Abstract
Enveloped viruses enter cells by inducing fusion of viral and cellular membranes, a process catalyzed by a specialized membrane-fusion protein expressed on their surface. This review focuses on recent structural studies of viral fusion proteins with an emphasis on their metastable prefusion form and on interactions with neutralizing antibodies. The fusion glycoproteins have been difficult to study because they are present in a labile, metastable form at the surface of infectious virions. Such metastability is a functional requirement, allowing these proteins to refold into a lower energy conformation while transferring the difference in energy to catalyze the membrane fusion reaction. Structural studies have shown that stable immunogens presenting the same antigenic sites as the labile wild-type proteins efficiently elicit potently neutralizing antibodies, providing a framework with which to engineer the antigens for stability, as well as identifying key vulnerability sites that can be used in next-generation subunit vaccine design.
A greater understanding of how viruses fuse with host cells, driven by recent advances in viral structural biology, is leading the way for new immunogen design and vaccine development.
Main Text
Introduction
Many of the circulating human pathogenic viruses are enveloped with a lipid bilayer, and infect their target cells by inducing the fusion of the viral envelope with the cell membrane. Although there are efficient vaccines against some of these viruses, for the majority there is no prophylactic or therapeutic treatment. The known vaccines typically work by eliciting antibodies that block entry of the pathogen into cells, which in the case of enveloped viruses involves antibody binding to the viral envelope proteins. The viral fusion protein is the key factor that induces the membrane fusion reaction that allows viral entry. A number of enveloped viruses display only a single protein on the particle surface, which necessarily mediates attachment to the cell surface as well as inducing the subsequent membrane fusion reaction. Understanding the interactions of antibodies with the viral envelope proteins—in particular with the fusion protein—and the antibody neutralization mechanism is paramount to develop potentially protective vaccines for HIV and many other human pathogenic enveloped viruses for which there are no vaccines available.
Three Structural Classes of Viral Membrane Fusion Proteins
The influenza A virus hemagglutinin (HA) protein binds sialic acid on host cells and is responsible for membrane fusion. Determination of its X-ray structure was a pioneering result that showed the way to study other viruses (Wilson et al., 1981). It then took 14 years until the structure of another fusion protein was determined, this time from a flavivirus, the tick-borne encephalitis virus (Rey et al., 1995), which revealed a completely different fold of the polypeptide chain compared to that of influenza virus HA. Later, as the X-ray structures of fusion proteins from a number of different viruses were determined, it became apparent that many otherwise unrelated viruses had related fusion proteins. This finding indicated that the corresponding gene had been either exchanged horizontally among viruses, or had been acquired from an ancient gene pool shared with cells—as suggested recently by the shared structural homology also observed in the ancient eukaryotic gamete fusion protein HAP2 (Doms, 2017)—evolving subsequently beyond the limits of homology detection by current amino acid sequence analysis tools. In total, there are currently three classes of structurally homologous proteins that have been characterized—termed class I, typified by influenza HA; class II, illustrated by the flavivirus envelope protein E; and class III, typified by the rhabdovirus glycoprotein G.
The Fusogenic Conformational Change
The accepted model for enveloped virus entry posits that interactions with a target cell trigger an exothermic fusogenic conformational change of the fusion protein, which irreversibly transits from a metastable, activated prefusion form to its lowest-energy, postfusion conformation. The three classes adopt a common postfusion hairpin-like arrangement, juxtaposing the target-membrane insertion element of the protein with its viral trans-membrane anchor, suggesting that in spite of their altogether different structures, they display a similar mechanism for catalyzing the membrane fusion reaction.
The energy released in the conformational transition is used to overcome the repulsive force between the two membranes generated as they approach each other by the required dehydration of the lipid heads to allow for direct interaction between the two bilayers (Leikin et al., 1993). This dehydration force, which is felt by the membranes as they approach to less than ∼3 nm from each other and increases very steeply as they become any closer, prevents direct membrane contacts and acts as an effective barrier for an overall exergonic (i.e., releasing free energy) membrane fusion reaction, which would otherwise occur spontaneously.
During the conformational change, the fusion protein adopts a transient, extended intermediate conformation in which a non-polar segment, termed the “fusion peptide” (FP) or “fusion loop” (FL), projects out to insert into the cell membrane, thereby bridging viral and cellular membranes at a distance in the order of 10–15 nm. This extended form then collapses into a “hairpin,” as illustrated in Figure 1 , in which the target membrane-inserted non-polar segment is relocated to the proximity of the viral trans-membrane anchor of the fusion protein, effectively bringing the two membranes to a distance within 1 nm (Harrison, 2015), overcoming the dehydration force and allowing direct membrane apposition. The two membranes then fuse via an initial hemifusion step in which the lipids of the outer leaflets of the membranes merge, followed by formation of an initial fusion pore (or multiple small pores) by merging of the inner leaflets, and then expansion of the fusion pore(s) to full fusion. The mechanistic details of the latter steps are, however, not completely understood (Nikolaus et al., 2011).
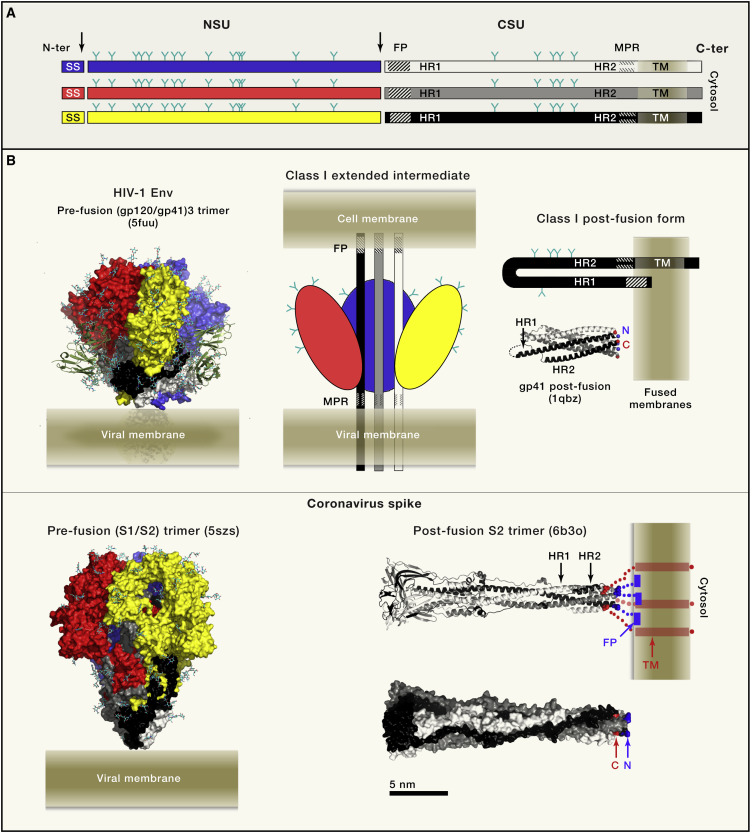
Figure 1.
Class I Viral Fusion Proteins
(A) Linear diagram illustrating the organization of the class I fusion protein polypeptide chain. A precursor glycoprotein is co-translationally translocated into the ER of the infected cell, where it is co- and post-translationally modified by glycosylation (Y symbols) and formation of disulfide bonds (not shown). An ER-resident signal peptidase generates the N terminus of the precursor upon cleavage (left vertical arrow) of the signal sequence (SS), while the precursor remains membrane-anchored through a C-terminal trans-membrane (TM) segment. The precursor folds as a trimer (represented by the three horizontal bars), which undergoes a subsequent proteolytic step (vertical arrow at the center) that generates two mature subunits NSU (N-terminal subunit, blue, red or yellow bars; the colors match the 3D diagrams in (B) and CSU (C-terminal subunit, white, gray or black bars). The two subunits remain associated in the mature trimer, which becomes metastable and primed for the irreversible conformational change that drives membrane fusion for infection of a new cell. The NSU is usually the receptor binding subunit, although in some viruses a separate protein carries this function. The CSU is the viral fusion protein, with a fusion peptide at its N-terminal end (symbolized by a dashed region) and an external membrane-proximal region (MPR) near the TM domain (symbolized by short dashes).
