Abstract
Background
TBX5 is a transcription factor that has an important role in development of heart. TBX5 variants in the region encoding the T-box domain have been shown to cause cardiac defects, such as atrial septal defect or ventricular septal defect, while TBX5 variants have also been identified in a few cardiomyopathy patients and considered causative. We identified a TBX5 variant (c.791G>A, p.Arg264Lys), that is over-represented in cardiomyopathy patients. This variant is located outside of the T-box domain, and its pathogenicity has not been confirmed by functional analyses.
Objective
To investigate whether the TBX5 R264K is deleterious and could contribute to the pathogenesis of cardiomyopathy.
Methods and results
We developed mice expressing Tbx5 R264K. Mice homozygous for this variant displayed compensated dilated cardiomyopathy; mild decreased fractional shortening, dilatation of the left ventricle, left ventricular wall thinning and increased heart weight without major heart structural disorders. There was no difference in activation of the ANF promotor, a transcriptional target of Tbx5, compared to wild-type. However, analysis of RNA isolated from left ventricular samples showed significant increases in the expression of Acta1 in left ventricle with concomitant increases in the protein level of ACTA1.
Conclusions
Mice homozygous for Tbx5 R264K showed compensated dilated cardiomyopathy. Thus, TBX5 R264K may have a significant pathogenic role in some cardiomyopathy patients independently of T-box domain pathway.
Introduction
Comprehensive genetic analysis of patients with genetic cardiomyopathies over the last decade have revealed significant genotype-phenotype associations. Most commonly, these genetic variants are single-nucleotide polymorphisms (SNPs), and may occur as familial or sporadic mutations. Identification of a genetic mutation in a patient with heart disease may provide useful information regarding prognosis, including the severity of the disease, morbidity or mortality and even therapeutic options. However, in many cases, the significance of the variant in pathogenesis is unknown and these are referred to as variants of unknown significance. Identification of a variant in multiple affected individuals may provide statistical evidence to support a causative role, but if a variant is rare it is difficult to establish a definitive role.
TBX5 was identified as the gene responsible for Holt-Oram syndrome, a syndrome that is clinically characterized by radial ray deficiencies and cardiac defects, such as atrial septal defects (ASD) and ventricular septal defects (VSD). However, multiple variants in TBX5 have been associated with Holt-Oram syndrome, with variations in cardiac morphology and severity of the defect [1–6]. Knockout or knockdown analysis of Tbx5 in mice has revealed that the protein has a central role in cardiac development, probably through regulation of downstream genes, including Anf, Gata4, Nkx2.5 and Brg1 [1,7,8].
Although the effect of mutations in the T-box domain of TBX5 have been widely studied, the mechanism of mutations outside of the T-box domain have not and their role in disease not clearly established. With the development of genome editing technology, it is now much simpler to create SNP knock-in animal models.
Here, we report the identification of a missense TBX5 variant R264K that is over-represented in patients with left ventricular non compaction (LVNC) or dilated cardiomyopathy (DCM) compared with controls. The variant is located outside of T-box domain. Therefore, the aim of this study is to analyze whether this TBX5 variant affects heart development and function using a knock-in mouse model.
Materials and methods
Collection of patient data
Basic clinical data were collected on patients at the time of enrollment. After the identification of genetic variants, detailed clinical data were retrospectively collected. The genetic testing of patients and use of the clinical data for the study were approved by the Ethics Committee of the University of Toyama and informed consent was obtained from all enrolled patients, or their parents.
Patient genetic testing
We designed a custom AmpliSeq panel of PCR primers (Life Technologies, Carlsbad, CA, USA) using Ion AmpliSeq designer software (www.ampliseq.com) to target all exons of 73 genes associated with cardiac disorders, including cardiomyopathies and channelopathies (S1 Table). This custom panel, which consisted of two separate PCR primer pools and produced a total of 1870 amplicons, was used to generate target amplicon libraries. Genomic DNA samples were PCR-amplified using the custom-designed panel and Ion AmpliSeq Library Kit Plus (Life Technologies) according to the manufacturer’s instructions. Samples were tagged using an Ion Xpress Barcode Adapters Kit (Life Technologies) and then pooled in equimolar concentrations. Emulsion PCR and Ion Sphere Particle (ISP) enrichment were performed using an Ion PGM Hi-Q View OT2 200 Kit (Life Technologies), according to the manufacturer’s instructions. ISPs were loaded on an Ion 318 Chip v2 and sequenced with the PGM™ system using an Ion PGM Hi-Q View Sequencing Kit (Life Technologies). All variants derived from PGM sequencing were prioritized and then confirmed by Sanger sequencing to validate the next-generation sequencing (NGS) results. For Sanger sequencing, the nucleotide sequences of the amplified fragments were analyzed by direct sequencing in both directions using a BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and an ABI 3130xl automated sequencer (Life Technologies).
Data analysis of genetic testing
Torrent Suite software and Ion Reporter Software 5.1 (Life Technologies) were used to perform primary to tertiary analyses, including optimized signal processing, base calling, sequence alignment, and variant analysis.
Pathogenic analysis in silico
SNP allelic frequencies were determined from the Human Genetic Variation Database of Kyoto University and the Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org), and multiple in silico algorithms were utilized to predict whether variants were likely deleterious and could contribute to disease pathogenesis.
Generation of Tbx5R264K/+ and Tbx5R264K/R264K mice
C57BL/6NJcl (CLEA Japan, Tokyo, Japan) strain mice were used in this study. Founder mice carrying a heterozygous R264K in Tbx5 (Tbx5R264K/+) were generated using CRISPR-Cas9 systems. The Slc:ICR (Japan SLC, Shizuoka, Japan) strain mouse was used as a surrogate mother, transplanted with an in vitro fertilized egg. Detailed methods are provided in the S1 File. Genotyping of offspring was performed by PCR using primers: 5’-AGGCTCAGCAAGGAGGTGAA-3’ and 5’-CATGGCAGCGAGCAGTAAGG-3’ followed by DNA sequencing. Homozygous mice (TBX5R264K/R264K) were obtained by mating of founder Tbx5R264K/+ mice. All animals were maintained under standard laboratory conditions (12–12 h/light–dark cycle with light on at 7:00 am; temperature, 22 ± 2°C; humidity, 50 ± 10%), and had free access to food and water, in the Laboratory Animal Resource Center of the University of Toyama. Mice were fed with a chow diet and housed in a barrier facility. All animal experiments were approved by the Animal Experiment Committee of the University of Toyama (Authorization No. A2017UH-2), and were carried out in accordance with the Guidelines for the Care and Use of Laboratory Animals of the University of Toyama.
Analysis of cardiac function and histological change
Echocardiography recordings
We used 10 week old Tbx5R264K/R264K mice as the young adult model and 6–9 month old Tbx5R264K/+ and Tbx5R264K/R264K mice as the mature to middle age model, based on the definition in The Mouse in Biomedical Research 2nd Edition (2007) [9]. Age-matched wild-type mice were used as controls. Sedation and echocardiography were performed as previously described [10]. Sedation was conducted using 5% isoflurane with 1L/min airflow rate in the induction chamber followed by maintenance of sedation using 1–2% isoflurane, maintaining a steady heart rate of between 400–500 beats per minutes.
