Abstract
The hydrolysis of triglycerides in triglyceride-rich lipoproteins by LPL is critical for the delivery of triglyceride-derived fatty acids to tissues, including heart, skeletal muscle, and adipose tissues. Physiologically active LPL is normally bound to the endothelial cell protein glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1 (GPIHBP1), which transports LPL across endothelial cells, anchors LPL to the vascular wall, and stabilizes LPL activity. Disruption of LPL-GPIHBP1 binding significantly alters triglyceride metabolism and lipid partitioning. In this study, we modified the NanoLuc® Binary Technology split-luciferase system to develop a novel assay that monitors the binding of LPL to GPIHBP1 on endothelial cells in real time. We validated the specificity and sensitivity of the assay using endothelial lipase and a mutant version of LPL and found that this assay reliably and specifically detected the interaction between LPL and GPIHBP1. We then interrogated various endogenous and exogenous inhibitors of LPL-mediated lipolysis for their ability to disrupt the binding of LPL to GPIHBP1. We found that angiopoietin-like (ANGPTL)4 and ANGPTL3-ANGPTL8 complexes disrupted the interactions of LPL and GPIHBP1, whereas the exogenous LPL blockers we tested (tyloxapol, poloxamer-407, and tetrahydrolipstatin) did not. We also found that chylomicrons could dissociate LPL from GPIHBP1 and found evidence that this dissociation was mediated in part by the fatty acids produced by lipolysis. These results demonstrate the ability of this assay to monitor LPL-GPIHBP1 binding and to probe how various agents influence this important complex.
Keywords: triglyceride, lipolysis, lipoprotein metabolism, chylomicrons, endothelial cell, glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1, NanoLuc® Binary Technology
Delivery of triglyceride-derived fatty acids to peripheral tissues such as heart, adipose tissue, and skeletal muscle is facilitated by the action of LPL, an extracellular triglyceride lipase that hydrolyzes the triglycerides of triglyceride-rich lipoproteins (i.e., VLDL and chylomicrons). In the vasculature, physiologically functional LPL is bound to the capillary wall by the endothelial cell protein glycosylphosphatidylinositol-anchored HDL binding protein 1 (GPIHBP1) (1, 2). GPIHBP1 facilitates LPL function by transporting LPL across endothelial cells (3, 4), anchoring LPL to the vascular wall (1, 2), and stabilizing LPL catalytic activity (5, 6). The interaction of LPL with GPIHBP1 is critical for efficient triglyceride clearance, as mutations that abolish LPL binding to GPIHBP1 lead to severe hypertriglyceridemia (7–12).
LPL activity is modulated by multiple endogenous factors including members of the angiopoietin-like (ANGPTL) family of proteins and apolipoproteins Apo-C2 and Apo-C3. Deficiency in ANGPTL3, ANGPTL4, ANGPTL8, or Apo-C3 results in decreased plasma triglyceride levels and, in the case of ANGPTL3, ANGPTL4, and Apo-C3, appears to protect against cardiovascular disease (13–22). Therapies targeting these endogenous inhibitors have gained traction in preclinical studies and clinical trials (13–15, 23–29). Conversely, for experimental studies, agents that block LPL-mediated triglyceride clearance, such as tyloxapol and poloxamer-407 (P-407), are used to analyze VLDL secretion and dietary lipid absorption (30–34).
Despite the common use of LPL blocking agents and the therapeutic importance of endogenous inhibitors, little study has been devoted to how these agents might perturb the functionally relevant LPL-GPIHBP1 complex. We have previously described evidence that inactivation of LPL by ANGPTL4 leads to the dissociation of LPL from GPIHBP1 (35). In this study, we describe a novel real-time potentially high-throughput assay for assessing the binding of LPL to GPIHBP1 on the surface of endothelial cells. This assay uses the NanoLuc® Binary Technology (NanoBiT) split-luciferase system (36). In our assay, two parts of the engineered luciferase enzyme nanoluciferase, the largeBiT and the smallBiT, were used to tag LPL and GPIHBP1 respectively. The binding of LPL to GPIHBP1 reconstituted luciferase activity and this activity could be monitored in real time. We used this assay to investigate the effects of common endogenous and exogenous LPL inhibitors on LPL-GPIHBP1 complex integrity.
MATERIALS AND METHODS
Endothelial cell line expressing smallBiT-GPIHBP1
Rat heart microvessel endothelial cells (RHMVECs; VEC Technologies) were grown in MCDB-131 base medium (Genedepot) supplemented with 10 mM l-glutamine, 1% PenStrep antibiotic solution (10,000 U/ml penicillin and 10,000 μg/ml streptomycin; Gibco), 5% FBS (Atlanta Biologicals), 1 μg/ml hydrocortisone (Sigma-Aldrich), 10 μg/ml human epidermal growth factor (Gibco, Life Technologies), and 12 μg/ml bovine brain extract (Lonza).
RHMVECs expressing smallBiT-tagged mouse GPIHBP1 were generated by transducing cells with lentivirus encoding smallBiT-GPIHBP1. A plasmid construct for expressing smallBiT-tagged mouse GPIHBP1 (pSS2) was generated by first replacing the S-tag of an S-tagged GPIHBP1 construct (pBD151) (3) with the smallBiT tag of the NanoBiT system (Promega) (36) using site-directed mutagenesis. Lentivirus encoding smallBiT-GPIHBP1 was generated by transfecting 293T cells with 4.35 μg pSS2 and 1.45 μg each of lentiviral packaging vectors pMD2.G (Addgene, #12259), pRSV-Rev (Addgene, #12253) (37), and pMDLg/pRRE (Addgene, #12251) (37). Viral packaging vectors were a gift from Didier Trono. The day after transfection, medium was changed to antibiotic-free DMEM medium (Gibco). Forty-eight hours later, virus-containing conditioned medium was collected and passed through a 0.45 μm filter, and then concentrated 10-fold using a Lenti-X™ concentrator (Takara, 631231). Transductions with lentivirus were carried out by adding 200 μl to 500 μl concentrated viral supernatant and 4 μg/ml Polybrene (#134220; Santa Cruz Biotechnology) in a total of 1 ml DMEM to ∼80% confluent RHMVECs in 6-well plates. Twenty-four hours posttransduction, cells were washed with PBS and incubated in MCDB-131 complete medium for 48 h. Cells were then passaged to new wells and grown in medium containing 6 μg/ml puromycin (Sigma) to select for transduced cells.
Production of LPL-conditioned medium
Generation of a construct expressing FLAG-tagged largeBiT-human LPL (pEB12) was described previously (38). A construct expressing largeBiT-human LPL-C445Y (pSS6) was generated by Phusion site-directed mutagenesis (Thermo Scientific). The C445Y mutation is sometimes referred to as C418Y when numbering according to the amino acids in mature LPL (39, 40). Concentrated lentiviruses containing this construct were produced by transfecting 293T cells with pSS6 and lentiviral packaging vectors as described above. 293T cells stably expressing largeBiT-LPL C445Y were generated by transducing 293T cells with lentivirus, followed by selection using 3 μg/ml puromycin (Sigma-Aldrich). To generate conditioned medium containing largeBiT-LPL or largeBiT-LPL C445Y, 293T cells stably expressing the respective LPL were grown to confluence and then switched to serum free DMEM medium containing 1× protease inhibitor (APExBIO protease inhibitor cocktail or GBiosciences TCM protease arrest). Conditioned medium was collected 48 h later and concentrated ∼10-fold using Amicon Ultra-15 centrifugal filter units (EMD Millipore, #UFC901024). The presence of LPL in the conditioned medium was assessed by Western blotting using a mouse antibody against the FLAG-tag (1:5,000; Sigma-Aldrich, #F1804). LPL was quantified using a human LPL ELISA kit (Aviva Systems Biology, #OKCD06285) and following the manufacturer’s instructions.
Production of endothelial lipase- conditioned medium
A construct generating largeBiT-endothelial lipase (EL) was generated by first cloning cDNA encoding human EL (Dharmacon, #MHS6278-202806078) into a pCDNA6-FLAG vector using In-fusion cloning (Clontech) to generate a human EL-FLAG construct (pVS1). Subsequently, the largeBiT was amplified from pEB12 (largeBiT LPL) and cloned into pVS1 using In-fusion cloning to generate FLAG-tagged largeBiT-EL (pAO3).
To generate conditioned medium containing FLAG-tagged largeBiT-EL, 293T, cells were transiently transfected with pAO3. After 24 h, the medium was switched to serum-free Opti-MEM medium (Gibco) containing 1× protease inhibitors (protease inhibitor cocktail; APExBIO) and 0.1 U/ml heparin. After an additional 24 h of incubation, conditioned medium was collected. The presence of EL in conditioned medium was assessed by Western blot using a rabbit polyclonal antibody against human EL (1:1,000; Invitrogen, PA1-16799). LargeBiT-EL was quantified by quantitative Western blot using an antibody against the FLAG-tag (1:5,000; Sigma-Aldrich, #F1804) and comparing band intensities to ELISA-quantified FLAG-tagged largeBiT-LPL.
Production of ANGPTL-conditioned media
Conditioned media containing ANGPTL4, ANGPTL3, ANGPTL8, or ANGPTL3 and ANGPTL8 were generated as described previously (35, 38). In brief, 293T cells were transiently transfected with constructs expressing V5-tagged human ANGPTL4 (pHS2), strep-tagged mouse ANGPTL3 (pHS18), or V5-tagged mouse ANGPTL8 (pWL1). Twenty-four hours posttransfection, media were switched to serum-free DMEM medium containing 1× protease inhibitors (TCM protease arrest, G-Biosciences). Forty-eight hours later, conditioned media were collected. The presence of ANGPTL proteins in the conditioned media was assessed by Western blot using a mouse monoclonal antibody against the V5-tag (1:5,000; Thermo Scientific, #PI MA5-15253) for ANGPTL4 and ANGPTL8 and a rabbit polyclonal antibody against the strep tag-II (1:3,000; Abcam, #ab76949) for ANGPTL3. The concentration of ANGPTL4-conditioned medium was determined using a human ANGPTL4 ELISA kit (RAB0017, Sigma-Aldrich). The concentrations of ANGPTL3 and ANGPTL8 were determined by quantitative Western blotting using previously quantified samples as standards (38). Conditioned medium collected from mock-transfected cells was used as a control for all experiments, and was referred to as control medium.
LPL-GPIHBP1 NanoBiT assay
RHMVECs expressing smallBiT-GPIHBP1 were grown to confluence in white clear-bottomed fibronectin-coated 96-well plates.
To detect LPL binding, cells were washed twice with 1× PBS and then 40 μl 2× NanoLuc Live Cell substrate (17.5 μl 5× substrate in 40 μl total control medium; Promega) were added. The plate was then placed in a multimode plate reader (Molecular Devices, SpectraMax i3) set at 37°C and luminescence was read every minute for 6 min. LargeBiT-tagged lipase (largeBiT-LPL, largeBiT-LPL (C445Y), or largeBiT-EL) was added to cells at a final concentration of 0–14 ng/ml and luminescence continued to be recorded every minute for an additional 30 min. Lipases were diluted using medium from untransduced cells (control medium) so that equal volumes were added to each well. To normalize across independent experiments, each luminescence recording was divided by the median luminescence of all reads for that particular experiment.
