Abstract
The last few decades have led to an explosion in our understanding of the major roles that small regulatory RNAs (sRNAs) play in regulatory circuits and the responses to stress in many bacterial species. Much of the foundational work was carried out with Escherichia coli and Salmonella enterica serovar Typhimurium. The studies of these organisms provided an overview of how the sRNAs function and their impact on bacterial physiology, serving as a blueprint for sRNA biology in many other prokaryotes. They also led to the development of new technologies. In this chapter, we first summarize how these sRNAs were identified, defining them in the process. We discuss how they are regulated and how they act and provide selected examples of their roles in regulatory circuits and the consequences of this regulation. Throughout, we summarize the methodologies that were developed to identify and study the regulatory RNAs, most of which are applicable to other bacteria. Newly updated databases of the known sRNAs in E. coli K-12 and S. enterica Typhimurium SL1344 serve as a reference point for much of the discussion and, hopefully, as a resource for readers and for future experiments to address open questions raised in this review.
INTRODUCTION
Historical Perspective
The first chromosomally encoded regulatory RNAs were, for the most part, found serendipitously, either in early studies of different types of RNAs in Escherichia coli (for example, 4.5S RNA, 6S RNA, 10Sa RNA = transfer-messenger RNA [tmRNA], 10Sb RNA = RNase P RNA, and Spot 42 RNA) or in in vitro and in vivo experiments originally aimed at nearby genes but that led to the identification of an unexpected transcript that proved to be a regulatory RNA (for example, MicF, DsrA, and OxyS) (reviewed in reference 1). The first of these small regulatory RNAs (sRNAs) that was studied in depth was MicF, encoded divergently to ompC (encoding one of two major porins), which was shown to downregulate the expression of the other porin by base pairing with the ompF mRNA (reviewed in reference 2). Studies of the functions of these initially identified RNAs, as well as regulatory RNAs controlling transposition and plasmid and phage replication (reviewed in references 3 and 4), led to an interest in more global approaches to finding these RNAs and, in conjunction, the need to define what one was looking for. Fully sequenced bacterial genomes were also just becoming available (5, 6), providing many options for new types of genomic analysis. Over the past 20 years, new technologies, in particular, RNA deep sequencing (RNA-seq), have continued to improve our ability to define these intriguing molecules and what they do. Parallel excitement about microRNAs (miRNAs) and, later, long noncoding RNAs (lncRNAs) in the eukaryotic world, developed at the same time but independently, contributed to both interest and the development of technologies for detecting and studying regulation by RNAs (reviewed in reference 7).
Finding sRNAs
A short discussion of the initial ways in which global searches for regulatory RNAs were carried out is useful for defining some of the characteristics of these molecules and considering how much we can depend upon the definitions. The more sensitive and higher-throughput approaches now available still depend, to some extent, on those characteristics to distinguish regulatory molecules from mRNAs, from ubiquitous low-level antisense transcripts, and from cis-acting RNA structures. Many of the findings were carried out simultaneously for both E. coli and Salmonella enterica and, if found for one organism, were true for the other. Thus, we will not differentiate between the species unless the findings only pertain to one organism.
Initial Approaches
The initial set of sRNAs that provided a basis for more global computational searches for sRNAs assumed the following properties: (i) expression from dedicated promoters and from regions previously defined as “intergenic,” meaning not part of an mRNA or operon, (ii) small size (100 to 200 nucleotides [nt]), (iii) the presence of a Rho-independent terminator, and (iv) the apparent lack of open reading frames. The initial searches also made use of conservation in related species. Though sRNA sequence conservation did not extend as broadly into different species as did protein conservation, it generally encompassed much of the RNA, and unlike that found between coding regions for the same species, did not show variation at third (wobble) positions. Expression of the predicted sRNAs was confirmed by Northern analysis. These initial combined computational and experimental screens (8–10) increased the set of likely sRNA candidates significantly and set the stage for their functional characterization. However, it is now known that not all of the characteristics listed above, which were critical for the initial searches, are necessary for defining these regulatory molecules. As we learn more, the sRNA definition may become broader yet.
As a second initial approach, a number of research groups also created cDNA libraries of RNAs selected for their small size, again with expression of predicted sRNAs confirmed by Northern analysis (11, 12). These studies provided the first hints that the sRNAs might not only be transcribed from intergenic regions but could also correspond to the 5′ and 3′ untranslated regions (UTRs) of mRNAs (Fig. 1).
Figure 1.
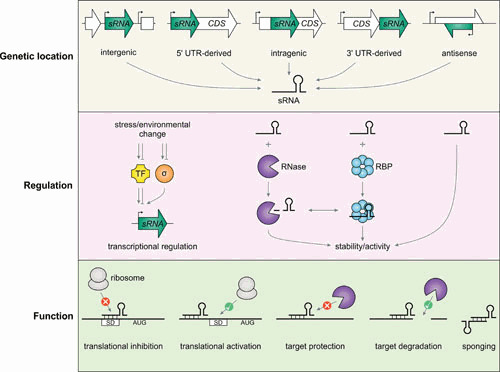
Overview of sRNA sources, mechanisms by which their levels and activities are regulated, and mechanisms of action.
Genome-wide RNA sequencing approaches, where sRNAs can be detected as distinct signals (13, 14), have largely superseded these cloning-based approaches but still rely on the detection of short transcripts and do not always readily distinguish functional RNAs from degradation products and transcriptional noise. Additional evidence of specific transcription, for instance, an increase in a short transcript in response to expression of a specific transcription factor, can provide further evidence of a likely functional sRNA (Fig. 1).
More Recent Approaches
A major step forward in identifying sRNAs and, subsequently, also defining their functions was the identification of protein cofactors, in particular, the RNA chaperone Hfq, which bound groups of sRNAs (8, 15, 16). These proteins will be described in more detail below, but recognition that many of the sRNAs bound to and were stabilized by Hfq provided a critical hook for further identification of additional Hfq-binding sRNAs. Thus, coimmunoprecipitation with Hfq and high-throughput analysis of the bound RNAs (17–19) and, more recently, cross-linking studies of RNAs associated with Hfq (20–22) have significantly increased the detection of sRNAs in this family and have simultaneously provided a strong indication of how these sRNAs are likely to act (Fig. 2).
Figure 2.

Approaches and methods for the characterization of base pairing sRNAs. Results of some of the approaches (in italics) are shown for the MicL sRNA (95).
The enrichment via Hfq binding allows the identification of sRNAs otherwise not easily detected due to poor expression of the sRNA under the growth conditions examined. Hfq binding also allows the relatively easy identification of sRNAs processed from mRNAs (primarily but not exclusively from the 3′ UTR of mRNAs) or expressed from transcripts that may have dedicated promoters internal to mRNAs. Enrichment by Hfq binding of these regions suggests that these are, in fact, sRNAs, usually accumulating because they are significantly more stable than the mRNAs from which they may be derived (20, 22). Copurification approaches also reinforced and extended information from crystal structures, allowing the identification of Hfq-binding motifs. Hfq-binding sRNAs, in particular, seem to require a factor-independent intrinsic terminator (stem-loop followed by a stretch of U residues). For the 3′ UTR-derived sRNAs, the 3′ UTR provides the terminator for both the sRNA and the mRNA. Such a factor-independent terminator may not necessarily be a requirement for sRNAs that do not bind Hfq, although it seems likely that the 3′ stem-loop of the terminator will protect the transcript from ribonucleases.
Other RNA-binding proteins are now known and have been used in parallel cross-linking or immunoprecipitation experiments. For example, ProQ is another RNA chaperone that also binds sRNAs and impacts their activities, and CsrA is a posttranscriptional regulator that is itself regulated by titrating sRNAs. Interactions with these proteins thus help to characterize the likely functions of even more RNA transcripts (22–25).
In addition to sRNA identification through enrichment with specific RNA-binding proteins, a number of more global approaches to identify RNAs binding to multiple proteins have recently been developed. These include Grad-seq, where all protein-RNA complexes are resolved on gradients; RNAs and proteins found in the same fractions, and thus possibly associating, are identified by RNA-seq and mass spectrometry, respectively (26). Other newly developed global approaches rely on cross-linking followed by extraction, such as organic phase separation, of the RNA-binding proteins and their cross-linked RNAs, again followed by transcriptomic and proteomic analyses (27–29).
Core and Specialized sRNAs in Pathogenic Strains
While most studies of sRNAs in E. coli have focused on the nonpathogenic laboratory K-12 strains, pathogenic E. coli share these sRNAs and, in addition, encode sRNAs in their pathogenicity islands. Microarrays and, more recently, RNA-seq, have improved the process of identifying potential new sRNAs. For example, multiple potential RNAs, including many encoded by lysogenic bacteriophages that are likely to vary from strain to strain, were identified in enterohemorrhagic E. coli upon cross-linking with Hfq (21). While these additional E. coli RNAs are not listed in Table S1, they are likely to play significant roles in pathogenesis, as well as in modifying the expression of core bacterial genes (reviewed in reference 30).
S. enterica and E. coli share many conserved sRNAs. Beyond these, the sum of several different dedicated searches has produced a detailed picture of S. enterica-specific sRNAs that are encoded on either Salmonella pathogenicity islands (SPIs) or other genomic regions not present in E. coli (31, 32). Many of these sRNAs show upregulation in media that reflect conditions that activate this bacterium’s major virulence regions or stresses encountered in the host environment (33). Examples of proven virulence-related functions of these sRNAs will be discussed further below. It should be mentioned that some of the seemingly virulence-associated sRNAs have homologs in many other bacteria and therefore have broadly conserved functions. For example, DapZ (STnc820) targets some of the same mRNAs as the GcvB sRNA, which is a global posttranscriptional regulator of amino acid synthesis and transport genes (19, 34). Similarly, IsrE (RyhB-2 or RfrA) is a functional paralog of the iron starvation-responsive RyhB sRNA (35). The overarching conclusion from these studies is that pathogenic and nonpathogenic bacteria of related species have the same core sRNAs with additional sRNAs from pathogenicity islands and other integrated DNA elements adding to the regulatory networks.
DEFINING sRNAs
Criteria for Annotation as an sRNA
Tables S1A and S2A show our current lists of regulatory RNAs, defined as those detected as a distinct band by Northern analysis and/or for which we detect clear signal in our RNA-seq datasets (most of these sRNAs have been detected as distinct bands by Northern analysis) for E. coli K-12 and S. enterica serovar Typhimurium SL1344, respectively. In addition, a number of newly detected sRNAs (studied in the authors’ labs) are included. Tables S1B and S2B provide what we consider “candidate sRNAs” in E. coli and S. enterica, respectively, detected by one means or another but not yet studied sufficiently to be added to the confirmed list or currently assigned a different function, such as a 5′ UTR. Some of these candidates may graduate to the confirmed list as studies proceed. For E. coli K-12, regulatory RNAs currently included in EcoCyc (36) are listed in either Table S1A or S1B.
