Abstract
Many plant species are able to regenerate adventitious roots either directly from aerial organs such as leaves or stems, in particularly after detachment (cutting), or indirectly, from over-proliferating tissue termed callus. In agriculture, this capacity of de novo root formation from cuttings can be used to clonally propagate several important crop plants including cassava, potato, sugar cane, banana and various fruit or timber trees. Direct and indirect de novo root regeneration (DNRR) originates from pluripotent cells of the pericycle tissue, from other root-competent cells or from non-root-competent cells that first dedifferentiate. Independently of their origin, the cells convert into root founder cells, which go through proliferation and differentiation subsequently forming functional root meristems, root primordia and the complete root. Recent studies in the model plants Arabidopsis thaliana and rice have identified several key regulators building in response to the phytohormone auxin transcriptional networks that are involved in both callus formation and DNRR. In both cases, epigenetic regulation seems essential for the dynamic reprogramming of cell fate, which is correlated with local and global changes of the chromatin states that might ensure the correct spatiotemporal expression pattern of the key regulators. Future approaches might investigate in greater detail whether and how the transcriptional key regulators and the writers, erasers, and readers of epigenetic modifications interact to control DNRR.
Keywords: root regeneration, adventitious roots, DNRR, callus, pluripotency, transcriptional networks, epigenetics
Introduction
During embryogenesis of higher plants, the shoot apical meristem (SAM) and the root apical meristem (RAM) are established at the opposite poles of the central axis. It is assumed that all other meristems derive from these two types of embryonic meristems, although all secondary SAMs or RAMs arise from partially differentiated cells, which need to go through reprogramming to become meristematic cells again. Root branching is based on these de novo RAMs that give rise to extensive root networks enabling plants to gain a stable hold in the ground, explore the soil and facilitate the uptake of water and mineral nutrients (Casimiro et al., 2003; Gonzali et al., 2005). Although the root system is genetically determined, they display a high plasticity in response to environmental variables such as water availability, nutrient levels, physical barriers or damage (Al-Ghazi et al., 2003; Sena and Birnbaum, 2010; Sugimoto et al., 2010; van Norman et al., 2013).
In dicotyledonous plants, such as Arabidopsis thaliana (Arabidopsis), the primary root grows as a thick central taproot (Bellini et al., 2014). Lateral roots (LRs) emerge post-embryonically and derive from pericycle cells close to the xylem pole cells of the primary root (De Smet et al., 2006); nonetheless, the morphology of primary root and LRs is basically identical (Birnbaum, 2016). Furthermore, adventitious roots (ARs) can be formed directly from various aerial organs (Fattorini et al., 2018) or indirectly from callus (Verstraeten et al., 2014). Depending on the status of the AR source cells, they can be directly fate-converted to AR root founder cells by a root-inducing signal or they first have to acquire root competence involving dedifferentiation (Druege et al., 2019). The natural ability of plants to regenerate is widely used in tissue culture, modern horticulture, and agriculture (Sussex, 2008; Druege and Franken, 2019). In rice and other cereals, ARs represent the main components of the root system as the primary root originating from the embryonic RAM is short-lived. During post-embryonic development, shoot-borne ARs form from nodes of the stem (Kawata et al., 1963; Liu et al., 2005). Some researchers prefer the term shoot-borne crown roots for these type of ARs, because they are part of the normal developmental program of cereals (Hochholdinger et al., 2004; Zhao et al., 2009). Nevertheless, ARs are also a common part of the regular root system of Arabidopsis under natural growth conditions in soil (Sheng et al., 2017). In recent years, research on plant root systems has made significant progress not only on natural root development but also on de novo root regeneration (DNRR). Besides classical DNRR research, which deals with the origin of ARs, some root regeneration studies have focused on reestablishment of the main RAM after pruning the root meristem tip (Efroni et al., 2016) or the replacement of single cells (Marhava et al., 2019). Here, we will give an overview of root regeneration systems and describe how recent breakthroughs in the model plants Arabidopsis and other plant species have changed our view of the molecular basis of cell fate reprogramming during DNRR focusing on transcriptional and epigenetic gene regulation. Furthermore, we will provide a short summary of the role of phytohormone signaling in DNRR, but the reviews of Lakehal and Bellini (2019) and Druege et al. (2019) provide a more comprehensive view on hormonal crosstalk and hormone-metabolic interactions in excision-induced AR formation.
The Role of Auxin in DNRR Systems
Adventitious shoots and roots, derived from isolated or injured tissues and organs, provide an important survival strategy for plants in natural conditions (Duclercq et al., 2011; Sugimoto et al., 2011; Chen et al., 2014). In 1957, Skoog and Miller made the breakthrough discovery of experimentally induced phytohormone-dependent de novo regeneration of shoot and roots (Skoog and Miller, 1957). This gave rise to tissue culture methods which are still in use today in agriculture, industry and research (Duclercq et al., 2011). Hence, we can distinguish two types of de novo shoot and root regeneration, one under tissue culture conditions and the other in natural surrounding (Chen et al., 2014; Yu et al., 2017). In tissue culture, isolated plant tissues or organs named explants are cultured on nutrient-rich media containing an appropriate ratio of the phytohormones auxin and cytokinin, which can promote root or shoot formation in a controlled manner (Skoog and Miller, 1957). Under natural conditions, isolated organs can produce adventitious shoots and roots, and in some cases form whole plants: for example, some species from the Crassulaceae family are able to regenerate shoots and roots from leaves placed on soil (Chen et al., 2014; Xu and Huang, 2014; Ikeuchi et al., 2016). Nevertheless, endogenous hormones are crucial to induce adventitious shoot and root formation under natural conditions, for example in petunia, polar auxin transport and early IAA accumulation are essential for AR formation (Ahkami et al., 2013; Xu and Huang, 2014).
Auxin plays an important role in root growth and development, especially in LR and AR initiation (Klerk et al., 1999; Overvoorde et al., 2010; Lavenus et al., 2013; Bellini et al., 2014). ARs initiate near the wounding site of detached organs, which likely depends on auxin accumulation in the area (Liu et al., 2014). ARs can form from young Arabidopsis leaves without application of exogenous auxin, whereas exogenous auxin can increase the chance of root regeneration from older leaves that have decreased levels of endogenous auxin (Shoji et al., 1951; Chen et al., 2014). In addition, DNRR from most trees and other hard-to-root plants requires the application of exogenous auxin to induce ARs (Klerk et al., 1999; Díaz-Sala, 2014). The earliest studies on DNRR were carried out by Zimmerman and Hitchcock (1935) using aerial roots of grapes (Vitis sp.). These did not develop LRs until touching the soil, however, several new roots occurred if the aerial roots were cut and placed in solutions containing “root-forming” substances (Zimmerman and Hitchcock, 1935). In the same year several growth substances were tested for their ability to promote ARs and alpha-naphthaleneacetic acid (NAA) and indolebutyric acid (IBA) were found to be the most effective root-forming substances (Zimmerman and Wilcoxon, 1935). Exogenous application of the natural auxin indole-acetic acid (IAA) can induce a large number of roots from tomato leaf explants (Coleman et al., 1980). In vitro, a supply of exogenous sucrose also supports root regeneration by providing carbohydrate for plant growth (Calamar and de Klerk, 2002) and leaf explants fail to induce roots when growing on media in the dark without sucrose (Chen et al., 2014). A low concentration of sugar promotes LR initiation, while a high concentration of sugar inhibits LR initiation (Malamy and Ryan, 2001).
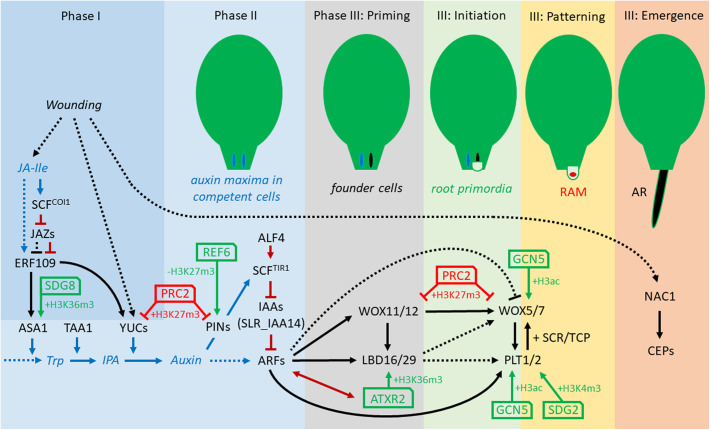
In tissue culture, the ratio of auxin to cytokinin can influence de novo organogenesis in nutrient-rich growth media. Callus-inducing medium (CIM) has a balanced ratio of auxin to cytokinin. The transfer of the callus to root-inducing medium (RIM) with a high ratio of auxin-to-cytokinin induces root regeneration, whereas transfer to media with a low ratio induces shoot regeneration (Skoog and Miller, 1957; Valvekens et al., 1988). Recently, it was shown that the transfer from CIM with high auxin levels to B5 medium without auxin triggers AR formation as well (Yu et al., 2017). Callus, which derives from pericycle or related cells in the vascular tissue, depends on several key transcriptional regulators, which are also involved in LR and AR formation (Che et al., 2007; Atta et al., 2009; Sugimoto et al., 2010; Kareem et al., 2015; Ikeuchi et al., 2016). Root regeneration from callus might be one of the simplest case of cell fate reprogramming in plants since callus itself has an identity resembling that of lateral root primordia (LRP) (Sugimoto et al., 2010). Interestingly, the induction of so-called endogenous callus, which is presumed to be the first required step for DNRR from leaf explants, can be achieved without the application of exogenous auxin (Bustillo-Avendaño et al., 2018). Recently, AR formation from Arabidopsis leaf explants was intensively studied and can be divided into three phases: (I) early after wounding, signaling pathways trigger (II) auxin production in so-called converter cells (0–4 hours after leaf explant detachment [HAD]), followed by auxin accumulation in the region of AR formation by directed auxin transport (around 12 HAD) and (III) fate transition from regeneration-competent cells into fully formed ARs (Xu, 2018). The fate transition phase can be subdivided into four steps: During the “priming” step (24–48 HAD), regeneration-competent cells became root founder cells by cell fate transition; in the “initiation” step, root founder cells start to divide to form a dome-shaped LRP (48–96 HAD); during the “patterning” step, continuous cell division and differentiation generate a well-organized RAM, whereas in the “emergence” step the new formed AR breaks through the epidermis of the leaf explant (Figure 1; Yu et al., 2017; Xu, 2018).