Echocardiography was performed using a SONIMAGE HS1 ultrasound system (KONICA MINOLTA, Inc.) and ultrawide waveband linear probe (L18-4) with mice in the supine position. The probe, with pre-warmed ultrasound gel, was gently placed on the left side parasternal thorax for the M-mode short axis view, including anterolateral and posteromedial papillary muscles, in order to measure systolic function and left ventricle diameter. In order to measure diastolic function, trans-mitral inflow data were obtained by pulse-wave Doppler on an apical four chamber view. The following measurements were recorded: left ventricle diameter at end diastole and systole (LVDd and LVDs), left ventricular anterior and posterior wall thickness at end diastole (AWD and PWD), fractional shortening (FS) and trans-mitral early wave (E) and atrial wave (A).
These measurements were performed by at least two individuals, including pediatric cardiologists, and measurements were taken over an average of three heart beats. After echocardiography, the mice were immediately euthanized by intraperitoneal injection of sodium pentobarbital (100 μg/g body weight) and their hearts were harvested. Heart weight (HW) was measured immediately. Although body size or heart size varied between individual mice, HW was standardized by dividing by body weight (BW). Cardiac defects were detected during echocardiography or during dissection for histological sections.
Isoproterenol stimulation to create myocardial injury
To examine cardiac tolerance in a stress environment, we used the isoproterenol-induced myocardial injury model as previously described [11–14]. Isoproterenol hydrochloride, a synthetic non-selective β-adrenergic agonist, was dissolved in physiological saline. Eight week old wild-type and Tbx5R264K/R264K mice were divided into two groups: isoproterenol (20 μg/g BW) group or isotonic saline group. Each group was injected subcutaneously once daily for 7 days. Seven days after the last injection, cardiac function was evaluated using echocardiography, as described above.
Histological analysis
Harvested hearts were stored at -80°C or fixed by immersion in 4% paraformaldehyde fixative. Fixed histological sections were stained with hematoxylin and eosin (HE) and Elastica van Gieson and Masson’s trichrome (Elastica-Masson) stains. Since it is known that the progression of myocardial fibrosis has two patterns of reactive fibrosis and replacement fibrosis, we performed semi-quantitatively scoring of Elastica-Masson stained sections according to the previously reported method [15], where the fibrosis score was defined as 0 = negative, 1 = perivascular, 2 = perivascular with extension to interstitium, which was finely classified; a = focal and b = diffuse, and 3 = encircling of individual myocytes, which was classified; a = without perivascular fibrosis and b = with perivascular fibrosis. Similarly, replacement fibrosis was scored 0 = negative, 1 = minimal foci, 2 = occasional foci and small scars, and 3 = extensive scarring (S2 Table). This semi-quantitative method has shown a high correlation between computerized quantitative measurements in cardiac fibrosis [15].
Electrocardiogram recordings
Electrocardiogram recordings were performed in 10 Tbx5R264K/+ mouse and 10 wild-type mice at 4 weeks of age. Anesthesia was provided by intraperitoneal injection of 0.02 ml/ g BW of a mixture containing 2,2,2-tribromoethanol (0.0175 g/ml), 2-methyl-2-butanol (1% v/v) and physiological saline. Needle electrodes were inserted into skin, and the electrocardiogram was recorded for 5 minutes in leads Ⅰ and Ⅱ on spine position using the LabChart 7 ECG software module (AD Instruments, Australia). The average waveform of 4 consecutive heart beats was recorded to remove baseline noise, then the average of 10 times measurements was used for analysis. The measurements were heart rate, interval of PR, QRS, QT, and amplitude of P, Q, R, ST and T wave. Arrhythmias, such as atrioventricular block or sinus node dysfunction, were automatically and visually detected.
Analysis of gene expression variation
Microarray and real time RT-PCR analysis of RNA expression.Total RNA was extracted from the left ventricular apex to the free wall of young adult TBX5R264K/R264K mice and wild-type littermates, using RNeasy Mini Kits (QIAGEN), according to the manufacturer’s instructions. The quality and quantity of RNA was measured using an Eppendorf BioPhotometer D30 (Eppendorf) and the RNA was stored at -80°C for future processing. For transcriptome analysis, Clariom™ D Arrays mouse (Thermo Fisher Scientific) was used, according to the manufacturer’s instruction. RNA expression was analysed using the Transcriptome Analysis Console software (v.4.0.1.36) (Thermo Fisher Scientific) and expressed as quantitative log2 metrics. Regarding the extracted genes, to explore the biological role of genes showing altered expression, functional enrichment analysis was performed using g:Profiler, a web server for functional enrichment analysis and conversions of gene lists [16]. For real time RT-PCR, cDNA synthesis was performed from 300 ng of RNA using Super Script Ⅳ Reverse Transcriptase (Invitrogen). Real time RT-PCR was performed using a THUNDERBIRD® SYBR qPCR Mix (TOYOBO CO., LTD. Life Science Department, OSAKA, JAPAN) and all real time RT-PCR experiments were performed in duplicate. Expression levels were measured using a Thermal Cycler Dice Real Time SystemⅡ(Takara Bio) and compared by threshold cycle difference from target gene to Gapdh (ΔCt). Real time RT-PCR primers are shown in S3 Table.
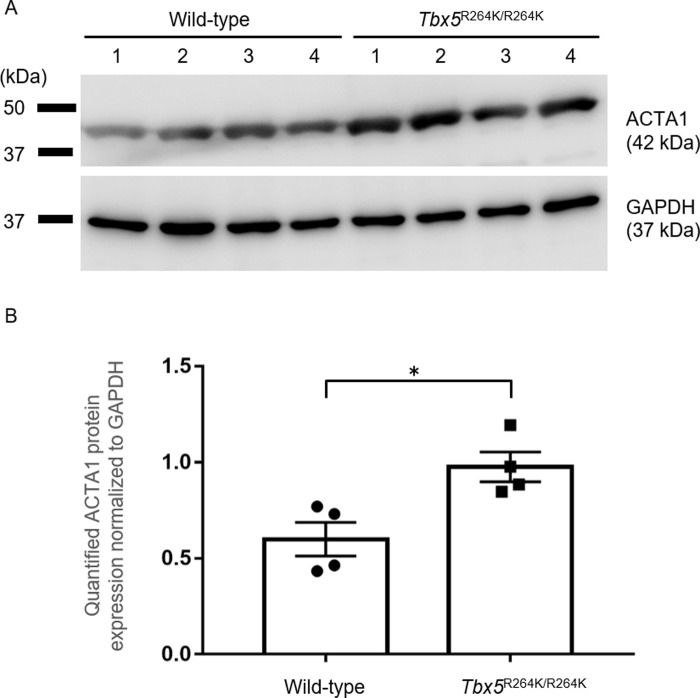
Western blotting.Total protein from left ventricle tissue was prepared using PRO-PREPTM Protein Extraction Solution (Cell/Tissue) (iNtRON Biotechnology, Inc), according to the manufacturer’s instruction. Protein extracts (10 μg) were separated by SDS-PAGE on 10% gel. Proteins were transferred to PVDF membranes, and the membranes were incubated with anti-skeletal muscle actin antibody (Mouse monoclonal LS-C413476 LifeSpan BioSciences, Inc) (1:5000) or anti-GAPDH antibody (Mouse monoclonal G8795 Sigma-Aldrich) (1:200) overnight at 4°C. The membranes were then incubated with Goat Anti-Mouse (H+L)-HRP conjugate (Bio-Rad) (1:5000) for 1 hour at room temperature. Protein bands were detected using Luminata Forte Western HRP Substrate (Merck Millipore) and a ImageQuant LAS 4000 mini (GE Healthcare) system. Protein levels of ACTA1 were normalized to those of GAPDH.