The primary criterion for defining a new regulatory RNA, even in the absence of any additional information, is the presence of an expressed transcript (Fig. 2). The increasing application of RNA-seq approaches together with widespread sequencing of microbial genomes means that, even for organisms without facile genetic analysis, transcriptome maps can be obtained and used to annotate sRNAs. In the initial studies, detection of a defined band by Northern analysis provided the strongest evidence of the size of the likely regulatory species and differentiated it from degradation products from mRNAs and other cellular RNAs less likely to be detected as defined bands (Fig. 2). Clearly, as depth of sequencing increases, the ability to detect an increasing number of transcripts provides ever more of a challenge for distinguishing functionally relevant RNAs from “noise” and degradation products. One example is understanding which of the extensive antisense transcripts detected in many studies (reviewed in reference 37) are functional sRNAs. Only the subset of antisense RNAs with demonstrated functions are listed in Tables S1A and S2A and will be discussed here.
General sRNA Categories
The sRNAs that have been characterized thus far fall into several general categories. The overwhelming majority of sRNAs act by base pairing with target mRNAs, leading to changes in the translation or stability of the target(s); these will be the major focus of this review. However, we first provide an overview of all the major categories.
sRNAs with Specialized Functions: 4.5S RNA, RNase P RNA, tmRNA, 6S RNA, and Y-Family RNAs
Four of the RNAs listed in Table S1A have rather specialized functions and are discussed only briefly here, with references provided for more extensive descriptions. All of these sRNAs are more broadly conserved than the other regulatory RNAs we will discuss. Whether still other, as yet uncharacterized, less broadly conserved sRNAs have similar specialized functions remains an open question.
Three of these abundant transcripts are key components of larger protein complexes. The 4.5S RNA is a component of the protein secretion apparatus (reviewed in reference 38). The RNA component of RNase P encoded by rnpB is a highly conserved ribozyme found in both eukaryotes and prokaryotes and is essential in E. coli, S. enterica, and other bacteria due to its role in tRNA processing (reviewed in reference 39). tmRNA, named for having properties of both tRNA and mRNA and encoded by ssrA, plays an interesting and important role in translational quality control and is also widely conserved in bacteria (reviewed in reference 40). When translation stops on mRNAs without a translation termination codon or stalls at rare codons, the tmRNA, with its cofactor protein SmpB, is recruited to the ribosome as a tRNA and then acts as a short mRNA, encoding an 11-amino acid sequence added to the end of a translating protein, which is then released from the ribosome. Since this sequence encodes a protein degradation signal, these stalled polypeptides are degraded upon completion. Thus, tmRNA relieves ribosome stalling and ensures that incomplete proteins are destroyed rather than accumulate. In some bacteria, tmRNA may also play a regulatory role. It is needed for optimal synthesis of the stress-induced sigma factor σS (RpoS) in E. coli (41).
6S RNA binds to and regulates the activity of RNA polymerase (reviewed in reference 42). Like the three RNAs discussed above, 6S is broadly conserved. The structure of folded 6S mimics that of the DNA at a promoter in an “open complex” and traps RNA polymerase with the vegetative σ70 (RpoD) promoter recognition subunit. This helps cells transition into stationary phase. When nutrients become abundant and cells transition from stationary phase back into exponential phase, RNA polymerase is able to transcribe a short segment of the 6S RNA, thus generating product RNAs (pRNAs). This leads to RNA polymerase release from the 6S RNA. Whether the ∼15- to 20-nt pRNAs themselves have a separate function is not known.
Two sRNAs with specialized functions, YrlA and YrlB, that are found in S. enterica (Table S2A) and other bacteria but not in E. coli, are members of the Y-family of RNAs (reviewed in reference 43). These RNAs all have secondary structures with similarities to tRNAs, including some tRNA nucleotide modifications, and bind the Rsr protein (related to the eukaryotic Ro60 protein). The Yrl RNAs together with Rsr associate with polynucleotide phosphorylase (PNPase), with the RNAs acting as both a tether between Rsr and PNPase and a gatekeeper to modulate the access of other RNAs to PNPase.
Antisense RNAs: Antitoxin sRNAs, GadY, ArrS, RyeA, SraG, and RyjB
Many antisense transcripts are seen in RNA-seq experiments; however, only a few have been shown to have regulatory consequences and are included here. Most of the antisense RNAs listed in Tables S1A and S2A downregulate expression from genes that encode proteins, frequently very small proteins, which are toxic at high levels (15 entries in Table S1A). Given that they are usually encoded antisense to the toxin gene, they have the potential for extended pairing with their target transcript and are likely to be, as far as is currently known, very specific in their function. They have not been shown to require chaperones to act, though a number of these RNAs have been shown to bind the protein ProQ (26). It is also worth noting that, for unknown reasons, there are multiple chromosomal copies of a number of these toxin-antitoxin systems, including the five Sib and four Ldr antitoxin RNAs. In many cases, the 5′ UTRs of the toxin mRNAs are quite long, with extensive secondary structures and potential RNase cleavage and alternative ribosome-binding sites, such that the regulation of toxin expression by the antitoxin sRNAs has the potential to be very complex (reviewed in reference 44). There are some antitoxin sRNAs encoded adjacent to the toxin gene that recognize their target mRNAs with limited complementarity but apparently also do not require chaperone proteins. One example is the IstR-tisAB pair, a well-characterized toxin-antitoxin system induced by DNA damage. Here, ∼25 nt of the ∼70-nt IstR sRNA base pairs with the tisAB toxin mRNA, transcribed divergently from the opposite strand (45, 46).
Five other antisense RNAs are listed for E. coli. Two of these, GadY and ArrS, strongly bind Hfq. While GadY has been shown to downregulate the antisense-encoded gadXW mRNA by directing RNase III-mediated cleavage (47, 48) and ArrS is required for RNase III-dependent processing of one form of the antisense-encoded gadE mRNA (49), it also appears that both of these sRNAs form pairs with trans-encoded targets (20). These observations suggest that they are not simply antisense RNAs but may be better considered Hfq-dependent base-pairing RNAs that happen to be antisense to one of their targets. The RyeA sRNA, encoded opposite the Hfq-binding SdsR sRNA, also shows some binding to Hfq. Given that RyeA seems to block SdsR activity and prevents the cell death caused by high levels of SdsR under some growth conditions, the RNA has been proposed to also be an antitoxin RNA, with SdsR RNA as the toxin (50). However, SdsR clearly has regulatory effects rather than simply functioning as a toxin; presumably, RyeA acts to modulate these effects as well (51, 52). Thus far, the only Hfq-independent antisense RNAs regulating something other than a toxin are SraG, encoded antisense to the pnp-rpsO mRNA and reported to downregulate levels of PNPase (53), and the uncharacterized RyjB sRNA, encoded antisense to sgcA.
sRNAs That Act by Limited Base Pairing
The group of sRNAs that have been characterized most extensively are the base-pairing sRNAs that bind to Hfq. On the order of 30 sRNAs clearly bind Hfq in either E. coli and S. enterica (column Q in Tables S1A and S2A). Base pairing with mRNAs usually leads to negative or, more rarely, to positive regulation, and some major well-characterized targets are indicated in Tables S1A and S2A. In the vast majority of cases, we would define these sRNAs as “trans-acting” that is, genes encoding the targeted mRNAs are generally located far from the gene encoding the sRNA. Many sRNAs and mRNAs meet on Hfq, though the less-well-characterized ProQ protein also binds sRNA-mRNA pairs, as discussed below. General approaches for characterizing these sRNAs are summarized in Fig. 2.
sRNA-BINDING PROTEINS
As already mentioned, important clues about possible functions of sRNAs often come from the identification of associated proteins. The three sRNA-binding proteins that have been studied most extensively in E. coli and S. enterica are Hfq, ProQ, and CsrA. Hfq is considered to be the main RNA chaperone that facilitates base pairing between the sRNAs and their targets. ProQ may have a similar chaperone role for some RNA pairs, but initial studies indicate that binding by this protein can have other consequences. Both Hfq and ProQ also stabilize sRNAs (Fig. 1) so that in many cases, sRNAs are destabilized in the corresponding mutant backgrounds. CsrA, on the other hand, is primarily a regulator of mRNA stability and translation, and sRNAs associated with CsrA modulate its activity, titrating the protein away from its mRNA targets.
A number of genome-wide approaches have been carried out to examine the set of RNAs that copurify with each of these proteins, initially using microarrays but subsequently using deep sequencing to identify the RNAs. Results from a subset of these studies are included in Tables S1A and S2A. A few caveats about interpreting these genome-wide data are warranted. The signal for RNAs is impacted by the sequencing coverage for an experiment, the abundance of the RNA, and the parameters used to analyze the data. For instance, these experiments indicate that some particularly abundant sRNAs, such as 6S RNA, are bound by many proteins in addition to RNA polymerase, but the signals may be due to nonspecific binding. Related to this, it should also be noted that binding does not necessarily suggest that there is a consequence for either the RNA or the protein. Additionally, protein binding and impact can change depending on growth conditions (25). The sRNAs for which there is the highest confidence for specific binding by Hfq and ProQ are indicated in Tables S1A and S2A (columns Q and R).
In this section, we describe the general properties of Hfq, ProQ, and CsrA as well as some of the RNases and other proteins that have been found to be associated with sRNAs (reviewed in references 54 and 55). Some of the sRNAs are bound by more than one protein, but the implications of this and possible competition between the proteins binding a given RNA are not well understood.
Sm-like Protein Hfq
Hfq, a homolog of eukaryotic Sm proteins and Lsm proteins that associate with splicing and RNA complexes, was first characterized as a host factor required for Qβ replication (56). Unlike the eukaryotic proteins, which commonly are hetero-heptamers, bacterial Hfq is a homo-hexamer. Structural and mutational studies have shown that the Hfq hexamer has a ring structure with multiple RNA-binding surfaces (reviewed in references 57 and 58). These include what is denoted the proximal face (where there is a binding site for a stretch of U residues around the ring opening), the opposite distal face (which binds repeats of an ARN sequence, where R = adenine/guanine and N = any nucleotide), the rim of the ring (which binds UA-rich sequences), and finally, an intrinsically disordered C-terminal tail which extends from the ring. All sRNAs bind to the proximal face of Hfq via the U-tail, which comprises the Rho-independent terminator of these sRNAs. The sRNAs additionally can interact with the rim or the distal face and are broadly classified as class I or class II sRNAs, respectively, based on their stabilities in strains expressing Hfq mutants defective in one or the other binding surface (59). However, it should be noted that there is a continuum of interactions, with some sRNAs affected by mutations on all surfaces (60). Differential interactions with the C-terminal tail also are likely to have an impact (61). Many Hfq-bound mRNAs have an ARN motif, which directs their binding to the distal face.
For sRNAs bound on the proximal and rim surfaces and mRNAs bound to the distal face, in vitro annealing assays for Hfq chaperone activity indicate that charged arginine residues on the Hfq rim promote base pairing (62). The C-terminal tail has been proposed to then facilitate release of the RNA duplex (63).
It is worth noting some caveats, however. The levels and activities of some sRNAs, such as DsrA, are less sensitive to the lack of Hfq, and there are some discrepancies between in vitro and in vivo results. For example, despite the proposed in vitro functions of the C-terminal tail, the lack of this sequence does not have strong functional consequences in vivo (61). While it is clear that there is competition between sRNAs for binding to Hfq and that RNAs must cycle on and off the chaperone, how this occurs is not fully elucidated (reviewed in reference 64). Similarly, there are conflicting reports about the subcellular localization of Hfq (65–67), a feature that is bound to have an impact on the function of this chaperone.
