Abstract
For over a decade, phage display has proven to be of immense value, allowing selection of a large variety of genes with novel functions from diverse libraries. However, the folding and modification requirements of complex proteins place a severe constraint on the type of protein that can be successfully displayed using this strategy, a restriction that could be resolved by similarly engineering a eukaryotic virus for display purposes. The quite recently established eukaryotic molecular biology tool, the baculovirus display vector system (BDVS), allows combination of genotype with phenotype and thereby enables presentation of eukaryotic proteins on the viral envelope or capsid. Data have shown that the baculovirus, Autographa californica multiple nucleopolyhedrovirus (AcMNPV), is a versatile tool for eukaryotic virus display. Insertion of heterologous peptides and/or proteins into the viral surface by utilizing the major envelope glycoprotein gp64, or foreign membrane‐derived counterparts, allows incorporation of the sequence of interest onto the surface of infected cells and virus particles. A number of strategies are being investigated in order to further develop the display capabilities of AcMNPV and improve the complexity of a library that may be accommodated. Numerous expression vectors for various approaches of surface display have already been developed. Further improvement of both insertion and selection strategies toward development of a refined tool for use in the creation of useful eukaryotic libraries is, however, needed. Here, the status of baculovirus display with respect to alteration of virus tropism, antigen presentation, transgene expression in mammalian cells, and development of eukaryotic libraries will be reviewed.
I. Introduction
In the era of genomics and proteomics, direct coupling of proteins to their DNA‐coding sequence is extremely valuable as it holds potential to derive functional information from unknown open reading frames. Proteins can be identified by virtue of their unique functional properties and their encoding gene subsequently isolated. Replicating nanoparticles, such as bacteriophages, have proven ideal for this type of application as they can be designed to display peptides or proteins of interest on their surface while encapsidating the gene of interest. Due to the exceptional titers that phages can achieve, the diversity of the resultant prokaryote‐based libraries is very high. Successful examples of such a display technology include isolation of antibodies from large combinatorial libraries displayed on the surface of bacteriophages. Phage display, however, has notable limitations due to the simple posttranslational machinery provided by the prokaryotic host.
The eukaryote‐based baculovirus expression vector system (BEVS), primarily based on the use of Autographa californica multiple nucleopolyhedrovirus (AcMNPV), was developed during the 1980s (Luckow 1988, Miller 1988a, Miller 1988b, Miller 1989, Sherman 1979, Smith 1983). Complex animal, human, and viral proteins, requiring folding, subunit assembly, and/or extensive posttranslational modification, can be successfully expressed using this system (Kost et al., 2005). The successful and wide adoption of BEVS benefits the choice of AcMNPV as a candidate for the development of a safe eukaryotic display system aimed at proper presentation of antigens, gene delivery to mammalian cells as well as development of eukaryotic libraries.
II. Targeting of Baculoviral Vectors by Surface Display
Surface glycoproteins of enveloped viruses are attractive candidates for control and manipulation of cellular recognition. The limitations of prokaryotic display systems regarding posttranslational modifications and folding of the displayed proteins has led to the development of alternative eukaryotic display systems. During the last decade, the expression of foreign peptides and proteins on the baculoviral surface has been quite extensively studied and display has already been employed in a number of applications. Although an insect virus, the tropism and transduction efficiency of AcMNPV with respect to mammalian cells or tissues can be manipulated by a variety of techniques including mutation of the major viral envelope protein (gp64), incorporation of targeting peptides or antibodies into virions, and vector pseudotyping (Fig. 1 ).
Fig 1.
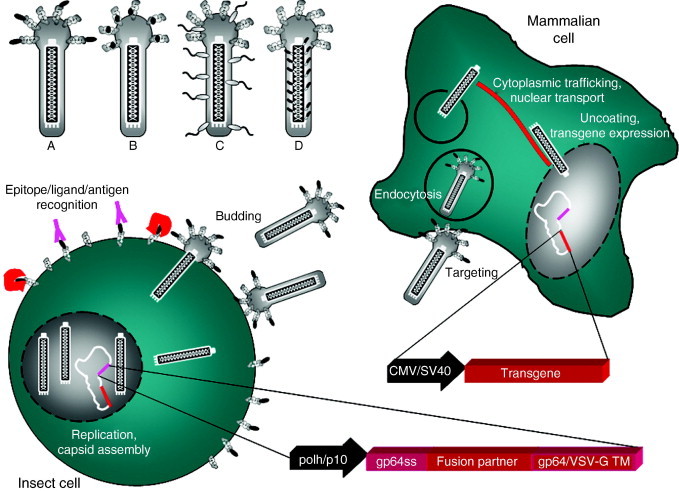
A generalized schematic outline of the multifunctional baculovirus display technology used for eukaryotic library development and mammalian gene delivery. Entry of baculovirus into mammalian cells is thought to be similar to that for insect cells. The receptor molecule(s) for baculovirus binding and entry are unknown, but they have been suggested to involve common cell surface components. The virus enters mammalian cells by endocytosis followed by low‐pH–induced membrane fusion with endosomes and capsid release into the cytoplasm. The capsid is then transported toward the nucleus along actin filaments and enters the nucleus presumably through nuclear pores followed by uncoating and release of the genome. Through incorporation of ligands with high and specific affinities into the virus envelope, it is possible to target baculoviral transduction to desired cell types expressing the receptor molecule for the displayed ligand. Foreign peptides or proteins can be displayed on the baculovirus envelope as N‐terminal (A) or internal (B) fusions to gp64, using a heterologous membrane anchor derived from VSV‐G (C) for example, or by fusion to the major capsid protein, vp39 (D). The displayed proteins are directed to the surface of the recombinant baculovirus‐infected insect cells using a signal sequence derived from the major AcMNPV envelope protein gp64 (gp64ss) for example. Genes encoding the fusion proteins are then coupled with a strong baculoviral promoter (e.g., polh or p10) for strong expression in insect cells. The vectors can be further equipped with an expression cassette encoding a reporter or a suicide gene under a mammalian promoter (e.g., CMV or SV40), enabling transduction monitoring in mammalian cells. Cell surface display can be applied in library screening for studying ligand–receptor interactions and antigen recognition. Display on the viral envelope provides possibilities for transductional targeting, whereas capsid display rather facilitates studies on intracellular trafficking as well as nuclear targeting of the virus.
The extensive diversity of mammalian cells that can be transduced by baculovirus vectors implies that the entry and uptake mechanisms of this insect virus by mammalian cells are universal (Kost and Condreay, 2002). Therefore, several strategies have been developed to restrict viral transduction to desired cell types. Baculovirus transduction has generally been considered as safe and nontoxic to mammalian cells, and cell growth has not been stalled even at notably high MOIs (Ho et al., 2004). Baculovirus enters insect cells by endocytosis followed by a low‐pH–induced fusion of the viral envelope with the endosomal membrane, consequently permitting viral entrance into the cytoplasm and nucleus (Dee and Shuler, 1997). Correspondingly, baculovirus is considered to enter mammalian cells via the same route (Fig. 1), as gene expression is inhibited by lysosomotropic agents that inhibit endosomal maturation (Boyce 1996, Hofmann 1995, van 2001). In addition to clathrin‐mediated endocytosis, baculovirus is presumably internalized by macropinocytosis (Matilainen et al., 2005). Although the receptor molecule(s) of AcMNPV are unknown, the cell surface molecules for the attachment and entry of the virus have been suggested to involve common constituents of the cell membrane including phospholipids or heparan sulfate proteoglycans (Duisit 1999, Tani 2001). Despite the somewhat limited knowledge regarding the molecular mechanism involved in baculovirus entry, functional alteration of baculovirus tropism has been achieved.
