Abstract
Enveloped viruses enclose their genomes inside a lipid bilayer which is decorated by membrane proteins that mediate virus entry. These viruses display a wide range of sizes, morphologies and symmetries. Spherical viruses are often isometric and their envelope proteins follow icosahedral symmetry. Filamentous and pleomorphic viruses lack such global symmetry but their surface proteins may display locally ordered assemblies. Determining the structures of enveloped viruses, including the envelope proteins and their protein-protein interactions on the viral surface, is of paramount importance. These structures can reveal how the virions are assembled and released by budding from the infected host cell, how the progeny virions infect new cells by membrane fusion, and how antibodies bind surface epitopes to block infection. In this chapter, we discuss the uses of cryogenic electron microscopy (cryo-EM) in elucidating structures of enveloped virions. Starting from a detailed outline of data collection and processing strategies, we highlight how cryo-EM has been successfully utilized to provide unique insights into enveloped virus entry, assembly, and neutralization.
Keywords: Cryogenic electron microscopy, Tomography, Virion, Enveloped virus, Membrane-containing virus, Single particle averaging, Subtomogram averaging, Viral glycoprotein, Membrane fusion, Virion budding
Abbreviations
- BDBV
Bundibugyo virus
- CCD
charge coupled device
- CHIKV
Chikungunya virus
- cryo-EM
cryogenic electron microscopy
- CTF
contrast transfer function
- DED
direct electron detector
- DENV
Dengue virus
- DNA
deoxyribonucleic acid
- EBOV
Ebola virus
- EEEV
eastern equine encephalitis virus
- ESCRT
endosomal sorting complex required for transport
- EV71
enterovirus 71
- FIB
focused ion beam
- GP
glycoprotein
- HA
hemagglutinin
- HAZV
Hazara virus
- HBV
hepatitis B virus
- HIV
human immunodeficiency virus
- HPeV3
human parechovirus 3
- HPIV3
human parainfluenza virus 3
- JEV
Japanese encephalitis virus
- LASV
Lassa virus
- MARV
Marburg virus
- MCP
major capsid protein
- MeV
measles virus
- MHV
mouse hepatitis virus
- MLD
mucin-like domain
- NA
neuraminidase
- NC
nucleocapsid
- NDV
Newcastle disease virus
- RNA
ribonucleic acid
- RVFV
Rift Valley fever virus
- SARS-CoV
severe acute respiratory syndrome-related coronavirus
- SeV
Sendai virus
- SFV
Semliki Forest virus
- SINV
Sindbis virus
- SPA
single particle averaging
- SSP
stable signal peptide
- STA
subtomogram averaging
- TBEV
tick borne encephalitis virus
- TULV
Tula virus
- UHV
University of Helsinki virus
- VACV
Vaccinia virus
- VEEV
Venezuelan equine encephalitis virus
- VSV
vesicular stomatitis virus
- VLP
virus-like particle
- VPP
volta phase plate
- WNV
West Nile virus
- ZIKV
Zika virus
1. Introduction
Enveloped viruses encompass a large group of viruses with different morphologies and genome types, spanning across different virus families and infecting host cells from all three domains of life (Eukaryota, Bacteria and Archaea). Common to all of these viruses is that their virions harbor a lipid bilayer which associates with integral and peripheral membrane proteins. Together, these structural components create a viral envelope that encloses the genome, or an internal protein–genome complex called the nucleocapsid (NC). The virion envelope is derived from the host cell membrane by fission in the budding process of progeny virions. During entry into a new host cell, the envelope must fuse to a host cell membrane for the enclosed genome or the NC to enter the cytoplasm. This process is catalyzed by membrane fusion proteins (Harrison, 2015; Kielian, 2014).
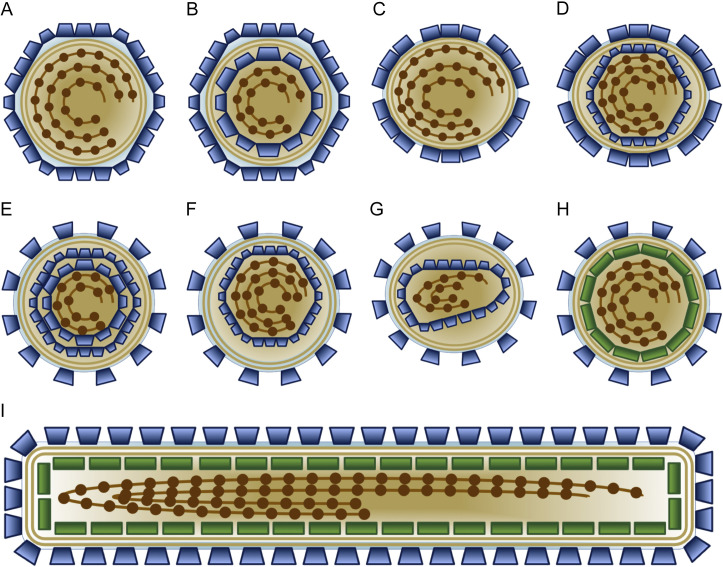
Understanding the structures of enveloped viruses is important for understanding their infection mechanisms, especially entry by membrane fusion in addition to assembly and budding by membrane fission. Various types of enveloped virus morphologies are schematized in Fig. 1 . The shapes of the enveloped virions range from spherical or icosahedral to ellipsoidal to pleomorphic and filamentous. The virion envelope surface is covered to varying degrees by membrane proteins that often form multimeric assemblies. Such morphological units on the virion surface are referred to as capsomers or spikes, as they often protrude from the surface and have pointed appearance. In animal viruses, these surface proteins are typically glycosylated (glycoproteins; GPs). In a seminal study on herpes simplex virus 1 (HSV-1), the GPs were visualized on the virion surface by cryo-electron tomography (cryo-ET) (Grünewald et al., 2003). The capsomers may further form higher order assemblies, which may have local symmetry. In many enveloped viruses the lipid bilayer is almost entirely covered by surface proteins, leaving hardly any naked membrane accessible from the virion exterior. This is the case for example in the members of Flaviviridae, such as dengue virus (DENV), where GPs form a continuous icosahedral protein shell on the envelope (Kuhn et al., 2002), and members of Hantaviridae, such as Tula virus (TULV), where GPs form locally ordered patches on the envelope (Huiskonen et al., 2010). In contrast, members of Retroviridae, such as human immunodeficiency virus (HIV), harbor very few glycoprotein spikes, leaving a large fraction of the membrane naked (Briggs et al., 2003). Some enveloped viruses harbor a symmetric nucleocapsid, which can have either icosahedral or helical symmetry. For example, members of Togaviridae, such as Semliki Forest virus (SFV), harbor an icosahedrally symmetric nucleocapsid (Fuller et al., 1995). Finally, many, but not all, enveloped viruses have a matrix protein directly under the envelope (Ke et al., 2018b; Li et al., 2016a).
Fig. 1.
Approximate schematic presentations of different types of enveloped virion morphologies. (A) Virion with an icosahedrally symmetric outer protein shell covering a lipid bilayer. Example structures include members of Flavivirus such as dengue virus (DENV) (Kuhn et al., 2002) and Phenuiviridae such as Rift Valley fever virus (RVFV) (Huiskonen et al., 2009) in addition to many membrane-containing prokaryotic viruses including members of Tectiviridae such as bacteriophage PRD1 (San Martín et al., 2002). (B) Virion with an icosahedrally symmetric outer protein shell covering a lipid bilayer and an additional icosahedrally symmetric inner protein shell. Example structures include members of Togaviridae such as SFV (Mancini et al., 2000). (C) A virion with a nonicosahedrally symmetric, but locally ordered outer protein shell covering most of the lipid bilayer and lacking a matrix layer. Example structures include members of order Bunyavirales such as Tula virus (TULV) (Hantaviridae) (Huiskonen et al., 2010), Hazara virus (HAZV) (Nairoviridae) (Punch et al., 2018) and Bunyamwera virus (BUNV) (Orthobunyaviridae) (Bowden et al., 2013). (D) A virion with an icosahedral inner capsid and a nonicosahedrally symmetric, but locally ordered outer protein shell. Example structures include members of Hepadnaviridae such as hepatitis B virus (HBV) (Dryden et al., 2006). (E) A virion with two internal icosahedrally symmetric protein shells surrounded by a lipid envelope with surface spikes. Example structures include members of Cystoviridae such as bacteriophage Φ6 (Jäälinoja et al., 2007a). (F) Members of Herpesviridae, such as HSV-1 (Grünewald et al., 2003), have an icosahedrally symmetric protein shell, enclosed by a tegument layer (not shown) and an external lipid envelope with surface glycoproteins. (G) Some members of Retroviridae, such as human immunodeficiency virus 1 (HIV-1) (Briggs et al., 2006), have mature virions with relatively few GPs on the virion envelope. (H) A virion with a lipid bilayer, decorated by glycoprotein spikes and an internal matrix protein layer is shown. Examples include members of Arenaviridae such as Lassa virus (LASV) (Li et al., 2016b), Coronaviridae such as severe acute respiratory syndrome-related coronavirus (SARS-CoV) (Neuman et al., 2006), and Paramyxoviridae such as measles virus (MeV) (Ke et al., 2018b). (I) A filamentous virion with envelope glycoprotein spikes and internal matrix layer. Examples include members of Filoviridae, such as Ebola virus (EBOV) (Bharat et al., 2012) and Pneumoviridae such as respiratory syncytial virus (RSV) (Ke et al., 2018a), in addition to filamentous forms of influenza A virus (Orthomyxoviridae) (Calder et al., 2010). Blue, viral structural protein; Light brown, lipid bilayer; Brown circles, nucleoprotein or other genome-associated protein; Brown line(s), viral genome segment(s); Green, matrix protein. Note that we have not attempted to accurately depict the genome type nor its organization or the symmetry and arrangement of different protein shells.
In this chapter, we review recent advances in understanding enveloped virus structures by cryogenic electron microscopy (cryo-EM). We will begin by an overview of different cryo-EM data collection and processing strategies relevant to the topic and then proceed to reviewing how cryo-EM structures of enveloped virions contribute to our understanding of the molecular interactions driving assembly, the dynamic nature of viral particles, their budding and membrane fusion mechanisms in addition to virus neutralization and furthermore how these structural biology studies are informing vaccine design.
2. Cryogenic electron microscopy in membrane virus research
Cryo-EM is a well-suited method for the structural analysis of enveloped virions (Subramaniam et al., 2007). As the structures are often pleomorphic (i.e., lacking a regular shape), they are often not amenable to X-ray crystallography, another structural biology technique that relies on crystallization of the sample of interest. In fact, in only a few cases has the structure of an enveloped virion been solved by X-ray crystallography (Abrescia et al., 2004, Abrescia et al., 2008). Cryo-EM methods, however, are applicable to both regular and pleomorphic virions, as no crystals are needed and structures can be determined from a relatively small amount of purified virions. The electric potential maps (or simply “cryo-EM density maps”) determined by cryo-EM have in many cases similar level of detail when compared to electron density maps determined by X-ray crystallography, although there are also subtle differences that become more significant at high resolution (Wang and Moore, 2017). Furthermore, cryo-EM allows structural investigations of more complex and rapid processes such as viral envelope–host membrane fusion and viral budding from the plasma membrane of infected cells.