FinO-Domain Protein ProQ
While the most attention was initially focused on the Hfq chaperone, it is clear that other proteins bind sRNAs and may also promote base pairing. One of these proteins, ProQ, contains an N-terminal FinO domain initially characterized for the plasmid-encoded FinO protein that promotes pairing of plasmid-expressed RNAs. In ProQ, the FinO domain is followed by a linker and C-terminal domain with structural similarity to the Tudor domains involved in binding to proteins with modified amino acids (reviewed in reference 68). ProQ is so named because mutations in the E. coli gene affect resistance to a toxic proline analog (69, 70). Cross-linking and coimmunoprecipitation experiments indicate that, generally, ProQ binds sRNAs distinct from those bound by Hfq (23, 25, 26). The protein has been suggested to recognize more structured RNA sequences than Hfq, and binding is frequently observed to be in the 3′ UTRs of mRNAs. However, given fewer in vivo and in vitro studies (71), many of the basic features of ProQ binding to RNA are not well understood, including definition of all of the RNA-binding surfaces and the recognition elements on RNAs.
Much also remains to be learned about the mechanism of ProQ action. The protein has been reported to promote pairing of RaiZ sRNA with hupA mRNA as well as STnc540 sRNA with mgtB mRNA in S. enterica (72, 73), but this activity has not been studied in detail. In E. coli, fewer RNA-RNA pairs are found to be bound by ProQ than Hfq, though interestingly, 30% of these pairs are associated with both proteins (25). However, for the one E. coli pair examined in detail, downregulation of the target RNA required Hfq, while high levels of ProQ blocked this regulation (25). Thus, competition between ProQ and Hfq as well as between ProQ and plasmid-encoded FinO-domain proteins for RNA binding and function are important topics for future studies.
RNA-Binding Protein CsrA
The gene for the mRNA-binding protein CsrA was identified in a genetic screen for mutants defective in glycogen biosynthesis. Further studies revealed that while CsrA clearly impacts carbon metabolism, it has a broad role in regulating biofilm formation, motility, quorum sensing, and virulence (reviewed in reference 74). CsrA binds a wide variety of mRNAs, predominantly in the 5′ UTR (24), where binding can have either negative effects, such as blocking translation or increasing transcription attenuation, or positive effects, such as increasing accessibility to a ribosome binding site. A range of structural and biochemical studies have shown that CsrA exists as a dimer, where each monomer binds a motif that has a GGA sequence in the loop of an RNA hairpin (75). However, aggregates, brought together by dimers binding multiple GGA motifs on the same RNA, can form.
Unlike hfq and proQ, the csrA gene is essential in E. coli (76). The activity of the CsrA protein is extensively regulated, in line with its central role in modulating critical cellular activities as well as a role in pathogenesis. Two sRNAs, CsrB and CsrC, which each have multiple GGA repeats, act by sequestering CsrA. Thus far, no other functions for these sRNAs have been discovered. However, several Hfq-binding sRNAs, such as McaS and GadY, also have been found to bind CsrA (22, 24). For McaS, studies showed that the sRNA impacts the expression of two targets through Hfq-mediated base pairing and activates expression of another target by sequestering CsrA (77). Interestingly, parts of mRNAs, such as the 5′ end of S. enterica fimAICDHF, additionally act to sequester the protein (78).
Ribonucleases and Other RNA-Binding Proteins
Ribonucleases can play critical roles for regulatory RNAs in both processing and degradation. Global RNA-seq experiments have documented that several sRNAs are bound as well as processed by the endoribonucleases RNase E and RNase III. In one study, cross-linking identified sRNAs associated with RNase E (79), while in other studies, the comparisons of the RNA-seq data for wild-type and mutant strains allowed the identification of cleavage sites in sRNAs for RNase E (80) and RNase III (81, 82). Other RNases that have not been studied as extensively but also impact sRNA-mediated actions are RNase II (23) and PNPase (83, 84), which is part of the RNase E degradosome complex. Another protein found to be associated with the GlmZ and GlmY sRNAs, RapZ, functions as an adaptor to target these sRNAs to RNase E (85). It is likely that other RNase adapter proteins remain to be found. The CsrD protein, which accelerates RNase-dependent decay of CsrA-interacting sRNAs, is another candidate for such molecular function (86).
Additionally, other types of RNA-binding proteins undoubtedly bind to and impact sRNAs. One such family is the cold shock proteins (Csps) (87), which have been proposed to affect the folding of RNAs, for instance, during cold shock (88). With the development of new approaches to globally identify proteins associated with RNAs (26–29), the study of these sRNA-binding proteins likely will be a fruitful area of research in the near future.
REGULATION OF sRNA LEVELS AND ACTIVITY
The levels of many sRNAs are highest under very specific growth conditions. All of the sRNAs characterized initially are encoded as separate transcripts, where their expression is strongly regulated by specific transcription factors. Later, it was found that sRNAs also correspond to the 3′ UTRs of some mRNAs (reviewed in reference 89). These can be transcribed from promoters internal to the coding sequence or cleaved from the longer mRNA. There also are hints of sRNAs being derived from 5′ UTRs (11, 20) and possibly even from within coding sequences (90). To generate the specific small transcript in these cases, there need to be promoters, terminators, or ribonuclease cleavage sites internal or adjacent to the coding sequence. Some sRNAs that are transcribed as defined RNAs also are cleaved to give derivatives that are more active or have altered activity (Fig. 1).
In addition to conditions and mechanisms that increase sRNA levels or activity, given that sRNAs are regulators, there also need to be ways to eliminate or turn off the sRNA when the inducing stress is no longer present. sRNAs were initially characterized as generally more stable than mRNAs, based on their slow turnover in the presence of inhibitors of transcription such as rifampicin (12). However, further work led to evidence for codegradation of sRNAs with mRNAs (59, 91). Regulation, both positive and negative, could also include other forms of processing such as RNA modification or degradation of the U tail; the latter is expected to greatly reduce Hfq binding (92).
Transcriptional Regulation
Many sRNAs are induced very highly as part of specific stress responses, frequently reflecting transcriptional regulation (see column S in Tables S1A and S2A). Thus, for example, RyhB is repressed by the Fur repressor and is only induced upon Fur deactivation under conditions of low iron (93); OxyS is strongly induced by oxidative stress by the OxyR activator (94). The expression of a number of other sRNAs is specifically controlled by alternate sigma factors. For instance, MicA, RybB, and MicL are all transcriptionally activated in response to cell envelope stress in a σE-dependent manner (95) (see Table S1A). Interestingly, many transcription or sigma factor-binding sites in the promoters of sRNA genes are close to the consensus, such that the sRNAs tend to be among the most rapidly and strongly induced transcripts in response to the different stress conditions (95).
All Hfq-binding sRNAs have a strong Rho-independent terminator with a stem-loop and stretch of U residues. While it might be assumed that the sRNAs end predominantly at the most 3′ U residue, examination of the 3′ ends in global RNA-seq sets indicated that the ends are far more variable. Additionally, it has been found that in the case of RyhB, the length of the 3′ end is dependent on the growth condition, with stronger transcription termination under stress conditions (96). There are also examples of sRNAs such as SroC, where the sRNA is derived from the intergenic region of an operon and must be generated by transcription termination that only occurs part of the time, such that the downstream genes, gltJKL, are also transcribed (97). Since the shorter 3′ U tails or readthrough past the U residues reduce or eliminate Hfq binding, changes in the sequence of the Rho-independent terminator or growth conditions that change transcription termination can impact sRNA activity (92).
RNA Modification
Generally, our knowledge of RNA modifications in bacterial transcripts other than rRNAs and tRNAs has greatly lagged behind what is known for eukaryotes (reviewed in references 98 and 99). However, there is some recent evidence that bacterial sRNAs also are modified. For example, sRNAs have been detected in genome-wide screens for RNAs with 5′ NAD caps (100) or N6-methyladenosine (101). At least in vitro, a 5′ NAD cap, which is introduced during transcription (102), impacts the ability of RNase E to cleave the transcript. While it is tempting to speculate that different 5′ caps, RNA methylation, or other modifications could serve additional regulatory roles, physiological evidence for this is still lacking.
RNA Processing
As already mentioned, several sRNAs are derived from the 3′ UTRs of longer mRNAs (Fig. 1). In this case, cleavage is critical for their generation. For most sRNAs where this has been examined, such as SdhX, the endoribonuclease RNase E is required for the cleavage (103, 104). Other sRNAs that are generated as defined short transcripts are also cleaved by RNase E. For some sRNAs, such as ArcZ, this cleavage leads to increased base-pairing activity (80, 105), whereas for other sRNAs, such as SdsN and SdsR, this cleavage can lead to different forms that have different sets of base-pairing targets (51, 106).
RNA Degradation
Cleavage is also a key mechanism to downregulate an sRNA-mediated regulatory response. Most sRNAs are relatively unstable when the transcription of the sRNA is specifically shut off, but their target mRNAs are still being transcribed. In contrast, sRNAs are very stable when cells are treated with rifampicin, which stops transcription of both the sRNA and target mRNAs. This observation was interpreted as coupled degradation of the sRNA with its target and was first shown for RyhB, where coupled RNase E-mediated degradation of the sRNA with its targets downregulates the RyhB response (91). Presumably, the sRNA degradation reflects loss of binding to Hfq after pairing. Further support for this model was provided by studies in which the ability of the sRNA to pair was impaired by mutation of the target mRNA-binding sites on Hfq; under these conditions, the unstable sRNA becomes significantly more stable (59).
While codegradation with the mRNA is a consequence for sRNA base pairing with a number of mRNAs, this is not always the case and can vary between targets of the same sRNA and among sRNAs. Generally speaking, class I sRNAs, which bind to the proximal face and rim of Hfq, are less stable and codegraded with their targets, while a subset of class II sRNAs, which bind to the proximal and distal faces of Hfq, are more stable and less likely to turn over upon mRNA pairing (59). For these sRNAs, other mechanisms must lead to turnover, and that is the case for ChiX, a stable sRNA that becomes unstable when paired with an RNA decoy (see below). In light of the impact of 3′ U tail length on Hfq binding (92), it is also conceivable that sRNAs could be “turned off” by selective trimming of the 3′ ends.
RNase E is not the only ribonuclease involved in the codegradation of sRNAs with targets. For example, the MicA sRNA was reported to be codegraded with the ompA mRNA by RNase III (107). Similarly, RybB is similarly degraded by this double strand-specific endoribonuclease in a manner that is dependent on Hfq but blocked by ProQ (25). Global maps of RNase III cleavage events in E. coli under different growth conditions are available (81, 82, 108) and lend themselves to interrogation of the impact of this enzyme on sRNA-mediated regulation.