Gp64 is the major baculoviral envelope (phospho)glycoprotein (Whitford et al., 1989) that is present on the surface of infected insect cells and on budded virions as homotrimers, forming typical peplomer structures at the pole of the virion (Markovic 1998, Oomens 1995). For the budded form of AcMNPV, it has been shown that gp64 determines the viral receptor preference in inhibition studies with a monoclonal anti‐gp64 antibody and, therefore, defines both the host range and the infection efficiency of the host (Hohmann 1983, Volkman 1985). Gp64 is necessary for the low‐pH–triggered membrane fusion activity (Blissard 1992, Jarvis 1994, Markovic 1998, Oomens 1995, Plonsky 1999) and is essential for viral budding from insect cells (Oomens and Blissard, 1999) as well as spreading the infection through cell‐to‐cell transmission (Monsma et al., 1996). Permissive epitope insertions into the gp64 have been achieved without altering or disturbing viral infectivity. One example of this approach is a study where Ernst et al. (2000) took advantage of the naturally occurring NotI restriction site of gp64 at amino acid position 278 and nondestructively inserted two short peptides, that is, ELDKVA of the human immunodeficiency virus type 1 (HIV‐1) gp41 and an eight amino acid streptavidin binding streptagII into the coding region of gp64. In a subsequent study, the ELDKVA peptide was also inserted into 17 different positions of gp64, where viral propagation was retained in as many as 13 cases, indicating that insertions of the affinity tags did not considerably affect the expression or function of gp64 (Spenger et al., 2002). Thus, small peptides with specific and high affinities to receptors on mammalian cells could be introduced into gp64 for targeting to desired cell types.
While direct modification of native gp64 may be advantageous (Ernst 1998, Ernst 2000, Spenger 2002), fusion of heterologous proteins and ligand‐binding moieties to an extra copy of the gp64 gene has generally been the method of choice for altering the baculovirus tropism. Boublik et al. (1995) were the first to demonstrate display of foreign proteins on the surface of baculovirus analogous to the established prokaryotic phage display systems. Glutathione‐S‐transferase was used to construct several fusion variants with the gp64 gene. In addition, the HIV‐1 major surface glycoprotein, gp120, was successfully incorporated to the amino terminus of gp64, illustrating functional ligand‐binding activity when displayed on the viral surface (Boublik et al., 1995). This report was followed by a study where another HIV‐1–derived protein, the ectodomain of gp41, was coupled with both the native and truncated (membrane anchor) forms of gp64 (Grabherr et al., 1997). To diversify the growing collection of heterologous gp64 fusion constructs, Mottershead et al. (1997) published a novel report where the green fluorescent protein (GFP) of Aequorea victoria or Rubella virus spike proteins E1 and E2 were displayed for the first time on the surface of an enveloped virus as N‐terminal fusions to gp64. Together, these initial reports illustrated that gp64 is a suitable fusion partner for functional display of foreign polypeptides on the viral surface, providing growing possibilities for viral vector targeting. Next, the gp64 display strategy was expanded to include presentation of targeting moieties, that is, murine and human single chain antibody fragments (scFv) for the hapten 2‐phenyloxazolone and carcinoembryonic antigen (CEA), respectively, and synthetic IgG‐binding Z/ZZ‐domains of protein A (Mottershead et al., 2000). The viruses exhibited strong binding ability to the corresponding antigens and intact antibodies in vitro, demonstrating that the characteristics of the displayed polypeptides were preserved when presented on the viral surface. To achieve targeted baculovirus transduction, the viruses displaying ZZ‐domains or scFv specific for the CEA were further modified to include a dual expression cassette containing GFP and enhanced green fluorescent protein (EGFP) under the transcriptional regulation of the polyhedrin and CMV promoters, respectively, allowing monitoring of transgene expression in both insect and mammalian cells (Ojala et al., 2001). Despite improved binding, enhanced transgene expression was not observed. A baculovirus displaying an integrin‐specific motif, RKK, as a part of two different loops of GFP fused with the gp64 was shown to bind a peptide representing the receptor‐binding site of an α2 integrin, the α2I domain, by ELISA. Again, this interaction was not strong enough to overcome binding of wild‐type gp64 to unknown cellular receptor(s) on the surface of α2 integrin‐expressing Chinese hamster ovary (CHO) cells or to improve virus uptake (Riikonen et al., 2005). Thus, although enhanced viral binding to desired targets has been achieved, this interaction has generally not led to improved internalization and gene transduction of the vectors. In a study, however, the avidin‐biotin technology was used for baculovirus targeting, resulting in both enhanced and targeted transduction (Raty et al., 2004). Due to its positive charge at physiological pH, avidin itself was demonstrated to enhance viral transduction of rat malignant glioma cells (BT4C) and rabbit aortic smooth muscle (RAASMC) cells by 5‐ and 26‐fold, respectively. Moreover, chimeric avidin–gp64 enabled viral targeting and efficient gene transfer to biotinylated cells, possibly providing a versatile tool for gene delivery. The display of αVβ3‐integrin–specific RGD motifs, derived from the C‐terminus of coxsackievirus A9 or human parechovirus 1 VP protein, on the viral surface, resulted in both improved binding and, thus, enhanced transduction of human lung carcinoma cells expressing αVβ3‐integrins (Ernst 2006, Matilainen 2006).
Although foreign protein sequences have mainly been displayed on the viral surface after fusion to gp64, heterologous viral glycoproteins are capable of serving in the same context. As a model system, a truncated form of the vesicular stomatitis virus glycoprotein (VSV‐G) was discovered to enhance display and enabled scattered distribution of the EGFP fusion proteins on the viral envelope (Chapple and Jones, 2002), whereas gp64 fusions normally accumulate only at the pole of the virion. To apply this fusion strategy to baculovirus targeting, the VSV‐G transmembrane anchor, comprising 29 amino acids of the cytoplasmic domain, the 20‐amino acid membrane spanning region in addition to the 21‐amino acid truncated ectodomain, was fused with the IgG‐binding ZZ domains of protein A (Ojala et al., 2004) and displayed on the surface of baculovirus vectors. The ZZ‐displaying viruses showed improved binding to IgG and, in principle, these vectors could be targeted to any desired cell type when a suitable IgG antibody is available, eliminating the need of preparing distinct vectors for each application. Improved transduction was not observed, however, when GFP was used as a reporter. To gain cancer cell‐selective tropism of baculovirus, the LyP‐1 (Laakkonen et al., 2002), F3 (Porkka et al., 2002), and CGKRK (Hoffman et al., 2003) tumor‐homing peptides were displayed on the surface of baculovirus by fusion to the membrane‐anchoring signal of VSV‐G (Mäkeläet al., 2006). To increase the specificity, the VSV‐G fusion strategy was further modified by excluding the 21‐amino acid VSV‐G ectodomain, known to mediate nonspecific binding and transduction of the baculovirus vectors (Kaikkonen 2005, Ojala 2004). These vectors exhibited significantly improved binding and transgene delivery to both human breast carcinoma and hepatocarcinoma cells, highlighting the potential of targeted baculovirus vectors in cancer gene therapy. In addition to VSV‐G, a class I transmembrane protein, the membrane‐spanning region of a class II membrane protein, neuraminidase A of influenza virus, is capable of serving as an N‐terminal anchor domain for efficient display of EGFP on the viral surface (Borg et al., 2004), providing an alternative display strategy to gp64 or VSV‐G membrane anchors.