FIGURE 1.
Concept of transcriptional and epigenetic regulation network during direct excision-induced DNRR in Arabidopsis. The three phases and four stages of phase III, priming, initiation, pattering, and emergence, are according to Yu et al. (2017) and Xu (2018). Note that several relations are not yet verified for DNRR but demonstrated for callus formation, LR initiation and/or regular root development. Black arrows, direct transcriptional regulation; dotted arrows, indirect regulation; blue arrows, JA and auxin synthesis and transport; dark-red arrows, protein-protein interaction; green arrows, positive epigenetic regulation; red arrows, negative epigenetic regulation. ALF4, ABERRANT LATERAL ROOT FORMATION4; ARFs, AUXIN RESPONSE FACTORs; ASA1, ANTHRANILATE SYNTHASE α1; ATXR2, ARABIDOPSIS THALIANA TRITHORAX-RELATED2; CEP, Cys endopeptidase; ERF109, ETHYLENE RESPONSE FACTOR109; GCN5, GENERAL CONTROL NON-REPRESSED5; H3K27me3, histone H3 tri-methylation of Lys27; IAAs, INDOLE-3-ACETIC ACID INDUCIBLEs; IPA, indole-3-pyruvic acid; JA-Ile, jasmonoyl-isoleucine; LBD16/29, LATERAL ORGAN BOUNDARIES DOMAIN16/29; NAC1, NAM/ATAF/CUC domain1; PINs, PIN-FORMED proteins; PLT1/2, PLETHORA1/2; PRC2, Polycomb Repressive Complex2; REF6, RELATIVE OF EARLY FLOWERING6; SDG2, SET-DOMAIN GROUP2; SLR_IAA14, SOLITARY ROOT_ INDOLE-3-ACETIC ACID INDUCIBLE 14; TAA1, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1; Trp, Tryptophan; WOX11/12, WUSCHEL-RELATED HOMEOBOX11/12; WOX5/7, WUSCHEL-RELATED HOMEOBOX5/7; YUCs, YUCCAs.
Root regeneration from detached organs relies on neo-biosynthesis of endogenous auxin, which is partly induced by wounding and/or the auxin accumulation resulting from the cutting off of the basal auxin drain (Cai et al., 2014; Liu et al., 2014; Chen X. et al., 2016; Druege et al., 2019; Zhang et al., 2019). Near the wounding sites, auxin level can increase rapidly in mesophyll cells, and then polar transport results in auxin accumulating in competent cells of procambium and vascular parenchyma to trigger cell fate transition (Liu et al., 2014; Chen L. et al., 2016). Loss of function of auxin influx carriers (AUX1 and LAX3) and auxin efflux carriers (ABCB19) reduces the regenerative potential of hypocotyl and leaf explants, demonstrating the importance of auxin transport for DNRR (Sukumar et al., 2013; Della Rovere et al., 2015; Bustillo-Avendaño et al., 2018). One of the main endogenous auxin biosynthesis pathways is conducted in two steps (Figure 1): first, the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family of aminotransferases converts the main precursor for IAA, tryptophan (Trp) to indole-3-pyruvic acid (IPA). Then, the YUCCA (YUC) family of flavin monooxygenases participate in the conversion of IPA to IAA (Mashiguchi et al., 2011; Won et al., 2011). During DNRR, the expression of YUC1 and YUC4 is up-regulated in response to wounding, which promotes auxin production in both mesophyll cells and competent cells resulting in cell fate transition (Table 1; Chen L. et al., 2016). The TAA1 mutation weak ethylene insensitive 8-1 (wei8-1) causes defects in AR formation, whereas the double mutant of wei8-1 tar2-2 (tryptophan aminotransferase related 2-2) was mostly unable to regenerate AR from leaf explants (Sun et al., 2016). Similarly, double mutations in YUC1/YUC4 and YUC2/YUC6 can partially block the rooting of leaf explants, while in yuc1246 quadruple mutants, rooting was severely blocked (Chen L. et al., 2016). In rice, OsYUC1 overexpression causes massive proliferation of ARs or crown roots, respectively, whereas loss of OsTAA1 reduces AR development (Zhang et al., 2018) confirming the central role of TAA and YUC mediated auxin biogenesis for AR formation that seems conserved between monocots and dicots.
TABLE 1.
Selection of transcription factors and components of phytohormone signaling evidently or putatively involved in DNRR further indicating their role in primary and lateral root development or callus formation.
| DNRR phases and stages | ||||||||||||
| Genes | PR | LR | Callus | I | II | III: | Pr | In | Pa | Em | Pro F | Comments |
| ABCB19 (MDR1) | ∙ | ∙Δ | Δ | IAA Tra | LFM display shorter LRs and reduced DNRR from hypocotyl explants, but DNRR is not impaired from intact hypocotyls (Wu et al., 2007; Sukumar et al., 2013) | |||||||
| ALF4 | ∙ | ∙Δ | Δ | Δ | Δ | IAA Sig | Regulator of SCF-TIR1 receptor, LFM acumulate IAAs, Exp in PR and LRs (Bagchi et al., 2018); LFM fail to produce LRs, callus and DNRR (Celenza et al., 1995; Sugimoto et al., 2010; Liu et al., 2014; Bustillo-Avendaño et al., 2018) | |||||
| ARF7,19 | ∙ | ∙Δ | Δ | nd | nd | nd | IAA Sig TFs | Directly activate LBD16/29 (Okushima et al., 2007), ARF7 LFM have less LRs, double mutants fail to produce any LRs, whereas DNRR is normal but callus formation is reduced, Exp in the root vasculature, LRPs and developing LRs (Okushima et al., 2005; 2007; Liu et al., 2014; Lee et al., 2017) | ||||
| ASA1 | ∙nd | ∙neΔ | Δ | IAA Syn | LFM have less DNRR (Zhang et al., 2019), less LRs in response to JA (Sun et al., 2009); Directly activated by ERF109 (Cai et al., 2014; Zhang et al., 2019) | |||||||
| AUX1, LAX3 | ∙ | ∙Δ | ∙Δ | IAA Tra | LFM have less LRs and detached mutant hypocotyls show a reduced rooting capacity for ARs, Exp in PR, LRs and ARP (Marchant et al., 2002; Swarup et al., 2008; Della Rovere et al., 2013; 2015; Bustillo-Avendaño et al., 2018) | |||||||
| COI1 | ∙(Δ) | Δ | Δ | JA Sig | JA receptor, LFM causes reduced DNRR (Zhang et al., 2019) and LR formation in response to JA (Raya-González et al., 2012); Exp in RAM, LFM are insensitive to root growth inhibition by JA (Chen et al., 2011) | |||||||
| ERF109 | nd | ∙Δ | ∙Δ | TF | JA induced after leaf detachment, LFM have reduced DNRR (Zhang et al., 2019); LFM have less LRs, ativates ASA1 and YUC2 (Cai et al., 2014) | |||||||
| ERF115 | ∙Δ | ∙Δ | (Δ) | ? | ? | TF | Induced by JA, IAA and ERF109 in protoxylem and QC cells, involved in root cell regeneration (Zhou et al., 2019); ERF115-SRDX blocks callus formation (Ikeuchi et al., 2017); controls QC cell devision (Heyman et al., 2013) | |||||
| FUS3, LEC2 | ∙Δ | ∙Δ | ? | TFs | Two homologous B3 TFs interact to activate directly YUC4 during LR formation (Tang et al., 2017); precocious growth of PR during embryogenesis (Vicente-Carbajosa and Carbonero, 2005) | |||||||
| GA1 (CPS1), GA5 (GA20OX1) | ∙Δ | GA Syn | Ent-Copalyl Diphosphate Synthetase and GA 20-Oxidase, respectively; involved in vascular proliferation in DNRR, LFM have less AR capacity (Ibáñez et al., 2019) | |||||||||
| GAI | ∙Δ | GA Sig | gai-1 is insensitive to GAs, involved in vascular proliferation during DNRR, LFM have reducted AR capacity (Ibáñez et al., 2019) | |||||||||
| IAA14 (SLR) | ∙(Δ) | Δ | Δ | Δ | Δ | IAA Sig TF | GFM, fail to produce LRs (Fukaki et al., 2002); less callus and DNRR (Shang et al., 2016; Bustillo-Avendaño et al., 2018); PR has less root hairs, Exp in RAM of PR and LRP (Fukaki et al., 2002; Vanneste et al., 2005) | |||||
| LBD16,29 | ∙ | ∙Δ | Δ | ∙Δ | ∙Δ | (∙) | TFs | OE enhances AR formation (Liu et al., 2014); Direct target of ARF7/19 (Okushima et al., 2007); LFM have less LRs and callus (Fan et al., 2012) | ||||
| NAC1 | ∙Δ | ∙Δ | ∙Δ | TF | Dominant-negative lines (NAC1-SRDX) have less LRs (Xie et al., 2000) and less ARs (Chen X. et al., 2016) | |||||||
| PIN 1,2,3,7 | ∙Δ | ∙Δ | ∙Δ | IAA Tra | Exp in leaf vasculature after excision, LFM have less ARs (Bustillo-Avendaño et al., 2018); Invoved PR and LR development (Petrásek and Friml, 2009) | |||||||
| PLT1,2 | ∙Δ | ∙Δ | Δ | Δ | TFs | Activate WOX5 (Shimotohno et al., 2018); double mutants: less DNRR (Bustillo-Avendaño et al., 2018), shorter PR but more LRs (Aida et al., 2004); Exp in LRPs and RAM of PR (Hofhuis et al., 2013; Du and Scheres, 2017) | ||||||
| PLT3,5,7 | ∙Δ | ∙ | ? | ? | ? | TFs | Promotes LR emergence, triple mutants have less LRs, Exp in a subset of pericycle cells requiring ARF7/19 as activators (Hofhuis et al., 2013) | |||||
| SCR | ∙Δ | ∙ | ∙ | ∙Δ | ∙ | ∙ | ∙ | TF | AR formation from hypocotyl is inhibited in LFM (Della Rovere et al., 2015); Involved in positioning the stem cell niche of RAMs, Exp in endodermis, QC and callus (Sabatini et al., 2003; Sugimoto et al., 2010; Kim et al., 2018) | |||