(B) Two representative class I fusion proteins in their prefusion conformation shown in surface representation (left panels), with NSU and CSU colored according to panel A. The glycan chains are shown as sticks with carbon atoms cyan and oxygen atoms red. The viral membrane is diagrammed to scale, with the aliphatic moiety in dark beige at the center, dingfa out as it enters the hydrophilic lipid head-group moiety. Left, top panel, structure of a fully-glycosylated, clade B native HIV Env trimer determined by cryo-EM to 4.4Å resolution (Lee et al., 2016) in the presence of the TM segment (empty arrow) and in complex with bnAb PGT151, which is displayed with the variable domains as green ribbons. The MPR segment and TM region appeared mostly disordered. The bottom left panel shows the cryo-EM structure of the NL63 α-coronavirus NL63 spike protein ectodomain, determined to 3.4Å resolution, and displaying clear density for many of the N-linked glycans (Walls et al., 2016b). The prefusion forms display the NSU subunit (S1 in coronaviruses and gp120 in HIV) making a crown (red/yellow/blue) around a spring-loaded CSU (black/gray/white; S2 in coronaviruses and gp41 in HIV). Interactions with the target cell trigger the release of the NSU crown, allowing the CSU to undergo a fusogenic conformational change going through an extended intermediate that bridges the two membranes (represented in the top-middle panel). The hairpin conformation adopted by the CSU (diagrammed at the top right) has the fusion peptide inserted into the membrane next to its TM region. A schematic fused membrane is shown to scale. Immediately below the hairpin is a ribbon representation of the postfusion CSU from SIV (gp41), forming a 6-helix bundle (Yang et al., 1999). The N and C-terminal ends of the gp41 subunit in black are marked. The two lower panels show a representative structure of the coronavirus postfusion CSU trimers displayed as ribbons (middle) and as surface representation (bottom) (Walls et al., 2017) with the N-and C-terminal ends visible in the structure of the ectodomain indicated in blue and red, respectively. A schematic on the ribbon diagram shows the expected location of the missing segments in the intact postfusion protein on membranes (not present in the structure; blue at the N-terminal end, connecting to the fusion peptide inserted superficially on the outer lipid leaflet of the fused membranes, represented as a full blue rectangle) and the C-terminal end in red, connecting to the TM helices and the cytosolic tail.
Membrane Fusion Triggers
The interactions with the target cell that trigger the fusogenic conformational change vary for different viruses, and can be divided into several broad categories (Yamauchi and Helenius, 2013). In the first one, fusion is triggered directly by interactions of the fusion protein (as in the case of HIV)—or by a companion viral glycoprotein at the particle surface (seen for paramyxoviruses and herpes viruses)—with a cellular receptor. In some cases, a sequential interaction with a primary receptor and then a co-receptor is required, as in HIV interacting with CD4, which triggers a first conformational change (Ozorowski et al., 2017) that then allows interaction with the chemokine receptors CCR5 or CXCR4 (Feng et al., 1996). In this case, the virus can fuse directly with the plasma membrane or can be incorporated into internal vesicles before fusion. In the second broad category, the interactions with receptor(s) at the cell surface lead to particle endocytosis, and binding of protons in the low pH endosomal environment triggers the fusogenic conformational change (seen for influenza viruses, alphaviruses, rhabdoviruses, flaviviruses, and bunyaviruses) (White and Whittaker, 2016). In the third category, the initial interactions of the virion at the cell surface trigger endocytosis, and then a second internal receptor, often found in late endosomes—and binding of which requires further proteolysis of the fusion protein by an endosomal protease in the case of some viruses (cf. Ebola virus)—triggers the final fusogenic conformational change (e.g, filoviruses and arenaviruses) (Jae and Brummelkamp, 2015). Finally, for certain viruses the fusion protein may require an activating redox reaction involving disulfide bonds to induce membrane fusion (Key et al., 2015).
With these broad classes defined, we focus this review on the structural biology of the viral fusion proteins, in particular on their activated metastable prefusion conformation that is recognized by neutralizing antibodies. We discuss the strategies that have been developed to stabilize them in their prefusion form enabling structural studies, and the role of antibodies in targeting and stabilizing relevant conformational epitopes. These studies also highlight the importance of engineering stabilized forms that preserve the antigencity of the metastable prefusion form on virions to use as immunogens for subunit vaccine design. In this regard, we discuss more extensively the class I and II fusion proteins. We mention at the end the members of class III, as well as the enveloped viruses that appear to fall outside these three classes and for which a full characterization of the membrane fusion machinery still awaits high resolution structural studies.
Class I Fusion Proteins
Class I fusion proteins form non-covalently linked homotrimers in their pre- and postfusion conformations. They are type I single pass trans-membrane proteins synthesized as a precursor, the N terminus of which is derived by signal peptidase cleavage shortly after translocation into the endoplasmic reticulum. This precursor undergoes subsequent proteolytic maturation to generate an N-terminal and a C-terminal subunit (NSU and CSU, Figure 1), which in some cases remain linked by a disulfide bond. The structural hallmark of class I fusion proteins is a parallel trimeric α-helical coiled-coil in the postfusion CSU. The long α-helix of the coiled-coil (HR1 in Figure 1) can be identified by bioinformatic analyses because of the presence of heptad repeats (HR), with non-polar amino acids at positions 1 and 4 of the repeats. The C-terminal HR2 helix, upstream of the external membrane-proximal region (MPR in Figure 1), runs antiparallel to HR1 along the grooves of the coiled-coil to complete the postfusion hairpin, thereby making a trimeric postfusion 6-helix bundle. In some viruses, the HR2 α-helix is absent, but the polypeptide chain still runs along the coiled-coil grooves antiparallel to HR1, albeit in an extended conformation (cf., influenza viruses). The fusion peptide is at—or near—the N-terminal end of the HR1 helix, which is spring-loaded in the prefusion form underneath an NSU trimer cap. The interactions with the target cell for entry destabilize the trimer contacts of the NSU cap, enabling HR1 to spring out and expose the fusion peptide for insertion into the cellular membrane.
The group of enveloped viruses carrying class I fusion proteins includes respiratory viruses such as the influenza viruses (four genera in the Orthomyxovirus family: influenza A, B, C and D), the respiratory syncytial virus (RSV, Pneumoviridae family) and the related measles, mumps and parainfluenza viruses in the Paramyxoviridae family, which also includes the recently emerged zoonotic Hendra and Nipah encephalitis viruses that cause serious disease in humans. Other respiratory virus members of the Class I group include the coronaviruses (CoVs) (Coronaviridae family) responsible for seasonal respiratory infections (NL73 CoV and HKU1 CoV, for instance), as well as the zoonotic severe acquired respiratory syndrome (SARS CoV) and Middle-Eastern respiratory syndrome coronaviruses (MERS CoV). The Retroviridae family, exemplified by HIV and the human T cell leukemia viruses (HTLVs), represent a very important subset of class I viruses. Last but not least, several important hemorrhagic fever agents have class I fusion proteins, the most notable ones being Lassa virus together with other members of the Arenaviridae family, and Ebola virus and relatives in the Filoviridae family.