As many enveloped viruses are human and animal pathogens, their production, purification and preparation for cryo-EM requires suitable containment facilities and bio-safety protocols (Sherman et al., 2013). To circumvent the need for decontaminating cryo-EM equipment, which may be in some cases impractical, purified virions can be inactivated for example by chemical fixation (Bharat et al., 2011; Halldorsson et al., 2018; Li et al., 2016b) or ultraviolet radiation (Park et al., 2011; Ye et al., 2018; Zhong et al., 2016) prior to cryo-EM sample preparation. Alternatively, virus-like particles (VLPs) that contain the relevant viral structural proteins with a lipid bilayer can be used as a model system instead of the native virion (Li et al., 2016b; Sun et al., 2013). Whether live or inactivated, virion or VLP, purified particles are prepared for cryo-EM similar to any other macromolecular complex (Thompson et al., 2016). A small aliquot of virus suspension, typically 3 μL, is pipetted on an EM sample grid, a circular metal mesh (3 mm in diameter), typically coated with a foil of holey carbon or gold. The grid, held by tweezers, is then blotted by a piece of filter paper to remove most of the sample in order to leave a very thin film of virus suspension on the grid (often not much thicker than the particle itself). The grid is then plunged in liquid ethane, which is cooled by liquid nitrogen. The extremely cold temperature of the cryogen (around –180 °C) leads to a very rapid cooling rate, and the formation of amorphous, glass-like ice, which is compatible with the vacuum of the electron microscope column and is also transparent to the electron beam.
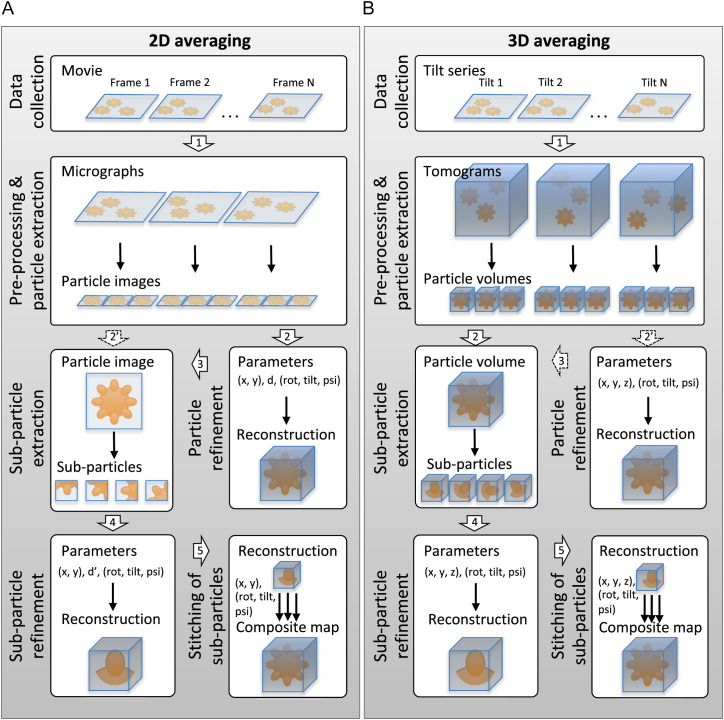
Typical cryo-EM data collection and processing workflows for enveloped viruses are outlined in Fig. 2 . Cryo-EM data can be collected either as 2D projection images (micrographs; Fig. 2A) or as a series of tilted images (a tomographic tilt series) from which a 3D tomographic volume (or a tomogram) can be calculated (Fig. 2B). To increase the inherently low signal-to-noise ratio (SNR), signal from multiple images (or volumes) needs to be averaged in a coherent manner to reach sufficiently high resolution in the average. The target resolution depends on the types of questions addressed and ranges from better than 3 Å (required to model the polypeptide chains of proteins and to see small bound molecules) to 30 Å and even lower (sufficient for addressing the organization of envelope proteins and visualizing the lipid bilayers; Table 1 ). Different approaches to averaging in the context of enveloped viruses are discussed below.
Fig. 2.
Cryo-EM data processing strategies for determining structures of enveloped virions and their glycoprotein spikes. (A) Here an enveloped virion is depicted as an orange sphere with protruding blobs that depict glycoprotein spikes. Defocus of the particle is d. Defocus of the subparticle is d’. The view direction of the particle or subparticle is defined by angles rot and tilt. The in-plane rotation of the particle projection is defined by psi. (B) Note that each image in a tomography tilt series is potentially recorded first as a movie. The range between tilt 1 and tilt N is typically [− 60,60°] and N is typically 41 (for 3-degree angular sampling) or 61 (for 2° angular sampling). Here and in the text we have assumed that the tomograms have been corrected for the effects of the contrast transfer function (CTF) during preprocessing but other approaches are possible. Here the three Euler angles (rot, tilt, psi) define the orientation of the particles and the subparticles. The block arrows refer to different data processing steps described in the text. The block arrows with dashed outlines depict less common data processing approaches.
Table 1.
Examples of enveloped virion structures and their membrane associated components determined by different cryogenic electron microscopy data processing strategies.
| Virus | Component | Method and stepsa | Resolution (Å) | Accession codes | Reference |
|---|---|---|---|---|---|
| Simian immunodeficiency virus 1 (Retroviridae) | Capsomer (Env) | 3D 1-2-4 |
28 | EMD-1216 PDB:2BF1 |
Zanetti et al. (2006) |
| Zika virus (Flaviviridae) | Virion | 2D 1-2 |
3.1 | EMD-7543 PDB:6CO8 |
Sevvana et al. (2018) |
| Sindbis virus (Togaviridae) | Virion | 2D 1-2-3-4-5 |
3.5 | EMD-9693 PDB:6IMM |
Chen et al. (2018) |
| Rift Valley fever virus (Phenuiviridae) | Capsomer (Gn–Gc) | 2D 1-2-3-4 |
7.7 | EMD-4201 PDB:6F9F | Halldorsson et al. (2018) |
| Hantaan virus (Hantaviridae) | Capsomer (Gn–Gc) | 2D 1-2′-4 |
25 | N/A | Battisti et al. (2011) |
| Tula virus (Hantaviridae) | Capsomer (Gn–Gc) | 3D 1-2-4 |
16 | EMD-3364 | Li et al. (2016a) |
| Bunyamwera virus (Orthobunyaviridae) | Capsomer (Gn–Gc) | 3D 1-2-4 |
30 | EMD-2352 | Bowden et al. (2013) |
| Hazara virus (Nairoviridae) | Capsomer (Gn–Gc) | 3D 1-2-4 |
25 | N/A | Punch et al. (2018) |
| Lassa virus (Arenaviridae) | Capsomer (GP1–GP2) | 3D 1-2-4 |
14.0 | EMD-3290 | Li et al. (2016b) |
| Measles virus (Paramyxoviridae) | Virion | 3D 1 |
N/A | N/A | Ke et al. (2018b) |
| Ebola virus (Filoviridae) | Capsomer (GP) | 2D 1-2 |
11 | EMD-8036 | Beniac and Booth (2017) |
| Influenza virus A (Orthomyxoviridae) | Virion | 3D 1 |
N/A | N/A | Calder et al. (2010) |
| Haloarcula hispanica SH1 virus (Sphaerolipoviridae) | Virion | 2D 1-2-3-4-5 |
3.8 | EMD-4633 PDB:6QT9 |
Colibus et al. (2019) |
2.1. Single particle averaging and localized reconstruction
Many enveloped viruses harbor an icosahedrally symmetric protein shell, which can be either external (Fig. 1A) or internal to the lipid bilayer (Fig. 1D and F). Some enveloped virions have two such shells sandwiching the lipid bilayer (Fig. 1B) and some have two internal shells (Fig. 1E). The structures of the icosahedrally symmetric protein shells in these types of virions can be determined by cryo-EM and single particle averaging (SPA). In standard SPA workflows, the virions are located in the micrographs (particle picking) and extracted in smaller images each containing one virion image in the middle (particle images). After determining the defocus value of each particle image, the exact location of the virion in the image (two coordinates) and orientation of the particle (three angles), the structure of the virion can be determined (or reconstructed) (Fig. 2A; steps 1 and 2). To increase the SNR and thus attainable resolution for the protein shells, icosahedral symmetry is normally applied at this stage. Several icosahedrally symmetric enveloped virus structures have been determined by this approach (see Section 3.1; Table 1). To obtain the highest possible resolution for large membrane viruses, it may also be crucial to take into account the thickness of the specimen in the 3D reconstruction process (Wolf et al., 2006).
The downside of this standard SPA workflow is that those components of the virion that are not organized in a strictly symmetric fashion will get incoherently averaged and this limits the attainable resolution, or hinders their reconstruction all together (Huiskonen, 2018). For example, if the structure of a virion presented in Fig. 1A is somewhat flexible, signal both between different particles and asymmetric units within each particle will be incoherently averaged. In order to improve the resolution for flexible enveloped virions and their symmetry-mismatched components, the localized reconstruction method can be used to divide the particle in several subparticles (Fig. 2A; subparticle extraction) (Huiskonen, 2018; Ilca et al., 2015). The orientation (three angles: rot, tilt and psi) of each subparticle and location in the particle image (two coordinates: x and y) can then be calculated and used as an initial estimate to further refine subparticles around their original positions and locations (Fig. 2A; steps 1, 2, 3, and 4). Furthermore, the defocus of each subparticle is calculated to take into account the defocus gradient across the specimen (Ilca et al., 2015). This approach has allowed improving the resolution of Rift Valley fever virus (RVFV) virions that have an icosahedrally ordered, but yet highly flexible, large protein shell (diameter ~ 110 nm), from 13 Å to 7.7 Å (Halldorsson et al., 2018). The resolution in this study and other similar studies may be limited due to overlaps between other subparticles, the membrane and the genome. In some cases, overlapping components can be subtracted from the particle image by partial signal subtraction to improve the accuracy of subparticle alignments (Bai et al., 2015; Huiskonen et al., 2007). Also, it is worth noting that in this approach only distortions in the image plane can be handled and any movement along the beam direction (corresponding to further changes in defocus) is ignored. After aligning subparticles in the image plane and reconstructing them, a composite 3D model of the virion can be created by “stitching” the entire virion from individual subparticle reconstructions (Fig. 2A, step 5). Recently the structure of the complete Sindbis virus (SINV) virion has been reconstructed this way from three separate subparticle reconstructions at 3.5 Å resolution (Chen et al., 2018).
Some virus particles are too pleomorphic for even rough orientational alignment rendering them challenging for SPA approaches. Several studies have attempted averaging glycoprotein spikes from the edge of particle images (Fig. 2A; steps 1 and 2′). This has been done for example for HNTV (Battisti et al., 2010), DENV at acidic pH (Zhang et al., 2015), and SARS (Neuman et al., 2006). In contrast to localized reconstruction where the three angles describing the orientation of the subparticle (rot, tilt and psi) can be estimated from the orientation parameters of the entire particle, in this approach only two angles of the three angles (tilt and psi) can be estimated (tilt can be assumed to be close to 90°; psi can be estimated from the normal of the membrane projection). Due to this limitation, it is not possible to calculate a 3D reconstruction without further exhaustive alignment to determine the third angle that is unknown (rot). Another limitation of this approach is that spikes at the edge of the particle can overlap other spikes and in the absence of an approximate 3D model of the particle, these overlaps cannot be removed by partial signal subtraction (Bai et al., 2015; Huiskonen et al., 2007). Due to these reasons, most studies have classified and averaged spike side-view projections only in 2D (Fig. 2A; steps 1 and 2′). At the 2D level, such studies have been informative. For example, two distinct conformations of the SARS M protein were observed by this approach (Neuman et al., 2006). In some cases 3D averaging has also been performed (Fig. 2A; step 4). In one example, the 3D structure of the tetrameric GP spike of HNTV was resolved to 25 Å from side-view subparticles (Battisti et al., 2010) (Fig. 2A; steps 1, 2′ and 4). This structure agreed well with the structure of TULV GP spike solved by cryo-ET and subtomogram averaging (STA) (Huiskonen et al., 2010). In another example, the structure of the trimeric GP spike of Ebola virus (EBOV) was determined at 11 Å resolution from side-view projections on the virion surface (Beniac and Booth, 2017) (Fig. 2A; steps 1, 2′ and 4). Consistent with a cryo-ET and subtomogram averaging (STA) investigation of GP from EBOV VLP (Tran et al., 2014), a mucin-like domain (MLD) was located to the apex and sides of each GP1 monomer, partially shielding the receptor-binding site, while the GP1 sits atop the GP2. In these side-view averaging approaches it is only possible to include spikes from the edge of the virion projection image so the packing of the spikes on the virion surface remains unattainable.