| SHR | ∙Δ | ∙Δ | ∙ | Δ | Δ | TF | LFM have reduced AR and LR formation as well as growth of the PR, Exp in the stelle, shr plt1,2 triple mutants fails to produce ARs (Helariutta et al., 2000; Lucas et al., 2011; Della Rovere et al., 2015; Bustillo-Avendaño et al., 2018) | |||||
| TAA1 (WEI8), TAR2 | ∙ | ∙Δ | ∙ | IAA Syn | Ubiquitously induced in leaf explants, double mutants are impaired in DNRR (Sun et al., 2016); Exp in RAM of PR (Stepanova et al., 2008) | |||||||
| TCP20,21 | (∙Δ) | ? | ? | TFs | Interact with PLT1/3 and SCR to bind and induce WOX5, Exp in precursor QC cells (in embryos) (Shimotohno et al., 2018) | |||||||
| WOX5,7 | ∙Δ | ∙Δ | ∙nd | ∙Δ | ∙Δ | ∙Δ | ∙ | TF | Activated by WOX11/12, WOX5 LFM have reduced DNRR, which is enhanced in double mutants (Hu and Xu, 2016); WOX5 maintains the stem cell niche of RAM, Exp in QC and callus (Sarkar et al., 2007; Sugimoto et al., 2010; Kim et al., 2018), whereas WOX7 is involved in LR initiation (Kong et al., 2016) | |||
| WOX11,12 | (Δ) | ∙Δ | ∙Δ | TF | LFM have less ARs, whereas OE inhibits AR and callus formation, Exp in AR founder cells, promotes LBD16/19 (Liu et al., 2014; Sheng et al., 2017) | |||||||
| YUC1,2,4,6 | ∙Δ | ∙Δ | IAA Syn | Induced in mesophyll cells of leaf explants, double mutants are partially, quadruple mutants severely impaired in DNRR (Chen L. et al., 2016), LFM of YUC4 reduces LR formation (Tang et al., 2017) | ||||||||
∙, expressed; ∆, mutant phenotype; ?, assumed involvement in DNRR; I, Phase I; II, Phase II; III:, Phase III:; Pr, Priming; In, Initiation; Pa, Patterning; En, Emergence; AR, adventitious root; ARP, adventitious root primordia; Exp, Expressed; GA sig, GA signaling; GA syn, GA biosynthesis; GFM, gain of function mutants; IAA sig, auxin signaling; IAA syn, auxin biosynthesis; IAA tra, auxin transport; JA sig, JA signaling; LFM, loss of function mutants; LR, lateral root formation; LRP, lateral root primordia; methylase, histone lysine methyl-transferase; nd, no defects observed; ne, not expressed; OE, overexpression; PR, primary (main) root; Pro F, protein function; QC, quiescent center; RAM, root apical meristem; TF(s), transcription factor(s).
Regulation of DNRR by Other Phytohormones
Beside the master player auxin, other phytohormones promote or antagonize DNRR: Cytokinin (CK), strigolactone (SL), and abscisic acid (ABA) suppress auxin production and/or AR formation whereas brassinosteroid (BR) and ethylene (ET) have positive effects (Table 1; Su and Zhang, 2014; Druege et al., 2019; Lakehal and Bellini, 2019). Although older studies suggest that gibberellin (GA) treatment inhibits AR formation (Busov et al., 2006; Mauriat et al., 2014), loss of components of GA synthesis (GA1 and GA5, GA REQUIRING) or GA signaling (GAI, GIBBERELLIC ACID INSENSITIVE) causes defective vascular proliferation and consequently delayed AR formation in leaf explants indicating a positive role for GA in DNRR (Ibáñez et al., 2019). The stress phytohormone jasmonic acid (JA) and its biologically active form jasmonoyl-isoleucine (JA-Ile) are derivates of the lipid α-linolenic acid (Huang et al., 2017). After wounding, JA and JA-Ile levels increase rapidly in local and undamaged distal plant tissue (Glauser et al., 2008). Until recently, it was a matter of dispute whether JA facilitates or inhibits DNRR (Lakehal and Bellini, 2019). Ahkami et al. (2009) suggested that JA is a positive regulator of AR formation since it rapidly accumulates at the wounding site before ARs emerge in petunia leafy stem explants. On the other hand, continuous JA treatment inhibits AR formation whereas it promotes LR formation (Sun et al., 2009; Lischweski et al., 2015). The latter is partially inhibited in yuc mutants indicating that JA triggered LR formation is dependent on auxin synthesis (Sun et al., 2009; Cai et al., 2014). Reviewing the experimental details of diverse studies on the role of JA in adventitious rooting and considering relations to auxin homeostasis and signaling, Druege et al. (2019) recently provided a coherent explanation for the different findings and postulated that early, particularly wound-induced, JA accumulation stimulates AR formation in cuttings via IAA accumulation in the stem base and/or canalization toward AR source cells, while induction of invertases as molecular drivers of sink activity may be further involved. According to these theories, it was recently shown by Zhang et al. (2019) that in detached leaf explants, JA concentration and expression of JA response genes rise very quickly, reach a maxima after 1 hour and then decline in Arabidopsis leaf explants whereas auxin synthesis and other auxin-related genes reach their expression maxima one hour later (2 h after leaf detachment) indicating that JA signaling, triggered by wounding, precedes auxin signaling dependent cell fate reprogramming during DNRR. Similarly, ET biosynthesis is also triggered by wounding and stimulates AR formation at the stem base of cuttings in petunia, whereas auxin controls the timing of different phases of DNRR (Druege et al., 2014). There is indication in the literature that the ET signal is important for the early reprogramming of the AR source cells and may act via enhanced auxin level and/or sensitivity (Druege et al., 2019; Lakehal and Bellini, 2019).
Key Transcriptional Regulators of DNRR
At the onset of DNRR in excised Arabidopsis leaves, wounding triggers the immediate accumulation of JA which activates the expression of ERF109 (ETHYLENE RESPONSE FACTOR109) encoding a transcription factor (TF) (Figure 1 and Table 1; Cai et al., 2014; Zhang et al., 2019). Loss-of-function mutants of ERF109 or the JA receptor COI1 (CORONATINE INSENSITIVE1) display defective rooting from leaf explants (Zhang et al., 2019). ERF109 upregulates directly ANTHRANILATE SYNTHASEα1 (ASA1) – a rate-limiting enzyme in tryptophan (Trp) biosynthesis – and YUC2 indicating that ERF109 mediates cross-talk between JA and auxin biosynthesis during DNRR (Cai et al., 2014; Zhang et al., 2019). To prevent hypersensitivity to wounding, JASMONATE ZIM-domain (JAZ) repressors bind physically to ERF109 proteins and so inhibit the activation of ASA1 by ERF109 (Zhang et al., 2019). Later, the accumulation of auxin at the wounding site triggers the expression of further key TFs forming a regulatory network (Figure 1) that is initially different but subsequently identical to the transcriptional network controlling regular root development. Thereby, the signaling pathway from auxin perception to transcriptional responses consists of only two steps: First, the binding of auxin to the receptor and F-box protein TRANSPORT INHIBITOR RESISTANT1 (TIR1) triggers the ubiquitin-mediated degradation of the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressors. Then, the degradation of the Aux/IAAs breaks the physical inhibition of the AUXIN RESPONSE FACTORs (ARFs), which bind as transcriptional activators to auxin response elements (AuxREs) in the promoters of auxin response genes (reviewed in more detail in Weijers and Wagner, 2016). In aberrant lateral root formation4-1 (alf4-1) mutants, the CULLIN1 subunit of the SCFTIR1 auxin receptor complex is destabilized leading to increases in the levels of Aux/IAA proteins, the repressors of ARFs (Bagchi et al., 2018). As the name implies, alf4-1 mutant plants are impaired in lateral root formation (Celenza et al., 1995) but they also fail to regenerate roots from leaf explants (Liu et al., 2014). Furthermore, the alf4-1 mutation blocks callus induction suggesting that callus and LR formation are under the same genetic control (Sugimoto et al., 2010). The reduction of auxin response in the alf4 mutant during LR and callus formation is also caused by the increasing level of IAA14 (Perez-Garcia and Moreno-Risueno, 2018). solitary root-1 (slr-1) mutants, which carry a dominant-negative version of IAA14, are not able to form regular LRs or callus on CIM (Shang et al., 2016) but can grow LRs after cutting the main root (root pruning) (Table 1; Xu et al., 2017).