The Early Structures of Class I Proteins in the Prefusion Form
Only recently have the three-dimensional structures of the fusion protein from representatives of each of the above virus families become available to near atomic resolution (Figure 2 ). Although the very first X-ray structures of viral fusion proteins were obtained using crystals of the ectodomain directly cleaved off the membrane surface (or cleaved after detergent solubilisation) by controlled proteolysis (Rey et al., 1995, Wilson et al., 1981), the structures that followed were determined by crystallizing the recombinant ectodomain, which in most cases required additional stabilization to maintain its trimeric prefusion form upon folding in the absence of the transmembrane segments. An early example is the structure of the HA ectodomain from the reconstituted influenza A virus (H1 serotype) that caused the 1918 pandemic (Stevens et al., 2004). The recombinant HA0 ectodomain (i.e., the uncleaved form obtained by mutating the cleavage site between NSU and CSU, termed HA1 and HA2, respectively) was monomeric unless engineered with a trimerization motif fused at its C terminus, replacing the trans-membrane segment and cytosolic tail. The motif used was derived from a bacteriophage trimeric fiber, termed “fibritin foldon.”
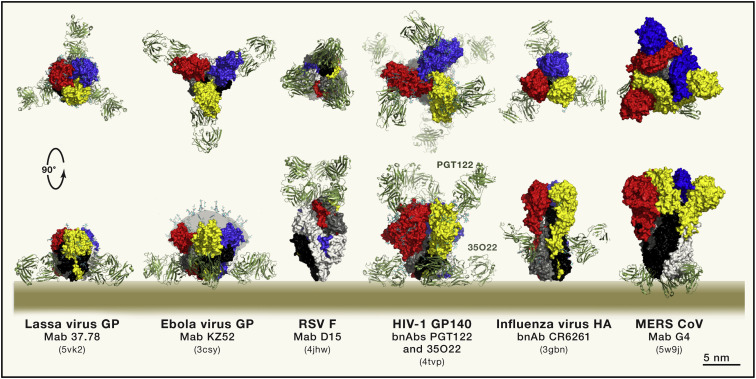
Figure 2.
Antibody Recognition of Class I Fusion Proteins
Representative structures of enveloped viruses with class I fusion proteins in complex with neutralizing antibodies (drawn at the same scale and aligned by height on the viral membrane): from left to right: Arenaviridae (Lassa virus GP); Filoviridae (Ebola virus GP), Retroviridae (HIV-1 Env), Pneumoviridae (Respiratory syncytial virus F), Orthomyxoviridae (Influenza virus HA), Coronaviridae (MERS CoV S). In the case of the Ebola virus GP, the mucin-like domain missing from the structure was diagrammed in gray with multiple glycans drawn as sticks, as a guide. The color coding is the same as in Figure 1; the top panel shows a top view down the 3-fold axis of the trimer, and the bottom panel displays a side view with the viral membrane represented to scale. Note that RSV F is different in the sense that the CSU is the bulkiest subunit, in contrast to the others, which have a large NSU crown. The PDB accession code of each of these structure is indicated in between parenthesis in each case.
In parallel, work on the paramyxovirus fusion protein F showed that—unexpectedly at the time—knocking out the cleavage site between NSU and CSU led to a trimeric recombinant ectodomain that had spontaneously adopted the postfusion form (Chen et al., 2001, Yin et al., 2005), indicating that in this case, it was not the maturation cleavage (NSU and CSU are called F2 and F1, respectively, and the precursor F0) that primed the protein to undergo the fusogenic conformational change, as in other class I proteins. On paramyxovirus particles, the fusogenic conformational change of F is triggered by a companion protein that binds a receptor present at the surface of target cells. Receptor binding alters the conformation of the companion protein, affecting its interactions with F at the virion surface, such that F is triggered to induce fusion (Bose et al., 2015). In the context of the isolated recombinant F ectodomain, this change occurs spontaneously, but engineering a trimer stabilizing motif at its C-terminal end—in this case a trimeric GCN4 element—resulted in stabilization of the prefusion F trimer. This approach allowed the determination of the X-ray structure of prefusion F of the parainfluenza 5 virus (Yin et al., 2006), followed by those of several other paramyxoviruses. Similarly, the structure of the F protein from the respiratory syncytial virus (RSV), which is closely related to the F protein of the paramyxoviruses but has two cleavage sites separating F2 from F1, was determined in its postfusion form, albeit cleaved and lacking the hydrophobic fusion peptide (McLellan et al., 2011, Swanson et al., 2011). In this case, addition of a trimerization foldon did not impede the conformational change, and only co-expression in the same cells with the antigen binding fragment (Fab) of a prefusion F specific, potently neutralizing antibody isolated from a patient (Figure 2), made it possible to maintain the trimeric prefusion form for structural studies (McLellan et al., 2013b).
For filoviruses, the determination of the X-ray structure of the Ebola virus glycoprotein (GP) in the prefusion form (at 3.4Å resolution) did not require trimer stabilization, but it did require the deletion of a large, glycosylated mucin-like domain, as well as genetic ablation of several N-glycosylation sites. Even after these manipulations, diffraction quality crystals were only obtained with the protein in complex with the Fab of a neutralizing antibody isolated from a patient who survived the disease (Figure 2) (Lee et al., 2008). This pioneering result was followed by several structures of the GP from several filoviruses in complex with various other neutralizing antibodies, obtained by X-ray crystallography to around 3.5Å resolution and by cryo-EM at lower resolutions, reviewed recently by (Kirchdoerfer et al., 2017). Subsequent protein engineering by fusion of the fibritin foldon sequence at the C-terminal end resulted in stabilization of the structure such that crystals of the unliganded GP ectodomain were obtained that diffracted to much higher resolution (2.2 Å resolution) with clear electron density for the HR2 region (not resolved in previous structures) forming additional trimer contacts in the membrane proximal region of the molecule (Zhao et al., 2016).
The HIV Env Trimer
The case of the retrovirus envelope (Env) protein trimer has been by far the most complex for structural studies due to its extensive glycosylation, conformational flexibility, and the tendency of the NSU and CSU to spontaneously dissociate. Initial attempts to generate stable, soluble trimers entailed knocking out the cleavage site between NSU and CSU, which are termed SU and TM for retroviruses in general, and gp120/gp41 for HIV, respectively. The resulting constructs were made trimeric by addition of a C-terminal foldon sequence, but it later became apparent that the resulting uncleaved Env trimer ectodomains did not display the same antigenicity pattern as native trimers on virions (Ringe et al., 2013). Other strategies to stabilize the trimer were therefore sought in order to preserve the native conformation as presented on infectious virions, which is the conformation recognized by neutralizing antibodies. Point mutations were designed to introduce cysteine residues strategically located for formation of inter-subunit disulfide bonds. This approach, inspired by the observation that some retroviruses have cysteines that cross-link NSU and CSU (Li et al., 2008a), involved a great deal of trial and error as the structure was not known at the time. It eventually led to an ectodomain variant termed “SOS” that was correctly processed into NSU and CSU, which remained covalently linked by the engineered disulfide bond (thereby avoiding NSU shedding), although this construct still did not form trimers (Binley et al., 2000). Further work was in part inspired by results on influenza virus HA introducing helix breaker residues (proline, but also glycine) in a region with strong α-helical propensity and displaying a heptad repeat pattern, but which forms a loop in between two consecutive helices in the structure of the prefusion form. This same segment is found as part of the long HR1 α-helical coiled-coil in the postfusion CSU. The proline mutants led to an HA molecule with identical antigenicity as wild-type, but which could not induce membrane fusion (Qiao et al., 1998). A similar strategy resulted in the HIV Env variant called “SOSIP,” which in addition to the SOS mutation has an I559P mutation in HR1 (Sanders et al., 2002). The SOSIP variant was trimeric and displayed an antigenic profile very similar to that of Env trimers exposed at the surface of virions. Additional modifications, such as removal of the amphipathic external membrane proximal region (MPR, Figure 1) by using a construct ending at position 664 (called SOSIP-664) avoided solubility issues of the initial constructs. Screening for orthologs from many strains—HIV-1 Env is highly variable in amino acid sequence and is phylogenetically divided into 9 clades (A, B, C, D, E, F, G, K and O)—with enhanced stability as examined in the electron microscope, further led to identifying HIV-1 clade A strain BG505 as a particularly stable trimer (Julien et al., 2013). The use of the BG505-SOSIP-664 trimer, together with key broadly neutralizing antibodies isolated from patients culminated in a number of structures (one of them illustrated in Figure 2). Readers are referred to recent reviews (Pancera et al., 2017, Ward and Wilson, 2017) that analyze the implications of the structures for understanding the fusogenic conformational change and for the design of better immunogens to use in developing HIV vaccines.
