters, 2006). Mutagenesis of the terminal heptad repeats and the predicted fusion peptides severely compromises SARS-CoV S protein-mediated cell-cell fusion (Petit et al., 2005). S protein-mediated cell-cell fusion is also dependent on a cysteine rich domain in the protein itself (Chang et al., 2000). Yet another point mutation, glutamine to leucine at position 294 of the IBV spike S1 subunit hampers processing of the protein into a matured protein capable of being translocated to the cell surface (Shen et al., 2004).
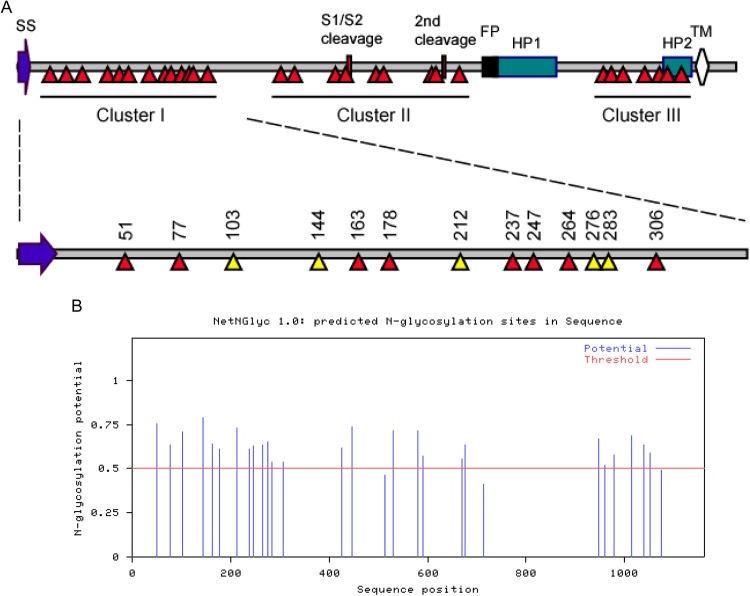
Fig. 1.
a Diagram showing the IBV spike protein with different functional domains indicated. Signal sequence(SS), amino acids 1–18; S1, amino acids 19–537; S2, amino acids 538–1162; Heptad Repeat 1 (HP1), amino acids 790–911; Heptad Repeat 2 (HP2), amino acids 1056–1089; Trans-membrane domain (TM), amino acids 1097–1118. Also indicated are the putative N-linked glycosylation sites in three clusters, and amino acid positions of the N-linked glycosylation sites in Cluster I. The relative importance of these N-linked glycosylation sites in Cluster I is indicated with colored triangles, with red indicating less importance and yellow indicating critical importance. b The 29 putative glycosylation sites on the IBV spike protein as predicted by NetNGlyc 1.0 software. The threshold and glycosylation potential are shown.
One aspect of the S protein that remains largely unexplored is the role of its glycans. Glycans are mainly involved in protein post-translational modification and folding. One of its most common forms is the N-linked glycosylation. This involves a high mannose core being attached to the amide nitrogen of asparagine (N), within a conserved motif Asn-X-Ser/Thr (where X is any amino acid except for proline). In the ER, this mannose core is added in the form of a block of fourteen sugars, Glc3Man9GlcNAc2 (Balzarini, 2007). The mannose oligosaccharide then moves through the ER and Golgi apparatus, during which it is altered to form different structures (Vigerust and Shepherd, 2007). Coronavirus S proteins typically contain 23–30 N-linked glycosylation sites, depending on the species in question. The protein is post-translationally glycosylated in the ER (Delmas and Laude, 1990), following which it is transported through the Golgi apparatus where high mannose oligosaccharides are trimmed and the protein further acylated.
The importance of glycans in S protein is likely to extend beyond protein modification and folding. They may play an important role in viral receptor binding, virus-cell and cell-cell fusion. Mutagenesis analyses of SARS-CoV S protein have identified seven glycosylation sites critical for DC/L-SIGN-mediated entry, an alternative site for SARS-CoV entry (Han et al., 2007). Interestingly, mutation of multiple sites together did not have a synergistic effect on entry (Han et al., 2007). On the other hand, mutation of a single N-linked glycosylation site (N330) abolishes the specific interaction between SARS-CoV S protein and mannose-binding lectin, a serum protein that serves an important function in host defenses during opsonization and complement activation (Zhou et al., 2010).
Mass spectrometry analysis has been used to determine the structure of the N-glycan on SARS-CoV S protein, and major glycans were shown to be high-mannose (Man5–9GlcNAc2), hybrid and bi-, tri- and tetra-antennary complex with and without bisecting GlcNAc and core fucose (Ritchie et al., 2010). Moreover, treatment with glucosidase inhibitor inhibits N-glycan processing and replication of SARS-CoV in Vero E6 cells (Ritchie et al., 2010).
Carbohydrate binding agents found in plants are able to inhibit coronavirus infections by targeting the S and membrane glycans of the mouse hepatitis virus (MHV) and feline infectious peritonitis virus (van der Meer et al., 2007). These results and those from other viruses show that the glycans on the S protein provide great avenues and potential for further exploration and understanding, particularly in the area of protein mediated fusion. Carbohydrate binding agents may be a useful tool in combating viral pathogenesis in future. As described by Balzarini in 2007, these agents may act via a dual mechanism of antiviral action, by firstly binding to the glycans and subsequently blocking virus entry, or by creating deletions in the “glycan shield” of a particular viral surface, inducing the immune system to act against otherwise hidden immunogenic epitopes. Before this becomes a full-fledged antiviral method, more work needs to be done to characterize the function of carbohydrates in these viruses, in particular, the S protein here.
In this study, 29 N-linked glycosylation sites on IBV S protein were predicted by NetNGlyc server 1.0 software. Among them, 8 sites were confidently confirmed by proteomics approaches. The functional importance of N-linked glycosylation in IBV S protein was further studied by mutagenesis of 13 predicted N-linked glycosylation sites in Cluster I. The impact of these mutations on S protein-mediated cell-cell fusion and viral infectivity was investigated.
2. Materials and methods
2.1. Cells and viruses
African Green Monkey kidney Vero cell line and a Vero-adapted IBV Beaudette strain (IBV p65) were used in this study (Ng and Liu, 1998). Confluent Vero cells were inoculated with IBV at a multiplicity of infection (moi) of approximately 1 plaque forming unit (PFU)/cell. Cells were then incubated for 24 h at 37 °C in 5% humidified CO2, until cytopathic effect (CPE) was observed. Both culture medium and cells were harvested and aliquot in appropriate amounts.
The recombinant vaccinia/virus (vTF7-3) was prepared by infection of confluent Vero cells with 0.1 PFU/cell of the virus for 24 h, before fresh medium was used to replace the inoculum. After 48 h, when cytopathic effects were observed, the infected cells were harvested. The viruses were released from cells via three rounds of freeze-thaw, before aliquot in 1.5 ml screw-cap vials and stored at –80 °C.
2.2. Construction and expression of plasmids containing mutant S genes
Forward and reverse primers were designed for the predicted N–linked glycosylation sites. Conventional PCR reactions were carried out. PCR products were treated with DpnI restriction endonuclease digestion for 2 h at 37 °C. For this study, overlapping PCR was done on PKT0-S plasmids (PKT0 vector containing S gene) to create the mutant constructs (Ng and Liu, 2002).