During DNRR, the newly formed auxin maximum induces quickly the expression of WUSCHEL-RELATED HOMEOBOX11 (WOX11) and its functional homolog WOX12 in procambium and parenchyma cells, which mediates cell fate transition toward the establishment of root founder cells (Figure 1; Liu et al., 2014; Xu, 2018). Notably, the current state of research cannot rule-out that root founder cells of ARs can also initiate from differentiated cells beside procambium and parenchyma tissue via cell fate reprogramming (Yu et al., 2017). Although loss of OsWOX11 causes severe growth defects including a near abolition of crown root production in rice, wox11 and wox12 single and double mutants show only slightly reduced AR formation in Arabidopsis (Zhao et al., 2009; Liu et al., 2014). The WOX11 promoter region carries several AuxREs, which are essential for the auxin response indicating direct binding and activation of WOX11 by ARFs (Liu et al., 2014). Although WOX11 promotes AR and callus formations, WOX11 is not involved in regular lateral root initiation (Liu et al., 2014; Sheng et al., 2017). The transition from root founder cells to root primordium cells is accompanied by decreasing WOX11 and WOX12 expression levels while those of WOX5 and WOX7 increase (Liu et al., 2014; Hu and Xu, 2016). This temporal succession of WOX11 and WOX5 expression is very similar to the temporal expression pattern during callus development supporting the idea that callus and AR initiation share the same genetic pathway at the cellular and molecular level (Liu et al., 2014). Recently, it has been shown that WOX11/12 directly activate WOX5/7 by binding to the promoters of WOX5 and WOX7 (Hu and Xu, 2016). WOX5, which encodes the functional homolog of the shoot stem cell promoting factor WUSCHEL (WUS), is expressed in the quiescent center (QC) of RAMs (Sarkar et al., 2007). Although wox5-1 mutants form roots with disorganized RAMs, wox5-1 mutant roots fail to maintain distal (columella) stem cells and, redundantly with the loss of other regulators, proximal stem cells during root development (Sarkar et al., 2007). In contrast to the activation of WOX11/12 by ARFs, ARF10 and ARF16 repress and restrict WOX5 to the QC (Ding and Friml, 2010). Interestingly, auxin is also required to activate WOX5/7 expression in root founder cells, which divide to form root primordia cells during DNRR (Hu and Xu, 2016). As WOX5 is expressed in callus as well as in RAMs of primary root, LRs and ARs, it is difficult to predict the stages of direct DNRR that would be affected by loss of WOX5 (Table 1).
WOX11/12 regulate at least partially the formation of AR and callus through activation of LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16) and LBD29 (Figure 1; Liu et al., 2014), whereby WOX11 directly binds to the WOX-binding sites in the LBD16 promoter region (Sheng et al., 2017). The relationship between WOX11 and LBD16 is also important for shoot regeneration because both promote the pluripotency acquisition in callus cells (Liu et al., 2018). However, the activation of LBD16 by WOX11 is not required for regular LRs that are also known as non-WOX11-mediated roots (Sheng et al., 2017). LBD16/29 are also direct targets of ARF7 and ARF19 during lateral root formation, and arf7-1 arf19-1 double mutants produce defective lateral roots (Okushima et al., 2005, 2007; Xu et al., 2017). Conversely, leaves of arf7-1 arf19-1 double mutant are still able to induce root regeneration and callus formation (Liu et al., 2014; Lee et al., 2017) and can recover LRs after cutting the tip of the primary root (Sheng et al., 2017; Xu et al., 2017).
Although LBD16 and WOX5 are regulated by WOX11/12 and auxin, their expression patterns are different: LBD16 is expressed in dividing root founder cells and the root primordia, but decreases during the formation of the root meristem whereas WOX5 is restricted to the stem cell niche in the new RAM (Hu and Xu, 2016). Overexpression of LBD16/29 can induce callus without exogenous auxin treatment, while LBD16-induced callus displays ectopic expression of WOX5 and PLETHORA (PLT1) (Fan et al., 2012). PLT1 and PLT2 genes, whose transcription requires auxin accumulation and ARFs, are essential for QC specification and stem cell activity in the RAM (Aida et al., 2004). During embryogenesis and LR formation, PLT proteins physically interact with SCARECROW (SCR) and TCP (teosinte-branched cycloidea PCNA) TFs to specify and maintain the new formed QC and stem cell niche (Shimotohno et al., 2018). PLT–TCP–SCR complexes assemble on PLT-binding sites in the WOX5 promoter to induce WOX5 expression (Shimotohno et al., 2018). In turn, WOX5 is needed for PLT1 expression in RAMs (Ding and Friml, 2010). Interestingly, the PLT genes PLT3/5/7, which are essential in shoot regeneration, facilitate pluripotency in callus tissues by activating the root-specific stem cell regulators PLT1 and PLT2 (Kareem et al., 2015). During callus formation, PLT1/2, WOX5, and SCR can act as major regulators in the establishment and maintenance of cell regeneration capacity and possible pluripotency by inhibiting factors that in turn promote differentiation (Kim et al., 2018). plt1 plt2 double mutants as well as short-root (shr) single mutants display significant reduction of leaf explants rooting and in rooting capacity (Bustillo-Avendaño et al., 2018). In shr plt1 plt2 triple mutants, AR primordia initiation is fully blocked, because the postembryonic root founder cells cannot form. These results demonstrate the importance of PLT1, PLT2, and SHR for DNRR (Table 1; Bustillo-Avendaño et al., 2018).
During DNRR, the emergence of ARs seems an easy task but leaf explants expressing a dominant-negative version of the NAC1 TF (NAC1-SRDX) fail to grow-out ARs, although they are unaffected in the earlier steps of auxin-mediated cell fate transition (Chen X. et al., 2016). NAC1 induces the expression of CEP (Cys endopeptidase) genes, which encode proteins that might be involved in programed cell death and in degradation of extensin proteins in the cell wall (Chen X. et al., 2016). Therefore, the NAC1 pathway controls auxin-independently the emerging of ARs by loosening of cell walls of the surrounding tissue (Table 1).
Callus is an organized tissue similar to LRP (Sugimoto et al., 2010), while LRP and adventitious root primordia (ARP) differ only in their early steps of formation (Sheng et al., 2017). All three developmental events largely share TFs, which control the morphological changes in similar hierarchical networks (see model for DNRR in Figure 1). Most components of the transcriptional network are direct targets of ARFs that might suggest simultaneous expression rather than the observed activation in a chronological order. Obviously, the reciprocal and hierarchical regulations of the TFs contribute to their distinct spatiotemporal expression pattern. Nevertheless, epigenetic regulation can stabilize gene networks, restrict gene expression to specific tissue and/or provide a time buffer, which allows delayed transcriptional response to an upstream TFs (Müller and Goodrich, 2011; Xiao et al., 2016).
Epigenetic Regulation of DNRR by the Repressive Mark H3K27me3
Epigenetic gene regulation here refers to mitotically or occasionally meiotically heritable changes in transcriptional activity that are not caused by changes in the DNA sequence but rather by covalent modifications to histone residues and DNA methylation (Bannister and Kouzarides, 2011; Smith and Meissner, 2013). The covalent nature of epigenetic chromatin modifications allows both stability through cell division as well as reversibility during development in response to extrinsic signals and endogenous clues (Bannister and Kouzarides, 2011; Müller-Xing et al., 2014b). For every chromatin modification like lysine methylation of histones exist both writers, which refers here to histone methyltransferases, and erasers, which refer here to histone demethylases, functioning as single proteins or protein complexes with enzyme activity (Xiao et al., 2016). Furthermore, chromatin modifications are specifically bound by so-called readers, proteins with domains which provide docking modules for the enzyme complexes (Andrews et al., 2016). While epigenetic regulation of callus formation and regular RAM development were subjects of several studies (Lee and Seo, 2018; Takatsuka and Umeda, 2015), the relationship of epigenetics and DNRR is a relatively new frontier, which was recently outlined as one of the hot topics for future research on adventitious rooting in cuttings (Chen L. et al., 2016; Lee et al., 2018; Druege et al., 2019; Zhang et al., 2019). To complete the current model of DNRR regulation and indicate future research directions, we also draw here parallels between the epigenetic control of callus formation, which is one of the earliest steps of cell fate reprogramming during AR initiation, and regular root development (Figure 1 and Table 2).
TABLE 2.