Competition, RNA Decoys, RNA Mimics, and RNA Sponges
Given that many sRNAs are unstable in the absence of association with the corresponding RNA-binding protein, such as Hfq or ProQ, competition for binding to these proteins can also have a profound effect on sRNA levels. Additionally, there are specific RNAs that base pair with sRNAs or compete with RNase adaptor proteins that impact sRNA levels (reviewed in reference 109). During the characterization of one sRNA, ChiX, whose expression is downregulated in the presence of chitosugars, it was found that the sRNA levels were not controlled by changes in transcription but, rather, were downregulated by base pairing with the chbBCARFG mRNA. Base pairing with this “decoy” mRNA target, which likely disrupts the terminator stem, led to degradation of ChiX; pairing with other targets, such as the chiP mRNA encoding the chitoporin, does not lead to degradation (110, 111). The GlmY sRNA, which is a “mimic” of the base-pairing sRNA GlmZ, represents yet another posttranscriptional regulatory mechanism to control sRNA levels, in this case, in response to low glucosamine-6-phosphate levels (reviewed in reference 112). In the absence of the inducer, GlmZ is destabilized by the binding of the ribonuclease adaptor protein RapZ, which leads to RNase E-dependent degradation. This degradation is prevented by GlmY, induced under low glucosamine-6-phosphate. GlmY, due to its similarity to GlmZ, also binds RapZ and thus titrates RapZ away from GlmZ (85).
Another sRNA, SroC, derived from the gltI-gltJ intergenic region, controls the levels of the base-pairing RNA GcvB. The GcvB-SroC base pairing directs RNase E-mediated cleavage of GcvB but not SroC, allowing SroC to recycle (97). The RNA-seq approaches are leading to the identification of sRNAs or even tRNA fragments, such as a 3′ fragment derived from tRNALeuZ (3′ETSLeuZ) processing, that base pair with sRNAs at regions overlapping the sequences involved in base pairing with targets. These “sponge” RNAs thus affect the levels of the sRNAs or block their activities (20, 113) (Fig. 1). Interestingly, the reduction of RyhB levels in a strain lacking poly(A) polymerase may be attributed to reduced degradation of the 3′ETSLeuZ RyhB sponge in this mutant background (114).
Ultimately, it is likely that the levels of a specific sRNA, especially over time and under changing conditions, is determined by the competition for binding among a very intricate network of mRNAs, sponge RNAs, and RNA-binding proteins. Understanding the actual intracellular levels of sRNAs under various conditions will be an important starting point for future research on these issues.
Complication of Translation for Dual-Function sRNAs
Although one initial criterion for the identification of sRNAs was that the transcripts are not translated, further inspection of several of the annotated sRNAs has revealed that some base-pairing sRNAs also serve as mRNAs encoding small proteins. In E. coli, the best-characterized example is SgrS, which encodes the 38-amino acid SgrT protein (reviewed in reference 115). Both SgrS and SgrT act to downregulate the PtsG transporter in response to sugar phosphate stress, SgrS at the level of ptsG mRNA translation and SgrT at the level of PtsG protein function. Undoubtedly, other base-pairing sRNAs similarly have dual functions. The presence of an adjacent or overlapping coding sequence likely will have consequences for the base-pairing activity of an sRNA, in part due to occlusion by the binding of the large ribosome complex. How translation affects base-pairing activity and what regulates whether one or the other function predominates is unexplored.
MECHANISMS OF BASE-PAIRING sRNA ACTION
The consequences of sRNA base-pairing interactions with their target mRNAs have been examined in relative detail for quite a number of sRNA-mRNA pairs. The most frequent outcome of the pairing is downregulation, though there are also multiple examples of positive regulation (reviewed in reference 116). The regulation can be at the level of translation or mRNA stability, and frequently both, but examples of pairing affecting transcription have also recently come to light. In all cases, the RNA duplex formation seems to involve one continuous stretch of base pairing of around 8 nt (denoted the seed region in the sRNA) but can extend beyond this initial region of pairing. The seed region typically is the most conserved part of an sRNA (117, 118). Similarly, the complementary recognition site in the target mRNA(s) generally shows high conservation (20, 119), a feature that can be used to improve in silico target predictions (120).
While there usually is only one region of pairing between the sRNA and mRNA, in a few cases, such as SgrS-manXYZ, MicF-lpxR, and CpxQ-nhaB, the same sRNA base pairs with two different regions of the same mRNA (121–123). Below, we describe the best-characterized mechanisms of base-pairing sRNA action as well as a few interesting variations.
Negative and Positive Regulation of Translation
Perhaps the most common way sRNAs act is by Hfq-mediated base pairing at or near the Shine-Dalgarno sequence, thereby occluding the ribosome entry (Fig. 1). Systematic assays of different positions of RybB-ompN pairing showed that occlusion can occur at sites as far as five codons into the coding sequence (124). There are, however, different permutations of sRNA-mediated translational repression. At many targets of GcvB, the sRNA downregulates translation by base pairing upstream of the Shine-Dalgarno sequence at C/A-rich sequences thought to be translational enhancers (117). Optimal translation of fepA and bamA requires stem-loop structures, and OmrA and OmrB repress these targets by disrupting these structures (125). In other examples, such as SgrS and DicF repression of manX, the sRNAs recruit or stabilize Hfq binding, and it is actually Hfq that occludes ribosome binding (126). For OmrA- and OmrB-mediated repression of dgcM, Hfq binding leads to a change in the dgcM 5′-UTR secondary structure, which then allows sRNA base pairing to block translation (127). Hfq also has been shown to repress translation without sRNAs, binding well upstream of the Shine-Dalgarno sequence and acting to remodel the RNA (128). These examples demonstrate the extent to which there is much still to be learned about ribosome entry and translation initiation. The detailed mechanistic studies of how sRNA and Hfq operate to inhibit repression continue to provide insight into how mRNA structure, “standby” ribosome-binding sites, and other proteins collaborate to regulate translation. In essence, while most sRNAs do inhibit translation of their targets, they can achieve this by base pairing within a much larger window than just the narrow region originally thought.
For the first characterized example of sRNA-mediated translational activation, the DsrA RNA base pairs with a region of the rpoS mRNA, which otherwise forms a secondary structure that blocks ribosome binding (129). Other examples of sRNAs that similarly activate translation by preventing the formation of inhibitory secondary structures, such as GlmZ-glmS and RyhB-shiA, have been found (130, 131). In a different permutation, Hfq binding blocks translation of the cirA mRNA, while RyhB base pairing with this mRNA changes the secondary structure, thereby displacing Hfq and facilitating ribosome binding (132). While not yet reported, it is conceivable that sRNA-Hfq-mRNA binding can have still other effects on translation, such as frameshifting or premature termination.
Negative and Positive Regulation of mRNA Stability
Decreased ribosome binding frequently leads to destabilization of the mRNA. This is the reason for the success of the initial studies of sRNA function that relied on the differences in RNA levels after a short pulse of overexpression of an sRNA to identify mRNA targets, even though the sRNAs primarily acted to block translation (Fig. 2). However, some examples where sRNA base pairing directly leads to target mRNA cleavage have also been found. For MicC base pairing internal to the S. enterica ompD coding sequence, this interaction was found to promote specific RNase E-mediated cleavage (133). In this case, the proximal 5′ monophosphate and other features provided by the sRNA serve to activate RNase E (134, 135). For sRNA-mediated downregulation of csgD, base pairing with McaS, RprA, or GcvB sRNAs, which was observed to be quite a distance upstream of the Shine-Dalgarno sequence, leads to mRNA cleavage in an AU-rich sequence in this long 5′ UTR (136).
sRNA-mRNA pairing can also impact target cleavage in a manner that results in stabilization of the mRNA. One example is SgrS-directed interference with RNase E-mediated decay of the pldB-yigL mRNA, resulting in a stabilized yigL product (137). In a different example, RydC pairing with the cfa mRNA blocks RNase E-mediated degradation (138). As for translation, other permutations of sRNA-Hfq-mRNA binding that promote or block the actions of RNases can be imagined.
Negative and Positive Regulation of Transcription Termination
There have been a few examples of direct sRNA-mediated regulation of transcription, specifically transcription termination (reviewed in reference 139). In two cases, sRNA binding affects the ability of Rho to access a rut (Rho-utilization site) on a transcript and bring about termination. For the chiPQ mRNA, the effect is negative. ChiX base pairing with the Shine-Dalgarno sequence of chiPQ prevents ribosome binding. In turn, there are fewer ribosomes blocking the rut site in the chiP coding sequence, resulting in increased transcription termination (140). For the rpoS mRNA, the effect is positive. DsrA, ArcZ, and RprA binding to the 5′ UTR blocks Rho access to rut sites, thus promoting transcription elongation (141). In a final example, the SraL RNA has been reported to increase synthesis of Rho itself by blocking premature termination (142). Again, one can envision still other mechanisms by which sRNAs can affect transcription termination.
IMPACT OF sRNAs ON REGULATORY CIRCUITS
As described above, much is now known about the class of Hfq-binding sRNAs, how their levels are regulated, and at least some of their targets, with the potential for defining the broader target sets. How does this regulation feed into cell physiology? Our expectation is that the regulation of targets helps the cell to adapt to or recover from the stress that led to induction of the sRNA and that the mRNAs targeted by the sRNA provide clues to its physiological role. In the selected set of examples below, we discuss some evidence that this is the case. However, for many other sRNAs, we do not currently understand the physiological relevance of the sRNA-mediated regulation, showing that there is still much to learn. We anticipate that as our understanding of the physiological significance of sRNA-mediated regulation increases, so will our broader appreciation of bacterial physiology.
General Principles of sRNAs in Regulatory Networks
Each regulatory network probably uses sRNAs in somewhat different ways, and new roles for these sRNAs continue to be discovered. Nonetheless, it is worth emphasizing some of the general properties of sRNA-based regulation (also discussed in references 143 and 144).
The first general concept is that sRNAs can significantly expand a regulon. In these cases, when a DNA-binding protein activates or represses transcription, it modulates expression not only from genes where the protein directly binds, but also from all those affected indirectly by the regulated sRNA or sRNAs. These sRNAs can be considered the “noncoding arm” of the regulon (with the protein-encoding genes regulated by the transcription factor being the “coding arm”), as discussed for the σE response below. The effects of the sRNAs can create a hierarchy of regulatory consequences. For instance, sRNA-mediated negative regulation, causing degradation of mRNAs, can rapidly overcome regulation at the level of transcription, turning off even well-expressed targets. This is further discussed below in the context of the RyhB regulatory network.
In a number of instances, sRNAs are critical components of positive feedback loops, reinforcing the transcriptional regulation of a set of genes by further regulating the same targets at the translational level. This has been best studied in the case of Spot 42, an sRNA whose expression is negatively regulated by cyclic AMP (cAMP) receptor protein (CRP) and cAMP, discussed here. Equally or possibly more common, however, are negative feedback loops, in which the sRNA carries out regulation that helps to restore homeostasis, leading to less activity of the upstream transcription factor. This negative feedback loop is indirect for the Fur-regulated RyhB RNA and the σE-regulated MicA, RybB, and MicL RNAs as discussed below but is direct for EnvZ-OmpR and the OmrA and OmrB sRNAs. OmrA and OmrB, whose expression is fully dependent upon EnvZ and OmpR for their synthesis, base pair directly with the envZ-ompR mRNA to downregulate this two-component system (145, 146). Intrinsic to many of these positive and negative feedback circuits is another principle of sRNA-based regulation, their capacity to change the sign of regulation for downstream genes. This is easiest to appreciate for transcriptional regulation by specialized sigma factors whose molecular function is limited to gene activation. If sigma factors positively regulate expression of sRNAs, those sRNAs can then carry out negative regulation (147).
Finally, and not surprisingly, sRNAs provide connections between different regulatory cascades. For critical regulatory hubs, multiple sRNAs, each made in response to a different environmental signal, can converge on a single target, at the same time providing complex combinatorial inputs, as discussed below for σS.
