Abstract
Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) are the most severe coronavirus (CoV)-associated diseases in humans. The causative agents, SARS-CoV and MERS-CoV, are of zoonotic origin but may be transmitted to humans, causing severe and often fatal respiratory disease in their new host. The two coronaviruses are thought to encode an unusually large number of factors that allow them to thrive and replicate in the presence of efficient host defense mechanisms, especially the antiviral interferon system. Here, we review the recent progress in our understanding of the strategies that highly pathogenic coronaviruses employ to escape, dampen, or block the antiviral interferon response in human cells.
Keywords: SARS-CoV, MERS-CoV, Interferon, RIG-I-like receptors, MDA5, PKR, Viral countermeasures
1. Introduction
Coronaviruses have made a remarkable career. Originally recognized as viral pathogens of veterinary importance but little medical (i.e., human) relevance, the appearance of SARS-CoV causing a worldwide epidemic with a large number of fatalities has changed everything. In 2003, the virus emerged in Chinese animal markets to circle the world in just a few weeks, teaching us important new lessons on perceived “differences” between animal and human pathogens. Just in case someone did not get the message, MERS-CoV repeated the coronavirus wake-up call 10 years later, providing yet another example for how easily animal viruses may be transmitted and adapt to new hosts including humans. Often, the tricks and strategies that viruses evolved to propagate in specific animal hosts may only need some fine-tuning (if at all) to enter the wide world of human crowds, air travel, and camel races. Here, we will summarize the insights gathered so far on an important aspect of virulence and host adaptation, the interactions of SARS-CoV and MERS-CoV with antiviral interferon (IFN) responses of human cells.
2. The Coronavirus Genome
The coronavirus genome is composed of a linear, single-stranded, monopartite RNA with a cap structure at its 5′ end and a polyA tail at the 3′ end (Fehr and Perlman, 2015). The 5′-terminal two-thirds of the CoV genome contain the open reading frames (ORF) 1a and 1b that together constitute the viral replicase gene. Translation is initiated at the start codon of ORF1a and may continue to ORF1b via a ribosomal frameshift mechanism, ultimately giving rise to two overlapping replicase polyproteins pp1a and pp1ab (Fehr and Perlman, 2015, Perlman and Netland, 2009, Snijder et al., 2003, Thiel et al., 2003). Virus-encoded proteinases, namely two papain-like cysteine proteases (PL1pro and PL2pro), residing in nonstructural protein (nsp) 3 and a 3C-like cysteine protease (3CLpro) associated with nsp5, proteolytically process the polyproteins into nsps 1–16 (Anand et al., 2003, Schiller et al., 1998, Thiel et al., 2003, Ziebuhr et al., 2007). A multitude of functions and enzymatic activities associated with specific nsps have been identified over the past years (for reviews, see Masters and Perlman, 2013, Ziebuhr, 2005). Moreover, ORF1b harbors several RNA-processing enzymes, including a 3′–5′ exonuclease and a guanosine N7-methyltransferase (associated with the N- and C-terminal domains, respectively, of nsp14), an endoribonuclease (nsp15) and a 2′-O-methyltransferase (nsp16) (Chen et al., 2009, Decroly et al., 2008, Fehr and Perlman, 2015, Ivanov et al., 2004, Kindler and Thiel, 2014, Minskaia et al., 2006, Perlman and Netland, 2009, Snijder et al., 2003, Thiel et al., 2003, Zust et al., 2011). The 3′ ORFs are translated from a set of subgenomic (sg) RNAs and yield on one hand four canonical structural proteins like the spike protein (S), the envelope (E), the membrane (M), and the nucleoprotein (N). Moreover, sgRNAs express accessory genes, which vary in function and number between different CoV strains and are interspersed between the structural genes (Fehr and Perlman, 2015, Perlman and Netland, 2009, Snijder et al., 2003, Thiel et al., 2003). Specifically, the genome of SARS-CoV expresses eight different accessory genes (3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b), while MERS-CoV encodes five accessory genes (3, 4a, 4b, 5, and 8b). The schematic overview of the genome organization of SARS-CoV and MERS-CoV is depicted in Fig. 1 .
Fig. 1.
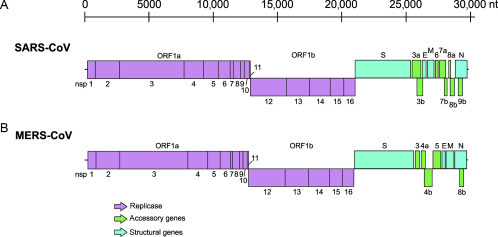
Coronavirus genomes. Schematic representation of the genome regions encoding nonstructural (nsp), structural, and accessory proteins of SARS-CoV (A) and MERS-CoV (B).
The CoV life cycle starts with the attachment of the viral spike protein to particular cellular receptors, subsequently leading to fusion between the viral envelope and the plasma membrane or the endosome membrane of the host. CoV uses a range of receptors, with SARS-CoV employing angiotensin-converting enzyme 2 (ACE2) and MERS-CoV employing dipeptyl peptidase 4 (DPP4) (Li et al., 2003, Raj et al., 2013). Following membrane fusion, the viral RNA genome is delivered into the host cytoplasm, where translation of the two 5′-terminal ORFs 1a and 1b is accomplished by the cellular translation machinery. Most of the newly synthesized nsps assemble with the N protein into a replicase–transcriptase complex (RTC) responsible for viral genome replication and transcription. At the site of replicative organelles (Knoops et al., 2008), the RTC initiates minus-strand synthesis using the full-length genome as template, thereby either copying the entire template to generate full-length minus strands or to move discontinuously along the template to produce a nested set of sgRNAs with negative polarity. The minus strands of genomic and sgRNAs are subsequently used as templates to synthesize positive sense strands (mRNAs), specifically the genomic RNA (genome replication) and sg mRNAs (transcription) (Sawicki et al., 2007). The N protein then encapsidates the newly synthetized RNA genome and thereby forms a helical nucleocapsid. Virion assembly is triggered by the action of the M protein, which assists in incorporating the nucleocapsid, the envelope and the spike into virus particles. Budding takes place between the endoplasmic reticulum and the Golgi and new viruses are released by exocytosis (McBride et al., 2014, Neuman et al., 2011, Ruch and Machamer, 2012, Ujike and Taguchi, 2015).