Analogous to surface display, a novel baculovirus capsid display method has been developed (Kukkonen 2003, Oker‐Blom 2003). This technique is based on the display of foreign proteins or peptides on the surface of the viral capsid as amino or carboxy terminal fusions to the major AcMNPV capsid protein vp39 (Thiem 1989a, Thiem 1989b). In the first capsid display report, vp39 was demonstrated to be compatible for incorporation of a foreign protein molecule, EGFP, in large quantities. EGFP was successfully fused either to the N‐ or C‐terminus of vp39 without compromising the viral titer or functionality (Kukkonen et al., 2003). In addition, it was proposed that the block in transduction of mammalian cells by baculovirus lies in the cytoplasmic trafficking or nuclear import instead of viral escape from the endosomes, as had previously been suggested. Thus, this new tool provides possibilities for specific intracellular and nuclear targeting of the viral capsids, and facilitates baculovirus entry and nuclear import studies in both insect and mammalian cells.
It is now evident that heterologous targeting moieties with specific and high avidities can be functionally displayed in large quantities on both the baculovirus envelope and capsid. Although targeting appears to be an attractive concept to enhance baculoviral transduction of specific cell types, its applicability in human disease could be partly limited by the fact that mammalian host cell nonpermissiveness cannot always be reversed simply by making baculoviral binding and entry possible. Therefore, tissue targeting and nuclear localization signals could be displayed in different combinations on the viral envelope and nucleocapsid, respectively, enabling both cellular and nuclear targeting. In addition to appropriate targeting molecules, introduction of tissue‐specific promoters and complement resistance (Huser et al., 2001) into the baculovirus vectors could enable targeting of this insect virus to desired cells and tissues in vivo and therefore provide potential applications in gene therapy.
III. Altering the Tropism of Baculoviral Vectors Through Pseudotyping
Pseudotyping, that is, phenotypic mixing, is a process in which the natural envelope glycoproteins of the virus are modified, replaced, or expressed with surface (glyco)proteins from a donor virus. In this way, the host range of virus vectors can be expanded or altered. If successful, such particles possess the tropism of the virus from which the protein was derived. The vesicular stomatitis virus (VSV) G‐protein (VSV‐G) is among the first and still most widely used glycoprotein for pseudotyping viral vectors due to the very extensive tropism and stability of the resulting pseudotypes. Generation of VSV‐G pseudotypes from a number of viruses has been described earlier. Both native and modified VSV‐G have been extensively used for pseudotyping retroviruses (Croyle 2004, Emi 1991, Guibinga 2004, Schnitzer 1977), adenoviruses (Yun et al., 2003), and herpesviruses (Anderson 2000, Tang 2001), for example. In the case of VSV‐G–pseudotyped viral vectors where tissue targeting through ligand incorporation into VSV‐G has been endeavored, several factors including the lack of three dimensional crystal structure of the glycoprotein, has rendered the tropism modification challenging. Regardless, permissive epitope/ligand insertion sites have been identified within native VSV‐G that allow modification of the protein without compromising folding or oligomerization (Guibinga et al., 2004). In addition, the use of different recombinant VSV vectors for vaccine production has been broadly studied (McKenna 2003, Schlehuber 2004) and different truncated forms of VSV‐G have served as partners for constructing chimeric fusion proteins to facilitate the study of the biological properties of viral or cellular membrane glycoproteins (Basu 2004, Buonocore 2002, Lagging 1998, Schnell 1996). While the VSV‐G–pseudotyped vectors are valuable for many diverse studies and even for some preliminary clinical applications, their promiscuous susceptibility for target cells and tissues may contribute to toxicity and serious adverse effects through transduction of nontarget cells (Burns 1993, Naldini 1999, VandenDriessche 2002).
Pseudotyped baculovirus vectors engineered to date represent viral particles bearing heterologous glycoproteins on their envelope, similar to other virus vectors, mainly the VSV‐G, expressed either alone or with the endogenous baculovirus surface glycoprotein, gp64 (Barsoum 1997, Facciabene 2004, Kitagawa 2005, Mangor 2001, Park 2001, Pieroni 2001, Tani 2001, Tani 2003). The primary objective of these studies was to engineer vectors possessing a wider tropism and improved transduction capacity of the target cells as compared to wild‐type baculovirus. Secondarily, the VSV‐G is expected to provide protection for baculovirus vectors against complement inactivation in potential in vivo gene therapy applications as has previously been demonstrated for VSV‐G–pseudotyped retroviral vectors (Ory et al., 1996).
Barsoum et al. (1997) demonstrated that AcMNPV can be pseudotyped with an envelope glycoprotein derived from another virus. The gene encoding VSV‐G was placed under the transcriptional control of the polyhedrin promoter, providing abundant expression in infected insect cells and subsequent incorporation into budded virions, which exhibited atypical oval‐shaped morphology and occasionally tail‐like structures. However, no further studies have been published where the effect of VSV‐G on the morphology of the budded form of AcMNPV has been described. These pseudotyped viruses improved transduction of HepG2 cells tenfold and also augmented transgene delivery to certain established as well as primary cell lines that are weakly or not susceptible to transduction by wild‐type baculovirus, thus broadening the tropism. In addition, it was speculated that the VSV‐G may augment the escape of the virus from intracellular vesicles via its membrane fusion activity rather than improve viral binding or entry into target cells, hence escalating transport of the viral genome into the nucleus (Barsoum et al., 1997). Later, VSV‐G and mouse hepatitis virus S protein (MHV‐S)‐pseudotyped baculovirus vectors were employed as a control system in a study where cell surface components involved in baculovirus infection of insect cells and entry into mammalian cells was explored using baculovirus displaying two copies of gp64 on the viral envelope (Tani et al., 2001). It was demonstrated that the virus overexpressing gp64, in addition to its endogenous copy of gp64, can incorporate ∼1.5‐ to 2‐fold the normal quantity of gp64 on the budded virion. These modified viruses mediated transduction resulting in 10‐ to 100‐fold increased reporter gene expression in a variety of cell lines as compared to the virus carrying an ordinary amount of gp64. It was also proposed that cell surface phospholipids provide a docking point for gp64, hence assisting viral entrance to mammalian cells (Tani et al., 2001). Park et al. (2001) combined tropism modification of baculovirus with transcriptional targeting, designed to be limited to cells of hepatic origin. Accordingly, a VSV‐G–pseudotyped virus, harboring an expression reporter (luciferase) gene placed under the control of a hepatocyte‐specific AFP (α‐fetoprotein) promoter/enhancer, was generated. The virus was able to transduce human hepatoma cells at an efficiency of approximately fivefold greater than the control virus lacking VSV‐G and transgene expression was restricted to cells of hepatic origin expressing AFP, of which concentration is elevated in hepatocellular carcinomas.
The VSV‐G is capable of complementing the function of gp64 by restoring the ability of a gp64‐null virus to assemble and produce infectious budded virions, although the kinetics of infection is somewhat delayed and viral titers reduced by 1 to 2 logs as compared to wild‐type AcMNPV (Mangor et al., 2001). However, these gp64‐null VSV‐G–pseudotyped virions were not tested for transduction of vertebrate cells, thus, whether they could enhance transduction analogous to recombinant vectors coexpressing gp64 and VSV‐G remains unanswered. In addition to VSV‐G, the function of a gp64‐deleted AcMNPV has been partially restored by inserting the recently identified F‐proteins from two group II nucleopolyhedroviruses (NPVs), Lymantria dispar MNPV and Spodoptera exigua MNPV, into the gp64 locus, demonstrating that F‐proteins derived from heterologous NPVs are functional analogs of gp64 (Lung et al., 2002). Parallel to the VSV‐G/gp64‐null virus (Mangor et al., 2001), infectious viral titers of the F‐protein pseudotypes were somewhat compromised as compared to the wild‐type counterpart, suggesting that the level of compatibility between the F‐proteins and other AcMNPV proteins may not be optimal. Further, the capacity of these F‐protein–pseudotyped vectors for gene transduction of mammalian cells remains to be explored.
