Abstract
E2f5 is a member of the E2f family of transcription factors that play essential roles during many cellular processes. E2f5 was initially characterized as a transcriptional repressor in cell proliferation studies through its interaction with the Retinoblastoma (Rb) protein for inhibition of target gene transcription. However, the precise roles of E2f5 during embryonic and post-embryonic development remain incompletely investigated. Here, we report that zebrafish E2f5 plays critical roles during spermatogenesis and multiciliated cell (MCC) differentiation. Zebrafish e2f5 mutants develop exclusively as infertile males. In the mutants, spermatogenesis is arrested at the zygotene stage due to homologous recombination (HR) defects, which finally leads to germ cell apoptosis. Inhibition of cell apoptosis in e2f5;tp53 double mutants rescued ovarian development, although oocytes generated from the double mutants were still abnormal, characterized by aberrant distribution of nucleoli. Using transcriptome analysis, we identified dmc1, which encodes an essential meiotic recombination protein, as the major target gene of E2f5 during spermatogenesis. E2f5 can bind to the promoter of dmc1 to promote HR, and overexpression of dmc1 significantly increased the fertilization rate of e2f5 mutant males. Besides gametogenesis defects, e2f5 mutants failed to develop MCCs in the nose and pronephric ducts during early embryonic stages, but these cells recovered later due to redundancy with E2f4. Moreover, we demonstrate that ion transporting principal cells in the pronephric ducts, which remain intercalated with the MCCs, do not contain motile cilia in wild-type embryos, while they generate single motile cilia in the absence of E2f5 activity. In line with this, we further show that E2f5 activates the Notch pathway gene jagged2b (jag2b) to inhibit the acquisition of MCC fate as well as motile cilia differentiation by the neighboring principal cells. Taken together, our data suggest that E2f5 can function as a versatile transcriptional activator and identify novel roles of the protein in spermatogenesis as well as MCC differentiation during zebrafish development.
Author summary
E2f family of transcription factors play essential roles during many cellular processes by activating or inhibiting target gene expression. E2F5 is recognized as a transcriptional repressor during cell-cycle progression. However, the role of E2f5 during embryonic development has not been fully investigated. Here, we show that E2f5 is required both for spermatogenesis and multiciliated cell differentiation in zebrafish. Specifically, E2f5 regulates homologous recombination, a central event in meiosis, by transactivating the expression of dmc1. Loss of E2f5 leads to spermatogenesis defects due to meiosis arrest. In addition, E2f5 plays an essential role during the differentiation of multiciliated cells and ion transporting principal cells in the pronephric ducts (embryonic zebrafish kidney). We showed that principal cells were devoid of cilia in the proximal straight tubule of pronephric ducts, while ectopic cilia developed in these cells due to lateral inhibition defects in the absence of E2f5 protein. Our data suggest that E2f5 functions as a versatile transcriptional activator during vertebrate development.
Introduction
E2f transcription factor family members are essential for cell proliferation, differentiation, apoptosis as well as many other cellular processes [1]. The roles of E2f proteins in cell-cycle control have been studied in great detail. In this context, interaction between Rb and E2f family members inhibits the transcription of E2f target genes at the G1 phase. Subsequently, phosphorylation of Rb by cyclin/cyclin-dependent kinases releases E2fs, which allows activation of their target genes and promotes G1 to S transition [2]. Consequently, unrestrained E2f activity due to inactivation of Rb, occurs in many types of cancers [2, 3].
Currently, eight different E2f proteins (E2f1-E2f8) have been identified, which can be generally classified into three categories according to their roles during cell cycle progression: transcription activators (E2f1, E2f2 and E2f3a), repressors (E2f3b, E2f4 and E2f5) and inhibitors (E2f6, E2f7 and E2f8) [4]. E2f1-3 are categorized as classic activators due to their transcriptional activity when binding to the promoter of target genes during cell-cycle entry. E2f4 and E2f5 function as transcription repressors through interactions with pocket proteins p107, p130, and Rb to inhibit target gene transcription during early G1 phase [4, 5]. Nevertheless, such classification is largely based on in vitro experiments probing their roles during the cell-cycle, which could be oversimplified, and may not necessarily reflect the complicated activities of E2f proteins in vivo [2].
Although being recognized as transcriptional repressors during cell cycle regulation, our current appreciation of the roles of E2f4 and E2f5 during embryonic development suggests alternative mechanisms of function. Mouse E2f4-/- mutants display defects in multiple tissues, and most die within the first week after birth due to increased susceptibility to opportunistic infections resulting from defects in the generation of MCCs within the airways [6–8]. E2f4 is also required for the development of MCCs within the male reproductive system as conditional knockout of E2f4 in E2f5+/- mice caused defects in the efferent ducts of the testes, where sperm are concentrated and transported into the epididymis [9]. By contrast, the role of E2f5 during embryonic development is relatively less well investigated. E2f5-/- mice have a shortened lifespan and develop hydrocephalus due to excessive cerebrospinal fluid (CSF) production [10]. Interestingly, mouse embryonic fibroblasts (MEFs) derived from both E2f4-/- and E2f5-/- mice display normal cell-cycle kinetics [6, 10]. All of these data suggest that the role of E2f4 and E2f5 during embryonic development is context-dependent, as opposed to these proteins functioning as general repressors in cell-cycle regulation.
MCCs are a specialized type of post-mitotic cells characterized by differentiation of hundreds of motile cilia that can beat unidirectionally to drive fluid flow over their epithelial surface. In mammals, MCCs are present in multiple organs, such as the spinal cord and brain ventricles, where they drive the flow of cerebrospinal fluid; the airways epithelia, where they help clearance of mucus; and in the fallopian tubes, where they are involved in ovum transport [11, 12]. The Notch signaling pathway is known to play a critical role during determination of the MCC fate [11]. Inhibition of Notch signaling leads to an expansion of MCC numbers at the expense of other cell types [13–17] in various tissues where this phenomenon has been examined. For example, in the zebrafish pronephric duct, the Notch ligand Jag2b is expressed in developing MCCs. Interaction between Jag2b and Notch1a/Notch3 receptors, a process called lateral inhibition, orchestrates the segregation of the “salt-and-pepper” pattern of MCCs and principal cells in the pronephros [13, 18].
Geminin coiled-coil domain-containing protein 1 (Gemc1, aka Gmnc) and Multicilin (Mci, aka Mcidas) are the known upstream activators of MCC differentiation downstream of Notch signals [19–22]. Interestingly, both proteins are devoid of an obvious DNA binding domain, although they both localize to the nucleus. Instead, Gemc1 and Mci associate with E2f4 and/or E2f5, together with Dp1 to activate gene expression for MCC specification and differentiation [20, 21, 23, 24]. Intriguingly, besides functioning as a transcription regulator, cytoplasmic E2f4 has also been shown to participate in the assembly of deuterosomes in the MCCs—electron-dense ring-like structures that are thought to seed the massive amplification of centrioles for multiciliation [25, 26].
Homologous recombination (HR) is an essential step during meiosis when homologous chromosomes pair and undergo reciprocal exchange of DNA during the first meiotic division. After homologous chromosome pairing, meiotic recombination is initiated by the formation of double-strand breaks (DSBs) at multiple positions through the actions of several factors including the topoisomerase-like enzyme Spo11. Later, binding of Dmc1 and Rad51 to the DSBs generates nucleoprotein filaments and initiates proper strand invasion to form D-loops. The DSBs are repaired either through a reciprocal exchange of chromosome arms by forming a crossover or non-crossover where no chromosome exchange happens [27, 28]. Both Rad51 and Dmc1 are recombinases that are required for strand invasion during meiotic HR, but they have overlapping and distinct functions. Rad51 regulates HR both in somatic cells and germ cells, whereas Dmc1 is expressed only in meiotic cells [29]. Several pieces of evidence suggest that Rad51 mainly performs an accessory function to promote Dmc1 foci formation during meiosis [30, 31]. Currently, the role of E2f5 during spermatogenesis, especially in meiosis, has not been reported.
We have generated loss-of-function alleles in zebrafish e2f5 and investigated the role of the gene during embryogenesis and adult development. Zebrafish e2f5 mutants display sex differentiation defects and develop exclusively as males. Interestingly, all male mutants are infertile due to meiotic arrest of spermatocytes. We have identified Dmc1 as the major factor accounting for HR defects in the mutants, and further show that E2f5 can bind to the promoter of dmc1, but not rad51. Overexpression of Dmc1 rescued spermatogenesis defects in e2f5 mutants. In addition, we found that E2f5 is not only required for MCC formation, but is also required cell-nonautonomously to orchestrate the differentiation of neighboring ion transporting principal cells by regulating the expression of jag2b in MCC precursors.
Results
Mutation of e2f5 results in male infertility in zebrafish
We first analyzed the expression pattern of e2f5 using whole-mount in situ hybridization. e2f5 showed tissue-specific expression at early developmental stages, with high levels of expression in the olfactory placode, otic vesicle and pronephric ducts at the 10-somite stage (Fig 1A) and 24 hours post-fertilization (hpf) (Fig 1B). At 24hpf, e2f5 was also strongly expressed in the posterior spinal cord (Fig 1B).