3. The Type I IFN System
3.1. Types of IFNs and Their Signaling Pathways
The antiviral IFN (IFN-alpha/beta) system confers an important part of the innate immune defense in chordates (tenOever, 2016). IFNs are cytokines that are produced and secreted by cells encountering viruses or parts thereof (Fig. 2 ). Humans are able to express one IFN-beta, 13 subtypes of IFN-alphas, and one each of IFN-kappa and IFN-omega (Schneider et al., 2014). All nucleated cells are able to respond to them as they express the IFN receptor (composed of the two subunits IFNAR-1 and IFNAR-2) on their surface (Bekisz et al., 2004). The docking of IFN-alpha/beta onto its cognate receptor activates the so-called JAK–STAT pathway. Thereby, the Janus kinases JAK1 and TYK2 are waiting to be activated on the cytoplasmic side of IFNAR2 and 1, respectively. The activated kinases then phosphorylate the signal transducers and transcription factors STAT1 and STAT2, which form a complex with IRF9 (ISGF3) that enters the nucleus to transactivate promoters of an antiviral gene expression program. Genes that are specifically upregulated by IFNs are collectively called ISGs (IFN-stimulated genes).
Fig. 2.
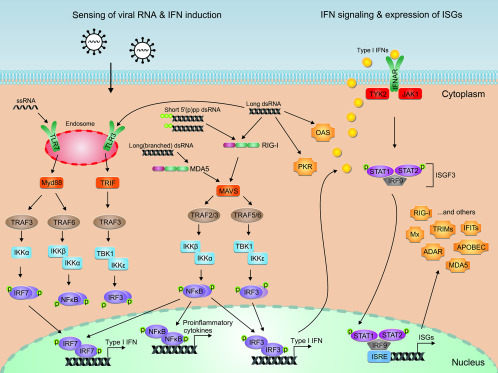
The antiviral IFN system. Induction of IFNs, IFN-dependent JAK/STAT signaling, and ISG expression is depicted. For details, see text.
The alpha/beta-IFNs are classified as type I IFNs, since they had been discovered first (Isaacs and Lindenmann, 1957). The type II IFNs use a different receptor and consist of only one member, IFN-gamma. IFN-gamma also confers some antiviral activity but is regarded more of an immunoregulator (produced by specialized immune cells) than a general antiviral mediator (Schneider et al., 2014). It signals through a JAK/STAT pathway that partially overlaps with one of the type I IFNs. IFN-gamma will not be further discussed here as it is not to the core of the antiviral IFN response to coronaviruses.
Recently, the IFN family was extended by the newly discovered type III IFNs, consisting of IFN-lambda 1–4 (Schneider et al., 2014). Type III IFNs resemble type I IFNs in that they also trigger STAT1/2 phosphorylation via JAK1 and TYK2. They employ however a different receptor which is only expressed by epithelial cells (Sommereyns et al., 2008). Thus, type I and type III IFNs trigger largely overlapping sets of ISGs, but while the former constitute a major, general antiviral cytokine system, the latter are mainly restricted to mucosal sites (Galani et al., 2015).
3.2. Induction of Type I IFNs
The molecular events leading to the upregulation of type I IFNs are well established. As indicated earlier, molecular structures that are specific for virus infections (often called PAMPs, for pathogen-associated molecular patterns) are sensed by pathogen recognition receptors (PRRs) of the host, that in turn are triggering the upregulation of IFN genes. A prototypical PAMP relevant for coronaviruses is double-stranded RNA (dsRNA), a by-product of genome replication and transcription (Weber et al., 2006, Zielecki et al., 2013). dsRNA can be sensed by toll-like receptor 3 (TLR3) in the endosome, and in the cytoplasm by the RNA helicases RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation antigen 5), as well as by the kinase PKR (protein kinase, RNA-activated) (Rasmussen et al., 2009, Yim and Williams, 2014, Yoneyama et al., 2016). RIG-I is thereby specific for long dsRNA molecules and short dsRNAs bearing a tri- or di-phosphorylated 5′ end, whereas MDA5 senses long dsRNAs, preferentially with a higher-order structure (Binder et al., 2011, Goubau et al., 2014, Pichlmair et al., 2009, Schlee, 2013). PKR is activated by dsRNA as well as by short stem-loop RNAs bearing a 5′ triphosphate end (Dabo and Meurs, 2012, Nallagatla et al., 2011). Also specific single-stranded RNAs (ssRNAs) can act as PAMPs, either if they are in the wrong location or if they display particular features. TLR7 senses GU-rich ssRNA in the endosome (Heil et al., 2004). RIG-I can be triggered by polyU/UC rich or 3′ monophosphorylated ssRNAs stretches, and MDA5 was found to bind ssRNA stretches of negative-sense RNA viruses and hypomethylated 5′ capped mRNAs (Luthra et al., 2011, Malathi et al., 2007, Malathi et al., 2010, Rasmussen et al., 2009, Runge et al., 2014, Saito et al., 2008, Zust et al., 2011).