2.2. Tomography and subtomogram averaging
Tomography is a method well suited for determining structures of flexible and truly pleomorphic virions from cryo-EM data that are challenging or unsuitable for the SPA approaches described above. When virions lack well defined shape, it is impossible to combine particles with different views extracted from 2D micrographs to reconstruct a correct 3D volume. Instead, different views must be collected for each virion as a series of tilted images (Fig. 2B). These views are then combined to calculate a tomographic 3D volume (a tomogram) of the specimen region under investigation. Due to the slab-shaped geometry of the cryo-EM specimen holders and the cryo-EM grid itself, the specimen cannot be tilted to 90° and thus the angular range in a typical tomographic tilt series is limited from − 60 to + 60°. This results in incomplete sampling of information in the 3D reconstruction, which can be described as a “missing wedge” in the 3D Fourier transform of the tomogram. Because of this limitation, features in tomograms are distorted and averaging of 3D particles in different orientations is required to fully sample the information in the final 3D reconstruction (Subramaniam et al., 2007).
Similar to the 2D processing workflow described in the previous section, it is often practical to extract smaller 3D volumes, each corresponding to a single virion, from the larger tomograms (Fig. 2B; particle extraction). If these 3D particles are homogenous enough it is then possible to align, classify and average them together. For example, the first low-resolution structure of a bunyavirus (Uukuniemi virus, UUKV; Phenuiviridae) was determined by aligning single 3D volumes of virions and by applying icosahedral symmetry (Överby et al., 2008) (Fig. 2B; steps 1 and 2′). Once the orientation (rot, tilt, psi) of each particle is known from an initial alignment, it is possible to deal with any possible flexibility of the virion by extracting 3D subparticles (also referred to as subvolumes or subtomograms; Fig. 2B; step 3) (Castaño-Díez et al., 2017). These subparticles, corresponding for instance to envelope GP spikes can then be refined further (Fig. 2B; step 4) and finally a composite map of the entire virion can be stitched from the 3D reconstruction of the subparticles (Fig. 2B; step 5) (Huiskonen et al., 2010).
In most cases where tomography is applied, however, the 3D particles are too dissimilar to be aligned and averaged in a coherent manner. This is the case with truly pleomorphic virions. In these cases, subparticles of GP spikes are extracted from unaligned 3D volumes of enveloped virions (Fig. 2B; steps 1 and 2). The locations of the spikes first need to be determined by a 3D search, which can be restricted close to the membrane surface (Castaño-Díez et al., 2017; Huiskonen et al., 2014). Also, the direction of the spike can be estimated from the membrane surface normals. Once the 3D-subvolumes, each corresponding to a centered spike, have been extracted, only the rotation around the spike long axis remains to be determined before a 3D reconstruction of the subparticle can be calculated (Fig. 2B, step 4) (Förster et al., 2005; Zanetti et al., 2006). For example, several studies have produced low resolution reconstructions of Env from native HIV virions using STA (Liu et al., 2008; Zanetti et al., 2006; Zhu et al., 2003, Zhu et al., 2006). As described above, these 3D subparticle reconstructions can then be plotted back onto the original particle volumes to stitch together composite models of entire virions. Here, the completeness of this stitching depends on the coverage of the 3D picking (Huiskonen et al., 2010).
2.3. Hybrid methods
As we have outlined in Fig. 2, averaging of 2D and 3D single particles follow highly analogous workflows. In some cases it may be beneficial to mix these two. In one example of such a hybrid approach, the structure of the GP spike of prototypic foamy virus (PFV) has been studied (Effantin et al., 2016). First STA was used to determine the structure of the trimeric GP at ~ 30 Å resolution (Fig. 2B; steps 1, 2, 3, 4). Plotting back the GP structure allowed visualizing hexagonal assemblies of six GPs on the envelope (Fig. 2B; step 5). A patch of six trimers was extracted, sixfold symmetry was imposed and this volume was used as a search model to pick and extract GP subparticles directly from untilted images (Fig. 2A; steps 1 and 2′). GP spike subparticles were then subjected to conventional SPA refinement that allowed determination of a threefold symmetrized GP density map at ~ 9 Å. The density map revealed a region interpreted as a coiled-coil of three α-helices, a hallmark of viral class I fusion proteins.
3. Structures of purified virions
3.1. Virions with icosahedrally symmetric protein shells
Structures of icosahedrally symmetric protein shells in enveloped virions have been a topic of several cryo-EM studies over the past two decades. The first 3D reconstructions of Semliki Forest virus (SFV; Togaviridae) and dengue virus (DENV; Flaviviridae) virions were determined at 22 Å resolution (Fuller et al., 1995) and at 24 Å resolution (Kuhn et al., 2002), respectively, by SPA with icosahedral symmetry applied (Fig. 2A; steps 1 and 2). At such limited resolution, only the rough morphology of the virions could be resolved. However, fitting of X-ray structures into cryo-EM maps has, in many cases, allowed the creation of so-called pseudo-atomic models of the glycoprotein shells. For example, fitting of the X-ray crystallographic structure of the E protein from another flavivirus, tick borne encephalitis virus (TBEV), (Rey et al., 1995) revealed the so-called herringbone arrangement of E-protein dimers on the DENV virion surface (Kuhn et al., 2002). The advent of direct electron detectors (DEDs) and other advances in electron microscope hardware and image processing software led to a “resolution revolution” in cryo-EM making it possible to derive atomic models from cryo-EM maps of single particles alone (Kühlbrandt, 2014). A notable exception from the pre-resolution revolution era of cryo-EM is the structure of mature DENV that was determined at 3.5 Å resolution from images collected on a charge coupled device (CCD) camera (Zhang et al., 2013a).
In the post-resolution revolution era, determining structures of icosahedrally symmetric protein shells in enveloped viruses from cryo-EM data alone has become routine. For instance, the structure of mature Zika virus (ZIKV; Flaviviridae) virion has been solved by cryo-EM and SPA by two research groups at ~ 3.8 Å resolution (Kostyuchenko et al., 2016; Sirohi et al., 2016). These studies revealed that despite the high level of structural similarity to other flaviviruses such as DENV, ZIKV particles displayed greater thermal stability at higher temperature (40 °C degrees) which could account for their survival in the semen and urine. The ZIKV protein shell also presents a unique amino acid region around the Asn154 glycosylation site which may explain the neurotropic nature of ZIKV similar to West Nile virus (WNV; Flaviviridae). A recent 4.3-Å cryo-EM structure of Japanese encephalitis virus (JEV; Flaviviridae) has allowed mapping of neurovirulence factors on the virus surface (Wang et al., 2017). The same strategy has been applied to several members of the Togaviridae family. A cryo-EM structure of Sindbis virus (SINV; Togaviridae) at 3.5 Å resolution has allowed the identification of a “pocket factor,” a 20-Å long molecule, possibly a phospholipid tail, projecting from the viral lipid bilayer into a membrane-proximal hydrophobic pocket of the GP shell (Chen et al., 2018). A chikungunya virus (CHIKV; Togaviridae) VLP structure has been determined at 5.3-Å resolution. This study shows that togavirus VLPs reflect the structures of mature virions and revealed that CHIKV E1 and E2 glycoproteins are not associated with the E3 glycoprotein unlike other alphaviruses such as Venezuelan equine encephalomyelitis virus (VEEV) (Sun et al., 2013).
Structures of several prokaryotic viruses with icosahedral capsids and an internal membrane have also been studied by cryo-EM and SPA. Typically these viruses harbor major capsid proteins (MCPs) that consist of upright beta-barrel folds and fully cover an internal membrane following different arrangements (described by the triangulation [T] number). These viruses include bacteriophages PRD1 (T = 25) (San Martín et al., 2002), Bam35 (T = 25) (Laurinmäki et al., 2005), PM2 (T = 21d) (Huiskonen et al., 2004) and FLiP (T = 21d) (Laanto et al., 2017), in addition to archaeal viruses SH1 (T = 28) (Colibus et al., 2019), STIV (T = 31d) (Veesler et al., 2013) and STIV2 (T = 31d) (Happonen et al., 2010). Similarity of the MCP fold and the arrangement MCPs of the virions surface have allowed grouping these viruses to a so-called “PRD1–adenovirus lineage,” based on the extended similarity to the nonenveloped adenovirus. Interestingly, cryo-EM and SPA have revealed that giant eukaryotic dsDNA viruses share the same basic MCP building block with aforementioned prokaryotic viruses and adenovirus. In these giant dsDNA viruses compelling evidence exists for an internal lipid bilayer (Xiao and Rossmann, 2011). Structures of several viruses belonging to this group have been determined by cryoEM and SPA, including CIV (T = 147) (Khayat et al., 2010), PBCV-1 (T = 169d) (Zhang et al., 2011), PpV01 (T = 219d) (Yan et al., 2005), CroV (T = 499) (Xiao et al., 2017) and mimivirus (972 ≥ T ≥ 1200) (Xiao et al., 2009; Klose et al., 2010). Cryo-EM has also been used in characterizing other types of enveloped bacteriophages belonging to the Cystoviridae family. Members of this family have an outer lipid envelope with surface proteins enclosing two internal icosahedral protein shells (with T = 1 and T = 13 L architecture) enclosing a segmented dsRNA genome. Structures including Φ6 (Jäälinoja et al., 2007b; Sun et al., 2017), Φ8 (Jäälinoja et al., 2007b) and Φ12 (Wei et al., 2009) have highlighted structural similarities in their protein shells to nonenveloped reoviruses. Taken together, these studies have started to exemplify possible distant evolutionary links between enveloped and nonenveloped viruses.
3.2. Dynamic nature of enveloped virions
In addition to the high-resolution cryo-EM structures of enveloped virions and their icosahedrally symmetric protein shells, several cryo-EM studies have highlighted the dynamic nature of these shells. One realization is that enveloped virions may assemble from a fixed number of GP capsomers (such as 12 pentamers and N hexamers) on a defined icosahedral lattice but the resulting virion structure may be flexible. The first representative structure for members of Phenuiviridae, and for the entire order of Bunyavirales, has been studied by cryo-ET of purified UUKV virions (Phlebovirus, Phenuiviridae) (Överby et al., 2008). This study revealed that the virion has icosahedral symmetry with T = 12 triangulation and should then in principle be amenable to SPA. Later the structure of RVFV (Phlebovirus, Phenuiviridae) virions has been studied by SPA but the resolution has been limited to 13 Å due to significant flexibility of the GP layer (Halldorsson et al., 2018). Despite this limitation, a localized reconstruction method (Ilca et al., 2015) has allowed partially dealing with flexibility to improve the resolution to 7.9 Å. This was sufficient for flexible fitting of Gn and Gc X-ray crystallographic structures, revealing how the Gn chaperone protein caps the fusion loops of the fusion protein Gc (Halldorsson et al., 2018).