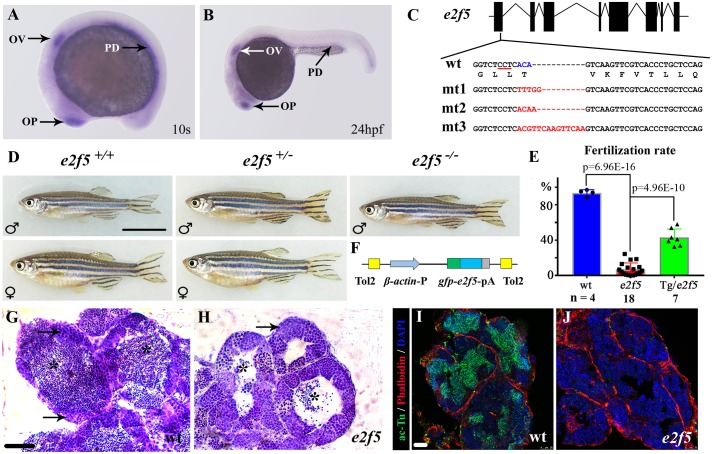
Fig 1. Mutation of e2f5 leads to male infertility.
(A-B) Whole-mount in situ hybridization showing expression of e2f5 at 10-somite stage (10s) and 24 hpf. OP, olfactory placode; OV, otic vesicle; PD, pronephric duct. (C) Genomic structure and sequences of wild- type (wt) and three e2f5 mutant alleles. The underlined sequence in wt indicates PAM sequence of sgRNA target. (D) Phenotypes of male and female wild-type and e2f5 heterozygotes. Only fish exhibiting the male phenotype were present among homozygous mutants. (E) Bar graph showing the percentage of fertilization rates of wild-type, e2f5 homozygote and e2f5 homozygotes carrying Tg(β-actin:gfp-e2f5) transgene as indicated. The numbers of adult males investigated are listed at the bottom. (F) Diagram of the constructs for making gfp-e2f5 transgene. (G-H) H&E staining results showing testes of wild-type (G) and e2f5 homozygous mutants (H). Arrows indicate primary spermatocytes. Asterisks point to mature spermatozoa which were substantially reduced in the mutants. (I-J) Confocal images showing the staining of sperm flagella in wild-type (I) and e2f5 mutant testis (J) visualized with acetylated alpha-tubulin (ac-Tu) antibody. Nuclei and actin filaments were counterstained with DAPI and phalloidin, respectively. Scale bars: 1cm in panel D, 50 μm in panel G, H and 25 μm in panel I, J.
To elucidate the function of E2f5, we generated three zebrafish e2f5 alleles, all of which contained frameshift mutations predicted to cause premature termination during translation and a complete loss-of-function condition (Fig 1C). Surprisingly, animals homozygous for all three mutant alleles are viable. However, all homozygous e2f5 mutants develop exclusively as males. Of a total of 723 adults from crosses between heterozygote mutants, we identified 245 wild-type, 330 heterozygote and 148 homozygous mutants, implying a slightly decreased survival rate of homozygous mutants. While both wild-type and heterozygote fish displayed a similar ratio between male and female, none of the 148 homozygous mutants were female (Fig 1D). When further crossed to wild-type females, all of the homozygous mutants from all three mutant alleles displayed severe fertility defects (the percentage of fertilization rates in mutants ca. 3%, n = 18; vs wild-type 94%, n = 4) (Fig 1E). To further explore this phenotype, we generated a stable transgenic line of E2f5 fused to the C-terminus of GFP, expression of which is driven by the constitutive and ubiquitously active β-actin promoter (Fig 1F). The fertilization rate was significantly rescued in e2f5 mutant males carrying this transgene (Fig 1E). Moreover, this transgene also allowed the recovery of female e2f5 homozygous mutants (S1A and S1B Fig). These data suggest that the fertility defects are due to a bona fide requirement of E2f5 in the germline.
In adult wild-type males, fully developed testes are plump and display milky white color, while testes from e2f5 mutants are slender and the color is also quite transparent (S1C and S1D Fig). Furthermore, histological sections through the seminiferous tubules showed a large number of mature spermatozoa and spermatocytes in the wild-type, while the number of mature spermatozoa were reduced substantially in the mutant testes (Fig 1G and 1H). Immunostaining with acetylated tubulin antibody, which labels the flagella of mature spermatozoa, also showed the absence of mature spermatozoa in the mutant testes (Fig 1I and 1J). These results demonstrate that spermatogenesis is strongly impaired in the absence of E2f5 activity.
Spermatogenesis is arrested in prophase I of meiosis in e2f5 mutants
After DNA replication and sister chromatid formation is complete, primary spermatocytes enter the first meiotic division (prophase I). Absence of mature spermatozoa is suggestive of defective meiosis in e2f5 mutants. To begin to understand the defect in greater detail, we analyzed the expression of Synaptonemal complex protein 3 (Sycp3), a marker of meiotic prophase I. Sycp3 is detected first in the lateral element of the synaptonemal complex at leptotene, and remains as part of this complex during homologous synapsis at zygotene and pachytene stages. At the later diplotene and diakinesis stages, Sycp3 signals become weakened and is maintained as short patches between the sister chromatids (Fig 2A–2E) [32]. In e2f5 mutants, we observed similar distribution of Sycp3 at early stages (Fig 2F–2H). However, the percentage of spermatocytes at leptotene and zygotene stages were increased dramatically, while those at later stages were barely seen (Fig 2I), confirming a meiotic arrest of spermatogenesis.
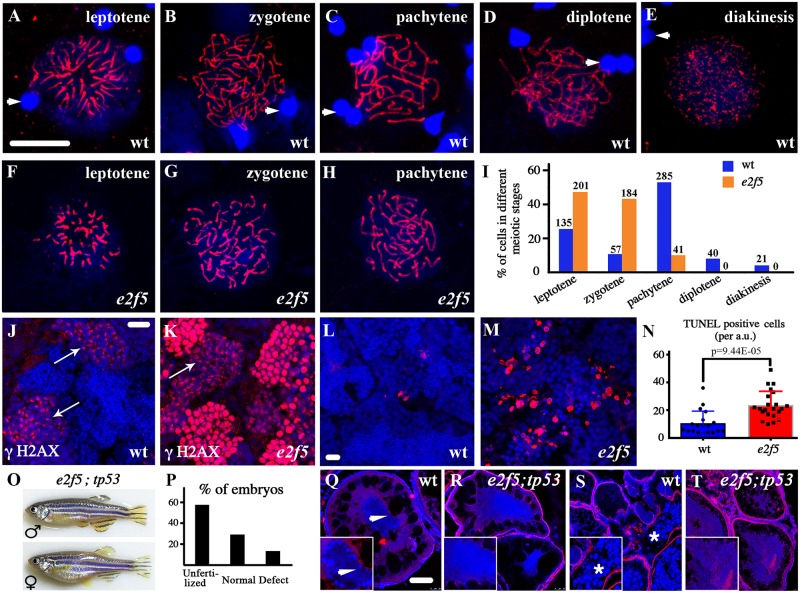
Fig 2. Spermatogenesis arrest in e2f5 mutants.
(A-H) Confocal images of primary spermatocytes at different prophase stages of meiosis I as indicated by anti-Sycp3 antibody staining (red). Arrowheads indicate spermatozoa nuclei from wild-type testis stained with DAPI in blue (A-E). (I) Bar graph showing the statistical results of the percentage of cells in different meiotic stages. The numbers of spermatocytes investigated are shown on top of each bar. (J-K) Staining of γH2AX (red) in the testes of wild-type and e2f5 mutant. Arrows point to the primary spermatocytes at leptotene and zygotene stages. (L-M) Confocal images showing apoptotic cells stained by TUNEL assay in red. (N) Bar graph with individual data points showing the number of TUNEL positive cells in wild-type and e2f5 mutant testes. (O) Phenotypes of e2f5;tp53 double mutants. (P) Bar graph showing the percentage of abnormal embryos produced from e2f5;tp53 double mutant females crossed with wild-type males. n = 441 from two double mutant females. (Q-T) Histological analysis of ovaries from wild-type and e2f5;tp53 double mutants. Inserted images are magnified views. Arrow in Q indicates peripheral localization of nucleoli stained by DAPI in stage II oocytes of wild-type ovary. The asterisk in S indicates cortical alveoli at later stage oocytes, which were visible under the confocal microscope due to autofluorescence. In panels J-M, Q-T, nuclei were counterstained with DAPI in blue. Scale bars: 10 μm in panel A-H, J-M and 50 μm in panel Q-T.
Phosphorylation of H2A histones is indispensable for the recruitment of repair factors to damaged DNA after DSB formation in leptotene and zygotene spermatocytes. In the wild-type, staining of phosphorylated histone H2AX (γ-H2AX) first appeared as clusters in the nuclear region at mid-leptotene stage, and then displayed a scattered pattern throughout the nucleus at late leptotene and early zygotene stages (Fig 2J). From mid-zygotene stage, γ-H2AX staining gradually disappeared and no staining was detected from late zygotene onward [33, 34]. In contrast, high levels of γ-H2AX staining was present in the mutant testes. In addition to the clusters of staining at mid-leptotene stage, most cells had strong γ-H2AX staining throughout their nuclei (Fig 2K), suggesting that spermatogenesis likely arrests at the zygotene stage in the absence of E2f5.