Confluent monolayers of Vero cells were infected with vTF7-3 for 2 h. Transfection was then performed using the Effectene® Transfection Kit. After incubation for 18 h, media was removed from cells. Cells were lysed with sample buffer comprising 2% SDS, 50 mM of Tris 6.8, 10% glycerol and 25 μl/ml of β–mercaptoethanol was added to each well.
2.3. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Separating gels of various acrylamide concentrations (10, 12.5, 15, or 17.5%) and 3% stacking gels were cast and run at a constant voltage for an appropriate length of time. Viral proteins were transferred to nitrocellulose membranes (Stratagene) by the semi-dry transfer cell (Bio-Rad Trans-Blot SD). The membranes were blocked overnight at 4 °C in blocking buffer (5% skim milk powder in PBST [20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween 20 (Sigma)), washed twice (5 min each with agitation) with PBST, and then subjected to primary antibody (rabbit anti-S antisera diluted in blocking buffer (1:6000)) for 1 h. This was then followed by incubation with anti-rabbit IgG conjugated with horseradish peroxidase (Dako) diluted in blocking buffer (1:2000) at room temperature for 1 h and subjected to chemiluminescent detection using detection reagents (ECL, Amersham Pharmacia Biotech) according to instructions of the manufacturer. Excess detection reagent was drained off from the membranes before exposure to X-ray film.
2.4. Indirect immunofluorescence
Vero cells grown on 24 well plates (Iwaki) were infected with IBV or transfected with plasmid DNAs. After washing twice with PBS, cells were fixed with 4% paraformaldehyde in PBS (Sigma) for 15 min at room temperature. For immunofluorescence of permeabilized cells, cells were incubated in 0.2% Triton X-100 (BDH) for 10 min before rinsing thrice with PBS. Anti-S antisera were diluted to 1:200 in a fluorescence dilution buffer (5% goat serum in PBS) and incubated for 2 h at room temperature. Cells were then subjected to three washes with PBS and incubated with anti-rabbit IgG conjugated to FITC (Sigma), diluted in fluorescence dilution buffer (1:200) for 2 h at 4 °C. After three washes with PBS, cells were mounted with glass coverslips using fluorescent mounting medium containing 15 mM NaN3 (Dako).
2.5. In vitro assembly of recombinant cDNA clones
Construction of an infectious IBV clone was done by the assembly of five cloned RT-PCR fragments from Vero-adapted IBV Beaudette strain, comprising of fragments A-E (Fang et al., 2007). Full-length transcripts were generated in vitro using the mMessage mMachine T7 kit (Ambion). The N transcripts were generated using a linearized pKT0-N containing IBV N gene and 3′-UTR as templates. The in vitro synthesized full-length and N transcripts were treated with DNase I and purified with phenol/chloroform. Vero cells were grown to 90% confluence, trypsinized, washed twice with PBS and resuspended in PBS. RNA transcripts were added to 400 μl of Vero cell suspension in an electroporation cuvette, and electroporated with one pulse at 450 V, 50 mF with a Bio-Rad Gene Pulser II electroporator. The transfected Vero cells were cultured overnight in 1% FBS containing DMEM in a 6 well plate and further cultured in DMEM without FBS.
2.6. In-solution deglycosylation
Purified IBV equivalent to 5 μg of protein was denatured by heating at 100 °C in the presence of 1x glycoprotein denaturing buffer (NEB), 5 μg of denatured proteins were split into two tubes prior to addition of 1x G7 reaction buffer (NEB) and 1% NP-40 (NEB). The two samples were subjected to SpeedVac to completely remove H2O16-water·H2O18-water (Cambridge Isotope Laboratories) was added and fully mixed with protein pellets. PNGase F (NEB P0704S) was added to one sample tube to induce deglycosylation reaction overnight at 37 °C and the other tube was mock-treated. After overnight incubation, two protein samples were dried by SpeedVac again to remove H2O18-water completely. This step was designed to limit and differentiate PNGase F deglycosylation and chemical deamidation. Two protein samples were subsequently analyzed by Western blot or subjected to in-gel digestion.
2.7. Protein sample in-gel digestion
The protein pellet was re-suspended in 14 μl 1x PBS. Polyacrylamide gel was formed with 14 μl of glycoprotein, 14 μl of 2% SDS in 25 mM NH4HCO3 (pH 8.0), 12 μl of acrylamide (40%, 29:1), 1.4 μl of ammonium persulfate and 0.8 μl of TEMED. Polymerization reaction was kept for 2 h at room temperature. The gel was removed, cut into small pieces and washed with 25 mM NH4HCO3 in 50% ACN for 15 min, three times with vortexing. The gel pieces were dried using a SpeedVac, kept in 10 mM DTT in 25 mM NH4HCO3 at 60 °C for 30 min, followed by alkylation by adding 55 mM IAA in 25 mM NH4HCO3 at room temperature in the dark for 30 min. The gel pieces were treated with 0.1 μg/μl trypsin (protein: trypsin=10:1, w/w) at 37 °C for 17 h. Trypsin was then heat-inactivated at 100 °C for 10 min. Chymotrypsin digestion (protein: chymotrypsin=30:1, w/w) was proceeded for 16 h at 37 °C. Peptides were extracted from gel using 100 μl of 25 mM NH4HCO3, 200 μl of 0.1% TFA in water, 300 μl of 0.1% TFA in 50% ACN and 100 μl of 100% ACN. The four steps were combined together, concentrated using SpeedVac and subjected to LC-MS/MS analysis.
2.8. LC-MS/MS analysis
Mass spectrometry analysis was performed using a nanoflow high-performance liquid chromatography (HPLC) system (Eksigent, Dublin, CA) connected to a hybrid LTQ-Orbitrap (Thermo Scientific, Bremen, Germany) equipped with a nanoelectrospray ion source (Thermo Scientific). The peptide mixture in each group was reconstituted in 8 μl of solution (0.1% formic acid in LC-MS water), and injected for LC separation. An autosampler (Dionex) with a switching valve (Dionex) and a trap column (C18 PepMap100, 5 µm, 100 µm × 2 cm, 100 Å, Dionex) were used for loading and desalting of peptides. Peptide separation was carried out using a nano-flow LC system (Ultimate plus, Dionex) with a nanoLC column (C18 PepMap100, 2 µm, 75 µm × 250 mm, 100 Å, Dionex). A solvent gradient with buffer B (0.1% formic acid / 100% acetonitrile) increasing from 0% to 40% in 90 min was used to elute the peptides followed by a 20-min wash with 80% buffer B and a 20-min equilibration with 100% buffer A. The flow rate was kept at 500 nl/min. The column was coupled to a Linear Trap Quadrupole (LTQ) Orbitrap (Thermo Fisher Scientific) having a T-union and a nanospray tip (20 µm ID tubing, 10 µm ID tip). The spray voltage of 1.8 kV was applied through the 22 T-union. The mass spectrometer was operated in data-dependent mode to switch between MS and MS/MS. Full scan MS spectra were acquired from m/z 300–2000 Da in the Orbitrap mass spectrometer with a resolution of 60,000 at m/z 400. The 5 most intense ions with ion intensity above 1000 counts and charged state ≥ 2 were sequentially isolated for fragmentation in the linear ion trap using collision induced dissociation (CID; normalized collision energy 35%, activation Q 0.250, and activation time 30 ms) at a target value of 10,000 ions. The dynamic exclusion list was restricted to a maximum retention period of 90 s and a relative mass window of 10 ppm. The MS and MS/MS spectra were recorded by the mass spectrometer as raw files using the Xcalibur software 2.0SR2 (Thermo Fisher Scientific).