Examples of sRNAs in Regulatory Networks
In the next section we describe some of the best-understood sRNA-mediated changes to cell physiology (Fig. 3). Most of the initial sRNA-mRNA pairs were identified during the characterization of individual sRNAs. This is now changing with the development of approaches that detect RNA-RNA proximity genome-wide (reviewed in reference 148).
Figure 3.
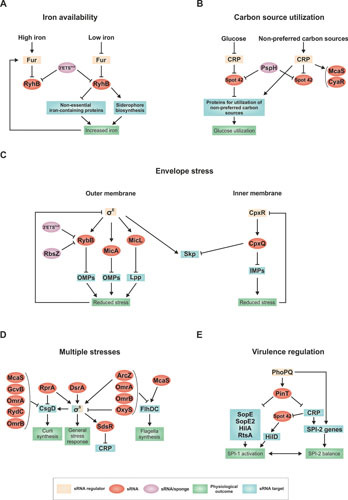
Examples of regulatory networks in E. coli and S. enterica. (A) Response to low iron regulated by RyhB. (B) Bias toward glucose utilization regulated by Spot 42. (C) Outer membrane protein (OMP) synthesis controlled by RybB, MicA, and MicL and inner membrane protein (IMP) synthesis controlled by CpxQ. (D) Regulation of the general stress response by multiple sRNAs. (E) Regulation of the transition between virulence programs in S. enterica by PinT.
Regulating Iron Acquisition and Homeostasis
Iron is a critical and often limiting nutrient for all organisms, particularly for bacterial pathogens that must acquire the metal ion from their hosts (reviewed in reference 149). When iron is abundant, the Fur repressor effectively shuts down expression of a large regulon that includes genes encoding iron acquisition systems. When iron is limiting, Fur no longer binds DNA, and the regulon is induced. RyhB, identified in the initial global searches for conserved sRNAs, is well conserved in enterobacteria and was quickly recognized to have a consensus Fur site overlapping its promoter (93). The Fur regulon has been extensively studied, and a number of genes apparently positively regulated by the Fur repressor have been identified in E. coli. The discovery of RyhB explained this conundrum; the positive regulation was indirect, due to Fur repression of RyhB expression relieving RyhB-mediated-negative regulation (93) (Fig. 3A).
Many RyhB targets have now been identified (reviewed in reference 150). Short-term overexpression of RyhB followed by microarray analysis demonstrated that a fairly extensive set of genes were negatively regulated, many encoding metabolic enzymes that use Fe-S clusters or otherwise make use of iron. Since RyhB is only synthesized when iron is limiting, it can rapidly decrease the demand for iron by downregulating synthesis of nonessential proteins that utilize iron. RyhB also contributes to the adaptation to iron starvation by positively regulating the expression of shiA, a gene encoding a permease for shikimate, needed for siderophore synthesis (130), as well as the enterobactin biosynthesis operon, and revamping cellular metabolism to ensure that serine is available for enterobactin synthesis (151). Therefore, RyhB helps the cell reestablish homeostasis. If enough iron becomes available due to the decrease in use and increased import, then Fur repression will be reestablished. RyhB is particularly critical for growth during severe iron limitation (152), though too much RyhB is also detrimental under some growth conditions. RyhB basal levels are kept low not only by Fur repression of synthesis but also by an RNA sponge, 3′ ETSleuZ, encoded by a processing product of leuZ tRNA. This processing product keeps the basal level of RyhB low and thus prevents regulation of targets in the absence of the inducing signal (113). Genes of succinate metabolism are repressed by RyhB, and cells mutant for the 3′ ETSleuZ sponge grow less well on succinate. Intriguingly, a RyhB paralog (RyhB-2) is encoded on a genetic island in S. enterica. While RyhB-1 and RyhB-2 have some overlapping targets, there also appear to be unique sets that are affected by only one or the other sRNA (reviewed in reference 153).
Reinforcing Carbon Usage Hierarchy
Spot 42, encoded by the spf gene, was first identified as a spot in a two-dimensional polyacrylamide gel of radioactively labeled sRNAs (154) and was found to be negatively regulated by cAMP and the transcriptional regulator CRP (155, 156) decades before its function as an Hfq-binding sRNA was defined (157, 158). cAMP sets cellular priorities for carbon metabolism; it is low when cells grow on glucose but increases when cells catabolize less favored carbon sources. cAMP-bound CRP binds DNA and positively regulates the synthesis of operons encoding proteins that metabolize these less favored carbon sources. At the same time, CRP-cAMP negatively regulates Spot 42 transcription, so the sRNA is abundant when cells grow on glucose but not when they grow on the less favored carbon sources whose metabolism requires positive regulation by CRP and cAMP (Fig. 3B).
While the first identified target of Spot 42 regulation was the gal operon, which has complex CRP and cAMP regulation (reviewed in reference 159), transient overexpression of Spot 42 identified many other regulated targets (160). Most of these downregulated genes were part of catabolic pathways, positively regulated by CRP-cAMP. Therefore, cAMP impacts these genes at two levels: by direct transcriptional regulation at the promoter and by repressing Spot 42 synthesis, thereby indirectly increasing translation and mRNA stability. This suggests that in glucose medium, Spot 42 prevents leaky expression of cAMP-CRP target genes by repressing the mRNAs. Consistent with this hypothesis, higher basal levels of Spot 42 targets in glucose-grown cells and changes in the kinetics of gene expression and repression upon changes in carbon sources were observed when spf was mutated (160). Spot 42 also has been found to be downregulated by an sRNA sponge, PspH, corresponding to the 3′ UTR of pspG (20), but the physiological consequences of this have not been explored.
Other sRNAs are either also regulated by CRP and cAMP or impact the utilization of carbon sources (reviewed in reference 161), indicating strong selection for sRNA-mediated regulation of carbon metabolism. While some regulators of these sRNAs as well as their targets are known, the impacts on metabolism are less well understood. For example, transcription of CyaR and McaS is positively regulated by CRP-cAMP in E. coli, such that these sRNAs have an expression pattern opposite that of Spot 42 (162, 163). The two sRNAs each have a set of targets, mostly distinct from those for Spot 42 and from each other, leading to significant specialization within the CRP regulon (162, 164). While there are possible explanations for CyaR downregulation of outer membrane proteins (OMPs) and McaS upregulation of flagella synthesis upon glucose starvation, the physiological consequences for carbon utilization have not been explored.
Responding to Cell Envelope Stress and Modulating Traffic to the Membrane
Many of the first examples of sRNA targets were OMPs. To some extent, negative regulation of OMPs may have been detected most easily because some initial experiments used changes in protein levels to identify possible targets, and many OMPs are very abundant and visible on protein gels (119). In addition, a major phenotype of cells deleted for the RNA chaperone Hfq is induction of the σE regulon (165). σE (encoded by the rpoE gene), a specialized sigma factor, which is the founding member of the ECF (extracytoplasmic function) sigma factor family, regulates the periplasmic proteins necessary for protein folding or degradation and the machinery necessary for moving proteins through the inner membrane and inserting them in the outer membrane (reviewed in reference 166).
σE was known to be induced by overproduction of misfolded or unfolded OMPs. Additionally, mRNAs for many OMPs are downregulated when σE is induced in a manner dependent upon Hfq. Given that sigma factors only act positively, stimulating transcription at a subset of promoters, the downregulation of OMPs suggested the existence of downstream regulators, and the dependence on Hfq suggested that these regulators might be sRNAs. That proved to be the case. The σE regulon contains at least three sRNAs, MicA and RybB, which downregulate OMPs (167–169), and MicL, which downregulates the major outer membrane lipoprotein, Lpp (95) (Fig. 3C). Thus, the current picture is that, in the absence of effective negative regulation of OMPs by the relevant sRNAs, robust transcription and translation of OMPs leads to more transport of these proteins into the periplasm than the uninduced machinery can handle. The resulting presence of unfolded OMPs in the periplasm is sensed, and the σE response is induced, increasing the synthesis of the OMP folding, secretion, and degradation machinery and, at the same time, increasing the levels of the repressing sRNAs.
Because induction of σE in the absence of Hfq occurs even without other inducing signals, and because deletion of the genes encoding MicA and RybB lead to activation of the σE response (169), basal levels of MicA, RybB, and MicL likely are critical for proper repression of the OMPs. Additionally, deletion of rpoE is lethal in E. coli, but lethality is suppressed if either MicA or RybB is overexpressed (147). Therefore, sRNAs are a critical part of the σE response, controlling trafficking to the membrane and providing a rapid and effective feedback mechanism both to limit basal expression and to restore homeostasis when the trafficking system is perturbed or stressed. The σE-regulated sRNAs also have additional, non-OMP targets; for the most part the role of the regulation of these other targets has not been studied extensively. As for RyhB, there seems to be a need to control RybB activity under some conditions, as both 3′ ETSleuZ and RbsZ (corresponding to the 3′ UTR of rbsB) sponge RybB (25, 113).
It is worth noting that there is a parallel sRNA-dependent regulatory pathway, which controls the levels of inner membrane proteins (IMPs) (123, 170). In this case, the cpxP mRNA and CpxQ, an sRNA derived from the 3′ end of cpxP, are induced in response to misfolded IMPs via the CpxA-CpxR two-component system (Fig. 3C). CpxQ downregulates synthesis of a number of IMPs, including the NhaB Na+/H+ antiporter, thus limiting the loss of membrane potential. CpxQ also cross-connects the inner and outer membrane stress responses by repressing synthesis of the σE-induced Skp. This periplasmic chaperone, which binds unfolded OMPs, may accidentally mistarget OMPs into the inner membrane, causing depolarization. CpxQ counteracts this potential toxicity by downregulating Skp production.
Regulating the General Stress Response
All of the examples discussed above are cases where the impact and activity of an upstream transcriptional regulator is expanded by the effects of one or more downstream sRNAs. However, sRNAs also can target mRNAs encoding transcriptional regulators, meaning that the whole regulon for that transcription factor can be affected by the sRNA activity. In E. coli, three transcription regulators, σS, CsgD, and FlhDC, play critical roles in transitions from rapid and planktonic growth to slower growth, frequently in a biofilm, and all are subject to complex levels of regulation, including regulation by multiple sRNAs. In these examples, the physiological significance of the regulation is not always apparent, but sRNAs provide opportunities for an abundance of signals to affect transcription factor levels, both positively and negatively (Fig. 3D).
The most extensively studied example of this sort of convergent regulation is positive regulation of the general stress response sigma factor σS (encoded by rpoS). σS plays a central role in the transition from exponential growth to stationary phase growth, with increased σS accumulation leading to induction of a large number of genes that help the cell cope with damage, extremes of temperature and pH, and dwindling energy and resources (reviewed in reference 171). As already discussed, translation of the rpoS mRNA is inhibited by a long 5′ UTR that folds to occlude ribosome entry. Induction of translation depends on any of at least three sRNAs, each of which can pair with a region of the 5′ UTR to open up the inhibitory RNA structure to allow translation. While the region of pairing within the rpoS 5′ UTR is the same for each sRNA (129, 172, 173), the sRNAs do not resemble each other and are each expressed under a different stress condition. The first to be found, DsrA, is expressed at low temperature (174, 175), as well as in response to increased levels of ppGpp (176), both conditions under which σS levels increase. The second sRNA, RprA, is positively regulated by the Rcs phosphorelay (172), which is activated when the cell surface is perturbed by antimicrobials such as polymyxin, by antibiotics such as ampicillin, and by interaction with a solid surface (reviewed in reference 177). ArcZ, the third activating sRNA, is negatively regulated by the two-component histidine kinase ArcB and response regulator ArcA under anaerobic growth conditions (178). Both DsrA and ArcZ contribute significantly to σS accumulation in growing cells and in cells entering stationary phase; loss of Hfq or all three sRNAs leads to cells that have extremely low levels of σS (173). Each of these sRNAs also regulates multiple additional targets, suggesting that the “global stress response” due to σS induction will be somewhat different under different growth conditions with different subprograms controlled by sRNAs. Thus, the regulation provides a complex combinatorial network, able to respond to multiple signals.