Apoptosis in e2f5 mutant spermatocytes
Germ cell apoptosis is a common consequence of meiotic arrest during spermatogenesis. The number of apoptotic cells increased significantly in the testes of e2f5 mutants (Fig 2L–2N). It has been shown earlier that mutation of the apoptotic gene tp53 (p53) can rescue the sex reversal defects caused by germ cell apoptosis [35, 36]. To test this, we generated e2f5-/-; tp53-/- double mutants to prevent cell apoptosis. Although double mutant males were still infertile, e2f5 female homozygous mutants could be recovered in the absence of p53 activity (4 female and 4 male double mutants were recovered among 128 adult fish derived from crossing between two e2f5+/-; tp53+/- heterozygotes) (Fig 2O), suggesting that p53 mediated germ cell apoptosis contributes to the absence of female e2f5 mutants. Interestingly, e2f5-/-; tp53-/- females were able to produce eggs when crossed to wild-type males, although the fertilization rates were low and most fertilized eggs developed abnormally at later stages (Fig 2P, S1E and S1F Fig). Further histological analysis showed that the symmetrical distribution of nucleoli near the nuclear periphery (stage IB oocyte) was consistently disrupted in oocytes from the double mutants (Fig 2Q and 2R). In addition, late-stage oocytes, normally distinguished by numerous cortical alveoli, were also barely seen in the mutant ovaries (Fig 2S and 2T). Thus, even though ovaries formed, oocyte development remained defective in the double mutant females.
Transcriptome analysis of e2f5 mutants
To further clarify the reason of meiotic arrest in e2f5 mutants, we performed RNA sequencing (RNA-seq) analysis on the testes of wild-type and e2f5 mutants. Sequencing results from two independent sample sets (in total from 10 wild-type and 10 mutants) revealed distinct gene expression patterns of e2f5 mutant testes relative to those of wild-type controls (Fig 3A). We found 1352 transcripts upregulated in e2f5 mutants compared to wild-type, whereas 763 transcripts were downregulated (FDR< = 0.05). Surprisingly, most genes associated with homologous recombination, including rad51, blm, ino80 and brca2 [37, 38], showed grossly normal expression levels in the mutants (Fig 3B, S2A Fig). However, the expression of dmc1, an essential gene involved in meiosis homologous recombination, was significantly downregulated in the mutants (Fig 3B, S2A Fig). To further validate this finding, we compared the expression levels of genes involved in meiotic HR via qRT-PCR. Of all the genes tested, dmc1 was again the only one significantly down regulated in e2f5 mutant testes (p = 4.21E-24), while other genes, including rad51, were expressed at similar levels to those of the wild-type controls (Fig 3C, S2B Fig). Expression level of tp53 was also increased, which is consistent with the TUNEL assay results (Fig 3C).
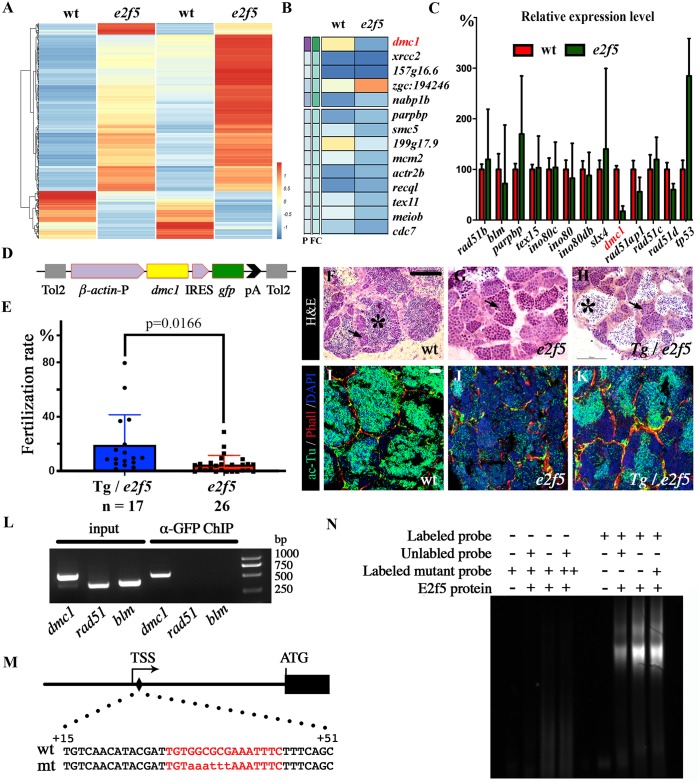
Fig 3. E2f5 binds to the promoter of dmc1.
(A) Heat map of RNA-seq transcriptome analysis from two sets of wild-type and mutant testes. (B) Expression heat map of genes involved in homologous recombination in wild-type and e2f5 mutants. The full list of genes analyzed is listed in S2A Fig. (C) qRT-PCR results showing the relative expression level of genes involved in homologous recombination in wild-type and e2f5 mutant testes. The expression of each gene in wild-type testes was set as 100%. The full list of genes analyzed is listed in S2B Fig. (D) Diagram of the construct used for generating Tg(β-actin:dmc1) transgenic fish. (E) Dot plot showing the fertilization rate of e2f5 mutants carrying the Tg(β-actin:dmc1) transgene. (F-H) H&E staining results showing testes from wild-type, e2f5 mutant and e2f5 mutant carrying Tg(β-actin:dmc1) transgene. Asterisks indicate mature spermatozoa. Arrows indicate spermatocytes. (I-K) Confocal images showing the staining of sperm flagella visualized with an anti-acetylated tubulin antibody in green in wild-type and mutant testes as indicated. Nuclei and actin filaments were counterstained with DAPI (blue) and phalloidin (red) respectively. (L) CHIP analysis of GFP-E2f5 binding to the promoter of dmc1. (M) Diagram showing the position and sequence of potential E2f5 binding site near the transcription start site (TSS) of the dmc1 gene. (N) EMSA analysis of the interaction between E2f5 protein and oligonucleotide probes corresponding to the wild-type and mutant E2F binding sites as indicated in panel M. “++” indicates that the amount of probes was doubled than those in other parallel experiments (+). Scale bars: 100 μm in panel F-H and 25 μm in panel I-K.
Overexpression of Dmc1 rescued spermatogenesis defects in e2f5 mutants
Mutation of DMC1 in humans and mice leads to male sterility due to defects in spermatogenesis [39]. In the medaka fish, loss-of-function of Dmc1 has been reported to produce abnormal sperm with multiple tails [40]. In line with this, we also observed abnormal sperm with two flagella in zebrafish e2f5 mutants (S3 Fig), suggesting that the decreased expression of Dmc1 could be the key issue underlying the defects in spermatogenesis in e2f5 mutants. To test this hypothesis directly, we generated a stable transgenic line expressing dmc1 under the control of the β-actin promoter (Fig 3D). With this transgene, the fertilization rates were significantly increased in e2f5 homozygous mutants (Fig 3E). The majority of e2f5 homozygous mutants (19 of 26 males) have fertilization rates lower than 5%, while 9 of 17 e2f5 mutants containing dmc1 transgene showed fertilization rates above 10%. Of note, in some of the transgenic lines, the fertilization rates increased to 77%, comparable to the rescue efficiency observed in Tg(β-actin:gfp-e2f5) transgenic e2f5 mutants (Figs 3E vs 1E). Histological analysis and immunostaining with acetylated tubulin antibody revealed that the number of mature spermatozoa was also substantially increased in the testes of the transgenic mutants (Fig 3F–3K).
E2f5 binds to the promoter of dmc1
To investigate whether E2f5 can regulate dmc1 gene expression directly, we performed chromatin immunoprecipitation (CHIP) analysis. We dissected testes from Tg(β-actin:gfp-e2f5) transgenic fish and incubated the lysate with anti-GFP antibody to pull down E2f5-binding DNA fragments. Following this, PCR analysis showed a –309 to +245 DNA fragment, near the transcription start site of dmc1, to be significantly enriched for E2f5-binding (Fig 3L). On the contrary, we failed to amplify any transcription elements of rad51 and blm genes, which are both involved in HR, but show normal expression levels in e2f5 mutants (S2A Fig). To confirm the binding activity of E2f5, we further performed electrophoretic mobility shift assays (EMSAs). By searching the promoter of dmc1, we identified one potential E2f5 binding site within the +29 to +44 region (Fig 3M). When incubated with recombinant zebrafish E2f5 protein, only FAM-labeled wild-type, but not mutant probe, showed band shift (Fig 3N). Together, these data suggest that E2f5 can transactivate the expression of dmc1 by directly binding to its promoter region.
Multiciliogenesis defects in e2f5 mutants
E2f5 plays an essential role during multicilia formation in MCCs by regulating the expression of genes required for centriole duplication and ciliary motility [23, 24]. Moreover, gene ontology analysis of biological functions of the transcriptome in e2f5 mutants also pointed to dramatic expression changes of genes required for ciliogenesis (S4 Fig). Whole-mount immunostaining results with anti-glycylated tubulin antibody showed that MCCs failed to develop in the mutant pronephric tubules and olfactory pits, while single cilia in spinal cord and ear hair cells developed normally (Fig 4A and 4B, S5A–S5H Fig). MCCs are enriched in the proximal tubule of zebrafish pronephros, which can be further subdivided into proximal convoluted tubule (PCT) and proximal straight tubule (PST) [41]. Interestingly, multicilia in the PST of pronephric tubules, but not PCT, recovered at 5dpf in the mutants (Fig 4C and 4D, S5G and S5H Fig). This could be due to the redundancy of function with E2f4. We therefore generated e2f4 mutants to address this issue (S5I Fig). Although MCCs developed completely normally in the e2f4 mutants, e2f4;e2f5 double mutants displayed a complete block in multiciliogeneis both in the pronephric tubules and the nose (Fig 4E–4H, S5J–S5M Fig). On the other hand, the recovery of MCCs in the PST was completely inhibited in the double mutants and only single cilia were present at 5dpf (Fig 4H).
