Abstract
This article focuses on the establishment of an accurate and sensitive quantitation method for the analysis of furan fatty acids. In particular, the sensitivity of GC/MS and UPLC/ESI/MS/MS was compared for the identification and quantification of furan fatty acids. Different methylation methods were tested with respect to GC/MS analysis. Special attention needs to be paid to the methylation of furan fatty acids, as acidic catalysts might lead to the degradation of the furan ring. GC/MS analysis in full-scan mode demonstrated that the limit of quantitation was 10 μM. UPLC/ESI/MS/MS in multiple reaction monitoring mode displayed a higher detection sensitivity than GC/MS. Moreover, the identification of furan fatty acids with charge-reversal derivatization was tested in the positive mode with two widely used pyridinium salts. Significant oxidation was unexpectedly observed using N-(4-aminomethylphenyl) pyridinium as a derivatization agent. The formed 3-acyl-oxymethyl-1-methylpyridinium iodide derivatized by 2-bromo-1-methylpyridinium iodide and 3-carbinol-1-methylpyridinium iodide improved the sensitivity more than 2,000-fold compared with nonderivatization in the negative mode by UPLC/ESI/MS/MS. This charge-reversal derivatization enabled the targeted quantitation of furan fatty acids in human plasma. Thus, it is anticipated that this protocol could greatly contribute to the clarification of pathological mechanisms related to furan fatty acids and their metabolites.
Keywords: charge-reversal derivatization, multiple reaction monitoring, precursor ion scan
The current state of the art in lipidomics and metabolomics enables the discovery of various lipids associated with the development and progression of T2D (1–3), cancer (4), and cardiovascular diseases (5, 6). Recent studies have shown that a dibasic urofuran acid, 3-carboxy-4-methyl-5-propyl-2-furanopropanoic acid (CMPF), is found in high concentrations in subjects diagnosed with prediabetics, gestational diabetes, and T2D (7, 8). The elevated CMPF has been recognized as a potential biomarker in diabetes development. CMPF is believed to increase oxidative stress and impair insulin granule maturation and secretion (9). It has been claimed that CMPF is a metabolite of furan fatty acids (Fig. 1); however, it has been documented that furan fatty acids have a line of homologs in nature. Intriguingly, very recent human studies demonstrated that CMPF formed as a significant metabolite of fish oil composed of only EPA and DHA ethyl esters without any furan fatty acids identified (10–12). Hence, the exact origin of CMPF remains controversial and related metabolic pathways remain elusive.
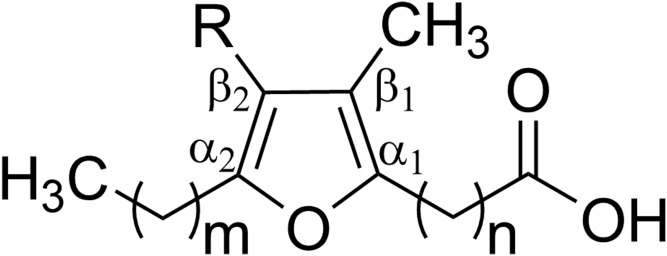
Fig. 1.
General structure of furan fatty acids. R = H or CH3. m = 0–4; n = 2–14.
An accurate and sensitive quantitation method of furan fatty acids in food and biological samples would greatly contribute to a clarification of the ongoing debate. GC-flame ion detection (FID) and GC/MS are generally considered as the most preferable methods for furan fatty acid profiling. For this, the methylation of the carboxylate group is inevitable (13–15). However, the similar chromatographic behavior of the furan ring with some common monounsaturated fatty acids at high concentrations renders the peak identification of furan fatty acids ambiguous and laborious (16). It has been documented that the hydrogenation of furan fatty acid methyl esters to more polar tetrahydrofurancarboxylic acid methyl esters could facilitate the chromatographic separations (17). Moreover, some improved analytical methods, including multidimensional GC/MS and HPLC coupled on-line with capillary gas chromatography with a photoionization detector mounted in series with a flame ionization detector, contribute greatly to the identification of furan fatty acids (18, 19). The quantification of furan fatty acids has also recently been achieved by 1H NMR (20). Accordingly, it has been reported that a variety of food sources, especially fish and fish oil, contain furan fatty acids below the mg/g level (21, 22). Nonetheless, little evidence for the existence of furan fatty acids in human tissues exists so far, and extremely low levels of less than 50 ng/ml have been reported (17, 23, 24). A complicated enrichment from large volumes of plasma/serum (e.g., >20 ml) is imperative before analysis (24). Additionally, a lack of commercial furan fatty acid standards and deuterated analogs represents further key limiting factors for the accurate quantitation in biological samples.
Generally, natural furan fatty acids can be divided into two major classes. The first class is represented by a propyl moiety, whereas the second is dominated by a pentyl moiety at the α2 position of the furan ring (19, 25). In addition to CMPF, 3-carboxy-4-methyl-5-pentyl-2-furanopropanoic acid has also been identified in human plasma and urine (10, 26). It has been suggested that the metabolic fate of furan fatty acids is associated with this difference (propyl or pentyl) (27). The most abundant furan fatty acid homologs detected in food matrices and human plasma are 11-(3,4-dimethyl-5-propylfuran-2-yl)undecanoic acid (11D3) and 11-(3,4-dimethyl-5-pentylfuran-2-yl)undecanoic acid (11D5), representing the aforementioned two classes (17, 28). Therefore, a targeted quantitation of these two classes of furan fatty acids, especially 11D3 and 11D5, in human diets and biological tissues represents a promising approach for clarifying the relationship of furan fatty acids with CMPF.
Shotgun lipidomics and LC/MS/MS-based lipidomics have also been well developed for fatty acid analysis (29, 30). However, an accurate and sensitive protocol for the quantitative analysis of furan fatty acids by shotgun lipidomics or LC/MS/MS has not been developed yet. To date, there is only one report on the qualitative analysis of furan fatty acids in fish lipids by HPLC/ESI/Q-TOF/MS (31).
Herein, we explored an approach for the identification and quantitation of furan fatty acids in biological samples by comparing the sensitivity of GC/MS and LC/MS/MS, with a special focus on the levels of 11D3 and 11D5. We paid particular attention to the derivatization of furan fatty acids. Notably, the abundant fragments derived from the derivatized moiety could be used for the quantitative analysis of individual furan fatty acid species. We believe this approach could facilitate the clarification of the metabolic precursor of CMPF and contribute to a greater understanding of the biological relevance of furan fatty acids.
EXPERIMENTAL METHODS
Reagents and materials
Acetonitrile and methanol of LC/MS grade were purchased from Merck. Ammonium acetate, 2-bromopyridine, 3-carbinolpyridine, Diazald®, formic acid, and triethylamine (TEA) were purchased from Sigma-Aldrich. 1,2-Dihenarachidoyl-sn-glycero-3-phosphocholine (PC-21:0/21:0) were obtained from Nu-Chek Prep, Inc. Diethylene glycol monoethyl ether was obtained from TCI (Shanghai) Development Co., Ltd. Iodomethane was purchased from Acros Organics. Water was prepared using a Milli-Q system (Millipore).