Epigenetic factors evidently or putatively involved in DNRR further indicating their role in primary and lateral root development or callus formation.
| DNRR phases and stages | |||||||||||
| Genes | PR | LR | Callus | I | II | III: | Pr | In | Pa | Em | Comments |
| SDG2 (ATXR3) | Δ | Δ | nd | ? | ? | H3K4me3 methylase involved in RAM organisation (Yao et al., 2013) | |||||
| ATX1 | Δ | Δ | nd | ? | H3K4me3 methylase required for resrtriction of QC markers and for LR initiation, morphogenesis, and emergence (Napsucialy-Mendivil et al., 2014) | ||||||
| SDG8 | Δ | nd | Δ | H3K36me3 methylase required for activation of ASI1 by ERF109 (Zhang et al., 2019), LOF causes less LR (Cazzonelli et al., 2009) | |||||||
| ATXR2 | Δ | Δ | Δ | Δ | H3K36me3 methylase, interacts with ARF7/9 and activates LBD16/29 (Lee et al., 2017, 2018) | ||||||
| CLF | Δ | Δ | nd | ? | ? | H3K27me3 methylase of PRC2, represses PIN1, LOF causes increased RAM, root size and the number of LRs (Gu et al., 2014; Wang et al., 2019) | |||||
| SWN | Δ | nd | ? | H3K27me3 methylase of PRC2, LOF causes decreased RAM and root size (Lucas et al., 2016) | |||||||
| CLF,SWN | Δ | Δ | Δ | ? | Δ | Double mutants fail to silence leaf identity genes, which prevent callus (He et al., 2012) and also AR formation (Liu et al., 2014) | |||||
| EMF2 | Δ | Δ | Δ | ? | ? | Component of PRC2, LOF inhibits PR growth, callus and LR formation (He et al., 2012; Gu et al., 2014) | |||||
| REF6 | Δ | nd | ? | ? | H3K27me3 demethylase, activates PIN1/3/7 that allows auxin transport and accumulation in LRP founder cells (Wang et al., 2019) | ||||||
| PKL | Δ | ? | CHD3 chromatin remodeler, conteracts as TrxG factor PRC2 function and maintains root stem cells (Aichinger et al., 2009, 2011) | ||||||||
| HAG1 (GCN5) | Δ | Δ! | nd | Histone acetyltransferase, activates PLT genes and maintains the stem cell niche (Kornet and Scheres, 2009), LOF causes faster callus growth (!) but does not impair indirect DNRR (Kim et al., 2018) | |||||||
∆, mutant phenotype; ?, assumed involvement in DNRR; I, Phase I; II, Phase II; III:, Phase III:; Pr, Priming; In, Initiation; Pa, Patterning; En, Emergence; AR, adventitious root; LOF, loss of function; LR, lateral root formation; LRP, lateral root primordia; methylase, histone lysine methyl-transferase; nd, no defects observed in callus formation (He et al.,2012) or indirect DNRR (Kim et al. 2018); PR, primary (main) root; RAM, root apical meristem; QC, quiescent center.
Reprogramming of cell fate requires the activation and repression not only of a few genes but whole transcriptional networks controlling developmental programs and it is accompanied by local and global changes in epigenetic modifications (Lee and Seo, 2018). The Polycomb repressive complex 2 (PRC2) is a key “writer” which deposits the repressive marks di- and tri-methylation of Lys27 on histone H3 (H3K27me2/3). It is assembled from four highly conserved core components as well as a more variable collection of associated proteins, some of which are DNA binding proteins that may guide the PRC2 to Polycomb (PcG) target genes (Xiao et al., 2017; Zhou et al., 2018). In Arabidopsis, three partially redundant homologs CURLY LEAF (CLF), MEDEA (MEA), and SWINGER (SWN) encode the catalytic subunit of the PRC2 histone methyltransferase (HMT), while EMBRYONIC FLOWER2 (EMF2) encodes a second core PRC2 component (Schmitges et al., 2011; Müller-Xing et al., 2014a). In contrast, RELATIVE OF EARLY FLOWERING6 (REF6) and its two close paralogs, EARLY FLOWERING6 and JMJ13, are three partially redundant H3K27me3 demethylases (erasers) with important functions in reprogramming during plant development (Table 2; Yan et al., 2018).
During callus formation from leaf explants, over 400 PcG targets lose H3K27me3, whereas less than 200 targets gain H3K27me3, indicating the significance of reactivation of former silenced genes (He et al., 2012). The levels of H3K27me3 decrease first at several genes of the auxin pathway including YUC4, NITRILASE2 (NIT2), IAA CARBOXYL METHYLTRANSFERASE1 (IAMT1), and PIN-FORMED1 (PIN1). Subsequently, H3K27me3 levels increase at leaf specific genes but decrease at genes involved in root development suggesting a central role for the repressive epigenetic mark H3K27me3 during regeneration (He et al., 2012). WOX11 expression, which is essential for establishing root founder cells during DNRR, depends on endogenic auxin synthesis by YUC proteins (Liu et al., 2014; Chen L. et al., 2016). The gene loci of YUC1 and YUC4, as well as WOX11, are H3K27me3-marked PcG targets whose levels decline in callus tissue cultured on CIM (He et al., 2012; Liu et al., 2014; Chen L. et al., 2016). Principally, reduction of H3K27me3 levels at target genes can be achieved either by active removal through H3K27me3 demethylases or by H3K27me3 dilution through cell divisions (Sun et al., 2014). During DNRR from leaf explants, the activation of YUC1 and YUC4 expression is accompanied by decreasing H3K27me3 levels after only a few hours on B5 medium (Chen L. et al., 2016) suggesting an active removal, e.g., by H3K27me3 demethylases as little or no cell division and DNA replication occurs in this time period.
In primary root and LRs, the polar auxin transport and local auxin maxima are widely achieved by the family of PIN-FORMED (PIN) efflux carriers, which includes PIN1, PIN2, PIN3, PIN4, and PIN7 (Petrásek and Friml, 2009). PRC2 inhibits LR formation and root growth by depositing the repressive H3K27me3 mark on chromatin at the PIN1 locus (Gu et al., 2014). Whereas the H3K27me3 demethylase REF6 binds to a specific DNA sequence (CTCTGYTY) and de-represses PIN1/3/7 during LR formation (Lu et al., 2011; Cui et al., 2016; Wang et al., 2019). Accordingly, ref6 mutants have fewer LRs than wild-type (Wang et al., 2019). In contrast, ref6 mutants are not impaired in callus formation from leaf explants (He et al., 2012), the first step of AR initiation (Sheng et al., 2017). As the contribution of REF6 to DNRR has not yet been tested, it remains unclear whether in ref6 mutant explants, AR formation is unaffected, as suggested by the result of callus formation, or delayed like LR formation in ref6. During rice shoot development, OsWOX11 recruits the REF6 homolog JUMONJI705 (JMJ705) to promote gene expression by H3K27me3 demethylation (Cheng et al., 2018). It would be interesting to test whether WOX11 and REF6 cooperate similarly during regular root development and DNRR in Arabidopsis.
PRC2 complexes containing either the HMT CLF or SWN are essential for post-embryonic plant development (Gutzat et al., 2012) and apparently play a role in DNRR. Although clf-50 swn-1 double mutants can form normal callus from root tissue, clf-50 swn-1 double and emf2 single mutants are defective in callus formation from leaf explants (He et al., 2012). In the latter case, the plants with reduced PRC2 activity fail to silence leaf identity genes such as SAWTOOTH1 (SAW1) and SAW2, whereas the root identity genes WOX5 and SHR are de-repressed as during normal callus formation (Kumar et al., 2007; He et al., 2012). Similarly, clf-50 swn-1 double mutants fail to form AR from leaf explants (Liu et al., 2014) but it is not yet clear whether impaired silencing of SAW1/2 or other leaf-regulatory genes plays a role. swn clf ± mutant explants, presumably harboring reductions in H3K27me3 levels, display an earlier re-activation of WUS enabling faster shoot regeneration (Zhang et al., 2017). This faster activation of WUS is likely achieved by easier access of the B-Type ARR and HD-ZIP class III TFs to the WUS chromatin (Zhang et al., 2017). Facilitated access to DNRR-related genes may also accelerate root regeneration in PcG mutants. Loss of CLF causes ectopic LR formation, a longer primary root (Gu et al., 2014), and higher root meristem activity as indicated by increased WOX5 expression and meristem size (Aichinger et al., 2011). On the other hand, the loss of the CLF homolog SWN causes shorter roots and decrease in the RAM size indicating that root growth defects varies in different PcG mutants (Lucas et al., 2016). Hence, alternative CLF-PRC2 and SWN-PRC2 complexes might target selectively different genes, which either promote or inhibit regular root development, but possibly also DNRR (Table 2).
Epigenetic Regulations of DNRR by Histone Modifications With Positive Effects on Transcription
Trithorax group (TrxG) proteins are a diverse group of antagonists of PcG-mediated gene repression that were originally defined genetically by their ability to suppress PcG mutant phenotypes (Kennison and Tamkun, 1992). Some TrxG proteins promote PcG target gene expression by catalyzing activation marks, others by removing the repressive H3K27me3 mark and yet others by chromatin remodeling to slide nucleosomes and facilitate access. The principle marks associated with transcriptional activation are di- and tri-methylation of histone H3 at lysine 4 and/or lysine 36 (H3K4me2/3 and H3K36me2/3) (Papp and Müller, 2006). In the early phase of DNRR, the upregulation of ASA1 by JA-activated ERF109 required H3K36me3 pre-deposition by SET DOMAIN GROUP8 (SDG8), which is also involved in LR formation (Table 2; Cazzonelli et al., 2009; Zhang et al., 2019). Another H3K36me3 HMTase, ARABIDOPSIS THALIANA TRITHORAX-RELATED2 (ATXR2) is involved in callus and AR formation by activating LBD16 and LBD29 expression (Lee et al., 2017, 2018). ATXR2 binds directly to the promoter region of LBD16 and LBD29 to depositing H3K36me3 at these loci (Lee et al., 2017). In addition, ATXR2 interacts physically with the known activators of LBD16 and LBD29 ARF7 and ARF19, suggesting that these ARF TFs could recruit ATXR2 to both LBD promoters (Lee et al., 2017). SDG2, also known as ATXR3, encodes the main H3K4me3 HMTase (Berr et al., 2010; Guo et al., 2010) and is required for the organization and function of stem cell niche in the RAM (Yao et al., 2013). sdg2-3 mutants have a shorter root and fewer LRs than wild type (Yao et al., 2013) but nonetheless are still able to induce callus formation (He et al., 2012), leaving the question unanswered whether DNRR initiation or only the later step of the emergence of ARs are impaired in sdg2 mutants. The preliminary data concerning other TrxG proteins such as ARABIDOPSIS THALIANA TRITHORAX1 (ATX1, encoding a H3K4me3 HMTase) are inconclusive or contradictory, for example inactivation causes reduced root length and LR number yet the mutants are able to form callus from leaf explant (He et al., 2012; Napsucialy-Mendivil et al., 2014). The chromatin remodeler PICKLE (PKL) counteract PcG function as pkl mutants display reduced expression of some PcG target genes (Aichinger et al., 2009) and have shorter roots and reduced root stem cell activity (Aichinger et al., 2011). It was proposed that PKL, together with PICKLE RELATED2 (PKR2), activate PcG targets outside of the RAM to promote cell differentiation, whereas PKL specifically maintains root stem cell (Aichinger et al., 2011). Plants carrying the slr-1 mutation are blocked in auxin signaling, and so the initiation of LRs is blocked (Shang et al., 2016). In slr-1 pkl double mutants, LR formation is rescued in an ARF7/19 function dependent manner revealing that chromatin remodeling by PKL regulates negatively auxin-mediated LR formation (Fukaki et al., 2006). It remains to be tested whether PKL or PKR2 have a similar function in AR formation.