Analysis of transcriptional activity by luciferase assay
Plasmid constructs
A human TBX5 expression vector (IRAK049F12) was provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan [17]. A series of TBX5 variant plasmids containing single-base substitutions at different positions were constructed using the PrimeSTAR Mutagenesis Basal Kit (Takara, Shiga, Japan). For the human NPPA (ANF) promoter reporter construct, the promoter region of NPPA gene, -20 base to -758 base from the ATG start codon, was amplified by PCR and cloned into the pGL3-basic vector (Promega) using an In-Fusion HD Cloning Kit (Takara). All plasmids containing the various mutations were validated by direct DNA sequencing.
Cell culture and DNA transfection
Human embryonic kidney (HEK) 293T cells (CRL-11268, American Type Culture Collection, VA, USA) were grown in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum and 0.5% penicillin-streptomycin, at 37°C and 5% CO2. Briefly, 2.0 × 105 HEK-293T cells were seeded into a 6-well plates. Twenty-four hours later, cells were transiently transfected with pCMV-SPORT6-TBX5 (1.0 μg/well) or pCMV-SPORT6-TBX5 mutant (R264K and P85T) (1.0 μg/well), and pGL3-vector (1.5 μg/well) or the pGL3-ANF promoter reporter (1.5 μg/well) vectors using TransIT-293 transfection reagent (Mirus, WI, USA). A previously reported TBX5 P85T mutant was included in the transactivation study as a positive control [18]. A β-galactosidase control vector (the RSV LTR/β-galactosidase plasmid DNA) (50 ng) was used as an internal control. The total amount of transfected plasmids was adjusted to 2.55 μg/well by adding the pCMV-SPORT6 plasmid. At 48 h post-transfection, cells were harvested for luciferase and β-galactosidase assays. Cell lysis and luciferase assays were performed using the Luciferase Assay System (Promega, WI, USA). Light emission was measured in a SpectraMax i3 (Molecular Devices, CA, USA). The values were obtained in relative light units (RLU). Variations in transfection efficiency were normalized to the activities of luciferase expressed from the co-transfected β-galactosidase control vector. β-Galactosidase activities were measured using the β-Galactosidase Enzyme Assay System (Promega). Activity of the pGL3 -ANF promoter vector was assigned an arbitrary value of 1.0.
Statistical analysis
Student's t-test, or one- or two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons as a post-hoc analysis, were used to determine significance of quantitative data. Statistical significance was set at P < 0.05. Data are expressed as the mean ± standard error.
Result
Patient characteristics
A total of 233 idiopathic patients (192 LVNC and 41 DCM) have been screened using NGS technologies at our institution. Among them, 5 patients (4 LVNC and 1 DCM) had a heterozygous missense variant (c.791G>A, R264K) in TBX5 (Table 1). The frequency of this variant in these patients (0.02) is significantly higher than in reported for Japanese individuals in the Human Genetic Variation Database (0.0075) and in gnomAD (0.0016). In addition, this variant was predicted to be likely pathogenic by multiple in silico algorithms (S4 and S5 Tables). None of these 5 patients had variants in any of the other genes previously associated with cardiomyopathy. LVNC was diagnosed in patients presenting with heart failure, heart murmur associated with a VSD or suspicion of anomalous cardiomyocyte maturation between the neonatal and infantile periods. The patient diagnosed with DCM presented with chest pain. Two patients presented with heart failure, one had severe heart failure and another complicated by an embolism that resulted in death at one month of age. One patient with LVNC had family history that included a paternal uncle who had died at 19 years old. The patient with the VSD had no other heart defects. Since cardiomyopathy due to TBX5 variant is rare, we made heterozygous and homozygous knock-in mice to confirm the genotype-phenotype correlation between TBX5 R264K variant and cardiomyopathies.
Table 1. Characteristics of the patients carrying the heterozygous TBX5 c.791G>A, R264K variant.
| patient | sex | diagnosis | Age | gene variant in NGS associated with cardiomyopathy | opportunity | familial or sporadic | congenital heart disease |
|---|---|---|---|---|---|---|---|
| 1 | F | LVNC | neonatal | screening | sporadic | VSD | |
| 2 | M | LVNC | 3m | heart failure | sporadic | none | |
| 3 | F | LVNC | neonatal | heart failure, embolus | familial | none | |
| 4 | M | LVNC | neonatal | fetal myocardial hypertrophy | sporadic | none | |
| 5 | F | DCM | 11y | - | chest pain | sporadic | none |
DCM: dilated cardiomyopathy, LVNC: left ventricular non compaction, VSD: ventricular septal defect, -: no relative gene variant was detected
Tbx5R264K/R264K mice developed compensated dilated cardiomyopathy
There was no significant difference in physique between Tbx5R264K/R264K mice and wild-type mice. Young adult Tbx5R264K/R264K mice displayed significantly reduced FS, AWD and PWD compared with wild-type littermates (Table 2A), as well as higher LVDd, LVDs and HW/BW ratios. No arrhythmias were detected during echocardiography and no cardiac defects, such as ASDs or VSDs, were detected during dissection or echocardiography. The changes in cardiac function were not related to gender. Furthermore, the isoproterenol administration test for assessment of myocardial tolerance resulted in no significant differences in FS between the two groups (Table 2B). From these data, it was concluded that the young adult Tbx5R264K/R264K mice developed dilated cardiomyopathy phenotype, but were within a compensable range.
Table 2.
A. Characteristics and echocardiographic data in young adult wild-type and Tbx5R264K/R264K mice. B. Characteristics and echocardiographic data of isoproterenol stimulation in young adult wild-type and Tbx5R264K/R264K mice.