Depending on the particular PRR, various-partially cross-talking-signaling pathways lead to the transactivation of promoters for antiviral genes (O’Neill et al., 2013). The endosomal PRR TLR3 engages the intracellular adapters TRIF (TIR domain-containing adapter protein inducing IFN-beta) and TRAF3 (TNF receptor-associated factor 3) to activate the kinases TBK1 (TANK-binding kinase 1) and IKKepsilon (inhibitor of NF-kappaB kinase epsilon). The kinases TBK1 and IKKepsilon then phosphorylate IRF3 (IFN regulatory factor 3), a transcription factor that activates genes for IFNs and other immunoregulatory cytokines. Signaling by TLR7, by contrast, requires the adaptor proteins MyD88 (myeloid differentiation primary-response protein 88) and TRAF3 to channel to the kinase IKKalpha. This kinase then phosphorylates IRF7, a transcription factor that covers a gene spectrum similar to IRF3. TLR7/MyD88 also recruits the adaptor protein TRAF6 that eventually activates the transcription factor NF-kappaB via the kinases IKKalpha and IKKbeta. NF-kappaB drives transcription of genes for proinflammatory cytokines but also enhances IFN gene expression.
In the cytoplasm, RNA sensing by the two PRRs RIG-I and MDA5 (collectively termed RIG-I-like receptors, RLRs) converges on the adaptor protein MAVS (mitochondrial antiviral signaling protein) that uses various TRAFs (TRAF 2, 3, 5, 6) to trigger TBK1/IKKepsilon and IKKalpha/IKKbeta. These kinases then activate IRF3 and NF-kappaB, respectively (Belgnaoui et al., 2011, Liu et al., 2013). Besides the RLRs, PKR contributes to IFN induction in the cytoplasm. PKR is a master regulator of mRNA translation (see later), but several lines of evidence indicate a role in activation of NF-kappaB and IRF3 via TRAF2/6, IKKalpha/beta, antiviral stress granule formation, and IFN-alpha/beta mRNA stability (Gil et al., 2004, Onomoto et al., 2012, Pfaller et al., 2011, Pham et al., 2016, Schulz et al., 2010, Zamanian-Daryoush et al., 2000).
Thus, several types of PRRs are constantly surveying the extracellular and intracellular space to detect virus infections in a timely and sensitive manner. Importantly, TLRs are preferentially expressed by immune cells, especially myeloid dendritic cells (mDCs) and plasmacytoid cells (pDCs), whereas RLRs and PKR are thought to be active in all nucleated cells. Detection of viral RNA in mDCs is mainly mediated by TLR3 (and some TLR7), and in pDCs by TLR7 (and TLR8 in human pDCs) (Schreibelt et al., 2010).
PAMP sensing by PRRs eventually culminates in activation of IRF3, IRF7, and NF-kappaB, as described earlier, the transcription factors driving the expression of genes for IFN-beta, IFN-alpha, and various proinflammatory and immunomodulatory cytokines (Belgnaoui et al., 2011).
3.3. IFN-Stimulated Gene Expression
Signaling by both type I and type III IFNs triggers the formation of ISGF3 (see Section 3.1), the heterotrimeric transcription factor complex consisting of phosphorylated STAT1 and STAT2, and IRF9 (Schneider et al., 2014). ISGF3 binds to the ISREs (for IFN-stimulated response element), specific promoter sequences of the so-called ISGs. Of note, there are actually several types of “ISREs” that are responding to different types of triggers and transcription factors. First, there are the ISREs that purely respond to IFN signaling and ISGF3, as it would be expected from the name. A prominent example is given by the promoter of the human antiviral protein MxA (Holzinger et al., 2007). Second, there are the—somewhat mislabeled—ISREs that do not respond to IFN at all, but only to the IRF3-, IRF7-, and NF-kappaB-related signal transduction that occurs much earlier, directly after virus infection has triggered a PRR. The IFN-beta promoter belongs to this class of ISREs (Freaney et al., 2013, Schmid et al., 2010). Third, there are mixed-type ISREs that can be activated by both virus infection and IFNs. An example is the promoter of the gene for the antiviral protein IFIT1 (also known as ISG56) (Fensterl and Sen, 2015). The different ISRE classes can be distinguished as IRF-specific ISREs (responding only to PRR signaling), ISGF3-specific ISREs (responding only to type I or type III IFNs), and universal ISREs (responding to both infection and IFNs) (Schmid et al., 2010). Many ISGs are controlled by additional promoter elements ensuring basal levels of expression already in the absence of IFN. Moreover, low levels of IFN itself are constitutively secreted by many tissues (tonic IFN), ensuring physiological homeostasis and priming of cells for a rapid response against pathogens (Gough et al., 2012).
It is estimated that, depending on the IFN subtype, dose, and cell type, IFNs regulate hundreds, if not thousands of genes (Rusinova et al., 2013). Many of the ISGs (i.e., those genes that are upregulated by IFNs) are known to have antiviral, immunomodulatory, or antiproliferative function (Samuel, 2001, Stark and Darnell, 2012). The broad antiviral activity of IFNs occurs on several levels, namely virus entry, viral polymerase function, host cell translation, RNA availability, RNA stability, particle budding, apoptosis, or general boosting of innate and adaptive immune responses.
4. Antiviral Action of IFNs Against Human Coronaviruses
High-dose IFN treatment (type I and type III) has clear effects against SARS-CoV and MERS-CoV in cell culture (Chan et al., 2013, Cinatl et al., 2003, Falzarano et al., 2013, Kindler et al., 2013, Spiegel et al., 2004, Stroher et al., 2004, Zielecki et al., 2013), in animal experiments (Channappanavar et al., 2016, Frieman et al., 2010, Haagmans et al., 2004, Mahlakoiv et al., 2012, Mordstein et al., 2008), and possibly also in patients (Loutfy et al., 2003, Omrani et al., 2014, Strayer et al., 2014). Remarkably, MERS-CoV was found to be substantially more IFN sensitive than SARS-CoV in cell culture (Zielecki et al., 2013).