Highly dynamic structural changes take place during virion maturation and entry (Hasan et al., 2018a). Cryo-EM studies have played a significant role in determining the structural changes exhibited by flaviviruses in their immature, fusogenic and mature forms. The first cryo-EM structure of a mature DENV showed that the mature infectious particles are icosahedral and ~ 500 Å in diameter. The surface is smooth and is comprised of 90 copies of an E protein dimer that is closely packed, suggesting that a major rearrangement is required before host cell fusion (Kuhn et al., 2002). Cryo-EM and crystallography of fusogenic virions has revealed how the parallel E protein dimers of the mature virion first rearrange into monomers and then into the E-protein fusogenic trimers with three-fold symmetry (Allison et al., 1995; Bressanelli et al., 2004; Modis et al., 2004; Zhang et al., 2015). The first cryo-EM studies of immature flavivirus particles (DENV and yellow fever virus) have shown striking differences in terms of the considerably larger diameter (600 Å) and the presence of 60 prominent trimeric spikes (Zhang et al., 2003b). More recently, the 9-Å resolution cryo-EM structure of the immature ZIKV has shown similar characteristics and spatial arrangement to DENV (Prasad et al., 2017). These studies demonstrate that flavivirus particles undergo a series of significant conformational changes during virion maturation and entry, reflecting the highly dynamic nature of the virions.
It is becoming increasingly clear that temperature may be a significant factor in the conformation of enveloped virions, yet typically samples are prepared for cryo-EM at ambient temperature or below. Two cryo-EM studies on the structure of the mature DENV virion serotype 2 (strains NGC and 16,681) have highlighted the structural changes to DENV-2 virion at elevated temperatures (Fibriansah et al., 2013; Zhang et al., 2013b). When heated to 37 °C, the envelopes of these particles changed their appearance from smooth to bumpy. This suggests that also mature DENV-2 virions involved in human infection may have bumpy structures and therefore optimal vaccines should consider also epitopes exposed on the bumpy form of the virus.
3.3. Inherently pleomorphic virions and their glycoproteins
Enveloped viruses whose structural protein shells do not exhibit icosahedral symmetry present a challenge for high resolution cryo-EM structure determination, as these virions are unsuitable to SPA processing. In these cases, cryo-ET has been used to study the overall virion ultrastructure. Examples of such studies include those on HSV-1 (Grünewald et al., 2003), vaccinia virus (Cyrklaff et al., 2005), HIV (Briggs et al., 2006), influenza virus (Calder et al., 2010; Harris et al., 2006), rabies virus (Guichard et al., 2011), EBOV (Bharat et al., 2012), MARV (Bharat et al., 2012) and baculovirus (Wang et al., 2016a). These studies have provided valuable insights into the virion morphology and high-level organization of the structural components. For example, cryo-ET of HIV has revealed that the virion is comprised of a protein core containing the viral genome surrounded by an envelope, in which the surface glycoprotein (Env) is embedded (Briggs et al., 2006). In mature virions, the HIV genome is contained within a conical capsid made up of the capsid protein CA arranged into hexamers and pentamers. Cryo-ET and STA have also been used to determine the structure of the capsid within intact mature virions (Mattei et al., 2016). Studies on influenza virions have revealed a capsular or a filamentous shape, with HA covering most of the virion surface and NA clustering in patches (Calder et al., 2010; Harris et al., 2006). A layer of matrix protein, M1, underneath the membrane and eight ribonucleoprotein (RNP) segments were also observed. Interestingly, NA and RNPs occupy opposite poles of the virion (Calder et al., 2010). Like the filamentous form of influenza, EBOV (Bharat et al., 2011) and MARV (Bharat et al., 2012) also present a strikingly filamentous morphology.
The first three-dimensional characterizations of arenavirus and their GP spikes were performed by cryo-ET and STA on University of Helsinki virus (UHV; reptarenavirus) (Hetzel et al., 2013) and on Lassa virus (LASV; mammarenavirus) (Li et al., 2016b). These studies have revealed the higher order assembly of the GP spikes, with each spike consisting of three protomers of GP1–GP2 heterodimers organized into a tripartite complex, distributed randomly over the whole virion surface. The improvement in resolution of the spike complex from 32 Å (UHV) to 14 Å (LASV) has allowed the fitting of a crystal structure of the LASV (GP1–GP2)3 trimeric ectodomain (Hastie et al., 2017). The resulting model places the GP1 receptor-binding glycoproteins to the membrane-distal region of the spike complex, where they sit atop the GP2 class I fusion glycoproteins that protrude from the virion membrane. Inspection of the tomographic slices shows that the spike complex penetrates the membrane and interacts with the underlying layer of the Z matrix protein that links to the genome (Li et al., 2016b). Although association between stable signal peptide (SSP) and GP2 has been reported (Bederka et al., 2014; Shankar et al., 2016), the structure and topology of SSP remain unresolved at 14 Å resolution.
Cryo-ET and STA have also started to reveal the striking differences in GP multimerization and higher level clustering on the viral envelopes, as shown by several studies focusing on bunyaviruses (Bunyavirales). The organization of hantavirus GP spikes on the virion has been studied by subtomogram averaging of the TULV (Hantaviridae) glycoprotein spikes, followed by placing them back to their correct positions on the virion envelope (Huiskonen et al., 2010; Li et al., 2016a). These studies showed how the tetrameric spikes, consisting of four copies of Gn–Gc heterodimers, cover the envelope surface in locally ordered patches. Improved resolution in later models allowed proposing a model where the globular domains of Gn are membrane distal with a tetrameric stalk that descends into the membrane and the Gc fusion proteins occupy the space between the spikes (Li et al., 2016a). Another prototypic bunyavirus, Bunyamwera virus (BUNV; Orthobunyaviridae) has been studied by cryo-ET and STA (Bowden et al., 2013). This study revealed the first low-resolution structure of the trimeric glycoprotein spikes and how these spikes create locally ordered lattices on the virion surface, analogous to TULV (Huiskonen et al., 2010). Nairoviruses are another example of a pleomorphic bunyavirus displaying an ordered lattice of glycoproteins on their envelope. The first structural insights into this lattice came from STA of HAZV, revealing a tetrameric array of spikes on the virion envelope (Punch et al., 2018).
The first representation of measles virus (MeV; Paramyxoviridae) ultrastructure (Liljeroos et al., 2011), derived by cryo-ET of purified virions, revealed that the virus particles are highly pleomorphic, heterogeneous in size (50–510 nm) and show no obvious glycoprotein ordering, agreeing with the observations of similar structural investigations of SeV (Loney et al., 2009), NDV (Battisti et al., 2012), and HPIV3 (Gui et al., 2015) virions. Although a recent crystallographic study revealed that the fusion glycoprotein is capable of forming a pseudo-hexameric arrangement (Xu et al., 2015), this organization has yet to be visualized on native virions. Human orthopneumovirus (formerly respiratory syncytial virus [RSV]; family Pneumoviridae) has also been studied by cryo-ET and STA (Liljeroos et al., 2013). These virions display a wide range of morphologies, including spherical and filamentous forms of different sizes. Interestingly, a matrix layer of the M protein can only be seen sporadically in spherical particles but is common in the filamentous form. This M layer is thought to provide the membrane curvature required for budding. On the virion surface, both the fusion protein F and the attachment glycoprotein G appear to be randomly distributed. The F protein can be seen in two different conformations, pre fusion and post-fusion, as determined by classification of subvolumes. This study notes that each virion typically has only one conformation of F protein on its surface, but differences can be seen between virions. Another study goes further to suggest that the F protein is exclusively in the prefusion form on filamentous particles but in the postfusion form on spherical particles (Ke et al., 2018a). This observation suggests that the infectious form of the virus is the filamentous form and highlights the need for studying virions in their most native form, free from purification artefacts, that is by cryo-ET of budding sites on the cell surface (see Section 5).
4. Enveloped virus membrane fusion
4.1. Fusion-triggered forms of purified virions
Entry of enveloped viruses into host cells can either occur by direct fusion at the plasma membrane or by endocytic pathways (i.e., macropinocytosis, clathrin-mediated and caveolin-mediated endocytosis), leading to the formation of endocytic vesicles and eventually fusion with endosomes or lysosomes (Yamauchi and Helenius, 2013). The harsh environment of these compartments, including low pH and unusually high or low ionic concentration, often results in significant changes in the morphologies of the virions, particularly the envelope-displayed fusion proteins (Harrison, 2015).
Cryo-EM has become the principal technique to visualize the structural transitions that enveloped viruses and their fusion proteins undergo during fusion. Virions from different viral families have been imaged by cryo-EM in acidic conditions by exposing the purified virions to low pH buffer. The results appear to agree that the fusion proteins, in spite of their class (I, II, III), undergo varying degrees of structural changes. DENV virions with a bound Fab and incubated at pH 5.5 buffer have been used to reconstruct a cryo-EM map at 26 Å resolution (Zhang et al., 2015). This map revealed extended E-protein spikes with consistent shape to that of the E-protein trimer in its postfusion conformation solved by X-ray crystallography earlier (Modis et al., 2004). Averaging of side-views of the spikes at the edge of the virion allowed calculating 2D class averages of spikes (Zhang et al., 2015). As these experiments were carried out in the absence of target membranes, these results suggested that the observed trimer is a transient prefusion trimeric state of the E fusion protein, stabilized by the bound Fab (Zhang et al., 2015). Similar side-view averaging of BUNV GP spikes from the surface of the virions, imaged at low pH, has suggested that the GP lattice loses some of its contacts at the tips of the spikes mainly formed by the class II fusion GPs (Bowden et al., 2013). Likewise, 2D averages of TULV side views has demonstrated the collapse of its ordered tetrameric lattice under low pH conditions (Rissanen et al., 2017). A recent cryo-ET and STA investigation of the LASV GP spike complex supports the sensitivity of GP1 attachment glycoprotein toward ambient pH (Cohen-Dvashi et al., 2015; Li et al., 2016b; Pryce et al., 2019). Conformational differences can be observed for GP1 structure as the pH drops from neutral (8–7) to endosomal or lysosomal pH (6.5–3.0), ultimately resulting in the shedding of the GP1 and the fusogenic rearrangement of the GP2 fusion protein (Li et al., 2016b).
Not just low pH, but other factors, either physiological or nonphysiological, can also be applied to trigger the fusion protein from its prefusion condition. HRPV5 is an enveloped archeal virus with a monomeric envelope fusion protein, which does not confer to any of the existing classes I–III (El Omari et al., 2019). HRPV5 virions have been triggered to their fusogenic conformation by exposure to high temperature (55 °C). This led to a conformational change in their monomeric fusion protein, allowing it to extend far enough to conceivably reach across the host cell S-layer to bridge the virion and host cell membranes (El Omari et al., 2019). Other factors such as the concentration of potassium ions (K+) has also been shown to be crucial for fusion events. An STA study of HAZV GP has demonstrated that the Gc fusion protein can be triggered by high K+ concentration into an extended conformation and subsequently embedded into the target membrane of co-purified vesicles (Punch et al., 2018).
4.2. Virus–liposome complexes
Liposomes have been used extensively as mimics of cellular plasma and endosomal membranes in cryo-EM studies of virus fusion. Liposomes serve as an accessible tool as their lipid compositions are well-defined and can be easily manipulated to suit the physiological fusion environment. As compared to a whole cell, the relatively small sizes of liposomes also enable cryo-EM imaging without complicated sample preparations, such as focused ion beam (FIB) milling or cryo-sectioning. The effect of various factors such as pH, temperature, lipid composition, ion concentration, in addition to antibodies or fusion inhibitors, have been investigated using virus–liposome complexes (Calder and Rosenthal, 2016; Chlanda et al., 2016; Halldorsson et al., 2018).