| A | ||||
| Wild-type | Tbx5R264K/R264K | |||
| (n = 5) | (n = 7) | |||
| male | 2 | 3 | ||
| BW (g) | 22.5±1.9 | 22.9±1.5 | ||
| HR (/min) | 413.2±12.7 | 431.2±34.2 | ||
| LVDd (mm) | 3.54±0.16 | 3.89±0.21* | ||
| LVDs (mm) | 2.14±0.23 | 2.56±0.17** | ||
| AWD (mm) | 0.68±0.12 | 0.56±0.05* | ||
| PWD (mm) | 0.82±0.07 | 0.66±0.1* | ||
| FS (%) | 39.6±5.4 | 34.2±1.7* | ||
| E (cm/s) | 51.9±7.0 | 61.2±14.1 | ||
| A (cm/s) | 31.2±6.3 | 34.1±5.0 | ||
| HW (mg) | 115.4±10.2 | 132.9±8.9* | ||
| HW/BW | 5.12±0.08 | 5.80±0.32** | ||
| B | ||||
| Wild-type | Tbx5R264K/R264K | |||
| Vehicle (n = 4) | Isoproterenol (n = 4) | Vehicle (n = 4) | Isoproterenol (n = 4) | |
| male | 2 | 2 | 2 | 2 |
| BW (g) | 24.5±1.0 | 24.7±0.9 | 22.8±1.5 | 23.4±0.8 |
| HR (/min) | 430.7±9.2 | 429.6±15.1 | 443.7±14.4 | 444.7±14.1 |
| LVDd (mm) | 3.63±0.22 | 3.83±0.11 | 4.10±0.25 | 4.18±0.09§ |
| LVDs (mm) | 2.33±0.17 | 2.66±0.11 | 2.90±0.2 | 3.18±0.09§ |
| AWD (mm) | 0.85±0.06 | 0.90±0.06 | 0.63±0.05† | 0.73±0.03§ |
| PWD (mm) | 0.95±0.09 | 0.88±0.05 | 0.58±0.05†† | 0.7±0.04§ |
| FS (%) | 36.0±1.1 | 30.4±1.7* | 29.4±0.8†† | 24.0±0.5‡§§ |
| E (cm/s) | 55.8±2.7 | 47.5±1.4* | 57.5±3.7 | 48.8±2.5 |
| A (cm/s) | 35.8±5.8 | 27.5±3.9 | 33.1±3.5 | 34.9±3.6 |
| HW (mg) | 117.0±5.5 | 119.0±4.3 | 116.3±5.9 | 125.5±3.9 |
| HW/BW | 4.45±0.11 | 4.82±0.18 | 5.11±0.10†† | 5.37±0.04§ |
BW: body weight, HR: heart rate, LVDd: left ventricle diameter at end diastole, LVDs: left ventricle diameter at end systole, AWD: left ventricular anterior wall thickness at end diastole, PWD: left ventricular posterior wall thickness at end diastole, FS: fractional shortening, E: trans-mitral early wave, A: trans-mitral atrial wave, HW: heart weight, P value *<0.05, versus wild-type vehicle,
†<0.05,
††<0.01, versus wild-type vehicle,
‡<0.05 versus young adult TBX5R264K/R264K vehicle, and.
§<0.05,
§§<0.01, versus wild-type isoproterenol. P value.
*<0.05,
**<0.01.
As the homozygous mutant mice aged, their FS remained significantly reduced compared to wild-type mice but the differences HW/BW, LVDd, LVDs, AWD, and PWD were no longer significant (S6 Table).
Heterozygous mutant mice, Tbx5R264K/+, had no significant changes on echocardiography or in heart weight. Further, electrocardiographic examination of heterozygous mice revealed no differences in HR, interval of PR, QRS, QT, and amplitude of P, Q, R, ST, and T waves (S7 Table).
Mild cardiac fibrosis in Tbx5R264K/R264K mice developing in an age-dependent manner
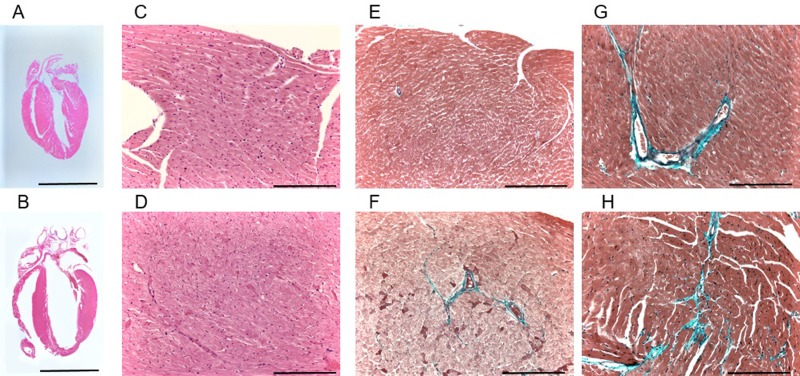
Young adult Tbx5R264K/R264K mice revealed slight fibrosis around vessels compared with wild-type on Elastica-Masson stain, with a reactive fibrosis score of 1. However, mature to middle aged Tbx5R264K/R264K mice had mild fibrosis from the endocardium to gaps between cardiomyocytes, and increased perivascular fibrosis with extension to the interstitium whereas mature to middle aged wild-type mice had only perivascular fibrosis. No other significant microscopic changes, such as hypertrophy of myocardial cells, difference in size or prolonged wavy appearance, were observed through histological analysis (Fig 1A–1H and Table 3).
Fig 1. Histological findings in wild-type (upper panels) and TBX5R264K/R264K mice (lower panels).
A and B. Longitudinal sections through the atria and ventricles with hematoxylin-eosin staining. Scale bars: 2 mm. C and D. Hematoxylin-eosin staining of high magnification sections of left ventricle myocardium. Scale bars: 100 μm.E and F. Elastica-Masson staining of sections from the left ventricular apex to free wall of hearts from young adult mouse. Slight fibrosis (blue signals) is seen around the vessels in young adult TBX5R264K/R264K mouse. Scale bars: 100 μm. G and H. Elastica-Masson staining of sections from the left ventricular apex to free wall of hearts from mature to middle age mice. Mild fibrosis (blue signals) from the endocardium to gaps between cardiomyocytes, and increased perivascular fibrosis with extension to the interstitium are shown in TBX5R264K/R264K compared with wild-type. Scale bars: 100 μm.
Table 3. Semi-quantitative evaluation of myocardial fibrosis based on S2 Table in wild-type and Tbx5R264K/R264K mice.
| Young adult | Mature to middle age | |||
|---|---|---|---|---|
| Wild-type | Tbx5R264K/R264K | Wild-type | Tbx5R264K/R264K | |
| Reactive fibrosis | 0 | 1 | 1 | 2a |
| Replacement fibrosis | 0 | 0 | 0 | 1 |
Tbx5R264K/R264K displayed significantly upregulated Acta1, specifically the isoform lacking exon 2
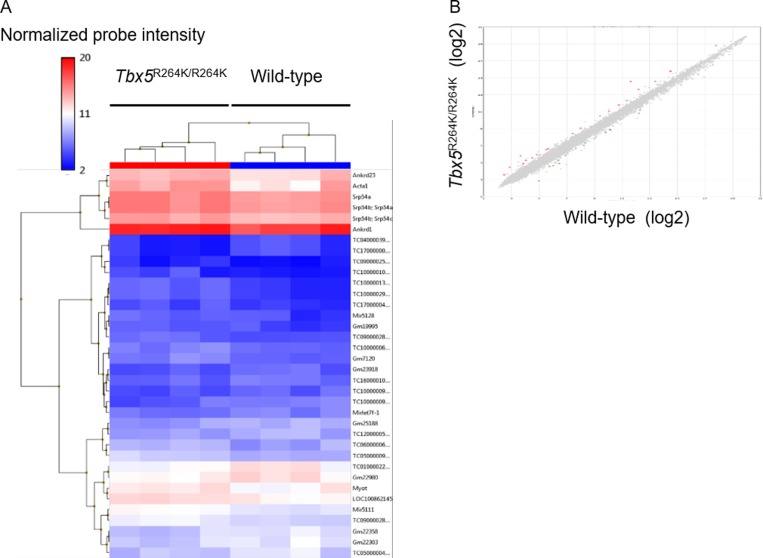
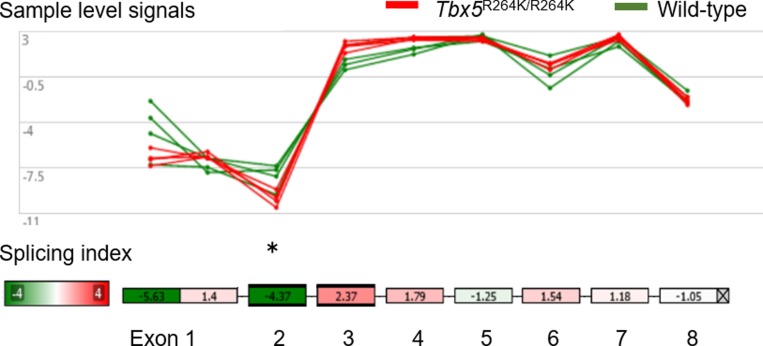
Microarray analysis of RNA isolated from the hearts of young adult Tbx5R264K/R264K mice revealed 36 genes (22 upregulated and 14 down regulated) differentially expressed (greater than 2.0 or less than -2.0 fold) by comparison with wild-type mice (Fig 2A and 2B and Table 4). After these were applied to g:Profiler analysis, noncoding genes were excluded, and as a result further analyses were limited to 8 coding genes, all of which were up-regulated genes in mutant mice (Table 4). Of the Biological Processes identified, there were two terms related to myocardium; response to muscle stretch and response to mechanical stimulus (S8 Table): this limited the relevant genes to Acta1, Myot, Ankrd1 and Ankrd23. Downregulated genes were non-coding genes, small RNAs or micro RNAs, the significance of which are unknown at this time. Exon splicing variant analysis of Acta1 showed that the significant variation derived from sample level signals was a 4.37-fold decrease in exon 2 in Tbx5R264K/R264K mice compared with wild-type mice (Fig 3).