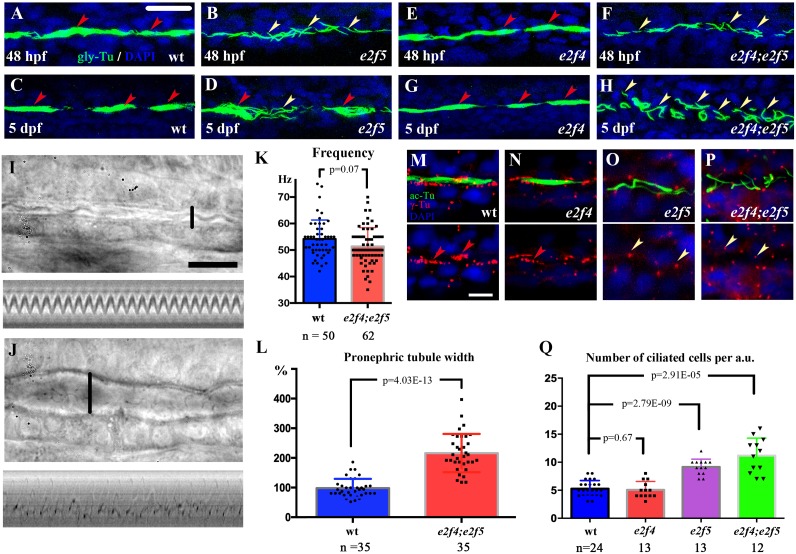
Fig 4. Multiciliogenesis defects in e2f5 mutants.
(A-H) Confocal images showing cilia in the PST region of the pronephros of wild-type and mutant larvae at different stages as indicated. Cilia were visualized with anti-glycylated tubulin antibody in green. Nuclei were labeled with DAPI in blue. Red arrowheads indicate multicilia bundles and yellow arrowheads indicate single cilia. (I-J) Still images from S1 and S2 Movies showing cilia in the PST of 5dpf wild-type (I) and e2f4;e2f5 (J) double mutants. Bottom images show kymographs of cilia movement. (K) Beating frequency of cilia in the pronephric tubules of wild-type and e2f4; e2f5 double mutants. (L) Dot plot showing the width of pronephric tubule lumen as indicated by vertical lines in panel I and J. (M-P) Confocal images showing cilia and basal bodies labeled with anti-acetylated tubulin (ac-Tu, green) and anti-γ tubulin (γ-Tu, red) antibodies in the PST of 48 hpf wild-type and mutant larvae as indicated. Staining of γ-tubulin is also indicated in the bottom to show the multiple basal bodies of MCCs (red arrowheads) and single ciliary basal bodies (yellow arrowheads). (Q) The number of cells bearing motile cilia per arbitrary unit (a.u.) in the PST region of wild-type and mutant larvae as indicated. Scale bars: 25 μm in panels A-H, 10 μm in panels I, J and 5μm in panels M-P.
To gauge whether the single cilia of the double mutants are motile or immotile, we assessed cilia motility in the pronephric tubules with high-speed video microscopy. In the PST of 5 dpf wild-type larvae, coordinated cilia beating formed a rhythmic sinusoidal wave, a characteristic pattern of multicilia motility (Fig 4I, S1 Movie) [42]. By contrast, in e2f4; e2f5 mutants, cilia movement appeared to be uncoordinated (Fig 4J, S2 Movie). Although ciliary beat frequency was similar to that of multicilia in wild-type embryos, the pronephric tubules were significantly dilated (Fig 4K–4L). Noticeably, all the cilia in the PST were single motile cilia, and the number of cells bearing cilia appeared to be increased in the mutants (S2 Movie). To further examine this, we labeled cilia and basal bodies with anti-acetylated alpha-tubulin and gamma-tubulin antibodies, respectively. In wild-type and e2f4 mutant larvae, each multicilia bundle arose from numerous basal bodies (Fig 4M and 4N). In contrast, each cilium was associated with a single basal body in e2f5 and e2f4;e2f5 double mutants (Fig 4O and 4P). We compared the number of cells bearing cilia in the PST region and found the number of ciliated cells increased significantly (Fig 4Q). Together, these data suggest that differentiation of the MCCs is interrupted in e2f4; e2f5 double mutants.
Cilia are not present in the PST principal cells
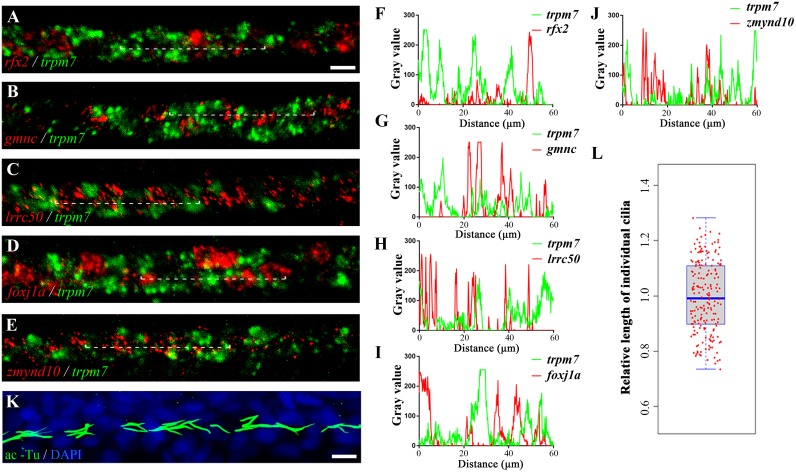
Principal cells have been suggested earlier to form cilia in the pronephric duct region [13, 18]. To clarify from where the extra motile cilia arose in the e2f4;e2f5 mutants, we further investigated ciliogenesis in the PST region. Using a stable Tg(β-actin:Arl13b-GFP) transgene which labels cilia in the pronephric duct, we observed that all the cilia in the PST region of wild-type embryos were motile at 24 hpf (S3 Movie). We further performed double in situ hybridization analysis using trpm7, a principal cell marker, together with several genes required for ciliary motility, including rfx2, foxj1a, zmynd10, lrrc50, as well as gmnc, a marker for multiciliated cells. The expression of trpm7 and all of these motile cilia related genes showed clearly non-overlapping expression pattern (Pearson’s Correlation Coefficient were all close to 0), suggesting that these motile cilia were not differentiated from principal cells (Fig 5A–5J, S6A Fig). Moreover, cilia were of similar length in this region when labeled with anti-acetylated tubulin antibody (Fig 5K and 5L). These findings suggest that contrary to previous reports [13, 18], motile cilia in the PST region do not differentiate on principal cells.
Fig 5. Cilia are not present in the PST principal cells.
(A-E) Double fluorescence in situ hybridization results showing the expression of trpm7 and different motile cilia related genes as indicated. (F-J) Line profile plots showing the pixel intensities of green and red channels along the dotted line in panels A-E. (K) Confocal image showing cilia in the PST region of a 24 hpf wild-type larva. Cilia were visualized with anti-acetylated tubulin antibody in green and nuclei were counterstained with DAPI in blue. (L) Box plot graph showing relative length of 198 individual cilia from 33 embryos. Each dot represents a single cilium. Scale bars: 10 μm.