Visualization of influenza virus–liposome fusion events at low pH has been achieved with cryo-ET and STA, showing a progression of sequential events from HA-liposome contact, membrane–membrane contact, full fusion, to redistribution of viral components and contents (Calder and Rosenthal, 2016). The M1 matrix layer has also been observed to undergo a conformational change at low pH prior to dissociating from the viral membrane, allowing membrane deformation and formation of a fusion pore (Fontana and Steven, 2013). In corroboration with crystal structures of HA obtained at neutral (Wilson et al., 1981) and fusion permissive pH (Bullough et al., 1994; Chen et al., 1999), cryo-EM investigations (Calder et al., 2010) have established that HA (class I fusion protein) undergoes a conformational change at low pH, exposing the fusion peptide for membrane fusion. Indeed, cryo-EM snapshots and tomographic slices of influenza virus–liposome interactions at low pH revealed an extended triple-stranded coiled coil HA structure, perpendicular to the membrane, that can be seen inserted into the target membrane. The membranes are pulled towards each other, first creating a dimple and then hemifusion of the two membranes. After the eventual formation of the fusion pore, the HA can be seen in the “foldback” conformation radiating from the membrane contact points (Calder and Rosenthal, 2016). Another cryo-ET study has proposed a hemifusion stalk-independent model of membrane fusion termed the “rupture-insertion” pathway utilized when the cholesterol content in the liposomes is low (Chlanda et al., 2016). Interestingly, the HA fusion peptide of influenza virus has been observed to insert into the viral membrane in cis in the absence of a target membrane (Calder et al., 2010; Calder and Rosenthal, 2016; Ruigrok et al., 1986; Skehel et al., 1982).
A recent cryo-ET and STA study of RVFV Gc (class II fusion protein) (Halldorsson et al., 2018) has shed more light on the prefusion conformation of Gc, showing how the hydrophobic fusion loop is protected prior to the fusion event, and how Gc embeds it into a target membrane upon acidification of the environment. Localized reconstructions of RVFV surface GP spikes have allowed their structures to be resolved at sufficiently high resolution (~ 8 Å) for flexible fitting of Gn and Gc X-ray crystallographic structures. This resulted in a model showing that the Gn glycoprotein shields the fusion loop by associating noncovalently with the Gc glycoprotein in the prefusion state at neutral pH (Halldorsson et al., 2018). Cryo-ET carried out at fusion permissive low pH and in the presence of liposomes showed that the Gn-shield shifts away to expose the fusion loop, allowing extension of the Gc molecule from a kinked, likely metastable conformation to a more straightened intermediate conformation. Extension of the Gc allows it to embed its fusion loops in the target membrane, with the aromatic side chains projected into the hydrophobic region of the lipid bilayer (Halldorsson et al., 2018). This Gn-fusion loop shielding mechanism resembles that of alphaviruses CHIKV (Sun et al., 2013) and SFV (Mancini et al., 2000), where the fusion peptide of the E1 fusion protein is shielded by the E2 receptor-binding protein but contrasts the homotypic shielding observed in E–E interactions of the flaviviruses DENV (Kuhn et al., 2002; Zhang et al., 2003a) and ZIKV (Sirohi et al., 2016).
5. Virus budding
Virus budding is a crucial step in the life cycle and propagation of enveloped viruses. Packaged with the newly synthesized genomic contents and viral proteins, virions escape from the host cell membrane prior to infecting more cells. Electron microscopy images show that budding virions can display different morphologies and often recruit viral matrix protein to the budding site. The assembly and organization of the matrix protein along with the spike GPs, NCs and in some cases also with the help of cellular ESCRT complexes induce a membrane curvature in the host cell membrane and form vesicles that are eventually pinched off to release the viral particles (Chen and Lamb, 2008). Other mechanisms that involve internal viral nonstructural proteins and lipid rafts have also been reported (Bavari et al., 2002; Hyatt et al., 1993; Leser and Lamb, 2005; Ono and Freed, 2001). Cryo-ET of infected cells has been proven to be a useful technique to study the morphologically variable process of virion budding (Bharat et al., 2011; Carlson et al., 2010; Ke et al., 2018a, Ke et al., 2018b).
The matrix protein (M) of MeV has been observed, using whole-cell cryo-ET and STA, to form a well-ordered lattice that lines the inner leaflet of the plasma or virion membrane of MeV-infected cells and released virus particles, respectively (Ke et al., 2018b). The M protein has also been shown to interact with the cytoplasmic or intravirionic tails of the envelope glycoproteins and the helical ribonucleoprotein (RNP) (Ke et al., 2018b). In contrast, purified MeV virions were observed to contain ~ 30 nm tubular structures composed of M protein tightly coating the inner RNP, but without M protein present at the membrane (Brown et al., 1987; Liljeroos et al., 2011). Matrix oligomerization at the membrane and association on the RNP suggest that the M protein plays an essential role in MeV assembly and budding by coordinating the M–glycoprotein and M–RNP interactions and that this is likely a common assembly mechanism utilized across the Paramyxoviridae family. These observations suggest that the methods of virus preparation and purification may result in alterations of the virus particles and subsequently impact our interpretation and understanding of the data (Kiss et al., 2014).
The roles of matrix protein and NC in virus budding have also been studied in filoviruses such as MARV. Cryo-ET and STA investigations of intact MARV virions (Bharat et al., 2011) have revealed that the membrane-associated VP40 matrix protein interacts with other viral proteins such as VP24, VP35, and NC, and that NC displays a strong structural disparity with characteristic “pointed” and “barbed” ends by analogy with actin. The analysis of MARV-infected cells by cryo-ET has shown that the helical NCs associates laterally with the inner leaflet of the host plasma membrane and are subsequently enveloped to form filopodia-like membrane protrusions prior to excision, releasing filamentous particles. The virion VP40 lattice has been observed to undergo structural rearrangement when associated with NC as compared to VP40 in VLP without NC, indicating that the VP40-NC interaction is likely required for NC envelopment and budding. Interestingly, the bullet-shaped rhabdovirus vesicular stomatitis virus (VSV), another member of Mononegavirales, appears to utilize different envelopment and budding mechanisms despite the morphological homology of its NC with that of filovirus (Ge et al., 2010). Evidently, the “barbed” ends of the VSV NC bud out first, whereas, all MARV NCs are oriented with their “pointed” ends outwards. This differential budding directionality is, however, reconciled by the authors as the requirement for the first base of the genome (3′) to be synthesized to bud first, placing the absolute RNA directionality a priority over the NC directionality.
A previous study of MARV budding has shown that filamentous particles constituted infectious virions while rounded particles of lower infectivity were released during late rounds of infection (Welsch et al., 2010). This morphological distinction has also been observed in the paramyxovirus RSV using cryo-ET in the presence of a fusion inhibitor to exclude the possibility of observing fusion events (Ke et al., 2018a). The particles are observed in various stages of the budding process including initiation, elongation, and scission. Interestingly, the particles bud in the filamentous form despite spherical forms of the virus having been also reported (Liljeroos et al., 2013). The authors suggest that this provides further evidence that the infectious form of RSV is the filamentous form.
In enveloped virions that lack a matrix protein, such as in the members of Bunyavirales order, organization of the spike GPs may become a key contributor to viral budding. For example, hantavirus TULV tetrameric GP forms locally ordered patches as shown by cryo-ET and STA. These patches have been proposed to create membrane curvature and contribute to the budding of hantaviruses (Huiskonen et al., 2010). Interestingly, nairovirus HAZV GP has also been observed to arrange in ordered tetrameric patches (Punch et al., 2018). Analogously, locally ordered patches of GP trimers have been observed also in BUNV, an orthobunyavirus, suggesting that such local order may be a generic driver of budding in pleomorphic bunyavirus virions (Bowden et al., 2013).
For retroviruses, the Gag polyprotein mediates many essential events including membrane binding, virion assembly, and genome packaging. Cryo-ET and STA analysis of native HIV-1 budding sites revealed a consistently continuous lattice of Gag polyprotein with the same organization and structure as seen in released immature virions, indicative that the organization of these particles is determined at their intracellular assembly point (Carlson et al., 2010). Cortical actin filaments were also visualized at these sites, particularly the filopodia-assisted buds, suggesting a role of actin filaments in retrovirus assembly. A lattice lacking the NC-RNA-p6 complex was also observed in some budding sites and virions, indicating that the viral genome was absent, an observation attributed to premature proteolytic maturation and failure to recruit the ESCRT machinery in some HIV-1 infected T-cells. Interestingly, the use of whole-cell cryo-ET permitted the direct visualization of HIV-1 virions and VLPs connected to each other and to the plasma membrane by highly dynamic and filamentous proteinaceous bodies, referred as tethers (Strauss et al., 2016). The localization of tethers at budding sites supported an established restriction model for HIV-1 release and dissemination. Indeed, observation of beads-on-a-string appearance on tomographic images suggested that virion budding had occurred sequentially through a tetherin-enriched microdomain.
6. Neutralization
The increasing number of outbreaks caused by arboviruses, such as DENV, ZIKV and CHIKV, has placed tremendous pressure on scientists worldwide to generate therapeutic agents or vaccines in response to the epidemics (Weaver et al., 2018). Cryo-EM has been applied extensively to study the structures of virion–antibody complexes which have provided insights into the molecular basis of antibody-mediated neutralization mechanisms and the identification of potential therapeutic immunogens.
The cryo-EM structures of DENV in complex with monoclonal antibodies (mAb) at elevated temperatures have provided a wealth of detailed information into neutralization of different DENV serotypes. These structures show that the bumpy form of DENV2 is able to bind an anti-DENV Fab 1A1D-2 (Lok et al., 2008) and E104 (Zhang et al., 2015) only at elevated temperatures. The cryo-EM structure of Fab 1A1D–DENV2 complex demonstrates that the complex formation was temperature dependent with a higher level of Fab binding at 37 °C than at ambient temperature due to the exposure of a hidden part of the Fab binding epitope on the E proteins (Lok et al., 2008). However, it has also been shown that this temperature-dependent transition from smooth to bumpy is strain-dependent. The cryo-EM structure of the DENV4 virion at 4.1-Å resolution suggests that this serotype has higher thermal stability than other DENV strains (Kostyuchenko et al., 2014). Furthermore, unlike other DENV strains, these virions do not expose the fusion loops to allow binding to flavivirus mAbs until the temperature is increased to 40 °C (Sukupolvi-Petty et al., 2013). A DENV2-specific human mAb (2D22) complex cryo-EM structure suggests that HMAb 2D22 neutralizes DENV2 by binding E dimers and prevents the E protein rearrangement required for viral entry (Fibriansah et al., 2015a). The cryo-EM structure of Fab HMAb 5J7-DENV3 complex shows that a single Fab molecule of HMAb 5J7 simultaneously binds to three functionally significant E protein domains, each on a different E protein molecule, to prevent virus attachment (Fibriansah et al., 2015b). The cryo-EM structure of a DENV1–HMAb 1F4 complex has revealed that the HMAb 1F4 binds to DI and DI–DII hinge region on an E protein monomer to prevent different stages of viral entry (Fibriansah et al., 2014) while the DENV1 specific HM14c10 targets an adjacent surface of E protein dimers to neutralize the virus by blocking virus attachment as shown by cryo-EM of the DENV1–HMAb 14c10 complex (Teoh et al., 2012). More recently, cryo-EM has been applied to study two highly neutralizing antibodies (2C8 and 3H5) against DENV E protein and to understand the difference in their capacity to promote antibody dependent enhancement (ADE) (Renner et al., 2018; Wirawan et al., 2019). It was proposed that antibody 3H5 promoted minimal ADE by its unusual binding to the viral surface resulting in poor access to the Fc region. Another study looked at the role of anti-prM antibodies in the pathogenesis of immature DENV by the cryo-EM structures of the immature DENV in complex with a Fab fragment of HMAb 1H10, at different pH values. This suggested the mechanism by which the Fab 1H10 enhanced attachment of immature DENV to liposomes by increasing dissociation of prM from E (Wirawan et al., 2019).
