Abstract
Coronaviruses (CoVs) have a remarkable potential to change tropism. This is particularly illustrated over the last 15 years by the emergence of two zoonotic CoVs, the severe acute respiratory syndrome (SARS)- and Middle East respiratory syndrome (MERS)-CoV. Due to their inherent genetic variability, it is inevitable that new cross-species transmission events of these enveloped, positive-stranded RNA viruses will occur. Research into these medical and veterinary important pathogens—sparked by the SARS and MERS outbreaks—revealed important principles of inter- and intraspecies tropism changes. The primary determinant of CoV tropism is the viral spike (S) entry protein. Trimers of the S glycoproteins on the virion surface accommodate binding to a cell surface receptor and fusion of the viral and cellular membrane. Recently, high-resolution structures of two CoV S proteins have been elucidated by single-particle cryo-electron microscopy. Using this new structural insight, we review the changes in the S protein that relate to changes in virus tropism. Different concepts underlie these tropism changes at the cellular, tissue, and host species level, including the promiscuity or adaptability of S proteins to orthologous receptors, alterations in the proteolytic cleavage activation as well as changes in the S protein metastability. A thorough understanding of the key role of the S protein in CoV entry is critical to further our understanding of virus cross-species transmission and pathogenesis and for development of intervention strategies.
Keywords: Coronavirus spike, Tropism, Cross-species transmission, Receptor interaction, Membrane fusion, Cryo-EM structure
1. Introduction
Coronaviruses (CoVs) (order Nidovirales, family Coronaviridae, subfamily Coronavirinae) are enveloped, positive-sense RNA viruses that contain the largest known RNA genomes with a length of up to 32 kb. The subfamily Coronavirinae, which contains viruses of both medical and veterinary importance, can be divided into the four genera alpha-, beta-, gamma- and deltacoronavirus (α-, β-, γ- and δ-CoV). The coronavirus particle comprises at least the four canonical structural proteins E (envelope protein), M (membrane protein), N (nucleocapsid protein), and S (spike protein). In addition, viruses belonging to lineage A of the betacoronaviruses express the membrane-anchored HE (hemagglutinin–esterase) protein. The S glycoprotein contains both the receptor-binding domain (RBD) and the domains involved in fusion, rendering it the pivotal protein in the CoV entry process.
Coronaviruses primarily infect the respiratory and gastrointestinal tract of a wide range of animal species including many mammals and birds. Although individual virus species mostly appear to be restricted to a narrow host range comprising a single animal species, genome sequencing and phylogenetic analyses testify that CoVs have crossed the host species barrier frequently (Chan et al., 2013; Woo et al., 2012). In fact most if not all human coronaviruses seem to originate from bat CoVs (BtCoVs) that transmitted to humans directly or indirectly through an intermediate host. It therefore appears inevitable that similar zoonotic infections will occur in the future.
In the past 15 years, the world witnessed two such zoonotic events. In 2002–2003 cross-species transmissions from bats and civet cats were at the base of the SARS (severe acute respiratory syndrome)-CoV epidemic that found its origin in the Chinese Guangdong province (Li et al., 2006, Song et al., 2005). The SARS-CoV nearly became a pandemic and led to over 700 deaths, before it disappeared when the appropriate hygiene and quarantine precautions were taken. In 2012, the MERS (Middle East respiratory syndrome)-CoV emerged in the human population on the Arabian Peninsula and currently continues to make a serious impact on the local but also global health system with 1800 laboratory confirmed cases and 640 deaths as of September 1, 2016 (WHO | Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia, 2016). The natural reservoir of MERS-CoV is presumed to be in dromedary camels from which zoonotic transmissions repeatedly give rise to infections of the lower respiratory tract in humans (Alagaili et al., 2014, Azhar et al., 2014, Briese et al., 2014, Reusken et al., 2013, Widagdo et al., 2016). Besides these two novel CoVs, four other CoVs were previously identified in humans which are found in either the alphacoronavirus (HCoV-NL63 and HCoV-229E) or the betacoronavirus genera (HCoV-OC43 and HCoV-HKU1). Phylogenetic analysis has shown that the bovine CoV (BCoV) has been the origin for HCoV-OC43 following a relatively recent cross-species transmission event (Vijgen et al., 2006). Moreover, HCoV-NL63, HCoV-229E, SARS-CoV, and MERS-CoV also have been predicted to originate from bats (Annan et al., 2013, Bolles et al., 2011, Corman et al., 2015, Hu et al., 2015, Huynh et al., 2012).
In general, four major criteria determine cross-species transmission of a particular virus (Racaniello et al., 2015). The cellular tropism of a virus is determined by the susceptibility of host cells (i.e., presence of the receptor needed for entry) as well as by the permissiveness of these host cells to allow the virus to replicate and to complete its life cycle. A third determinant consists of the accessibility of susceptible and permissive cells in the host. Finally, the innate immune response may restrict viral replication in a host species-specific manner. The above-mentioned criteria may play a critical role in the success of a cross-species transmission event. However, for CoVs, it seems that host tropism and changes therein are particularly determined by the susceptibility of host cells to infection. While CoV accessory genes, including the HE proteins, are thought to play a role in host tropism and adaptation to a new host, the S glycoprotein appears to be the main determinant for the success of initial cross-species infection events. In this review, we focus on the molecular changes in the S protein that underlie tropism changes at the cellular, tissue, and host species level and put these in perspective of the recently published cryo-EM structures.
2. Structure of the Coronavirus S Protein
The CoV S protein is a class I viral fusion protein (Bosch et al., 2003) similar to the fusion proteins of influenza, retro-, filo-, and paramyxoviruses (Baker et al., 1999, Bartesaghi et al., 2013, Lee et al., 2008, Lin et al., 2014). Like other class I viral fusion proteins, the S protein folds into a metastable prefusion conformation following translation. The size of the abundantly N-glycosylated S protein varies greatly between CoV species ranging from approximately 1100 to 1600 residues in length, with an estimated molecular mass of up to 220 kDa. Trimers of the S protein form the 18–23-nm long, club-shaped spikes that decorate the membrane surface of the CoV particle. Besides being the primary determinant in CoV host tropism and pathogenesis, the S protein is also the main target for neutralizing antibodies elicited by the immune system of the infected host (Hofmann et al., 2004).
The S protein can be divided into two functionally distinct subunits: the globular S1 subunit is involved in receptor recognition, whereas the S2 subunit facilitates membrane fusion and anchors S into the viral membrane (Fig. 1A). The S1 and S2 domains may be separated by a cleavage site that is recognized by furin-like proteases during S protein biogenesis in the infected cell. X-ray crystal structures of several S domains have furthered our understanding of the S protein in the past. In addition, recent elucidation of the high-resolution structures of the spike ectodomain of two betacoronaviruses—MHV and HCoV-HKU1—by single-particle cryo-electron microscopy (Kirchdoerfer et al., 2016, Walls et al., 2016) has provided novel insights into the architecture of the S trimer in its prefusion state (Fig. 1B and C).
Fig. 1.