E2f5 determines principal cell differentiation by regulating jag2b expression in MCCs
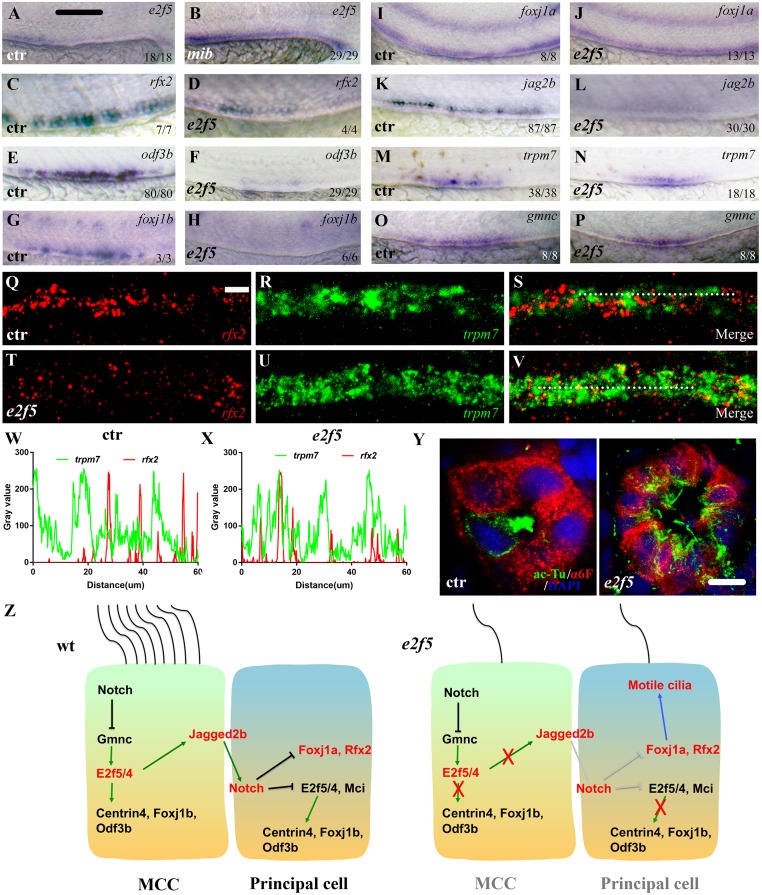
As mentioned before, during zebrafish pronephros development, the fate of MCCs and principal cells is determined through the Notch signaling pathway. Zebrafish mind bomb (mib) mutants are deficient in a ubiquitin ligase essential for Notch signaling, and thus, develop supernumerary MCCs in the pronephric ducts (S7A–S7J Fig) [13]. Consistent with this and the requirement of e2f5 in MCC formation, the expression of e2f5 was expanded in mib mutants (Fig 6A and 6B). We next examined the expression of a suite of genes associated with MCC development in the e2f5 mutants to uncover the exact defect in the MCC developmental program. We observed that the expression of rfx2, foxj1b, odf3b and cetn4, which are highly expressed in the MCCs, was strongly reduced in e2f5 mutants, further confirming the defect in multiciliogenesis (Fig 6C–6H, S7K and S7L Fig). On the other hand, the expression of foxj1a, a master regulator for motile cilia formation, was maintained in the mutants, suggesting different roles of foxj1a and foxj1b during MCC differentiation (Fig 6I and 6J).
Fig 6. Ectopic motile cilia developed in the principal cells of e2f5 mutants.
(A-P) Whole-mount in situ hybridization results showing the expression of e2f5 and other marker genes in the pronephric duct of 24 hpf control and mutant larvae as indicated. The numbers in the bottom right-hand corners indicate the numbers of embryos with similar staining results (left) and total numbers of embryos analyzed (right). (Q-V) Fluorescence in situ hybridization results showing the expression of rfx2 (red) and trpm7 (green) in the pronephric tubule of 36 hpf wild-type control (Q-S) and e2f5 mutant larvae (T-V). (W-X) Line profile plots showing the pixel intensities of green and red channels along the dotted line in panels S and V. (Y) Immunofluorescence results showing the staining of anti-acetylated tubulin (ac-Tu) and α6F antibodies on cross-sections through the pronephric tubules of 5dpf control and e2f5 mutant larvae. (Z) Model illustrating dual roles of E2f5 during multiciliogenesis. In the absence of E2f5 (maybe also E2f4), both MCC and principal progenitor cells developed a single motile cilium. Scale bars: 500 μm in panels A-P, 10 μm in panels Q-V and 5 μm in panel Y.
Unexpectedly, the expression of jag2b was also dramatically diminished in e2f5 mutants (Fig 6K and 6L). Inhibition of Jag2b expression leads to MCC hyperplasia at the expense of principal cells [13, 18]. To test whether the number of MCCs and principal cells changed in e2f5 mutants, we further stained the mutants with several MCC and principal cells markers. The expression of trpm7, together with other markers for transporting epithelial cells were unchanged in the mutants (Fig 6M and 6N, S7M–S7T Fig). Furthermore, the expression of gmnc, an early MCC differentiation marker was also unchanged (Fig 6O and 6P). These results suggest that although lateral inhibition was diminished, the number of the progenitor cells of MCCs and principal cells was maintained in the mutants. We hypothesized that extra motile cilia in the PST region of pronephric tubules of the e2f5 mutants may come from principal cells. To test this, we performed double fluorescence in situ hybridization assay with trpm7 and rfx2 genes. Rfx2 is a central regulator for ciliogenesis and also a marker for MCCs [18]. In wild-type larvae, rfx2 and trpm7 displayed a mutually exclusive expression pattern (Fig 6Q–6S and 6W). By contrast, rfx2 and trpm7 transcripts partially colocalized in the mutant pronephric ducts (Fig 6T–6V and 6X). Pearson’s Correlation Coefficient analysis also suggested a positive correlation in the expression of these two genes in the mutants (S6B Fig). Together, these data suggest that some of the principal cells expressed rfx2 in the mutants. Indeed, double immunostaining with α6F (antibody against chick alpha-1 subunit of the Na+/K+ ATPase) and acetylated-alpha tubulin antibody showed that cilia developed in the principal cells of e2f5 mutants, but not in those of wild-type larvae (Fig 6Y).
Together, all of these findings suggest that E2f5, possibly together with E2f4, is required both for multiciliated and principal cell differentiation. In e2f5 single or e2f4; e2f5 double mutants, the progenitor cells of MCCs and principal cells developed but failed to further undergo their specific differentiation programs. Instead, both MCCs and principal cells developed single motile cilia (Fig 6Z).
Discussion
During cell-cycle regulation, E2F5 has been recognized as a transcriptional repressor that inhibits gene expression at G1 phase. In this study, we provide data to show that E2f5 also functions as a transcription activator. In particular, E2f5 activates the transcription of dmc1 to promote homologous recombination during meiosis. Our data, together with previous reports, suggest that E2f5 also functions as a transactivator to control the expression of genes required for centriole duplication and ciliogenesis during MCC differentiation. Interestingly, when investigating the role of E2f5 through luciferase reporter assay, we found that E2f5 can function as a transcriptional repressor in cultured cells (S8A and S8B Fig). Furthermore, we generated an e2f5 mutant line carrying the inducible transgene Tg(fabp10:rtTA2s-M2;TRE2:EGFP-krasG12V), which contains a liver-specific double transgene to induce the expression of EGFP- KrasG12V specifically in the liver that leads to excessive cell proliferation and subsequent carcinogenesis [43, 44]. e2f5 mutant fish developed significantly enlarged and hyperplastic livers than wild-type siblings, providing corroborating evidence that E2f5 does play an inhibitory role during cell proliferation (S8C and S8D Fig). Thus, our present data together with earlier studies suggest that E2f5 is an ambivalent transcription factor, acting as an activator as well as repressor during development, in a context-dependent manner.
During sexual maturation, juvenile zebrafish usually first develop presumptive ovaries that contain gonocytes, early meiotic oocytes, and perinucleolar oocytes. Later, oocytes in animals that will become females continue to mature. By contrast, those fish with oocyte degeneration and spermatogonia and spermatocyte development develop as males [45]. Whole-mount in situ hybridization with vasa, a germ cell marker, showed that the number of primordial germ cells (PGCs) appeared to be normal in e2f5 mutants at early stages. Recovery of females among e2f5; tp53 double mutants indicates that oocyte apoptosis is the main reason for the loss of females among e2f5 mutants. These findings also suggest that similar to spermatocyte apoptosis, E2f5 may also regulate homologous recombination during oocyte meiosis. Meiosis arrest in e2f5 mutants is similar to that reported for brca2 and fancl mutants [35, 36, 46]. Both Brca2 and Fancl function to repair Spo11-induced double strand breaks to initiate meiotic recombination by forming complex with Rad51 and Dmc1 [28]. These data indicate that mutations affecting factors involved in DSB repair will result in meiosis arrest and germ cell apoptosis, which finally causes the male infertility phenotype. We have shown here that E2f5 binds directly to dmc1 promoter and that the downregulation of dmc1 expression is the major factor accounting for HR defects in e2f5 mutants. Although overexpression of Dmc1 increased male fertility in e2f5 mutants, we failed to recover mutant females, suggesting that other genes regulated by E2f5 also contributes to proper oocyte development.
We have also extended and further clarified the role of E2f5 in the development of the MCCs. Our data are consistent with a model of E2f5 playing cell autonomous and non-cell autonomous functions for the differentiation of the MCCs and the principal cells. e2f5 expression is regulated by Notch signaling, the major signaling pathway regulating MCC differentiation. E2f5, by interacting with Gmnc, activates the expression of genes required for basal body amplification and multiciliogenesis in the MCCs. This may also include the expression of Jag2b, which activates Notch signaling in the neighboring principal cells and prevents them from adopting the MCC fate (Fig 6Z). In e2f5 mutants, MCCs failed to form due to the absence of key genes required for centriole duplication. Interestingly, expression of gmnc, an early marker for MCCs, is unaffected in e2f5 mutants (Fig 6O and 6P). Gmnc functions both in the initial step for MCC precursor specification and is also required for later multiciliogenesis [47]. These results suggest that multiciliated progenitor cells develop in the mutants, and the numbers of these cells are unchanged. In the neighboring principal cells, the expression of several ciliary genes is de-repressed due to the absence of inhibitory Notch signaling from the loss of Jag2b in the MCCs. Consequently, the expression of foxj1a is activated in these cells, which promotes the formation of a single motile cilium. Interestingly, our data also suggest a functional difference of foxj1a and foxj1b during multicilia formation. Foxj1b likely contributes to multiciliogenesis in MCCs, while Foxj1a is a general factor for motile cilia differentiation. In gmnc mutants, the expression of foxj1b is absent while foxj1a remains unchanged, which further confirms their functional diversity [19]. Finally, principal cell markers, including trpm7, appear to be expressed normally in the e2f5 mutants. Together, these results suggest that both MCCs and principal cell precursors are specified in the mutants, while multiciliation and lateral inhibition is interrupted due to the loss of E2f5, and this finally results in the formation of single motile cilia in both cell types.