11-(3,4-Dimethyl-5-propylfuran-2-yl)undecanoic acid (purity >98%) and 11-(3,4-dimethyl-5-pentylfuran-2-yl)undecanoic acid (purity >98%) were synthesized by Shanghai Medicilon Inc. 11-(3,4-Dimethyl-5-(propyl-2,2,3,3,3-d5)furan-2-yl)undecanoic acid (chemical purity >99.5%; deuterium purity >99.0%) and 11-(3,4-dimethyl-5-(pentyl-4,4,5,5,5-d5)furan-2-yl)undecanoic acid (chemical purity >99.5%; deuterium purity >99.0%) were synthesized by Syncom. The AMP+ MaxSpec® Kit (50 tests, Catalog #710000) containing N-(4-aminomethylphenyl) pyridinium (AMPP), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride aqueous solution, N-hydroxybenzotriazole, and acetonitrile/N,N-dimethylformamide (4:1; v/v) solution was purchased from Cayman Chemical Company.
Nomenclature
Furan fatty acids are abbreviated according to the simplified number-letter-number notation (21). The first number indicates the length of the carboxyalkyl chain at the α1 position. The letter “M” or “D” represents one or two methyl moieties at β positions. The second letter donates the length of the alkyl chain at the α2 position. Accordingly, 11-(3,4-dimethyl-5-propylfuran-2-yl)undecanoic acid and 11-(3,4-dimethyl-5-pentylfuran-2-yl)undecanoic acid are denoted as 11D3 and 11D5, respectively. The corresponding deuterated analogs, 11-(3,4-dimethyl-5-(propyl-2,2,3,3,3-d5)furan-2-yl)undecanoic acid and 11-(3,4-dimethyl-5-(pentyl-4,4,5,5,5-d5)furan-2-yl)undecanoic acid, are abbreviated as 11D3-d5 and 11D5-d5, respectively.
Preparation of furan fatty acid mixtures
The stock solutions of furan fatty acids and deuterated internal standards (ISs) were prepared in pure acetonitrile with a concentration of 10 μg/ml. The concentration of each stock solution was determined gravimetrically. The mixtures of furan fatty acids were prepared from individual stock solutions.
Preparation of human plasma samples
This study was approved by the Ethics Committee of Shanghai General Hospital, which is affiliated with Shanghai Jiao Tong University School of Medicine. Fasting blood samples were collected from healthy subjects and stored at −80°C according to a standardized procedure as previously described (32).
Methylation of furan fatty acids
The methylation of furan fatty acids was conducted according to three different methods: ethereal diazomethane, methanolic sulfuric acid (H2SO4-MeOH), and boron trifluoride-methanol (BF3-MeOH).
First, 5 μg PC-21:0/21:0 was mixed with furan fatty acid standards (11D3 and 11D5) and dried under N2. The methyl esters were prepared according to the method described by Uchida et al. (31) with slight modifications. The mixed standards were dissolved in toluene (0.2 ml) and mixed with 0.5 M sodium methoxide in methanol (0.4 ml). The solution was maintained at 50°C for 10 min. Glacial acetic acid (20 μl) was then added, followed by ddH2O (1 ml). The required methyl esters and nonesterified free furan fatty acids were extracted into n-hexane (3 × 1 ml) and dried under N2. The following methylation of free fatty acids was performed by ethereal diazomethane. The final products were resuspended in 50 μl n-hexane and then subjected to GC/MS analysis.
Second, furan fatty acid standards (11D3 and 11D5) with 5 μg PC-21:0/21:0 as ISs were derivatized with 2 ml methanol containing 2% sulfuric acid for 2 h at 80°C in sealed borosilicate glass tubes. A saturated aqueous solution of NaCl (1 ml) was then added, and the generated furan fatty acid methyl esters were extracted three times using 1 ml n-hexane. The combined organic layer was dried under N2 and resuspended in 50 μl n-hexane and then subjected to GC/MS analysis.
Finally, furan fatty acid standards (11D3 and 11D5) with 5 μg PC-21:0/21:0 as ISs were derivatized with 2 ml of ∼15% boron trifluoride in methanol for 1 h at 90°C in sealed borosilicate glass tubes. A saturated aqueous solution of NaCl (1 ml) was then added, and the generated furan fatty acid methyl esters were extracted three times using 1 ml n-hexane. The combined organic layer was dried under N2 and redissolved in 50 μl n-hexane and then subjected to GC/MS analysis.
Synthesis of 2-bromo-1-methylpyridinium iodide (BMP) and 3-carbinol-1-methylpyridinium iodide (CMP)
Iodomethane (50 mmol, 3.11 ml) was added to 2-bromopyridine (10 mmol, 0.97 ml) or 3-carbinolpyridine (10 mmol, 0.96 ml) and stirred at room temperature for 1 h. The yellow crystals were washed with 5 ml cold acetone three times and dried in a vacuum.
Derivatization of furan fatty acids with AMPP and BMP and CMP
The derivatization of furan fatty acids with AMPP was carried out according to the manufacturer’s instructions. Briefly, 100 μl 11D3 and 11D5 (1 μg/ml in acetonitrile) was dried under N2 and resuspended in 20 μl cold acetonitrile/N,N-dimethylformamide (4:1; v/v). Cold 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (20 μl) and N-hydroxybenzotriazole (10 μl) were sequentially added to the solution. The vial was briefly mixed on a vortex mixer and placed on ice. Finally, the AMP+ solution (30 μl) was added, mixed, and heated at 60°C for 30 min.
The derivatization of furan fatty acids with BMP and CMP was conducted according to a previously described method (33) with slight modifications. 11D3 and 11D5 (10 μg/ml in acetone, 10 μl), BMP (7.5 mg/ml in acetonitrile, 20 μl), and CMP (10 mg/ml in acetonitrile, 20 μl) were added and briefly mixed on a vortex mixer. TEA (1 μl) was then added to the solution, mixed, and heated at 50°C for 30 min. After derivatization, the solution was dried under N2 and resuspended in 100 μl acetonitrile-H2O (7:3; v/v). The formed 3-acyl-oxymethyl-1-methylpyridinium iodides (AMMPs) are abbreviated as 11D3-AMMP and 11D5-AMMP. The scheme of furan fatty acid derivatization is shown in supplemental Fig. S1.
GC/MS analysis
Furan fatty acids were analyzed as fatty acid methyl esters, which were separated on an Agilent HP-INNOWax column [30 m × 0.25 mm inner diameter (ID), 0.25 μm film thickness] and analyzed by triple quadrupole GC/MS (Agilent Technologies) based on EI. EI at 70 eV was performed with a full scan range of m/z 50–500. All measurements were carried out according to the following oven temperature program: the initial temperature of 60°C was held for 2 min and raised to 160°C with a ramp of 20°C/min; the temperature was then increased to 240°C with a ramp of 5°C/min and held for 7 min, resulting in a total run time of 30 min.