2.9. Database searching
The acquired MS raw spectra were processed with PAVAw7.exe into ms2 file and searched against IBV (strain Beaudette) S protein (P11223) by The Global Proteome Machine (http://www.thegpm.org/) searching engine for peptide identifications. A reversed-concatenated database was used to determine the false discovery rate (FDR). The search criteria included trypsin as protease and additional cleavage sites induced by chymotrypsin at Tyr, Trp, and Phe were added. Carbamidomethylation at Cys was set as a fixed modification, while deamidation at N and Q with incorporation of O16 and O18, and oxidation at methionine, were set as variable modifications. 2.98 Da at Asn was also set as variable modification. The maximum number of missed cleavages was set at 2 with the mass tolerance for precursor ions +/− 0.4 Da and for fragment ions +/− 8 ppm. Identifications with less than 1% FDR were considered for further data analysis and MS/MS spectra of all the peptide ions were manually inspected.
3. Results
3.1. Prediction of N-linked glycosylation sites in IBV S protein by NetNGlyc server
The amino acid sequence of S protein from the Vero-adapted IBV Beaudette strain (p65) was subjected to analysis by NetNGlyc server 1.0 software (Blom et al., 2004). This analysis predicts a total of 29 potential N-linked glycosylation sites in the IBV S protein (Fig. 1b and Table 1). These sites were distributed in the S protein as three clusters, with 13 sites in Cluster I, 9 sites in cluster II and 7 sites in Cluster III (Fig. 1a).
Table 1.
NetNGlyc 1.0 Server - prediction results (Threshold+0.5).
| Position | Sequence | Potential | Agreement | N-Glyc result |
|---|---|---|---|---|
| 51 | NISS | 0.7530 | (9/9) | +++ |
| 77 | NASS | 0.6344 | (7/9) | + |
| 103 | NFSD | 0.7078 | (8/9) | + |
| 144 | NLTV | 0.7907 | (9/9) | +++ |
| 163 | NLTS | 0.6421 | (9/9) | ++ |
| 178 | NETI | 0.6101 | (6/9) | + |
| 212 | NGTA | 0.7300 | (9/9) | ++ |
| 237 | NFSD | 0.6116 | (8/9) | + |
| 247 | NSSL | 0.6270 | (8/9) | + |
| 264 | NTTC | 0.6341 | (8/9) | + |
| 276 | NETG | 0.6528 | (8/9) | + |
| 283a | NPSG | 0.5366 | (8/9) | + |
| 306 | NFSF | 0.5353 | (4/9) | + |
| 425 | NITL | 0.6154 | (7/9) | + |
| 447 | NVTD | 0.7393 | (9/9) | ++ |
| 513 | NETG | 0.4646 | (5/9) | – |
| 530 | NGTR | 0.7150 | (9/9) | ++ |
| 579 | NVTE | 0.7121 | (9/9) | ++ |
| 591 | NLTV | 0.5723 | (8/9) | + |
| 669 | NVST | 0.5542 | (5/9) | + |
| 676 | NISL | 0.6323 | (8/9) | + |
| 714 | NCTA | 0.4121 | (8/9) | – |
| 947 | NVTA | 0.6689 | (9/9) | ++ |
| 960 | NASQ | 0.5179 | (6/9) | + |
| 979 | NGSY | 0.5787 | (9/9) | ++ |
| 1014 | NKTV | 0.6853 | (8/9) | + |
| 1038 | NDTK | 0.6343 | (8/9) | + |
| 1051 | NYTV | 0.5906 | (7/9) | + |
| 1074 | NDSL | 0.4930 | (5/9) | – |
WARNING: PRO-X1.
3.2. Determination of N-linked glycosylation sites on IBV S protein by proteomic analysis of PNGase F-mediated deglycosylation
PGNase F mediated Asn to Asp conversion has been used to specifically identify N-glycosylation sites (Bailey et al., 2012). The PNGase F digestion results in the release of the N-linked glycans into the solvent and the glycosylated N within the N-linked glycosylation sequence motif N-X-S/T/C could be converted to D by incorporating extra oxygen derived from the solvent ( Fig. 2a). In addition, deglycosylation occurring in H2O18 environment instead of H2O16 enables incorporation of O18 to Asp which results in a mass increment of 2.98 Da, leading to more robust identification of glycosylated sites (Fig. 2a). However, Asn deamidation also occurs during sample preparation. Asn is converted to a succinimide intermediate that could further hydrolyze rapidly to produce L-Aspartic acid and L-iso Aspartic acid, typically in a 1:3 ratio (Geiger and Clarke, 1987). It is thus possible that Asn deamidation in H2O18 will lead to the mass increment of 2.98 Da. To avoid false positives, a control without addition of PNGase F would be set up for direct comparison with PNGase F mediated deglycosylation.
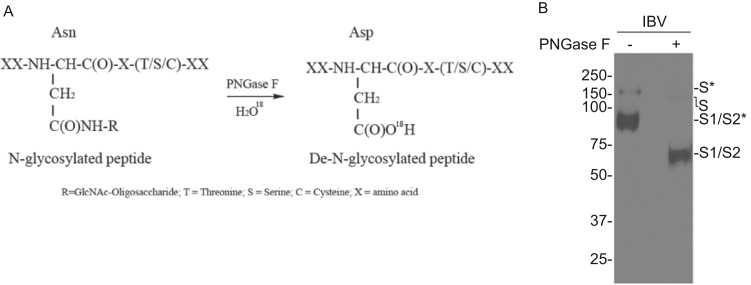
Fig. 2.
a Enzymatic reaction mechanism of PNGase F-mediated Asn to Asp conversion in H2O18 solvent. b Western blot analysis of IBV S protein from partially purified IBV particles before and after PNGase F digestion. Polypeptides were separated on a 10% SDS-PAGE gel and analyzed by Western blot with anti-IBV S antiserum. The full-length glycosylated S (S*), full-length unglycosylated S (S), glycosylated S1/S2 (S1/S2*) and de-glycosylated S1/S2 (S1/S2) are indicated. Numbers on the left indicate molecular masses in kilodalton.
As shown in Fig. 2b, purified IBV was subjected to enzymatic deglycosylation with PNGase F, resulting in remarkable molecular weight shift of IBV S protein detected by Western blot.