The cellular basis for the relatively low (SARS-CoV) and high (MERS-CoV) IFN sensitivity is currently unknown. Several prominent (i.e., potent) ISG products were studied in the context of human pathogenic coronaviruses, but only some of them were found to have an effect. The IFN-induced transmembrane (IFITMs) proteins 1, 2, and 3 restrict the entry of many enveloped viruses including SARS-CoV (Huang et al., 2011b) as well as reoviruses (Bailey et al., 2014). They act by altering the site of membrane fusion, but the exact mechanism remains to be elucidated (Bailey et al., 2014). Strikingly, while IFITMs are inhibitory for the highly pathogenic SARS-CoV, they appear to boost infection with the related, low pathogenic coronavirus HCoV-OC43 (Zhao et al., 2014). In particular, IFITM2 or IFITM3 acts as entry factor for HCoV-OC43 by facilitating—rather than impeding—membrane fusion. Human MxA (for Myxovirus resistance protein A) is a well-known antiviral host factor with activity against a wide range of (mostly) RNA viruses (Haller et al., 2015). It blocks early replication steps of influenza viruses but was found have no effect on SARS-CoV (Spiegel et al., 2004). The kinase PKR is an ISG product acting as a signaling PRR on one hand (see earlier), but its main function in antiviral defense is the inhibition of protein synthesis. After binding viral dsRNA, PKR undergoes autophosphorylation to activate itself, and subsequently phosphorylates eIF-2alpha that is thereby converted from a translation initiation factor to a translation inhibitor (Yim and Williams, 2014). PKR has a broad antiviral spectrum. Nonetheless, PKR has no bearing on the replication of SARS-CoV, although it is involved in virally induced apoptosis (Krahling et al., 2009). Also the 2′–5′ oligoadenylate synthetase (OAS) family members are triggered by viral dsRNA (Chakrabarti et al., 2011). In the dsRNA-bound state they synthesize short chains of 2′–5′ oligoadenylates that activate the latent RNase L. RNase L then cleaves virus and host ssRNAs, predominantly at single-stranded UA and UU dinucleotides (Wreschner et al., 1981). Interestingly, the small 3′-monophosphorylated cleavage products of RNase L are recognized by the PRRs RIG-I and MDA5, thus amplifying the IFN response in an infection-dependent manner (Malathi et al., 2007). Polymorphisms of the OAS-1 gene might affect susceptibility to SARS-CoV (Hamano et al., 2005), but to our knowledge, there is no direct data on antiviral effects of the OAS/RNase L system on human coronaviruses. For the mouse coronavirus MHV-A59, however, it was shown that mutants deficient in the ns2 gene are highly sensitive against RNase L (Zhao et al., 2012) (see also later).
As mentioned, there are several hundreds of ISGs, of which about 40 were characterized as being antiviral (Schneider et al., 2014). It is in a way remarkable that relatively little is known about ISGs that impede human pathogenic coronaviruses. Most likely, active and passive evasion mechanisms such as the ones described later are responsible for the relative insensitivity of at least SARS-CoV against IFN and potent antiviral ISGs. Although our review will focus on the human pathogenic coronaviruses SARS-CoV and MERS-CoV, we will draw additional conclusions from well investigated other coronaviruses whenever adequate.
5. Evasion Strategies of Coronaviruses
Viral evasion strategies against the IFN response can act on several levels, namely the induction of IFN, IFN signaling, or antiviral action of individual ISG products (Gack, 2014, Kindler and Thiel, 2014, Vijay and Perlman, 2016, Weber and Weber, 2014, Wong et al., 2016, Zinzula and Tramontano, 2013). The viruses can thereby actively sequester or destroy key regulators, or otherwise interfere with the IFN system. Moreover, several aspects of the viral replication cycle can be regarded as a passive IFN evasion. The strategies described later are also summarized in three tables.
5.1. Inhibition of IFN Induction
Both SARS-CoV and MERS-CoV induce very little—if any—IFN in most cell types (Chan et al., 2013, Cheung et al., 2005, Kindler et al., 2013, Lau et al., 2013, Menachery et al., 2014a, Spiegel et al., 2005, Zhou et al., 2014, Ziegler et al., 2005, Zielecki et al., 2013). In fact, it was recently shown in a mouse model of SARS that the delay in IFN induction is responsible for the activation of proinflammatory monocyte-macrophages and cytokines in the lung, resulting in vascular leakage and impaired adaptive immune responses (Channappanavar et al., 2016). Thus, the high levels of dsRNA that are produced during replication (Weber et al., 2006, Zielecki et al., 2013) do not result in an adequate IFN induction. One of the reasons (besides the active measures described later) is certainly the storage of coronaviral dsRNA inside double-membrane vesicles (Knoops et al., 2008, van Hemert et al., 2008, Versteeg et al., 2007). Moreover, the N protein sequesters IFN-inducing RNA PAMPs (Kopecky-Bromberg et al., 2007, Lu et al., 2011). However, the fact that infection with coronaviruses activates the cytosolic dsRNA-sensing host factors PKR and OAS (Birdwell et al., 2016, Krahling et al., 2009, Zhao et al., 2012), as well as the existence of numerous mechanisms dedicated to suppress dsRNA-dependent IFN induction (see later) strongly suggest that dsRNA stashing alone is not sufficient and that some dsRNA or other PAMPs are exposed to PRRs, thus necessitating the presence of additional, active mechanisms.
While most cell types remain IFN-silent after infection, a notable exception are pDCs, which express high levels of IFN-alpha/beta in response to infection with both SARS-CoV and MERS-CoV (Cervantes-Barragan et al., 2007, Channappanavar et al., 2016, Scheuplein et al., 2015). For the mouse coronavirus MHV-A59 it was shown that IFN induction in pDCs occurs through TLR7 (Cervantes-Barragan et al., 2007), suggesting the same to be true for SARS-CoV and MERS-CoV. Indeed, GU-rich ssRNAs from the SARS-CoV genome were shown to activate an excessive innate immune response via TLR7 (Li et al., 2013). Moreover, the membrane (M) protein and the envelope (E) protein of SARS-CoV are able to activate a TLR-like pathway and NF-kappaB signaling, respectively (DeDiego et al., 2014, Wang and Liu, 2016).