The antibody fragments bound to the protein were essential for the structural studies as they provided packing contacts to form the crystal lattice, since the extensive Env glycosylation (around 30 glycans attached per protomer, depending on the strain) interfered with crystal formation. The bNAb fragments also helped with structure determination by cryo-EM by providing a larger size complex to facilitate image analysis. The concomitant advent of direct electron counting detectors allowed the application of single particle cryo-EM to reach resolutions approaching 3Å, which in turn made possible the visualization of natively glycosylated trimers. A fully glycosylated trans-membrane clade B native Env trimer (lacking the cytosolic tail but containing the MPR and the TM segments) was recently visualized to 4.2Å resolution by cryo-EM (Figure 1B, left) (Lee et al., 2016). This detergent-solubilized Env trimer lacks the cytosolic tail, but contains the external MPR and TM segment, and has an ectodomain conformation highly similar to that of the soluble SOSIP trimer (Figure 2), confirming the native conformation of the latter. The cryo-EM reconstruction also showed that the external MPR and TM segments do not form a rigid structure relative to the ectodomain, as they appear ill-defined and mobile within a detergent micelle in the structure.
Overall, one of the important observations from these studies was the importance of glycan recognition by some of the most potent bNAbs, such as those targeting the Env trimer apex (Andrabi et al., 2017) or the region of the Asn332-linked glycan (Landais et al., 2016). In turn, these structures have guided further mutagenesis approaches to make stable trimers belonging not only to HIV-1 clade A, but also to the other HIV-1 clades, reviewed by (Medina-Ramírez et al., 2017b). This approach now offers hope of developing new approaches to obtain immunogens capable of eliciting such antibodies in humans.
The Coronavirus Spike
In addition to the retrovirus Env trimer, the coronavirus spike (S) protein is another class I fusion protein that proved difficult to crystallize due to its extensive glycosylation (around 40 N-linked glycans attached per protomer, depending on the CoV) and very large size, as each protomer in the trimer is 1,300 amino acids long. The recent advances in cryo-EM enabled the determination of the structures of the prefusion S ectodomain from several CoVs, beginning with the β-coronaviruses mouse hepatitis virus (MHV) (Walls et al., 2016a) and the HKU1 human coronavirus (Kirchdoerfer et al., 2016) to around 4Å resolution, where the polypeptide chains could be traced. These structures used the S0 precursor ectodomain (i.e, with ablation of cleavage site between NSU and CSU) fused at the C-terminal end to a trimer-stabilizing element in order to maintain its trimeric conformation. In spite of the presence of the added trimerization motif, the HR2 region is disordered in all the structures, suggesting that it may not contribute to interactions stabilizing the trimer as seen for other class I proteins. In several coronaviruses S0 undergoes more than one proteolytic maturation step: an initial cleavage generating the S1 and S2 subunits in the producer cell (a cleavage event that is not conserved across coronaviruses) is followed by further cleavage of S2 to generate an S2′ subunit within an endosomal compartment of the target cell (Burkard et al., 2014). As S2′ has the fusion peptide at its N-terminal end, it corresponds better to the CSU indicated in Figure 1, and it is rather this proteolytic event that would be expected to be universal across coronaviruses. Cryo-EM studies also showed that the receptor binding domain (RBD) of the SARS CoV and the MERS CoV adopts different conformations on the spike, with a closed and an open conformation in which the surface that binds receptor is occluded and exposed, respectively, thereby regulating attachment to host cells (Pallesen et al., 2017, Yuan et al., 2017).
The available structures further enabled an immunogen design strategy, inspired by the HIV SOSIP trimer, that involved stabilization of the CoV S0 prefusion trimer by introduction of two consecutive proline residues in a region of HR1 that forms a loop between two short helices in the spring-loaded prefusion form. This modification allowed a 50-fold increase in immunogen production yields, and the resulting molecule elicited strongly neutralizing antibodies in immunized animals. It also led to a structure in complex with a neutralzing antibody targeting the stem region (Figure 2) (Pallesen et al., 2017). The possibility to now evaluate S0 immunogens of MERS and SARS CoV locked in an RBD “closed” conformation is a further window of opportunity opened by these structures.
Engineering the RSV F Prefusion Trimer
Similar to the coronavirus S protein, the initial structures of RSV F suggested approaches to engineer mutations to stabilize the prefusion trimer for its use as immunogen, since the prefusion conformation contains epitopes that are the target of the most potently neutralizing antibodies (Ngwuta et al., 2015). The first stabilized constructs contained an engineered intra-CSU (F1 in RSV) disulfide bond and two cavity-filling mutations in addition to the fibritin foldon fused at the C-terminal end (McLellan et al., 2013a). Further work identified proline mutations again in the HR1 region of F1, which allowed much higher yields of recombinant prefusion trimer (Krarup et al., 2015). This work also identified the role of p27 (the segment between the two maturation cleavage sites in F), the presence of which blocks F trimerization. Eliminating the p27 segment altogether was found to facilitate trimerization with the resulting trimers retaining the wild-type antigenicity (Krarup et al., 2015). In parallel, additional studies have introduced a “cysteine zipper” in the HR2 region in prefusion F0, which preserves the native conformation of the protein (Stewart-Jones et al., 2015). The enhanced trimer stabilization provided by the cysteine zipper in HR2 renders the foldon unnecessary, which is useful as its presence is undesirable when using the recombinant molecule for vaccine development.
The Lassa Virus Glycoprotein Prefusion Trimer
The above trimer-stabilization strategies were also successful in enabling the determination of the X-ray structure of the prefusion form of the envelope protein of Lassa virus (Hastie et al., 2017). The arenavirus envelope glycoprotein complex (GPC) differs from other class I fusion proteins because it has a Zn2+ binding domain at the C-terminal cytosolic side and has an unusually long “stable signal peptide” (SSP) that remains associated to the complex after signal peptidase cleavage. The maturation cleavage to generate the NSU (called GP1) and the CSU (GP2) is carried out by the subtilase SKI-1/S1P in the early Golgi compartment (Burri et al., 2012). The strategy used to obtain the structure combined engineering an “SOS” disulfide bond between GP1 and GP2, introducing a proline residue in the metastable HR1 region of GP2, and mutating the SKI-1/S1P cleavage site between GP1 and GP2 to make it efficiently cleavable by the furin protease present in the producing cells, in this case Schneider 2 Drosophila melanogaster cells. This form of the protein retained the native antigenicity and was recognized by strongly neutralizing antibodies against Lassa virus isolated from survivors of the infection, the majority of which do not recognize the individual GPC subunits expressed separately. The crystals were obtained in complex with the Fab of one of these neutralizing antibodies (Mab 37.38, Figure 2) and yielded diffraction to 3Å resolution, which is remarkable for this heavily glycosylated protein (27 predicted N-linked glycosylation sites per protomer). As most of the neutralizing antibodies have overlapping epitopes on the GPC trimer and compete with Mab 37.38 for binding, the structure also identifies a main antigenic determinant of the virus. Furthermore, the observed conformation of the Lassa virus prefusion trimer ectodomain observed in the X-ray structure matches that of authentic spikes at the virion surface visualized by cryo-electron tomography and sub-tomogram averaging to about 14 Å resolution (Li et al., 2016b). The stabilized Lassa virus trimer will thus guide next generation vaccine design against arenaviruses, which constitute a large group of rodent-transmitted hemorrhagic disease viruses distributed worldwide.