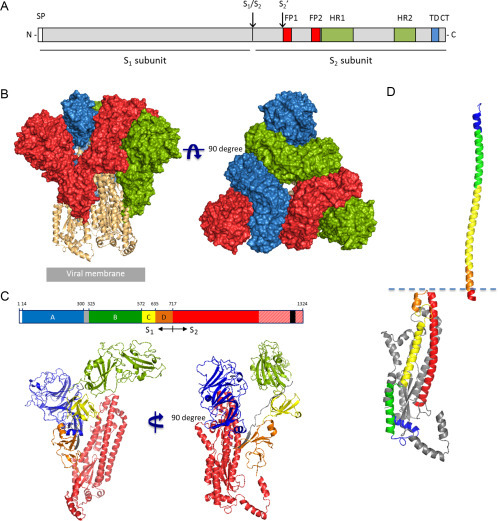
Spike protein features and structure of the mouse hepatitis coronavirus spike glycoprotein trimer. (A) Schematic linear representation of the coronavirus S protein with relevant domains/sites indicated: signal peptide (SP), two proteolytic cleavage sites (S1/S2 and S2′), two proposed fusion peptides (FP1 and FP2), two heptad repeat regions (HR1 and HR2), transmembrane domain (TD), and cytoplasmic tail (CT). (B) Front and top view of the trimeric mouse hepatitis coronavirus (strain A59) spike glycoprotein ectodomain obtained by cryo-electron microscopy analysis (Walls et al., 2016; PDB: 3JCL). Three S1 protomers (surface presentation) are colored in red, blue, and green. The S2 trimer (cartoon presentation) is colored in light orange. (C) Schematic representation of MHV spike protein sequence (drawn to scale), the S1 domains A, B, C, and D are colored in blue, green, yellow, and orange, respectively, and the linker region connecting domains A and B in gray, the S2 region is colored in red, and the TM region is indicated as a black box. Red-shaded region indicates spike region that was not resolved in the cryo-EM structure. (Lower panel) Two views on the structure of the mouse hepatitis virus spike glycoprotein protomer (cartoon representation); domains are colored as depicted earlier. (D) Comparison of the S2 HR1 region in its pre- and postfusion conformation. (Lower left) Structure of the MHV S2 protomer (cartoon presentation) with four helices of the HR1 region (and consecutive linker region) and the downstream central helix colored in blue, green, yellow, orange, and red, respectively. (Upper right) The structure of a single SARS-CoV S HR1 helix of the postfusion six-helix bundle structure (PDB: 1WYY) is colored according to the homologous HR1 region in the MHV S2 prefusion structure shown in the lower left panel. Structures are aligned based on the N-terminal segment of the central helix (in red). Figures were generated with PyMOL.
2.1. Structure of the S1 Subunit
The S1 subunit of the betacoronavirus spike proteins displays a multidomain architecture and is structurally organized in four distinct domains A–D of which domains A and B may serve as a RBD (Fig. 1C). The core structure of domain A displays a galectin-like β-sandwich fold, whereas domain B contains a structurally conserved core subdomain of antiparallel β-sheets (Kirchdoerfer et al., 2016, Li et al., 2005a, Walls et al., 2016, Wang et al., 2013). Importantly, domain B is decorated with an extended loop on the viral membrane-distal side. This loop may differ greatly in size and structure between virus species of the betacoronavirus genus and is therefore also referred to as hypervariable region (HVR). The cryo-EM structures of the MHV-A59 and HCoV-HKU1 S trimers show an intricate interlocking of the three S1 subunits (Fig. 1B). Oligomerization of the S protomers results in a closely clustered trimer of the individual B domains close to the threefold axis of the spike on top of the S2 trimer, whereas the three A domains are ordered more distally of the center. In contrast to domains A and B, the S1 C-terminal domains C and D are made up of discontinuous parts of the primary protein sequence and form β-sheet-rich structures directly adjacent to the S2 stalk core, while the separate S1 domains are interconnected by loops covering the S2 surface. Compared to the S2 subunit, the S1 subunit displays low level of sequence conversation among species of different CoV genera. Moreover, S1 subunits vary considerably in sequence length ranging from 544 (infectious bronchitis virus (IBV) S) to 944 (229-related bat coronavirus S) residues in length (Fig. 2 ), indicating differences in architecture of the spikes of species from different CoV genera. Structural information from the spikes of gamma- and deltacoronavirus species is currently lacking. Two independently folding domains have been assigned in the S1 subunit of alphacoronavirus spikes, that can interact with host cell surface molecules, an N-terminal domain (in transmissible gastroenteritis virus (TGEV) S residues 1–245) and a more C-terminal domain (in TGEV S residues 506–655). Contrary to betacoronaviruses, these two receptor-interacting domains in alphacoronavirus spikes are separated in sequence by some 275 residues, which may fold into one or more separate domains. Structural information is only available for the C-terminal S1 RBD of two α-CoV S proteins, which differs notably from that of betacoronaviruses. The RBD in the S1 CTR of alphacoronaviruses displays a β-sandwich core structure, whereas a β-sheet core structure is seen for betacoronaviruses (Reguera et al., 2012, Wu et al., 2009).
Fig. 2.
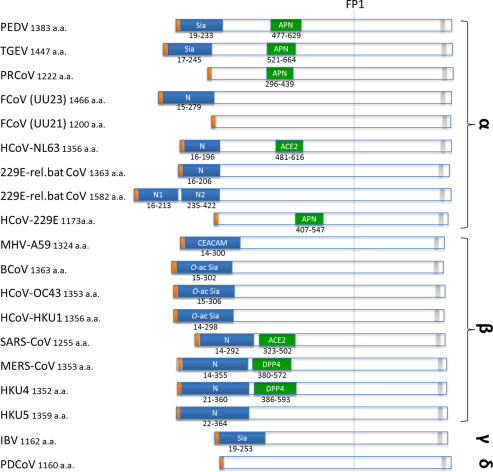
Overview of currently known receptors and their binding domains within S1. Schematic representation of coronavirus spike proteins drawn to scale. Yellow boxes indicate signal peptides. Blue boxes indicate the N-terminal regions in alpha- and betacoronavirus spike proteins, which were mapped based on sequence homology between viruses within the same genus. Green boxes indicate known receptor-binding domains in the C-terminal region of S1. Known receptors are indicated in the boxes: APN, aminopeptidase N; ACE2, angiotensin-converting enzyme 2; CEACAM, carcinoembryonic antigen-related cell adhesion molecule 1; Sia, sialic acid; O-ac Sia, O-acetylated sialic acid; DPP4, dipeptidyl peptidase-4. Gray boxes indicate transmembrane domains. Spikes proteins are shown of PEDV strain CV777 (GB: AAK38656.1), TGEV strain Purdue P115 (GB: ABG89325.1), PRCoV strain ISU-1 (GB: ABG89317.1), Feline CoV strain UU23 (GB: ADC35472.1), Feline CoV strain UU21 (GB: ADL71466.1), Human CoV NL63 (GB: YP_003767.1), 229E-related bat CoV with one N domains (GB: ALK28775.1), 229E-related bat CoV with two N domains (GB: ALK28765.1), Human CoV 229E strain inf-1 (GB: NP_073551.1), MHV strain A59 (GB: ACO72893), BCoV strain KWD1 (GB: AAX38489), HCoV-OC43 strain Paris (GB: AAT84362), HCoV-HKU1 (GB: AAT98580), SARS-CoV strain Urbani (GB: AAP13441), MERS-CoV strain EMC/2012 (GB: YP_009047204), HKU4 (GB: AGP04928), HKU5 (GB: AGP04943), IBV strain Beaudette (GB: ADP06471), and PDCoV strain USA/Ohio137/2014 (GB: AIB07807). PSI-BLAST analysis using the NTR of the HCoV-NL63 S protein (residues 16–196) as a query detected two homologous regions in the first 425 residues of the 229E-related bat coronavirus spike protein (GB: ALK28765.1)—designated N1 (residues 32–213) and N2 (residues 246–422) with 32% and 35% amino acid sequence identity, respectively, suggesting a duplication of the NTR. Spike proteins are drawn to scale and aligned at the position of the conserved fusion peptide (FP1).