The mouse coronavirus MHV-A59 also naturally induces IFN in brain macrophages/microglia, with MDA5 being the responsible PRR (Birdwell et al., 2016, Roth-Cross et al., 2008). Also in oligodendrocytes IFN induction by MHV occurs through both MDA5 and RIG-I (Li et al., 2010). Interestingly, a general (i.e., not restricted to particular cell types) MDA5-dependent IFN induction can be obtained by ablating the ribose 2′-O-methylation activity of the nsp16. As it was shown for MHV-A59, SARS-CoV, and the mildly human pathogenic coronavirus HCoV-229E, nsp16-mediated 2′-O-methylation of viral mRNA cap structures prevents recognition by MDA5 (Menachery et al., 2014b, Zust et al., 2011).
Besides these “hiding” or “disguising” strategies, active mechanisms targeting specific host factors are in place (Table 1 ). SARS-CoV was shown to inhibit IRF3 by preventing its hyperphosphorylation, dimerization, and interaction with the cofactor CBP (Spiegel et al., 2005). Curiously, IRF3 initially enters the nucleus of infected cells, but later returns to the cytoplasm. SARS-CoV also inhibits the nuclear import of the related transcription factor IRF7 (Kuri et al., 2009). In this context, the papain-like protease (PLpro) domain of nsp3 (the largest coronaviral protein) of SARS-CoV and the mildly pathogenic HCoV-NL63 both interact with IRF3 and block its activation (Devaraj et al., 2007, Frieman et al., 2009). Moreover, PLpro was shown to drive the deubiquitination (or inhibit ubiquitination) of RIG-I, TBK1, and IRF3 (Clementz et al., 2010, Devaraj et al., 2007, Frieman et al., 2009, Sun et al., 2012). IRF3 activation is also prevented by the M protein of SARS-CoV through inhibiting complex formation between TRAF3 and TBK1 (Siu et al., 2009). Since M was also found to activate a TLR-like signaling pathway (Wang and Liu, 2016), a final picture of M protein function in the context of IFN induction/inhibition remains to be provided. IFN induction is also disturbed by the SARS-CoV nsp1, nsp7, nsp15, ORF3b, ORF6, and ORF9b proteins, respectively (Frieman et al., 2009, Kopecky-Bromberg et al., 2007, Shi et al., 2014, Zust et al., 2007). The anti-IFN function of nsp1 is based on its ability to mediate host mRNA degradation, while sparing viral mRNAs at the same time, and to block host mRNA translation (Huang et al., 2011a, Narayanan et al., 2008, Tanaka et al., 2012). Nsp1 also has a function in evasion from IFN signaling (see later), providing a possible reason why nsp1 mutants are particularly IFN sensitive (Wathelet et al., 2007, Zust et al., 2007). While the mechanisms of other SARS-CoV IFN induction antagonists like nsp7, nsp15, ORF3b, and ORF6 proteins remain to be characterized, for the ORF9b protein it was shown that it drives degradation of MAVS, TRAF3, and TRAF6 by interacting with the host factors PCBP2 and the E3 ubiquitin ligase AIP4 (Shi et al., 2014).
Table 1.
Mechanisms and Factors of Human Coronaviruses to Counteract IFN Induction
| Virus | Viral Protein or Function | Mechanism | References |
|---|---|---|---|
| SARS-CoV (MHV-A59) | Storage of dsRNA inside double-membrane vesicles | Prevents exposure of dsRNA to PRRs | Knoops et al. (2008), van Hemert et al. (2008), and Versteeg et al. (2007) |
| SARS-CoV | N | Sequesters IFN-inducing RNA PAMPs | Kopecky-Bromberg et al. (2007) and Lu et al. (2011) |
| SARS-CoV, HCoV-229E (MHV-A59) | nsp16 | Ribose 2′-O-methylation of viral mRNA cap structures prevents recognition by MDA5 | Menachery et al. (2014b) and Zust et al. (2011) |
| SARS-CoV, NL63 | PLpro | Interacts with IRF3, inhibits IRF3 activation, deubiquitinates RIG-I, TBK1, IRF3 | Clementz et al. (2010), Devaraj et al. (2007), Frieman et al. (2009), and Sun et al. (2012) |
| SARS-CoV | M | Inhibits TRAF3/TBK1 complex formation | Siu et al. (2009) |
| SARS-CoV | nsp7, nsp15, ORF3b, ORF6 | Mechanism unclear | Frieman et al. (2009) and Kopecky-Bromberg et al. (2007) |
| SARS-CoV | nsp1 | Mediates host mRNA degradation | Huang et al. (2011a) and Narayanan et al. (2008) |
| SARS-CoV | nsp1 | Blocks host mRNA translation | Narayanan et al. (2008) and Tanaka et al. (2012) |
| SARS-CoV | ORF9b protein | Proteasomal degradation of MAVS, TRAF3, and TRAF6 | Shi et al. (2014) |
| MERS-CoV | ORF4a protein | Interacts with dsRNA and the RLR cofactor PACT | Niemeyer et al. (2013) |
| MERS-CoV | ORF4a protein | Interacts with the RLR cofactor PACT | Siu et al. (2014) |
| MERS-CoV | ORF4a, 4b, and ORF5 proteins, M | Prevent IRF3 translocation | Yang et al. (2013) |
| MERS-CoV | ORF4b protein | Binds TBK1 and IKKepsilon | Matthews et al. (2014) and Yang et al. (2015) |
| MERS-CoV | PLpro | Deubiquitination | Bailey-Elkin et al. (2014) and Mielech et al. (2014) |
| MERS-CoV | nsp1 | Degrades host mRNAs | Lokugamage et al. (2015) |
| MERS-CoV | Unknown | Repressive histone modifications | Menachery et al. (2014a) |
Also for MERS-CoV, the reason for the low levels of IFN produced by infected cells (Chan et al., 2013, Kindler et al., 2013, Lau et al., 2013, Menachery et al., 2014a, Zhou et al., 2014, Zielecki et al., 2013) was further investigated. The ORF4a protein inhibits IFN induction by interaction with dsRNA and the RLR cofactor PACT (Niemeyer et al., 2013, Siu et al., 2014). Like the ORF4a, the ORF4b, 5, and M proteins of MERS-CoV were shown to prevent IRF3 translocation (Yang et al., 2013). The ORF4b protein, in particular, inhibits IFN induction by binding to TBK1 and IKKepsilon (Matthews et al., 2014, Yang et al., 2015). In agreement with the data on SARS-CoV, the PLpro of MERS-CoV has deubiquitinating activity and inhibits IFN induction (Bailey-Elkin et al., 2014, Mielech et al., 2014), and the nsp1 mediates host mRNA degradation (Lokugamage et al., 2015). In contrast to SARS-CoV, however, infection with MERS-CoV additionally activates repressive histone modifications that downregulate ISG expression (Menachery et al., 2014a).