The efficiency of gene delivery in vivo has also been explored using VSV‐G–pseudotyped baculovirus vectors. The modified virus enhanced transgene delivery by five‐ to tenfold when mouse myoblasts and myotubes were transduced in vitro (Pieroni et al., 2001). Similarly, the same increase in reporter gene (β‐galactosidase) expression was detected in vivo after injection of the VSV‐G–pseudotyped vector in the quadriceps of BALB/c and C57BL/6 mice. Moreover, expression of the transgene, mouse erythropoietin, was monitored to last for 35 and 178 days in the skeletal muscle of BALB/c or C57BL/6, and DBA/2J mice, respectively (Pieroni et al., 2001). The VSV‐G–coated baculovirus also exhibited improved resistance to inactivation by human, rabbit, guinea pig, hamster, and mouse, but not rat sera (Tani et al., 2003). This modified virus could also be used for transduction of the cerebral cortex and testis of mice by direct inoculation in vivo. No comparisons were conducted, however, with the unmodified virus to evaluate putative enhancement in transduction efficiency. A truncated form of VSV‐G (VSV‐GED), composed of the cytoplasmic and membrane‐spanning domains in addition to the 21‐amino acid ectodomain, was shown to enhance transduction by the VSV‐GED–pseudotyped baculovirus both in vitro and in vivo (Kaikkonen et al., 2005). Thus, the enhancement of virus transduction, which is characteristic of full‐length VSV‐G, was retained by the truncated form. It was speculated that the improved gene delivery was due to possible augmentation of gp64‐mediated release from endosomes during viral entry into the target cells. Moreover, induction of humoral and cell‐mediated immune response has been studied with a recombinant baculovirus vector displaying VSV‐G on the viral surface and expressing hepatitis C virus glycoprotein, E2, under the CMV promoter. The results demonstrated that cell‐mediated immunity to the E2 antigen can be elicited in mice by injecting recombinant baculovirus vectors expressing the target antigen and that the display of VSV‐G on the viral surface increases the immunogenic efficiency tenfold leading to greater induction of E2 antigen‐specific CD8+ T cells (Facciabene et al., 2004). Ligand‐directed gene delivery was achieved by pseudotyped gp64‐deleted baculovirus vectors carrying measles virus receptors, CD46 and SLAM, on their surface (Kitagawa et al., 2005). The viruses were able to replicate and spread infection in gp64‐complementing Sf9 cells, whereas virus propagation was strongly reduced in cells not expressing gp64. However, after three rounds of passage of the pseudotyped viruses, the gp64‐coding gene was integrated into the baculovirus genome probably through nonhomologous recombination. The corresponding viruses were able to target gene delivery to BHK cells expressing the measles virus H and F envelope glycoproteins and the transduction could be inhibited by pretreatment with specific monoclonal antibodies for the displayed ligands. A short hairpin RNA (shRNA) delivery system mediated by a VSV‐G–displaying baculovirus vector was generated, resulting in knock down of an endogenous reporter gene, EGFP, and suppression of porcine reproductive and respiratory syndrome virus replication in tissue culture (Lu et al., 2006), highlighting the potential of recombinant baculovirus as an alternative vehicle for antivirus shRNA delivery.
Overall, the AcMNPV‐pseudotyping system provides an efficient and powerful method for examining the functions and compatibilities of heterologous viral or cellular membrane proteins as well as enabling diversification or constraint of the viral tropism. The selection of cell surface components during virus assembly in infected insect cells is flexible enough to allow incorporation of unrelated membrane proteins into baculovirus particles, yet specific enough to exclude the bulk of host proteins. The first proofs of the principle were the VSV‐G–pseudotyped (gp64‐null) baculovirus vectors, which retained their ability to replicate in insect cells and transduce a large collection of mammalian cells. VSV‐G may use common cell surface determinants as putative receptor molecules, rendering the VSV‐G–pseudotyped baculovirus vectors inappropriate for cell‐specific gene delivery, but ideal in applications where a limited tropism is not required. Thus, such pseudotyped vectors would be particularly suitable for ex vivo gene therapeutic applications where there is no risk of transducing nontarget cell populations. The introduction of ligands with high and specific avidity into the viral envelope, as demonstrated by Kitagawa et al. (2005), could also enable baculovirus targeting in vivo.
IV. Baculovirus Display of Immunogens
For generation of antibodies by traditional procedures, the protein or peptide is produced in a system of choice and subsequently purified before immunization. This is often cumbersome, and more importantly, the final product may not be correctly folded—an essential requirement for an adequate immune response in the host, and thereby, for generation of functional antibodies. In addition to recombinant proteins, several other systems including phage display, DNA‐based immunization, as well as recombinant viral infections and/or fusions to viral proteins are available for generation of antibodies. Here, examples from the literature are presented where baculovirus surface display has been employed for generation of functional monoclonal antibodies against proteins of different origin. Several reports also show clear evidence that display of the immunogen on the viral surface can elicit protective immune responses against viral or parasite infections by using animal models.
To produce monoclonal antibodies against the human nuclear receptors LXRβ and FXR, the N‐terminal domains of these antigens were displayed on the baculoviral surface by inserting the corresponding coding sequences between the signal sequence and the mature domain of gp64 of AcMNPV (Lindley et al., 2000). This study illustrated that baculovirus display is a versatile tool applicable for antigen presentation and for rapid production of functional monoclonal antibodies once the antigen‐coding sequence is available (Lindley et al., 2000). Similarly, monoclonal antibodies against human peroxisome proliferator‐activated receptors (PPARs) using baculovirus display have been generated. The amino terminal sequences of human PPARd and PPARg2 were placed at the N‐terminus of gp64 and antibodies were raised by immunization with whole virus without prior purification of the immunogens (Tanaka et al., 2002). The antibodies generated by this method were functional in a variety of techniques such as immunohistochemistry, immunoblotting, and electrophoretic mobility shift assays. Antigenic epitopes of Theileria parva, an intracellular protozoan parasite that causes East Coast fever, a severe lymphoproliferative disease in cattle, were also successfully presented on the surface of AcMNPV by adopting the gp64 N‐terminal fusion strategy (Kaba et al., 2003). This approach was applied because previous attempts to produce recombinant sporozoite surface antigen (p67) in bacterial or insect cells for vaccine purposes had not resulted in correctly folded protein molecules. Further, a small, immunodominant antigenic site (site A) and the large polyprotein (P1) coding for the four structural proteins of foot‐and‐mouth disease virus (FMDV) have been displayed on the membrane of infected insect cells and consequently on the baculoviral surface by fusion to the N‐terminus of gp64 (Tami et al., 2000). Later, the investigators have shown that these FMVD antigens were able to elicit a specific immune response against FMVD in mice (Tami et al., 2004). Similarly, Yoshida et al. (2003) have shown that the rodent malaria Plasmodium berghei circumsporozoite protein (PbCSP) displayed on the surface of baculovirus as a fusion to gp64 protects mice against a malaria sporozoite infection.