To test LPL binding after phosphatidylinositol-specific phospholipase C (PIPLC) treatment, cells were washed with 1× PBS and then treated with 5 U/ml PIPLC for 30 min at 37°C or water (in 22.5 μl total volume of control medium). Seventeen and one-half microliters of 5× NanoLuc Live Cell substrate were added directly to the treated cells or, in some cases, cells were washed twice with 1× PBS before adding 22.5 μl control medium and 17.5 μl 5× NanoLuc Live Cell substrate. After addition of substrate, luminescence was recorded every minute. After 6 min, largeBiT-tagged LPL was added to a final concentration of 14 ng/ml and luminescence continued to be recorded for an additional 45 min. To normalize across independent experiments, each luminescence recording was divided by the median luminescence of all reads for that particular experiment.
To detect LPL dissociation, endothelial cells were incubated with largeBiT-LPL at 4°C for 3–4 h to allow LPL to bind to GPIHBP1. After washing off the unbound LPL, 62.5 μl 1× NanoLuc Live Cell substrate (12.5 μl 5× substrate diluted in 62.5 μl control medium) were added and luminescence was recorded every 3 min for 15 min. The agents being tested were then added and luminescence was recorded every 3 min for an additional 9–45 min. In this study, the agents used were tetrahydrolipstatin (THL) (APExBIO; 10–80 μM), tyloxapol (Sigma-Aldrich; 0.15–0.6 mg/ml), P-407 (Sigma-Aldrich; 2–8 mg/ml), ANGPTL4 (300 ng/ml), ANGPTL3 (3 μg/ml), ANGPTL8 (30 ng/ml), coexpressed ANGPTL3 and ANGPTL8 (3 μg/ml and 30 ng/ml, respectively), chylomicrons (1–10 μg/ml), sodium oleate (Sigma-Aldrich; 0.375–3 mM), heparin (Hospira; 100 U/ml), and the appropriate vehicles (control medium for ANGPTLs; ethanol for THL; 1× PBS for tyloxapol, P-407, and chylomicrons; water for heparin and sodium oleate). In some cases, after reading luminescence for 9–30 min, cells were washed and fresh substrate (50 μl control medium and 12.5 μl 5× NanoLuc Live Cell substrate) was added to the cells and luminescence was read for another 15 min. To normalize dissociation assays across independent experiments, the luminescence of each sample was normalized as a percentage of the maximum luminescence of that particular sample. Except for Fig. 2C, the control for each experiment was set at 100% at each time point, and the test samples were expressed as a percentage of the control. For experiments testing the combination of THL and chylomicrons, experiments were performed as above, but after washing off unbound LPL, cells were treated with 80 μM THL or vehicle (0.32% ethanol) for 30 min at 37°C, and then washed before adding substrate. Cells were then treated with 80 μM THL or vehicle for 30 min at 37°C.
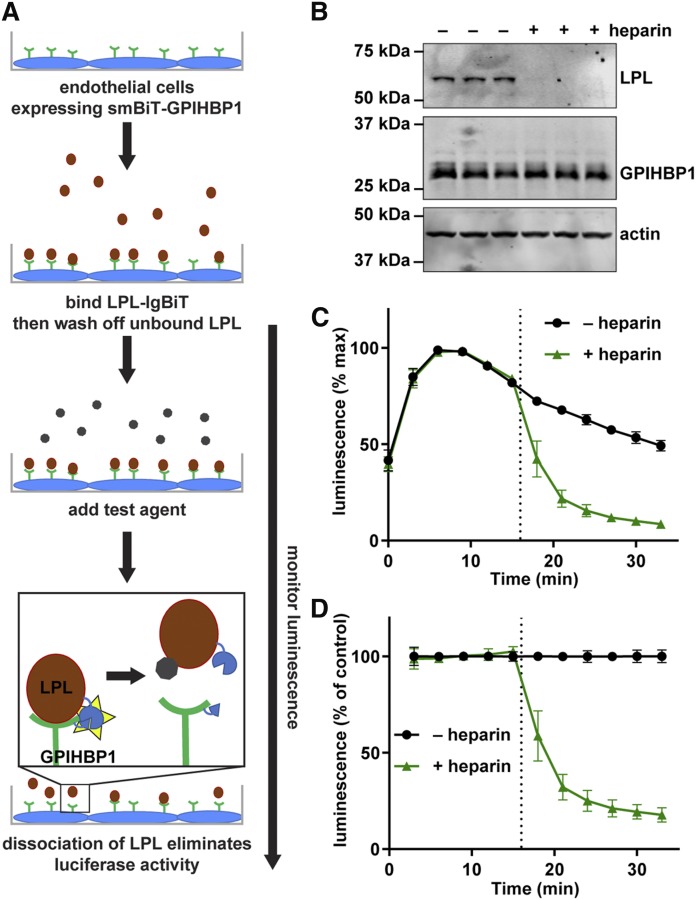
Fig. 2.
Disruption of LPL-GPIHBP1 binding by heparin. A: Schematic of the NanoBiT LPL dissociation assay. B: Western blot of lysates of smallBiT-GPIHBP1-expressing RHMVECs incubated with LargeBiT-LPL for 3.5 h at 4°C, washed, and then treated with or without 100 U/ml heparin for 30 min. C, D: SmallBiT-GPIHBP1-expressing endothelial cells were incubated with LargeBiT-LPL for 2 h at 4°C and washed. Luminescent substrate was added to cells and luminescence read every 3 min for 15 min. After 15 min 100 U/ml heparin (+heparin) or water (–heparin) were added to the samples and luminescence was read every 3 min for an additional 15 min. C: Luminescent signal over time (maximum signal for each sample set to 100%). D: Luminescence signal over time normalized to the –heparin control at each time point. Points represent mean ± 95% CI of three independent experiments (n = 3–6 per group per experiment).
An endpoint-only version of the dissociation assay was used for Fig. 7D. In this case, after binding largeBiT-LPL to smallBiT-GPIHBP1 expressing RHMVECs as before, 20 μg/ml chylomicrons mixed with 2 or 8 mg/ml tyloxapol were added to cells and incubated at 37°C for 30 min. Cells were washed, luminescent substrate was added, and luminescence was read every minute for 15 min at 37°C. The maximum luminescence of each sample was plotted as a percentage of the untreated LPL control.
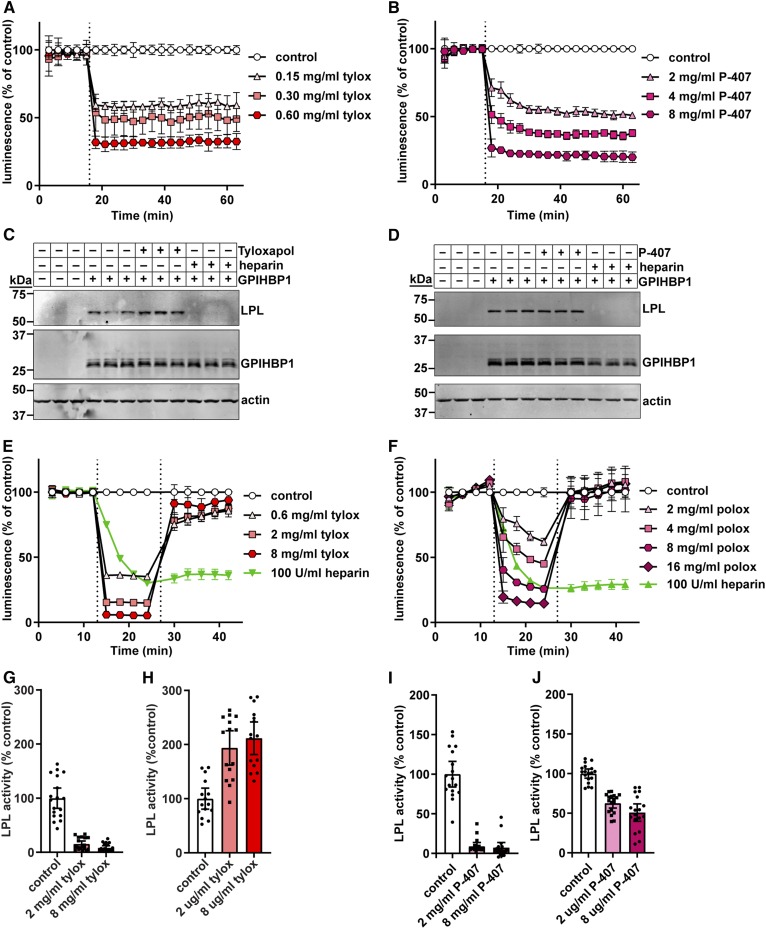
Fig. 7.
Effect of tyloxapol and P-407 on GPIHBP1-LPL binding and LPL activity. A, B: RHMVECs expressing smallBiT-GPIHBP1 were incubated with largeBiT-LPL for 2 h at 4°C and washed. Luminescent substrate was added to cells and luminescence read every 3 min for 15 min. After 15 min (dotted line), 150–600 μg/ml of tyloxapol (tylox) (A) or 2–8 mg/ml of P-407 (B) were added and luminescence continued to be measured for 45 min. Points represent mean ± 95% CI of luminescent signal over time normalized to the control at each time point. Data represent three independent experiments, each with three biological triplicates per group. C, D: Western blot of cell lysates from cells incubated with LargeBiT-LPL for 3.5 h at 4°C, washed, and then treated with 8 mg/ml tyloxapol (C) or 8 mg/ml P-407 (D) for 30 min at 37°C. Bands show LPL (using an antibody against the FLAG tag), GPIHBP1, and actin for biological triplicates. E, F: Luminescence of smallBiT-GPIHBP1-expressing RHMVECs incubated with LgBiT-LPL. After washing off unbound LPL, luminescence was measured for 12 min (3 min intervals). Tyloxapol (E) or P-407 (F) was added (first dotted line) at the indicated concentrations and luminescence was measured for an additional 12 min. Cells were then washed, fresh substrate was added (second dotted line), and luminescence was measured for another 15 min. Points represent luminescent signal over time normalized to the LPL only control at each time point (mean ± 95% CI of three independent experiments; n = 3–6 per group per experiment). G–J: LPL activity of LPL bound to smallBiT-GPIHBP1-expressing RHMVECs washed and treated with 0 (control), 2, or 8 mg/ml tyloxapol (G, H) or 0 (control), 2, or 8 mg/ml poloxamer (I, J) for 30 min at 37°C. For G and I, heparin was added directly to the reaction to release LPL. For H and J, cells were washed extensively and then LPL was released from the cells with heparin. In all cases, LPL activity assays were performed on heparin-released LPL. Bars represent normalized LPL activity (mean ± 95% CI of three independent experiments; n = 3–6 per experiment).