Class II Fusion Proteins
In contrast to class I, the class II proteins in their prefusion form do not form independent trimers on the viral membrane, but rather form a multimeric assembly that encases the whole viral membrane (Figure 3 ). They do not have long α helices and are essentially folded as β sheets, featuring three structured domains termed I, II, and III. The central domain I is a β sandwich with up-and-down topology with two of the connections between adjacent β strands making long excursions at one end to form domain II, which carries an internal fusion loop at the distal end of the first excursion (see diagram of Figure 3C). A linker connects domain I at its C terminus to domain III, which has an immunoglobulin superfamily fold. An additional segment called the “stem” connects domain III to the trans-membrane region at the C-terminal end of the protein. The ectodomain is organized as a long rod with domain I at the center and domains II and III at either end. The postfusion form features the rod bent in half with domains I and II packing centrally about the 3-fold molecular axis (analogous to HR1 in class I proteins) and domain III with the stem packing externally (like HR2 in class I) to reach the fusion loop, forming a trimeric hairpin analogous to the 6-helix bundle in the class I proteins—albeit with a radically different structure (Figure 4 ).
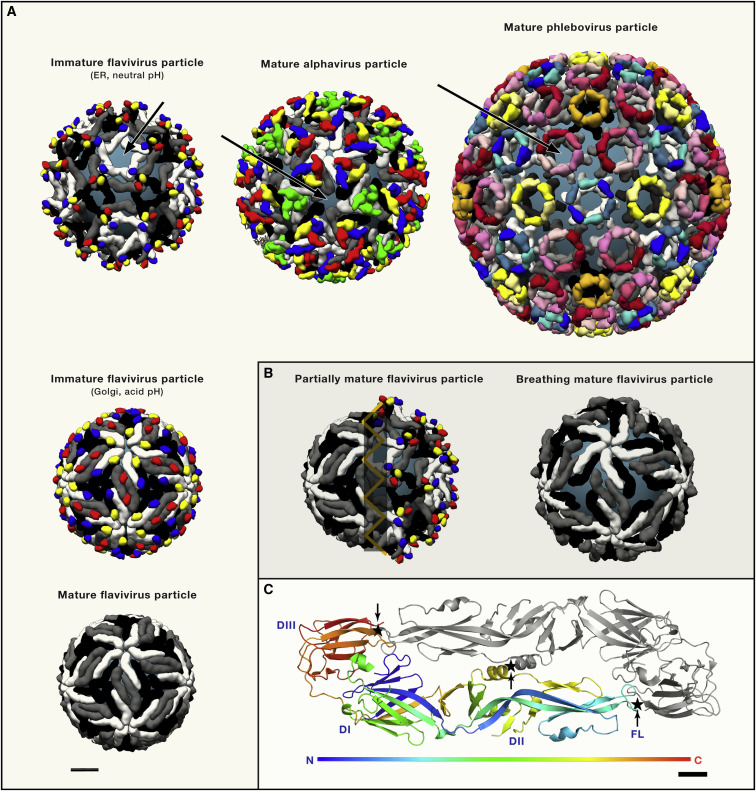
Figure 3.
Class II Viruses: Cryo-EM Structures of Flavivirus, Alphavirus and Phlebovirus Particles
(A) The top row shows the organization of an immature flavivirus particle (left, icosahedral non quasi-equivalent lattice with 3 protomers per asymmetric unit, 180 UGP/DGP heterodimers) (PDB 5u4w), the mature alphavirus (middle, icosahedral T = 4 quasi equivalent lattice, 240 UGP/DGP heterodimers)(PDB 5vU2) and phlebovirus particles (icosahedral T = 12 quasi-equivalent lattice, i.e., 720 UGP/DGP heterodimers) (PDB 6f9b) The three structures represented display a similar organization in which the DGP (shown in white, gray and black) makes the lateral contacts between spikes (trimeric in flaviviruses and alphaviruses, hexameric and pentameric in phleboviruses), and the UGP (in various colors) caps the fusion loop of the DGP at the spike apices. The viral membrane is shown in steel-grey (arrows). Left column: Flavivirus particle maturation: Immature flavivirus particles undergo a conformational change in response to exposure to low pH in the Golgi, in which the 60 (prM/E)3 spikes (top panel) reorganize into 90 (prM/E)2 dimers (middle panel) (PDB 3c6R). In this conformation, prM exposes a cleavage site specific for the cellular furin proteinase. Upon cleavage, the peripheral “pr” domain (red, blue and yellow) stays bound as long as the pH is acidic, but is shed from the particle in the neutral pH external environment. Mature flavivirus particles (bottom left) expose the E protein in a herringbone arrangement, completely covering the viral membrane (PDB 5iz7). All structures are shown at the same scale (bar bottom left, 10nm).
(B) Flavivirus particle heterogeneity (left panel) arising from incomplete furin maturation. In the case of dengue viruses, the conformational change between the top left and middle left panels in (A) is reversible with pH, whereas the one from the middle to the bottom panel is irreversible. Because furin is membrane bound, the particles are often processed only on the side that is closest to the TGN membrane with the opposite side uncleaved. Upon release into the neutral external environment, the non-processed side returns to the immature conformation, whereas the processed side adopts the mature conformation, giving rise to mosaic particles exposing the fusion loop in the immature patches. A representation of a “breathing” mature particle (right) shows an expanded size and exposure of the membrane underneath. These particles also expose epitopes normally buried by dimer contacts on virions.
(C) Organization of the flavivirus E dimer as present on mature virions as a representative class II DGP (PDB code 5lbv). One subunit is rainbow colored along the polypeptide chain (from N to C terminus, as indicated in the color key bar below), the other is gray. The three structured domains DI, DII and DIII are indicated as well as the fusion loop (FL). Sites where disulfide bridges have been engineered to stabilize the dimer in the prefusion conformation are marked (star and arrow). Scale bar (bottom, right): 1 nm.