UPLC/ESI/MS/MS analysis
The analysis of free and derivatized furan fatty acids, including furan fatty acids-AMPP and furan fatty acids-AMMP, was performed on a Shimadzu UPLC/MS/MS 8050 system composed of a 30AD LC system (an LC-30 A binary pump, an SIL-30AC autosampler, and a CTO-30AC column oven) and an 8050 triple quadrupole mass spectrometer equipped with a heated ESI source.
The analysis of free furan fatty acids was performed in the negative mode. Analytes were separated with an Agilent ZORBAX Eclipse Plus C18 column (2.1 mm × 100 mm, 1.8 μm ID) at a flow rate of 0.3 ml/min. The mobile phase consisted of solvent A (H2O with 10 mM CH3COONH4) and solvent B (MeOH with 10 mM CH3COONH4). Samples were applied to the column at 60% B and eluted with a linear increase in B (60–80% B from 1.0 to 6.5 min) that reached 100% at 7.5 min and was held for 6 min. The elution was then decreased to 80% in B at 14.0 min and held for 3.5 min. Finally, the elution was returned to the initial condition at 18.0 min for 2 min of equilibration. The MS parameters were as follows: nebulizing gas (N2) flow rate, 3 l/min; drying gas (N2) flow rate, 15 l/min; desolvation line temperature, 250°C; heat block temperature, 400°C; and interface temperature, 350°C.
The analysis of derivatized furan fatty acids was performed in the positive mode. Analytes were separated with a Waters ACQUITY UPLC CSH C18 column (2.1 mm × 100 mm, 1.7 μm ID) at a flow rate of 0.4 ml/min. The mobile phase consisted of solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Samples were applied to the column at 10% B and then raised to 30% B from 1.0 to 2.0 min. The elution was increased linearly in B (30–80% from 2.0 to 5.0 min) that reached 100% at 6.0 min and was held for 1 min before returning to initial conditions for 3 min of equilibration.
Validation procedure
The peak area ratios of furan fatty acid standards to corresponding ISs against the concentration ratios were plotted to construct the standard curves in pooled plasma. The concentrations of standards are shown in supplemental Table S1.
A seven-point dilution series of each furan fatty acid species was prepared from the stock solution for external calibration. The above calibrator solutions were selected according to the range of physiological levels. Three aliquots of one low-concentrated plasma sample spiked with different furan fatty acid concentrations were used to determine precision and accuracy. Intra- and interassay precision was evaluated by the relative standard deviation (RSD) of triplicate injections on the same day and on three consecutive days, respectively. The accuracy was assessed according to the following formula:
Additionally, five standard solutions with a concentration range of 0.005 to 50 ng/ml were prepared to evaluate the LOD and lower limit of quantitation (LLOQ). The LOD was determined as the lowest concentration with a single-to-noise ratio >3, while the LLOQ was determined according to the calculated precision (coefficient of variation <20%) and accuracy (80–120%) by measuring each concentration five times.
Analysis of furan fatty acids in human plasma
The furan fatty acid extraction was carried out by the following protocol. 11D3-d5 and 11D5-d5 (each 10 ng) and plasma (20 μl) were added to a glass vial. The vial was then sealed with a Teflon/silicone disk and incubated at 60°C for 2 h after 1 M KOH in 95% ethanol (500 μl) was mixed with the plasma. The pH was then adjusted to between 3 and 4 with 1 M HCl when the mixture was cooled to room temperature. Free fatty acids were extracted by 300 μl n-hexane three times and dried under N2. The following derivatization with BMP and CMP was carried out according to the above protocol.
Statistical analysis
Significant differences of furan fatty acid concentrations between female and male subjects were evaluated by Student’s unpaired t-tests. Data were analyzed using GraphPad Prism version 8.0. P < 0.05 was considered to be statistically significant.
RESULTS AND DISCUSSION
Comparison of different methylation methods of furan fatty acids for GC/MS analysis
Generally, fatty acids are analyzed by GC-FID or GC/MS as their volatile nonpolar derivatives. Methyl esters are the preferred derivates and can be derived by acid- or base- catalyzed methylation or using diazomethane and related reagents (34). Acid- and base- catalyzed methylation are suitable for the most common fatty acids such as straight-chain and branched-chain fatty acids. However, special attention must be paid to some unusual fatty acids (e.g., fatty acids containing cyclopropene, cyclopropane, or epoxy groups), as they are susceptible to chemical attack by acidic catalysts (34, 35).
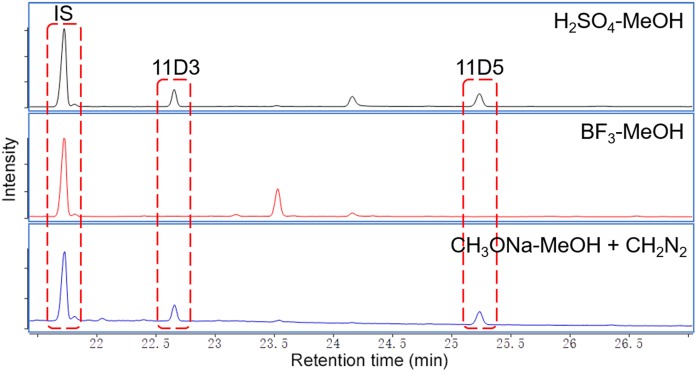
To compare the efficiency of different methylation protocols for furan fatty acids, 11D3 and 11D5 standards were derivatized with H2SO4-MeOH, BF3-MeOH, and ethereal diazomethane using PC-21:0/21:0 as the IS. The typical GC/MS chromatograms are shown in Fig. 2. While BF3-MeOH was inefficient for the methylation of 11D3 and 11D5 (no methyl ester peaks were observed), both H2SO4-MeOH and CH3ONa-MeOH + CH2N2 yielded the desired methyl products at approximately the same levels. BF3 possibly induced the hydrolytic degradation of the furan moiety (36). It should be noted that also using H2SO4-MeOH can lead to degreased yields of furan fatty acid methyl esters if performed at elevated reaction temperatures (>90°C) or in the presence of elevated concentrations (>2%) (data not shown). These findings cast some doubt on the accuracy of some furan fatty acid determinations in food samples using the BF3-MeOH method (21). Overall, our results strongly indicate that basic derivatization protocols should be used for the quantification of furan fatty acids.
Fig. 2.
Furan fatty acid methyl esters derivatized by different methylation protocols. IS: PC-21:0/21:0; CH2N2: ethereal diazomethane.
The LOQ and LOD for the diazomethane and methanolic sulfuric acid method were 10 μM and 3.33 μM, respectively. The level of furan fatty acids within the human blood is less than 0.30 μM (17). As a consequence, tedious isolation and concentration of large volumes of plasma/serum would be necessary, which means that GC/MS in full-scan mode is not ideally suited for the identification of furan fatty acids in human plasma or serum.