Fig 2. Microarray analysis of RNAs isolated from the left ventricle myocardium of young adult Tbx5R264K/R264K mice compared with wild-type.
A Clustering map shows 36 RNAs were significantly (P value <0.05) changed with greater than 2.0 or less than -2.0 fold expression changes. B Scatter plot shows those 36 RNAs indicated as red (increased) and blue (decreased) points.
Table 4. Significant 8 mRNAs detected with microarray analysis, all of which were increased in young adult Tbx5R264K/R264K mice, remaining after excluding non-coding genes from 36 RNAs.
| Gene Symbol | Fold Change |
|---|---|
| Gm7120 | 2.05 |
| Myot | 2.08 |
| Ankrd1 | 2.12 |
| Srp54b; Srp54c | 2.32 |
| Srp54a | 2.44 |
| Srp54b; Srp54a | 2.47 |
| Ankrd23 | 2.82 |
| Acta1 | 7.92 |
Fig 3. Exon splicing variant analysis in Acta1 by Transcriptome Analysis Console software.
Transcriptome Analysis Console software shows the sample level signals and schema of Acta1 for each exon from 1 to 8 of each 4 wild-type mice (green) and Tbx5R264K/R264K mice (red) in the young adult group. Of these, the only significant variation derived from sample level signals was a 4.37-fold decrease in exon 2 in the Tbx5R264K/R264K mice group. P value *<0.05, versus wild-type.
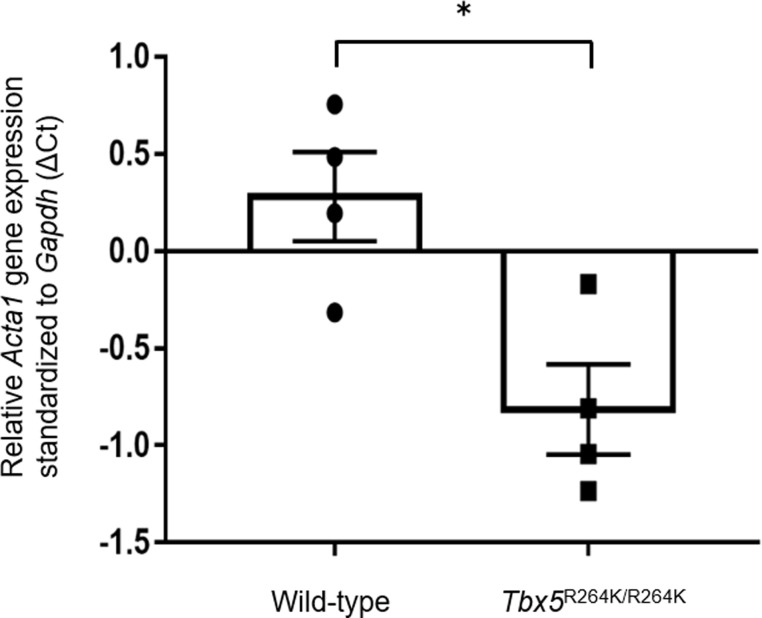
By real time RT-PCR, the expression of 7 of the 8 genes was not significantly different (S1 Fig). However, the expression level of Acta1 by real time RT-PCR was about 2 times higher in Tbx5264K/R264K mice than in the wild-type mice and ACTA1 protein expression was significantly higher by western blot analysis (Figs 4, 5A and 5B, and S2 Fig). No significant changes in the expression of Tbx5 itself and Nppa as a load marker for the heart, were detected (S3 Fig). Since there were no significant changes other than Acta1, we also performed real time RT-PCR on other genes related to sarcomere or calcium ion kinetics. However, no differences in the following genes were observed; myosin heavy chain 7 (Myh7), myosin binding protein C3 (Mybpc3), actin alpha cardiac muscle 1 (Actc1), sarcolipin (Sln) and myosin light polypeptide 7 regulatory (Myl7) (S4 Fig).
Fig 4. Relative gene expression of Acta1 in hearts of young adult Tbx5R264K/R264K and wild-type mice by real time RT-PCR analysis.
In the young adult Tbx5R264K/R264K mice group, the expression of Acta1 mRNA was approximately 2 times higher (lower ΔCt value) than in the wild-type mice group by real time RT-PCR. The number of individuals is four in each group. Average value of ΔCt (mean ± SEM) in each group is plotted. P value *<0.05, versus wild-type.
Fig 5. Comparison of ACTA1 protein expression in hearts of young adult Tbx5R264K/R264K and wild-type mice.
A Western blotting shows expression of ACTA1 protein (upper) in young adult Tbx5R264K/R264K and wild-type mice. GAPDH (lower) is shown as loading controls. The number of samples is four respectively. B In the young adult Tbx5R264K/R264K mice, the amount of ACTA1, normalized to GAPDH was about 1.6 times higher than that of in the wild-type mice. Quantified ACTA1 value normalized to GAPDH (mean ± SEM) is plotted. P value *<0.05, versus wild-type.
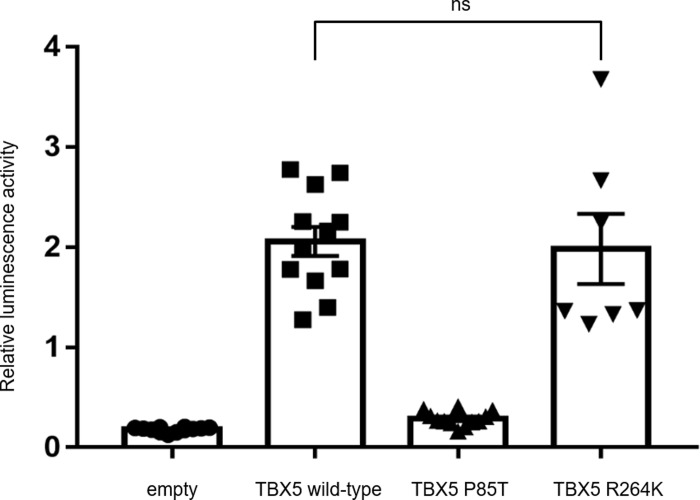
Transactivation of the ANF promoter was unaffected by the TBX5 variant
There were no significant changes in activation of the ANF promoter by the R264K mutation compared to wild-type TBX5: the positive control P85T mutant showed significantly reduced transcriptional activity (Fig 6).