2.2. Structure of the S2 Subunit
The highly conserved S2 subunit contains the key protein segments that facilitate virus-cell fusion. These include the fusion peptide, two heptad repeat regions (HR1 and HR2) and the transmembrane domains which are well conserved among CoV species across different genera. In the MHV and HKU1 S prefusion structures, the S2 domain consists of multiple α-helical segments and a three-stranded antiparallel β-sheet at the viral membrane-proximal end. A 75 Å long central helix located immediately downstream of the HR1 region stretches along the threefold axis over the entire length of the S2 trimer. The HR1 motif itself folds as four individual α-helices along the length of the S2 subunit, in contrast to the 120 Å long α-helix formed by this region in postfusion structures (Duquerroy et al., 2005, Gao et al., 2013, Xu et al., 2004). A 55 Å long helix upstream of the S2′ cleavage site runs parallel to and is packed against the central helix via hydrophobic interactions (Fig. 1C). The fusion peptide forms a short helix of which the strictly conserved hydrophobic residues are buried in an interface with other elements of S2. Unlike other class I fusion proteins, this conserved fusion peptide (FP1) is not directly upstream of HR1 but located some 65 residues upstream of this region (Fig. 1A). Intriguingly, a recent published report provided experimental evidence for the existence of another fusion peptide (FP2) immediately upstream of the HR1 region (Ou et al., 2016), that had been predicted earlier based on the position, hydrophobicity profile and amino acid composition canonical for class I viral fusion peptides (Bosch and Rottier, 2008, Bosch et al., 2004, Chambers et al., 1990). The HR2 region locates closely to the C-terminal end of the S ectodomain, but it appeared to be disordered in both cryo-EM structures and therefore its prefusion conformation remains unknown.
The metastable prefusion conformation of S2 is locked by the cap formed by the intertwined S1 protomers. The distal tip of the S2 trimer connects via hydrophobic interactions with domains B. This distal tip of the S2 trimer consists of the C-terminal region of HR1 in the prefusion conformation, while the entire HR1 rearranges to form a central three-helix coiled coil in the postfusion structure (Duquerroy et al., 2005, Lu et al., 2014, Supekar et al., 2004). Interactions between this region of the S2 trimer and domain B may therefore prevent premature conformational changes resulting in the conversion of the prefusion S protein into the very stable postfusion structure. Also domains C and D of the betacoronavirus S1 subunit and the linker region connecting domain A and B interact with the surface of the adjacent S2 protomer and may hence play a role in stabilizing the prefusion S2 trimer. Domain A appears to play a minor role in this respect in view of its relatively small a surface area that interacts with the S2 trimer.
3. Spike–Receptor Interactions
3.1. Different Domains Within S1 May Act as RBD
Over the past decades, molecular studies on the CoV S glycoprotein have shown that both the N-terminal region (NTR, domain A in β-CoV) and the C-terminal region of S1 (CTR, comprising domain B, C, and D in β-CoV) can bind host receptors and hence function as RBDs (Fig. 2) (Li, 2015). The CTR of alpha- and betacoronaviruses appears to bind proteinaceous receptors exclusively. The α-CoV HCoV-229E, serotype II feline CoV (FCoV), TGEV, and porcine respiratory coronavirus use the human aminopeptidase N (APN) of their respective hosts as receptors (Bonavia et al., 2003, Delmas et al., 1992, Reguera et al., 2012). The HCoV-NL63 (α-CoV) and SARS-CoV (β-CoV) both utilize angiotensin-converting enzyme 2 (ACE2) as a functional receptor (Li et al., 2005b, Wu et al., 2009), whereas the β-CoVs MERS-CoV and BtCoV-HKU4 recruit dipeptidyl peptidase-4 (DPP4) as a functional receptor (Lu et al., 2013, Mou et al., 2013, Raj et al., 2013, Wang et al., 2014, Yang et al., 2014).
The receptor-binding motifs (RBMs) in the S1 CTRs of alpha- and betacoronavirus spike proteins are presented on one or more loops extending from the β-sheet core structure. Within alpha- and betacoronavirus genera the RBD core is structurally conserved yet the RBM(s) that determine receptor specificity may vary extensively. For instance, the CTR of the α-CoVs PRCoV and HCoV-NL63 has a similar core structure suggesting common evolutionary origin but diverged in their RBMs recruiting different receptors (APN and ACE2, respectively). A similar situation is seen for the CTRs of β-CoVs SARS-CoV and MERS-CoV that bind ACE2 and DPP4, respectively (Li, 2015). Conversely, the CTRs of the α-CoV HCoV-NL63 and β-CoV SARS-CoV both recognize ACE2, yet via distinct molecular interactions (ACE2 recognition via three vs one RBM, respectively), which suggested a convergent evolution pathway for these viruses in recruiting the ACE2 receptor (Li, 2015). The core structures of the CTRs in α- and β-CoVs provide a scaffold to present RBMs from extending loop(s), which may accommodate facile receptor switching by subtle alterations in or exchange of the RBMs via mutation/recombination.
Contrary to the CTR, the NTR appears to mainly bind glycans. The NTR of the α-CoV TGEV and of the γ-CoV IBV S proteins binds to sialic acids (Promkuntod et al., 2014, Schultze et al., 1996), while the NTR of betacoronaviruses including BCoV and HCoV-OC43 was shown to bind to O-acetylated sialic acids (Künkel and Herrler, 1993, Peng et al., 2012, Schultze et al., 1991, Vlasak et al., 1988). Only the NTR of MHV (domain A) is known to interact with a protein receptor, being mCEACAM1a (Peng et al., 2011), while lacking any detectable sialic acid-binding activity (Langereis et al., 2010). However, as the NTR of MHV displays the β-sandwich fold of the galectins, a family of sugar-binding proteins, it probably has evolved from a sugar-binding domain (Li, 2012).
The presence of RBDs in different domains of the S protein that can bind either proteinaceous or glycan receptors illustrates a functional modularity of this glycoprotein in which different domains may fulfill the role of binding to cellular attachment or entry receptors. The CoV S protein is thought to have evolved from a more basic structure in which receptor recognition was confined to the CTR within S1 (Li, 2015). The observed deletions of the NTR in some CoV species in nature are indicative of a less stringent requirement and integration of this domain with other regions of the spike trimer compared to the more C-terminally located domains of S1 and support a scenario in which the NTR has been acquired at a later time point in CoV evolutionary history. For example, the NTR of MHV, which displays a human galectin-like fold, was suggested to originate from a cellular lectin acquired early on in CoV evolution (Peng et al., 2011). Acquisition of glycan-binding domains and fusion thereof to the ancestral S protein may have resulted in a great extension of CoV host range and may have caused an increase in CoV diversity. The general preference of the NTR and CTR to bind to, respectively, glycan or protein receptors may be related to their arrangement in the S protein trimer. In contrast to the CTR, which is located in the center of the S trimer, the NTR is more distally oriented (Fig. 1B). As protein–glycan interactions are often of low affinity, the more distal orientation of domain A may allow multivalent receptor interactions, thereby increasing avidity. Interestingly, some CoVs appear to have a dual receptor usage as they may bind via their NTR and CTR to glycan and protein receptors, respectively (Fig. 2).