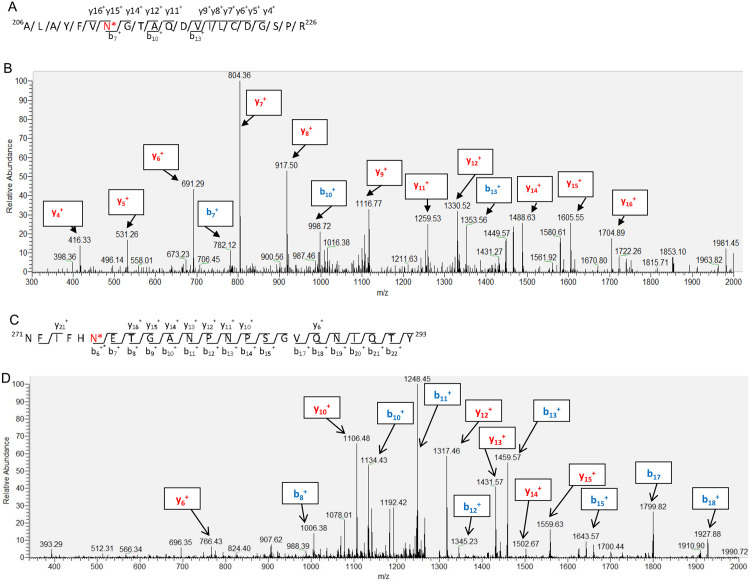
Analysis of the deglycosylated S protein by LC-MS/MS identified 8 N-linked gylcosylation sites, including N212, N237, N247, N276, N513, N591, N1051 and N1074 ( Table 2). These sites are within the N-linked glycosylation sequence motif N-X-S/T/C and the y ions and b ions generated matched well to their theoretical m/z scores (Supplementary Fig.). As examples, two tandem mass spectrometric data of N212 and N276 glycosylation were shown in Fig. 3, respectively. In all these identified N-glycosylation sites, none of them was present in the control sample without PNGase F treatment, suggesting that these identified glycosylation sites are derived from PNGase F mediated de-glycosylation but not Asn deamidation.
Table 2.
Proteomic identification of the N-linked glycosylation sites on IBV S protein.
| N-glycosylation sites | Identified peptides | Measured [M+H]+ | Calc. [M+H]+ with no glycosylation | Δm |
|---|---|---|---|---|
| N212 | 206ALAYFVN*GTAQDVILCDGSPR226 | 2270.112 | 2267.1124 | 2.9996 |
| N237 | 234NTGN*FSDGFYPFTN*SSLVK252 | 2100.9767 | 2094.9767 | 6.0000 |
| N247 | 234NTGN*FSDGFYPFTN*SSLVK252 | 2100.9767 | 2094.9767 | 6.0000 |
| N276 | 271NFIFHN*ETGANPNPSGVQNIQTY293 | 2565.2008 | 2562.2008 | 3.0000 |
| N513 | 513N*ETGSQLLENQFY525 | 1545.7071 | 1542.7071 | 3.0000 |
| N591 | 591N*LTVTDEYIQTR602 | 1455.7329 | 1452.7329 | 3.0000 |
| N1051 | 1051N*YTVPILDIDSEIDR1065 | 1765.8858 | 1762.8857 | 3.0001 |
| N1074 | 1066IQGVIQGLN*DSLIDLEK1082 | 1858.0171 | 1855.0171 | 3.0000 |
Fig. 3.
a Glycosylation peptide (ALAYFVN*GTAQDVILCDGSPR) of IBV S protein following PNGase F treatment of purified IBV. N* represents N-glycosylation site. Identities of the b-ions and y-ions in the spectrum shown in (b). b LC-MS/MS spectrum of the glycosylation peptide in (a) with a [M+2 H]2+ precursor ion at 1136.08 m/z. c Another glycosylation peptide (NFIFHN*ETGANPNPSGVQNIQTY) of IBV S protein following PNGase F treatment of purified IBV. N* represents N-glycosylation site. Identities of the b-ions and y-ions in the spectrum shown in (d). d LC-MS/MS spectrum of the glycosylation peptide in (c) with a [M+2 H]2+ precursor ion at 1283.61 m/z.
3.3. Functional impact of single and double mutations in the N-linked glycosylaltion sites on IBV S protein-mediated cell-cell fusion
To analyze the functions of N-linked glycosylation on IBV replication, the 13 predicted N-linked glycosylation sites in Cluster I were mutated from asparagine (N) to aspartic acid (D), respectively, by site-directed mutagenesis (Fig. 1a and Table 3). Asparagine to aspartate mutation represents the smallest change in molecular mass, and both amino acids have similar polar side chains as well as a hydropathy index of –3.5. As our previous studies showed that expression of IBV S protein in Vero cells using the vaccinia/T7 recombinant virus system induced formation of extensive cell-cell fusion (Xiao et al., 2008), this expression system was then used to assess the impacts of these point mutations on S protein-mediated cell-cell fusion. As summarized in Table 3 and representative results shown in Fig. 4a, N–D point mutations at positions 51, 77, 103, 163, 178, 237, 264 and 306 did not render significant impact on S protein-mediated cell-cell fusion. Significant reduction of cell-cell fusion was observed in cells expressing N144D and N247D and no induction of cell-cell fusion in cells expressing N212D, N276D and N283D mutant constructs (Table 3 and Fig. 4a). Among them, N212, N247 and N276 are confirmed N-linked glycosylation sites as shown in our proteomics analysis.
Table 3.
Summary of the functional impact of point mutation in the cluster 1 glycosylation sites on IBV S protein-mediated cell-cell fusion and viral infectivity.
| Position | Distribution in IBV strains | Glycosylation | Cell-cell fusion |
Virus recovery |
||
|---|---|---|---|---|---|---|
| N→D | N→Q | N→D | N→Q | |||
| N51 | 98 / 129 | + | ++ | ND | + | ND |
| N77 | 111 / 129 | + | ++ | ND | + | ND |
| N103 | 128 / 129 | + | ++ | ND | + | ND |
| N144 | 128 / 129 | + | + | + | + | + |
| N163 | 128 / 129 | + | ++ | ND | + | ND |
| N178 | 128 / 129 | + | ++ | ND | + | ND |
| N212 | 129 / 129 | + | – | – | – | – |
| N237 | 126 / 129 | + | ++ | ND | + | ND |
| N247 | 125 / 129 | + | + | + | + | + |
| N264 | 124 / 129 | + | ++ | ND | + | ND |
| N276 | 128 / 129 | + | – | – | – | – |
| N283 | 77 / 129 | – | – | + | – | + |
| N306 | 127 / 129 | + | ++ | ND | + | ND |
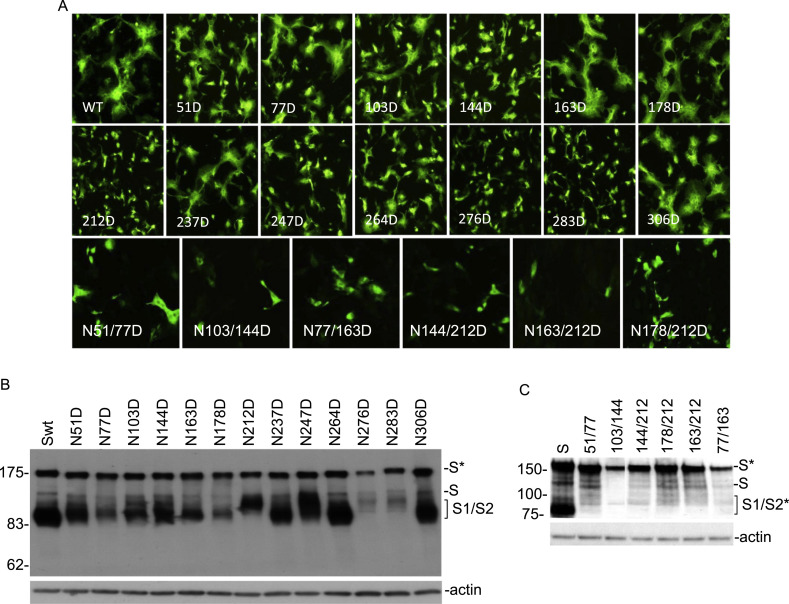
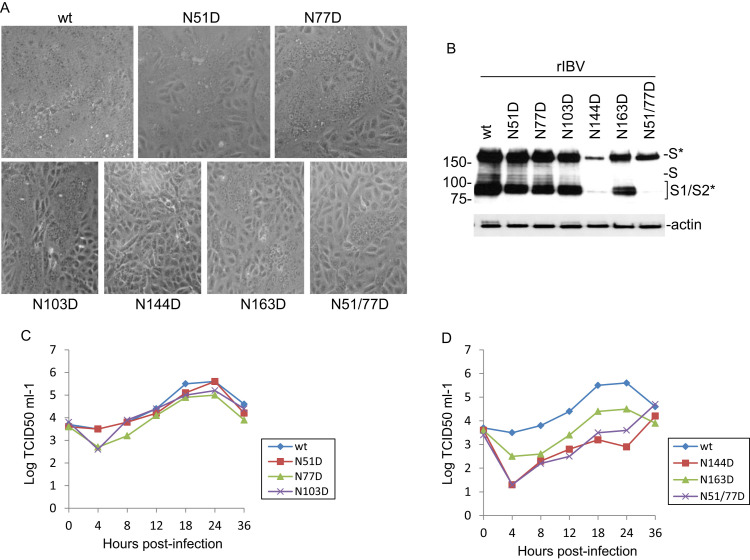
Fig. 4.