Fig 6. Assays of TBX5 transcriptional activity.
TBX5 activates transcription from the ANF promoter. HEK293T cells were cotransfected with the ANF reporter plasmid and plasmids overexpressing TBX5, TBX5P85T, TBX5R264K. The TBX5R264K mutant activated the ANF promoter in a manner similar to the wild-type TBX5. Relative luminescence activity (mean ± SEM) is plotted.
Discussion
In this study, we found that homozygous Tbx5R264K/R264K mice exhibited cardiac dysfunction mimicking dilated cardiomyopathy. In particular, young adult Tbx5R264K/R264K mice had decreased FS, dilatation of the left ventricle, left ventricular wall thinning, and heart weight increases. We also showed that young adult Tbx5R264K/R264K mice had significant up-regulation of expression of Acta1.
TBX5 has been shown to be an essential transcription factor for cardiac development. Tbx5 heterozygous knockout mice develop cardiac defects, such as ASDs or VSDs, due to heart developmental patterning defects [19,20]. These result from significantly decreased expression of Cx40 and mis-expression of ANF into the right ventricle and the intraventricular groove rather than limited to the left ventricle, as seen in wild-type mice. Tbx5 homozygous hypomorphs develop hypoplastic left ventricles, resulting in embryonic lethality at E10.5 [19,20]. These structural defects often cause conduction diseases, such as arrhythmia and atrioventricular block [3,21,22]. In addition, TBX5 is considered to play a role in diastolic function, through Ca2+ handling modulation [23–25]. TBX5 variants have been identified in patients with congenital cardiac defects, with most located in the T-box domain (S5 Fig) [26–29]. However, there are only few reports of the identification of TBX5 variants in patients with LVNC or DCM unlike congenital heart defects [2, 5,30]. There is not a significant relationship between the cardiac phenotype and whether the variant is in the T-box or non T-box domain. A recent study revealed that a non T-box domain (amino acids 255–264) interacts with the nucleosome remodeling and deacetylase (NuRD) repressor complex (S5 Fig). This NuRD interaction pathway was considered to repress incompatible gene programs involved in cardiac development and, as a result, cardiac defects are inhibited [28].
In this context, we describe the analysis of a heterozygous TBX5 R264K variant that is over-represented in cardiomyopathy patients. This variant lies within the TBX5-NuRD interaction domain and is predicted to be likely pathogenic by multiple in silico algorithms. Therefore, we aimed to investigate whether this variant affects in cardiac development and/or cardiac function.
We found that Tbx5R264K/R264K mice develop DCM phenotype, without a major heart structural disorder. DCM is a chronic disease presenting left ventricular dilatation and systolic dysfunction, except for secondary reasons such as hypertension, valvular disease and coronary artery disease [31]. LVNC is defined by the feature of prominent trabeculae, intratrabecular recesses, and two distinct myocardial wall layers: a thin compacted epicardial layer and a thickened endocardial non-compacted layer [31]. In most patients with LVNC heart failure, the clinical characteristics are often similar to DCM in terms of left ventricular dilatation and systolic dysfunction. The findings in Tbx5R264K/R264K mice were characteristic of compensable DCM without heart structural defects or arrhythmias.
As the mice aged, the deposition of fibrosis increased in the endocardium and interstitium. The reactive fibrosis occurs with angiogenesis against the load on cardiomyocytes, and the replacement fibrosis occurs to fill the gap of cardiomyocytes when the cells fall into ischemia or inflammation leading to necrosis [32]. These fibrosis events were observed in mature to middle age Tbx5R264K/R264K mice. The slow progression of these histological changes is dependent on time with increasing contractile dysfunction, according to a mouse model of severe DCM [33]. Thus, the lack of changes, except for systolic dysfunction on echocardiography, in mature to middle aged Tbx5R264K/R264K mice was due to myocardial remodeling that occurred slowly as the mice aged.
Microarray analysis showed significant changes in the expression of 36 genes in the hearts of young adult Tbx5R264K/R264K mice compared with wild-type mice, and functional enrichment analysis based on these genes showed specific terms related to myocardium. However, there were no significant changes in expression detected by RT-PCR except for Acta1. Although the relationship between real time RT-PCR results and functional enrichment analysis could not be determined, it is presumed that the number of candidate genes was too small to investigate biological process. Moreover, the function of noncoding RNAs has not been fully elucidated. However, as there is a study describing noncoding RNA profiles in DCM patients [34], the noncoding RNAs identified in our study may also have important implications for cardiomyopathy pathogenesis.
ACTA1 is regulated by TBX5 during heart development, although it is expressed more highly in skeletal muscle than heart muscle [35]. ACTA1 is the causal gene of nemaline myopathy, characterized by an abnormal distribution of muscle filaments resulting in myopathy [36,37]. There have been reports of DCM patients with ACTA1 mutations [38]. Elevated ACTA1 expression has been reported in human heart failure samples and in a model mouse of DCM [39]. It has been concluded that increasing ACTA1 in heart failure is to compensate for contraction failure, irrespective of cause [39]. Transcriptome Analysis Console software identified a splicing variant of Acta1, with significantly increased splicing of exon 2. The function of exon 2 of Acta1 is unclear, though the exon encodes part of the 5’ untranslated region, and, in general, untranslated regions are speculated to regulate protein translation, translation activity or mRNA expression [40–42]. The transcription factor Tbx5 R264K was functionally and quantitatively preserved showing ANF expression via T-box domain pathway. However, it has been previously reported that the non-T-box domain pathway associated with amino acids 255–264 has a role in DCM pathogenesis in mice [28].
Since we were unable to identify the underlying mechanism of DCM in Tbx5R264K/R264K mice by gene expression analysis, we investigated the effects of isoproterenol stimulation on phenotype. Excessive β-adrenergic receptor stimulation activates the mitogen-activated protein kinase cascade, causing remodeling of myocardial hypertrophy resulting in systolic or diastolic dysfunction. The role of specific gene mutations in diastolic dysfunction induced by isoproterenol stimulation has been reported [43]. If isoproterenol stimulation caused a significant cardiac dysfunction in the Tbx5R264K/R264K mice, it is likely that the Tbx5R264K/R264K caused fragility in cardiomyocytes associated with mutation-specific changes in gene expression. However, no phenotypic differences were observed on isoproterenol stimulation, so we were unable to investigate possible changes in gene expression.
The patients with variants in the TBX5 gene showed severe cardiomyopathy phenotypes despite heterozygosity, while the mice with homozygous variant barely presented with compensatory chronic, decreasing cardiac contractility. Therefore, the cause of cardiomyopathy in these patients is not likely to be TBX5 R264K alone and it is likely that other unknown genes or environmental factors might have triggered those patients’ diseases [44]. In other words, this variant could act as modifier of the phenotype.