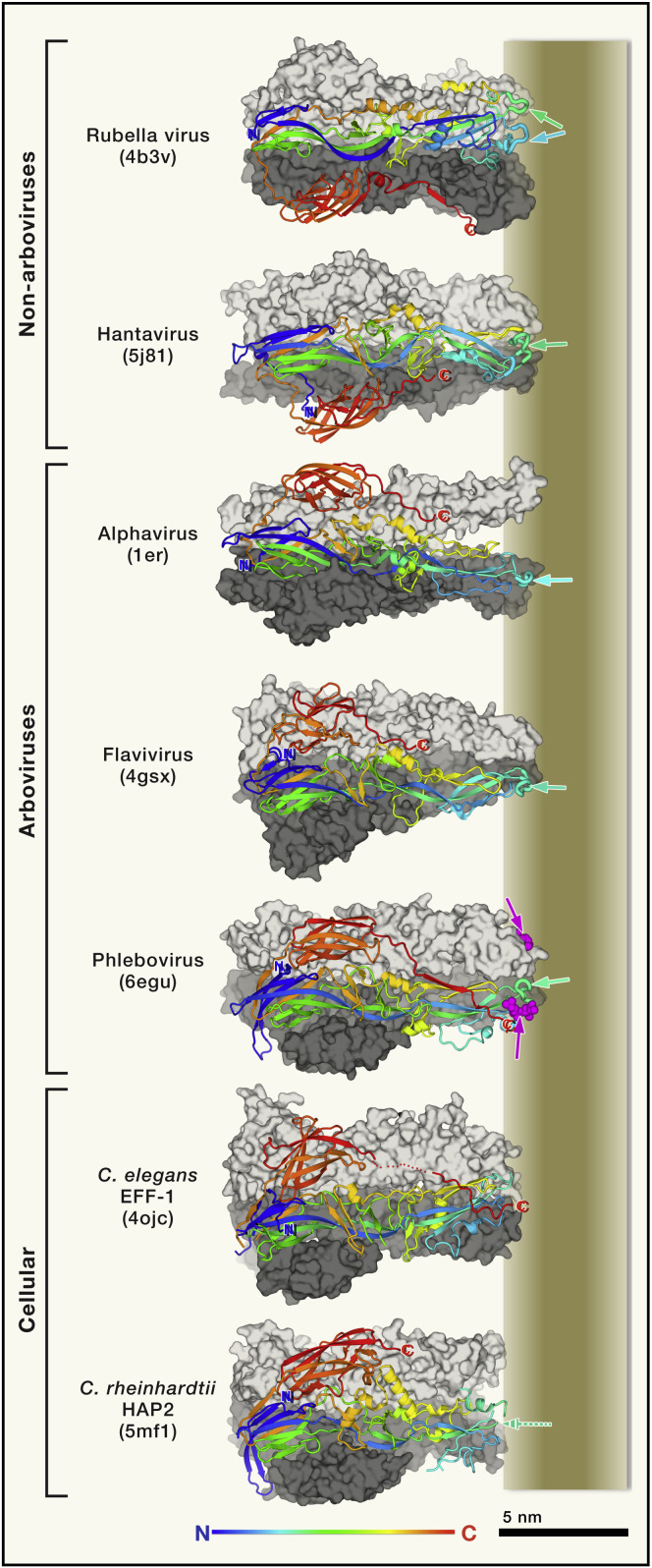
Figure 4.
Postfusion Class II Fusion Proteins
The viruses are listed in the left with the corresponding PDB accession code indicated in parenthesis. Each postfusion trimer is displayed with two protomers in surface representation colored light and dark gray, and with the third protomer (in the foreground) shown in ribbons ramp colored from N to C terminus (compare to the pre-fusion form in Figure 3C), highlighting the common fold of the polyeptide chains. The membrane is represented on the right, roughly to scale, and the N and C-terminal ends of the crystallized ectodomain are labeled in blue and red, respectively. Domain III (red) swaps protomers in the non-arboviruses packing against the bottom one (dark gray) instead of the top one (light gray) in the others. The fusion loop in the front protomer is shown as thicker tubes and marked by green/cyan arrows in the schematized fused membrane represented to the right (note that the Rubella virus protomer has two fusion loops). In the case of the phlebovirus protein, a glycerophospholipid head group present in the structure is shown in magenta spheres and marked by magenta arrows. In the cellular proteins, C. elegans EFF-1 does not have a non-polar fusion loop, but the three C-terminal ends converge toward the 3-fold axis at the level of the aliphatic moiety of the membrane (marked by a red “C”, the C-terminal end of the ectodomain). In C. rheinhadtii HAP2, the fusion loops were disordered in the crystal structure (indicated by the broken arrow).
The Class II Viruses
This group encompasses several important human pathogenic viruses, the majority of them arthropod-borne (called “arboviruses”). They include flaviviruses such as the mosquito-borne hemorrhagic fever agents dengue and yellow fever viruses, the encephalitic mosquito-borne West Nile and Japanese encephalitis viruses, as well as tick-borne encephalitis viruses. They also include the neurotropic and teratogenic Zika virus. Alphaviruses (Togaviridae family) such as Chikungunya virus, which causes severe arthralgia with joint pain and fever, and the equine encephalitic viruses that spread zoonotically to humans, such as the Venezuelan equine encephalitis virus, are also mosquito-borne class II viruses. Viruses in several families of the recently defined Bunyavirales Order (previously known as Bunyaviridae family), and generically called bunyaviruses, also belong to the class II group. The majority of the bunyaviruses are zoonotic arboviruses, the most studied being the mosquito transmitted Rift Valley fever virus (RVFV), which causes hemorrhagic fever and belongs to the Phlebovirus genus (Phenuiviridae family). The Orthobunyavirus genus (Peribunyaviridae family) also includes zoonotic viruses distributed worldwide, the most known being California encephalitis and La Crosse viruses, transmitted by mosquitoes in North America. The Nairoviridae family includes the highly pathogenic tick-borne Crimean-Congo hemorrhagic fever virus, classified as a biosafety level 4 pathogen. In addition to the arboviruses, the class II group includes hantaviruses (bunyaviruses of the Hantaviridae family), zoonotic viruses carried by chronically infected rodents and causing severe renal or respiratory syndromes (depending on the virus) in humans.
Another example of class II virus is the teratogenic rubella virus (Rubivirus genus in the Togaviridae family), an airborne, highly contagious strictly human virus. Finally, structural and functional homologs of the class II fusion proteins have been found in eukaryotic organisms. The somatic cell-cell fusion protein EFF-1 from C. elegans (Pérez-Vargas et al., 2014) makes syncytia necessary for skin formation during embryogenesis, and glycoprotein HAP2 is responsible for gamete fusion (or sperm/egg fusion) found in almost all the main branches of eukaryotes (Fedry et al., 2017).
Organization of the Class II Genes
Viral class II proteins are expressed within a polyprotein precursor, which contains at least two proteins (hantaviruses), and often more as in the case of the flaviviruses where all 10 virus-encoded proteins are derived from a single precursor polyprotein. The fusion protein folds as a heterodimer with a companion glycoprotein present upstream in the precursor. For ease of description, we term the companion glycoprotein UGP, for “upstream glycoprotein,” and the fusion protein as DGP (“downstream glycoprotein”). UGP and DGP form a heterodimer in the ER of the infected cell, most likely co-translationally as the UGP is already present in the ER when the DGP emerges from the translocon. Both UGP and DGP are anchored in the viral membrane by their C-terminal ends. Their topology is similar to the type I single transmembrane proteins, except that in some viruses they have two C-terminal trans-membrane segments. They are in general released by signal peptidase cleavage from the polyprotein precursor (Elliott and Schmaljohn, 2013, Kuhn, 2013, Lindenbach et al., 2013), although for some class II viruses other enzymes are also involved, with the most complex processing being that of the nairovirus mature glycoproteins (Altamura et al., 2007)). The UGP/DGP heterodimers interact laterally on the cell membrane to make a large assembly, in many cases with icosahedral symmetry. During morphogenesis in the infected cell, lateral interactions between glycoprotein spikes generate strong curvature in the underlying lipid bilayer, driving particle budding across the cell membrane (Vaney and Rey, 2011). They are thus reminiscent of the COP proteins involved in formation of intracellular vesicles, albeit with reversed topology. The main difference from class I viruses, in which the prefusion trimer assembles independently of the other trimers at the virus surface and its structure can be studied in isolation, is that in class II proteins it is the whole assembly that is important to understand in interaction with antibodies.