a Detection of membrane fusion in cells overexpressing wild type and N-D mutant S proteins at the predicted N-linked glycosylation sites in Cluster I by indirect immunofluorescence. Vero cells were infected with the vaccinia/T7 recombinant virus and transfected with the indicated constructs. At 18 h post-transfection, cells were fixed with 4% paraformaldehyde and stained with rabbit anti-IBV S polyclonal antibodies and anti-rabbit IgG conjugated to FITC. b Western Blot analysis of cells expressing wild type and N-D mutant S proteins at the predicted N-linked glycosylation sites in Cluster I. Vero cells were infected with the vaccinia/T7 recombinant virus and transfected with the indicated constructs. At 18 h post-transfection, cells were harvested and lysates prepared. Polypeptides were separated on a 10% SDS-PAGE gel and analyzed by Western blot with anti-IBV S antiserum. The full-length glycosylated S (S*), full-length unglycosylated S (S) and glycosylated S1/S2 (S1/S2*) are indicated. Numbers on the left indicate molecular masses in kilodalton. c Western Blot analysis of cells expressing wild type and double N-D mutant S proteins at selected N-linked glycosylation sites in Cluster I. Vero cells were infected with the vaccinia/T7 recombinant virus and transfected with the indicated constructs. At 18 h post-transfection, cells were harvested and lysates prepared. Polypeptides were separated on a 10% SDS-PAGE gel and analyzed by Western blot with anti-IBV S and anti-actin antibodies, respectively. The full-length glycosylated S (S*), full-length unglycosylated S (S) and glycosylated S1/S2 (S1/S2*) are indicated. Numbers on the left indicate molecular masses in kilodalton.
Western blot analysis of protein samples collected at 18 h post-transfection detected the full-length glycosylated S protein from cells transfected with wild type and all mutant S constructs (Fig. 4b), but with varied expression efficiency. Among them, wild type, N51D, N237D, N264D and N306D were highly expressed, and N77D, N178D, N276D and N283D were among the lowest expressed (Fig. 4b). The expression levels appeared not to be related to the cell-cell fusion induced by the constructs. In addition, expression of most constructs led to the detection of glycosylated S1/S2 cleavage products, but the level of S1/S2 cleavage was variable among constructs (Fig. 4b). Interestingly, very limited cleavage of S protein to S1/S2 was observed in cells transfected with N212D, N276D and N283D constructs (Fig. 4b). As almost no cell-cell fusion was observed in cells transfected with these three constructs, it suggests that cleavage of S protein to S1/S2 might either be the cause or the consequence of cell-cell fusion mediated by coronavirus S protein.
A few more mutant S constructs with double glycosylation site mutations, i.e., N51/77D, N103/144D, N77/163D, N144/212D, N163/212D and N178/212D, were created. Expression of these mutant constructs showed reduced cell-cell fusion in cells expressing these constructs. Except N51/77D which showed reduced induction of cell-cell fusion, no apparent cell-cell fusion was observed in cells expressing all other double mutants (Fig. 4a). Once again, the expression of these structures led to the detection of the glycosylated full-length S protein, but cleavage to S1/S2 was much reduced in cells expressing most of these constructs (Fig. 4c).
3.4. Functional impact of mutations in the N-linked glycosylation sites on recovery of infectious IBV
These mutations were then introduced into an infectious IBV cDNA cloning system (Fang et al., 2007), and their impact on the recovery of infectious virus was assessed. Recombinant viruses were recovered at 72 h post-electroporation from cells transfected with mutant full-length transcripts N51D, N77D, N103D, N144D, N163D, N178D, N237D, N247D, N264D and N306D (Table 3). Interestingly, no infectious virus was recovered from cells electroporated with N212D, N276D and N283D full-length transcripts (Table 3), correlating well with the cell-cell fusion data that expression of mutant S protein harboring point mutation at these positions induced limited or no cell-cell fusion (Fig. 4a). These observations prompted further characterization of the replication, infectivity and growth kinetics of a selected set of recovered mutant IBV, including N51D, N77D, N103D, N144D and N163D.
3.5. Correlation between cell-cell fusion and viral replication/infectivity among mutant S proteins
The selected set of mutant viruses were then analyzed by examining the syncytium formation in infected cells. After passage of the recovered mutant IBV in Vero cells twice, TCID50 assay was performed to determine the viral titers and growth kinetics, and Western blot analysis was done to check the viral S protein expression in cells infected with these recombinant viruses. As shown in Fig. 5a, in cells infected with N51D and N77D mutant viruses, very similar syncytium formation with cells infected with wild type IBV was observed. Interestingly, a mutant virus harboring N-D mutations at both N51 and N77 was also recovered. This virus showed much reduced syncytium formation in infected cells (Fig. 5a). Mild reduction in cells infected with N103D and N163D mutant viruses, and much reduced formation of syncytia was found in cells infected with N144D mutant virus (Fig. 5a).