Urano et al. (2003) used an alternative approach of exploiting the baculovirus for monoclonal antibody production by displaying an integral ER membrane protein SCAP on the extracellular, budded form of the virus. Thus, SCAP was not displayed as a fusion to baculovirus specific proteins. SCAP is known to be involved in cleavage of sterol element‐binding protein‐2, hence its function is tightly coupled to cholesterol regulation (Urano et al., 2003). Other membrane receptors, such as the β‐adrenergic receptor (Loisel et al., 1997) and the leukotriene B4 receptor (BLT1) (Masuda et al., 2003) residing on the plasma membrane have also been functionally displayed in the same context.
In addition to AcMNPV, Bombyx mori NPV (BmNPV) has been modified to display immunogens with similar aims as described earlier. Rahman et al. (2003) displayed the immunodominant ectodomains of the fusion glycoprotein (F) of peste‐des‐petitis‐ruminants virus (PPRV) and the hemagglutinin protein (H) of rinderpest virus (RPV), on budded virus particles. The strategy was identical, in that the antigens were fused to gp64 of BmNPV and expressed under transcriptional regulation of the polyhedrin promoter. The investigators showed that the antigenic epitopes were properly displayed and that the recombinant virions were able to induce an immune response in mice against both PPRV and RPV. Finally, Chang et al. (2004) aimed to produce a recombinant baculovirus that mimics severe acute respiratory syndrome corona virus (SARS‐CoV) in its host range and infection mechanism. A baculovirus displaying a 688‐amino acid fragment of SARS‐CoV S glycoprotein as a gp64 fusion was generated and then used to examine the effect on the IL‐8 release in A549, NCI‐H520, HFL‐1, and MRC‐5 cells (Chang et al., 2004).
Together, these reports provide convincing evidence that baculovirus can be used for the functional display of heterologous proteins on its surface through budding from the infected insect cell. Consequently, the baculovirus‐displayed immunogens have been used to elicit immune responses needed for production of monoclonal antibodies and/or to protect the animal host against a viral or parasite infection.
V. Generation of Display Libraries
Display on the surface of bacteriophage is currently the most widespread method for display and selection of large collections of antibodies. This approach is robust, simple to use and, in addition, highly versatile. The selection procedures can be adapted to many specific conditions including selections on whole cells, tissues, and even animals. Originally, generation of eukaryotic cDNA libraries was based on plasmid vectors capable of replicating in particular eukaryotic cell types. During the last decade, however, a variety of display methods and other library‐screening techniques have been under study for isolating monoclonal antibodies from collections of recombinant antibody fragments. The development of virus‐based cDNA expression libraries has offered several advantages over nonviral vectors regarding host cell tropism, transduction efficiency, stability of transgene expression, and production of the vector in high quantities.
In 1997, Granziero et al. (1997) aimed to develop a rapid method for generating baculovirus‐based cDNA expression libraries for screening cell surface molecules, for which antibodies are available beforehand and whose expression pattern is restricted to particular cell types. The first proof of principle was gained by cloning a cDNA pool, reverse transcribed from human placenta, into the baculovirus genome and sorting the virus‐infected insect cells by flow cytometry using monoclonal antibodies of an unknown specificity as probes. By this method, single positive cells could be sorted and viruses carrying the cDNAs encoding the cell surface epitopes isolated. The first demonstration of using baculovirus display for generation and screening of expression was described by Ernst et al. (1998). An HIV‐1 gp41 epitope (ELDKWA), specific for the neutralizing human mAb 2F5, was inserted into the antigenic site B of influenza virus A hemagglutinin, and expressed on the surface of baculovirus‐infected insect cells. The epitope was displayed in a library form, such that each clone contained different amino acids adjacent to the epitope. Thus, the purpose of the experiment was to alter the structural environment so that the corresponding epitope would be presented in the most accessible way, leading to an increased binding capacity of the mAb. The library consisted of 8000 variants out of which one clone showed an increased specific binding capacity when screened by fluorescence activated cell sorting (Ernst et al., 1998). Later, the group also described a system where the same epitope as well as the biotin mimic streptag II were inserted at position 278 of gp64 (Ernst et al., 2000). The fact that the insertions into the coding sequence of the major envelope protein of the virus did not alter virus propagation may be of value in further development of display libraries.
Crawford et al. (2004) have described the use of baculovirus‐infected insect cells as a display platform for class II major histocompatibility complex (MHCII) molecules covalently bound to a library of potential peptide mimotopes. The sequence encoding the peptide was embedded within the genes for the MHC molecule in the viral genome. Thereby, each insect cell infected with a virus particle from a library coding for different peptides, displayed a unique peptide–MHC complex on its cellular membrane. Crawford et al. (2004) were able to identify such peptide mimotope–MHC complexes that bound to the soluble receptors and stimulating T cells bearing the same receptors by “fishing” with fluorescent, soluble T cell receptors. These findings should, therefore, have implications for the relative importance of peptide and MHC in T cell receptor‐ligand recognition. Later, the same group used this baculovirus‐based display system for identification of antigen mimotopes for MHC class I‐specific T cells (Wang et al., 2005). Here, a mouse MHC class I molecule was displayed on the surface of baculovirus‐infected insect cells with a 9‐ to 10‐mer peptide library tethered to the N‐terminus of beta2 microglobulin via a flexible linker. Although there are relatively few studies on libraries generated by using baculovirus/insect cell technology, the present examples clearly show that this technology has potential and interest in further development and utilization of this technology will likely increase.
VI. Summary
In this chapter, we have given a “state of the art” overview of strategies and technologies developed for display of foreign peptides and/or proteins on the surface of baculovirus‐infected insect cells and budded baculovirus particles. Data on virus targeting and transgene expression in mammalian cells, and on the generation of libraries for studying molecular recognition and protein–protein interactions using these techniques were summarized. Production of monoclonal antibodies by utilization of these techniques and the benefits of using baculovirus display to elicit protective immune responses in animal models were reviewed. Together, these studies show the potential for baculovirus within these areas of research and illustrate that further development and broadening of the interdisciplinary applications of this versatile and unique insect virus are justified.