One of the genes dependent on σS is csgD, which encodes a master regulator for curli synthesis and thus one pathway of biofilm formation. While CsgD is regulated in a complex transcriptional manner, it is also regulated by multiple sRNAs (reviewed in reference 179). Most of the sRNAs that negatively regulate csgD translation are not implicated in direct regulation of rpoS. The exception is RprA, which positively regulates rpoS translation and negatively regulates csgD (180). Therefore, under conditions of high expression of the Rcs phosphorelay, RprA may allow induction of the σS response but block the branch of the response that leads to curli-dependent biofilm formation.
Another mRNA that is affected by multiple sRNAs encodes the FlhDC transcription regulators, which sit at the top of a cascade of genes necessary for motility. Negative regulation by at least three sRNAs and positive regulation by another were found for the flhDC transcript (162, 181). One of the sRNAs that downregulates flhDC is ArcZ, which activates rpoS. Thus, in situations where ArcZ is well expressed, motility may be downregulated while σS is induced.
As if these regulatory networks were not sufficiently complex, the levels of a number of sRNAs increase in stationary phase. One of them, SdsR, is clearly dependent on σS. Among other targets, SdsR downregulates CRP and OmpD, the latter of which is not present in E. coli (51, 182). As still more sRNAs are characterized, the web of connections between the key stress transcription factors and sRNA undoubtedly will be found to be even more intricate.
Controlling Pathogenesis
The success of pathogens such as E. coli and S. enterica depends not only on their ability to quickly adapt to changing and often harsh environments during host infections; it also requires the precise timing of the expression of their virulence factors and coordination of these processes with general gene expression. The observations that inactivation of the hfq (reviewed in reference 183) or proQ (73) genes attenuate bacterial virulence have been taken as evidence that sRNAs are involved in bacterial pathogenesis. sRNAs with direct and indirect functions in controlling virulence and host survival have been discovered and characterized in both E. coli and S. enterica (reviewed in reference 30 and 184). Arguably, our knowledge is most advanced for the Hfq-associated sRNAs in S. enterica, as we will illustrate with three examples showing sRNAs encoded in virulence regions targeting both virulence factors and genes from the core genome, as well as a core genome-encoded sRNA targeting virulence factors.
One complex regulatory network is associated with PinT, the most highly upregulated sRNA in the intracellular phase of S. enterica (185) (Fig. 3E). PinT is encoded on a horizontally acquired locus and is activated by the PhoPQ two-component system together with the physically unlinked SPI-2 locus, which encodes a type 3 secretion system required for intracellular survival. The sRNA downregulates virulence factors (SopE and SopE2) from the initial SPI-1 invasion gene program (185). At the same time, PinT downregulates the two major SPI-1-encoded transcription factors, HilA and RtsA, thus globally inhibiting invasion gene expression (186). Finally, as the sRNA acts to shut off SPI-1 functions, PinT delays the full expression of SPI-2 indirectly by inhibiting synthesis of the general transcription factor CRP (185). Overall, PinT acts as a posttranscriptional timer, shaping the transition from one virulence program (SPI-1, invasion) to the other (SPI-2, intracellular lifestyle). Interestingly, in S. enterica, CRP-repressed Spot 42 acquired an additional function of positively regulating hilD encoding the master regulator of virulence (187), providing an example of a conserved sRNA influencing species-specific regulons.
InvR sRNA is encoded by SPI-1 and is coactivated with these SPI-1 genes for host cell invasion. Surprisingly, however, the main target of InvR was found to be the ompD mRNA, encoding S. enterica’s most abundant outer membrane porin. This led to a model whereby InvR acts to limit the synthesis of an abundant membrane protein to support the insertion of the bulky SPI-1 type 3 secretion system into the bacterial envelope (32). This hypothesis has been bolstered by an independent observation that a synthetic minimal SPI-1 locus can only be functional in S. enterica if it also carries the invR gene (188).
Finally, SgrS provides another example of how a core genome-encoded sRNA was recruited to regulate a horizontally acquired, S. enterica-specific virulence factor (189). SgrS base pairs with the mRNA of the secreted effector protein, SopD, via the same seed region used in E. coli and S. enterica for downregulation of the major glucose importer PtsG and other targets. While the biological meaning of this regulation has remained somewhat unclear, these studies revealed the intriguing ability of sRNAs to discriminate between a G-C pair (in the productive SgrS-sopD duplex) and a G-U pair (in the nonproductive G-U pair in the potential SgrS-sopD2 RNA). In general, Spot 42 and SgrS illustrate how pathogens utilize conserved Hfq-associated sRNAs to integrate horizontally acquired genes into existing posttranscriptional networks, just as conserved transcription factors are recruited for transcriptional networks.
The major hurdle for gauging the importance of sRNAs in virulence regulation has been a lack of phenotypes of sRNA deletion strains in standard virulence assays or animal infection experiments. However, recently developed molecular methods promise a much broader assessment of their functions (reviewed in reference 190). For example, dual RNA-seq, providing simultaneous transcriptomes of pathogen and host showed that PinT dramatically impacts host cells, with ∼10% of all host genes showing altered expression using infection with a ΔpinT strain versus wild-type bacteria (185). Molecular readouts, including changes in the host transcriptome, might provide more sensitive approaches to understanding sRNA functions in host-pathogen interactions.
PERSPECTIVES
This review gives an overview of what has been learned about sRNAs in E. coli and S. enterica and provides resources for future studies of these regulators. Given the relatively young age of the field of bacterial sRNAs, it is not surprising that, while much has been discovered, many questions remain. These include, but are not limited to, the impact of subcellular localization and competition among sRNAs, mRNAs, mRNA and tRNA fragments, and multiple RNA-binding proteins; novel mechanisms of regulating sRNA activity and sRNA action; the physiological processes regulated by sRNAs; and processes in which sRNAs still remain to be identified. We look forward to seeing what answers, as well as unexpected new mechanisms and new questions, will come from continued studies of sRNAs in the next 20 years.
SUPPLEMENTARY MATERIAL
ACKNOWLEDGMENTS
We thank P. Adams, E. Holmqvist, S. Melamed, and K. Papenfort for comments on the review.
Research in the Vogel laboratory is funded in part by Deutsche Forschungsgemeinschaft (DFG) grants Vo875/14-1, Vo875/18-1, and Vo875/19-1, research in the Gottesman laboratory is funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research, and research in the Storz laboratory is funded by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development.
No potential conflicts of interest relevant to this review were reported.
Contributor Information
Jens Hör, Institute of Molecular Infection Biology, University of Würzburg, 97080 Würzburg, Germany.
Gianluca Matera, Institute of Molecular Infection Biology, University of Würzburg, 97080 Würzburg, Germany.
Jörg Vogel, Institute of Molecular Infection Biology, University of Würzburg, 97080 Würzburg, Germany; Helmholtz Institute for RNA-based Infection Research (HIRI), 97080 Würzburg, Germany.
Susan Gottesman, Laboratory of Molecular Biology, National Cancer Institute, Bethesda, MD 20892.
Gisela Storz, Division of Molecular and Cellular Biology, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD 20892.
Susan T. Lovett, Brandeis University, Waltham, MA
Deborah Hinton, Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD.