In conclusion, our study shows that besides its function as a transcription repressor during cell division, E2f5 also functions as a transcription activator both in spermatogenesis and multiciliogenesis. This information will help us to further appreciate the genetic complexity in the function of the E2f factors during embryonic and post-embryonic development as well as in adult physiology.
Materials and methods
Ethics statement
All zebrafish study was conducted according standard animal guidelines and approved by the Animal Care Committee of Ocean University of China (Animal protocol number: OUC2012316).
Zebrafish strains and mutants
All zebrafish strains were maintained at 14-hour light / 10-hour dark cycle at 28.5°C. mibta52b mutant line was obtained from Dr. Anming Meng. tp53zdf1 mutant line was obtained from Dr. Jianfeng Zhou. The Tg (fabp10:rtTA2s-M2;TRE2:EGFP-krasG12V) transgenic line was a gift from Dr. Zhiyuan Gong. Zebrafish e2f4 and e2f5 mutants were generated with CRISPR/Cas9 system with the following target sequences: e2f4: 5’- AGATCTCAAGTTGGAACTGG -3’, e2f5: 5’-GGGTGACGAACTTGACTGTG -3’. The single guide RNA and Cas9 mRNA were synthesized using Ambion’s MEGAshortscript T7 Transcription Kit (AM1354). Guide sgRNAs and Cas9 mRNA were co-injected into zebrafish embryos at the one-cell stage.
Transgenic lines
The transgenic fish expressing e2f5 or dmc1 were generated using Multisite Gateway technology. Briefly, the open reading frame of e2f5 and dmc1 was amplified from zebrafish cDNA library and cloned into pDONR vector using BP reaction (Invitrogen). For e2f5 gene, the ORF was cloned into pDONRP2R-P3 vector, then multisite gateway recombination was performed with the following constructs: pDEST vector, p5E-β-actin, pME-EGFP and pDONRP2R-P3-e2f5. The dmc1 final construct was generated through multisite gateway with pDEST vector, p5E-β-actin, pDONR221-dmc1 and p3E-IRES-EGFP. Multisite gateway cloning was performed according to the standard protocol from Invitrogen (Life Technologies).
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed according to the standard protocol. The primers used to amplify genes involved in the development of MCCs and principal cells are listed in S1 Table. The clcnk, slc12a1 and slc4a4a genes were obtained from Dr. Ying Cao. Double fluorescent in situ hybridization was carried out using TSA-plus Fluorescein System (Perkin Elmer) according to manufacturer’s protocol. Images were captured with Leica M165FC microscope or Leica TCS SP8 confocal microscope.
Immunohistochemical staining, immunofluorescence and TUNEL assay
Zebrafish testes were dissected and fixed in 4% paraformaldehyde (PFA) (w/v) in PBST overnight at 4°C. After gradual dehydration through 30%, 50%, 75%, 95% and 100% ethanol, the testes were embedded in JB4 embedding medium (Polysciences Inc.). Sections and H&E staining were performed as previously described [44]. For immunofluorescence on whole-mount larvae or cryosections through the testis, the following antibodies were used: mouse anti-acetylated tubulin (sigma, T6793), rabbit anti-acetylated tubulin (Cell Signaling Technology, 5335S), anti-glycylated tubulin (EMD, TAP952), anti- γ tubulin (sigma,T5326), anti-γ-H2Ax (Genetex), and a6F (Developmental Studies Hybridoma Bank). TUNEL assay was performed using the in situ cell death detection kit (Roche) according to standard protocols from the manufacturer.
Quantitative PCR
Testes from e2f5 mutants or control siblings were dissected to isolate total RNA. cDNA was synthesized using PrimeScript 1st strand cDNA Synthesis Kit (Takara). qPCR was performed on the Step One real-time PCR system (Thermo Scientific) using the Eva-Green Master Mix (ABM). The primers used for qPCR are listed in Supplementary S2 Table. Relative gene expression levels were quantified using the comparative Ct method (2−ΔΔCt method) based on Ct values for target genes and zebrafish β-actin.
Chromatin-immunoprecipitation assay
Testes from Tg(β-actin:GFP-e2f5) transgenic fish were first crosslinked in 1.85% PFA for 15 min at room temperature, and subsequently quenched with 0.125M glycine for 5 min with gentle shake, then homogenized with nuclei lysis buffer (50 mM Tris-HCl pH8.0, 10 mM EDTA, 1% SDS, 1 mM PMSF and protease inhibitor APL). The sample was sonicated using Bioruptor sonicator (Diagenode) for 15 min with 30s ON and 30s OFF at high power. 50 μl sonicated samples were saved as input control. Then, 25 μl GFP-Trap-A (Chromotek) was added to the sonicated samples and incubated overnight at 4°C with rotation. After 1 min centrifuge at 1000g, the beads were washed sequentially with wash buffer I (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 1% TritonX-100, 150 mM NaCl and 0.1% SDS), wash buffer II (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 1% TritonX-100, 500 mM NaCl and 0.1% SDS), wash buffer III (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% NP-40 and 1% Deoxycholate Sodium Salt) and TE buffer (10 mM Tris-HCl pH 8.0 and 1mM EDTA). Finally, 500 μl elution buffer (25 mM Tris-HCl, 10 mM EDTA and 0.5% SDS) were added to elute the DNA-protein complex at 65° for 15 min. The supernatant was transferred to a new tube and 20 μl 5M NaCl was added, then incubated at 65° for 4 hours to reverse crosslinks. Finally, the immunoprecipitated DNA were purified using Phenol/Chloroform/Isoamyl alcohol and further analyzed with regular PCR. The following primers were used for PCR analysis: dmc1 forward: 5’-GCTGCAAAGCTGAAGTATTC-3’, reverse 5’- CACGTAATTTGGTAACAAG-3’; rad51 forward 5’- TGCCAGTAGTTTGAATGAGC -3’, reverse 5’- TCACTCACCCGCTAAGCTAC -3’ and blm forward 5’- TAGTCCTATTATTAGCGCCG -3’, reverse 5’- AACCAAATAACACAACAAAG -3’.
Electrophoretic mobility shift assay (EMSA)
The coding region of e2f5 was amplified from cDNA library and ligated into pDONR221 Gateway entry vector. After LR reaction, full-length coding sequence was further cloned into pET30A vector. The expression of E2f5 was induced by 0.5 mM IPTG and purified with nickel-nitrilotriacetic acid resin column (GE Healthcare). All the synthetic oligonucleotides were FAM-labelled at the 5-terminal (Beijing Genomics Institute). The oligonucleotides were first dissolved in distilled water to a final concentration of 100 μM and annealed with their complementary oligonucleotides to a final concentration of 25 μM. The binding reaction was carried out with 3 μg purified protein, 1μl annealed oligonucleotide in binding buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT and 4% glycerol). After 40 min incubation at room temperature, the samples were analyzed by 6.5% native polyacrylamide gel electrophoresis and detected using ChemiDoc MP Imaging System (Bio-Rad).
Chromosome spreading
To determine the stages of meiosis, testes were dissected from wild-type and mutant zebrafish and incubated in 50 μl 1 X PBS. After gently tearing up using a pair of tweezers, 10 μl liquid was transferred into 300 μl hypotonic solution (100 μl 1xPBS plus 200 μl double distilled water) onto an adhesive slide. After 25 min incubation, the cells were fixed in 4% PFA for 5 min, washed three times with PBS, then blocked 30 min with blocking solution (10% goat serum, 3% BSA, 0.05% TritonX-100). The slides were further incubated with Anti-Sycp3 antibody (Abcam, ab150292) for 4–5 hours at room temperature. Then after a brief wash, the secondary antibody was added and the slides were incubated at 37°C for 2 hours. Images were collected with a Leica Sp8 confocal microscope.
RNA-seq transcriptome analysis
Testes from five wild-type or e2f5 mutants were dissected and total RNA was isolated using Trizol according to standard protocol. RNA sequencing was performed by Novogene using Illumina HiSeq X Ten (Novogene Bioinformatics Technology Co., Ltd., Tianjin, China). To reduce potential sequencing errors, we repeated this experiment with Annoroad (Annoroad Gene Technology Co., Ltd, Beijing, China). These two sample sets displayed similar changes in gene expression pattern. Both of these RNA-seq data have been deposited to the Sequence Read Archive (SRA) database under accession numbers SRR10854654, SRR10854655, SRR10854656 and SRR10854657 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA599540). We have mainly shown the data that we obtained from Novogene. For data processing, the reads were mapped to the reference genome index constructed by hisat2 (version 2.1.0) using Danio_rerio.GRCz10.90.gtf and Danio_rerio.GRCz10.dna.chromosome.1.fa. The raw counts of the genes were obtained using featureCounts in the subread version 1.6.2 package. Sample normalization and differential expression analysis was performed using edgeR Bioconductor package. Then, the relative expression level RPKM of the differential genes was calculated according to the traditional formula, and the gene expression heat map was obtained using the pheatmap Bioconductor package.