Analysis of furan fatty acids by UPLC/ESI/MS/MS in the negative mode
LC/MS/MS is widely used in lipidomics due to its high sensitivity and accuracy. We therefore also analyzed free furan fatty acids (11D3 and 11D5, both at 1 μg/ml) with LC/MS/MS in multiple reaction monitoring mode (supplemental Fig. S2). To optimize the detection of 11D3 and 11D5, parameters such as dwell time, Q1 voltage, collision energy, and Q3 voltage were investigated in detail (supplemental Table S2). The main fragmentations were as follows: 11D3 (m/z 321→71, 321→99, 321→141) and 11D5 (m/z 349→71, 349→127, 349→141) (supplemental Fig. S2). The most intensive fragments for both 11D3 and 11D5 derived from the cleavage of the alkyl carboxyl chains. It is worth noting that the abundance of fragments in the MS2 spectra was 1,000-fold less than the molecular ion in MS1. The fragmentation of furan fatty acids in the collision cell seemed to be relatively random rather than specific. Results showed that the LOQ and LOD were 50 ng/ml and 16.67 ng/ml, respectively.
Analysis of furan fatty acids by UPLC/ESI/MS/MS in the positive mode
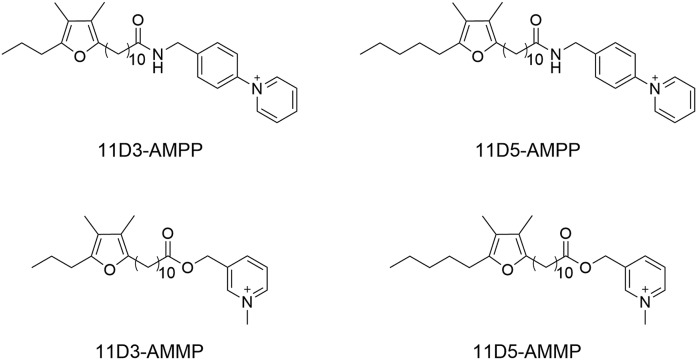
It is generally acknowledged that in the ESI source cations exhibit a better ionization efficiency than anions (37). Therefore, charge-reversal derivatization is a powerful protocol for improving the sensitivity of the mass spectrometric detection of fatty acids (29). In particular, pyridinium salts such as N-[4-(aminomethyl)phenyl]pyridinium, N-[4-(aminomethyl)benzyl]2,4,6-trimethylpyridinium, N-(benzenemethylamine)-2,4,6-trimethylpyridinium, BMP, and CMP can result in remarkable enhancements of the detection sensitivity of fatty acids (29, 33). Among these pyridinium salts, AMMP and AMPP show the highest derivatization efficiency and therefore enhance the detection sensitivity most efficiently (29, 33). In addition, fragmentation patterns of most of the other pyridinium derivates are more complex (29). Therefore, in the current study, derivatizations of furan fatty acids with AMMP and AMPP were investigated. The chemical structures of the derivatives (-AMPP and -AMMP) are illustrated in Fig. 3.
Fig. 3.
The derivative products of furan fatty acids. A: 11D3-AMPP. B: 11D5-AMPP. C: 11D3-AMMP. D: 11D5-AMMP.
In the case of the AMPP derivatization of furan fatty acids, oxidation products ([M + 16]+ and [M + 32]+) were detected even if the derivates had been stored at −80°C overnight. m/z 367 was especially prominent in the MS/MS spectra of [11D3-AMPP + 32]+ (m/z 521) and [11D5-AMPP + 32]+ (m/z 549) (supplemental Fig. S3). In MS1 scanning, the oxidation product peaks exhibit a low abundance and therefore can easily be overlooked.
We hypothesize on the basis of the fragmentation pattern that the activated methylene groups at the furan moiety are oxidized. [M − 18]+ was observed in the MS/MS spectra of both 11D3-AMPP and 11D5-AMPP. Presumably, [M − H2O]+ stems from the dehydration of the aromatic side chain of the oxidation product. Interestingly, similar hydroxylated furan fatty acids have been tentatively identified at very low concentrations in fish lipids (31). In light of our findings, however, this interpretation is questionable, and further analyses will be necessary to confirm or disprove the natural occurrence of these products. In any case, we conclude that AMPP derivatization is not ideally suited for the derivatization of furan fatty acids.
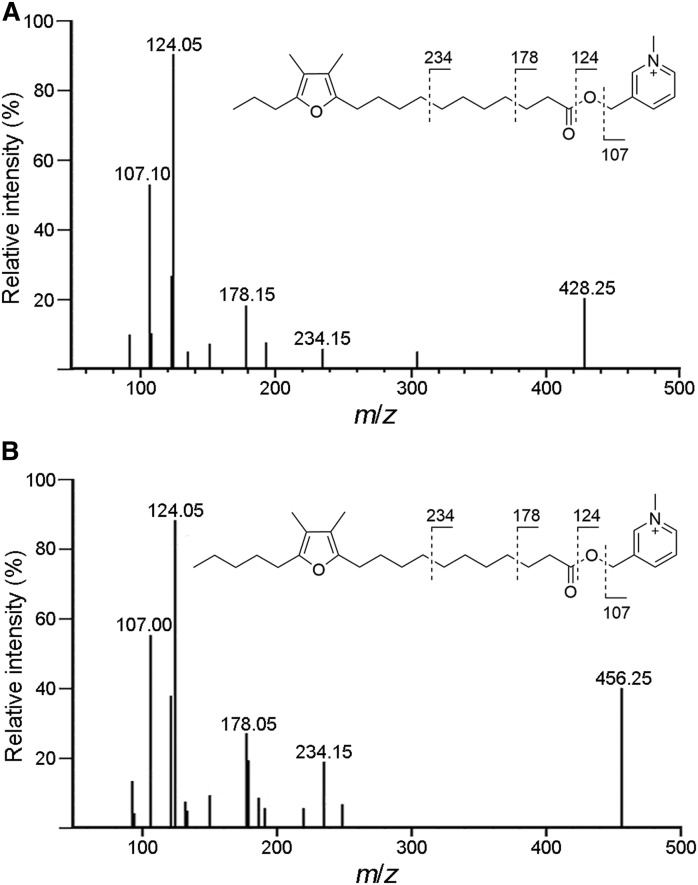
Fortunately, no such artifacts (oxidation of the methylene group) were observed upon derivatization with BMP and CMP. Charge-remote fragmentation could be clearly observed in the MS/MS spectra of both 11D3-AMMP and 11D5-AMMP (Fig. 4). The fragmentation of the N-pyridylcarbinol moiety yielded product ions with m/z 107 and m/z 124. The fragment at m/z 178 was derived from the heterolytic cleavage between the β- and γ-C in the alkylcarboxyl chain (33). Additionally, a series of fragments at m/z 192, 206, 220, 234, and 248 were observed in the MS/MS spectra, indicating a difference of —CH2— (14 amu). A transition ion of M+→124 was selected for the quantitative analysis of 11D3-AMMP and 11D5-AMMP, with M+→107 and M+→178 as qualitative references. Key parameters such as dwell time, Q1 voltage, collision energy, and Q3 voltage for monitoring the above three transition ions were optimized. The results are shown in supplemental Table S3.
Fig. 4.
MS/MS spectra of furan fatty acids-AMMP. A: [11D3-AMMP]+. B: [11D5-AMMP]+.