Detection of LPL activity on the cell surface
To detect LPL activity on the cell surface, RHMVECs expressing smallBiT-GPIHBP1 were incubated with largeBiT-LPL at 4°C for 3–4 h in a 12-well plate. After washing off unbound LPL, cells were treated with THL (80 μM), P-407 (2 or 8 mg/ml), tyloxapol (2 or 8 mg/ml), ANGPTL4 (300 ng/ml), ANGPTL3 (3 μg/ml), ANGPTL8 (30 ng/ml), coexpressed ANGPTL3 (3 μg/ml) and ANGPTL8 (30 ng/ml), or control medium at 37°C for 30 min. After again washing three times with 1× PBS, cells were treated with heparin (100 U/ml in water) for 10 min at 4°C to release surface-bound LPL. Released LPL was collected and immediately assayed for lipase activity. In some cases, after incubation with P-407 or tyloxapol, cells were not washed. Instead, 50% (150 μl) of the detergent medium was removed (from 300 μl total medium volume) and 16.5 μl 1,000 U/ml heparin (100 U/ml final concentration) were added to cells. After incubation for 10 min at 4°C to release surface-bound LPL, LPL was collected and immediately assayed for lipase activity. Lipase activity was assayed using EnzChek lipase fluorescent substrate (Molecular Probes) as described previously (41). Briefly, 50 μl sample was mixed with 25 μl 4× assay buffer [0.6 M NaCl, 80 mM Tris-HCl (pH 8.0), and 6% fatty acid-free BSA]. Twenty-five microliters of substrate solution containing 2.48 μM EnzChek lipase fluorescent substrate and 0.05% 3-(N,N-dimethylmyristylammonio) propanesulfonate zwittergent detergent (Acros Organics) in 1% methanol was then added to each sample. Samples were then incubated at 37°C with fluorescence (485 nm excitation/528 nm emission) read every minute for 30 min with a Synergy Neo multimode plate reader (BioTek). Relative lipase activity was calculated by first subtracting background (calculated by reading fluorescence of a sample with no LPL) and then calculating the slope of the curve between the 10 and 15 min reads.
LPL cell binding assays
RHMVECs expressing smallBiT-GPIHBP1 grown on fibronectin-coated 12-well plates were incubated with LPL at 4°C for 3.5 h. Following incubation, unbound LPL was removed by washing extensively with PBS and the cells were incubated with THL (80 μM), P-407 (8 mg/ml), tyloxapol (8 mg/ml), ANGPTL4 (300 ng/ml), ANGPTL3 (3 μg/ml), ANGPTL8 (30 ng/ml), coexpressed ANGPTL3 (3 μg/ml) and ANGPTL8 (30 ng/ml), heparin (100 U/ml), or control medium at 37°C for 30 min. Cells were then lysed with radioimmunoprecipitation assay buffer (1% NP-40 substitute, 0.5% sodium deoxycholate, and 0.1% SDS in PBS), clarified by centrifugation, and subjected to Western blotting.
Western blot
Equal volumes of cell lysates or conditioned media were size fractionated on 12% SDS-PAGE gels and then electrophoretically transferred to a nitrocellulose membrane (Bio-Rad). Membranes were blocked with casein buffer (1% casein; Fisher Science Education) at room temperature for 1 h. Primary antibodies were diluted in casein buffer + 0.2% Tween, and incubations with primary antibody were carried out overnight at 4°C. Primary antibody dilutions were 1:5,000 for a mouse monoclonal antibody against FLAG-tag (F1804; Sigma-Aldrich), 1:2,000 for a rat monoclonal antibody against GPIHBP1 [11A12 (42); a kind gift from Loren Fong], and 1:1,000 for a goat antibody against actin (SC-1615; Santa Cruz). After washing with PBS + 0.1% Tween (PBS-T), membranes were incubated with Dylight680- or Dylight800-labeled secondary antibodies (Thermo Scientific) diluted 1:5,000 in casein buffer + 0.2% Tween. After washing with PBS-T, antibody binding was detected using an Odyssey Clx infrared scanner (LI-COR).
Mice
All animal procedures conformed to the Public Health Service Policy on Humane Care and Use of Laboratory Animals and were carried out according to guidelines approved by the Institutional Animal Care and Use Committee at the University of Iowa. Mice were group housed (up to five per cage) in a controlled environment with a 12/12 h light/dark cycle, with food and water provided ad libitum. Mice were fed a normal chow diet (Envigo, 7913). Gpihbp1−/− mice were originally obtained from the Mutant Mouse Resource and Research Center (https://www.mmrrc.org/; strain name: B6;129S5-Gpihbp1tm1Lex/Mmucd) (43, 44) and were maintained by breeding on a mixed C57Bl/6J-129S5 background.
Preparation of chylomicrons
Chylomicrons were prepared as described previously (45). Gpihbp1−/− mice were anesthetized and blood was collected by cardiac puncture. Blood was diluted 1:10 with 0.5 M EDTA (pH 8.0) and centrifuged 1,500 g for 15 min at 4°C to pellet blood cells. The plasma was then transferred to ultracentrifuge tubes and mixed 1:1 with sterile PBS. After centrifugation at 424,000 g for 2 h at 10°C, the chylomicrons form an upper layer. The chylomicron layer was resuspended in sterile PBS and the centrifugation was repeated. Following the second centrifugation, the resulting chylomicron layer was resuspended in a volume of sterile PBS equal to that of the original plasma sample. The concentration of protein present in the chylomicron preparation was assayed using the Bio-Rad DC protein assay.
Fluorescently labeled chylomicrons for immunofluorescence were prepared using the Alexa Fluor® 488 protein labeling kit (Thermo Fisher Scientific). Chylomicrons were diluted to 2 mg/ml (protein concentration) using 100 mM bicarbonate (pH ∼8.3) and incubated with fluorescent dye Alexa Fluor® 488 for 1 h at room temperature with constant rotation. Labeled chylomicrons were separated from unincorporated dye using Econo-Pac 10DG columns (Bio-Rad).
Analysis of chylomicron particles
Chylomicron diameters were measured by dynamic laser light-scattering analysis by using an ultrafine particle analyzer with a laser probe tip (UPA-250; Microtrac, Clearwater, FL) and appropriate software (Microtrac, Honeywell, Washington, PA). Sample characteristics were set to irregular transparent particles with a refractive index of 1.46 and density of <0.98 g/ml. Each measurement set comprised three 40 s measurements. The laser probe was gently placed into the top 5 mm of the prepared sample to prevent air bubbles at the probe-liquid interface. After the initial reading, tyloxapol was added to a subset of chylomicrons to a final concentration of 8 mg/ml, mixed by inversion, and incubated at 37°C for 30 min. In parallel, untreated chylomicrons were stored at 4°C for 30 min. The results of primary data collection can be expressed as a distribution of particle number, particle area, or particle volume; particle area distribution is reported here albeit results from other distributions produced similar conclusions. Raw particle diameter distributions were converted to population percentiles to facilitate comparisons.
Immunofluorescence
RHMVECs expressing smallBiT-GPIHBP1 were grown on fibronectin-coated glass coverslips. Cells were incubated with control medium or LPL for 3 h at 4°C. Cells were then washed three times with PBS to remove unbound LPL. For THL experiments, cells were then incubated with THL (80 μM) or vehicle (0.32% ethanol) for 30 min at 37°C. Cells were then washed three times with PBS to remove unbound THL and incubated with fluorescently labeled chylomicrons (20 μg/ml) or PBS for 30 min at 37°C. For tyloxapol experiments, after removing unbound LPL, cells were incubated with either PBS or fluorescently labeled chylomicrons (20 μg/ml) for 30 min at 37°C. After washing three times with PBS to remove unbound chylomicrons, cells were incubated with 8 mg/ml tyloxapol (or PBS) for 30 min at 37°C. For all experiments, cells were washed and then fixed with cold 100% methanol for 5 min. After washing twice with PBS, cells were blocked with 0.2% BSA and 10% FBS in PBS for 1 h at room temperature and then incubated with a monoclonal anti-FLAG antibody (F1804; Sigma-Aldrich) diluted 1:1,000 in blocking buffer containing 0.2% Triton X-100 overnight at 4°C. Following another set of PBS washes, cells were incubated with AlexaFluor-555 donkey anti-mouse IgG (Life Technologies) diluted 1:1,000 in blocking buffer containing 0.2% Triton X-100. Coverslips were then mounted on glass slides with ProLong Gold antifade reagent with DAPI (Molecular Probes).
RESULTS
Real-time detection of LPL-GPIHBP1 binding
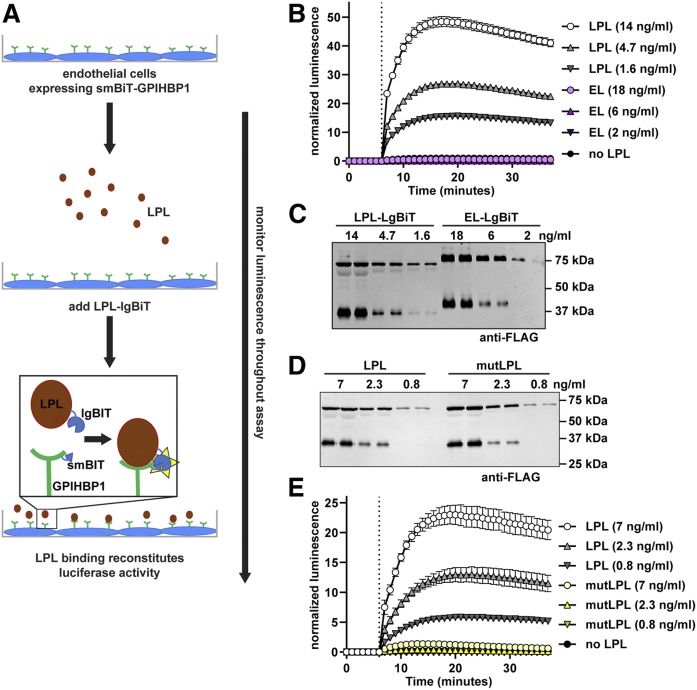
GPIHBP1 binds LPL, anchoring LPL to the surface of vascular endothelial cells (1). We developed an assay to assess the binding of LPL to GPIHBP1 on the surface of endothelial cells using the NanoBiT split-luciferase system (36). In the NanoBiT system, two parts of the engineered luciferase enzyme NanoLuciferase, the largeBiT and the smallBiT, are used to tag proteins of interest. Luciferase activity is reconstituted when the tagged proteins interact. For our assay, we used LPL tagged with largeBiT (38) and generated GPIHBP1 tagged with smallBiT. We then asked whether the binding of LPL to GPIHBP1 on the surface of endothelial cells could be detected in real time by monitoring luminescence (Fig. 1A). We grew RHMVECs stably expressing smallBiT-GPIHBP1 in 96-well plates. Luciferase substrate was added to each well and luminescence was read every minute. After 6 min, different concentrations of largeBiT-LPL were added to cells, and we continued reading luminescence for 30 min. As a negative control, we added a largeBiT-tagged version of EL. EL belongs to the same lipase family as LPL, but does not bind GPIHBP1 (46). We observed a dose-dependent increase in luminescent signal upon LPL addition (Fig. 1B), indicating that luminescent signal accurately reflected binding of LPL to GPIHBP1. No luminescent signal was observed upon addition of largeBiT-EL despite similar protein levels (Fig. 1B, C). We also performed a similar experiment using a wider range of LPL concentrations. Our assay was able to differentiate LPL binding to GPIHBP1 across an 80-fold range of LPL concentrations (supplemental Fig. S1A). When we used this data to plot a binding curve (supplemental Fig. S1B), we calculated a Kd value of approximately 10 nM, a value in line with previous reports (1, 5).