High-speed video microscopy
Cilia motility in the PST of the zebrafish pronephric tubules was recorded at 5dpf. Briefly, wild-type or mutant embryos were first treated with 30 μg/ml 1-phenyl 2-thiourea (PTU) to inhibit pigmentation from 24 hpf. At 5dpf, the larvae were anesthetized with 0.01% tricaine, and then placed on top of a cover glass. After removing most of the embryo medium, the cover glass was placed upside down on the center of a depression slide containing 50 μl embryo medium. Cilia movement in the pronephric tubules was recorded with 100 X oil objective on a Leica Sp8 confocal microscope equipped with a high-speed camera (Motion- BLITZ EoSens mini1; Mikrotron, Germany). Cilia movement was captured at rates of 500 frames per second, and playback was set at 25 frames per second.
To record cilia motility in the Tg(β-actin:Arl13b-GFP) embryos, 24 hpf zebrafish embryos were embedded in the same way as 5dpf larvae and movement of beating cilia was recorded using an Olympus IX83 microscope equipped with a 60X, 1.3 NA objective lens, an EMCCD camera (iXon+ DU-897D-C00-#BV-500; Andor Technology), and a spinning disk confocal scan head (CSU-X1 Spinning Disk Unit; Yokogawa Electric Corporation). Ciliary motility movies were acquired using μManager (https://www.micro-manager.org) at an exposure time of 25 ms, and playback speed of 10 fps. Image processing was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Luciferase assay
Partial dmc1 promoter containing wild-type or mutant E2f5 binding site was cloned into the pGL3 Luciferase Reporter Vector. For the effector vector, full length zebrafish e2f5 was first cloned into pDONR221vector using BP reaction, then further cloned into pCS2-6xMyc destination vector using LR reaction. We co-transfected the effector and reporter vectors, along with the Renilla luciferase vector, into HEK293 cells using Lipofectamine 2000 (Invitrogen) with standard protocol. Luciferase activity was measured using the Dual Luciferase reporter assay kit (Promega) at 48 hours after transfection.
Pharmacological treatments
To test the role of E2f5 during cell proliferation in vivo, we generated e2f5 heterozygotes carrying the Tg(fabp10:rtTA2s-M2;TRE2:EGFP-krasG12V) transgene, which drives liver-specific expression of an activating mutation of KRAS to induce excessive cell proliferation after drug treatment. We crossed e2f5 heterozygotes carrying the EGFP-krasG12V transgene with e2f5 heterozygotes, and compared the liver size between e2f5 homozygotes carrying the EGFP-krasG12V transgene with wild-type carrying EGFP-krasG12V transgene. The embryos were treated with 60 μg/mL doxycycline hydrochloride (Sangon Biotech, Shanghai, China) or DMSO starting from 60 hours post fertilization and harvested at 1, 2 and 3 days after treatment. Image capture and statistical analysis were as described previously[44].
Statistical analysis
Statistical analysis was performed using Microsoft Excel or GraphPad Prism 6 software. All data were presented as mean ±S.D. as indicated in the figure legends. Statistical significance was evaluated by means of the two-tailed Student’s t-test for unpaired data. Pearson Coefficient analysis were performed using the Leica Sp8 confocal colocalization software. A value of p<0.05 was considered statistically significant. All the p values are indicated in the figures.
Supporting information
(A-B) External phenotypes of male and female e2f5 mutants rescued by gfp-e2f5 transgene under the regulation of the ubiquitously expressed β-actin promoter. (C-D) Dissected testes from wild-type (C) and e2f5 adult mutant (D). (E-F) External phenotypes of 72 hpf embryos collected from crosses between wild-type male and e2f5;tp53 double heterozygous female (E) or e2f5;tp53 homozygous female (F). Scale bars: 1 cm in panel A, B and 1mm in panel C-F.
(TIF)
(A) Heat map showing relative expression of genes involved in homologous recombination from RNA-seq transcriptome analysis. The genes are listed according to fold change (FC) and p-value. (B) qPCR results showing the relative expression level of genes involved in homologous recombination in wild-type and e2f5 mutant testes.
(TIF)
(A-D) Confocal images showing the phenotypes of mature spermatozoa in wild-type (A) and e2f5 mutants (B-D). Flagella were labeled with anti-acetylated tubulin antibody in green. Nuclei were stained with DAPI in blue. Scale bar: 5 μm.
(TIF)
The genes were clustered according to biological processes. The colors of the bars indicate p adjust value of different GO terms.
(TIF)
(A-H) Confocal images showing cilia in the cristae (A-B), spinal canal (SC) (C-D), olfactory pit (OP) (E-F) and PCT of the pronephros (G-H) in 5dpf wild-type and e2f5 mutants. Cilia were visualized with anti-glycylated tubulin antibodies in green and nuclei were counterstained with DAPI in blue. Arrow in (E) points to cilia bundle of MCCs and asterisk indicates single primary cilia. (I) Diagram showing the genomic structure of e2f4 locus. The sequences of the wild-type and e2f4 mutant alleles generated with CRISPR/Cas9 method is shown at the bottom. The sgRNA target sequence and corresponding PAM region are also labeled. (J-M) Confocal images showing the localization of basal bodies visualized with anti-γ tubulin (green) in the olfactory pits of wild-type and mutant larvae as indicated. Arrows point to MCCs characterized by multiple basal bodies. Inserted images are magnified views. Nuclei were stained with DAPI in blue and F-actin was counterstained with phalloidin in red. Scale bars: 10 μm.
(TIF)
(A) Colocalization analysis of different genes as indicated in 24 hpf wild-type embryos. (B) Colocalization analysis of rfx2 and trpm7 expression in the PST of 36 hpf wild-type or e2f5 mutants as indicated. In panels A and B, each dot represents one zebrafish embryo analyzed.
(TIF)
Whole mount in situ hybridization results showing the expression of ciliary genes (A-H, K-L) and marker genes for transporter cells (I-J, M-T) in the pronephric duct of 24 hpf control and mutant embryos as indicated. The numbers of positive/total analyzed embryos are shown in the bottom right-hand corner of each panels.
(TIF)
(A) Diagram showing the constructs used for reporter assays. Part of the promoter region of dmc1 was used to drive the expression of the luciferase gene. The E2f5 binding site is also indicated. The mutant sequence of E2f5 binding site is the same as used for EMSA assay. (B) Bar graph showing the relative luciferase activity in the different combinations as indicated. Increase in the amount of E2f5 constructs further inhibited luciferase activity. (C) Representative images showing the liver of control and e2f5 mutants as highlighted by EGFP-KrasG12V expression at different time points after doxycycline treatment. dpt: days post treatment. (D) Dot plot showing the average liver size in wild-type or e2f5 mutants at different time points after treatment. Scale bar: 200 μm.
(TIF)
(MOV)
(MOV)
Cilia were visualized using Tg(β-actin:Arl13b-GFP) transgene. Scale bar: 5 μm.
(AVI)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. A. Meng for providing mind bomb mutants, Dr. J. Zhou for providing tp53 mutants, Dr. Z. Gong for providing the EGFP-krasG12V transgenic line and Dr. G. Ou for helping with the spinning disk confocal. We also thank Dr. Y. Cao for providing probes for in situ hybridization analysis.