References
- Anderson D.B., Laquerre S., Ghosh K., Ghosh H.P., Goins W.F., Cohen J.B., Glorioso J.C. Pseudotyping of glycoprotein D‐deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables mutant virus attachment and entry. J. Virol. 2000;74:2481–2487. doi: 10.1128/jvi.74.5.2481-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Brown R., McKee M., Boyce F.M. Efficient transduction of mammalian cells by a recombinant baculovirus having the vesicular stomatitis virus G glycoprotein. Hum. Gene Ther. 1997;8:2011–2018. doi: 10.1089/hum.1997.8.17-2011. [DOI] [PubMed] [Google Scholar]
- Basu A., Beyene A., Meyer K., Ray R. The hypervariable region 1 of the E2 glycoprotein of hepatitis C virus binds to glycosaminoglycans, but this binding does not lead to infection in a pseudotype system. J. Virol. 2004;78:4478–4486. doi: 10.1128/JVI.78.9.4478-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blissard G.W., Wenz J.R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH‐dependent membrane fusion. J. Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J., Nevsten P., Wallenberg R., Stenstrom M., Cardell S., Falkenberg C., Holm C. Amino‐terminal anchored surface display in insect cells and budded baculovirus using the amino‐terminal end of neuraminidase. J. Biotechnol. 2004;114:21–30. doi: 10.1016/j.jbiotec.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Boublik Y., Di Bonito P., Jones I.M. Eukaryotic virus display: Engineering the major surface glycoprotein of the Autographa californica nuclear polyhedrosis virus (AcNPV) for the presentation of foreign proteins on the virus surface. Biotechnology (NY) 1995;13:1079–1084. doi: 10.1038/nbt1095-1079. [DOI] [PubMed] [Google Scholar]
- Boyce F.M., Bucher N.L. Baculovirus‐mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore L., Blight K.J., Rice C.M., Rose J.K. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 2002;76:6865–6872. doi: 10.1128/JVI.76.14.6865-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: Concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.J., Liu C.Y., Chiang B.L., Chao Y.C., Chen C.C. Induction of IL‐8 release in lung cells via activator protein‐1 by recombinant baculovirus displaying severe acute respiratory syndrome‐coronavirus spike proteins: Identification of two functional regions. J. Immunol. 2004;173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
- Chapple S.D., Jones I.M. Non‐polar distribution of green fluorescent protein on the surface of Autographa californica nucleopolyhedrovirus using a heterologous membrane anchor. J. Biotechnol. 2002;95:269–275. doi: 10.1016/s0168-1656(02)00023-8. [DOI] [PubMed] [Google Scholar]
- Crawford F., Huseby E., White J., Marrack P., Kappler J.W. Mimotopes for alloreactive and conventional T cells in a peptide‐MHC display library. PLoS Biol. 2004;2:E90. doi: 10.1371/journal.pbio.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croyle M.A., Callahan S.M., Auricchio A., Schumer G., Linse K.D., Wilson J.M., Brunner L.J., Kobinger G.P. PEGylation of a vesicular stomatitis virus G pseudotyped lentivirus vector prevents inactivation in serum. J. Virol. 2004;78:912–921. doi: 10.1128/JVI.78.2.912-921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee K.U., Shuler M.L. Optimization of an assay for baculovirus titer and design of regimens for the synchronous infection of insect cells. Biotechnol. Prog. 1997;13:14–24. doi: 10.1021/bp960086t. [DOI] [PubMed] [Google Scholar]
- Duisit G., Saleun S., Douthe S., Barsoum J., Chadeuf G., Moullier P. Baculovirus vector requires electrostatic interactions including heparan sulfate for efficient gene transfer in mammalian cells. J. Gene Med. 1999;1:93–102. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<93::AID-JGM19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Emi N., Friedmann T., Yee J.K. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst W., Grabherr R., Wegner D., Borth N., Grassauer A., Katinger H. Baculovirus surface display: Construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 1998;26:1718–1723. doi: 10.1093/nar/26.7.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst W., Schinko T., Spenger A., Oker‐Blom C., Grabherr R. Improving baculovirus transduction of mammalian cells by surface display of a RGD‐motif. J. Biotechnol. 2006 doi: 10.1016/j.jbiotec.2006.04.012. (in press) [DOI] [PubMed] [Google Scholar]
- Ernst W.J., Spenger A., Toellner L., Katinger H., Grabherr R.M. Expanding baculovirus surface display. Modification of the native coat protein gp64 of Autographa californica NPV. Eur. J. Biochem. 2000;267:4033–4039. doi: 10.1046/j.1432-1327.2000.01439.x. [DOI] [PubMed] [Google Scholar]
- Facciabene A., Aurisicchio L., La Monica N. Baculovirus vectors elicit antigen‐specific immune responses in mice. J. Virol. 2004;78:8663–8672. doi: 10.1128/JVI.78.16.8663-8672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr R., Ernst W., Doblhoff‐Dier O., Sara M., Katinger H. Expression of foreign proteins on the surface of Autographa californica nuclear polyhedrosis virus. Biotechniques. 1997;22:730–735. doi: 10.2144/97224rr02. [DOI] [PubMed] [Google Scholar]
- Granziero L., Nelboeck P., Bedoucha M., Lanzavecchia A., Reid H.H. Baculovirus cDNA libraries for expression cloning of genes encoding cell‐surface antigens. J. Immunol. Methods. 1997;203:131–139. doi: 10.1016/s0022-1759(97)00018-5. [DOI] [PubMed] [Google Scholar]
- Guibinga G.H., Hall F.L., Gordon E.M., Ruoslahti E., Friedmann T. Ligand‐modified vesicular stomatitis virus glycoprotein displays a temperature‐sensitive intracellular trafficking and virus assembly phenotype. Mol. Ther. 2004;9:76–84. doi: 10.1016/j.ymthe.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Ho Y.C., Chen H.C., Wang K.C., Hu Y.C. Highly efficient baculovirus‐mediated gene transfer into rat chondrocytes. Biotechnol. Bioeng. 2004;88:643–651. doi: 10.1002/bit.20239. [DOI] [PubMed] [Google Scholar]
- Hofmann C., Sandig V., Jennings G., Rudolph M., Schlag P., Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J.A., Giraudo E., Singh M., Zhang L., Inoue M., Porkka K., Hanahan D., Ruoslahti E. Progressive vascular changes in a transgenic mouse model of squamous cell carcinoma. Cancer Cell. 2003;4:383–391. doi: 10.1016/s1535-6108(03)00273-3. [DOI] [PubMed] [Google Scholar]
- Hohmann A.W., Faulkner P. Monoclonal antibodies to baculovirus structural proteins: Determination of specificities by Western blot analysis. Virology. 1983;125:432–444. doi: 10.1016/0042-6822(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Huser A., Rudolph M., Hofmann C. Incorporation of decay‐accelerating factor into the baculovirus envelope generates complement‐resistant gene transfer vectors. Nat. Biotechnol. 2001;19:451–455. doi: 10.1038/88122. [DOI] [PubMed] [Google Scholar]
- Jarvis D.L., Garcia A., Jr. Biosynthesis and processing of the Autographa californica nuclear polyhedrosis virus gp64 protein. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- Kaba S.A., Hemmes J.C., van Lent J.W., Vlak J.M., Nene V., Musoke A.J., van Oers M.M. Baculovirus surface display of Theileria parva p67 antigen preserves the conformation of sporozoite‐neutralizing epitopes. Protein Eng. 2003;16:73–78. doi: 10.1093/proeng/gzg004. [DOI] [PubMed] [Google Scholar]