Fig. 1.
Association of largeBiT-LPL with smallBiT-GPIHBP1. A: Schematic of the NanoBiT LPL binding assay. B: SmallBiT-GPIHBP1-expressing RHMVECs were grown to confluence. Luminescent substrate (Nano-Glo® live cell substrate) was added to cells and luminescence was read every minute. After 6 min (dotted line), the indicated concentrations of largeBiT-LPL or largeBiT-EL were added to each well and luminescence continued to be measured each minute for an additional 30 min. Points represent mean ± 95% CI of four independent experiments (n = 6 per group per experiment). C: Western blot of the LargeBiT LPL (LPL-LgBiT)- and LargeBiT-EL (EL-LgBiT)-conditioned media used in B. D: Western blot of the LargeBiT LPL (LPL)- and LargeBiT-LPL C445Y (mutLPL)-conditioned media used in E. E: SmallBiT-GPIHBP1-expressing RHMVECs were grown to confluence. Luminescent substrate was added to cells and luminescence was read every minute. After 6 min (dotted line), the indicated concentrations of largeBiT-LPL (LPL) or largeBiT-LPL C445Y (mutLPL) were added to each well and luminescence continued to be measured each minute for an additional 30 min. Points represent mean ± 95% CI of three independent experiments (n = 6 per group per experiment).
Detection of LPL binding to GPIHBP1 did not require the association of GPIHBP1 with the plasma membrane. We treated GPIHBP1-expressing cells with PIPLC to cleave the GPI anchor and release GPIHBP1 into the media. Addition of LPL to cells where GPIHBP1 had been released into the surrounding media produced just as much luminescence as when LPL was added to cells where GPIHBP1 had not been released from the plasma membrane (supplemental Fig. S2A). In contrast, if we used PIPLC to release GPIHBP1 from the plasma membrane and then washed away the released GPIHBP1 before adding LPL, luminescent signal was greatly reduced compared with control-treated cells (supplemental Fig. S2B), indicating that PIPLC did indeed release GPIHBP1 from the cells in these experiments.
A cysteine to tyrosine mutation in residue 445 (C445Y) of human LPL has been shown to result in chylomicronemia despite retaining catalytic activity (39). Additional studies showed that the C445Y mutation disrupts the binding of LPL to GPIHBP1 (40). Recent structural studies confirm that this region of LPL is important for the binding of LPL to GPIHBP1 (47). To validate our binding assay, we generated a mutant construct containing the C445Y mutation in largeBiT-LPL and tested the ability of the resultant protein to bind smallBiT-GPIHBP1. As expected, the mutant protein was expressed and secreted at similar levels to wild-type largeBiT-LPL (Fig. 1D). However, binding of mutant LPL to GPIHBP1 was greatly reduced (Fig. 1E). Interestingly, the NanoBiT assay was sensitive enough to detect some binding of mutant LPL to GPIHBP1, but this binding was less than 10% of that observed with wild-type largeBiT-LPL (Fig. 1E).
We next expanded our assay to detect the dissociation of LPL from GPIHBP1 (Fig. 2A). Heparin is known to release LPL from GPIHBP1, likely by competing with the acidic domain of GPIHBP1 for binding to the heparin binding domain of LPL (1, 48). Consistent with previous studies, when cells expressing smallBiT-GPIHBP1 were incubated with largeBiT-LPL and then washed, LPL attached to cells could easily be detected by Western blot (Fig. 2B). Treatment of cells with heparin released almost all of the LPL from cells (Fig. 2B). This heparin-induced dissociation of LPL from GPIHBP1 could be detected in real time using our NanoBiT assay. After incubating smallBiT-GPIHBP1 expressing endothelial cells with LPL at 4°C to bind LPL to the surface, cells were incubated with luciferase substrate for 15 min. A strong luminescent signal indicated that LPL was bound to GPIHBP1. Heparin was then added to a subset of samples and luminescence continued to be read for an additional 18 min. Luminescent signal rapidly decreased in samples treated with heparin, indicating the rapid release of LPL from GPIHBP1 (Fig. 2C). This was especially apparent when we corrected for the natural decay of luminescent signal by normalizing to the untreated samples (Fig. 2D). These data indicate that our NanoBiT assay could detect both the binding and release of LPL from GPIHBP1.
Disruption of LPL-GPIHBP1 binding by ANGPTL proteins
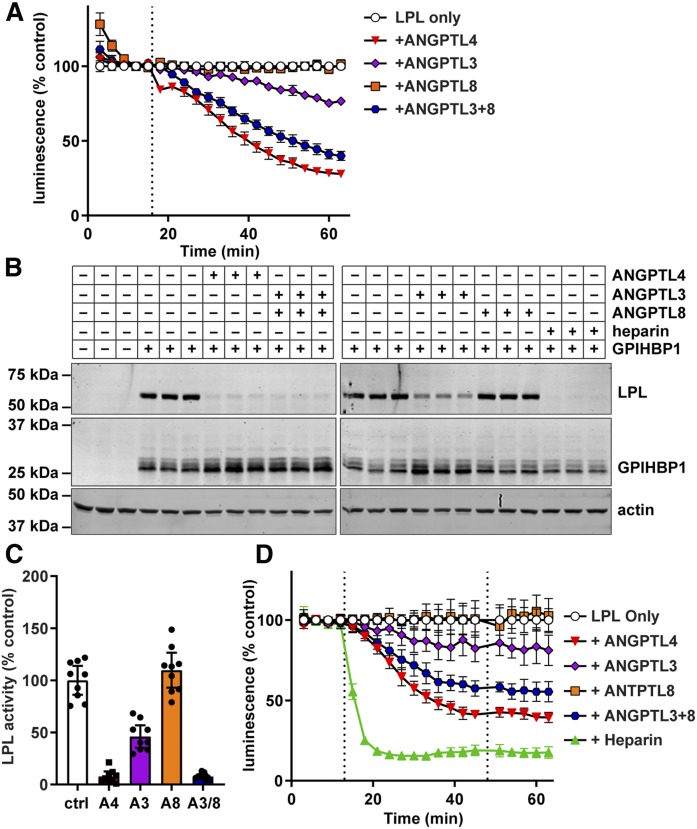
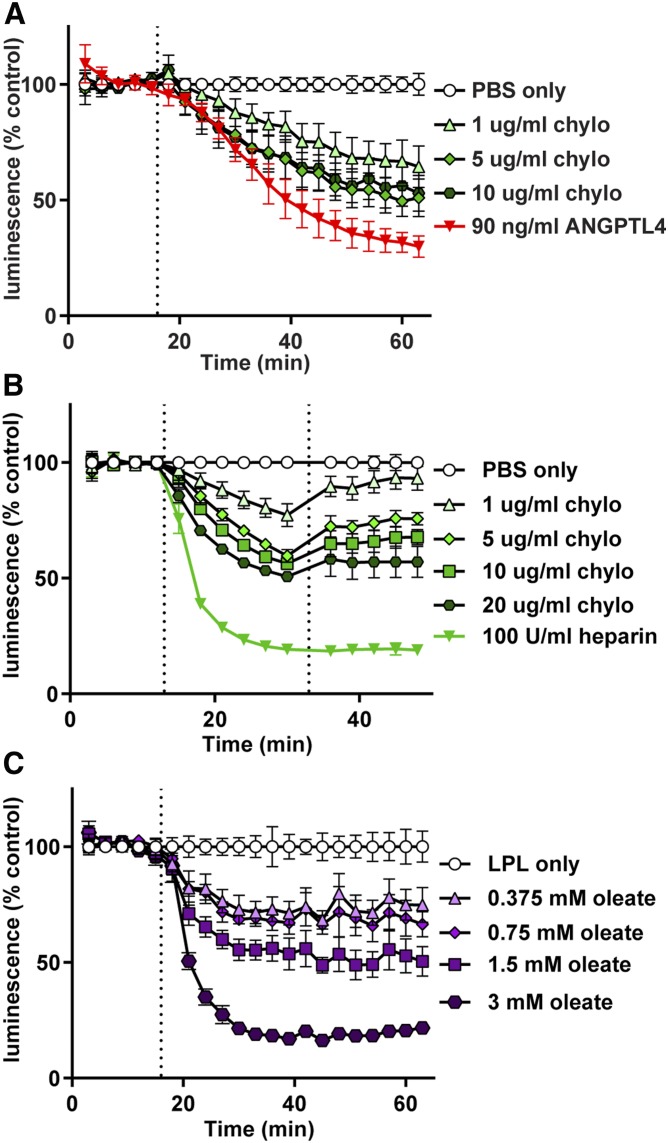
We have previously shown that inactivation of LPL by ANGPTL4 results in the dissociation of LPL from GPIHBP1 (35). We asked whether a similar dissociation of LPL from GPIHBP1 could be observed in our NanoBiT assay. We also tested the ability of ANGPTL3, ANGPTL8, and ANGPTL3-ANGPTL8 complexes to mediate the dissociation of LPL from GPIHBP1. We have previously shown that neither ANGPTL3 nor ANGPTL8 alone bind well to LPL, but that ANGPTL3-ANGPTL8 complexes effectively bind and inhibit LPL (38). Again, largeBiT-LPL was bound to smallBiT-GPIHBP1 on the surface of endothelial cells. After washing off unbound LPL, luminescent substrate was added. Then, after 15 min, ANGPTL3, ANGPTL4, ANGPTL8, and ANGPTL3-8 complexes were added to the cells. Treatment with either ANGPTL4 or ANGPTL3-8 complexes caused dissociation of LPL from GPIHBP1 (Fig. 3A). Treatment with ANGPTL3 also caused dissociation, but to a lesser extent, whereas ANGPTL8 treatment did not result in any observable dissociation (Fig. 3A). These results correlated well with traditional Western blot-based cell binding assays (Fig. 3B). However, our real-time assay provided additional kinetic information. Unlike heparin, where treatment quickly released LPL from GPIHBP1 (see Fig. 2B, C), incubation of LPL-GPIHBP1 complexes with ANGPTL proteins led to a slower sustained dissociation of LPL from GPIHBP1 (see Fig. 3A). It is also of note that when we ran parallel experiments analyzing the activity of LPL, rather than the binding of LPL to GPIHBP1, the inhibition of LPL activity by ANGPTL proteins tracked closely to the level of dissociation of LPL from GPIHBP1 (Fig. 3C).
Fig. 3.