Particle Morphogenesis
Depending on the virus, class II virions assemble and bud internally in the infected cell into the ER lumen (flaviviruses) or into the Golgi apparatus, (bunyaviruses, rubella virus [Elliott and Schmaljohn, 2013, Kuhn, 2013]), and are then transported via the secretory pathway of the cell to reach the external environment. They can also assemble at the cell surface and bud directly across the plasma membrane into the external milieu (alphaviruses, [Kuhn, 2013]). Class II virions undergo a maturation process that primes the viral particle for acid activation for fusion within an endosome of a target cell. In the case of rubella virus and of the bunyaviruses, this maturation has been observed by electron microscopy with clear morphological differences between immature and mature particles (Mangala Prasad et al., 2017, Salanueva et al., 2003), but the actual maturation process is not understood. In the case of flaviviruses and alphaviruses, maturation is proteolytic, via the trans-Golgi network (TGN)-resident furin protease of the infected cell, which cleaves the UGP such that the N-terminal portion is later shed, priming the particle to react upon exposure to acidic pH in the endosome of the target cell to induce fusion (Kuhn, 2013, Lindenbach et al., 2013). This maturation cleavage is analogous to that taking place in class I proteins, priming the fusion machinery by cleavage to form the NSU and CSU, except that in class II viruses it takes place on the companion glycoprotein.
Alphaviruses
The organization of alphavirus particles has been reviewed earlier (Vaney et al., 2013). The UGP and DGP are called p62 and E1, respectively. The heterodimer trimerizes to make (p62/E1)3 spikes, which are transported to the cell surface where they accumulate for subsequent assembly of a glycoprotein shell and budding. Proteolytic maturation in the TGN converts p62 into the mature proteins E3 (N-terminal fragment, a small protein that remains peripherally attached to the spike) and E2. E3 is then lost in the neutral pH of the extracellular environment, priming the spike complex to react to subsequent exposure to acidic pH in the endosome of a target cell. The surface glycoprotein shell of infectious virions contain 80 (E2/E1)3 spikes interacting laterally to make a T = 4 icosahedral lattice encasing the viral membrane (Kuhn, 2013) (Figure 3A, top-middle panel). High-resolution structural studies by X-ray crystallography have provided the structure of the ectodomains of the p62/E1 and E3/E2/E1 complexes of Chikungunya virus at neutral pH (Voss et al., 2010), and the E2/E1 complex of Sindbis virus at acidic pH, in which E1 and E2 remain partially bound and in which the fusion loop of E1 is exposed (Li et al., 2010). There are also sub-nanometer resolution structures by single particle cryo-EM of several alphaviruses, and the combination with the X-ray structures has provided a detailed model of the interactions that form the glycoprotein shell (Sun et al., 2013, Tang et al., 2011, Zhang et al., 2011), showing that the E1 DGP forms the icosahedral scaffold, with E2 capping E1 at the top of each of the spikes and being ideally positioned to interact with cellular receptors. Most of the alphavirus neutralizing antibodies target the exposed E2 cap (Fox et al., 2015, Long et al., 2015, Sun et al., 2013), but some also target E1.
Flaviviruses
The structural biology of flaviviruses has been reviewed extensively (see Hasan et al., 2018 for a recent review). In contrast to alphaviruses, the flavivirus particle assembles and buds into the ER lumen of the infected cell in the form of an immature particle with 60 trimeric spikes ((prM/E)3) containing trimers of the precursor-membrane protein (prM, the UGP) and the envelope protein (E, the membrane fusion DGP) (Figure 3A, top-left panel). These immature particles are then transported across the secretory pathway, trafficking from the neutral pH environment in the ER into the acidic milieu of the Golgi apparatus. Exposure to this mildly acidic environment triggers a radical conformational change of the particle, such that the 60 trimeric spikes dissociate and reassociate as 90 (prM/E)2 dimers, with prM bound at the E dimer interface (Li et al., 2008b, Yu et al., 2008) (Figure 3A, middle left). In this conformation, a furin specific site in prM becomes exposed, giving rise to proteolytic maturation by furin to yield protein M (the C-terminal, viral-membrane anchor portion) and the peripheral “pr” portion. Mature virions are then released into the extracellular milieu where pr is shed as its affinity for E at neutral pH is weak, thus priming the particle to become infectious upon subsequent exposure to acid pH in the endosome of a target cell. The mature flavivirus particle has a smooth aspect, featuring 90 head-to-tail E dimers organized in a herringbone pattern (Figure 3A), with the fusion loop buried at the E dimer interface (Figure 3C) (Kuhn et al., 2002, Zhang et al., 2013).
Bunyaviruses
The bunyavirus genome is composed of three segments of negative–sense single stranded RNA. The medium segment (M) encodes a polyprotein precursor containing glycoprotein GN in the amino-terminal side (UGP) and GC (the membrane fusion DGP) in the C-terminal side. The first X-ray structure of a bunyavirus fusion protein corresponded to RVFV GC in the prefusion form, which showed that it is a class II protein (Dessau and Modis, 2013). The structure of postfusion GC was later determined for several phleboviruses (Guardado-Calvo et al., 2017, Halldorsson et al., 2016, Zhu et al., 2017. In particular, the structure of RVFV GC in the postfusion form revealed the path of the stem, which was unresolved in the previous structures of arbovirus DGPs, extending from domain III to the fusion loop. The end of the stem interacts with the fusion loop and completes an interaction pocket specific for a glycerophospholipid head group (Guardado-Calvo et al., 2017) at the site of membrane insertion (magenta spheres and arrows in Figure 4). This pocket appears to be also present in the flavivirus and alphavirus DGPs, although these proteins have not yet been visualized in complex with a lipid head group. Of note, a key residue in direct interaction with the lipid head group in RVFV GC occupies the same location as residue 226 in the fusion protein E1 of Chikugunya virus. The A226V mutation in E1 affects the lipid dependence for fusion and coincided with a change of mosquito vector transmitting the virus, in turn causing an explosive outbreak of Chikungunya disease in Reunion Island (Indian Ocean) in 2005 and 2006 (Tsetsarkin et al., 2007), although the mechanistic explanation for this effect remained elusive (Tsetsarkin et al., 2011). The current view is that the lipid composition of the endosomal membranes may vary for different mosquito species, and that minor differences in affinity of the DGP for the lipid head groups may have a big impact in the overall outcome of the infection and the transmission capacity of the infected mosquito.
Very recently, the X-ray structure of a phlebovirus GN was also reported, crystallized on its own (Halldorsson et al., 2018) and in complex with a human neutralizing antibody, thereby defining an important epitope exposed at the virion surface (Wu et al., 2017). Moreover, a cryo-EM reconstruction of the T = 12 icosahedral RVFV particle to about 13Å resolution was reported recently (Halldorsson et al., 2018). These authors performed a localized 3D reconstruction centered on individual hexamers and pentamers of the T = 12 lattice to extend the resolution locally to about 8Å. The resulting cryo-EM map made possible a clear docking of the available atomic models for GN and GC, thereby revealing the organization of the GC shell on the virion, which provides particle cohesion by forming the contacts between adjacent pentamers and hexamers. It also revealed the capping of the fusion loop by GN at the hexamer and pentamer apices (Figure 3), although the detailed interactions between GN and GC could not be visualized at this resolution. In addition, by using cryo-ET, this study also captured the interaction of virions with liposomes at acid pH (mimicking the pH in the endosomes) at low resolution (around 20 Å). This early stage of the fusogenic conformational change featured a subset of the DGPs on the virion (the GC proteins located around the 5-fold axes of the T = 12 icosahedral particles) which had lost the GN cap and were in an extended intermediate conformation bridging the two membranes, while those at around quasi-6-fold axes of the particle still maintained the GN cap (Halldorsson et al., 2018). This study therefore reported the first structural visualization of a class II fusion protein in the elusive extended intermediate conformation (schematized in the middle panel of Figure 1A for the class I proteins) occurring during the process of membrane fusion.