Recent epidemics of ZIKV in the Americas have accelerated structural studies into the neutralization mechanism of ZIKV virion–mAb complexes (Cauchemez et al., 2016; Fauci and Morens, 2016; Mlakar et al., 2016). The first cryo-EM structure of a ZIKV–mAb complex used a cross-reactive DENV human mAb (C10) which inhibited viral fusion to acidified endosomes by locking of the E proteins (Zhang et al., 2016). This was followed by the 9-Å resolution cryo-EM complex structure of ZIKV and the ZIKV specific human mAb (Fab Z23) which demonstrated the binding of Fab Z23 to DIII domain of the E protein (Wang et al., 2016b). Another study showed that the neutralizing human mAb (ZIKV-117) cross-links monomers with E dimers as well as between neighboring E dimers to prevent the conformational changes of E dimers into fusogenic trimers in acidified endosomes (Hasan et al., 2017). More recently, a 4-Å resolution cryo-EM structure of ZIKV in complex with Fab fragments of the highly specific human mAb (ZIKV-195) suggested that Fab ZIKV-195 neutralizes ZIKV by binding to two adjacent E dimers to prevent the structural reorganization to trimers required for membrane fusion (Long et al., 2019). Cryo-EM has been applied also on other flavivirus–mAb complexes. TBEV, in complex with the neutralizing mouse mAb Fab fragment (19/1786) has been resolved to a resolution of 3.9 Å revealing that this Fab neutralizes by inhibiting virus-induced membrane fusion (Füzik et al., 2018). The 14.5 Å resolution cryo-EM structure of WNV in complex with the Fab of the strongly neutralizing antibody E16 has revealed that it binds to DIII and neutralizes by preventing the conformational change of E prior to membrane fusion (Kaufmann et al., 2006).
Many alphavirus neutralizing antibodies in complex with virions have been studied by cryo-EM. These studies have shown that most of these neutralizing mAbs target the exposed E2 protein cap (Fox et al., 2015; Hasan et al., 2018b; Long et al., 2015; Porta et al., 2016; Sun et al., 2013). One of the first studies produced four ~ 15-Å resolution cryo-EM structures of CHIKV VLPs complexed with the Fab fragments of neutralizing mouse MAbs (Sun et al., 2013). This study shows that the CHK-152 mAb neutralizes by stabilizing the viral surface and preventing the exposure of fusion-loop and thereby the fusion. The three mAbs CHK-9, m10 and m242 antibodies have been suggested to block the receptor-attachment site (Sun et al., 2013). More recently, two cryo-EM studies of CHIKV VLPs complexed with Fabs of highly neutralizing human mAbs have been described providing insights into the structural basis of neutralization (Long et al., 2015; Porta et al., 2016). Furthermore, it has been suggested that the Fabs 4J21 and 5M16 bind to domains B on E2 blocking virus fusion (Long et al., 2015). The Fab 8B10 inhibits attachment by covering the receptor binding site whereas 5F10 inhibits fusion by restricting B domain movement (Porta et al., 2016). Another cryo-EM study of Fab fragments of two human mAbs has revealed a mechanism for inhibiting membrane fusion and also identified E2-W64 as a key neutralizing epitope of CHIKV E protein (Jin et al., 2015). The 16-Å resolution cryo-EM structure of CHIKV in complex with CHK-265 Fab fragments has shown that the Fab binds to domain B of E2 and induces significant conformational changes in domain A and cross-links adjacent E2 spikes to block viral entry and egress steps (Fox et al., 2015). The cryo-EM study of another alphavirus, VEEV, in complex with the highly neutralizing mAb Fab fragments (F5 and 3B4C-4) at resolution of ~ 17.5 Å has revealed the difference in their neutralization mechanisms. While both Fab fragments neutralize by stabilizing E2 trimeric spikes and preventing the viral fusion, the F5 cross-links E2 within a trimeric spike to block the receptor binding site, whereas the 3B4C-4 Fab cross links E2 from neighboring spikes to prevent the exposure of the fusion loop by steric hindrance (Porta et al., 2014). Recent cryo-EM studies looking at the SINV-EEEV chimera in complex with five different Fabs have revealed that three Fab fragments of neutralizing mAbs (EEEV-5, >EEEV-42 and EEEV-58) bound to domain A of E2 protein, which cause intraspike cross-linking, while the other two (EEEV-3 and EEEV-69) interact only with domain B favoring interspike cross-linking (Hasan et al., 2018b).
In addition to extensive cryo-EM studies of the arboviruses belonging to Flaviviridae and Togaviridae families described above, cryo-EM has been applied to study the structures of virion–antibody complexes for several viruses from other families, notably HIV and EBOV. STA has been used to study the HIV Env trimer on native virions in complex with a broadly neutralizing antibody Fab fragment, b12, and the cell surface receptor, CD4 (Liu et al., 2008). This study shows that both the Fab and the receptor cause a large conformational change that “opens” the trimer, but this change is more dramatic with the receptor CD4. It has therefore been proposed that the b12 antibody exerts its neutralizating effect by locking Env in a confirmation that prevents a further CD4 induced conformational change that is required to continue the virus life cycle. To understand the structural basis of ZMapp (a cocktail of 3 mAbs: c24G4, c4G7 and C13C6), a promising anti-EBOV therapeutic, cryo-EM structures of c2G4, c4G7 and c13C6 IgGs bound to EBOV VLPs have been studied by STA (Tran et al., 2016). This study suggests that two of these antibodies (c2G4 and c4G7) remain bound through the endocytic pathway and exert their neutralizing effect by preventing conformational changes required for fusion.
7. Conclusions
Cryo-EM has always been the method of choice to study the structures of enveloped virions. Due to technological developments in both microscope hardware and image processing software, the rate of discoveries has been accelerating in the past 5 years. Structures of regular enveloped virions whose protein shell(s) follow icosahedral symmetry can now be determined in a matter of days by SPA. Pleomorphic virions and virions in complex with receptors or antibodies remain a challenge for structure determination. Those virions or complexes that show slight deviations from perfect icosahedral symmetry may be tractable by improved computational 2D averaging approaches that deal with structural deformations accurately. When deviations are more severe or when the virion structure is truly pleomorphic or filamentous, STA remains the method of choice and is becoming more widely utilized. In the near future, more studies are expected to utilize various hybrid data processing approaches to tackle the most challenging targets for which resolution has remained limited or which have remained entirely unattainable. The 2D and 3D averaging workflows depicted in Fig. 2 will likely merge into one framework within the same software package in the future. This should facilitate routinely resolving structural components in pleomorphic, enveloped virions to near-atomic resolution, and to further address mechanisms of virus infection and neutralization of enveloped viruses in general.
References
- Abrescia N.G.A., Cockburn J.J.B., Grimes J.M., Sutton G.C., Diprose J.M., Butcher S.J., Fuller S.D., San Martín C., Burnett R.M., Stuart D.I. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- Abrescia N.G.A., Grimes J.M., Kivelä H.M., Assenberg R., Sutton G.C., Butcher S.J., Bamford J.K.H., Bamford D.H., Stuart D.I. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Allison S.L., Schalich J., Stiasny K., Mandl C.W., Kunz C., Heinz F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X.-C., Rajendra E., Yang G., Shi Y., Scheres S.H. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti A.J., Chu Y.K., Chipman P.R., Kaufmann B., Jonsson C.B., Rossmann M.G. Structural studies of Hantaan virus. J. Virol. 2010;85:835–841. doi: 10.1128/JVI.01847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti A.J., Chu Y.-K., Chipman P.R., Kaufmann B., Jonsson C.B., Rossmann M.G. Structural studies of Hantaan virus. J. Virol. 2011;85:835–841. doi: 10.1128/JVI.01847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti A.J., Meng G., Winkler D.C., McGinnes L.W., Plevka P., Steven A.C., Morrison T.G., Rossmann M.G. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S., Bosio C.M., Wiegand E., Ruthel G., Will A.B., Geisbert T.W., Hevey M., Schmaljohn C., Schmaljohn A., Aman M.J. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bederka L.H., Bonhomme C.J., Ling E.L., Buchmeier M.J. Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization. MBio. 2014;5 doi: 10.1128/mBio.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Booth T.F. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci. Rep. 2017;7:46374. doi: 10.1038/srep46374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat T.A.M., Riches J.D., Kolesnikova L., Welsch S., Krähling V., Davey N., Parsy M.-L., Becker S., Briggs J.A.G. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat T.A.M., Noda T., Riches J.D., Kraehling V., Kolesnikova L., Becker S., Kawaoka Y., Briggs J.A.G. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden T.A., Bitto D., McLees A., Yeromonahos C., Elliott R.M., Huiskonen J.T. Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S., Stiasny K., Allison S.L., Stura E.A., Duquerroy S., Lescar J., Heinz F.X., Rey F.A. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J.A.G., Wilk T., Welker R., Kräusslich H.-G., Fuller S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J.A.G., Grünewald K., Glass B., Förster F., Kräusslich H.-G., Fuller S.D. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Brown H.R., Goller N., Thormar H., Norrby E. Fuzzy material surrounding measles virus nucleocapsids identified as matrix protein. Brief report. Arch. Virol. 1987;94:163–168. doi: 10.1007/BF01313735. [DOI] [PubMed] [Google Scholar]
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Calder L.J., Rosenthal P.B. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat. Struct. Mol. Biol. 2016;23:853–858. doi: 10.1038/nsmb.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder L.J., Wasilewski S., Berriman J.A., Rosenthal P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L.-A., de Marco A., Oberwinkler H., Habermann A., Briggs J.A.G., Kräusslich H.-G., Grünewald K. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Díez D., Kudryashev M., Stahlberg H. Dynamo catalogue: geometrical tools and data management for particle picking in subtomogram averaging of cryo-electron tomograms. J. Struct. Biol. 2017;197:135–144. doi: 10.1016/j.jsb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Cauchemez S., Besnard M., Bompard P., Dub T., Guillemette-Artur P., Eyrolle-Guignot D., Salje H., Van Kerkhove M.D., Abadie V., Garel C. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet. 2016;387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.J., Lamb R.A. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Skehel J.J., Wiley D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang M., Zhu D., Sun Z., Ma J., Wang J., Kong L., Wang S., Liu Z., Wei L. Implication for alphavirus host-cell entry and assembly indicated by a 3.5 Å resolution cryo-EM structure. Nat. Commun. 2018;9:5326. doi: 10.1038/s41467-018-07704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P., Mekhedov E., Waters H., Schwartz C.L., Fischer E.R., Ryham R.J., Cohen F.S., Blank P.S., Zimmerberg J. The hemifusion structure induced by influenza virus haemagglutinin is determined by physical properties of the target membranes. Nat. Microbiol. 2016;1:16050. doi: 10.1038/nmicrobiol.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Dvashi H., Cohen N., Israeli H., Diskin R. Molecular mechanism for LAMP1 recognition by Lassa virus. J. Virol. 2015;89:7584–7592. doi: 10.1128/JVI.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colibus L.D., Roine E., Walter T.S., Ilca S.L., Wang X., Wang N., Roseman A.M., Bamford D., Huiskonen J.T., Stuart D.I. Assembly of complex viruses exemplified by a halophilic euryarchaeal virus. Nat. Commun. 