Separation efficiency
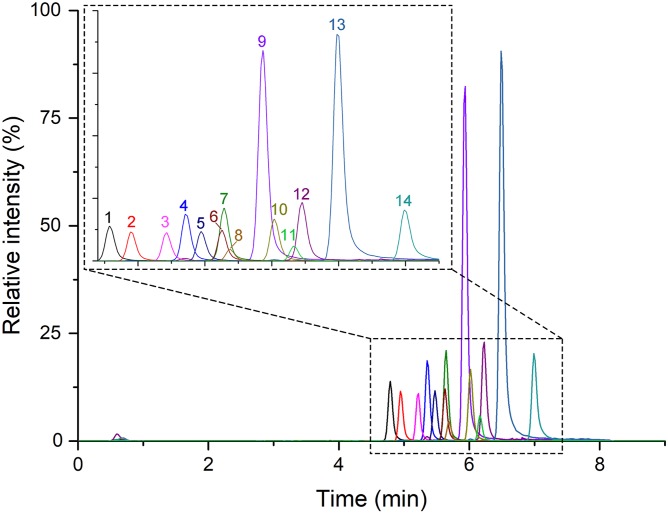
The introduction of the pyridinium group obviously contributes to the polarity of fatty acids. Fatty acids derivatized with AMMP (from C12:0-AMMP to C20:0-AMMP) could be separated well on a C18 column (Fig. 5). Although the peak of 11D3-AMMP (m/z 428) overlapped to some extent with the peaks of C18:2-AMMP (m/z 386) and C15:0-AMMP (m/z 348), the first quadruple (Q1) could discriminate the parent molecular ions clearly due to the different m/z (differences of mass-to-charge ratios >>0.7).
Fig. 5.
LC/MS chromatograms of derivatized fatty acid standards. 1) Lauric acid(C12:0)-AMMP, RT = 4.79 min, MW = 306; 2) myristoleic acid(C14:1)-AMMP, RT = 4.95 min, MW = 332; 3) 10-pentadecenoic acid(C15:1)-AMMP, RT = 5.22 min, MW = 346; 4) myristic acid(C14:0)-AMMP, RT = 5.35 min, MW = 334; 5) palmitoleic acid(C16:1)-AMMP, RT = 5.47 min, MW = 360; 6) linoleic acid(C18:2)-AMMP, RT = 5.63 min, MW = 386; 7) pentadecanoic acid(C15:0)-AMMP, RT = 5.65 min, MW = 348; 8) 11D3-AMMP, RT = 5.69 min, MW = 428; 9) palmitic acid(C16:0)-AMMP, RT = 5.94 min, MW = 362; 10) oleic acid(C18:1)-AMMP, RT = 6.02 min, MW = 388; 11) 11D5-AMMP, RT = 6.16 min, MW = 456; 12) heptadecanoic acid(C17:0)-AMMP, RT = 6.23 min, MW = 376; 13) stearic acid(C18:0)-AMMP, RT = 6.49 min, MW = 390; and 14) eicosanoic acid(C20:0)-AMMP, RT = 6.99 min, MW = 418.
Method development and validation
The esterification of free fatty acids with AMMP and derivative yields has been studied extensively (33, 38). Mechanistically, the final 3-acyloxymethyl-1-methylpyridinum iodide could be formed via a nucleophilic attack by CMP on the 2-acyloxy1-methylpyridinum iodide (the esterification product of free fatty acids and BMP). Using equimolar amounts of starting materials and a 2-fold molar excess of TEA, the desired products have been obtained in good yields (>80%) (38). Based on these observations, derivatization was carried out in a one-pot strategy by simply mixing 11D3/11D5, BMP, CMP, and TEA. The derivatized 11D3 and 11D5 were quantified by UPLC/ESI/MS/MS with deuterium labeled 11D3 and 11D5, respectively, as ISs. Standard curves were established based on the peak area ratios of derivatized furan fatty acids and the corresponding ISs with transition ions (M+→124). As shown in Table 1, good linearities were obtained in dynamic ranges between 0.5 and 320 ng/ml and 2.5 and 1,600 ng/ml for 11D3-AMMP and 11D5-AMMP, respectively.
TABLE 1.
Linearity range of furan fatty acid derivatives
| Furan Fatty Acid Derivatives | IS | R2 | Linearity (ng/ml) |
| 11D3-AMMP | 11D3-d5-AMMP | 0.9995 | 0.5–320 |
| 11D5-AMMP | 11D5-d5-AMMP | 0.9991 | 2.5–1,600 |
The precision and accuracy of this UPLC/ESI/MS/MS approach were analyzed on seven physiological concentrations covering the range of measured levels in subjects. As can be seen from Tables 2 and 3, most intraday and interday RSDs were lower than 12%, while the accuracy was between 84% and 115%. Moreover, our results demonstrated that 11D3-AMMP and 11D5-AMMP had an LLOQ of 0.05 ng/ml. The detection sensitivity was 1,000-fold higher than in the negative mode of ionization used with free 11D3 and 11D5. Therefore, this derivatization method is accurate for the quantitation of furan fatty acids in plasma or other biological samples.
TABLE 2.
Precision of furan fatty acid derivatives
| Furan Fatty Acid Derivatives | STD 1 | STD 2 | STD 3 | STD 4 | STD 5 | STD 6 | STD 7 | |||||||
| Intra-assay | Interassay | Intra-assay | Interassay | Intra-assay | Interassay | Intra-assay | Inter-assay | Intra-assay | Interassay | Intra-assay | Interassay | Intra-assay | Interassay | |
| 11D3-AMMP | 6.31 | 12.01 | 3.49 | 6.17 | 4.35 | 3.19 | 3.78 | 7.61 | 3.37 | 4.08 | 5.73 | 4.90 | 2.12 | 2.65 |
| 11D5-AMMP | 1.99 | 7.86 | 2.92 | 4.82 | 1.14 | 6.74 | 3.87 | 3.35 | 4.16 | 4.60 | 3.88 | 1.71 | 2.17 | 2.61 |
Data shown are RSDs. To determine the precision of 11D3, aliquots of one plasma sample with a low level of 5.01 ± 0.17 ng/ml were spiked with seven different concentrations (STDs 1–7: 5, 10, 15, 20, 25, 30, and 35 ng/ml). To determine the precision of 11D5, aliquots of one plasma sample with a low level of 104.41 ± 0.62 ng/ml were also spiked with seven different concentrations (STDs 1–7: 20, 40, 60, 80, 100, 120, 140 ng/ml). STD, standard.
TABLE 3.
Accuracy of furan fatty acid derivatives
| STD 1 | STD 2 | STD 3 | STD 4 | STD 5 | STD 6 | STD 7 | Range | |
| Furan Fatty Acid Derivatives | % | |||||||
| 11D3-AMMP | 102.51 ± 10.98 | 104.11 ± 3.86 | 98.13 ± 4.80 | 100.57 ± 5.31 | 102.07 ± 4.72 | 90.34 ± 6.28 | 93.42 ± 2.37 | 84–115 |
| 11D5-AMMP | 104.37 ± 4.27 | 90.72 ± 5.23 | 99.52 ± 3.85 | 102.77 ± 4.47 | 101.25 ± 2.93 | 102.25 ± 4.30 | 96.36 ± 2.70 | 86–108 |
Data shown are means ± SDs. To determine the accuracy of 11D3, aliquots of one plasma sample with a low level (5.01 ± 0.17 ng/ml) were spiked with seven different concentrations (STDs 1–7: 5, 10, 15, 20, 25, 30, and 35 ng/ml). To determine the accuracy of 11D5, aliquots of one plasma sample with a low level (104.41 ± 0.62 ng/ml) were also spiked with seven different concentrations (STDs 1–7: 20, 40, 60, 80, 100, 120, and 140 ng/ml). STD, standard.