3.2. CoV Protein Receptor Preference
Although the number of currently known CoV receptors is limited, receptor usage does not appear to be necessarily conserved between closely related virus species such as HCoV-229E (APN) and HCoV-NL63 (ACE2), whereas identical receptors (ACE2) can be targeted by virus species from different genera such as HCoV-NL63 and SARS-CoV. It seems that CoVs prefer certain types of host proteins as their entry receptor, with three out of four of the so far identified proteinaceous receptors being ectopeptidases (APN, ACE2, and DPP4), although enzymatic activity of these proteins was shown not to be required for infection by their respective viruses (Bosch et al., 2014). Possibly, the localization to certain membrane microdomains and efficient internalization of two of these proteins in polarized cells (APN and DPP4) may contribute to their suitability to function as entry receptors (Aït-Slimane et al., 2009). In the case of MERS-CoV, the region of DPP4 that is bound by the S protein coincides with the binding site for its physiological ligand adenosine deaminase (Raj et al., 2014). Employment of conserved epitopes such as these may also contribute to the cross-species transmission potential of viruses (Bosch et al., 2014), as is exemplified by MERS-CoV being able to use goat, camelid, cow, sheep, horse, pig, monkey, marmoset, and human DPP4 as entry receptor (Barlan et al., 2014, Eckerle et al., 2014, Falzarano et al., 2014, Müller et al., 2012, van Doremalen et al., 2014). Similarly, this may apply for the ability of feline, canine, porcine, and human CoVs to use fAPN as entry receptor, at least in vitro (Tresnan et al., 1996).
4. S Protein Proteolytic Cleavage and Conformational Changes
Coronavirus entry is a tightly regulated process that appears to be orchestrated by multiple triggers that include receptor binding and proteolytic processing of the S protein and that ultimately results in virus-cell fusion. It is initiated by virion attachment mediated through interaction of either the NTR or CTR (or both) in the S1 subunit of the spike protein with host receptors. Upon attachment, the virus is taken up via receptor-mediated endocytosis by clathrin- or caveolin-dependent pathways (Burkard et al., 2014, Eifart et al., 2007, Inoue et al., 2007, Nomura et al., 2004) although other entry routes have also been reported (Wang et al., 2008). Prior to and/or during endocytic uptake the CoV S protein is proteolytically processed. The spike protein may contain two proteolytic cleavage sites. One of the cleavage sites is located at the boundary between the S1 and S2 subunits (S1/S2 cleavage site), while the other cleavage site is located immediately upstream of the first fusion peptide (S2′ cleavage site). Although not irrevocably proven, it is expected that all CoVs depend on proteolytic cleavage on or close to S2′ for fusion to occur. Virus-cell fusion thus not only critically depends on the conformational changes following spike–receptor engagement, and perhaps on acidification of endosomal vesicles (Eifart et al., 2007, Matsuyama and Taguchi, 2009, Zelus et al., 2003), but also on proteolytic activation of the S protein by proteases along the endocytic route (Burkard et al., 2014, Simmons et al., 2005). Indeed, inhibition of intracellular proteases has been shown to block virus entry and virus-cell fusion (Burkard et al., 2014, Frana et al., 1985, Simmons et al., 2005, Yamada and Liu, 2009). The specific proteolytic cleavage requirements of the S protein at the S1/S2 boundary and particularly at the S2′ site may furthermore determine the intracellular site of fusion (Burkard et al., 2014). In agreement herewith, it has become evident that the protease expression profile of host cells may form an additional determinant of the host cell tropism of coronaviruses (Millet and Whittaker, 2015).
Analysis of the CoV S prefusion conformation suggests that relocation (or shedding) of the S1 subunits that cap the S2 subunit is a prerequisite for the conformational changes in S2 that ultimately result in fusion. Shedding of S1 probably requires receptor binding as well as proteolytic processing at S1/S2. The cryo-EM structure indicates that the S1/S2 proteolytic cleavage site is accessible to proteases prior to spike–receptor interaction, and depending on the particular cleavage site present may already be processed in the cell in which the virions are produced. As indicated earlier, the conformational changes in the S protein that result in virus-cell fusion most likely also require cleavage at the S2′ site immediately upstream of the fusion peptide. Interestingly, the S2′ cleavage site is located within an α-helix exposed on the prefusion S structure which prevents efficient proteolytic cleavage (Robertson et al., 2016). This indicates the necessity for preceding conformational changes induced by receptor binding and subsequent shedding of S1, upon which the secondary structure of the S2′ site transforms into a cleavable flexible loop. Following proteolytic cleavage activation at the S2′ site, hydrophobic interactions between the fusion peptide and the adjacent S2 helices are disturbed which allows the four α-helices and the connecting regions that make up the HR1 region in the prefusion S protein to refold into a long trimeric coiled coil (Fig. 1D). This coiled coil forms an N-terminal extension of the central helix projecting the fusion peptide(s) toward the target membrane. Successively, the fusion peptide(s) will be inserted into the limiting membrane of the host cell endocytic compartment. Next, as a consequence of S2 rearrangements, the two HR regions will interact to form an antiparallel energetically stable six-helix bundle (Bosch et al., 2003, Bosch et al., 2004), enabling the close apposition and subsequent fusion of the viral and host lipid bilayers.
5. Tropism Changes Associated with S Protein Mutations
Changes in the S protein may result in an altered host, tissue, or cellular tropism of the virus. This is clearly exemplified by genomic recombination events that result in exchange of (part of) the S protein and in a concomitant change in tropism. The propensity of CoVs to undergo homologous genomic recombination has been exploited for the genetic manipulation of these viruses (de Haan et al., 2008, Haijema et al., 2003, Kuo et al., 2000). To this end, interspecies chimeric coronaviruses were generated, which carried the spike ectodomain of another CoV and which could be selected based on their altered requirement for an entry receptor. Exchange of S protein genes may also occur in vivo, resulting in altered tropism as is illustrated by the occurrence of serotype II feline infectious peritonitis virus (FIPV). This virus results from a naturally occurring recombination event between feline and canine CoVs (CCoVs) in which the feline virus acquires a CCoV spike gene (Herrewegh et al., 1995, Terada et al., 2014). As a result of the acquisition of this new S protein, the rather harmless enteric feline CoV (FECV) turns into a systemically replicating and deadly FIPV. As FECV has a strict feline tropism (Myrrha et al., 2011), while CCoV has been shown to infect feline cells (Levis et al., 1995), it is likely that serotype II FIPVs arise in cats coinfected with serotype I FECV and CCoV. Furthermore, as different recombination sites have been observed for each serotype II FIPV, while serotype II FECVs have not been observed, it appears that serotype II FIPVs exclusively result of reoccurring recombination events (Terada et al., 2014). In addition to these feline–CCoV recombinants, a chimeric porcine coronavirus with a TGEV backbone and a spike of the porcine epidemic diarrhea virus (PEDV) was recently isolated from swine fecal samples in Italy and Germany, likely also resulting from a recombination event (Akimkin et al., 2016, Boniotti et al., 2016). Moreover, the α-CoV HKU2 BtCoV probably resulted from genomic recombination as it encodes an S protein that resembles a betacoronavirus S protein except for its N-terminal region that is similar to that of alphacoronaviruses (Lau et al., 2007). Thus, such genomic recombination events are not necessarily restricted to occur between viruses of the same genus.