2019;10:1456. doi: 10.1038/s41467-019-09451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M., Risco C., Fernández J.J., Jiménez M.V., Estéban M., Baumeister W., Carrascosa J.L. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K.A., Wieland S.F., Whitten-Bauer C., Gerin J.L., Chisari F.V., Yeager M. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Effantin G., Estrozi L.F., Aschman N., Renesto P., Stanke N., Lindemann D., Schoehn G., Weissenhorn W. Cryo-electron microscopy structure of the native prototype foamy virus glycoprotein and virus architecture. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Li S., Kotecha A., Walter T.S., Bignon E.A., Harlos K., Somerharju P., De Haas F., Clare D.K., Molin M. The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nat. Commun. 2019;10:846. doi: 10.1038/s41467-019-08728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. Zika virus in the Americas—yet another arbovirus threat. N. Engl. J. Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- Fibriansah G., Ng T.-S., Kostyuchenko V.A., Lee J., Lee S., Wang J., Lok S.-M. Structural changes in dengue virus when exposed to a temperature of 37 °C. J. Virol. 2013;87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Tan J.L., Smith S.A., de Alwis A.R., Ng T.-S., Kostyuchenko V.A., Ibarra K.D., Wang J., Harris E., de Silva A. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Ibarra K.D., Ng T.-S., Smith S.A., Tan J.L., Lim X.-N., Ooi J.S.G., Kostyuchenko V.A., Wang J., de Silva A.M. Dengue virus. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Tan J.L., Smith S.A., de Alwis R., Ng T.-S., Kostyuchenko V.A., Jadi R.S., Kukkaro P., de Silva A.M., Crowe J.E. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J., Steven A.C. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J. Virol. 2013;87:5621–5628. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster F., Medalia O., Zauberman N., Baumeister W., Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Long F., Edeling M.A., Lin H., van Duijl-Richter M.K.S., Fong R.H., Kahle K.M., Smit J.M., Jin J., Simmons G. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell. 2015;163:1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.D., Berriman J.A., Butcher S.J., Gowen B.E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- Füzik T., Formanová P., Růžek D., Yoshii K., Niedrig M., Plevka P. Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody. Nat. Commun. 2018;9:436. doi: 10.1038/s41467-018-02882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P., Tsao J., Schein S., Green T.J., Luo M., Zhou Z.H. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald K., Desai P., Winkler D., Heymann J., Belnap D., Baumeister W., Steven A. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Gui L., Jurgens E.M., Ebner J.L., Porotto M., Moscona A., Lee K.K. Electron tomography imaging of surface glycoproteins on human parainfluenza virus 3: association of receptor binding and fusion proteins before receptor engagement. MBio. 2015;6 doi: 10.1128/mBio.02393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Krell T., Chevalier M., Vaysse C., Adam O., Ronzon F., Marco S. Three dimensional morphology of rabies virus studied by cryo-electron tomography. J. Struct. Biol. 2011;176:32–40. doi: 10.1016/j.jsb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Halldorsson S., Li S., Li M., Harlos K., Bowden T.A., Huiskonen J.T. Shielding and activation of a viral membrane fusion protein. Nat. Commun. 2018;9:349. doi: 10.1038/s41467-017-02789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happonen L.J., Redder P., Peng X., Reigstad L.J., Prangishvili D., Butcher S.J. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J. Virol. 2010;84:4747–4754. doi: 10.1128/JVI.02156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A., Cardone G., Winkler D.C., Heymann J.B., Brecher M., White J.M., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.S., Saif Hasan S., Miller A., Sapparapu G., Fernandez E., Klose T., Long F., Fokine A., Porta J.C., Jiang W. A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun. 2017;8:14722. doi: 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.S., Sevvana M., Kuhn R.J., Rossmann M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018;25:13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- Hasan S.S., Sun C., Kim A.S., Watanabe Y., Chen C.-L., Klose T., Buda G., Crispin M., Diamond M.S., Klimstra W.B. Cryo-EM structures of eastern equine encephalitis virus reveal mechanisms of virus disassembly and antibody neutralization. Cell Rep. 2018;25:3136–3147.e5. doi: 10.1016/j.celrep.2018.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Zandonatti M.A., Kleinfelter L.M., Heinrich M.L., Rowland M.M., Chandran K., Branco L.M., Robinson J.E., Garry R.F., Saphire E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel U., Sironen T., Laurinmäki P., Liljeroos L., Patjas A., Henttonen H., Vaheri A., Artelt A., Kipar A., Butcher S.J. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J. Virol. 2013;87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen J.T. Image processing for cryogenic transmission electron microscopy of symmetry-mismatched complexes. Biosci. Rep. 2018;38 doi: 10.1042/BSR20170203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen J.T., Kivelä H.M., Bamford D.H., Butcher S.J. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., Jäälinoja H.T., Briggs J.A.G., Fuller S.D., Butcher S.J. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J. Struct. Biol. 2007;158:156–164. doi: 10.1016/j.jsb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., Overby A.K., Weber F., Grünewald K. Electron cryo-microscopy and single-particle averaging of Rift Valley fever virus: evidence for GN-GC glycoprotein heterodimers. J. Virol. 2009;83:3762–3769. doi: 10.1128/JVI.02483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen J.T., Hepojoki J., Laurinmäki P., Vaheri A., Lankinen H., Butcher S.J., Grünewald K. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 2010;84:4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen J.T., Parsy M.-L., Li S., Bitto D., Renner M., Bowden T.A. Averaging of viral envelope glycoprotein spikes from electron cryotomography reconstructions using Jsubtomo. J. Vis. Exp. 2014;92 doi: 10.3791/51714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt A.D., Zhao Y., Roy P. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology. 1993;193:592–603. doi: 10.1006/viro.1993.1167. [DOI] [PubMed] [Google Scholar]
- Ilca S.L., Kotecha A., Sun X., Poranen M.M., Stuart D.I., Huiskonen J.T. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 2015;6:8843. doi: 10.1038/ncomms9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäälinoja H.T., Huiskonen J.T., Butcher S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jäälinoja H.T., Huiskonen J.T., Butcher S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages ϕ6 and ϕ8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jin J., Liss N.M., Chen D.-H., Liao M., Fox J.M., Shimak R.M., Fong R.H., Chafets D., Bakkour S., Keating S. Neutralizing monoclonal antibodies block Chikungunya virus entry and release by targeting an epitope critical to viral pathogenesis. Cell Rep. 2015;13:2553–2564. doi: 10.1016/j.celrep.2015.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B., Nybakken G.E., Chipman P.R., Zhang W., Diamond M.S., Fremont D.H., Kuhn R.J., Rossmann M.G. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Strauss J.D., Hampton C.M., Brindley M.A., Dillard R.S., Leon F., Lamb K.M., Plemper R.K., Wright E.R. Promotion of virus assembly and organization by the measles virus matrix protein. Nat. Commun. 2018;9:1736. doi: 10.1038/s41467-018-04058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Dillard R.S., Chirkova T., Leon F., Stobart C.C., Hampton C.M., Strauss J.D., Rajan D., Rostad C.A., Taylor J.V. The morphology and assembly of respiratory syncytial virus revealed by cryo-electron tomography. Viruses. 2018;10:446. doi: 10.3390/v10080446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat R., Fu C.-Y., Ortmann A.C., Young M.J., Johnson J.E. The architecture and chemical stability of the archaeal Sulfolobus turreted icosahedral virus. J. Virol. 2010;84:9575–9583. doi: 10.1128/JVI.00708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M. Mechanisms of virus membrane fusion proteins. Annu. Rev. Virol. 2014;1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- Kiss G., Chen X., Brindley M.A., Campbell P., Afonso C.L., Ke Z., Holl J.M., Guerrero-Ferreira R.C., Byrd-Leotis L.A., Steel J. Capturing enveloped viruses on affinity grids for downstream cryo-electron microscopy applications. Microsc. Microanal. 2014;20:164–174. doi: 10.1017/S1431927613013937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T., Kuznetsov Y.G., Xiao C., Sun S., McPherson A., Rossmann M.G. The three-dimensional structure of Mimivirus. Intervirology. 2010;53:268–273. doi: 10.1159/000312911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko V.A., Chew P.L., Ng T.-S., Lok S.-M. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J. Virol. 2014;88:477–482. doi: 10.1128/JVI.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko V.A., Lim E.X.Y., Zhang S., Fibriansah G., Ng T.-S., Ooi J.S.G., Shi J., Lok S.-M. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanto E., Mäntynen S., De Colibus L., Marjakangas J., Gillum A., Stuart D.I., Ravantti J.J., Huiskonen J.T., Sundberg L.-R. Virus found in a boreal lake links ssDNA and dsDNA viruses. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8378–8383. doi: 10.1073/pnas.1703834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurinmäki P.A., Huiskonen J.T., Bamford D.H., Butcher S.J. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure. 2005;13:1819–1828. doi: 10.1016/j.str.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Leser G.P., Lamb R.A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Li S., Sun Z., Pryce R., Parsy M.-L., Fehling S.K., Schlie K., Siebert C.A., Garten W., Bowden T.A., Strecker T. Acidic pH-Induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rissanen I., Zeltina A., Hepojoki J., Raghwani J., Harlos K., Pybus O.G., Huiskonen J.T., Bowden T.A. A molecular-level account of the antigenic hantaviral surface. Cell Rep. 2016;15:959–967. doi: 10.1016/j.celrep.2016.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeroos L., Huiskonen J.T., Ora A., Susi P., Butcher S.J. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeroos L., Krzyzaniak M.A., Helenius A., Butcher S.J. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bartesaghi A., Borgnia M.J., Sapiro G., Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok S.-M., Kostyuchenko V., Nybakken G.E., Holdaway H.A., Battisti A.J., Sukupolvi-Petty S., Sedlak D., Fremont D.H., Chipman P.R., Roehrig J.T. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Loney C., Mottet-Osman G., Roux L., Bhella D. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of Sendai virus. J. Virol. 2009;83:8191–8197. doi: 10.1128/JVI.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Fong R.H., Austin S.K., Chen Z., Klose T., Fokine A., Liu Y., Porta J., Sapparapu G., Akahata W. Cryo-EM structures elucidate neutralizing mechanisms of anti-chikungunya human monoclonal antibodies with therapeutic activity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13898–13903. doi: 10.1073/pnas.1515558112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Doyle M., Fernandez E., Miller A.S., Klose T., Sevvana M., Bryan A., Davidson E., Doranz B.J., Kuhn R.J. Structural basis of a potent human monoclonal antibody against Zika virus targeting a quaternary epitope. Proc. Natl. Acad. Sci. U. S. A. 2019;116:1591–1596. doi: 10.1073/pnas.1815432116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E.J., Clarke M., Gowen B.E., Rutten T., Fuller S.D. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Mattei S., Glass B., Hagen W.J.H., Kräusslich H.-G., Briggs J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science. 2016;354:1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V. Zika virus associated with microcephaly. N. Engl. J. Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the Dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Överby A.K., Pettersson R.F., Grünewald K., Huiskonen J.T. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2375–2379. doi: 10.1073/pnas.0708738105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.W., Linden K.G., Sobsey M.D. Inactivation of murine norovirus, feline calicivirus and echovirus 12 as surrogates for human norovirus (NoV) and coliphage (F) MS2 by ultraviolet light (254 nm) and the effect of cell association on UV inactivation. Lett. Appl. Microbiol. 2011;52:162–167. doi: 10.1111/j.1472-765X.2010.02982.x. [DOI] [PubMed] [Google Scholar]