Targeted quantification of furan fatty acids in human plasma
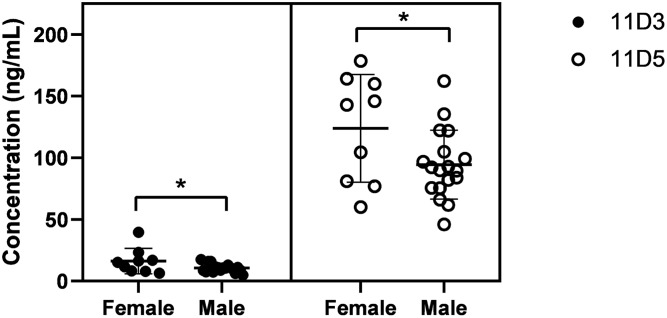
11D3 and 11D5 were quantified in the plasma of 27 healthy Chinese subjects (18 males and 9 females) using the AMMP derivatization methods. The UPLC/ESI/MS/MS chromatograms of 11D3 and 11D5 in human plasma and corresponding ISs are shown in Fig. 6. Although some unidentified peaks were observed in these samples, the detection and identification of furan fatty acids were not impaired. Therefore, targeted quantitation of 11D3 and 11D5 in human plasma could be achieved. The concentrations of 11D3 and 11D5 in the individuals are shown in Fig. 7. The concentration of 11D3 was generally low, usually below 20 ng/ml. In contrast, 11D5 was found at concentrations above 100 ng/ml. Interestingly, in female subjects, the average level of furan fatty acids was significantly (P < 0.05) higher than in male subjects. In addition, the contents of furan fatty acids in female cohorts showed larger deviations than in male cohorts.
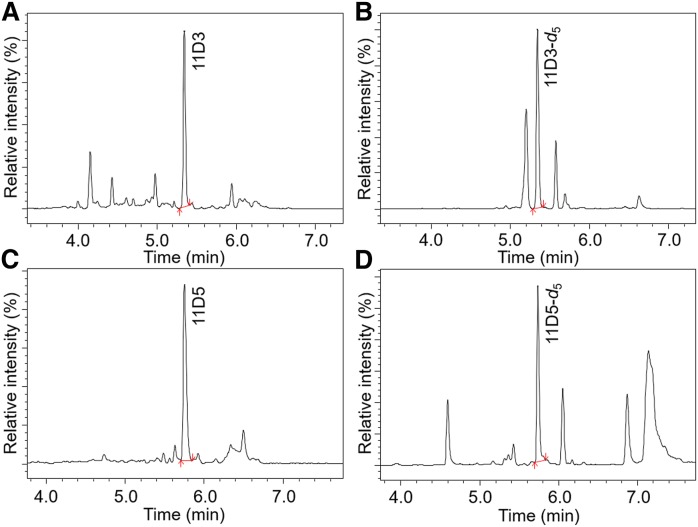
Fig. 6.
Multiple reaction monitoring chromatography of (A) 11D3, (B) 11D3-d5, (C) 11D5, and (D) 11D5-d5.
Fig. 7.
The concentration of furan fatty acids in the plasma of healthy subjects.
Fatty acid profiles in human plasma have been extensively investigated by GC/MS (39), multidimensional MS-based shotgun lipidomics (29), and LC/MS/MS (33). However, to the best of our knowledge, furan fatty acids have been reported in three publications so far investigating the fatty acid content of German individuals (17, 23, 24). In these studies, unusually high sample volumes (>20 ml plasma or 30 ml whole blood) were used and concentrated before analysis. This explains why furan fatty acids have not been reported since then. In addition, the “unusual” fragmentation pattern of furan fatty acids may have prevented their identification.
Comparing our results on furan fatty acids in human blood with a previous study (17), we found parallel levels of 11D3 and significantly higher levels of 11D5. Furthermore, it is interesting to note that the sex-specific differences had not been reported previously. Future investigations will need to clarify these striking differences. Overall, it can be asserted that the sensitivity of our new UPLC/ESI/MS/MS protocol (not necessitating isolation or enrichment) is approximately 1,000-fold higher than the previously established GC/MS protocols.
The metabolic precursor of CMPF still remains to be identified. According to previous findings, β-oxidation does not occur on the alkyl chain at the α2 position of furan fatty acids (40). Therefore, it was proposed that furan fatty acids with a propyl group at α2 positions serve as precursors of CMPF. Accordingly, homologs bearing a pentyl group at the α2 position serve as precursors of 3-carboxy-4-methyl-5-pentyl-2-furanopropanoic acid (27). It is well documented that the concentrations of CMPF in males is generally higher than in females (7, 10). Our results and previous findings indicate that higher levels of CMPF coincide with lower levels of 11D3. Therefore, this sex-specific difference in 11D3 and CMPF concentrations in human plasma may indicate that 11D3 serves as a metabolic precursor for CMPF. This assumption also warrants further specific studies.
Nontargeted analysis of furan fatty acid analogs
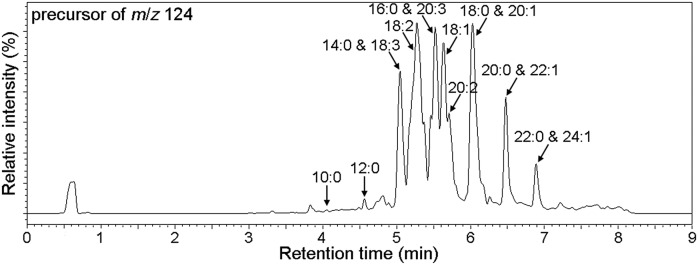
The charge-reversal derivatization method described above enables the identification of furan fatty acids (11D3 and 11D5) according to the transition ions of M+→107, M+→124, and M+→178. Several saturated and unsaturated furan fatty acids have previously been identified in other biological samples (27). To investigate the occurrence of 11D3 and 11D5 analogs in human plasma, precursor ion scan (PIS) was used for the identification of furan fatty acids in pooled samples based on the product ions of m/z 107, 124, and 178 at the collision energy of −48, −40 and −52 eV, respectively. The total ion chromatogram of PIS (precursor of m/z 124) is shown in Fig. 8. Precursors of m/z 107, 124, and 178 were compared for comprehensive identification of all fatty acids in human plasma (supplemental Figs. S4–S13). However, some homologs, including 7-(3,4-dimethyl-5-pentylfuran-2-yl)heptanoic acid, 9-(3,4-dimethyl-5-propylfuran-2-yl)nonanoic acid, 9-(3-methyl-5-pentylfuran-2-yl)nonanoic acid, and 9-(3,4-dimethyl-5-pentylfuran-2-yl)nonanoic acid (former three: <0.5 ng/ml; the last: ∼4.0 ng/ml), which had been identified in German individuals (17), were not found in our subjects. These differences might be related to race, diet, and pathophysiological conditions. Therefore, future investigations will be necessary to confirm this observation.