- Kaikkonen M.U., Raty J.K., Airenne K.J., Wirth T., Heikura T., Yla‐Herttuala S. Truncated vesicular stomatitis virus G protein improves baculovirus transduction efficiency in vitro and in vivo. Gene Ther. 2005 doi: 10.1038/sj.gt.3302657. Online publication November 3. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y., Tani H., Limn C.K., Matsunaga T.M., Moriishi K., Matsuura Y. Ligand‐directed gene targeting to mammalian cells by pseudotype baculoviruses. J. Virol. 2005;79:3639–3652. doi: 10.1128/JVI.79.6.3639-3652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost T.A., Condreay J.P. Recombinant baculoviruses as mammalian cell gene‐delivery vectors. Trends Biotechnol. 2002;20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- Kost T.A., Condreay J.P., Jarvis D.L. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 2005;23(5):567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen S.P., Airenne K.J., Marjomaki V., Laitinen O.H., Lehtolainen P., Kankaanpaa P., Mahonen A.J., Raty J.K., Nordlund H.R., Oker‐Blom C., Kulomaa M.S., Yla‐Herttuala S. Baculovirus capsid display: A novel tool for transduction imaging. Mol. Ther. 2003;8:853–862. doi: 10.1016/j.ymthe.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Laakkonen P., Porkka K., Hoffman J.A., Ruoslahti E. A tumor‐homing peptide with a targeting specificity related to lymphatic vessels. Nat. Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- Lagging L.M., Meyer K., Owens R.J., Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J. Virol. 1998;72:3539–3546. doi: 10.1128/jvi.72.5.3539-3546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindley K.M., Su J.L., Hodges P.K., Wisely G.B., Bledsoe R.K., Condreay J.P., Winegar D.A., Hutchins J.T., Kost T.A. Production of monoclonal antibodies using recombinant baculovirus displaying gp64‐fusion proteins. J. Immunol. Methods. 2000;234:123–135. doi: 10.1016/s0022-1759(99)00133-7. [DOI] [PubMed] [Google Scholar]
- Loisel T.P., Ansanay H., St‐Onge S., Gay B., Boulanger P., Strosberg A.D., Marullo S., Bouvier M. Recovery of homogeneous and functional beta 2‐adrenergic receptors from extracellular baculovirus particles. Nat. Biotechnol. 1997;15:1300–1304. doi: 10.1038/nbt1197-1300. [DOI] [PubMed] [Google Scholar]
- Lu L., Ho Y., Kwang J. Suppression of porcine arterivirus replication by baculovirus‐delivered shRNA targeting nucleoprotein. Biochem. Biophys. Res. Commun. 2006;340:1178–1183. doi: 10.1016/j.bbrc.2005.12.133. [DOI] [PubMed] [Google Scholar]
- Luckow V.A., Summers M.A. Trends in the development of baculovirus expression vectors. Bio/Technology. 1988;6:47–55. [Google Scholar]
- Lung O., Westenberg M., Vlak J.M., Zuidema D., Blissard G.W. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä A.R., Matilainen H., White D.J., Rouslahti E., Oker‐Blom C. Enhanced baculovirus‐mediated transduction of human cancer cells by tumor‐homing peptides. J. Virol. 2006;80:6603–6611. doi: 10.1128/JVI.00528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangor J.T., Monsma S.A., Johnson M.C., Blissard G.W. A GP64‐null baculovirus pseudotyped with vesicular stomatitis virus G protein. J. Virol. 2001;75:2544–2556. doi: 10.1128/JVI.75.6.2544-2556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic I., Pulyaeva H., Sokoloff A., Chernomordik L.V. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 1998;143:1155–1166. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Itoh H., Sakihama T., Akiyama C., Takahashi K., Fukuda R., Yokomizo T., Shimizu T., Kodama T., Hamakubo T. A combinatorial G protein‐coupled receptor reconstitution system on budded baculovirus. Evidence for Galpha and Galphao coupling to a human leukotriene B4 receptor. J. Biol. Chem. 2003;278:24552–24562. doi: 10.1074/jbc.M302801200. [DOI] [PubMed] [Google Scholar]
- Matilainen H., Rinne J., Gilbert L., Marjomaki V., Reunanen H., Oker‐Blom C. Baculovirus entry into human hepatoma cells. J. Virol. 2005;79:15452–15459. doi: 10.1128/JVI.79.24.15452-15459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen H., Mäkelä A.R., Riikonen R., Saloniemi T., Korhonen E., Hyypiä T., Heino J., Grabherr R., Oker‐Blom C. RGD motifs on the surface of baculovirus enhance transduction of human lung carcinoma cells. J. Biotechnol. 2006;125(1):114–126. doi: 10.1016/j.jbiotec.2006.02.002. [DOI] [PubMed] [Google Scholar]
- McKenna P.M., McGettigan J.P., Pomerantz R.J., Dietzschold B., Schnell M.J. Recombinant rhabdoviruses as potential vaccines for HIV‐1 and other diseases. Curr. HIV Res. 2003;1:229–237. doi: 10.2174/1570162033485320. [DOI] [PubMed] [Google Scholar]
- Miller L.K. Baculoviruses as gene expression vectors. Annu. Rev. Microbiol. 1988;42:177–199. doi: 10.1146/annurev.mi.42.100188.001141. [DOI] [PubMed] [Google Scholar]
- Miller L.K. Baculoviruses for foreign gene expression in insect cells. Biotechnology. 1988;10:457–465. doi: 10.1016/b978-0-409-90042-2.50029-5. [DOI] [PubMed] [Google Scholar]
- Miller L.K. Insect baculoviruses: Powerful gene expression vectors. Bioessays. 1989;11:91–95. doi: 10.1002/bies.950110404. [DOI] [PubMed] [Google Scholar]
- Monsma S.A., Oomens A.G., Blissard G.W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell‐to‐cell transmission of infection. J. Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottershead D., van der Linden I., von Bonsdorff C.H., Keinanen K., Oker‐Blom C. Baculoviral display of the green fluorescent protein and rubella virus envelope proteins. Biochem. Biophys. Res. Commun. 1997;238:717–722. doi: 10.1006/bbrc.1997.7372. [DOI] [PubMed] [Google Scholar]
- Mottershead D.G., Alfthan K., Ojala K., Takkinen K., Oker‐Blom C. Baculoviral display of functional scFv and synthetic IgG‐binding domains. Biochem. Biophys. Res. Commun. 2000;275:84–90. doi: 10.1006/bbrc.2000.3264. [DOI] [PubMed] [Google Scholar]
- Naldini L. In vivo gene delivery by lentiviral vectors. Thromb. Haemost. 1999;82:552–554. [PubMed] [Google Scholar]
- Ojala K., Mottershead D.G., Suokko A., Oker‐Blom C. Specific binding of baculoviruses displaying gp64 fusion proteins to mammalian cells. Biochem. Biophys. Res. Commun. 2001;284:777–784. doi: 10.1006/bbrc.2001.5048. [DOI] [PubMed] [Google Scholar]
- Ojala K., Koski J., Ernst W., Grabherr R., Jones I., Oker‐Blom C. Improved display of synthetic IgG‐binding domains on the baculovirus surface. Technol. Cancer Res. Treat. 2004;3:77–84. doi: 10.1177/153303460400300109. [DOI] [PubMed] [Google Scholar]
- Oker‐Blom C., Airenne K.J., Grabherr R. Baculovirus display strategies: Emerging tools for eukaryotic libraries and gene delivery. Brief. Funct. Genomic. Proteomic. 2003;2:244–253. doi: 10.1093/bfgp/2.3.244. [DOI] [PubMed] [Google Scholar]
- Oomens A.G., Blissard G.W. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology. 1999;254:297–314. doi: 10.1006/viro.1998.9523. [DOI] [PubMed] [Google Scholar]
- Oomens A.G., Monsma S.A., Blissard G.W. The baculovirus GP64 envelope fusion protein: Synthesis, oligomerization, and processing. Virology. 1995;209:592–603. doi: 10.1006/viro.1995.1291. [DOI] [PubMed] [Google Scholar]
- Ory D.S., Neugeboren B.A., Mulligan R.C. A stable human‐derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., Lee H.K., Kim T.G., Yoon S.K., Paik S.Y. Hepatocyte‐specific gene expression by baculovirus pseudotyped with vesicular stomatitis virus envelope glycoprotein. Biochem. Biophys. Res. Commun. 2001;289:444–450. doi: 10.1006/bbrc.2001.6009. [DOI] [PubMed] [Google Scholar]