Histone lysine acetylation (Kac) may have more direct links to transcriptional control than most other chromatin modifications, as Kac promotes transcription not only by recruiting reader proteins but also by neutralizing the positive charge on the lysine side chain, which directly facilitates RNA synthesis by making the DNA more accessible to the transcriptional machinery (Zhao et al., 2018). Kac can overcome repressive histone marks such as H3K27me3 or can be replaced by such epigenetic marks. The histone acetyltransferase HISTONE ACETYLTRANSFERASE OF THE GNAT FAMILY1 (HAG1), also known as A. thaliana GENERAL CONTROL NON-REPRESSED5 (AtGCN5) (Vlachonasios et al., 2003; Servet et al., 2010), plays a key role in the establishment of callus pluripotency and subsequent shoot regeneration (Kim et al., 2018). The expression of PLT genes is positively regulated by AtGCN5 to maintain the stem cell niche in roots (Kornet and Scheres, 2009). hag1-6 mutants display very short roots and a smaller root meristem size (Kornet and Scheres, 2009) but faster-growing callus on CIM, which is associated with decreasing expression of the root key regulators WOX5, SCR, PLT1, and PLT2 (Kim et al., 2018). Although hag1-6 mutant callus fails to induce shoot regeneration on shoot inducing medium (SIM), it is surprisingly able to regenerate roots after transferring from CIM to RIM (Table 2; Kim et al., 2018). In rice, OsWOX11 interacts with the ADA2-GCN5 histone acetyltransferase complex to activate downstream target genes during AR formation (Zhou et al., 2017) giving a textbook example of how TFs acts as recruiter of epigenetic factors (EFs) for long-term transcription regulation.
Conclusion and Outlook
The integration of genes and their protein products in transcriptional networks, such as the one controlling DNRR, often start with the description of their mutant phenotypes and their expression patterns. Nevertheless, the components of transcriptional-epigenetic networks have to be interconnected through direct binding of one component to the promoter of another component or by direct protein-protein interaction or by phytohormone or other signaling pathways. These physical interactions need to be tested by yeast-two-hybrid (Y-2-H) or proteomics assays, chromatin immunoprecipitation (ChIP), electrophoretic mobility shift assay (EMSA), DNA-affinity precipitation assay (DAPA) and other approaches. Recent progress in DNRR research allowed us to develop a model on the transcriptional and epigenetic reprogramming network controlling DNRR (Figure 1). However, several links of the model are not yet verified in DNRR systems and several gaps remain obvious, for example: (1) Taking into account that ERF109, which functions upstream of ERF115 (Zhou et al., 2019), is JA inducible, it would be interesting to test whether MYC2, the main JA response factor, can directly bind to the ERF109 promoter or whether the loss of other JA signaling components impairs DNRR, like for mutations in the JA receptor COI1 (Zhang et al., 2019). (2) Although several publications showed that auxin can induce NAC1 (Xie et al., 2000; Guo et al., 2005; Wang et al., 2006), a recent publication suggests that NAC1 expression near the wounding side does not require auxin (Chen X. et al., 2016) raising the question whether JA or another signal could be the missing link. (3) Considering the linkages between ET signaling, the WIND (WOUND INDUCED DEDIFFERENTIATION) TF gene family, micro RNAs and epigenetic processes recently outlined by Druege et al. (2019), these relationships should additionally be taken into account. (4) It is also unclear whether WOX5/7 or PLT1/2 are direct targets of LBD16/29. Another open question is whether ARs only originates from cells, which are root regeneration-competent from the beginning or is it possible that ARs also derivate from cells, which have first to acquire root regeneration competence.
Writing and erasing of histone marks facilitates and stabilizes long-term changes in transcriptional programs. Therefore, the recruitment of EFs by long non-coding RNAs and TFs is highly important for DNRR yet our current knowledge is very limited and only a few interactions between TFs and EFs are known. Using DNRR-related TFs as bait, approaches, like Y-2-H screen or immunoaffinity-purification with subsequent mass-spectrometry, can identify further TF/EF protein complexes. On the other hand, if the up-stream TFs are unknown, they can be identified in yeast-one-hybrid screens using promoter sequences of DNRR-related genes.
DNRR itself is an inducible system that can give us temporal resolution of gene expression and changes of epigenetic marks during the reprogramming of cell fate. Nevertheless, most DNRR studies, involving ChIPs and other chromatin-related approaches, used callus, mixed callus/explant tissue or whole leaf explants. At least after accumulation of auxin in the competent cells, using the mixed tissue cannot provide the needed spatiotemporal resolution. An approach combining DNRR and INTACT (isolation of nuclei tagged in specific cell types) system (Deal and Henikoff, 2011) or classical protoplast sorting (Birnbaum et al., 2003) will provide more specific and accurate data.
The question of transferability of DNRR studies in Arabidopsis to crop plants and trees seems to be a big issue as Arabidopsis is still the main genetic and molecular biological tool in plant research. Arabidopsis belongs to the Brassicaceae family, which include many important agricultural crop varieties, such as canola and cabbage. Therefore, the knowledge gained from Arabidopsis research has also agricultural significance (Paulraj and Yeung, 2012). Interestingly, several key regulators of DNRR were first described in rice including (Os)WOX11 and ARL1/LOB29 (Liu et al., 2005; Zhao et al., 2009) with similar functions in AR development indicating a high degree of conservation of the DNRR transcriptional networks between monocots and dicots and therefore likely between most of the crop plants. Nevertheless, that has to be confirmed by further efforts to study the molecular mechanisms of DNRR in crops.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Justin Goodrich and Lin Xu for critical reading and comments on the manuscript and all members of our group for helpful discussions and apologize for references not cited due to space limitations.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Project Nos. 31640054 and 31771602), the Fundamental Research Funds for the Central Universities, China (Grant No. 2572016DA03), and the Natural Science Foundation of Heilongjiang Province of China, General Program (Grant No. C2016007) to QXi and RM. We are also grateful for financial support from the Northeast Forestry University Starting Grant for Distinguished Young Scholar to RM.