Fig. 8.
Total ion chromatogram of precursor ion scan (precursor of m/z 124). 1) RT = 4.05 min, MW = 278, C10:0; 2) RT = 4.56 min, MW = 306, C12:0; 3) RT = 5.04 min, MW = 334, C14:0; 4) RT = 5.04 min, MW = 384, C18:3; 5) RT = 5.38 min, MW = 386, C18:2; 6) RT = 5.46 min, MW = 412, C20:3; 7) RT = 5.50 min, MW = 362, C16:0; 8) RT = 5.64 min, MW = 388, C18:1; 9) RT = 5.77 min, MW = 414, C20:2; 10) RT = 6.01 min, MW = 390, C18:0; 11) RT = 6.08 min, MW = 416, C20:1; 12) RT = 6.48 min, MW = 418, C20:0; 13) RT = 6.48 min, MW = 444, C22:1; 14) RT = 6.89 min, MW = 446, C22:0; and 15) RT = 6.89 min, MW = 472, C24:1.
CONCLUSIONS
We have demonstrated the superiority of UPLC/ESI/MS/MS over the established GC/MS methods. An acid-catalyzed ring opening of the furan moiety and oxidation of the activated methylene moieties were identified as major side reactions during sample preparation and derivatization. The optimized protocol reported here largely avoid these side reactions, thereby allowing for a more accurate, quantitative determination of furan fatty acids from biological samples. Moreover, the nontargeted strategy using PIS shows great potential for the detection and identification of furan fatty acid analogs.
To the best of our knowledge, this is the first study using this strategy for the characterization and quantification of furan fatty acids in human tissues. We believe that the UPLC/ESI/MS/MS approach will greatly contribute to the illumination of the CMPF precursor and related metabolic pathway.
Supplementary Material
Acknowledgments
The authors would like to thank Mingjiang Zhu (Zhejiang Hisun Pharmaceutical Co., Ltd.), Shuyuan Guo (SCIEX, Analytical Instrument Trading Corporation), Zheng Yan (Chinese Academy of Sciences), Xia Shen (ShanghaiTech University), Ningning Liang (Chinese Academy of Sciences), Shanshan Zhong (Chinese Academy of Sciences) and Luxiao Li (Chinese Academy of Sciences) for their contributions to the mass spectrometric analysis.
Footnotes
Abbreviations:
- AMMP
- 3-acyl-oxymethyl-1-methylpyridinium iodide
- AMPP
- N-(4-aminomethylphenyl) pyridinium
- BMP
- 2-bromo-1-methylpyridinium iodide
- CMP
- 3-carbinol-1-methylpyridinium iodide
- CMPF
- 3-carboxy-4-methyl-5-propyl-2-furanopropanoic acid
- FID
- flame ion detection
- ID
- inner diameter
- IS
- internal standard
- LLOQ
- lower limit of quantification
- LOQ
- limit of quantification
- PC-21:0/21:0
- 1,2-dihenarachidoyl-sn-glycero-3-phosphocholine
- PIS
- precursor ion scan
- RSD
- relative standard deviation
- TEA
- triethylamine
- 11D3
- 11-(3,4-dimethyl-5-propylfuran-2-yl)undecanoic acid
- 11D3-d5
- 11-(3,4-dimethyl-5-(propyl-2,2,3,3,3-d5)furan-2-yl)undecanoic acid
- 11D5
- 11-(3,4-dimethyl-5-pentylfuran-2-yl)undecanoic acid
- 11D5-d5
- 11-(3,4-dimethyl-5-(pentyl-4,4,5,5,5-d5)furan-2-yl)undecanoic acid
This work was supported by National Science Fund for Distinguished Young Scholars of China Grant 31725022; National Natural Science Foundation of China Grant 31601398; Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University Grant 20171049; and “Blue Ocean Talent Plan” Innovation and Entrepreneurship Team Grant 201811070001. The authors declare that they have no conflicts of interest with the contents of the article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Razquin C., Toledo E., Clish C. B., Ruiz-Canela M., Dennis C., Corella D., Papandreou C., Ros E., Estruch R., and Guasch-Ferré M.. 2018. Plasma lipidomic profiling and risk of type 2 diabetes in the PREDIMED trial. Diabetes Care. 41: 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balgoma D., Pettersson C., and Hedeland M.. 2019. Common fatty markers in diseases with dysregulated lipogenesis. Trends Endocrinol. Metab. 30: 283–285. [DOI] [PubMed] [Google Scholar]
- 3.Allalou A., Nalla A., Prentice K. J., Liu Y., Zhang M., Dai F. F., Ning X., Osborne L. R., Cox B. J., and Gunderson E. P.. 2016. A predictive metabolic signature for the transition from gestational diabetes mellitus to type 2 diabetes. Diabetes. 65: 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandu R., Mok H. J., and Kim K. P.. 2018. Phospholipids as cancer biomarkers: mass spectrometry-based analysis. Mass Spectrom. Rev. 37: 107–138. [DOI] [PubMed] [Google Scholar]
- 5.Newgard C. B. 2017. Metabolomics and metabolic diseases: where do we stand? Cell Metab. 25: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah S. H., Kraus W. E., and Newgard C. B.. 2012. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 126: 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prentice K. J., Luu L., Allister E. M., Liu Y., Jun L. S., Sloop K. W., Hardy A. B., Wei L., Jia W., and Fantus I. G.. 2014. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 19: 653–666. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Prentice K. J., Eversley J. A., Hu C., Batchuluun B., Leavey K., Hansen J. B., Wei D. W., Cox B., Dai F. F., et al. 2016. Rapid elevation in CMPF may act as a tipping point in diabetes development. Cell Reports. 14: 2889–2900. [DOI] [PubMed] [Google Scholar]
- 9.Koppe L., and Poitout V.. 2016. CMPF: a biomarker for type 2 diabetes mellitus progression? Trends Endocrinol. Metab. 27: 439–440. [DOI] [PubMed] [Google Scholar]
- 10.Prentice K. J., Wendell S. G., Liu Y., Eversley J. A., Salvatore S. R., Mohan H., Brandt S. L., Adams A. C., Wang X. S., and Wei D.. 2018. CMPF, a metabolite formed upon prescription omega-3-acid ethyl ester supplementation, prevents and reverses steatosis. EBioMedicine. 27: 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan H., Brandt S. L., Kim J. H., Wong F., Lai M., Prentice K. J., Al Rijjal D., Magomedova L., Batchuluun B., Burdett E., et al. 2019. 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) prevents high fat diet-induced insulin resistance via maintenance of hepatic lipid homeostasis. Diabetes Obes. Metab. 21: 61–72. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair A., Xu L., and Wang Y.. 2018. 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF): a metabolite identified after consumption of fish oil and fish. Nutr. Bull. 43: 153–157. [Google Scholar]