5.2. Inhibition of IFN Signaling
Several proteins of SARS-CoV and MERS-CoV were found to interfere with the signal transduction chain that leads from IFN docking onto its receptor to the upregulation of ISGs by ISGF3, the STAT1/STAT2/IRF9 complex (Table 2 ). The ORF3a protein was shown to decrease levels of IFNAR, most probably by ubiquitination and proteolytic degradation (Minakshi et al., 2009). The ORF6 protein was the first factor described for SARS-CoV that affects IFN signaling in infected cells, disrupting nuclear import of STAT1 (Frieman et al., 2007, Kopecky-Bromberg et al., 2007). The ORF6 protein binds to the nuclear import factor karyopherin alpha 2 and tethers it (together with karyopherin beta 1) to intracellular membranes (Frieman et al., 2007). There, they become unavailable for their normal cellular function, the import of, e.g., STAT1. The phosphorylation of STAT1 is impeded by the multifunctional nsp1 protein of SARS-CoV, which otherwise drives degradation of host mRNAs and inhibits translation (see earlier) (Wathelet et al., 2007). For MERS-CoV, the ORF4a, 4b, and M proteins inhibit ISRE activation after stimulation with IFN (Yang et al., 2013). The mechanisms are currently unknown. The ORF4a protein, which also acts as an inhibitor of IFN induction (see earlier), had the strongest activity. Lastly, the repressive modifications that are imposed by MERS-CoV onto the cellular histones are also a strategy to dampen ISG expression (Menachery et al., 2014a).
Table 2.
Mechanisms and Factors of Human Coronaviruses to Counteract IFN-Stimulated Gene Expression
| Virus | Viral Protein or Function | Mechanism | References |
|---|---|---|---|
| SARS-CoV | ORF3a protein | Proteolytic degradation of IFNAR | Minakshi et al. (2009) |
| ORF6 protein | Inhibits STAT1 nuclear import by sequestering karyopherin alpha 2 to intracellular membranes | Frieman et al. (2007) and Kopecky-Bromberg et al. (2007) | |
| SARS-CoV | nsp1 | Decreases phosphorylation of STAT1 | Wathelet et al. (2007) |
| MERS-CoV | ORF4a, and ORF4b proteins, M | Inhibit ISRE activation after stimulation with IFN, mechanism unknown | Yang et al. (2013) |
| MERS-CoV | Unknown | Repressive histone modifications | Menachery et al. (2014a) |
5.3. Increasing IFN Resistance
Despite having some sensitivity toward IFN, especially MERS-CoV (see Section 4), viral strategies to increase IFN resistance are also in place (Table 3 ). The sequestration of viral dsRNA in DMVs (Knoops et al., 2008, van Hemert et al., 2008, Versteeg et al., 2007) not only reduces cytoplasmic exposure to PRRs and hence IFN induction but also limits activation of antiviral dsRNA-responsive ISG products like PKR. However, PKR is eventually activated by SARS-CoV infection, but has no effect on viral replication (Krahling et al., 2009). Interestingly, other coronaviruses cope differently with PKR. The avian infectious bronchitis virus (IBV) expresses a weak inhibitor or PKR (nsp2) and additionally upregulates the phosphatase subunit GADD34 to reduce phosphorylation of the PKR downstream target eIF-2alpha (Wang et al., 2009). By contrast, the porcine reproductive and respiratory syndrome virus (PRRSV; a member of the Arteriviridae that are related to the Coronaviridae and other nidoviruses) does not inhibit but rather requires PKR for optimal replication and gene expression (Wang et al., 2016). Thus, the interactions and interdependencies of coronaviruses with PKR are complex and far from being understood. With respect to the antiviral OAS/RNase L system that is also activated by dsRNA, the mouse coronavirus MHV-A59 was shown to expresses an ns2 protein that antagonizes by degrading the product of the OAS enzyme, 2′–5′ oligoadenylate that would activate RNase L (Zhao et al., 2012). SARS-CoV and MERS-CoV do not possess an ns2 homolog (Silverman and Weiss, 2014), but the MERS-CoV ns4b was recently demonstrated to cleave 2′–5′ oligoadenylate (Thornbrough et al., 2016). Although ns4b-mutated MERS-CoV was not attenuated in cell culture, it provoked increased RNAse L activity in infected cells (Thornbrough et al., 2016). A critical factor for IFN resistance of SARS-CoV (and of the low pathogenic HCoV-229E) is the ADP-ribose-1″-monophosphatase (ADRP) domain that is contained within the nsp3 protein (Kuri et al., 2011). Virus mutants lacking a functional ADRP domain (also called macrodomain) display an increased IFN sensitivity. ADRP-like macrodomains are encoded by other coronaviruses and several other positive-strand RNA viruses (Gorbalenya et al., 1991). Also for MHV-A59, a role of the ADRP domain in pathogenesis was shown (Eriksson et al., 2008, Fehr et al., 2015), but this seems not be related to IFN sensitivity.