Fig. 5.
a Cytopathic effect of Vero cells transfected with wild type and selected N-D mutant full-length transcripts. Cells were transfected with RNA and syncytium formation was observed by microscopy at 24–36 h post-transfection. b Western blot analysis of cells infected with wild type and a set of recombinant IBV with N-D mutation(S) in N-linked glycosylation sites. Vero cells were infected with virus, harvested at 24 h post-infection and lysates prepared. Polypeptides were separated on a 10% SDS-PAGE gel and analyzed by Western blot with anti-IBV S and anti-actin antibodies, respectively. The full-length glycosylated S (S*), full-length unglycosylated S (S) and glycosylated S1/S2 (S1/S2*) are indicated. Numbers on the left indicate molecular masses in kilodalton. c Growth kinetics of wild type and N51D, N77D and N103D recombinant IBV. Confluent Vero cells were infected with IBV, harvested at 0, 4, 8, 12, 18, 24 and 36 h post-infection, respectively. Viral stocks were prepared by freezing/thawing of the cells three times and TCID50 was determined. d Growth kinetics of wild type and N144D, N163D and N51-77D recombinant IBV. Confluent Vero cells were infected with IBV, harvested at 0, 4, 8, 12, 18, 24 and 36 h post-infection, respectively. Viral stocks were prepared by freezing/thawing of the cells three times and TCID50 was determined.
Western blot analysis of protein samples collected at 18 h post-infection detected similar levels of S protein expression in cells infected with wild type and N51D, N77D and N103D mutants (Fig. 5b). Mild reduction of S protein expression was observed in cells infected with N163 mutant, and severe reduction of S protein expression was observed in cells infected with N51/77D and N144D mutant viruses (Fig. 5b). These results are largely correlated with syncytium formation data shown in Fig. 5a.
TCID50 assay of the growth kinetics of wild type and mutant viruses was carried out with samples harvested at 0, 4, 8, 12, 18, 24 and 36 h post-infection. The results showed that N51D mutant virus displayed very similar growth kinetics as wild type virus, both reaching the peak titer at 24 h post-infection (Fig. 5c). Mutants N77D and N103D also showed similar growth kinetics as wild type virus, but with 2–5 fold reduction in the peak titers (Fig. 5c). Mutant N144D, N163D and N51/77D grew much slower than wild type virus. At 24 h post-infection, the titers of these mutant viruses showed 10–25 fold reduction compared to wild type virus (Fig. 5d).
The stability of these mutant viruses was checked by DNA sequencing using a series of six primers covering the S gene, confirming the presence of the mutation(s) introduced into the recombinant viruses. No other insertions, deletions or mutations were found to be present in the S gene of these mutant viruses.
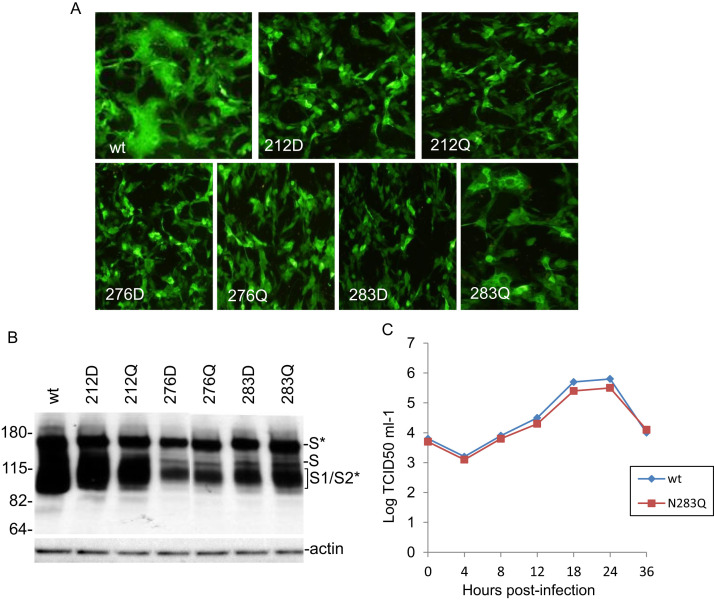
3.6. N-linked glycosylation-independent role of N283 in IBV replication and infectivity
Although asparagine to aspartate mutation represents the smallest change in molecular mass, the negatively charged aspartate may have additional effects on the S protein independent of N-linked glycosylation. To address this concern, asparagine to glutamine (Q) mutation was made in N144, N212, N247, N276 and N283 (Table 3). As summarized in Table 3, mutations of N to Q at positions N144, N212, N247 and N276 in IBV S protein render same effects on cell-cell fusion and viral recovery as N to D mutations. Mutation of N283 to Q, however, generated a mutant S protein similar to wild type protein (Table 3 and Fig. 6). As can be seen, expression of N283Q in Vero cells showed induction of extensive cell-cell fusion (Fig. 6a), and efficient glycosylation and processing of the mutant S protein (Fig. 6b). The recovered recombinant virus containing this mutation displayed similar growth kinetics as wild type IBV, with a 2-fold reduction in the peak titers at 24 h post-infection (Fig. 6c). These results suggest that N283 plays an essential role in viral replication and infectivity independent of N-linked glycosylation.
Fig. 6.
a Detection of membrane fusion in cells overexpressing wild type and N-D/Q mutant S proteins at three predicted N-linked glycosylation sites in Cluster I by indirect immunofluorescence. Vero cells were infected with the vaccinia/T7 recombinant virus and transfected with the indicated constructs. At 18 h post-transfection, cells were fixed with 4% paraformaldehyde and stained with rabbit anti-IBV S polyclonal antibodies and anti-rabbit IgG conjugated to FITC. b Western Blot analysis of cells expressing wild type and N-D/Q mutant S proteins at three predicted N-linked glycosylation sites in Cluster I. Vero cells were infected with the vaccinia/T7 recombinant virus and transfected with the indicated constructs. At 18 h post-transfection, cells were harvested and lysates prepared. Polypeptides were separated on a 10% SDS-PAGE gel and analyzed by Western blot with anti-IBV S antiserum. The full-length glycosylated S (S*), full-length unglycosylated S (S) and glycosylated S1/S2 (S1/S2*) are indicated. Numbers on the left indicate molecular masses in kilodalton. c Growth kinetics of wild type and N283Q recombinant IBV. Confluent Vero cells were infected with IBV, harvested at 0, 4, 8, 12, 18, 24 and 36 h post-infection, respectively. Viral stocks were prepared by freezing/thawing of the cells three times and TCID50 was determined.
4. Discussion
In this study, the S glycoprotein of IBV was deglycosylated by PNGase F in H2O18 solvent which induces Asn-Asp conversion that results in a mass increment of 2.98 Da. Analysis was done by proteomics to identify the N-linked glycosylation sites in the IBV S glycoprotein. A control was used to exclude any false positive caused by chemical deamination that can happen spontaneously during sample preparation. The functional importance of 13 N-glycosylation sites in Cluster I on IBV S protein-mediated membrane fusion, infectivity and replication was characterized using mutagenesis analysis via both transfection of mutant S proteins and construction of recombinant viruses. In SARS-CoV S protein, certain positions are suggested to be of importance for binding to cellular components, such as L-SIGN and DC-SIGN (Han et al., 2007). IBV S protein-mediated fusion is not well understood, and the importance of the S1 region of the protein in fusion is largely unknown. With the postulation that glycans may play an important function, the presence of a large number of glycosylation sites in this region suggests that it may play a role in this process. This forms the motivation behind this study.