5.1. S1 Receptor Interactions Determining Tropism
5.1.1. S1 NTR Changes
Several changes in the amino-terminal domain of S1 have been associated with changes in the tropism of the virus. For example, for several α-CoVs, loss of NTR of the S protein appears to be accompanied with a loss of enteric tropism. While the porcine CoV TGEV displays a tropism for both the gastrointestinal and respiratory tract, the closely related PRCoV, which lacks the sialic acid-binding N-terminal region (Krempl et al., 1997), only replicates in the respiratory tract. The loss of sialic acid-binding activity by four-amino acid changes in the NTR of its S protein resulted in an almost complete loss of enteric tropism (Krempl et al., 1997). Similar to TGEV, enteric serotype I FCoVs also have been reported to bind to sialic acids (Desmarets et al., 2014). Large deletions within the S1 subunit corresponding to the N-terminal region have been found in variants of the systemically replicating FIPV (strains UU16, UU21, and C3663) after intrahost emergence from enteric FECV (Chang et al., 2012, Terada et al., 2012). Also FIPVs seem to have lost the ability to replicate in the enteric tract (Pedersen, 2014). Clinical isolates of human coronavirus 229E as well as of the related alpaca coronavirus, both of which cause respiratory infections, encode relatively short spike proteins that lack the NTR (Crossley et al., 2012, Farsani et al., 2012). In contrast, closely related bat coronaviruses with intestinal tropism contain S proteins with a NTR or sometimes even two copies of the NTR (Corman et al., 2015) (Fig. 2). Overall, these observations suggest that the alphacoronavirus spike NTR—in particular its sialic acid-binding activity—may contribute to the enteric tropism of these alphacoronaviruses, while it is not required for replication in the respiratory tract or in other extraintestinal organs. It has been hypothesized that the sialic acid-binding activity of the spike protein can allow virus binding to (i) soluble sialoglycoconjugates that may protect the virus from hostile conditions in the stomach or (ii) to mucins that may prevent the loss of viruses by intestinal peristalsis and allow the virus to pass the thick mucus barrier, thereby gaining access to the intestinal cells to initiate infection (Schwegmann-Wessels et al., 2003).
Besides deletions of entire domains of the S protein, more subtle changes consisting of amino acid substitutions in S1 NTR may also suffice to alter the virus’ tropism. For example, MHV variants have been observed that acquired the ability to use the human homologue of their murine CEACAM1a receptor to enter cells as a result of mutations in their RBD that is located in S1 NTR (Baric et al., 1999).
5.1.2. S1 CTR Changes
As the CTR of the S1 subunit contains the protein RBD for most CoVs, also mutations in this part of S have been associated with changes in the virus’ tropism. Perhaps the most well-known example of viral cross-species transmission involves the SARS-CoV. Studies support a transmission model in which a SARS-like CoV was transmitted from Rhinolophus bats to palm civets, which subsequently transmitted the palm civet-adapted virus to humans at local food markets in southern China (Li et al., 2006). According to this model, SARS-like viruses adapted to both the palm civet and human host, which was reflected in the rapid viral evolution observed for these viruses within these species (Song et al., 2005). Two-amino acid substitutions within the RBD were elucidated that are of relevance for binding to the ACE2 proteins of palm civets and humans (Li et al., 2005b, Li et al., 2006, Qu et al., 2005). From these studies it appears that due to strong conservation of ACE2 between mammalian species only a few amino acid alterations within the RBD are needed to change coronavirus host species tropism. Indeed serial passage of SARS-CoVs in vitro or in vivo can rapidly lead to adaptation to new host species (Roberts et al., 2007). SARS-like viruses isolated from bats displayed major differences including a deletion in the ACE2 RBM compared to human SARS-CoV (Drexler et al., 2010, Ren et al., 2008) and as a consequence were unable of using human ACE2 as an entry receptor (Becker et al., 2008). However, recently a novel SARS-like BtCoV was identified, which could use ACE2 of Rhinolophus bats, palm civets as well as of humans as a functional receptor (Ge et al., 2013). These findings not only provide further evidence that bats are indeed the natural reservoir for SARS-like CoVs, but also that these bat coronaviruses can directly include human ACE2 in their receptor repertoire. The detection of sequences of SARS-CoV-like viruses in palm civets and raccoon dogs (Guan et al., 2003, Tu et al., 2004) therefore probably reflects the unusually wide host range of these viruses. A similar promiscuous receptor usage is also observed for MERS-CoV which binds to DPP4 of many species (Barlan et al., 2014, Eckerle et al., 2014, Falzarano et al., 2014, Müller et al., 2012, van Doremalen et al., 2014) as indicated earlier.
Just as SARS like and MERS-CoVs are able to use entry receptors of different host species, also several α-CoVs display promiscuity to orthologous receptors. For example, the feline APN molecule can be used as a receptor by feline (serotype II FIPV), canine (CCoV), porcine (TGEV), and human (HCoV-229E) α-CoVs in cell culture (Tresnan and Holmes, 1998, Tresnan et al., 1996). Conversely, serotype II FIPV can only enter cells expressing feline APN (Tresnan and Holmes, 1998). The ability of TGEV and CCoV to use feline APN as a receptor probably results from strong conservation of the viral-binding motif (VBM) among APN orthologs in combination with the RBDs recognizing APN in a similar fashion (Reguera et al., 2012). Though recruiting the same receptor, HCoV-229E binds another domain within APN, which apparently is also conserved in feline APN (Kolb et al., 1997, Tusell et al., 2007). Conservation of the VBM obviates the need for large adaptations within the RBD of these viruses to orthologous receptors allowing more facile cross-species transmission.
Other mutations in the S1 CTR associated with altered tropism have been described for the β-CoV MHV. Similar to the humanized CEACAM1a-recognizing MHV variant, serial passaging of virus-infected cells resulted in the selection of viruses with an extended host range, which were subsequently shown to be able to enter cells in a heparan sulfate-dependent and CEACAM1a-independent manner (de Haan et al., 2005, Schickli et al., 1997). Two sets of mutations in the S protein were shown to be critically required for this phenotype, both of which resulted in the occurrence of multibasic heparan sulfate-binding sites. While one heparan sulfate-binding site was located in the S2 subunit immediately upstream of the fusion peptide, the other was located in the S1 CTR. The presence of this latter, but not of the former, domain resulted in MHV that depended on both heparan sulfate and CEACAM1a for entry. Additional introduction of the second heparan sulfate-binding site enabled the virus to become mCEACAM1a independent (de Haan et al., 2006). In addition, a mutation of the HVR of S1 may affect CoV tropism as was demonstrated for the MHV strain JHM (MHV-JHM). The spike protein of MHV-JHM may induce receptor-independent fusion (Gallagher et al., 1992, Gallagher et al., 1993). However, deletion of residues in HVR of MHV-JHM resulted in the spike protein being entirely dependent on CEACAM1a binding for fusion (Dalziel et al., 1986, Gallagher and Buchmeier, 2001, Phillips and Weiss, 2001).