Disruption of LPL-GPIHBP1 binding by ANGPTL proteins. A: RHMVECs expressing smallBiT-GPIHBP1 were incubated with largeBiT-LPL for 2 h at 4°C and washed. Luminescent substrate was added to cells and luminescence read every 3 min for 15 min. After 15 min (dotted line), the indicated ANGPTL proteins were added and luminescence continued to be measured every 3 min for 45 min. Points represent mean ± 95% CI of luminescent signal over time normalized to the “LPL only” control at each time point. Data represent seven independent experiments with three biological replicates per group. B: Western blot of cell lysates from cells incubated with largeBiT-LPL for 3.5 h at 4°C, washed, and then treated with ANGPTL4, ANGPTL3, ANGPTL8, or heparin for 30 min at 37°C. Bands show LPL (using an antibody against the FLAG tag), GPIHBP1, and actin for biological triplicates. C: LPL activity of LPL bound to smallBiT-GPIHBP1-expressing RHMVECs washed and treated with ANGPTL4 (A4), ANGPTL3 (A3), ANGPTL8 (A8), or ANGPTL3 and ANGPTL8 (A3/8) for 30 min at 37°C. After incubation, cells were washed, and LPL was released from the cells with heparin before performing LPL activity assays. Bars represent normalized LPL activity (mean ± 95% CI of three independent experiments; n = 3 per experiment). D: Luminescence of smallBiT-GPIHBP1-expressing RHMVECs incubated with largeBiT-LPL. After washing off unbound LPL, luminescence was measured for 15 min at 3 min intervals. The indicated ANGPTL proteins were added (first dotted line) and luminescence was measured for an additional 30 min. Cells were then washed, fresh substrate was added (second dotted line), and luminescence was measured for another 15 min. Points represent luminescent signal normalized to the LPL only control at each time point (mean ± 95% CI of four independent experiments; n = 3 per group per experiment).
We considered the possibility that the decrease in luminescence we observed in our NanoBiT assay was not a result of actual dissociation of LPL from GPIBHP1, but rather the result of some kind of direct effect of ANGPTL proteins on the reconstituted luciferase or the luminescence substrate. To investigate this possibility, we performed a washout experiment. As before, LPL was bound to cells and substrate was added. After the subsequent addition of ANGPTL proteins or heparin, we again observed a decrease in luminescence. After 30 min of incubation with ANGPTL proteins or heparin, we washed the cells to remove substrate, unbound ANGPTL proteins, heparin, and any dissociated LPL. We then added fresh substrate and continued to measure luminescence. We reasoned that if loss in luminescent signal was the result of interference by ANGPTL proteins on some aspect of the luciferase system rather than dissociation of LPL, luminescent signal would be restored after washout. On the other hand, if decreased luminescent signal was the result of actual dissociation, signal would remain low after the removal of dissociated LPL and the addition of fresh substrate. We observed that after washout and addition of new substrate, the level of luminescence remained lower relative to the untreated control, indicating that reduced luminescence was indeed indicative of LPL dissociation (Fig. 3D).
Effect of THL on LPL-GPIHBP1 binding
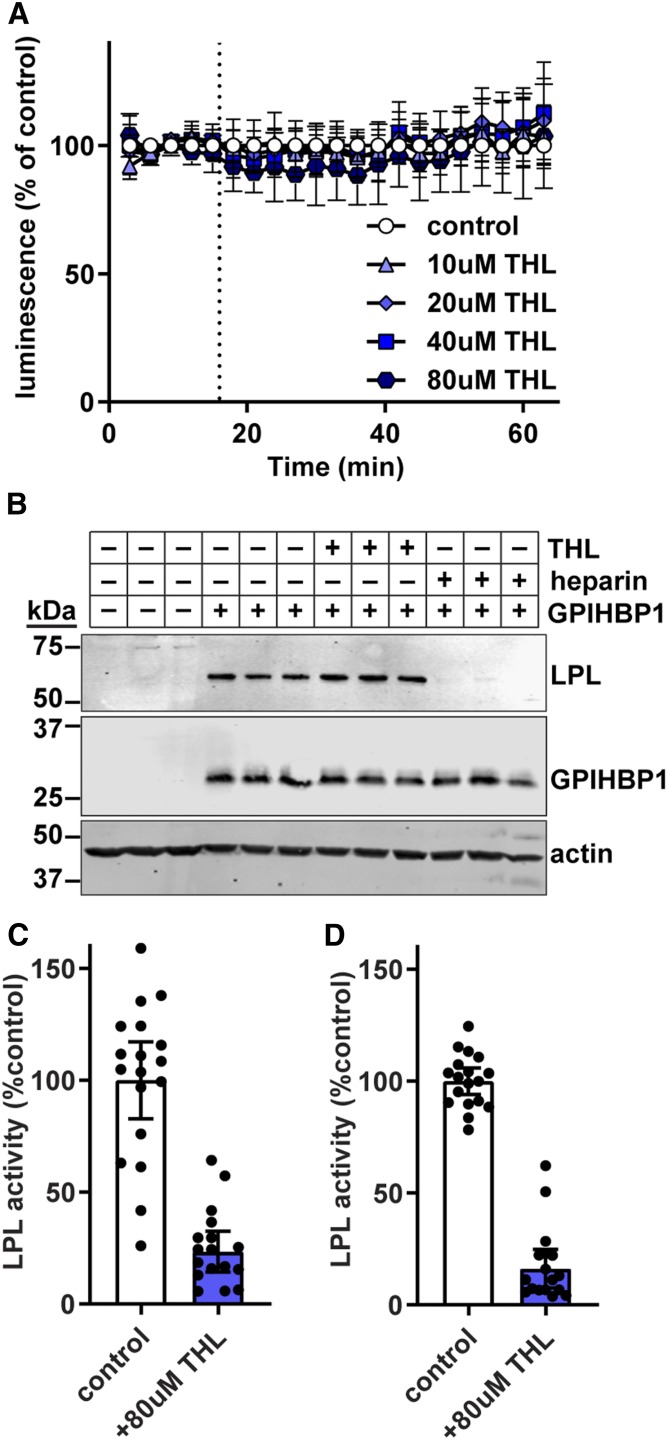
We next tested how the commonly used LPL inhibitors, THL, tyloxapol, and P-407, affect LPL-GPIHBP1 binding. THL (also known as Orlistat) is an active-site lipase inhibitor and has been used to inhibit LPL in the vasculature (2, 49, 50). As it has been reported that THL may alter the conformation of LPL or lead to LPL tetramerization (49), we tested whether THL disrupts the binding of LPL to GPIHBP1. Using our NanoBiT assay, we found that THL did not disrupt the binding of LPL to GPIHBP1 at any concentration, though the vehicle, DMSO, does partially reduce luminescent signal (Fig. 4A). Likewise, we saw no disruption of binding when we used a Western blot cell-binding assay (Fig. 4B). The lack of dissociation was not due to an inability of THL to inhibit GPIHBP1-bound LPL, as we found that THL was able to inhibit the activity of both largeBiT-LPL bound to smallBiT-GPIHBP1 and FLAG-tagged LPL bound to S-tagged GPIHBP1 on the surface of endothelial cells (Fig. 4C, D). Thus, we conclude that THL does not disrupt LPL-GPIHBP1 binding.
Fig. 4.
Effect of THL on GPIHBP1-LPL binding. A: RHMVECs expressing smallBiT-GPIHBP1 were incubated with largeBiT-LPL for 2 h at 4°C and washed. Luminescent substrate was added to the cells, and luminescence was read every 3 min for 15 min. After 15 min (dotted line), 0–80 μM of THL were added and luminescence continued to be measured every 3 min for 45 min. Points represent mean ± 95% CI of luminescent signal over time normalized to the vehicle (ethanol). Data represent three independent experiments each with three biological replicates per group. B: Western blot of cell lysates from cells incubated with largeBiT-LPL for 3.5 h at 4°C, washed, and then treated with 80 μM of THL or 100 U/ml heparin for 30 min. Bands show LPL (using an antibody against the FLAG tag), GPIHBP1, and actin for biological triplicates. C, D: LPL activity of largeBiT-LPL bound to smallBiT-GPIHBP1-expressing RHMVECs (C) or FLAG-LPL bound to S tag-GPIHBP1 (D) after treatment with 80 μM of THL for 30 min at 37°C. After incubation, the cells were washed, and LPL was released from the cells with heparin before performing LPL activity assays. Bars represent normalized LPL activity (mean ± 95% CI of three independent experiments; n = 6 per experiment).
Effect of chylomicrons and fatty acids on LPL-GPIHBP1 binding
Triglyceride emulsions, and presumably triglyceride-rich lipoproteins, can strip off LPL bound to the vascular wall (51). We asked whether we could reproduce this finding with our assay. Indeed, we found that chylomicrons could dissociate some LPL from GPIHBP1 (Fig. 5A). Again, we considered the possibility that chylomicrons might interfere with the reconstituted luciferase or the luminescence substrate. Therefore, we performed a washout experiment to test this possibility and found that after washout and addition of new substrate, luminescence signal was not restored, indicating that LPL had indeed dissociated from GPIHBP1 (Fig. 5B).
Fig. 5.
Effect of chylomicrons and fatty acids on GPIHBP1-LPL binding. RHMVECs expressing smBiT-GPIHBP1 were incubated with LargeBiT-LPL for 2 h at 4°C and washed. Luminescent substrate was added to cells and luminescence read every 3 min for 12–15 min. A: After 15 min (dotted line), chylomicrons (chylo) (1–10 μg/ml by protein) were added and luminescence continued to be measured for 45 min. ANGPTL4 (90 ng/ml) was also added as a control. B: After 12 min, chylomicrons or heparin were added (first dotted line) at the indicated concentrations and luminescence was measured for an additional 12 min. Cells were then washed, fresh substrate was added (second dotted line), and luminescence was measured for another 15 min. C: After 15 min (dotted line), sodium oleate (0.375–3 mM) was added and luminescence continued to be measured for 45 min. For all panels, points represent luminescent signal over time normalized to control at each time point (mean ± 95% CI of three independent experiments; n = 3–6 per group per experiment).