References
- Ahkami A. H., Lischewski S., Haensch K.-T., Porfirova S., Hofmann J., Rolletschek H., et al. (2009). Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol. 181 613–625. 10.1111/j.1469-8137.2008.02704.x [DOI] [PubMed] [Google Scholar]
- Ahkami A. H., Melzer M., Ghaffari M. R., Pollmann S., Ghorbani Javid M., Shahinnia F., et al. (2013). Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta 238 499–517. 10.1007/s00425-013-1907-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C. B. R., Di Mambro R., Sabatini S., Köhler C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23 1047–1060. 10.1105/tpc.111.083352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C. B. R., Farrona S., Reyes J. C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:e1000605. 10.1371/journal.pgen.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Al-Ghazi Y., Muller B., Pinloche S., Tranbarger T. J., Nacry P., Rossignol M., et al. (2003). Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signalling. Plant Cell Environ. 26 1053–1066. 10.1046/j.1365-3040.2003.01030.x [DOI] [Google Scholar]
- Andrews F. H., Strahl B. D., Kutateladze T. G. (2016). Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol. 12 662–668. 10.1038/nchembio.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., et al. (2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57 626–644. 10.1111/j.1365-313X.2008.03715.x [DOI] [PubMed] [Google Scholar]
- Bagchi R., Melnyk C. W., Christ G., Winkler M., Kirchsteiner K., Salehin M., et al. (2018). The Arabidopsis ALF4 protein is a regulator of SCF E3 ligases. EMBO J. 37 255–268. 10.15252/embj.201797159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C., Pacurar D. I., Perrone I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65 639–666. 10.1146/annurev-arplant-050213-035645 [DOI] [PubMed] [Google Scholar]
- Berr A., McCallum E. J., Ménard R., Meyer D., Fuchs J., Dong A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22 3232–3248. 10.1105/tpc.110.079962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D. E., Wang J. Y., Jung J. W., Lambert G. M., Galbraith D. W., et al. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. 10.1126/science.1090022 [DOI] [PubMed] [Google Scholar]
- Birnbaum K. D. (2016). How many ways are there to make a root? Curr. Opin. Plant Biol. 34 61–67. 10.1016/j.pbi.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Busov V., Meilan R., Pearce D. W., Rood S. B., Ma C., Tschaplinski T. J., et al. (2006). Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 224 288–299. 10.1007/s00425-005-0213-9 [DOI] [PubMed] [Google Scholar]
- Bustillo-Avendaño E., Ibáñez S., Sanz O., Sousa Barros J. A., Gude I., Perianez-Rodriguez J., et al. (2018). Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis. Plant Physiol. 176 1709–1727. 10.1104/pp.17.00980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.-T., Xu P., Zhao P.-X., Liu R., Yu L.-H., Xiang C.-B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5:5833. 10.1038/ncomms6833 [DOI] [PubMed] [Google Scholar]
- Calamar A., de Klerk G.-J. (2002). Effect of sucrose on adventitious root regeneration in apple. Plant Cell Tissue Organ Cult. 70 207–212. 10.1023/A:1016356123367 [DOI] [Google Scholar]
- Casimiro I., Beeckman T., Graham N., Bhalerao R., Zhang H., Casero P., et al. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. 10.1016/S1360-1385(03)00051-7 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C. I., Yin K., Pogson B. J. (2009). Potential implications for epigenetic regulation of carotenoid biosynthesis during root and shoot development. Plant Signal. Behav. 4 339–341. 10.4161/psb.4.4.8193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Grisafi P. L., Fink G. R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. 10.1101/gad.9.17.2131 [DOI] [PubMed] [Google Scholar]
- Che P., Lall S., Howell S. H. (2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226 1183–1194. 10.1007/s00425-007-0565-4 [DOI] [PubMed] [Google Scholar]
- Chen L., Tong J., Xiao L., Ruan Y., Liu J., Zeng M., et al. (2016). YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 67 4273–4284. 10.1093/jxb/erw213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sun J., Zhai Q., Zhou W., Qi L., Xu L., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23 3335–3352. 10.1105/tpc.111.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cheng J., Chen L., Zhang G., Huang H., Zhang Y., et al. (2016). Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 170 2136–2145. 10.1104/pp.15.01733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Qu Y., Sheng L., Liu J., Huang H., Xu L. (2014). A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 5:208. 10.3389/fpls.2014.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Tan F., Lu Y., Liu X., Li T., Yuan W., et al. (2018). WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 46 2356–2369. 10.1093/nar/gky017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman W. K., Huxter T. J., Reid D. M., Thorpe T. A. (1980). Ethylene as an endogenous inhibitor of root regeneration in tomato leaf discs cultured in vitro. Physiol. Plant. 48 519–525. 10.1111/j.1399-3054.1980.tb03298.x [DOI] [Google Scholar]
- Cui X., Lu F., Qiu Q., Zhou B., Gu L., Zhang S., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48 694–699. 10.1038/ng.3556 [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60 871–887. 10.1007/s11103-005-4547-2 [DOI] [PubMed] [Google Scholar]
- Deal R. B., Henikoff S. (2011). The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6 56–68. 10.1038/nprot.2010.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rovere F., Fattorini L., D’Angeli S., Veloccia A., Del Duca S., Cai G., et al. (2015). Arabidopsis SHR and SCR transcription factors and AUX1 auxin influx carrier control the switch between adventitious rooting and xylogenesis in planta and in in vitro cultured thin cell layers. Ann. Bot. 115 617–628. 10.1093/aob/mcu258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rovere F., Fattorini L., D’Angeli S., Veloccia A., Falasca G., Altamura M. M. (2013). Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 112 1395–1407. 10.1093/aob/mct215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Sala C. (2014). Direct reprogramming of adult somatic cells toward adventitious root formation in forest tree species: the effect of the juvenile-adult transition. Front. Plant Sci. 5:310. 10.3389/fpls.2014.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 107 12046–12051. 10.1073/pnas.1000672107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege U., Franken P. (2019). Petunia as model for elucidating adventitious root formation and mycorrhizal symbiosis: at the nexus of physiology, genetics, microbiology and horticulture. Physiol. Plant. 165 58–72. 10.1111/ppl.12762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege U., Franken P., Lischewski S., Ahkami A. H., Zerche S., Hause B., et al. (2014). Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in Petunia cuttings. Front. Plant Sci. 5:494. 10.3389/fpls.2014.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege U., Hilo A., Pérez-Pérez J. M., Klopotek Y., Acosta M., Shahinnia F., et al. (2019). Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann. Bot. 123 929–949. 10.1093/aob/mcy234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Scheres B. (2017). PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc. Natl. Acad. Sci. U.S.A. 114 11709–11714. 10.1073/pnas.1714410114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J., Sangwan-Norreel B., Catterou M., Sangwan R. S. (2011). De novo shoot organogenesis: from art to science. Trends Plant Sci. 16 597–606. 10.1016/j.tplants.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Efroni I., Mello A., Nawy T., Ip P.-L., Rahni R., DelRose N., et al. (2016). Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165 1721–1733. 10.1016/j.cell.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Xu C., Xu K., Hu Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22 1169–1180. 10.1038/cr.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattorini L., Hause B., Gutierrez L., Veloccia A., Della Rovere F., Piacentini D., et al. (2018). Jasmonate promotes auxin-induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 18:182. 10.1186/s12870-018-1392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Fukaki H., Taniguchi N., Tasaka M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48 380–389. 10.1111/j.1365-313X.2006.02882.x [DOI] [PubMed] [Google Scholar]
- Glauser G., Grata E., Dubugnon L., Rudaz S., Farmer E. E., Wolfender J.-L. (2008). Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 283 16400–16407. 10.1074/jbc.M801760200 [DOI] [PubMed] [Google Scholar]
- Gonzali S., Novi G., Loreti E., Paolicchi F., Poggi A., Alpi A., et al. (2005). A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J. 44 633–645. 10.1111/j.1365-313X.2005.02555.x [DOI] [PubMed] [Google Scholar]
- Gu X., Xu T., He Y. (2014). A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant 7 977–988. 10.1093/mp/ssu035 [DOI] [PubMed] [Google Scholar]
- Guo H.-S., Xie Q., Fei J.-F., Chua N.-H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17 1376–1386. 10.1105/tpc.105.030841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu Y., Law J. A., Zhang X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine corrected 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107 18557–18562. 10.1073/pnas.1010478107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R., Borghi L., Gruissem W. (2012). Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 17 139–148. 10.1016/j.tplants.2011.12.001 [DOI] [PubMed] [Google Scholar]
- He C., Chen X., Huang H., Xu L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8:e1002911. 10.1371/journal.pgen.1002911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567. 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K. S., van Leene J., Vercauteren I., et al. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342 860–863. 10.1126/science.1240667 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Park W. J., Sauer M., Woll K. (2004). From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci. 9 42–48. 10.1016/j.tplants.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Hofhuis H., Laskowski M., Du Y., Prasad K., Grigg S., Pinon V., et al. (2013). Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 23 956–962. 10.1016/j.cub.2013.04.048 [DOI] [PubMed] [Google Scholar]
- Hu X., Xu L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172 2363–2373. 10.1104/pp.16.01067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Liu B., Liu L., Song S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68 1349–1359. 10.1093/jxb/erw495 [DOI] [PubMed] [Google Scholar]
- Ibáñez S., Ruiz-Cano H., Fernández M. Á, Sánchez-García A. B., Villanova J., Micol J. L., et al. (2019). A network-guided genetic approach to identify novel regulators of adventitious root formation in Arabidopsis thaliana. Front. Plant Sci. 10:461. 10.3389/fpls.2019.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Iwase A., Rymen B., Lambolez A., Kojima M., Takebayashi Y., et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175 1158–1174. 10.1104/pp.17.01035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143 1442–1451. 10.1242/dev.134668 [DOI] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A. J., Trivedi Z. B., et al. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25 1017–1030. 10.1016/j.cub.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata S.-I., Yamazaki K., Ishihara K., Shibayama H., Lai K.-L. (1963). Studies on root system formation in rice plants in a paddy. Jpn. J. Crop Sci. 32 163–180. 10.1626/jcs.32.163 [DOI] [Google Scholar]
- Kennison J. A., Tamkun J. W. (1992). Trans-regulation of homeotic genes in Drosophila. New Biol. 4 91–96. [PubMed] [Google Scholar]
- Kim J.-Y., Yang W., Forner J., Lohmann J. U., Noh B., Noh Y.-S. (2018). Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 37:e98726. 10.15252/embj.201798726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klerk G.-J., de, van der Krieken W., de Jong J. C. (1999). Review the formation of adventitious roots: new concepts, new possibilities. In Vitro Cell. Dev. Biol. Plant 35 189–199. 10.1007/s11627-999-0076-z [DOI] [Google Scholar]
- Kong D., Hao Y., Cui H. (2016). The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol. Plant 9 261–270. 10.1016/j.molp.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21 1070–1079. 10.1105/tpc.108.065300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Kushalappa K., Godt D., Pidkowich M. S., Pastorelli S., Hepworth S. R., et al. (2007). The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19 2719–2735. 10.1105/tpc.106.048769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakehal A., Bellini C. (2019). Control of adventitious root formation: insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 165 90–100. 10.1111/ppl.12823 [DOI] [PubMed] [Google Scholar]
- Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., de Smet I., et al. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18 450–458. 10.1016/j.tplants.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Lee K., Park O.-S., Seo P. J. (2017). Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci. Signal. 10:eaan0316. 10.1126/scisignal.aan0316 [DOI] [PubMed] [Google Scholar]
- Lee K., Park O.-S., Seo P. J. (2018). ATXR2 as a core regulator of de novo root organogenesis. Plant Signal. Behav. 13:e1449543. 10.1080/15592324.2018.1449543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Seo P. J. (2018). Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 23 235–247. 10.1016/j.tplants.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Lischweski S., Muchow A., Guthörl D., Hause B. (2015). Jasmonates act positively in adventitious root formation in Petunia cuttings. BMC Plant Biol. 15:229. 10.1186/s12870-015-0615-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S., Yu X., Yu J., He X., Zhang S., et al. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43 47–56. 10.1111/j.1365-313X.2005.02434.x [DOI] [PubMed] [Google Scholar]
- Liu J., Hu X., Qin P., Prasad K., Hu Y., Xu L. (2018). The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 59 734–743. 10.1093/pcp/pcy010 [DOI] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26 1081–1093. 10.1105/tpc.114.122887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43 715–719. 10.1038/ng.854 [DOI] [PubMed] [Google Scholar]
- Lucas M., de Pu L., Turco G., Gaudinier A., Morao A. K., Harashima H., et al. (2016). Transcriptional regulation of Arabidopsis polycomb repressive complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28 2616–2631. 10.1105/tpc.15.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Swarup R., Paponov I. A., Swarup K., Casimiro I., Lake D., et al. (2011). Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155 384–398. 10.1104/pp.110.165126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J. E., Ryan K. S. (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127 899–909. 10.1104/pp.010406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A., Bhalerao R., Casimiro I., Eklöf J., Casero P. J., Bennett M., et al. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597. 10.1105/tpc.010354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P., Hoermayer L., Yoshida S., Marhavı P., Benková E., Friml J. (2019). Re-activation of stem cell pathways for pattern restoration in plant wound healing. Cell 177 957–969.e13. 10.1016/j.cell.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriat M., Petterle A., Bellini C., Moritz T. (2014). Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 78 372–384. 10.1111/tpj.12478 [DOI] [PubMed] [Google Scholar]
- Müller R., Goodrich J. (2011). Sweet memories: epigenetic control in flowering. F1000 Biol. Rep. 3:13. 10.3410/B3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Xing R., Clarenz O., Pokorny L., Goodrich J., Schubert D. (2014a). Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell 26 2457–2471. 10.1105/tpc.114.123323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Xing R., Xing Q., Goodrich J. (2014b). Footprints of the sun: memory of UV and light stress in plants. Front. Plant Sci. 5:474. 10.3389/fpls.2014.00474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napsucialy-Mendivil S., Alvarez-Venegas R., Shishkova S., Dubrovsky J. G. (2014). Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65 6373–6384. 10.1093/jxb/eru355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2:a001537. 10.1101/cshperspect.a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Müller J. (2006). Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20 2041–2054. 10.1101/gad.388706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulraj S., Yeung E. C. (2012). Improved shoot regeneration from root explants using an abscisic acid-containing medium. Methods Mol. Biol. 877 183–189. 10.1007/978-1-61779-818-4_15 [DOI] [PubMed] [Google Scholar]
- Perez-Garcia P., Moreno-Risueno M. A. (2018). Stem cells and plant regeneration. Dev. Biol. 442 3–12. 10.1016/j.ydbio.2018.06.021 [DOI] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136 2675–2688. 10.1242/dev.030353 [DOI] [PubMed] [Google Scholar]
- Raya-González J., Pelagio-Flores R., López-Bucio J. (2012). The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J. Plant Physiol. 169 1348–1358. 10.1016/j.jplph.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17 354–358. 10.1101/gad.252503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. 10.1038/nature05703 [DOI] [PubMed] [Google Scholar]
- Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42 330–341. 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Sena G., Birnbaum K. D. (2010). Built to rebuild: in search of organizing principles in plant regeneration. Curr. Opin. Genet. Dev. 20 460–465. 10.1016/j.gde.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servet C., Conde e Silva N., Zhou D.-X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3 670–677. 10.1093/mp/ssq018 [DOI] [PubMed] [Google Scholar]
- Shang B., Xu C., Zhang X., Cao H., Xin W., Hu Y. (2016). Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113 5101–5106. 10.1073/pnas.1522466113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L., Hu X., Du Y., Zhang G., Huang H., Scheres B., et al. (2017). Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 144 3126–3133. 10.1242/dev.152132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Heidstra R., Blilou I., Scheres B. (2018). Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 32 1085–1100. 10.1101/gad.314096.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji K., Addicott F. T., Swets W. A. (1951). Auxin in relation to leaf blade abscission. Plant Physiol. 26 189–191. 10.1104/pp.26.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C. O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11 118–130. [PubMed] [Google Scholar]
- Smith Z. D., Meissner A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14 204–220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D.-Y., Dolezal K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Su Y. H., Zhang X. S. (2014). The hormonal control of regeneration in plants. Curr. Top. Dev. Biol. 108 35–69. 10.1016/B978-0-12-391498-9.00010-3 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Gordon S. P., Meyerowitz E. M. (2011). Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 21 212–218. 10.1016/j.tcb.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18 463–471. 10.1016/j.devcel.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Sukumar P., Maloney G. S., Muday G. K. (2013). Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 162 1392–1405. 10.1104/pp.113.217174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Chen L., Liu J., Zhang X., Yang Z., Liu W., et al. (2016). TAA family contributes to auxin production during de novo regeneration of adventitious roots from Arabidopsis leaf explants. Sci. Bull. 61 1728–1731. 10.1007/s11434-016-1185-9 [DOI] [Google Scholar]
- Sun B., Looi L.-S., Guo S., He Z., Gan E.-S., Huang J., et al. (2014). Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science 343:1248559. 10.1126/science.1248559 [DOI] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21 1495–1511. 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex I. M. (2008). The scientific roots of modern plant biotechnology. Plant Cell 20 1189–1198. 10.1105/tpc.108.058735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Benková E., Swarup R., Casimiro I., Péret B., Yang Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10 946–954. 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- Takatsuka H., Umeda M. (2015). Epigenetic control of cell division and cell differentiation in the root apex. Front. Plant Sci. 6:1178. 10.3389/fpls.2015.01178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. P., Zhou C., Wang S. S., Yuan J., Zhang X. S., Su Y. H. (2017). FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 213 1740–1754. 10.1111/nph.14313 [DOI] [PubMed] [Google Scholar]
- Valvekens D., van Montagu M., van Lijsebettens M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. U.S.A. 85 5536–5540. 10.1073/pnas.85.15.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Norman J. M., Xuan W., Beeckman T., Benfey P. N. (2013). To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140 4301–4310. 10.1242/dev.090548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Rybel B., de Beemster G. T. S., Ljung K., de Smet I., van Isterdael G., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17 3035–3050. 10.1105/tpc.105.035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten I., Schotte S., Geelen D. (2014). Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 5:495. 10.3389/fpls.2014.00495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Carbonero P. (2005). Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49 645–651. 10.1387/ijdb.052046jc [DOI] [PubMed] [Google Scholar]
- Vlachonasios K. E., Thomashow M. F., Triezenberg S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15 626–638. 10.1105/tpc.007922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gao J., Gao S., Li Z., Kuai B., Ren G. (2019). REF6 promotes lateral root formation through de-repression of PIN1/3/7 genes. J. Integr. Plant Biol. 61 383–387. 10.1111/jipb.12726 [DOI] [PubMed] [Google Scholar]
- Wang Y., Duan L., Lu M., Li Z., Wang M., Zhai Z. (2006). Expression of NAC1 up-stream regulatory region and its relationship to the lateral root initiation induced by gibberellins and auxins. Sci. China C Life Sci. 49 429–435. 10.1007/s11427-006-2021-2 [DOI] [PubMed] [Google Scholar]
- Weijers D., Wagner D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lewis D. R., Spalding E. P. (2007). Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19 1826–1837. 10.1105/tpc.106.048777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Jin R., Yu X., Shen M., Wagner J. D., Pai A., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49 1546–1552. 10.1038/ng.3937 [DOI] [PubMed] [Google Scholar]
- Xiao J., Lee U.-S., Wagner D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34 41–53. 10.1016/j.pbi.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036. 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Miao J., Yumoto E., Yokota T., Asahina M., Watahiki M. (2017). YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant Cell Physiol. 58 1710–1723. 10.1093/pcp/pcx107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. (2018). De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 41 39–45. 10.1016/j.pbi.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Xu L., Huang H. (2014). Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 108 1–33. 10.1016/B978-0-12-391498-9.00009-7 [DOI] [PubMed] [Google Scholar]
- Yan W., Chen D., Smaczniak C., Engelhorn J., Liu H., Yang W., et al. (2018). Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 4 681–689. 10.1038/s41477-018-0219-5 [DOI] [PubMed] [Google Scholar]
- Yao X., Feng H., Yu Y., Dong A., Shen W.-H. (2013). SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS One 8:e56537. 10.1371/journal.pone.0056537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Liu W., Liu J., Qin P., Xu L. (2017). Auxin control of root organogenesis from callus in tissue culture. Front. Plant Sci. 8:1385. 10.3389/fpls.2017.01385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zhao F., Chen L., Pan Y., Sun L., Bao N., et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 5 491–497. 10.1038/s41477-019-0408-x [DOI] [PubMed] [Google Scholar]
- Zhang T., Li R., Xing J., Yan L., Wang R., Zhao Y. (2018). The YUCCA-auxin-WOX11 module controls crown root development in rice. Front. Plant Sci. 9:523. 10.3389/fpls.2018.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.-Q., Lian H., Zhou C.-M., Xu L., Jiao Y., Wang J.-W. (2017). A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29 1073–1087. 10.1105/tpc.16.00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang X., Li H. (2018). Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol. 53 169–177. 10.1016/j.sbi.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.-X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21 736–748. 10.1105/tpc.108.061655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Jiang W., Long F., Cheng S., Yang W., Zhao Y., et al. (2017). Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 29 1088–1104. 10.1105/tpc.16.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lozano-Torres J. L., Blilou I., Zhang X., Zhai Q., Smant G., et al. (2019). A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177 942–956.e14. 10.1016/j.cell.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang Y., Krause K., Yang T., Dongus J. A., Zhang Y., et al. (2018). Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 50 638–644. 10.1038/s41588-018-0109-9 [DOI] [PubMed] [Google Scholar]
- Zimmerman P. W., Hitchcock A. E. (1935). The response of roots to “Root- Forming” substances. Contrib. Boyce Thompson Inst. 7 439–455. [Google Scholar]
- Zimmerman P. W., Wilcoxon F. (1935). Several chemical growth substances which cause initiation of roots and other responses in plants. Contrib. Boyce Thompson Inst. 7 209–229. [Google Scholar]