- 13.Müller M., Hogg M., Ulms K., and Vetter W.. 2017. Concentrations, stability and isolation of the furan fatty acid 9-(3-methyl-5-pentylfuran-2-yl)-nonanoic acid from disposable latex gloves. J. Agric. Food Chem. 65: 7919–7925. [DOI] [PubMed] [Google Scholar]
- 14.Chvalová D., and Špička J.. 2016. Identification of furan fatty acids in the lipids of common carp (Cyprinus carpio L.). Food Chem. 200: 183–188. [DOI] [PubMed] [Google Scholar]
- 15.Wendlinger C., and Vetter W.. 2014. High concentrations of furan fatty acids in organic butter samples from the german market. J. Agric. Food Chem. 62: 8740–8744. [DOI] [PubMed] [Google Scholar]
- 16.Wahl H. G., Liebich H. M., and Hoffmann A.. 1994. Identification of fatty acid methyl esters as minor components of fish oil by multidimensional GC-MSD: new furan fatty acids. J. High Resolut. Chromatogr. 17: 308–311. [Google Scholar]
- 17.Puchta V., Spiteller G., and Weidinger H.. 1988. F-Säuren: Eine bisher unbekannte Komponente der Phospholipide des Humanblutes. Liebigs Ann. Chem. 1988: 25–28. German. [Google Scholar]
- 18.Boselli E., Grob K., and Lercker G.. 2000. Determination of furan fatty acids in extra virgin olive oil. J. Agric. Food Chem. 48: 2868–2873. [DOI] [PubMed] [Google Scholar]
- 19.Wahl H. G., Chrzanowski A., Liebich H. M., and Hoffmann A.. 1994. Identification of furan fatty acids in nutritional oils and fats by multi-dimensional GC-MSD. Accessed February 10, 2020, at https://www.gerstel.com/pdf/p-gc-an-1994-06-ar.pdf. [Google Scholar]
- 20.Gottstein V., Mueller M., Günther J., Kuballa T., and Vetter W.. 2019. Direct 1H NMR quantitation of valuable furan fatty acids in fish oils and fish oil fractions. J. Agric. Food Chem . 67: 11788–11795. [DOI] [PubMed] [Google Scholar]
- 21.Vetter W., Laure S., Wendlinger C., Mattes A., Smith A. W. T., and Knight D. W.. 2012. Determination of furan fatty acids in food samples. J. Am. Oil Chem. Soc. 89: 1501–1508. [Google Scholar]
- 22.Kirres C., and Vetter W.. 2018. Furan fatty acid content and homologue patterns of fresh green matrices. J. Food Compos. Anal. 67: 63–69. [Google Scholar]
- 23.Puchta V., and Spiteller G.. 1988. Struktur der F-Säuren enthaltenden Plasmalipide. Liebigs Ann. Chem. 1988: 1145–1147. German. [Google Scholar]
- 24.Wahl H. G., Chrzanowski A., Müller C., Liebich H. M., and Hoffmann A.. 1995. Identification of furan fatty acids in human blood cells and plasma by multi-dimensional gas chromatography-mass spectrometry. J. Chromatogr. A. 697: 453–459. [Google Scholar]
- 25.Spiteller G. 2005. Furan fatty acids: occurrence, synthesis, and reactions. Are furan fatty acids responsible for the cardioprotective effects of a fish diet? Lipids. 40: 755–771. [DOI] [PubMed] [Google Scholar]
- 26.Tovar J., Mello V. D., Nilsson A., Johansson M., Paananen J., Lehtonen M., Hanhineva K., and Björck I.. 2017. Reduction in cardiometabolic risk factors by a multifunctional diet is mediated via several branches of metabolism as evidenced by nontargeted metabolite profiling approach. Mol. Nutr. Food Res. 61: 1600552. [DOI] [PubMed] [Google Scholar]
- 27.Xu L., Sinclair A. J., Faiza M., Li D., Han X., Yin H., and Wang Y.. 2017. Furan fatty acids – beneficial or harmful to health? Prog. Lipid Res. 68: 119–137. [DOI] [PubMed] [Google Scholar]
- 28.Vetter W., and Wendlinger C.. 2013. Furan fatty acids - valuable minor fatty acids in food. Lipid Technol. 25: 7–10. [Google Scholar]
- 29.Wang M., Han R. H., and Han X.. 2013. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal. Chem. 85: 9312–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bian X., Sun B., Zheng P., Li N., and Wu J-L.. 2017. Derivatization enhanced separation and sensitivity of long chain-free fatty acids: application to asthma using targeted and non-targeted liquid chromatography-mass spectrometry approach. Anal. Chim. Acta. 989: 59–70. [DOI] [PubMed] [Google Scholar]
- 31.Uchida H., Itabashi Y., Watanabe R., Matsushima R., Oikawa H., Suzuki T., Hosokawa M., Tsutsumi N., Ura K., Romanazzi D., et al. 2018. Detection and identification of furan fatty acids from fish lipids by high-performance liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Food Chem. 252: 84–91. [DOI] [PubMed] [Google Scholar]
- 32.Han X., Rozen S., Boyle S. H., Hellegers C., Cheng H., Burke J. R., Welsh-Bohmer K. A., Doraiswamy P. M., and Kaddurah-Daouk R.. 2011. Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One. 6: e21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W-C., Adamec J., and Regnier F. E.. 2007. Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding. Anal. Chem. 79: 5150–5157. [DOI] [PubMed] [Google Scholar]
- 34.Christie W. W. 1993. Preparation of ester derivatives of fatty acids for chromatographic analysis. Adv. Lipid. Method. 2: e111. [Google Scholar]
- 35.Christie W. W., and Han X.. 2010. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. 4th edition Woodhead Publishing, Philadelphia, PA. [Google Scholar]
- 36.Jie M. L. K., and Sinha S.. 1981. Fatty acids, part 21: ring opening reactions of synthetic and natural furanoid fatty esters. Chem. Phys. Lipids. 28: 99–109. [Google Scholar]
- 37.Bollinger J. G., Thompson W., Lai Y., Oslund R. C., Hallstrand T. S., Sadilek M., Turecek F., and Gelb M. H.. 2010. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal. Chem. 82: 6790–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukaiyama T., Usui M., Shimada E., and Saigo K.. 1975. A convenient method for the synthesis of carboxylic esters. Chem. Lett. 4: 1045–1048. [Google Scholar]
- 39.Han L-D., Xia J-F., Liang Q-L., Wang Y., Wang Y-M., Hu P., Li P., and Luo G-A.. 2011. Plasma esterified and non-esterified fatty acids metabolic profiling using gas chromatography-mass spectrometry and its application in the study of diabetic mellitus and diabetic nephropathy. Anal. Chim. Acta. 689: 85–91. [DOI] [PubMed] [Google Scholar]
- 40.Sand D. M., Schlenk H., Thoma H., and Spiteller G.. 1983. Catabolism of fish furan fatty acids to urofuran acids in the rat. Biochim. Biophys. Acta. 751: 455–461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.