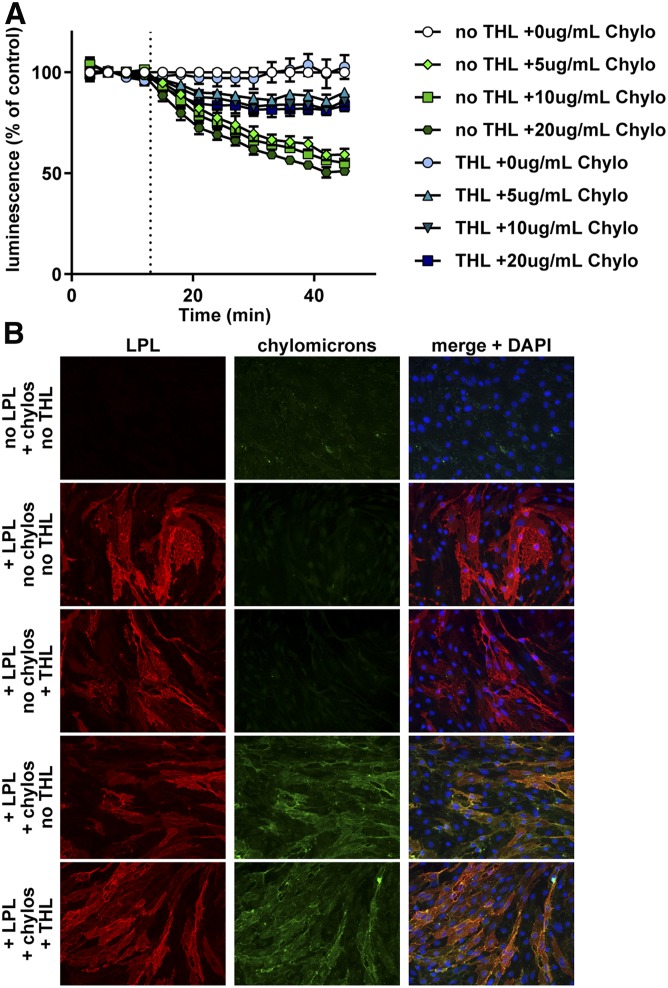
The mechanism by which triglyceride-rich lipoproteins strip LPL from GPIHBP1 is not known. One possibility is that the fatty acids released by LPL-mediated lipolysis facilitate the dissociation of LPL from GPIHBP1. To test this possibility, we first asked whether oleate could dissociate LPL from GPIHBP1. We observed that oleate did, in fact, dissociate LPL from GPIHBP1 in a dose-dependent manner (Fig. 5C). To further explore this issue, we asked whether chylomicrons could dissociate LPL from GPIHBP1 in the presence of THL. We reasoned that if fatty acids released from chylomicrons are the true causative agent, preventing the production of fatty acids from chylomicrons by inhibition of LPL catalytic activity would prevent the dissociation of the LPL-GPIHBP1 complex. As before, we found that THL itself did not interfere with the binding of LPL to GPIHBP1 (Fig. 6A). However, when THL was added in combination with chylomicrons, the ability of chylomicrons to dissociate LPL from GPIHBP1 was greatly reduced (Fig. 6A). THL did not interfere with the binding of chylomicrons to LPL, as judged by fluorescence microscopy. When fluorescent chylomicrons were incubated with endothelial cells expressing GPIHBP1, we could easily detect chylomicrons bound to cells, but only in the presence of LPL (Fig. 6B, supplemental Fig. S3). The level of chylomicron binding was not reduced when THL was also added (Fig. 6B, supplemental Fig. S3), indicating that THL does not prevent chylomicrons from binding LPL. Together, these data suggest that fatty acid products are at least partially responsible for the ability of chylomicrons to dissociate LPL from GPIHBP1.
Fig. 6.
Effect of THL on chylomicron-LPL interactions. A: RHMVECs expressing smBiT-GPIHBP1 were incubated with LargeBiT-LPL for 2 h at 4°C and washed. Cells were then treated with 80 μM of THL or vehicle for 30 min at 37°C. After washing, luminescent substrate was added to cells and luminescence read every 3 min for 12 min. After 12 min, chylomicrons (Chylo) (0–20 μg/ml by protein) were added and luminescence continued to be measured for 33 min. Points represent mean ± 95% CI of luminescent signal over time normalized to control at each time point. Data represent three independent experiments, each with 3–6 biological replicates per group. B: Immunofluorescence showing binding of LPL and chylomicrons to smallBiT-GPIHBP1-expressing RHMVECs. RHMVECs were incubated with largeBiT-LPL for 3 h at 4°C. After washing away unbound LPL, cells were incubated with or without THL (80 μM) for 30 min at 37°C. Cells were then washed and incubated with or without fluorescently labeled chylomicrons (green) for 30 min at 37°C. Cells were then stained for LPL (red) using an antibody against the FLAG tag and with DAPI (blue).
Effect of detergent inhibitors on LPL-GPIHBP1 binding
Tyloxapol and P-407 are detergents often used to block LPL-mediated triglyceride clearance in VLDL secretion and fat absorption assays (30–34). We asked whether these agents could disrupt LPL-GPIHBP1 binding. Initially, our NanoBiT assay indicated that both tyloxapol and P-407 disrupted LPL-GPIHBP1 binding in a dose-dependent manner (Fig. 7A, B). However, these results were inconsistent with Western blot cell-binding assays, which indicated no dissociation of LPL from GPIBHP1 in the presence of tyloxapol or P-407 (Fig. 7C, D). We reasoned that the presence of these detergents might interfere with luminescent signal rather than actually disrupting binding. Therefore, we performed a washout experiment. After treating LPL-GPIHBP1 complexes with detergent and observing a decrease in luminescent signal, we washed off the detergent and all unbound proteins. We then added fresh substrate to the cells. If LPL had actually dissociated from GPIHBP1, it would have been removed in the wash and, after the addition of fresh substrate, luminescent signal would remain lower than that in the control. Indeed that was the case with heparin-treated control cells (Fig. 7E, F; green line). However, in the tyloxapol- and P-407-treated cells, luminescence returned to levels indistinguishable from untreated cells (Fig. 7E, F), indicating that LPL remained bound to GPIHBP1. These data suggest that neither tyloxapol nor P-407 disrupted LPL-GPIHBP1 binding, but rather disrupted luminescent signal.
Tyloxapol and P-407 are used to block LPL-mediated triglyceride clearance in vivo. Under these circumstances, LPL is bound to GPIHBP1 on the surface of endothelial cells (1–3). Therefore, we next measured the ability of tyloxapol and P-407 to block LPL-activity when LPL is bound to endothelial cells by GPIHBP1. We found that LPL activity was strongly suppressed in the presence of either tyloxapol or P-407 (Fig. 7G, I). However, once either detergent was washed away, LPL activity was largely restored (Fig. 7H, J). These data suggest that either the interactions of tyloxapol and P-407 with LPL are easily reversible or that they do not act by interacting with LPL per se, but by interacting with triglyceride-rich lipoproteins, preventing these substrates from interacting with LPL.
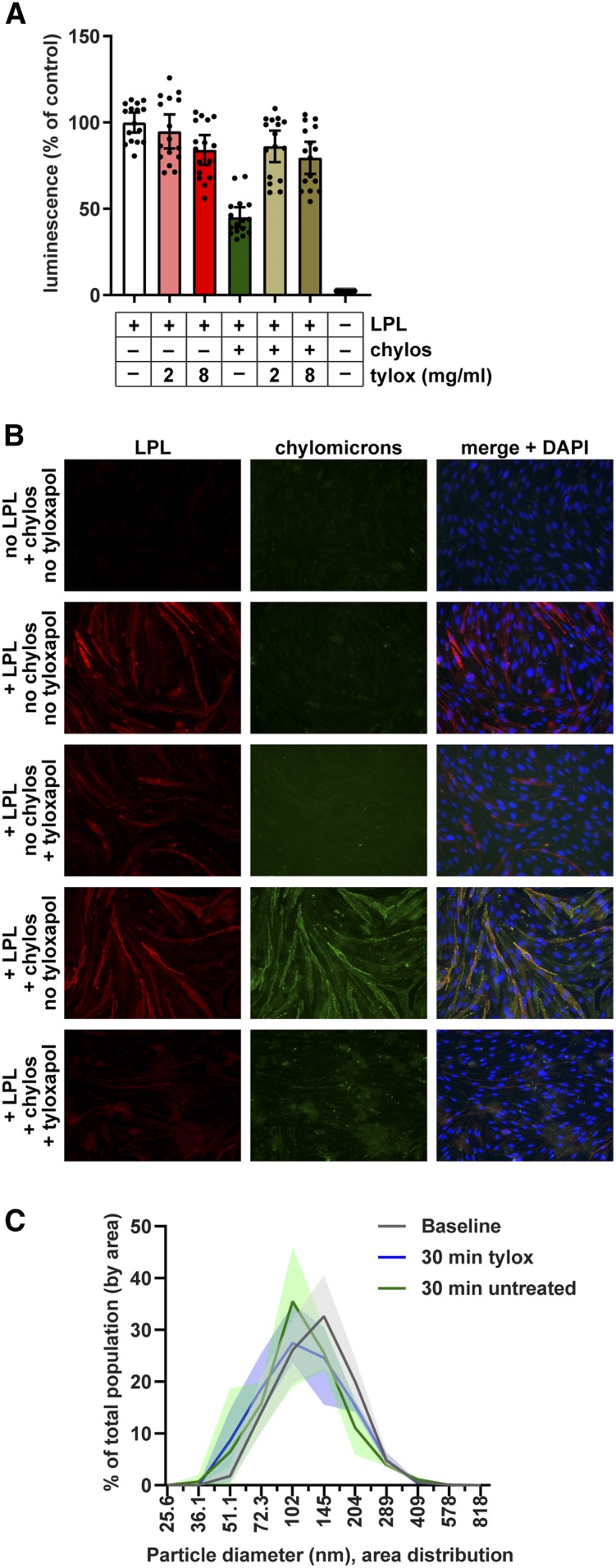
To further explore this issue, we asked whether tyloxapol could prevent chylomicrons from binding LPL and thus from dissociating LPL from GPIHBP1. Chylomicrons were mixed with tyloxapol and then added to LPL bound to GPIHBP1 on the surface of endothelial cells. Because of our finding that tyloxapol interferes with luminescent signal, we did not perform a kinetic experiment. Instead, we incubated the chylomicron-tyloxapol mixture with cells for 30 min and then washed the cells. Substrate was then added and luminescence assessed. As before, we found that treatment with chylomicrons reduced the amount of LPL bound to GPIHBP1, while treatment with tyloxapol on its own did not (Fig. 8A). However, when chylomicrons were pretreated with tyloxapol, they were unable to promote the dissociation of LPL from GPIHBP1 (Fig. 8A). These data support the idea that tyloxapol prevents triglyceride-rich lipoproteins from binding LPL. We next asked whether tyloxapol could release chylomicrons from LPL using immunofluorescence microscopy. When fluorescent chylomicrons were incubated with endothelial cells expressing smallBiT-GPIHBP1, we could easily detect chylomicrons bound to cells, but only in the presence of LPL (Fig. 8B, supplemental Fig. S4). Treatment with tyloxapol strikingly decreased the number of fluorescent chylomicrons bound to cells (Fig. 8B, supplemental Fig. S4), indicating that tyloxapol releases chylomicrons from LPL.
Fig. 8.
Effect of tyloxapol on binding of chylomicrons to LPL. A: Chylomicrons (20 μg/ml) were mixed with 2 or 8 mg/ml tyloxapol (tylox) and then incubated with largeBiT-LPL bound to smallBiT-GPIHBP1-expressing RHMVECs at 37°C. After 30 min, cells were washed, substrate was added, and luminescence was read. Bars represent luminescent signal (mean ± 95% CI of four independent experiments; n = 4 per group per experiment) normalized to the no chylomicron no tyloxapol control. B: Immunofluorescence showing binding of LPL and chylomicrons (chylos) to smallBiT-GPIHBP1-expressing RHMVECs. RHMVECs were incubated with largeBiT-LPL for 3.5 h at 4°C. After washing away unbound LPL, cells were incubated with or without with 20 µg/ml fluorescently labeled chylomicrons (green) for 30 min at 37°C, then with or without 8 mg/ml tyloxapol for 30 min at 37°C. Cells were then stained for LPL (red) using an antibody against the Flag tag and with DAPI (blue). C: Distribution of chylomicron diameters before (baseline) and after 30 min treatment with 8 mg/ml tyloxapol as measured by dynamic laser light scattering. Untreated chylomicrons stored at 4°C for 30 min after baseline reading were also analyzed (30 min untreated). Lines represent mean ± range (shaded area) particle distribution of three different reads.