Data Availability
RNA-Seq data are available from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra), under accession numbers SRR10854654, SRR10854655, SRR10854656 and SRR10854657.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31991194), the Marine S&T Fund of Shandong Province for the Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0406-2) and by grants from the Fundamental Research Funds for Central Universities (Grants 201941004) to C.Z. and funds from the Agency for Science, Technology and Research (A*STAR) of Singapore to S.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DeGregori J., Johnson D. G., Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Current molecular medicine 6, 739–748 (2006). 10.2174/1566524010606070739 [DOI] [PubMed] [Google Scholar]
- 2.Chen H. Z., Tsai S. Y., Leone G., Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature reviews. Cancer 9, 785–797 (2009). 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick F. A., Goodrich D. W., Sage J., Dyson N. J., Non-canonical functions of the RB protein in cancer. Nature reviews. Cancer 18, 442–451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ertosun M. G., Hapil F. Z., Osman Nidai O., E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine & growth factor reviews 31, 17–25 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Attwooll C., Lazzerini Denchi E., Helin K., The E2F family: specific functions and overlapping interests. The EMBO journal 23, 4709–4716 (2004). 10.1038/sj.emboj.7600481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humbert P. O. et al. , E2F4 is essential for normal erythrocyte maturation and neonatal viability. Molecular cell 6, 281–291 (2000). 10.1016/s1097-2765(00)00029-0 [DOI] [PubMed] [Google Scholar]
- 7.Rempel R. E. et al. , Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Molecular cell 6, 293–306 (2000). 10.1016/s1097-2765(00)00030-7 [DOI] [PubMed] [Google Scholar]
- 8.Ruzhynsky V. A. et al. , Cell cycle regulator E2F4 is essential for the development of the ventral telencephalon. The Journal of neuroscience: the official journal of the Society for Neuroscience 27, 5926–5935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danielian P. S., Hess R. A., Lees J. A., E2f4 and E2f5 are essential for the development of the male reproductive system. Cell cycle 15, 250–260 (2016). 10.1080/15384101.2015.1121350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindeman G. J. et al. , A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes & development 12, 1092–1098 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spassky N., Meunier A., The development and functions of multiciliated epithelia. Nature reviews. Molecular cell biology 18, 423–436 (2017). 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- 12.Brooks E. R., Wallingford J. B., Multiciliated cells. Current biology: CB 24, R973–982 (2014). 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Pathak N., Kramer-Zucker A., Drummond I. A., Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development 134, 1111–1122 (2007). 10.1242/dev.02806 [DOI] [PubMed] [Google Scholar]
- 14.Mori M. et al. , Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development 142, 258–267 (2015). 10.1242/dev.116855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsao P. N. et al. , Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 (2009). 10.1242/dev.034884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcet B. et al. , Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nature cell biology 13, 693–699 (2011). 10.1038/ncb2241 [DOI] [PubMed] [Google Scholar]
- 17.Deblandre G. A., Wettstein D. A., Koyano-Nakagawa N., Kintner C., A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126, 4715–4728 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Ma M., Jiang Y. J., Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS genetics 3, e18 (2007). 10.1371/journal.pgen.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou F. et al. , Gmnc Is a Master Regulator of the Multiciliated Cell Differentiation Program. Current biology: CB 25, 3267–3273 (2015). 10.1016/j.cub.2015.10.062 [DOI] [PubMed] [Google Scholar]
- 20.Terre B. et al. , GEMC1 is a critical regulator of multiciliated cell differentiation. The EMBO journal 35, 942–960 (2016). 10.15252/embj.201592821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbi M. et al. , GemC1 controls multiciliogenesis in the airway epithelium. EMBO reports 17, 400–413 (2016). 10.15252/embr.201540882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stubbs J. L., Vladar E. K., Axelrod J. D., Kintner C., Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nature cell biology 14, 140–147 (2012). 10.1038/ncb2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong Y. L., Zhang Y., Zhou F., Roy S., Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo. Developmental biology 443, 165–172 (2018). 10.1016/j.ydbio.2018.09.013 [DOI] [PubMed] [Google Scholar]
- 24.Ma L., Quigley I., Omran H., Kintner C., Multicilin drives centriole biogenesis via E2f proteins. Genes & development 28, 1461–1471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori M. et al. , Cytoplasmic E2f4 forms organizing centres for initiation of centriole amplification during multiciliogenesis. Nature communications 8, 15857 (2017). 10.1038/ncomms15857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H. et al. , The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nature cell biology 15, 1434–1444 (2013). 10.1038/ncb2880 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Copenhaver G. P., Meiotic Recombination: Mixing It Up in Plants. Annu Rev Plant Biol 69, 577–609 (2018). 10.1146/annurev-arplant-042817-040431 [DOI] [PubMed] [Google Scholar]
- 28.Neale M. J., Keeney S., Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 (2006). 10.1038/nature04885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sehorn M. G., Sigurdsson S., Bussen W., Unger V. M., Sung P., Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429, 433–437 (2004). 10.1038/nature02563 [DOI] [PubMed] [Google Scholar]
- 30.Cloud V., Chan Y. L., Grubb J., Budke B., Bishop D. K., Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012). 10.1126/science.1219379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Ines O. et al. , Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS genetics 9, e1003787 (2013). 10.1371/journal.pgen.1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bisig C. G. et al. , Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS genetics 8, e1002701 (2012). 10.1371/journal.pgen.1002701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito K., Siegfried K. R., Nusslein-Volhard C., Sakai N., Isolation and cytogenetic characterization of zebrafish meiotic prophase I mutants. Dev Dyn 240, 1779–1792 (2011). 10.1002/dvdy.22661 [DOI] [PubMed] [Google Scholar]
- 34.Mahadevaiah S. K. et al. , Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27, 271–276 (2001). 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Mari A. et al. , Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS genetics 7, e1001357 (2011). 10.1371/journal.pgen.1001357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shive H. R. et al. , brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc Natl Acad Sci U S A 107, 19350–19355 (2010). 10.1073/pnas.1011630107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao W., Wiese C., Kwon Y., Hromas R., Sung P., The BRCA Tumor Suppressor Network in Chromosome Damage Repair by Homologous Recombination. Annu Rev Biochem 88, 221–245 (2019). 10.1146/annurev-biochem-013118-111058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., McCorvie T. J., Yates L. A., Zhang X., Structural basis of homologous recombination. Cell Mol Life Sci 77, 3–18 (2020). 10.1007/s00018-019-03365-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagawa W., Kurumizaka H., From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J 277, 590–598 (2010). 10.1111/j.1742-4658.2009.07503.x [DOI] [PubMed] [Google Scholar]
- 40.Chen J. et al. , Disruption of dmc1 Produces Abnormal Sperm in Medaka (Oryzias latipes). Sci Rep 6, 30912 (2016). 10.1038/srep30912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond I. A., Davidson A. J., Zebrafish kidney development. Methods Cell Biol 100, 233–260 (2010). 10.1016/B978-0-12-384892-5.00009-8 [DOI] [PubMed] [Google Scholar]
- 42.Zhao C., Malicki J., Genetic defects of pronephric cilia in zebrafish. Mech Dev 124, 605–616 (2007). 10.1016/j.mod.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 43.Nguyen A. T. et al. , An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Disease models & mechanisms 5, 63–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang Y., Xie H., Zhao C., Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation. Genes (Basel) 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kossack M. E., Draper B. W., Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr Top Dev Biol 134, 119–149 (2019). 10.1016/bs.ctdb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Mari A. et al. , Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS genetics 6, e1001034 (2010). 10.1371/journal.pgen.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu H. et al. , Mcidas mutant mice reveal a two-step process for the specification and differentiation of multiciliated cells in mammals. Development 146 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-B) External phenotypes of male and female e2f5 mutants rescued by gfp-e2f5 transgene under the regulation of the ubiquitously expressed β-actin promoter. (C-D) Dissected testes from wild-type (C) and e2f5 adult mutant (D). (E-F) External phenotypes of 72 hpf embryos collected from crosses between wild-type male and e2f5;tp53 double heterozygous female (E) or e2f5;tp53 homozygous female (F). Scale bars: 1 cm in panel A, B and 1mm in panel C-F.
(TIF)
(A) Heat map showing relative expression of genes involved in homologous recombination from RNA-seq transcriptome analysis. The genes are listed according to fold change (FC) and p-value. (B) qPCR results showing the relative expression level of genes involved in homologous recombination in wild-type and e2f5 mutant testes.
(TIF)
(A-D) Confocal images showing the phenotypes of mature spermatozoa in wild-type (A) and e2f5 mutants (B-D). Flagella were labeled with anti-acetylated tubulin antibody in green. Nuclei were stained with DAPI in blue. Scale bar: 5 μm.
(TIF)
The genes were clustered according to biological processes. The colors of the bars indicate p adjust value of different GO terms.
(TIF)
(A-H) Confocal images showing cilia in the cristae (A-B), spinal canal (SC) (C-D), olfactory pit (OP) (E-F) and PCT of the pronephros (G-H) in 5dpf wild-type and e2f5 mutants. Cilia were visualized with anti-glycylated tubulin antibodies in green and nuclei were counterstained with DAPI in blue. Arrow in (E) points to cilia bundle of MCCs and asterisk indicates single primary cilia. (I) Diagram showing the genomic structure of e2f4 locus. The sequences of the wild-type and e2f4 mutant alleles generated with CRISPR/Cas9 method is shown at the bottom. The sgRNA target sequence and corresponding PAM region are also labeled. (J-M) Confocal images showing the localization of basal bodies visualized with anti-γ tubulin (green) in the olfactory pits of wild-type and mutant larvae as indicated. Arrows point to MCCs characterized by multiple basal bodies. Inserted images are magnified views. Nuclei were stained with DAPI in blue and F-actin was counterstained with phalloidin in red. Scale bars: 10 μm.
(TIF)
(A) Colocalization analysis of different genes as indicated in 24 hpf wild-type embryos. (B) Colocalization analysis of rfx2 and trpm7 expression in the PST of 36 hpf wild-type or e2f5 mutants as indicated. In panels A and B, each dot represents one zebrafish embryo analyzed.
(TIF)
Whole mount in situ hybridization results showing the expression of ciliary genes (A-H, K-L) and marker genes for transporter cells (I-J, M-T) in the pronephric duct of 24 hpf control and mutant embryos as indicated. The numbers of positive/total analyzed embryos are shown in the bottom right-hand corner of each panels.
(TIF)
(A) Diagram showing the constructs used for reporter assays. Part of the promoter region of dmc1 was used to drive the expression of the luciferase gene. The E2f5 binding site is also indicated. The mutant sequence of E2f5 binding site is the same as used for EMSA assay. (B) Bar graph showing the relative luciferase activity in the different combinations as indicated. Increase in the amount of E2f5 constructs further inhibited luciferase activity. (C) Representative images showing the liver of control and e2f5 mutants as highlighted by EGFP-KrasG12V expression at different time points after doxycycline treatment. dpt: days post treatment. (D) Dot plot showing the average liver size in wild-type or e2f5 mutants at different time points after treatment. Scale bar: 200 μm.
(TIF)
(MOV)
(MOV)
Cilia were visualized using Tg(β-actin:Arl13b-GFP) transgene. Scale bar: 5 μm.
(AVI)
(DOCX)
(DOCX)
Data Availability Statement
RNA-Seq data are available from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra), under accession numbers SRR10854654, SRR10854655, SRR10854656 and SRR10854657.