There is a limitation to this study. We have not confirmed that the non-Tbox domain pathway: NuRD interaction pathway is actually impaired. Therefore, the genetic pathway that led to DCM in Tbx5R264K/R264K mice has not been clarified.
Conclusion
We developed knock-in mice expressing the R264K Tbx5 variant that is frequently detected in cardiomyopathy patients. The homozygous variant of R264K in Tbx5 leads to compensatory DCM, without major heart structural disorders in mice, and ACTA1 expression was increased in cardiomyocytes in association with decreased contractility. Since the influence of other genes and environmental factors cannot be ignored, the TBX5 R264K variant in the non-Tbox domain involving NuRD interaction pathway may play a role in the pathogenesis of human cardiomyopathy.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
The numbers below indicate the positions of previously reported amino acid substitutions associated with congenital heart disease (35 variants in all). The variants described in this study, R264K, is located within the NuRD Interaction Domain.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors would like to thank Dr. Hideki Origasa, division of Biostatistics and Clinical Epidemiology, University of Toyama, Toyama, Japan for advice of statistical methods. The authors also would like to thank Mr.Hitoshi Moriuchi, technical assistant in department of pediatrics, University of Toyama for preparation and proxy purchase of laboratory equipment. We thank Dr. Yoshihide Hayashizaki at RIKEN and Dr. Sumio Sugano at University of Tokyo for providing us the human TBX5 expression vector (IRAK049F12). The authors are grateful to Dr.Neil E Bowles for assistance with editing.
Non-standard abbreviations and acronyms
- A
trans-mitral atrial wave
- ACTA1
actin, alpha 1, skeletal muscle
- ANF
Atrial natriuretic factor
- ANKRD1
ankyrin repeat domain 1
- ANKRD23
ankyrin repeat domain 23
- ANOVA
analysis of variance
- ASD
atrial septal defect
- AWD
anterior wall thickness at end diastole
- BRG1
Brahma-related gene-1
- BW
body weight
- DCM
dilated cardiomyopathy
- E
trans-mitral early wave
- FS
fractional shortening
- GATA4
GATA binding protein 4
- gnomAD
Genome Aggregation Database
- HW
heart weight
- LMNA
lamin A/C
- LVDd
left ventricle diameter at end diastole
- LVDs
left ventricle diameter at end systole
- LVNC
left ventricular non compaction
- MYBPC
myosin binding protein C
- MYH
myosin heavy chain
- MYL
myosin light chain
- MYOT
myotilin
- NGS
next generation sequencing
- Nkx2.5
NK2 homeobox 5
- NPPA
natriuretic peptide type A
- NuRD
nucleosome remodeling and deacetylase
- PWD
posterior wall thickness at end diastole
- SNP
single-nucleotide polymorphisms
- TBX5
T-box transcription factor 5
- VSD
ventricular septal defect
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Steimle JD, Moskowitz IP. TBX5: A Key Regulator of Heart Development. Curr Top Dev Biol. 2017; 122: 195–221. 10.1016/bs.ctdb.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross SB, Bagnall RD, Yeates L, Sy RW, Semsarian C. Holt-Oram syndrome in two families diagnosed with left ventricular noncompaction and conduction disease. Hear Rhythm Case Reports 2018; 4(4): 146–151. 10.1016/j.hrcr.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo DF, Li RG, Yuan F, Shi HY, Hou XM, Qu XK, et al. TBX5 loss-of-function mutation contributes to atrial fibrillation and atypical Holt-Oram syndrome. Mol Med Rep. 2016; 13(5): 4349–4356. 10.3892/mmr.2016.5043 [DOI] [PubMed] [Google Scholar]
- 4.Mcdermott DA, Bressan MC, He J, Lee JS, Aftimos S, Brueckner M, et al. TBX5 genetic testing validates strict clinical criteria for Holt-Oram syndrome. Pediatr Res. 2005; 58(5): 981–986. 10.1203/01.PDR.0000182593.95441.64 [DOI] [PubMed] [Google Scholar]
- 5.Brassington AE, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, et al. Expressivity of Holt-Oram Syndrome is not predicted by TBX5 genotype. Am J Hum Genet. 2003; 73: 74–85. 10.1086/376436 PMCID:PMC1180592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA, et al. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012; 21(14): 3255–3263. 10.1093/hmg/dds165 PMCID:PMC3384386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu T, Qiao L, Wang Q, Mi R, Chen J, Lu Y, et al. T-box family of transcription factor-TBX5, insights in development and disease. Am J Transl Res. 2017; 9(2): 442–453. PMCID:PMC5340680 [PMC free article] [PubMed] [Google Scholar]
- 8.Su W, Zhu P, Wang R, Wu Q, Wang M, Zhang X, et al. Congenital heart diseases and their association with the variant distribution features on susceptibility genes. Clin Genet. 2017; 91(3): 349–354. 10.1111/cge.12835 [DOI] [PubMed] [Google Scholar]
- 9.Flurkey K, Currer JM, Harrison DE. The Mouse in Aging Research Mouse in Biomed Res 2nd Edition 2007: 637–672 [Google Scholar]
- 10.Lindsey ML, Kassiri Z, Virag JAI, de Castro Brás LE, Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol. 2018; 314(4): H733–H752. 10.1152/ajpheart.00339.2017 PMCID:PMC5966769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grune J, Beyhoff N, Smeir E, Chudek R, Blumrich A, Ban Z, et al. Selective Mineralocorticoid Receptor Cofactor Modulation as Molecular Basis for Finerenone’s Antifibrotic Activity. Hypertension 2018; 71(4): 599–608. 10.1161/HYPERTENSIONAHA.117.10360 [DOI] [PubMed] [Google Scholar]
- 12.Wang L-X, Yang X, Yue Y, Fan T, Hou J, Chen G-X, et al. Imatinib attenuates cardiac fibrosis by inhibiting platelet-derived growth factor receptors activation in isoproterenol induced model. PLoS One 2017; 12(6): 1–19. 10.1371/journal.pone.0178619 PMCID:PMC5453565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks WW, Conrad CH. Isoproterenol-induced myocardial injury and diastolic dysfunction in mice: structural and functional correlates. Comp Med. 2009; 59(4): 339–343. PMCID:PMC2779208 [PMC free article] [PubMed] [Google Scholar]
- 14.Ma XW, Song Y, Chen C, Fu YN, Shen Q, Li Z, et al. Distinct actions of intermittent and sustained β-adrenoceptor stimulation on cardiac remodeling. Sci China Life Sci. 2011; 54(6): 493–501. 10.1007/s11427-011-4183-9 [DOI] [PubMed] [Google Scholar]
- 15.Segura AM, Frazier OH, Demirozu Z, Buja LM. Histopathologic correlates of myocardial improvement in patients supported by a left ventricular assist device. Cardiovasc Pathol. 2011; 20: 139–145. 10.1016/j.carpath.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 16.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019. July 2; 47(W1): W191–W198 10.1093/nar/gkz369 PMCID: PMC6602461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004; 36(1): 40–45. 10.1038/ng1285 [DOI] [PubMed] [Google Scholar]