To determine whether treatment of tyloxapol grossly altered the structure of chylomicrons, perhaps by solubilizing lipid within the chylomicron, we performed dynamic laser-light scattering analysis of chylomicrons before and after tyloxapol treatment. We found that after 30 min of treatment with tyloxapol, there was a small shift toward smaller particle size (Fig. 8C). However, this shift appeared to be primarily time dependent, as we observed a similar shift in untreated chylomicrons stored at 4°C for 30 min (Fig. 8C). We therefore concluded that tyloxapol had little effect on the gross structure of chylomicrons.
DISCUSSION
In this study, we described a new assay for assessing the binding of LPL to GPIHBP1 on the surface of endothelial cells. Using this assay, we found that ANGPTL4 and ANGPTL3-ANGPTL8 complexes, endogenous inhibitors of LPL, promote the dissociation of LPL from GPIHBP1; whereas commonly used exogenous agents, THL, tyloxapol, and P-407, block LPL activity but do not promote dissociation from GPIHBP1. We also describe evidence that fatty acids liberated from chylomicrons by LPL activity dissociate LPL from GPIHBP1 and that tyloxapol prevents the binding of chylomicrons to LPL.
Several methods for assessing the binding of LPL to GPIHBP1 have been described previously. These include immunofluorescence, Western blot, and ELISA based experiments that measure the LPL bound to GPIHBP1-expressing cells (1) as well as cell-free antibody-based pull-down assays using a soluble version of GPIHBP1 (42). Unquestionably, these methods have been critical in analyzing the interactions of LPL with GPIHBP1 and in assessing LPL and GPIHBP1 mutants. Nonetheless, the advantages of the assay described in this study are manyfold: 1) Our assay can easily be performed in a 96-well plate, allowing higher throughput, and thus more replication, than Western blot- and immunofluorescence-based methods. 2) Our assay detects the direct interaction of LPL with GPIHBP1 rather than the interaction of LPL with GPIHBP1-expressing cells. 3) Our assay is quantitative and very sensitive. We were able to distinguish differential binding of LPL over nearly two orders of magnitude of LPL concentrations. 4) In most situations, our assay can be used to monitor binding and dissociation in real time.
Although we performed our assay using RHMVECs, there is no reason to believe it would not work in other cell lines. While different vascular beds and endothelial cell types definitely vary greatly in their expression of GPIHBP1 (1, 52, 53), when GPIHBP1 is present on the cell surface, we know of no evidence that GPIHBP1-LPL interactions differ according to cell type.
The current assay is not without limitations. Most notably, the assay requires the smallBiT and largeBiT tags, thus necessitating the use of recombinant protein. In addition, when testing the effects of detergents such as tyloxapol and P-407 on LPL binding, the detergents can interfere with the generation of luminescence. In these cases, as demonstrated in this study, the assay could still be used as an endpoint assay after washing out detergent.
In this study, we found that ANGPTL4 caused dissociation of LPL from GPIHBP1. These data are consistent with our previous observations (35). In addition, we also found that ANGPTL3-ANGPTL8 complexes also promoted dissociation of LPL from GPIHBP1. For both ANGPTL4 and ANGPTL3-ANGPTL8 complexes, dissociation of LPL from GPIHBP1 proceeded at a slow, steady rate, contrasting with heparin, which mediated LPL dissociation very quickly (see Fig. 3). This steady rate of dissociation could reflect catalytic time-dependent inactivation of LPL by ANGPTL proteins and is consistent with the catalytic unfolding mechanism of ANGPTL4-mediated inhibition (54, 55). The mechanism by which ANGPTL3-ANGPTL8 complexes inhibit LPL has not yet been clearly elucidated, but it could be similar to that of ANGPTL4 (56). Alternatively, it is possible that inhibition or inactivation of LPL by ANGPTL proteins does not lead to immediate dissociation of LPL from GPIHBP1, but rather alters the binding affinity of LPL such that it gradually dissociates from GPIHBP1.
We found that inhibition of LPL by THL (Orlistat) does not cause LPL to dissociate from GPIHBP1. THL has been used previously to probe the noncatalytic functions of LPL, such as bridging of lipoproteins to receptors (57, 58). The use of THL in these studies assumes that THL blocks only LPL catalytic function and not the ability of LPL to bind receptors. The role of GPIHBP1 in facilitating the noncatalytic functions of LPL has not been studied in depth. However, given the critical role of GPIHBP1 in transporting and anchoring LPL, it is likely that the binding of LPL to GPIHBP1 helps to mediate the noncatalytic functions of LPL. By showing that THL does not disrupt LPL binding, our study supports the continued use of THL to distinguish between the catalytic and noncatalytic functions of LPL.
Although tyloxapol and P-407 are commonly used to block the lipolytic action of LPL, the mechanism by which they do this remains unclear. Suggested mechanisms include the coating of substrate (triglyceride-rich lipoproteins) thereby preventing the interaction of triglycerides with LPL or direct inhibition of LPL (59–61). Although complete determination of the mechanism of action of these two nonionic detergents is beyond the scope of the current study, our data do provide some insight. We found that after treating LPL with tyloxapol or P-407, inhibition was easily reversible by washing the cells, suggesting that the interactions of the detergents with LPL, if any, are transient. We also found that in the presence of tyloxapol, chylomicrons could not dissociate LPL from GPIHBP1. Moreover, tyloxapol could be used to release chylomicrons bound to LPL, suggesting that in the presence of tyloxapol, chylomicrons cannot bind LPL. Thus, our data support a model in which detergents prevent the interaction of triglyceride-rich lipoproteins with LPL and likely do so by interacting with the lipoprotein rather than with LPL.
The ability of triglyceride emulsions to strip LPL from the vascular wall and, in particular, from GPIHBP1 has been documented previously (51). In our study, we confirm that chylomicron particles can indeed dissociate LPL from GPIHBP1. Furthermore, we found evidence that this dissociation is mediated by the fatty acid products of lipolysis. Free oleate was able to dissociate LPL from GPIHBP1, and treating LPL with THL to prevent lipolysis greatly reduced dissociation of LPL-GPIHBP1 complexes by chylomicrons. The mechanism by which fatty acids dissociate LPL from GPIHBP1 is not clear, and it remains to be determined whether the ability of chylomicrons to remove LPL from GPIHBP1 serves a physiological purpose.
In conclusion, the current study demonstrates the utility of a new real-time assay for assessing the binding of LPL to GPIHBP1. In the future, this assay could be used for variety of studies. For example, recently, it has been shown that autoantibodies against GPIHBP1 can block LPL binding and cause hypertriglyceridemia (62). Our assay could also be used to test for the presence of these blocking autoantibodies. Moreover, our assay could be used to assess how novel mutations in LPL or GPIHBP1 affect complex formation, or to investigate how additional proteins alter binding of LPL to GPIHBP1. As the study of LPL-mediated triglyceride metabolism continues, our assay provides another tool to interrogate this important pathway. Moreover, this assay could be adapted to probe the interaction of other extracellular proteins with their cognate membrane receptors.
Supplementary Material
Acknowledgments
The authors would like to thank Ashley Segura-Roman for providing largeBiT EL and Kathryn Spitler and Kelli Sylvers-Davie for their careful edits of the written manuscript.
Footnotes
Abbreviations:
- ANGPTL
- angiopoietin-like
- EL
- endothelial lipase
- GPIHBP1
- glycosylphosphatidylinositol-anchored HDL binding protein 1
- NanoBiT
- NanoLuc® Binary Technology
- P-407
- poloxamer-407
- PIPLC
- phosphatidylinositol-specific phospholipase C
- THL
- tetrahydrolipstatin
This work was supported by National Institutes of Health Grant R01HL130146 (B.S.J.D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Beigneux A. P., Davies B. S. J., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., et al. 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulbourne C. N., Gin P., Tatar A., Nobumori C., Hoenger A., Jiang H., Grovenor C. R. M., Adeyo O., Esko J. D., Goldberg I. J., et al. 2014. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies B. S. J., Beigneux A. P., Barnes R. H. II, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyrén R., Goldberg I., Olivecrona G., et al. 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies B. S. J., Goulbourne C. N., Barnes R. H., Turlo K. A., Gin P., Vaughan S., Vaux D. J., Bensadoun A., Beigneux A. P., Fong L. G., et al. 2012. Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J. Lipid Res. 53: 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen K. K., Midtgaard S. R., Mysling S., Kovrov O., Hansen L. B., Skar-Gislinge N., Beigneux A. P., Kragelund B. B., Olivecrona G., Young S. G., et al. 2018. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 115: E6020–E6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mysling S., Kristensen K. K., Larsson M., Beigneux A. P., Gårdsvoll H., Fong L. G., Bensadouen A., Jørgensen T. J., Young S. G., and Ploug M.. 2016. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 5: e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S. J., Kuivenhoven J. A., et al. 2009. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler. Thromb. Vasc. Biol. 29: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivecrona G., Ehrenborg E., Semb H., Makoveichuk E., Lindberg A., Hayden M. R., Gin P., Davies B. S. J., Weinstein M. M., Fong L. G., et al. 2010. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J. Lipid Res. 51: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franssen R., Young S. G., Peelman F., Hertecant J., Sierts J. A., Schimmel A. W. M., Bensadoun A., Kastelein J. J. P., Fong L. G., Dallinga-Thie G. M., et al. 2010. Chylomicronemia with low postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ Cardiovasc Genet. 3: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coca-Prieto I., Kroupa O., Gonzalez-Santos P., Magne J., Olivecrona G., Ehrenborg E., and Valdivielso P.. 2011. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: identification of a novel GPIHBP1 mutation. J. Intern. Med. 270: 224–228. [DOI] [PubMed] [Google Scholar]
- 11.Charrière S., Peretti N., Bernard S., Di Filippo M., Sassolas A., Merlin M., Delay M., Debard C., Lefai E., Lachaux A., et al. 2011. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J. Clin. Endocrinol. Metab. 96: E1675–E1679. [DOI] [PubMed] [Google Scholar]
- 12.Rios J. J., Shastry S., Jasso J., Hauser N., Garg A., Bensadoun A., Cohen J. C., and Hobbs H. H.. 2012. Deletion of GPIHBP1 causing severe chylomicronemia. J. Inherit. Metab. Dis. 35: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewey F. E., Gusarova V., O’Dushlaine C., Gottesman O., Trejos J., Hunt C., Van Hout C. V., Habegger L., Buckler D., Lai K-M. V., et al. 2016. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N. Engl. J. Med. 374: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewey F. E., Gusarova V., Dunbar R. L., O’Dushlaine C., Schurmann C., Gottesman O., McCarthy S., Van Hout C. V., Bruse S., Dansky H. M., et al. 2017. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N. Engl. J. Med. 377: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham M. J., Lee R. G., Brandt T. A., Tai L-J., Fu W., Peralta R., Yu R., Hurh E., Paz E., McEvoy B. W., et al. 2017. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N. Engl. J. Med. 377: 222–232. [DOI] [PubMed] [Google Scholar]