Table 3.
Mechanisms and Factors of Human Coronaviruses to Increase IFN Resistance
| Virus | Viral Protein or Function | Mechanism | References |
|---|---|---|---|
| SARS-CoV (MHV-1) | Storage of dsRNA inside double-membrane vesicles | Prevents exposure of dsRNA to PKR and OAS | Knoops et al. (2008), van Hemert et al. (2008), and Versteeg et al. (2007) |
| SARS-CoV | Unknown | Insensitivity to activated PKR | Krahling et al. (2009) |
| SARS-CoV | ADP-ribose-1″-monophosphatase domain of nsp3 | Unknown | Kuri et al. (2011) |
6. Conclusions and Outlook
The last 10 + years have seen tremendous progress toward the identification of IFN antagonists of human coronaviruses (De Diego et al., 2014, Gralinski and Baric, 2015, Kindler and Thiel, 2014, Perlman and Netland, 2009, Thiel and Weber, 2008, Totura and Baric, 2012, Vijay and Perlman, 2016, Wong et al., 2016). For SARS-CoV, the catalogue of IFN antagonists may be nearly complete by now and that of MERS-CoV may follow soon. Nonetheless, we are still far from comprehensively understanding the manifold interactions of human pathogenic coronaviruses with the IFN system. Many of the factors described here were identified by overexpression studies, and still lack the final biological assessment through generation and characterization of adequate virus mutants. It would also be interesting to see at which infections stage, in which subcellular compartment, and with which comparative intensity the IFN antagonists act, and whether and how they interact with each other. It is however safe to state that coronaviruses, which have the largest RNA genome known to date, do not rely on single virulence factors but employ several layers of anti-IFN strategies. Otherwise they would not be able to exist, thrive, and even expand to new hosts in the presence of powerful antiviral IFN responses.
Acknowledgments
F.W. is supported by the SFB 1021 and Grant We 2616/7-1 (SPP 1596) of the Deutsche Forschungsgemeinschaft. E.K. and V.T. were supported by the Swiss National Science Foundation (SNF Grant 149784).
Disclosures: No conflicts of interest declared.
References
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Bailey C.C., Zhong G.C., Huang I.C., Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu. Rev. Virol. 2014;1(1):261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C.M., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B. Crystal structure of the middle east respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekisz J., Schmeisser H., Hernandez J., Goldman N.D., Zoon K.C. Human interferons alpha, beta and omega. Growth Factors. 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- Belgnaoui S.M., Paz S., Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Binder M., Eberle F., Seitz S., Mucke N., Huber C.M., Kiani N. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J. Biol. Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell L.D., Zalinger Z.B., Li Y., Wright P.W., Elliott R., Rose K.M. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal OAS gene expression and independent of virus-induced interferon. J. Virol. 2016;90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Zust R., Weber F., Spiegel M., Lang K.S., Akira S. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.W.Y., Chan M.C.W., Agnihothram S., Chan L.L.Y., Kuok D.I.T., Fong J.H.M. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabo S., Meurs E.F. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses. 2012;4:2598–2635. doi: 10.3390/v4112598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Castano-Rodriguez C., Fernandez-Delgado R. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M.L., Selisko B., Alvarez K. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2'O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Tseng M., Barretto N., Lin R. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K.K., Cervantes-Barragan L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci. Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V., Sen G.C. Interferon-induced IFIT proteins: their role in viral pathogenesis. J. Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freaney J.E., Kim R., Mandhana R., Horvath C.M. Extensive cooperation of immune master regulators IRF3 and NFkappaB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M.U. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J. Virol. 2014;88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Koltsida O., Andreakos E. Type III interferons (IFNs): Emerging Master Regulators of Immunity. vol. 850. Springer International Publishing; 2015. Crossroads between innate and adaptive immunity V; pp. 1–15. (Advances in Experimental Medicine and Biology). [DOI] [PubMed] [Google Scholar]
- Gil J., Garcia M.A., Gomez-Puertas P., Guerra S., Rullas J., Nakano H. TRAF family proteins link PKR with NF-kappa B activation. Mol. Cell. Biol. 2004;24:4502–4512. doi: 10.1128/MCB.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M.C. Putative papain-related thiol proteases of positive-strand RNA viruses—identification of rubivirus and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubivirus, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough D.J., Messina N.L., Clarke C.J.P., Johnstone R.W., Levy D.E. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Baric R.S. Molecular pathology of emerging coronavirus infections. J. Pathol. 2015;235:185–195. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A.M., Rimmelzwaan G.F., van Amerongen G. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Holzinger D., Jorns C., Stertz S., Boisson-Dupuis S., Thimme R., Weidmann M. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J. Virol. 2007;81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V. To sense or not to sense viral RNA—essentials of coronavirus innate immune evasion. Curr. Opin. Microbiol. 2014;20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Jonsdottir H.R., Muth D., Hamming O.J., Hartmann R., Rodriguez R. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio. 2013;4:e00611–e00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahling V., Stein D.A., Spiegel M., Weber F., Muhlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J. Virol. 2009;83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri T., Zhang X., Habjan M., Martinez-Sobrido L., Garcia-Sastre A., Yuan Z. Interferon priming enables cells to partially overturn the SARS coronavirus-induced block in innate immune activation. J. Gen. Virol. 2009;90:2686–2694. doi: 10.1099/vir.0.013599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri T., Eriksson K.K., Putics A., Zust R., Snijder E.J., Davidson A.D. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Lau C.C.Y., Chan K.H., Li C.P.Y., Chen H.L., Jin D.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J. Gen. Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Y., Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen M., Cao H.W., Zhu Y.F., Zheng J., Zhou H. Extraordinary GU-rich single-strand RNA identified from SARS coronavinis contributes an excessive innate immune response. Microbes Infect. 2013;15:88–95. doi: 10.1016/j.micinf.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y.T. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. elife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.T. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutfy M.R., Blatt L.M., Siminovitch K.A., Ward S., Wolff B., Lho H. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome—a preliminary study. J. Am. Med. Assoc. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P., Sun D.Y., Silverman R.H., He B.A. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakoiv T., Ritz D., Mordstein M., DeDiego M.L., Enjuanes L., Muller M.A. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 2012;93:2601–2605. doi: 10.1099/vir.0.046284-0. [DOI] [PubMed] [Google Scholar]