5.2. Changes in Proteolytic Cleavage Site and Other S2 Mutations Associated with Altered Tropism
5.2.1. Changes in Proteolytic Cleavage Sites
Although the S2 subunit does not appear to contain any RBDs, several mutations in this subunit have been associated with changes in the virus’ tropism. Some of these changes affect the cleavage sites in the S protein that are located at the S1/S2 boundary or immediately upstream of the fusion peptide (S2′ cleavage site). As these cleavages appear to be essential for virus-cell fusion, the availability of host proteases to process the S protein is of critical importance for the virus’ tropism. The importance of S protein cleavage at the S1/S2 boundary for the tropism of the virus is exemplified by the BtCoV HKU4, which is closely related to the MERS-CoV. Although domain B of the HKU4 S protein can interact with both bat and human DPP4, it is only in the context of bat cells, but not human cells, that the virus can utilize these molecules as entry receptors (Yang et al., 2014). In contrast, MERS-CoV can enter cells of human and bat origin via both DPP4 orthologues. This difference results from host restriction factors at the level of proteolytic cleavage activation. Two-amino acid substitutions (S746R and N762A) in the S1/S2 boundary of the S protein were shown to be crucial for the adaptation of bat MERS-like CoV to the proteolytic environment of the human cells (Yang et al., 2015).
Although probably not directly responsible for the tropism change associated with the enterically replicating FECV evolving into the systemically replicating FIPV, loss of a furin cleavage site at S1/S2 junction is observed in the majority of the FIPVs, whereas this furin cleavage site is strictly conserved in the parental FECV strains (Licitra et al., 2013). Apparently, conservation of this furin cleavage site is not required for efficient systemic replication. However, as FIPV is generally not found in the feces of cats, it may well be that loss of the furin cleavage site at S1/S2—as well as mutations in other parts of the genome, such as the accessory genes—may prevent efficient replication of FIPV in the enteric tracts.
Besides the influence of the S1/S2 cleavage site, virus tropism may also depend on the S2′ cleavage site upstream of FP1. In contrast to wild-type MHV strain A59, a recombinant MHV carrying a furin cleavage site at this position was shown to no longer depend on lysosomal proteases for efficient entry to occur (Burkard et al., 2014). As a consequence, this virus was able to infect cells in which trafficking to lysosomes was inhibited. Cleavage at the S2′ site may also be important for the tropism of PEDV, which causes major damage to the biofood industry in Asia and the Americas (Lee, 2015, Song et al., 2015). PEDV replication in cell culture is strictly dependent on trypsin-like proteases, a requirement which is expected to limit its tropism in vivo to the enteric tract. The trypsin dependency of PEDV entry was shown, however, to be lifted after introduction of a furin cleavage site at the S2′ cleavage site by a single-amino acid substitution. Such mutations may potentially affect the spread of this virus in the pig by allowing it to replicate in nonenteric tissues in the absence of trypsin-like proteases (Li et al., 2015).
5.2.2. Other S2 Mutations Associated with Altered Tropism
Mutations in other parts of the S2 subunit than those affecting the proteolytic cleavage sites may also influence the tropism of different CoVs. Several studies report a correlation between mutations in the HR1 region of FCoVs and the conversion of FECV into FIPV (Bank-Wolf et al., 2014, Desmarets et al., 2016, Lewis et al., 2015). Such a correlation appeared even more convincing for mutations found in the recently identified FP2 (Chang et al., 2012, Ou et al., 2016). While these correlations suggest an important role for the S protein in the transition of FECV into FIPV, the causal relationship between these mutations in S and FIP remains to be determined. It is plausible, however, that such mutations may play a role in the acquired ability of FIPVs to infect macrophages. Indeed, for serotype II FCoV, the ability to replicate in macrophages was shown to be determined by residues located in the C-terminal part of the S2 subunit, although the responsible residues were not identified (Rottier et al., 2005).
Also for other CoVs, mutations in the S2 subunit have been linked to changes in the virus’ tropism. A serially passaged MHV-A59 virus was shown to obtain mutations (M936V, P939L, F948L, and S949I) in and adjacent to the HR1 region which conveyed host range expansion of the mutant virus to normally nonpermissive mammalian cell types in vitro (Baric et al., 1999, McRoy and Baric, 2008). Contrary, Krueger et al. reported three mutations in the S2 subunit of MHV-JHM (V870A located upstream of the S2′ cleavage site and A994V and A1046V located in the HR1 region) all of which reduced the CEACAM1a-independent fusogenicity of this virus (Krueger et al., 2001). Many studies on MHV-JHM point to a crucial role of a leucine at amino acid position 1114 in S protein fusogenicity. The MHV S cryo-EM structure demonstrates that the L1114 residue is located in the central helix and contributes to interprotomer interactions. A L1114F substitution in the MHV-JHM S protein was observed in a mutant strain of JHM and correlated with an increased S1–S2 stability and the loss of the ability to induce CEACAM1a-independent fusion (Taguchi and Matsuyama, 2002), while a substitution of the same residue to an Arg (L1114R) reduced the neurotropism of this virus (Tsai et al., 2003). Mutants resistant to a monoclonal antibody (Wang et al., 1992) and soluble receptor (Saeki et al., 1997) also correlate with substitutions at this specific residue, illustrating the importance of this residue in S fusogenicity. For the MERS-CoV, mutations in HR1 have been identified that are thought to be associated with its adaptive evolution (Forni et al., 2015). Among these sites, position 1060 is particularly interesting, as it appears to correspond to substitutions found in MHV and IBV that modify the tropism of these viruses (MHV: E1035D; IBV: L857F; Navas-Martin et al., 2005, Yamada et al., 2009). Substitution E1035D in HR1 of MHV was shown to restore the hepatotropism of an otherwise nonhepatotropic MHV, the latter resulting from mutations in the S1 NTR and the S1/S2 cleavage site. These studies collectively indicate that mutations in and close to the HR regions may affect CoV tropism, possibly by affecting the metastability and consequently fusogenicity of the S protein and/or the formation of the postfusion six-helix bundle.
6. Concluding Remarks
It appears that changes in the S protein associated with altered tropism can be found in several regions of the spike protein. These regions obviously include the NTR and CTR of S1 that are involved in the interaction with attachment and/or entry receptors. Substitutions within the S1 RBDs may convey an altered viral tropism by adaptation of the virus to new or orthologous entry receptors. In addition, the S protein cleavage sites are important for host tropism as the processing of these sites by host proteases will critically affect the removal of the S1-mediated locking of the S2 prefusion conformation by shedding of S1 (S1/S2 cleavage site) and the release of the fusion peptide(s) (S2′ cleavage site). Finally, changes in S2 (particularly in the HR regions) may compensate for yet suboptimal spike binding to orthologous receptors by which low relative affinity interactions suffice to induce the required conformational changes of the S protein that ultimately result in the formation of the postfusion six-helix bundle and virus-cell fusion.
The observation that the different domains of the S protein all contribute to the tropism of CoVs is indicative of a coordinated interplay between these domains. This interplay has also been inferred from several studies, which reported changes in one S protein subunit often to be accompanied by adaptations in the other subunit (Saeki et al., 1997, Wang et al., 1992). In addition, the interplay between S1 and S2 has also been shown to be important for changes in the tropism of the virus as indicated earlier (de Haan et al., 2006, Navas-Martin et al., 2005). The recently published cryo-EM structures of CoV spike proteins (Kirchdoerfer et al., 2016, Walls et al., 2016) now provide structural evidence for the complex interplay between the subunits and domains of the S protein.
From all these studies, a picture arises in which the S protein is progressively destabilized through receptor engagement and proteolytic activation. In this process the S1 subunits serve as a safety pin that stabilizes the fusogenic S2 trimer. The safety pin is discharged upon interaction with a specific receptor and processing by host cell proteases and thereby gives way to conformational changes of the instable S2 subunit. Subsequent release of the fusion peptide may resemble the pulling of the trigger which inevitably results in fusion of viral and host membranes through interaction of the heptad repeats regions.