Our results demonstrated that N-linked glycans at different positions may differentially affect S protein-mediated fusion and viral infectivity and replication. Positions N212 and N276, which are confirmed by LC–MS/MS, in particular show high influence when analyzed both for cell-cell fusion and recovery of infectious virus. Although the targeted sites were mutated to remove glycosylation, there is a possibility though that the changes present were down to the mutations in amino acids, rather than the absence of glycans themselves. As mentioned earlier the change in molecular mass is minimal, both amino acids also having similar polar side chains and hydropathy index. Taking the various protein folding forces into consideration, it is thus unlikely that any conformational and binding changes as proposed earlier are due to the amino acids themselves. Given that glycans modify protein structures and assist in folding and interaction, it is likely that they are the factors that determine the phenotypic changes.
On the other hand, among the 29 software-predicted N-linked glycosylation sites, only 8 sites could be validated using LC-MS/MS analysis. This seemingly under-representation could be partially attributed to the massive amount of IBV S protein produced during IBV infection and the limited capacities of the host cell glycosylation enzymes. It is possible that due to their better spatial availability or other unknown advantages, the asparagine residues at these 8 sites were preferentially modified by glycosylation. The remaining sites were either inefficiently glycosylated, or they might miss the chance of modification because the partially glycosylated IBV S protein has already been transported to the next compartment where glycosylation enzymes are no longer available. It is also possible that there exists a detection limit for LC-MS/MS set up during the time for sample analysis. When one or more of the confirmed glycosylation sites in S protein was mutated, the cellular glycosylation enzymes might be “freed up”, so that N-linked glycosylation of the remaining sites could be observed in theory. This could be studied by determining the N-linked glycosylation of virus particles from mutant recombinant IBVs using LC-MS/MS analysis. Glycosylation of these “redundant” sites might be able to functionally complement the mutations on the “canonical” sites, explaining why several of the rIBV with these mutations could still be successfully recovered and grew well in cell culture.
Interestingly, the asparagine at 283 (N283PSG) with the proline at ×1 position should not be glycosylated in theory, but its mutation to aspartate abolishes cell-cell fusion and recovery of recombinant virus. To this end, the same residue was mutated to glutamine, and it was found that the N283Q mutant supports both cell-cell fusion and virus recovery. Therefore, it is concluded that the critical function of N283 is independent of N-linked glycosylation. While N to D mutation involves a change in charge, there is no net change in charge but only a small size increase of side chain for N to Q mutation. Thus, N283 might be positioned in a crucial domain in IBV S protein, in which even minor charge (but not size) changes would disrupt delicate internal electrostatic interactions and lead to drastic modifications of protein structure and function.
Proteolytic cleavage of the S protein into S1 and S2 subunits is essential for initiating fusion (Burkard et al., 2014, Yamada and Liu, 2009). Each peplomer consists of two to three spike molecules already cleaved into S1 and S2 subunits. It is possible that the two subunits need to interact in a certain way to form a proper peplomer as well as to facilitate binding to cellular receptors as the virus approaches the cell. Once again, the presence or absence of glycans on the S1 region may affect its interaction with the S2 region, thus affecting binding, or even the formation of proper peplomers on complete viral particles. This may help to explain why several of the mutant S proteins cannot induce cell-cell fusion, as well as failing to recover recombinant mutants. The observations that very low cleavage bands were seen when some mutant S proteins were expressed suggest that mutations at some glycosylation sites may also affect cleavage efficiency, although the glycosylation sites near the cleavage site were not mutated.
Coronavirus S protein is mainly synthesized in the ER, and N-linked glycosylation occurs mainly in the ER lumen and Golgi apparatus. N-linked glycosylation significantly increases the size of S protein, and has been shown to promote its folding and trimerization (Delmas and Laude, 1990). Therefore, it is reasonable to propose that changes of N-linked glycosylation status in the cluster I mutants may modify the proper folding, processing and/or trimerization of the S protein, thus affecting its proteolytic cleavage pattern and compromising its fusogenic functions. In the schematic diagram of Fig. 1a, Cluster I glycans are shown to be in the N-terminus region of IBV S protein and do not cover the S1/S2 cleavage site and the fusion peptide. However, in the fully folded and trimerized coronavirus S protein, the S1 subunits are closely associated with the S2 subunits. In fact, in a recent publication showing the cryo-electron microscopy structure of MHV S trimer, the fusion peptides were exposed in the periphery, and positioned very closed to the major antigenicity determinant in the S1 subunit (Walls et al., 2016). If the cluster I N-linked glycosylation sites were located closed to critical regions of the S2 subunit, the presence/absence of N-linked glycans could affect the cleavage and/or fusion ability. This may explain why some N to D or N to Q mutants in the cluster I sites could affect the S1/S2 ratio or cell-cell fusion of the overexpressed S protein.
In this study, we have identified 13 potential N-linked glycosylation sites in cluster I of the IBV S protein. Among them, four (N212, N237, N247 and N276) have been confirmed to be N-linked glycosylated in S protein originated from partially purified virions by proteomics studies. N to D/Q mutations in N212 and N276 were found to abolish the infectivity of the recombinant IBV. The receptor binding domain (RBD) on the S1 subunit of each coronavirus may vary, with RBD of MHV located at the N-terminus (Kubo et al., 1994), SARS-CoV at the center (Wong et al., 2004), and TGEV and HCoV-229E at the C-terminus (Bonavia et al., 2003, Godet et al., 1994) of the S1 region. For the M41 strain of IBV, the RBD has been mapped to the N-terminal 253 residues of the S protein, which was both necessary and sufficient for 2,3-sialic acid-dependent binding to chicken respiratory tract (Promkuntod et al., 2014). Interestingly, N212 was located within this putative RBD, while N276 was also very closely positioned. However, the involvement of N-linked glycans at these two sites for IBV (strain Beaudette) receptor binding, both in cell culture and in vivo, requires further investigations. Nonetheless, the importance of N-linked glycosylation on receptor binding and viral infectivity has been well documented for SARS-CoV. For example, seven glycosylation sites (N109, N118, N119, N158, N227, N589 and N699) in SARS-CoV S protein have been shown to be critical for virus entry mediated by the C-type lectin DC-SIGN and L-SIGN (Han et al., 2007). Furthermore, mannose-binding lectin (MBL) has been shown to interact with SARS-CoV S-pseudotyped virus and block viral binding to DC-SIGN, with another glycosylation site (N330) on the SARS-CoV S protein found to be critical for MBL binding (Zhou et al., 2010). It is intriguing that even if these N-link glycans may not directly participate in receptor binding, they may be recognized by different lectin proteins and modulate the immune response during viral infection.
Viruses are constantly needing new ways and mechanisms to evade antiviral mechanisms a host cell or system might impose. Since glycosylation patterns in different host cells differ, viruses such as the West Nile Virus have been shown to be able to exploit different host glycosylation profiles to escape such mechanisms, via exerting a specific cytokine blocking effect (Arjona et al., 2007). Other studies also showed that type I interferon and TNF-α are further examples of substances differentially induced by varied glycosylation profiles. It is possible that depriving the IBV S of certain glycans may alter its profile and make it more susceptible to antiviral actions by the host cell. The possibility that recombinant IBV with glycosylation-defective at certain positions recovered in this study may be used to study such virus-host interaction would be worth exploring.