- Pieroni L., Maione D., La Monica N. In vivo gene transfer in mouse skeletal muscle mediated by baculovirus vectors. Hum. Gene Ther. 2001;12:871–881. doi: 10.1089/104303401750195845. [DOI] [PubMed] [Google Scholar]
- Plonsky I., Cho M.S., Oomens A.G., Blissard G., Zimmerberg J. An analysis of the role of the target membrane on the Gp64‐induced fusion pore. Virology. 1999;253:65–76. doi: 10.1006/viro.1998.9493. [DOI] [PubMed] [Google Scholar]
- Porkka K., Laakkonen P., Hoffman J.A., Bernasconi M., Ruoslahti E. A fragment of the HMGN2 protein homes to the nuclei of tumor cells and tumor endothelial cells in vivo. Proc. Natl. Acad. Sci. USA. 2002;99:7444–7449. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.M., Shaila M.S., Gopinathan K.P. Baculovirus display of fusion protein of Peste des petits ruminants virus and hemagglutination protein of Rinderpest virus and immunogenicity of the displayed proteins in mouse model. Virology. 2003;317:36–49. doi: 10.1016/j.virol.2003.08.022. [DOI] [PubMed] [Google Scholar]
- Raty J.K., Airenne K.J., Marttila A.T., Marjomaki V., Hytonen V.P., Lehtolainen P., Laitinen O.H., Mahonen A.J., Kulomaa M.S., Yla‐Herttuala S. Enhanced gene delivery by avidin‐displaying baculovirus. Mol. Ther. 2004;9:282–291. doi: 10.1016/j.ymthe.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Riikonen R., Matilainen H., Rajala N., Pentikainen O., Johnson M., Heino J., Oker‐Blom C. Functional display of an alpha2 integrin‐specific motif (RKK) on the surface of baculovirus particles. Technol. Cancer Res. Treat. 2005;4:437–445. doi: 10.1177/153303460500400411. [DOI] [PubMed] [Google Scholar]
- Schlehuber L.D., Rose J.K. Prediction and identification of a permissive epitope insertion site in the vesicular stomatitis virus glycoprotein. J. Virol. 2004;78:5079–5087. doi: 10.1128/JVI.78.10.5079-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell M.J., Buonocore L., Kretzschmar E., Johnson E., Rose J.K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc. Natl. Acad. Sci. USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer T.J., Weiss R.A., Zavada J. Pseudotypes of vesicular stomatitis virus with the envelope properties of mammalian and primate retroviruses. J. Virol. 1977;23:449–454. doi: 10.1128/jvi.23.3.449-454.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman K.E., McIntosh A.H. Baculovirus replication in a mosquito (dipteran) cell line. Infect. Immun. 1979;26:232–234. doi: 10.1128/iai.26.1.232-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.E., Summers M.D., Fraser M.J. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 1983;3:2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenger A., Grabherr R., Tollner L., Katinger H., Ernst W. Altering the surface properties of baculovirus Autographa californica NPV by insertional mutagenesis of the envelope protein gp64. Eur. J. Biochem. 2002;269:4458–4467. doi: 10.1046/j.1432-1033.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- Tami C., Farber M., Palma E.L., Taboga O. Presentation of antigenic sites from foot‐and‐mouth disease virus on the surface of baculovirus and in the membrane of infected cells. Arch. Virol. 2000;145:1815–1828. doi: 10.1007/s007050070058. [DOI] [PubMed] [Google Scholar]
- Tami C., Peralta A., Barbieri R., Berinstein A., Carrillo E., Taboga O. Immunological properties of FMDV‐gP64 fusion proteins expressed on SF9 cell and baculovirus surfaces. Vaccine. 2004;23:840–845. doi: 10.1016/j.vaccine.2004.03.070. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takeno T., Watanabe Y., Uchiyama Y., Murakami T., Yamashita H., Suzuki A., Aoi R., Iwanari H., Jiang S.Y., Naito M., Tachibana K., The generation of monoclonal antibodies against human peroxisome proliferator‐activated receptors (PPARs) J. Atheroscler. Thromb. 2002;9:233–242. doi: 10.5551/jat.9.233. [DOI] [PubMed] [Google Scholar]
- Tang J., Yang T., Ghosh H.P., Geller A.I. Helper virus‐free HSV‐1 vectors packaged both in the presence of VSV G protein and in the absence of HSV‐1 glycoprotein B support gene transfer into neurons in the rat striatum. J. Neurovirol. 2001;7:548–555. doi: 10.1080/135502801753248132. [DOI] [PubMed] [Google Scholar]
- Tani H., Nishijima M., Ushijima H., Miyamura T., Matsuura Y. Characterization of cell‐surface determinants important for baculovirus infection. Virology. 2001;279:343–353. doi: 10.1006/viro.2000.0699. [DOI] [PubMed] [Google Scholar]
- Tani H., Limn C.K., Yap C.C., Onishi M., Nozaki M., Nishimune Y., Okahashi N., Kitagawa Y., Watanabe R., Mochizuki R., Moriishi K., Matsuura Y. In vitro and in vivo gene delivery by recombinant baculoviruses. J. Virol. 2003;77:9799–9808. doi: 10.1128/JVI.77.18.9799-9808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem S.M., Miller L.K. A baculovirus gene with a novel transcription pattern encodes a polypeptide with a zinc finger and a leucine zipper. J. Virol. 1989;63:4489–4497. doi: 10.1128/jvi.63.11.4489-4497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem S.M., Miller L.K. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 1989;63:2008–2018. doi: 10.1128/jvi.63.5.2008-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Yamaguchi M., Fukuda R., Masuda K., Takahashi K., Uchiyama Y., Iwanari H., Jiang S.Y., Naito M., Kodama T., Hamakubo T. A novel method for viral display of ER membrane proteins on budded baculovirus. Biochem. Biophys. Res. Commun. 2003;308:191–196. doi: 10.1016/s0006-291x(03)01355-x. [DOI] [PubMed] [Google Scholar]
- van Loo N.D., Fortunati E., Ehlert E., Rabelink M., Grosveld F., Scholte B.J. Baculovirus infection of nondividing mammalian cells: Mechanisms of entry and nuclear transport of capsids. J. Virol. 2001;75:961–970. doi: 10.1128/JVI.75.2.961-970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenDriessche T., Naldini L., Collen D., Chuah M.K. Oncoretroviral and lentiviral vector‐mediated gene therapy. Methods Enzymol. 2002;346:573–589. doi: 10.1016/s0076-6879(02)46078-8. [DOI] [PubMed] [Google Scholar]
- Wang Y., Rubtsov A., Heiser R., White J., Crawford F., Marrack P., Kappler J.W. Using a baculovirus display library to identify MHC class I mimotopes. Proc. Natl. Acad. Sci. USA. 2005;102:2476–2481. doi: 10.1073/pnas.0409798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford M., Stewart S., Kuzio J., Faulkner P. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 1989;63:1393–1399. doi: 10.1128/jvi.63.3.1393-1399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman L.E., Goldsmith P.A. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: Inhibition of entry by adsorptive endocytosis. Virology. 1985;143:185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Kondoh D., Arai E., Matsuoka H., Seki C., Tanaka T., Okada M., Ishii A. Baculovirus virions displaying Plasmodium berghei circumsporozoite protein protect mice against malaria sporozoite infection. Virology. 2003;316:161–170. doi: 10.1016/j.virol.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yun C.O., Cho E.A., Song J.J., Kang D.B., Kim E., Sohn J.H., Kim J.H. dl‐VSVG‐LacZ, a vesicular stomatitis virus glycoprotein epitope‐incorporated adenovirus, exhibits marked enhancement in gene transduction efficiency. Hum. Gene Ther. 2003;14:1643–1652. doi: 10.1089/104303403322542284. [DOI] [PubMed] [Google Scholar]