Based on the presented data we propose a model in which the ability of a CoV to cross the host species barrier is critically dependent on the interplay between the different regions of the S proteins. In this model, the probable low affinity of the S1 RBD for a novel receptor must be compensated by sufficiently low S2 metastability, which depends on both proteolytic cleavage of the S protein and the S2 interprotomer interactions. These required S protein characteristics may be generated during naturally occurring quasispecies variation and may result in the ability of the virus to replicate in and adapt to a new host.
Acknowledgments
This study is supported by TOP Project Grant (91213066) funded by ZonMW and as part of the Zoonotic Anticipation and Preparedness Initiative (ZAPI project; IMI Grant Agreement No. 115760), with the assistance and financial support of IMI and the European Commission. We thank Mark Bakkers for his help in preparing figures.
Contributor Information
C.A.M. de Haan, Email: c.a.m.dehaan@uu.nl.
B.-J. Bosch, Email: b.j.bosch@uu.nl.
References
- Aït-Slimane T., Galmes R., Trugnan G., Maurice M. Basolateral internalization of GPI-anchored proteins occurs via a clathrin-independent flotillin-dependent pathway in polarized hepatic cells. Mol. Biol. Cell. 2009;20(17):3792–3800. doi: 10.1091/mbc.E09-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimkin V., Beer M., Blome S., Hanke D., Höper D., Jenckel M., Pohlmann A. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg. Infect. Dis. 2016;22(7):1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., Burbelo P.D. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5(2) doi: 10.1128/mBio.00884-14. e00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19(3):456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;26(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Baker K.A., Dutch R.E., Lamb R.A., Jardetzky T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3(3):309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- Bank-Wolf B.R., Stallkamp I., Wiese S., Moritz A., Tekes G., Thiel H.J. Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis. Vet. Microbiol. 2014;173(3–4):177–188. doi: 10.1016/j.vetmic.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Sullivan E., Hensley L., Yount B., Chen W. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 1999;73(1):638–649. doi: 10.1128/jvi.73.1.638-649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Perlman S., Gallagher T. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88(9):4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A., Merk A., Borgnia M.J., Milne J.L.S., Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U.S.A. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Donaldson E., Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr. Opin. Virol. 2011;1(6):624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. Dis. 2016;22(1):83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Rottier P.J.M. Nidovirus entry into cells. In: Perlman S., Gallagher T., Snijder E., editors. Nidoviruses. American Society of Microbiology; Washington, DC: 2008. pp. 157–178. [DOI] [Google Scholar]
- Bosch B.J., Van Der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Smits S.L., Haagmans B.L. Membrane ectopeptidases targeted by human coronaviruses. Curr. Opin. Virol. 2014;6(1):55–60. doi: 10.1016/j.coviro.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Mishra N., Jain K., East M., Syndrome R., Quasispecies C. Dromedary camels in Saudi Arabia include homologues of human isolates revealed through whole-genome analysis etc. mBio. 2014;5(3):1–5. doi: 10.1128/mBio.01146-14. Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10(11):e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71(12):3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- Chan F.J., To K.K., Tse H., Jin D.-Y., Yuen K.-Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.W., Egberink H.F., Halpin R., Spiro D.J., Rottier P.J.M. Spike protein fusion peptide and feline coronavirus virulence. Emerg. Infect. Dis. 2012;18(7):1089–1095. doi: 10.3201/eid1807.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89(23):11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley B.M., Mock R.E., Callison S.A., Hietala S.K. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses. 2012;4(12):3689–3700. doi: 10.3390/v4123689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel R.G., Lampert P.W., Talbot P.J., Buchmeier M.J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J. Virol. 1986;59(2):463–471. doi: 10.1128/jvi.59.2.463-471.1986. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3016306 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Li Z., te Lintelo E., Bosch B.J., Haijema B.J., Rottier P.J.M. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005;79(22):14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., te Lintelo E., Li Z., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J.M. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80(22):10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Haijema B.J., Masters P.S., Rottier P.J.M. Manipulation of the coronavirus genome using targeted RNA recombination with interspecies chimeric coronaviruses. Methods Mol. Biol. 2008;454:229–236. doi: 10.1007/978-1-59745-181-9_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357(6377):417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarets L.M.B., Theuns S., Roukaerts I.D.M., Acar D.D., Nauwynck H.J. Role of sialic acids in feline enteric coronavirus infections. J. Gen. Virol. 2014;95(9):1911–1918. doi: 10.1099/vir.0.064717-0. [DOI] [PubMed] [Google Scholar]
- Desmarets L.M.B., Vermeulen B.L., Theuns S., Conceição-Neto N., Zeller M., Roukaerts I.D.M. Experimental feline enteric coronavirus infection reveals an aberrant infection pattern and shedding of mutants with impaired infectivity in enterocyte cultures. Sci. Rep. 2016;6:20022. doi: 10.1038/srep20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84(21):11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquerroy B.S., Vigouroux A., Rottier P.J.M., Rey F.A., Berend T., Bosch J. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335(2):276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle I., Corman V.M., Müller M.A., Lenk M., Ulrich R.G., Drosten C. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg. Infect. Dis. 2014;20(2):276–279. doi: 10.3201/eid2002.131182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifart P., Ludwig K., Böttcher C., de Haan C.A.M., Rottier P.J.M., Korte T., Herrmann A. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 2007;81(19):10758–10768. doi: 10.1128/JVI.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10(8):e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsani S.M.J., Dijkman R., Jebbink M.F., Goossens H., Ieven M., Deijs M. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes. 2012;45(3):433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Filippi G., Cagliani R., De Gioia L., Pozzoli U., Al-Daghri N. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci. Rep. 2015;5:14480. doi: 10.1038/srep14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56(3):912–920. doi: 10.1128/jvi.56.3.912-920.1985. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=252664&tool=pmcentrez&rendertype=abstract Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J., Perlman S. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology. 1992;19(1):517–522. doi: 10.1016/0042-6822(92)90223-C. http://www.ncbi.nlm.nih.gov/pubmed/1413526 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J., Perlman S. Dissemination of MHV4 (strain JHM) infection does not require specific coronavirus receptors. Adv. Exp. Med. Biol. 1993;342:279–284. doi: 10.1007/978-1-4615-2996-5_43. http://www.ncbi.nlm.nih.gov/pubmed/8209743 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87(24):13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J.M. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77(8):4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Vennema H., Horzinek M.C., Rottier P.J.M., de Groot R.J. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212(2):622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78(12):6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Ge X., Wang L.-F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12(1):221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86(23):12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81(16):8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A.F., Hegyi A., Siddell S.G. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. Gen. Virol. 1997;78(11):2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71(4):3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger D.K., Kelly S.M., Lewicki D.N., Ruffolo R., Gallagher T.M. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 2001;75(6):2792–2802. doi: 10.1128/JVI.75.6.2792-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkel F., Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195(1):195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74(3):1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., van Vliet A.L.W., Boot W., de Groot R.J. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J. Virol. 2010;84(17):8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Wang M., Lam C.S.F. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367(2):428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12(1):193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454(7201):177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Cardellichio C.B., Scanga C.A., Compton S.R., Holmes K.V. Multiple receptor-dependent steps determine the species specificity of HCV-229E infection. Adv. Exp. Med. Biol. 1995;380:337–343. doi: 10.1007/978-1-4615-1899-0_55. http://www.ncbi.nlm.nih.gov/pubmed/8830504 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Lewis C.S., Porter E., Matthews D., Kipar A., Tasker S., Helps C.R., Siddell S.G. Genotyping coronaviruses associated with feline infectious peritonitis. J. Gen. Virol. 