REFERENCES
- 1.Wassarman KM, Zhang A, Storz G. 1999. Small RNAs in Escherichia coli. Trends Microbiol 7:37–45 10.1016/S0966-842X(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 2.Inouye M, Delihas N. 1988. Small RNAs in the prokaryotes: a growing list of diverse roles. Cell 53:5–7 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- 3.Eguchi Y, Itoh T, Tomizawa J. 1991. Antisense RNA. Annu Rev Biochem 60:631–652 10.1146/annurev.bi.60.070191.003215. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Wagner EGH, Simons RW. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 48:713–742 10.1146/annurev.mi.48.100194.003433. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512 10.1126/science.7542800. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94 10.1016/j.cell.2014.03.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651 10.1101/gad.901001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157–177 10.1016/S0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 10.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11:941–950 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 11.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 33:1040–1050 10.1093/nar/gki256. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EGH. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31:6435–6443 10.1093/nar/gkg867. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hébrard M, Händler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA 109:E1277–E1286 10.1073/pnas.1201061109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomason MK, Bischler T, Eisenbart SK, Förstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197:18–28 10.1128/JB.02096-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J 17:6061–6068 10.1093/emboj/17.20.6061. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sledjeski DD, Whitman C, Zhang A. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol 183:1997–2005 10.1128/JB.183.6.1997-2005.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124 10.1046/j.1365-2958.2003.03734.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4:e1000163 10.1371/journal.pgen.1000163. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019 10.1038/emboj.2012.229. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H. 2016. Global mapping of small RNA-target interactions in bacteria. Mol Cell 63:884–897 10.1016/j.molcel.2016.07.026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. 2014. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell 55:199–213 10.1016/j.molcel.2014.05.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35:991–1011 10.15252/embj.201593360. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmqvist E, Li L, Bischler T, Barquist L, Vogel J. 2018. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol Cell 70:971–982.e6 10.1016/j.molcel.2018.04.017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Potts AH, Vakulskas CA, Pannuri A, Yakhnin H, Babitzke P, Romeo T. 2017. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat Commun 8:1596 10.1038/s41467-017-01613-1. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melamed S, Adams PP, Zhang A, Zhang H, Storz G. 2020. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol Cell 77:411–425.e7 10.1016/j.molcel.2019.10.022. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA 113:11591–11596 10.1073/pnas.1609981113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Queiroz RML, Smith T, Villanueva E, Marti-Solano M, Monti M, Pizzinga M, Mirea DM, Ramakrishna M, Harvey RF, Dezi V, Thomas GH, Willis AE, Lilley KS. 2019. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat Biotechnol 37:169–178 10.1038/s41587-018-0001-2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shchepachev V, Bresson S, Spanos C, Petfalski E, Fischer L, Rappsilber J, Tollervey D. 2019. Defining the RNA interactome by total RNA-associated protein purification. Mol Syst Biol 15:e8689 10.15252/msb.20188689. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urdaneta EC, Vieira-Vieira CH, Hick T, Wessels HH, Figini D, Moschall R, Medenbach J, Ohler U, Granneman S, Selbach M, Beckmann BM. 2019. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat Commun 10:990 10.1038/s41467-019-08942-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatt S, Egan M, Jenkins V, Muche S, El-Fenej J. 2016. The tip of the iceberg: on the roles of regulatory small RNAs in the virulence of enterohemorrhagic and enteropathogenic Escherichia coli. Front Cell Infect Microbiol 6:105 10.3389/fcimb.2016.00105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. 2008. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36:1913–1927 10.1093/nar/gkn050. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66:1174–1191 10.1111/j.1365-2958.2007.05991.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695 10.1016/j.chom.2013.11.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81:1144–1165 10.1111/j.1365-2958.2011.07751.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Ellermeier JR, Slauch JM. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol 190:476–486 10.1128/JB.00926-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karp PD, Ong WK, Paley S, Billington R, Caspi R, Fulcher C, Kothari A, Krummenacker M, Latendresse M, Midford PE, Subhraveti P, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Santos-Zavaleta A, Mackie A, Collado-Vides J, Keseler IM, Paulsen I. 12 November 2018, posting date. The EcoCyc Database. Ecosal Plus 2015 10.1128/ecosalplus.ESP-0006-2018. 10.1128/ecosalplus.ESP-0006-2018. [PubMed] [DOI] [Google Scholar]
- 37.Georg J, Hess WR. 2018. Widespread antisense transcription in prokaryotes. Microbiol Spectr 6(4):RWR-0029-2018. 10.1128/microbiolspec.RWR-0029-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akopian D, Shen K, Zhang X, Shan SO. 2013. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82:693–721 10.1146/annurev-biochem-072711-164732. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondragón A. 2013. Structural studies of RNase P. Annu Rev Biophys 42:537–557 10.1146/annurev-biophys-083012-130406. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Keiler KC. 2015. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol 13:285–297 10.1038/nrmicro3438. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Ranquet C, Gottesman S. 2007. Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J Bacteriol 189:4872–4879 10.1128/JB.01838-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassarman KM. 2018. 6S RNA, a global regulator of transcription. Microbiol Spectr 6(3):RWR-0019-2018. 10.1128/microbiolspec.RWR-0019-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sim S, Wolin SL. 2018. Bacterial Y RNAs: gates, tethers, and tRNA mimics. Microbiol Spectr 6(4):RWR-0023-2018. 10.1128/microbiolspec.RWR-0023-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masachis S, Darfeuille F. 2018. Type I toxin-antitoxin systems: regulating toxin expression via Shine-Dalgarno sequence sequestration and small RNA binding. Microbiol Spectr 6(4):RWR-0030-2018. 10.1128/microbiolspec.RWR-0030-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel J, Argaman L, Wagner EGH, Altuvia S. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol 14:2271–2276 10.1016/j.cub.2004.12.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Darfeuille F, Unoson C, Vogel J, Wagner EGH. 2007. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell 26:381–392 10.1016/j.molcel.2007.04.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Opdyke JA, Kang JG, Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186:6698–6705 10.1128/JB.186.20.6698-6705.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opdyke JA, Fozo EM, Hemm MR, Storz G. 2011. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol 406:29–43 10.1016/j.jmb.2010.12.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiso T, Kamiya S, Yonezawa H, Gamou S. 2014. Overexpression of an antisense RNA, ArrS, increases the acid resistance of Escherichia coli. Microbiology 160:954–961 10.1099/mic.0.075994-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Choi JS, Kim W, Suk S, Park H, Bak G, Yoon J, Lee Y. 2018. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol 15:1319–1335 10.1080/15476286.2018.1532252. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fröhlich KS, Haneke K, Papenfort K, Vogel J. 2016. The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res 44:10406–10422. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker A, Gottesman S. 2016. Small RNA regulation of TolC, the outer membrane component of bacterial multidrug transporters. J Bacteriol 198:1101–1113 10.1128/JB.00971-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fontaine F, Gasiorowski E, Gracia C, Ballouche M, Caillet J, Marchais A, Hajnsdorf E. 2016. The small RNA SraG participates in PNPase homeostasis. RNA 22:1560–1573 10.1261/rna.055236.115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babitzke P, Lai YJ, Renda AJ, Romeo T. 2019. Posttranscription initiation control of gene expression mediated by bacterial RNA-binding proteins. Annu Rev Microbiol 73:43–67 10.1146/annurev-micro-020518-115907. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmqvist E, Vogel J. 2018. RNA-binding proteins in bacteria. Nat Rev Microbiol 16:601–615 10.1038/s41579-018-0049-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Franze de Fernandez MT, Eoyang L, August JT. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588–590 10.1038/219588a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138 10.1016/j.mib.2016.02.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woodson SA, Panja S, Santiago-Frangos A. 2018. Proteins that chaperone RNA regulation. Microbiol Spectr 6(4):RWR-0026-2018. 10.1128/microbiolspec.RWR-0026-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schu DJ, Zhang A, Gottesman S, Storz G. 2015. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34:2557–2573 10.15252/embj.201591569. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. 2013. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425:3678–3697 10.1016/j.jmb.2013.01.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santiago-Frangos A, Kavita K, Schu DJ, Gottesman S, Woodson SA. 2016. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc Natl Acad Sci USA 113:E6089–E6096 10.1073/pnas.1613053113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panja S, Santiago-Frangos A, Schu DJ, Gottesman S, Woodson SA. 2015. Acidic residues in the Hfq chaperone increase the selectivity of sRNA binding and annealing. J Mol Biol 427:3491–3500 10.1016/j.jmb.2015.07.010. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santiago-Frangos A, Jeliazkov JR, Gray JJ, Woodson SA. 2017. Acidic C-terminal domains autoregulate the RNA chaperone Hfq. eLife 6:e27049 10.7554/eLife.27049. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner EGH. 2013. Cycling of RNAs on Hfq. RNA Biol 10:619–626 10.4161/rna.24044. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malabirade A, Morgado-Brajones J, Trépout S, Wien F, Marquez I, Seguin J, Marco S, Velez M, Arluison V. 2017. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci Rep 7:10724 10.1038/s41598-017-11157-5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persson F, Lindén M, Unoson C, Elf J. 2013. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat Methods 10:265–269 10.1038/nmeth.2367. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Kannaiah S, Livny J, Amster-Choder O. 2019. Spatiotemporal organization of the E. coli transcriptome: translation independence and engagement in regulation. Mol Cell 76:574–589.e7 10.1016/j.molcel.2019.08.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Olejniczak M, Storz G. 2017. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 104:905–915 10.1111/mmi.13679. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stalmach ME, Grothe S, Wood JM. 1983. Two proline porters in Escherichia coli K-12. J Bacteriol 156:481–486 10.1128/JB.156.2.481-486.1983. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milner JL, Wood JM. 1989. Insertion proQ220:Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J Bacteriol 171:947–951 10.1128/JB.171.2.947-951.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gonzalez GM, Hardwick SW, Maslen SL, Skehel JM, Holmqvist E, Vogel J, Bateman A, Luisi BF, Broadhurst RW. 2017. Structure of the Escherichia coli ProQ RNA-binding protein. RNA 23:696–711 10.1261/rna.060343.116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36:1029–1045 10.15252/embj.201696127. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt WD, Vogel J. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar Typhimurium. MBio 10:e02504-18 10.1128/mBio.02504-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6(2):RWR-0009-2017. 10.1128/microbiolspec.RWR-0009-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gutiérrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol 187:3496–3501 10.1128/JB.187.10.3496-3501.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Timmermans J, Van Melderen L. 2009. Conditional essentiality of the csrA gene in Escherichia coli. J Bacteriol 191:1722–1724 10.1128/JB.01573-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–1145 10.1101/gad.214734.113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sterzenbach T, Nguyen KT, Nuccio SP, Winter MG, Vakulskas CA, Clegg S, Romeo T, Bäumler AJ. 2013. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J 32:2872–2883 10.1038/emboj.2013.206. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ. 2017. Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J 36:374–387 10.15252/embj.201694639. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, Śmiga M, Papenfort K, Reinhardt R, Wieden HJ, Luisi BF, Vogel J. 2017. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell 65:39–51 10.1016/j.molcel.2016.11.002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altuvia Y, Bar A, Reiss N, Karavani E, Argaman L, Margalit H. 2018. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli. Nucleic Acids Res 46:10530–10531 10.1093/nar/gky816. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon GC, Cameron JC, Pfleger BF. 2017. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. MBio 8:e00128-17 10.1128/mBio.00128-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Lay N, Gottesman S. 2011. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17:1172–1189 10.1261/rna.2531211. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cameron TA, Matz LM, Sinha D, De Lay NR. 2019. Polynucleotide phosphorylase promotes the stability and function of Hfq-binding sRNAs by degrading target mRNA-derived fragments. Nucleic Acids Res 47:8821–8837 10.1093/nar/gkz616. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. 2013. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev 27:552–564 10.1101/gad.210112.112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20:2605–2617 10.1101/gad.1461606. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michaux C, Holmqvist E, Vasicek E, Sharan M, Barquist L, Westermann AJ, Gunn JS, Vogel J. 2017. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc Natl Acad Sci USA 114:6824–6829 10.1073/pnas.1620772114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, Gross CA. 2018. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol Cell 70:274–286.e7 10.1016/j.molcel.2018.02.035. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyakoshi M, Chao Y, Vogel J. 2015. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol 24:132–139 10.1016/j.mib.2015.01.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Dar D, Sorek R. 2018. Bacterial noncoding RNAs excised from within protein-coding transcripts. MBio 9:e01730-18 10.1128/mBio.01730-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Massé E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383 10.1101/gad.1127103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morita T, Nishino R, Aiba H. 2017. Role of the terminator hairpin in the biogenesis of functional Hfq-binding sRNAs. RNA 23:1419–1431 10.1261/rna.060756.117. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99:4620–4625 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43–53 10.1016/S0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 95.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. 2014. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28:1620–1634 10.1101/gad.243485.114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morita T, Ueda M, Kubo K, Aiba H. 2015. Insights into transcription termination of Hfq-binding sRNAs of Escherichia coli and characterization of readthrough products. RNA 21:1490–1501 10.1261/rna.051870.115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyakoshi M, Chao Y, Vogel J. 2015. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J 34:1478–1492 10.15252/embj.201490546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Höfer K, Jäschke A. 2018. Epitranscriptomics: RNA modifications in bacteria and archaea. Microbiol Spectr 6(3):RWR-0015-2017. 10.1128/microbiolspec.RWR-0015-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marbaniang CN, Vogel J. 2016. Emerging roles of RNA modifications in bacteria. Curr Opin Microbiol 30:50–57 10.1016/j.mib.2016.01.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Cahová H, Winz ML, Höfer K, Nübel G, Jäschke A. 2015. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519:374–377 10.1038/nature14020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C. 2015. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res 43:6557–6567 10.1093/nar/gkv596. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bird JG, Zhang Y, Tian Y, Panova N, Barvík I, Greene L, Liu M, Buckley B, Krásný L, Lee JK, Kaplan CD, Ebright RH, Nickels BE. 2016. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535:444–447 10.1038/nature18622. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Mets F, Van Melderen L, Gottesman S. 2019. Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc Natl Acad Sci USA 116:1043–1052 10.1073/pnas.1815288116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miyakoshi M, Matera G, Maki K, Sone Y, Vogel J. 2019. Functional expansion of a TCA cycle operon mRNA by a 3′ end-derived small RNA. Nucleic Acids Res 47:2075–2088 10.1093/nar/gky1243. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. 2010. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA 107:9602–9607 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hao Y, Updegrove TB, Livingston NN, Storz G. 2016. Protection against deleterious nitrogen compounds: role of σS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res 44:6935–6948 10.1093/nar/gkw404. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. 2011. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res 39:2918–2930 10.1093/nar/gkq1239. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. 2014. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci USA 111:3134–3139 10.1073/pnas.1315974111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Figueroa-Bossi N, Bossi L. 2018. Sponges and predators in the small RNA world. Microbiol Spectr 6(4):RWR-0021-2018. 10.1128/microbiolspec.RWR-0021-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. 2009. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23:2004–2015 10.1101/gad.541609. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. 2009. Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73:790–800 10.1111/j.1365-2958.2009.06807.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Göpel Y, Khan MA, Görke B. 2014. Ménage à trois: post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol 11:433–442 10.4161/rna.28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Massé E. 2015. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell 58:393–405 10.1016/j.molcel.2015.03.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Sinha D, Matz LM, Cameron TA, De Lay NR. 2018. Poly(A) polymerase is required for RyhB sRNA stability and function in Escherichia coli. RNA 24:1496–1511 10.1261/rna.067181.118. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Raina M, King A, Bianco C, Vanderpool CK. 2018. Dual-function RNAs. Microbiol Spectr 6(5):RWR-0032-2018. 10.1128/microbiolspec.RWR-0032-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Papenfort K, Vanderpool CK. 2015. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39:362–378 10.1093/femsre/fuv016. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. 2007. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 21:2804–2817 10.1101/gad.447207. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54:1076–1089 10.1111/j.1365-2958.2004.04348.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. 2010. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA 107:20435–20440 10.1073/pnas.1009784107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. 2013. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci USA 110:E3487–E3496 10.1073/pnas.1303248110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rice JB, Balasubramanian D, Vanderpool CK. 2012. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc Natl Acad Sci USA 109:E2691–E2698 10.1073/pnas.1207927109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84:428–445 10.1111/j.1365-2958.2012.08031.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Chao Y, Vogel J. 2016. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–363 10.1016/j.molcel.2015.12.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32:827–837 10.1016/j.molcel.2008.10.027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Jagodnik J, Chiaruttini C, Guillier M. 2017. Stem-loop structures within mRNA coding sequences activate translation initiation and mediate control by small regulatory RNAs. Mol Cell 68:158–170.e3 10.1016/j.molcel.2017.08.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Azam MS, Vanderpool CK. 2018. Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res 46:2585–2599 10.1093/nar/gkx1286. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoekzema M, Romilly C, Holmqvist E, Wagner EGH. 2019. Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation. EMBO J 38:e101199 10.15252/embj.2018101199. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen J, Gottesman S. 2017. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev 31:1382–1395 10.1101/gad.302547.117. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA 95:12462–12467 10.1073/pnas.95.21.12462. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64:1260–1273 10.1111/j.1365-2958.2007.05733.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Urban JH, Vogel J. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6:e64 10.1371/journal.pbio.0060064. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Salvail H, Caron MP, Bélanger J, Massé E. 2013. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J 32:2764–2778 10.1038/emboj.2013.205. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16:840–846 10.1038/nsmb.1631. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. 2012. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47:943–953 10.1016/j.molcel.2012.07.015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Göpel Y, Khan MA, Görke B. 2016. Domain swapping between homologous bacterial small RNAs dissects processing and Hfq binding determinants and uncovers an aptamer for conditional RNase E cleavage. Nucleic Acids Res 44:824–837 10.1093/nar/gkv1161. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andreassen PR, Pettersen JS, Szczerba M, Valentin-Hansen P, Møller-Jensen J, Jørgensen MG. 2018. sRNA-dependent control of curli biosynthesis in Escherichia coli: McaS directs endonucleolytic cleavage of csgD mRNA. Nucleic Acids Res 46:6746–6760 10.1093/nar/gky479. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153:426–437 10.1016/j.cell.2013.03.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fröhlich KS, Papenfort K, Fekete A, Vogel J. 2013. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J 32:2963–2979 10.1038/emboj.2013.222. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen J, Morita T, Gottesman S. 2019. Regulation of transcription termination of small RNAs and by small RNAs: molecular mechanisms and biological functions. Front Cell Infect Microbiol 9:201 10.3389/fcimb.2019.00201. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. 2012. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev 26:1864–1873 10.1101/gad.195412.112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. 2016. sRNA-mediated control of transcription termination in E. coli. Cell 167:111–121.e13 10.1016/j.cell.2016.09.004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Silva IJ, Barahona S, Eyraud A, Lalaouna D, Figueroa-Bossi N, Massé E, Arraiano CM. 2019. SraL sRNA interaction regulates the terminator by preventing premature transcription termination of rho mRNA. Proc Natl Acad Sci USA 116:3042–3051 10.1073/pnas.1811589116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brosse A, Guillier M. 2018. Bacterial small RNAs in mixed regulatory networks. Microbiol Spectr 6(3):RWR-0014-2017. 10.1128/microbiolspec.RWR-0014-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nitzan M, Rehani R, Margalit H. 2017. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys 46:131–148 10.1146/annurev-biophys-070816-034058. [PubMed] [DOI] [PubMed] [Google Scholar]
- 145.Brosse A, Korobeinikova A, Gottesman S, Guillier M. 2016. Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res 44:9650–9666 10.1093/nar/gkw642. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guillier M, Gottesman S. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36:6781–6794 10.1093/nar/gkn742. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. 2011. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci USA 108:12875–12880 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hör J, Gorski SA, Vogel J. 2018. Bacterial RNA biology on a genome scale. Mol Cell 70:785–799 10.1016/j.molcel.2017.12.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Lopez CA, Skaar EP. 2018. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe 23:737–748 10.1016/j.chom.2018.05.008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chareyre S, Mandin P. 2018. Bacterial iron homeostasis regulation by sRNAs. Microbiol Spectr 6(2):RWR-0010-2017. 10.1128/microbiolspec.RWR-0010-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lépine F, Dozois CM, Imlay J, Massé E. 2010. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci USA 107:15223–15228 10.1073/pnas.1007805107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jacques JF, Jang S, Prévost K, Desnoyers G, Desmarais M, Imlay J, Massé E. 2006. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol 62:1181–1190 10.1111/j.1365-2958.2006.05439.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Kim JN. 2016. Roles of two RyhB paralogs in the physiology of Salmonella enterica. Microbiol Res 186-187:146–152 10.1016/j.micres.2016.04.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Ikemura T, Dahlberg JE. 1973. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem 248:5033–5041. [PubMed] [Google Scholar]
- 155.Sahagan BG, Dahlberg JE. 1979. A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. II. Accumulation and distribution. J Mol Biol 131:593–605 10.1016/0022-2836(79)90009-3. [DOI] [PubMed] [Google Scholar]
- 156.Polayes DA, Rice PW, Garner MM, Dahlberg JE. 1988. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol 170:3110–3114 10.1128/JB.170.7.3110-3114.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16:1696–1706 10.1101/gad.231702. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9:23–30 10.1016/S1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 159.Lewis DE, Adhya S. 2015. Molecular mechanisms of transcription initiation at gal promoters and their multi-level regulation by GalR, CRP and DNA loop. Biomolecules 5:2782–2807 10.3390/biom5042782. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Beisel CL, Storz G. 2011. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41:286–297 10.1016/j.molcel.2010.12.027. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Durica-Mitic S, Göpel Y, Görke B. 2018. Carbohydrate utilization in bacteria: making the most out of sugars with the help of small regulatory RNAs. Microbiol Spectr 6(2):RWR-0013-2017. 10.1128/microbiolspec.RWR-0013-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35 10.1111/j.1365-2958.2012.07965.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68:890–906 10.1111/j.1365-2958.2008.06189.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 164.De Lay N, Gottesman S. 2009. The Crp-activated sRNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191:461–476 10.1128/JB.01157-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. 2007. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol 189:1963–1973 10.1128/JB.01243-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lonetto MA, Donohue TJ, Gross CA, Buttner MJ. 2019. Discovery of the extracytoplasmic function σ factors. Mol Microbiol 112:348–355 10.1111/mmi.14307. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. 2006. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J Mol Biol 364:1–8 10.1016/j.jmb.2006.09.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Thompson KM, Rhodius VA, Gottesman S. 2007. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol 189:4243–4256 10.1128/JB.00020-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. 2006. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62:1674–1688 10.1111/j.1365-2958.2006.05524.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Grabowicz M, Koren D, Silhavy TJ. 2016. The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. MBio 7:e00312-16 10.1128/mBio.00312-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gottesman S. 2019. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294:11685–11700 10.1074/jbc.REV119.005593. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Majdalani N, Hernandez D, Gottesman S. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 46:813–826 10.1046/j.1365-2958.2002.03203.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Mandin P, Gottesman S. 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29:3094–3107 10.1038/emboj.2010.179. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sledjeski DD, Gupta A, Gottesman S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15:3993–4000 10.1002/j.1460-2075.1996.tb00773.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Repoila F, Gottesman S. 2003. Temperature sensing by the dsrA promoter. J Bacteriol 185:6609–6614 10.1128/JB.185.22.6609-6614.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. 2017. DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J Bacteriol 200:e00463-17 10.1128/JB.00463-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wall E, Majdalani N, Gottesman S. 2018. The complex Rcs regulatory cascade. Annu Rev Microbiol 72:111–139 10.1146/annurev-micro-090817-062640. [PubMed] [DOI] [PubMed] [Google Scholar]
- 178.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JCD, Vogel J. 2009. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74:139–158 10.1111/j.1365-2958.2009.06857.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507 10.4161/rna.28867. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. 2012. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol 84:51–65 10.1111/j.1365-2958.2012.08002.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538 10.1111/j.1365-2958.2012.08209.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fröhlich KS, Papenfort K, Berger AA, Vogel J. 2012. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 40:3623–3640 10.1093/nar/gkr1156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33 10.1016/j.mib.2010.01.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 184.Westermann AJ. 2018. Regulatory RNAs in virulence and host-microbe interactions. Microbiol Spectr 6(4):RWR-0002-2017. 10.1128/microbiolspec.RWR-0002-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501 10.1038/nature16547. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Kim K, Palmer AD, Vanderpool CK, Slauch JM. 2019. The small RNA PinT contributes to PhoP-mediated regulation of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J Bacteriol 201:e00312-19 10.1128/JB.00312-19. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.El Mouali Y, Gaviria-Cantin T, Sánchez-Romero MA, Gibert M, Westermann AJ, Vogel J, Balsalobre C. 2018. CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet 14:e1007401 10.1371/journal.pgen.1007401. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Song M, Sukovich DJ, Ciccarelli L, Mayr J, Fernandez-Rodriguez J, Mirsky EA, Tucker AC, Gordon DB, Marlovits TC, Voigt CA. 2017. Control of type III protein secretion using a minimal genetic system. Nat Commun 8:14737 10.1038/ncomms14737. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Papenfort K, Podkaminski D, Hinton JC, Vogel J. 2012. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci USA 109:E757–E764 10.1073/pnas.1119414109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Barquist L, Westermann AJ, Vogel J. 2016. Molecular phenotyping of infection-associated small non-coding RNAs. Philos Trans R Soc Lond B Biol Sci 371:20160081 10.1098/rstb.2016.0081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