- 16.Romeo S., Yin W., Kozlitina J., Pennacchio L. A., Boerwinkle E., Hobbs H. H., and Cohen J. C.. 2009. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 119: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musunuru K., Pirruccello J. P., Do R., Peloso G. M., Guiducci C., Sougnez C., Garimella K. V., Fisher S., Abreu J., Barry A. J., et al. 2010. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 363: 2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeo S., Pennacchio L. A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H. H., and Cohen J. C.. 2007. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 39: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel N. O., Stirrups K. E., Masca N. G., Erdmann J., Ferrario P. G., König I. R., Weeke P. E., Webb T. R., Auer P. L., et al. 2016. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 374: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollin T. I., Damcott C. M., Shen H., Ott S. H., Shelton J., Horenstein R. B., Post W., McLenithan J. C., Bielak L. F., Peyser P. A., et al. 2008. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 322: 1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jørgensen A. B., Frikke-Schmidt R., Nordestgaard B. G., and Tybjærg-Hansen A.. 2014. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 371: 32–41. [DOI] [PubMed] [Google Scholar]
- 22.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute, Crosby J., Peloso G. M., Auer P. L., Crosslin D. R., Stitziel N. O., Lange L. A., Lu Y., Tang Z., Zhang H., et al. 2014. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 371: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gusarova V., Alexa C. A., Wang Y., Rafique A., Kim J. H., Buckler D., Mintah I. J., Shihanian L. M., Cohen J. C., Hobbs H. H., et al. 2015. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J. Lipid Res. 56: 1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusarova V., Banfi S., Alexa-Braun C. A., Shihanian L. M., Mintah I. J., Lee J. S., Xin Y., Su Q., Kamat V., Cohen J. C., et al. 2017. ANGPTL8 blockade with a monoclonal antibody promotes triglyceride clearance, energy expenditure, and weight loss in mice. Endocrinology. 158: 1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Z., Abou-Samra A. B., and Zhang R.. 2015. A lipasin/Angptl8 monoclonal antibody lowers mouse serum triglycerides involving increased postprandial activity of the cardiac lipoprotein lipase. Sci. Rep. 5: 18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham M. J., Lee R. G., Bell T. A., Fu W., Mullick A. E., Alexander V. J., Walter S., Nick V., Richard G., John S., et al. 2013. Antisense oligonucleotide inhibition of apolipoprotein C-III reduces plasma triglycerides in rodents, nonhuman primates, and humans. Circ. Res. 112: 1479–1490. [DOI] [PubMed] [Google Scholar]
- 27.Gaudet D., Brisson D., Tremblay K., Alexander V. J., Singleton W., Hughes S. G., Geary R. S., Baker B. F., Graham M. J., Crooke R. M., et al. 2014. Targeting APOC3 in the familial chylomicronemia syndrome. N. Engl. J. Med. 371: 2200–2206. [DOI] [PubMed] [Google Scholar]
- 28.Gaudet D., Alexander V. J., Baker B. F., Brisson D., Tremblay K., Singleton W., Geary R. S., Hughes S. G., Viney N. J., Graham M. J., et al. 2015. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N. Engl. J. Med. 373: 438–447. [DOI] [PubMed] [Google Scholar]
- 29.Desai U., Lee E-C., Chung K., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K. A., Sands A. T., et al. 2007. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. USA. 104: 11766–11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otway S., and Robinson D. S.. 1967. The effect of a non-ionic detergent (Triton WR 1339) on the removal of triglyceride fatty acids from the blood of the rat. J. Physiol. 190: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otway S., and Robinson D. S.. 1967. The use of a non-ionic detergent (Triton WR 1339) to determine rates of triglyceride entry into the circulation of the rat under different physiological conditions. J. Physiol. 190: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Catalina F., Grundy S. M., and Patel S.. 1996. Method to measure apolipoprotein B-48 and B-100 secretion rates in an individual mouse: evidence for a very rapid turnover of VLDL and preferential removal of B-48- relative to B-100-containing lipoproteins. J. Lipid Res. 37: 210–220. [PubMed] [Google Scholar]
- 33.Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., and Billheimer J. T.. 2005. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J. Lipid Res. 46: 2023–2028. [DOI] [PubMed] [Google Scholar]
- 34.Voshol P. J., Minich D. M., Havinga R., Elferink R. P., Verkade H. J., Groen A. K., and Kuipers F.. 2000. Postprandial chylomicron formation and fat absorption in multidrug resistance gene 2 P-glycoprotein-deficient mice. Gastroenterology. 118: 173–182. [DOI] [PubMed] [Google Scholar]
- 35.Chi X., Shetty S. K., Shows H. W., Hjelmaas A. J., Malcolm E. K., and Davies B. S. J.. 2015. Angiopoietin-like 4 modifies the interactions between lipoprotein lipase and its endothelial cell transporter GPIHBP1. J. Biol. Chem. 290: 11865–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon A. S., Schwinn M. K., Hall M. P., Zimmerman K., Otto P., Lubben T. H., Butler B. L., Binkowski B. F., Machleidt T., Kirkland T. A., et al. 2016. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11: 400–408. [DOI] [PubMed] [Google Scholar]
- 37.Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., and Naldini L.. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72: 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi X., Britt E. C., Shows H. W., Hjelmaas A. J., Shetty S. K., Cushing E. M., Li W., Dou A., Zhang R., and Davies B. S. J.. 2017. ANGPTL8 promotes the ability of ANGPTL3 to bind and inhibit lipoprotein lipase. Mol. Metab. 6: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson H. E., Hassan F., Marais D., and Hayden M. R.. 1996. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem. Biophys. Res. Commun. 227: 189–194. [DOI] [PubMed] [Google Scholar]
- 40.Voss C. V., Davies B. S. J., Tat S., Gin P., Fong L. G., Pelletier C., Mottler C. D., Bensadoun A., Beigneux A. P., and Young S. G.. 2011. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl. Acad. Sci. USA. 108: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basu D., Manjur J., and Jin W.. 2011. Determination of lipoprotein lipase activity using a novel fluorescent lipase assay. J. Lipid Res. 52: 826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beigneux A. P., Gin P., Davies B. S. J., Weinstein M. M., Bensadoun A., Fong L. G., and Young S. G.. 2009. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 284: 30240–30247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang T., Li L., Tang J., Li Y., Lin W. Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. 2010. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28: 749–755. [DOI] [PubMed] [Google Scholar]
- 44.Young S. G., Davies B. S. J., Voss C. V., Gin P., Weinstein M. M., Tontonoz P., Reue K., Bensadoun A., Fong L. G., and Beigneux A. P.. 2011. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J. Lipid Res. 52: 1869–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cushing E. M., Chi X., Sylvers K. L., Shetty S. K., Potthoff M. J., and Davies B. S. J.. 2017. Angiopoietin-like 4 directs uptake of dietary fat away from adipose during fasting. Mol. Metab. 6: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gin P., Beigneux A. P., Voss C., Davies B. S. J., Beckstead J. A., Ryan R. O., Bensadoun A., Fong L. G., and Young S. G.. 2011. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31: 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birrane G., Beigneux A. P., Dwyer B., Strack-Logue B., Kristensen K. K., Francone O. L., Fong L. G., Mertens H. D. T., Pan C. Q., Ploug M., et al. 2019. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc. Natl. Acad. Sci. USA. 116: 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gin P., Yin L., Davies B. S. J., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., and Beigneux A. P.. 2008. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 283: 29554–29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lookene A., Skottova N., and Olivecrona G.. 1994. Interactions of lipoprotein lipase with the active-site inhibitor tetrahydrolipstatin (Orlistat)R. Eur. J. Biochem. 222: 395–403. [DOI] [PubMed] [Google Scholar]
- 50.Merkel M., Loeffler B., Kluger M., Fabig N., Geppert G., Pennacchio L. A., Laatsch A., and Heeren J.. 2005. Apolipoprotein AV accelerates plasma hydrolysis of triglyceriderich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. J. Biol. Chem. 280: 21553–21560. [DOI] [PubMed] [Google Scholar]
- 51.Weinstein M. M., Yin L., Beigneux A. P., Davies B. S. J., Gin P., Estrada K., Melford K., Bishop J. R., Esko J. D., Dallinga-Thie G. M., et al. 2008. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J. Biol. Chem. 283: 34511–34518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies B. S. J., Waki H., Beigneux A. P., Farber E., Weinstein M. M., Wilpitz D. C., Tai L-J., Evans R. M., Fong L. G., Tontonoz P., et al. 2008. The expression of GPIHBP1, an endothelial cell binding site for lipoprotein lipase and chylomicrons, is induced by peroxisome proliferator-activated receptor-gamma. Mol. Endocrinol. 22: 2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olafsen T., Young S. G., Davies B. S. J., Beigneux A. P., Kenanova V. E., Voss C., Young G., Wong K-P., Barnes R. H., Tu Y., et al. 2010. Unexpected expression pattern for glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1) in mouse tissues revealed by positron emission tomography scanning. J. Biol. Chem. 285: 39239–39248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sukonina V., Lookene A., Olivecrona T., and Olivecrona G.. 2006. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc. Natl. Acad. Sci. USA. 103: 17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mysling S., Kristensen K. K., Larsson M., Kovrov O., Bensadouen A., Jørgensen T. J., Olivecrona G., Young S. G., and Ploug M.. 2016. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 5: e20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovrov O., Kristensen K. K., Larsson E., Ploug M., and Olivecrona G.. 2019. On the mechanism of angiopoietin-like protein 8 for control of lipoprotein lipase activity. J. Lipid Res. 60: 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nykjaer A., Bengtsson-Olivecrona G., Lookene A., Moestrup S. K., Petersen C. M., Weber W., Beisiegel U., and Gliemann J.. 1993. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds lipoprotein lipase and beta-migrating very low density lipoprotein associated with the lipase. J. Biol. Chem. 268: 15048–15055. [PubMed] [Google Scholar]
- 58.Goti D., Balazs Z., Panzenboeck U., Hrzenjak A., Reicher H., Wagner E., Zechner R., Malle E., and Sattler W.. 2002. Effects of lipoprotein lipase on uptake and transcytosis of low density lipoprotein (LDL) and LDL-associated α-tocopherol in a porcine in vitro blood-brain barrier model. J. Biol. Chem. 277: 28537–28544. [DOI] [PubMed] [Google Scholar]
- 59.Johnston T. P., and Palmer W. K.. 1993. Mechanism of poloxamer 407-induced hypertriglyceridemia in the rat. Biochem. Pharmacol. 46: 1037–1042. [DOI] [PubMed] [Google Scholar]
- 60.Borensztajn J., Rone M. S., and Kotlar T. J.. 1976. The inhibition in vivo of lipoprotein lipase (clearing-factor lipase) activity by triton WR-1339. Biochem. J. 156: 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schotz M. C., Scanu A., and Page I. H.. 1957. Effect of triton on lipoprotein lipase of rat plasma. Am. J. Physiol. 188: 399–402. [DOI] [PubMed] [Google Scholar]
- 62.Beigneux A. P., Miyashita K., Ploug M., Blom D. J., Ai M., Linton M. R. F., Khovidhunkit W., Dufour R., Garg A., McMahon M. A., et al. 2017. Autoantibodies against GPIHBP1 as a cause of hypertriglyceridemia. N. Engl. J. Med. 376: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.