There are many forms of carbohydrate binding agents, such as lectins. Carbohydrate binding agents have been shown to elicit favorable responses in HIV infected cells, able to inhibit fusion of cell free HIV particles with susceptible cells and also the formation of syncytia between persistently infected and uninfected cells (Balzarini et al., 1991). They are also able to prevent capture of viruses by DC-SIGN and the subsequent transmission of viruses to T-cells (Wang et al., 2008). Carbohydrate binding agents are unique as they will interact directly with the glycans present on the glycoproteins, thus need not be taken up by cells. Also multiple binding of these agents to the glycoprotein exerts a high genetic barrier, which means that if the virus was to mutate and evolve resistance by deletion of its N-glycans, several mutations need to accumulate in the protein before a significant phenotypic drug resistance occurs. Whereas other drugs only require a couple of mutations to evolve resistance, this method of antiviral treatment offers a more stable effect (Balzarini, 2007). With the gradual improvement and increase in significance of carbohydrate binding agents as antivirals, the IBV S glycoprotein should also come into the picture as one of the targets. This would also lead to similar uses in other coronaviruses such as the SARS-CoV. Identifying regions of glycans that exert the most influence on viral infectivity and pathogenesis may facilitate design of such binding agents.
Notably, a recent study has evaluated the antigenicity of recombinant IBV S1 protein expressed in mammalian cells (Andoh et al., 2015). Result shows that although the recombination S1 protein is highly glycosylated and is able to induce antibodies against S1 in immunized chickens, the antibodies have lower neutralizing activity compared to those in chickens immunized with inactivated virus (Andoh et al., 2015). Therefore, the glycosylation patterns on IBV S proteins synthesized in mammalian cells might differ from those produced in avian cells, thus accounting for the lacking of neutralizing epitopes.
During coronavirus infection, the massive production of major structural proteins, in particular the large and heavily glycosylated spike protein, has been shown to impose enormous burden to the protein synthetic machinery of ER, leading to the ER stress and induction of unfolded protein response (UPR) (Fung et al., 2014a, Fung et al., 2016). In fact, overexpression of the S protein of SARS-CoV and MHV induces potent ER stress in cell culture (Chan et al., 2006, Versteeg et al., 2007), while one or more of the three UPR branches are activated in cells infected with SARS-CoV, MHV, TGEV and IBV (Fung et al., 2014b, Fung and Liu, 2014, Liao et al., 2013). However, the involvement of N-glycan in spike-induced ER stress has not been fully investigated. In a previous study, overexpression of SARS-CoV and HCoV-HKU1 spike proteins with multiple putative N-glycosylation site mutated was shown to activate UPR reporter similarly to the wild type protein (Siu et al., 2014). Nevertheless, the implication of spike protein N-glycosylation on cellular stress responses and other aspects of virus-host interactions should be further investigated under the context of actual infection.
Acknowledgements
This work was partially supported by an Academic Research Fund (AcRF) Tier 2 grant (ACR47/14), Ministry of Education, Singapore. We thank Yeo Jia chi, Zhi wei for technical assistance.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.10.003.
Appendix A. Supplementary material
Supplementary material
References
- Andoh K., Suenaga K., Sakaguchi M., Yamazaki K., Honda T. Decreased neutralizing antigenicity in IBV S1 protein expressed from mammalian cells. Virus Res. 2015;208:164–170. doi: 10.1016/j.virusres.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A., Ledizet M., Anthony K., Bonafé N., Modis Y., Town T., Fikrig E. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J. Immunol. Baltim. Md. 2007;1950(179):8403–8409. doi: 10.4049/jimmunol.179.12.8403. [DOI] [PubMed] [Google Scholar]
- Bailey U.-M., Jamaluddin M.F., Schulz B.L. Analysis of congenital disorder of glycosylation-Id in a yeast model system shows diverse site-specific under-glycosylation of glycoproteins. J. Proteome Res. 2012;11:5376–5383. doi: 10.1021/pr300599f. [DOI] [PubMed] [Google Scholar]
- Balzarini J. Carbohydrate-binding agents: a potential future cornerstone for the chemotherapy of enveloped viruses?? Antivir. Chem. Chemother. 2007;18:1–11. doi: 10.1177/095632020701800101. [DOI] [PubMed] [Google Scholar]
- Balzarini J., Schols D., Neyts J., Van Damme E., Peumans W., De Clercq E. Alpha-(1-3)- and alpha-(1-6)-D-mannose-specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob. Agents Chemother. 1991;35:410–416. doi: 10.1128/aac.35.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a Receptor-Binding Domain of the Spike Glycoprotein of Human Coronavirus HCoV-229E. J. Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J.M., Bosch B.J., de Haan C.A.M. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J. Gen. Virol. 1983;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Chan C.P., Siu K.L., Chin K.T., Yuen K.Y., Zheng B., Jin D.Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80:9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.W., Sheng Y., Gombold J.L. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology. 2000;269:212–224. doi: 10.1006/viro.2000.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Chen B., Tay F.P.L., Ng B.S., Liu D.X. An. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus–host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. Regulation of stress responses and translational control by coronavirus. Viruses. 2016:8. doi: 10.3390/v8070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. The ER stress sensor IRE1α protects cells from apoptosis induced by coronavirus infectious bronchitis virus. J. Virol. 2014;88:12752–12764. doi: 10.1128/JVI.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:1–13. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus ((coronavirus)) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN-and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Fung T.S., Huang M., Fang S.G., Zhong Y., Liu D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013;87:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F., Liu D.X. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998;243:388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341:215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promkuntod N., van Eijndhoven R.E.W., de Vrieze G., Gröne A., Verheije M.H. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie G., Harvey D.J., Feldmann F., Stroeher U., Feldmann H., Royle L., Dwek R.A., Rudd P.M. Identification of N-linked carbohydrates from severe acute respiratory syndrome ((SARS)) spike glycoprotein. Virology. 2010;399:257–269. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Law Y.C., Liu D.X. A single amino acid mutation in the spike protein of coronavirus infectious bronchitis virus hampers its maturation and incorporation into virions at the nonpermissive temperature. Virology. 2004;326:288–298. doi: 10.1016/j.virol.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Chan C.-P., Kok K.-H., Woo P.C., Jin D.-Y. Comparative analysis of the activation of unfolded protein response by spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus. HKU1. 2014;4:1–9. doi: 10.1186/2045-3701-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer F.J.U.M., de Haan C. a.M., Schuurman N.M.P., Haijema B.J., Verheije M.H., Bosch B.J., Balzarini J., Egberink H.F. The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin A target the glycans of the coronavirus envelope glycoproteins. J. Antimicrob. Chemother. 2007;60:741–749. doi: 10.1093/jac/dkm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Van De Nes P.S., Bredenbeek P.J., Spaan W.J.M. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81:10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-K., Liang P.-H., Astronomo R.D., Hsu T.-L., Hsieh S.-L., Burton D.R., Wong C.-H. Targeting the carbohydrates on HIV-1: interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc. Natl. Acad. Sci. USA. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Xu L.H., Yamada Y., Liu D.X. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One. 2008;3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lu K., Pfefferle S., Bertram S., Glowacka I., Drosten C., Pöhlmann S., Simmons G. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84:8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material