- 18.Dreßen M, Lahm H, Lahm A, Wolf K, Doppler S, Deutsch M-A, et al. A novel de novo TBX5 mutation in a patient with Holt–Oram syndrome leading to a dramatically reduced biological function. Mol Genet Genomic Med.2016; 4(5): 557–567. 10.1002/mgg3.234 PMCID:PMC5023941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-Box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001; 106(6): 709–721. 10.1016/s0092-8674(01)00493-7 [DOI] [PubMed] [Google Scholar]
- 20.Hatcher CJ, Goldstein MM, Mah CS, Delia CS, Basson CT. Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Dev Dyn. 2000;219(1):90–95. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R, Tian X, Gao L, Li H, Yin X, Dong Y, et al. Common Variants in the TBX5 gene associated with atrial fibrillation in a Chinese Han population. PLoS One 2006; 11(8): 1–8. 10.1371/journal.pone.0160467 PMCID:PMC4968840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadadur RD, Broman MT, Boukens B, Mazurek SR, Yang X, van den Boogaard M et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci. Transl. Med. 2016; 8(354): 1–11. 10.1126/scitranslmed.aaf4891 PMCID:PMC5266594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Gramolini AO, Walsh MA, Zhou Y-Q, Slorach C, Friedberg MK, et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Nati Acad Sci U S A 2008; 105(14): 5519–5524. 10.1073/pnas.0801779105 PMCID:PMC2291114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y-Q, Zhu Y, Bishop J, Davidson L, Henkelman RM, Bruneau BG, et al. Abnormal cardiac inflow patterns during postnatal development in a mouse model of Holt-Oram syndrome. Am J Physiol Circ Physiol. 2005; 289(3): H992–H1001. 10.1152/ajpheart.00027.2005 [DOI] [PubMed] [Google Scholar]
- 25.Mikhailov AT, Torrado M. Myocardial transcription factors in diastolic dysfunction: clues for model systems and disease. Heart Fail. Rev. 2016; 21(6): 783–794. 10.1007/s10741-016-9569-0 [DOI] [PubMed] [Google Scholar]
- 26.Su W, Zhu P, Wang R, Wu Q, Wang M, Zhang X, et al. Congenital heart diseases and their association with the variant distribution features on susceptibility genes. Clin Genet. 2017; 91(3): 349–354. 10.1111/cge.12835 [DOI] [PubMed] [Google Scholar]
- 27.Yoshida A, Morisaki H, Nakaji M, Kitano M, Kim K-S, Sagawa K, et al. Genetic mutation analysis in Japanese patients with non-syndromic congenital heart disease. J Hum Genet. 2016;. 61(2): 157–162. 10.1038/jhg.2015.126 [DOI] [PubMed] [Google Scholar]
- 28.Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, et al. The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation Article The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation. Dev Cell 2016; 36(3): 262–275. 10.1016/j.devcel.2016.01.009 PMCID:PMC4920128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Qattan MM, Al-Shaar HA. Molecular basis of the clinical features of Holt-Oram syndrome resulting from missense and extended protein mutations of the TBX5 gene as well as TBX5 intragenic duplications. Gene 2015; 560(2): 129–136. 10.1016/j.gene.2015.02.017 [DOI] [PubMed] [Google Scholar]
- 30.Zhang X-L, Qiu X-B, Yuan F, Wang J, Zhao C-M, Li R-G, et al. TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2015; 459(1): 166–171. 10.1016/j.bbrc.2015.02.094 [DOI] [PubMed] [Google Scholar]
- 31.Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies:a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2008; 29: 270–276. 10.1093/eurheartj/ehm342 [DOI] [PubMed] [Google Scholar]
- 32.Segura AM, Frazier OH, Buja LM. Fibrosis and heart failure. Heart Fail Rev. 2014. March; 19(2): 173–85. 10.1007/s10741-012-9365-4 [DOI] [PubMed] [Google Scholar]
- 33.Arimura T, Sato R, Machida N, Bando H, Zhan D-Y, Morimoto S, et al. Improvement of Left Ventricular Dysfunction and of Survival Prognosis of Dilated Cardiomyopathy by Administration of Calcium Sensitizer SCH00013 in a Mouse Model. J Am Col. Cardiol. 2010; 55(14): 1503–1505. 10.1016/j.jacc.2009.10.065 [DOI] [PubMed] [Google Scholar]
- 34.Lin F, Gong X, Yu P, Yue A, Meng Q, Zheng L, et al. Distinct Circulating Expression Profiles of Long Noncoding RNAs in Heart Failure Patients With Ischemic and Nonischemic Dilated Cardiomyopathy. Front Genet. 2019. November 12; 10: 1116 10.3389/fgene.2019.01116 PMCID: PMC6861296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plageman TF, Yutzey KE. Microarray Analysis of Tbx5-Induced Genes Expressed in the Developing Heart. Dev Dyn. 2006; 235(10): 2868–2880. 10.1002/dvdy.20923 [DOI] [PubMed] [Google Scholar]
- 36.Ravenscroft G, McNamara E, Griffiths LM, Papadimitriou JM, Hardeman EC, Bakker AJ, et al. Cardiac α-actin over-expression therapy in dominant ACTA1 disease. Hum Mol Genet. 2013; 22(19): 3987–3997. 10.1093/hmg/ddt252 [DOI] [PubMed] [Google Scholar]
- 37.Laing NG, Dye DE, Wallgren-Pettersson C, Richard G, Monnier N, Lillis S, et al. Mutations and Polymorphisms of the Skeletal Muscle α-Actin Gene (ACTA1). Hum Mutat. 2009; 30(9): 1267–1277. 10.1002/humu.21059 PMCID:PMC2784950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reza N, Garg A, Merrill SL, Chowns JL, Rao S, Owens AT. ACTA1 Novel Likely Pathogenic Variant in a Family With Dilated Cardiomyopathy. Circ Genom Precis Med. 2018; 11(10): e002243 10.1161/CIRCGEN.118.002243 PMCID:PMC6208136[Available on 2019-10-01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copeland O, Nowak KJ, Laing NG, Ravenscroft G, Messer AE, Bayliss CR. et al. Investigation of changes in skeletal muscle a-actin expression in normal and pathological human and mouse hearts. J Muscle Res Cell Motil. 2010; 31(3): 207–214. 10.1007/s10974-010-9224-7 [DOI] [PubMed] [Google Scholar]
- 40.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006. October; 12(10): 1755–85. 10.1261/rna.157806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta. 2009. Sep-Oct;1789(9–10):584–91. 10.1016/j.bbagrm.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wethmar K, Smink JJ, Leutz A. Upstream open reading frames: molecular switches in (patho)physiology. Bioessays. 2010. October;32(10):885–93. 10.1002/bies.201000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Huertas-Vazquez A, Wang Y, Lusis AJ. Isoproterenol-Induced Cardiac Diastolic Dysfunction in Mice: A Systems Genetics Analysis. Front Cardiovasc Med. 2019. July 31;6:100 eCollection 2019. 10.3389/fcvm.2019.00100 PMCID: PMC6684968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka R, Minamisawa S, Akimoto K, Ichida F, Ohta Y, Ogata S, et al. Cardiovascular Disease Epidemiology Committee of Japanese Society of Pediatric Cardiology and Cardiac Surgery: Strong association between single-gene etiology and the intrauterine environment. Proceedings of the 2nd IREIIMS Open Symposium. Tokyo Women’s Medical University. Tokyo Japan, December 3–5, 2006. 43: 151–154