2015;96(Pt. 6):1358–1368. doi: 10.1099/vir.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86(5):2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wong S.-K., Li F., Kuhn J.H., Huang I.-C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 2006;80(9):4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wicht O., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.-J. A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea coronavirus trypsin-independent for cell entry and fusion. J. Virol. 2015;89(15):8077–8081. doi: 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19(7):1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Eddy N.R., Noel J.K., Whitford P.C., Wang Q., Ma J., Onuchic J.N. Order and disorder control the functional rearrangement of influenza hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12049–12054. doi: 10.1073/pnas.1412849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.-H., Qin L., Li Y. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009;83(21):11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRoy W.C., Baric R.S. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J. Virol. 2008;82(3):1414–1424. doi: 10.1128/JVI.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Raj V.S., van Kuppeveld F.J.M., Rottier P.J.M., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87(16):9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.A., Raj V.S., Muth D., Meyer B., Kallies S., Smits S.L. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3(6) doi: 10.1128/mBio.00515-12. e00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrrha L.W., Silva F.M.F., de Oliveira Peternelli E.F., Junior A.S., Resende M., de Almeida M.R. The paradox of feline coronavirus pathogenesis: a review. Adv. Virol. 2011;2011:109849. doi: 10.1155/2011/109849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Martin S., Hingley S.T., Weiss S.R. Murine coronavirus evolution in vivo: functional compensation of a detrimental amino acid substitution in the receptor binding domain of the spike glycoprotein. J. Virol. 2005;79(12):7629–7640. doi: 10.1128/JVI.79.12.7629-7640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 2004;78(16):8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. 78/16/8701 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Zheng W., Shan Y., Mu Z., Dominguez S.R., Holmes K.V., Qian Z. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J. Virol. 2016;90(12):5586–5600. doi: 10.1128/JVI.00015-16. JVI.00015–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. An update on feline infectious peritonitis: virology and immunopathogenesis. Vet. J. 2014;201(2):123–132. doi: 10.1016/j.tvjl.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108(26):10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Xu L., Lin Y.L., Chen L., Pasquarella J.R., Holmes K.V., Li F. Crystal structure of bovine coronavirus spike protein lectin domain. J. Biol. Chem. 2012;287(50):41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.J., Weiss S.R. MHV neuropathogenesis: the study of chimeric S genes and mutations in the hypervariable region. Adv. Exp. Med. Biol. 2001;494:115–119. doi: 10.1007/978-1-4615-1325-4_18. http://www.ncbi.nlm.nih.gov/pubmed/11774454 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Promkuntod N., van Eijndhoven R., de Vrieze G., Gröne A., Verheije M. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V.R., Skalka A.M., Flint J., Rall G.F. American Society of Microbiology; Washington, DC: 2015. Principles of Virology, Bundle. [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Smits S.L., Provacia L.B., van den Brand J.M.A., Wiersma L., Ouwendijk W.J.D. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88(3):1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordoño D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8(8):e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Qu X., Li W., Han Z., Yu M., Zhou P. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;82(4):1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.-J., Meyer B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.L., Headey S.J., Ng N.M., Wijeyewickrema L.C., Scanlon M.J., Pike R.N., Bottomley S.P. Protein unfolding is essential for cleavage within the α-helix of a model protein substrate by the serine protease, thrombin. Biochimie. 2016;122:227–234. doi: 10.1016/j.biochi.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Rottier P.J.M., Nakamura K., Schellen P., Volders H., Haijema B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005;79(22):14122–14130. doi: 10.1128/JVI.79.22.14122-14130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki K., Ohtsuka N., Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 1997;71(12):9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. http://www.ncbi.nlm.nih.gov/pubmed/9371559 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickli J.H., Zelus B.D., Wentworth D.E., Sawicki S.G., Holmes K.V. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 1997;71(12):9499–9507. doi: 10.1128/jvi.71.12.9499-9507.1997. http://www.ncbi.nlm.nih.gov/pubmed/9371612 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Gross H.-J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-0-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65(11):6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Krempl C., Ballesteros M.L., Shaw L., Schauer R., Enjuanes L., Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996;70(8):5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. http://www.ncbi.nlm.nih.gov/pubmed/8764078 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Zimmer G., Schroder B., Breves G., Herrler G. Binding of transmissible gastroenteritis coronavirus to brush border membrane sialoglycoproteins. J. Virol. 2003;77(21):11846–11848. doi: 10.1128/JVI.77.21.11846-11848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.-D., Tu C.-C., Zhang G.-W., Wang S.-Y., Zheng K., Lei L.-C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015;4(2):166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfí A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101(52):17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Matsuyama S. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J. Virol. 2002;76(3):950–958. doi: 10.1128/JVI.76.3.950-958.2002. http://www.ncbi.nlm.nih.gov/pubmed/11773370 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Shiozaki Y., Shimoda H., Mahmoud H.Y.A.H., Noguchi K., Nagao Y. Feline infectious peritonitis virus with a large deletion in the 5′-terminal region of the spike gene retains its virulence for cats. J. Gen. Virol. 2012;93(Pt. 9):1930–1934. doi: 10.1099/vir.0.043992-0. [DOI] [PubMed] [Google Scholar]
- Terada Y., Matsui N., Noguchi K., Kuwata R., Shimoda H., Soma T. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One. 2014;9(9):e106534. doi: 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan D.B., Holmes K.V. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. http://www.ncbi.nlm.nih.gov/pubmed/9782266 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70(12):8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=190961&tool=pmcentrez&rendertype=abstract Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.C., De Groot L., Pinon J.D., Iacono K.T., Phillips J.J., Seo S.H. Amino acid substitutions within the heptad repeat domain 1 of murine coronavirus spike protein restrict viral antigen spread in the central nervous system. Virology. 2003;312(2):369–380. doi: 10.1016/S0042-6822(03)00248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10(12):2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S.M., Schittone S.A., Holmes K.V. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J. Virol. 2007;81(3):1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88(16):9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80(14):7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 1988;85(12):4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., DiMaio F. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.I., Fleming J.O., Lai M.M. Sequence analysis of the spike protein gene of murine coronavirus variants: study of genetic sites affecting neuropathogenicity. Virology. 1992;186(2):742–749. doi: 10.1016/0042-6822(92)90041-M. http://www.ncbi.nlm.nih.gov/pubmed/1310195 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO | Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia . 2016. WHO. [Google Scholar]
- Widagdo W., Raj V.S., Schipper D., Kolijn K., van Leenders G.J.L.H., Bosch B.J., Bensaid A. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J. Virol. 2016;90(9):4838–4842. doi: 10.1128/JVI.02994-15. Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 2009;106(47):19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279(47):49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83(17):8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu X.B., Fang S.G., Tay F.P.L., Liu D.X. Acquisition of cell-cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS One. 2009;4(7):e6130. doi: 10.1371/journal.pone.0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Du L., Liu C., Wang L., Ma C., Tang J. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2014;111(34):12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R.S., Li F. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89(17):9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelus B.D., Schickli J.H., Blau D.M., Weiss S.R., Holmes K.V. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 degrees C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 2003;77(2):830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