- Porta J., Jose J., Roehrig J.T., Blair C.D., Kuhn R.J., Rossmann M.G. Locking and blocking the viral landscape of an alphavirus with neutralizing antibodies. J. Virol. 2014;88:9616–9623. doi: 10.1128/JVI.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta J., Mangala Prasad V., Wang C.-I., Akahata W., Ng L.F.P., Rossmann M.G. Structural studies of Chikungunya virus-like particles complexed with human antibodies: neutralization and cell-to-cell transmission. J. Virol. 2016;90:1169–1177. doi: 10.1128/JVI.02364-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V.M., Miller A.S., Klose T., Sirohi D., Buda G., Jiang W., Kuhn R.J., Rossmann M.G. Structure of the immature Zika virus at 9 Å resolution. Nat. Struct. Mol. Biol. 2017;24:184–186. doi: 10.1038/nsmb.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce R., Ng W.M., Zeltina A., Watanabe Y., El Omari K., Wagner A., Bowden T.A. Structure-based classification defines the discrete conformational classes adopted by the arenaviral GP1. J. Virol. 2019;93 doi: 10.1128/JVI.01048-18. e01048-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punch E.K., Hover S., Blest H.T.W., Fuller J., Hewson R., Fontana J., Mankouri J., Barr J.N. Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus. J. Biol. Chem. 2018;293:9937–9944. doi: 10.1074/jbc.RA118.002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Flanagan A., Dejnirattisai W., Puttikhunt C., Kasinrerk W., Supasa P., Wongwiwat W., Chawansuntati K., Duangchinda T., Cowper A. Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus. Nat. Immunol. 2018;19:1248–1256. doi: 10.1038/s41590-018-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rissanen I., Stass R., Zeltina A., Li S., Hepojoki J., Harlos K., Gilbert R.J.C., Huiskonen J.T., Bowden T.A. Structural transitions of the conserved and metastable hantaviral glycoprotein envelope. J. Virol. 2017;91 doi: 10.1128/JVI.00378-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok R.W., Wrigley N.G., Calder L.J., Cusack S., Wharton S.A., Brown E.B., Skehel J.J. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5:41–49. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martín C., Huiskonen J.T., Bamford J.K.H., Butcher S.J., Fuller S.D., Bamford D.H., Burnett R.M. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 2002;9:756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- Sevvana M., Long F., Miller A.S., Klose T., Buda G., Sun L., Kuhn R.J., Rossmann M.G. Refinement and analysis of the mature Zika virus Cryo-EM structure at 3.1 Å resolution. Structure. 2018;26:1169–1177.e3. doi: 10.1016/j.str.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S., Whitby L.R., Casquilho-Gray H.E., York J., Boger D.L., Nunberg J.H. Small-molecule fusion inhibitors bind the pH-sensing stable signal peptide-GP2 subunit interface of the Lassa virus envelope glycoprotein. J. Virol. 2016;90:6799–6807. doi: 10.1128/JVI.00597-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.B., Trujillo J., Leahy I., Razmus D., Dehate R., Lorcheim P., Czarneski M.A., Zimmerman D., Newton J.T.M., Haddow A.D. Construction and organization of a BSL-3 cryo-electron microscopy laboratory at UTMB. J. Struct. Biol. 2013;181:223–233. doi: 10.1016/j.jsb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D., Chen Z., Sun L., Klose T., Pierson T.C., Rossmann M.G., Kuhn R.J. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Bayley P.M., Brown E.B., Martin S.R., Waterfield M.D., White J.M., Wilson I.A., Wiley D.C. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.D., Hammonds J.E., Yi H., Ding L., Spearman P., Wright E.R. Three-dimensional structural characterization of HIV-1 tethered to human cells. J. Virol. 2016;90:1507–1521. doi: 10.1128/JVI.01880-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Bartesaghi A., Liu J., Bennett A.E., Sougrat R. Electron tomography of viruses. Curr. Opin. Struct. Biol. 2007;17:596–602. doi: 10.1016/j.sbi.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S., Brien J.D., Austin S.K., Shrestha B., Swayne S., Kahle K., Doranz B.J., Johnson S., Pierson T.C., Fremont D.H. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J. Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Xiang Y., Akahata W., Holdaway H., Pal P., Zhang X., Diamond M.S., Nabel G.J., Rossmann M.G. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. Elife. 2013;2 doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., El Omari K., Sun X., Ilca S.L., Kotecha A., Stuart D.I., Poranen M.M., Huiskonen J.T. Double-stranded RNA virus outer shell assembly by bona fide domain-swapping. Nat. Commun. 2017;8 doi: 10.1038/ncomms14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh E.P., Kukkaro P., Teo E.W., Lim A.P.C., Tan T.T., Yip A., Schul W., Aung M., Kostyuchenko V.A., Leo Y.S. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 2012;4:139ra83. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- Thompson R.F., Walker M., Siebert C.A., Muench S.P., Ranson N.A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods. 2016;100:3–15. doi: 10.1016/j.ymeth.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.E.H., Simmons J.A., Bartesaghi A., Shoemaker C.J., Nelson E., White J.M., Subramaniam S. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J. Virol. 2014;88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.E.H., Nelson E.A., Bonagiri P., Simmons J.A., Shoemaker C.J., Schmaljohn C.S., Kobinger G.P., Zeitlin L., Subramaniam S., White J.M. Mapping of ebolavirus neutralization by monoclonal antibodies in the ZMapp cocktail using cryo-electron tomography and studies of cellular entry. J. Virol. 2016;90:7618–7627. doi: 10.1128/JVI.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D., Ng T.-S., Sendamarai A.K., Eilers B.J., Lawrence C.M., Lok S.-M., Young M.J., Johnson J.E., Fu C.-Y. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Moore P.B. On the interpretation of electron microscopic maps of biological macromolecules. Protein Sci. 2017;26:122–129. doi: 10.1002/pro.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Bosch B.-J., Vlak J.M., van Oers M.M., Rottier P.J., van Lent J.W.M. Budded baculovirus particle structure revisited. J. Invertebr. Pathol. 2016;134:15–22. doi: 10.1016/j.jip.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yang H., Liu X., Dai L., Ma T., Qi J., Wong G., Peng R., Liu S., Li J. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci. Transl. Med. 2016;8:369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- Wang X., Li S.-H., Zhu L., Nian Q.-G., Yuan S., Gao Q., Hu Z., Ye Q., Li X.-F., Xie D.-Y. Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat. Commun. 2017;8:14. doi: 10.1038/s41467-017-00024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Charlier C., Vasilakis N., Lecuit M. Zika, Chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 2018;69:395–408. doi: 10.1146/annurev-med-050715-105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Holland Cheng R., Berriman J., Rice W.J., Stokes D.L., Katz A., Morgan D.G., Gottlieb P. Three-dimensional structure of the enveloped bacteriophage Φ12: an incomplete T = 13 lattice is superposed on an enclosed T = 1 shell. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Kolesnikova L., Krähling V., Riches J.D., Becker S., Briggs J.A.G. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wirawan M., Fibriansah G., Marzinek J.K., Lim X.X., Ng T.-S., Sim A.Y.L., Zhang Q., Kostyuchenko V.A., Shi J., Smith S.A., Verma C.S., Anand G., Crowe J.E., Bond P.J., Lok S.-M. Mechanism of enhanced immature dengue virus attachment to endosomal membrane induced by prM antibody. Structure. 2019;27:253–267. doi: 10.1016/j.str.2018.10.009. e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M., DeRosier D.J., Grigorieff N. Ewald sphere correction for single-particle electron microscopy. Ultramicroscopy. 2006;106:376–382. doi: 10.1016/j.ultramic.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Xiao C., Rossmann M.G. Structures of giant icosahedral eukaryotic dsDNA viruses. Curr. Opin. Virol. 2011;1:101–109. doi: 10.1016/j.coviro.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Kuznetsov Y.G., Sun S., Hafenstein S.L., Kostyuchenko V.A., Chipman P.R., Suzan-Monti M., Raoult D., McPherson A., Rossmann M.G. Structural studies of the giant mimivirus. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Fischer M.G., Bolotaulo D.M., Ulloa-Rondeau N., Avila G.A., Suttle C.A. Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses. Sci. Rep. 2017;7:5484. doi: 10.1038/s41598-017-05824-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Chan Y.-P., Bradel-Tretheway B., Akyol-Ataman Z., Zhu Y., Dutta S., Yan L., Feng Y., Wang L.-F., Skiniotis G. Crystal structure of the pre-fusion Nipah virus fusion glycoprotein reveals a novel hexamer-of-trimers assembly. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Helenius A. Virus entry at a glance. J. Cell Sci. 2013;126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- Yan X., Chipman P.R., Castberg T., Bratbak G., Baker T.S. The marine algal virus PpV01 has an icosahedral capsid with T = 219 quasisymmetry. J. Virol. 2005;79:9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Chang P.H., Hartert J., Wigginton K.R. Reactivity of enveloped virus genome, proteins, and lipids with free chlorine and UV254. Environ. Sci. Technol. 2018;52:7698–7708. doi: 10.1021/acs.est.8b00824. [DOI] [PubMed] [Google Scholar]
- Zanetti G., Briggs J.A.G., Grünewald K., Sattentau Q.J., Fuller S.D. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2 doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chipman P.R., Corver J., Johnson P.R., Zhang Y., Mukhopadhyay S., Baker T.S., Strauss J.H., Rossmann M.G., Kuhn R.J. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Corver J., Chipman P.R., Zhang W., Pletnev S.V., Sedlak D., Baker T.S., Strauss J.H., Kuhn R.J., Rossmann M.G. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xiang Y., Dunigan D.D., Klose T., Chipman P.R., Van Etten J.L., Rossmann M.G. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., Zhou Z.H. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Sheng J., Plevka P., Kuhn R.J., Diamond M.S., Rossmann M.G. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Sheng J., Austin S.K., Hoornweg T.E., Smit J.M., Kuhn R.J., Diamond M.S., Rossmann M.G. Structure of acidic pH dengue virus showing the fusogenic glycoprotein trimers. J. Virol. 2015;89:743–750. doi: 10.1128/JVI.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Kostyuchenko V.A., Ng T.-S., Lim X.-N., Ooi J.S.G., Lambert S., Tan T.Y., Widman D.G., Shi J., Baric R.S. Neutralization mechanism of a highly potent antibody against Zika virus. Nat. Commun. 2016;7 doi: 10.1038/ncomms13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Carratalà A., Nazarov S., Guerrero-Ferreira R.C., Piccinini L., Bachmann V., Leiman P.G., Kohn T. Genetic, structural, and phenotypic properties of MS2 coliphage with resistance to ClO disinfection. Environ. Sci. Technol. 2016;50:13520–13528. doi: 10.1021/acs.est.6b04170. [DOI] [PubMed] [Google Scholar]
- Zhu P., Chertova E., Bess J., Jr., Lifson J.D., Arthur L.O., Liu J., Taylor K.A., Roux K.H. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J.D., Grisé H., Ofek G.A., Taylor K.A., Roux K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]