- Malathi K., Dong B., Gale M., Jr., Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., Saito T., Crochet N., Barton D.J., Gale M., Silverman R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. vol. 1. Wolters Kluwer; Philadelphia: 2013. pp. 825–858. (Fields Virology). [Google Scholar]
- Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schafer A., Josset L., Sims A.C., Proll S. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5:e01174–01114. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M., Kochs G., Dumoutier L., Renauld J.C., Paludan S.R., Klucher K. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla S.R., Toroney R., Bevilacqua P.C. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Golenbock D., Bowie A.G. The history of Toll-like receptors—redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- Onomoto K., Jogi M., Yoo J.S., Narita R., Morimoto S., Takemura A. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller C.K., Li Z., George C.X., Samuel C.E. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A.M., Santa Maria F.G., Lahiri T., Friedman E., Marie I.J., Levy D.E. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog. 2016;12:e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.B., Reinert L.S., Paludan S.R. Innate recognition of intracellular pathogens: detection and activation of the first line of defense. APMIS. 2009;117:323–337. doi: 10.1111/j.1600-0463.2009.02456.x. [DOI] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S., Sparrer K.M.J., Lassig C., Hembach K., Baum A., Garcia-Sastre A. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Owen D.M., Jiang F.G., Marcotrigiano J., Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuplein V.A., Seifried J., Malczyk A.H., Miller L., Hocker L., Vergara-Alert J. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89:3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J.J., Kanjanahaluethai A., Baker S.C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M. Master sensors of pathogenic RNA—RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S., Mordstein M., Kochs G., Garcia-Sastre A., Tenoever B.R. Transcription factor redundancy ensures induction of the antiviral state. J. Biol. Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreibelt G., Tel J., Sliepen K.H.E.W.J., Benitez-Ribas D., Figdor C.G., Adema G.J. Toll-like receptor expression and function in human dendritic cell subsets: implications for dendritic cell-based anti-cancer immunotherapy. Cancer Immunol. Immunother. 2010;59:1573–1582. doi: 10.1007/s00262-010-0833-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., Pichlmair A., Rehwinkel J., Rogers N.C., Scheuner D., Kato H. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R.H., Weiss S.R. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J. Interferon Cytokine Res. 2014;34:455–463. doi: 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Yeung M.L., Kok K.H., Yuen K.S., Kew C., Lui P.Y. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Muhlberger E., Haller O., Weber F. The antiviral effect of interferon-beta against SARS-coronavirus is not mediated by MxA protein. J. Clin. Virol. 2004;30:211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Martinez-Sobrido L., Cros J., Garcia-Sastre A., Haller O. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Darnell J.E. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer D.R., Dickey R., Carter W.A. Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect. Disord. Drug Targets. 2014;14:37–43. doi: 10.2174/1871526514666140713152858. [DOI] [PubMed] [Google Scholar]
- Stroher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F. Severe acute respiratory syndrome related coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Thiel V., Weber F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 2008;19:121–132. doi: 10.1016/j.cytogfr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7:e00258. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Bredenbeek P.J., van den Worm S.H., Spaan W.J. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R., Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr. Opin. Virol. 2016;16:70–76. doi: 10.1016/j.coviro.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu L. The membrane protein of severe acute respiratory syndrome coronavirus functions as a novel cytosolic pathogen-associated molecular pattern to promote beta interferon induction via a toll-like-receptor-related TRAF3-independent mechanism. mBio. 2016;7:e01872-15. doi: 10.1128/mBio.01872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.X., Liao Y., Yap P.L., Png K.J., Tam J.P., Liu D.X. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 2009;83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Q., Zhang H.M., Abel A., Nelson E. Protein kinase R (PKR) plays a pro-viral role in porcine reproductive and respiratory syndrome virus (PRRSV) replication by modulating viral gene transcription. Arch. Virol. 2016;161:327–333. doi: 10.1007/s00705-015-2671-0. [DOI] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Weber F. Segmented negative-strand RNA viruses and RIG-I: divide (your genome) and rule. Curr. Opin. Microbiol. 2014;20:96–102. doi: 10.1016/j.mib.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.-Y.R., Lui P.-Y., Jin D.-Y. A molecular arms race between host innate antiviral response and emerging human coronaviruses. Virol. Sin. 2016;31:12–23. doi: 10.1007/s12250-015-3683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner D.H., McCauley J.W., Skehel J.J., Kerr I.M. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H.C.H., Williams B.R.G. Protein kinase R and the inflammasome. J. Interferon Cytokine Res. 2014;34:447–454. doi: 10.1089/jir.2014.0008. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Jogi M., Onomoto K. Regulation of antiviral innate immune signaling by stress-induced RNA granules. J. Biochem. 2016;159:279–286. doi: 10.1093/jb/mvv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Daryoush M., Mogensen T.H., DiDonato J.A., Williams B.R. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol. Cell. Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A.E. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.S., Guo F., Liu F., Cuconati A., Chang J.H., Block T.M. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H.Y., Cheng Z.S., Poon V.K.M. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Schelle B., Karl N., Minskaia E., Bayer S., Siddell S.G. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007;81:3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T., Matikainen S., Ronkko E., Osterlund P., Sillanpaa M., Siren J. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 2005;79:13800–13805. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013;87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L., Tramontano E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antivir. Res. 2013;100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