Pleomorphic Class II Viruses
Structural studies are also available for hantavirus Gc in both pre-and postfusion forms (Guardado-Calvo et al., 2016, Willensky et al., 2016). In addition, the X-ray structure of hantavirus UGP (glycoprotein GN) was used to interpret a 3D reconstruction obtained by cryo-ET and subtomogram averaging to about 14Å resolution, as the particles are pleomorphic (Li et al., 2016a), showing that the spikes have a square outline with pseudo quasi-4-fold symmetry incompatible with icosahedral assembly. As for RVFV, these studies showed that the hantavirus DGP is responsible for the inter-spike contacts on the virion, while the UGPs cap the fusion loop at the spike apices.
Finally, the rubella virus particles are also pleomorphic but display locally ordered glycoprotein arrays, as visualized by cryo-ET at 11Å resolution (Mangala Prasad et al., 2017). The glycoprotein spikes are composed of heterodimers of glycoproteins E2 and E1 (the UGP and DGP, respectively), and the cryo-ET 3D reconstruction was interpreted using the available X-ray structure of the postfusion form of the rubella virus DGP (DuBois et al., 2013) (Figure 4), showing that the E1 spikes form rows surrounding the particles in a helical fashion. It also showed that the ribonucleoprotein complex of the capsid protein with the genomic RNA forms an array underneath the viral membrane in which the nucleocapsid subunits are directly beneath each spike, probably interacting with the E2 cytosolic tail.
Emerging Concepts
Overall, the picture emerging from the structural studies of class II proteins provides interesting parallels with the class I proteins. UGP capping the DGP, and the interactions with the target cell resulting in the release of the UGP cap in order to project the fusion loop of the DGP into the cellular membrane, function analogously to the functions of the NSU and CSU in the class I proteins. An exception is seen with the flaviviruses, however, where the UGP only partially caps the DGP in the immature form, and the fusion loop then rearranges to become buried the DGP dimer interface in mature particles. This particular organization of the particle has important implications in flavivirus biology and for vaccine design, as discussed below.
Class III Fusion Proteins
Class III fusion proteins (Baquero et al., 2015) are trimeric in both their pre and postfusion conformations, although the presence of monomers has also been detected at the virus surface in the case of the Vesicular Stomatitis virus, the class III prototype (Libersou et al., 2010). Class III proteins appear as a combination of classes I and II, with a central coiled-coil in the postfusion form, which bears at its N-terminal end a β sheet-rich fusion domain that is reminiscent of domain II in class II fusion proteins. However, the connectivity of the secondary structure elements in the respective domains are unrelated. Another difference is that class III proteins do not undergo activating maturation cleavage like the one occurring in class I proteins. But they behave as class I proteins in the sense that they do not make a glycoprotein assembly encasing the viral membrane as class II proteins. Class III proteins were originally identified in the rhabdovirus vesicular stomatitis virus (VSV) glycoprotein G (Roche et al., 2006) and in the herpes simplex virus glycoprotein B (gB) (Heldwein et al., 2006) and were later also identified in baculoviruses (Kadlec et al., 2008). Very recently, the structure of class III fusion protein from two thogotoviruses were reported (Peng et al., 2017). Thogotoviruses are a group of arboviruses forming a genus in the Orthomyxoviridae family that also includes the influenza virus genera, which can cause serious disease when infecting humans. Except for VSV G, which was reported in both in prefusion and postfusion forms (Roche et al., 2007), the other class III fusion proteins were crystallized in their postfusion conformation only. The envelope glycoprotein of rabies virus, the type species in the Rhabdoviridae family, is also expected have a class III fold, and structural studies of its prefusion form would be important to understand the way antibodies neutralize this important human pathogen. Similarly, the surface glycoprotein of members of the Bornaviridae family has been suggested to have a class III fusion fold, although this has not been experimentally confirmed. As these viruses have been associated with fatal cases of zoonotic encephalitis (Hoffmann et al., 2015), it would also important to undertake structural studies on members of this family.
Viral Fusion Proteins outside the Three Characterized Structural Classes
The family Flaviviridae has three other genera in addition to the Flavivirus genus (the Pestivirus, Hepacivirus, and Pegivirus genera). Because the genomic organization is the same as that of the flaviviruses, displaying two envelope glycoproteins in tandem (E1 and E2) within the polyprotein precursor, it was initially thought that they would also belong to class II, and would correspond to the UGP and DGP, respectively, and that therefore E2 would be the fusion protein. The structure of E2 from representatives of the Pestivirus genus (El Omari et al., 2013, Li et al., 2013) and also the structures of a core fragment of glycoprotein E2 from the hepatitis C virus (HCV) in the Hepacivirus genus (Khan et al., 2014, Kong et al., 2013) showed that they do not have a class II fusion protein fold, and it is not clear how the fusion machinery of these viruses work. Clarification of this issue awaits the structures of the E1/E2 heterodimers of these viruses, and also a clear cryo-EM 3D reconstruction of the virus particles.
Another virus for which the fusion protein does not appear to belong to any of the classes described above is the hepatitis B virus (HBV). In this case, the envelope gene appears to have emerged de novo, overlaid on a shifted reading frame on the reverse transcriptase gene of an evolutionary precursor non-enveloped virus (Lauber et al., 2017). It therefore cannot have the same origin as any of the proteins of the structurally characterized classes of homologous fusion proteins. Indeed, HBV features an envelope glycoprotein with three isoforms (large, medium and small protein), sharing the same C-terminal trans-membrane domain and displaying different N-terminal extensions. The common C-terminal domain crosses the membrane 3 or 4 times—depending on the maturation state of the particle (Seitz et al., 2016)—and has only a relatively small domain facing the exterior of the virion, in an arrangement that does not match any of the three classes described above.
Finally, the Poxviridae, a large family of enveloped DNA viruses (with vaccinia virus as type species), have a complex of eleven proteins that is embedded in the membrane of the mature virion and is activated for membrane fusion by the acidic pH of the endosomes (Moss, 2016). This complex also appears to fall outside the three structurally characterized classes of membrane fusion proteins, and is also awaiting structural studies to shed light on the poxvirus fusion machinery.
Common Features and Implications for Vaccine Design
Taken together, and in spite of the vast variation among viruses, the structures reviewed here reveal a number of common features to be taken into account for the design of efficient prophylactic vaccines against enveloped viruses in general. One aspect highlighted by the studies on HIV Env is the importance of conformational dynamics of the glycoprotein, presenting open and closed forms in equilibrium (Munro et al., 2014). This equilibrium between different forms is a functional requirement as the protein must react to the interactions with the receptors by changing its conformation to induce membrane fusion to infect a cell. This dynamism is valid not only for HIV, but is common to all enveloped viruses. Specific to HIV-1 and to HCV is an extraordinary strain diversity. In addition, cell-culture adapted HIV-1 strains are easily neutralizable (termed “tier 1 viruses”) in contrast to the difficult to neutralize “tier 2” circulating clinical strains (Seaman et al., 2010). The challenge is to develop bNAbs targeting the tier 2 strains. Several structure-based studies have shown that it is possible to engineer the Env trimer to obtain a stable closed form, which although non-functional, maintains the overall antigenicity of authentic Env trimers on infectious virions. A whole body of research indicates that it is this closed form that is recognized by the known bNAbs, whereas the epitopes exposed in the open form are not.
Undesirable Antibodies Directed against the V3 Loop of HIV Env
The hypervariable V3 loop of gp120, which is concealed in a closed trimer, is immunodominant and elicits antibodies that can only neutralize autologous tier 1 viruses. Several very recent studies have revealed ways of eliminating elicitation of such antibodies, which distract the immune system from the production of the relevant bNAbs, by further stabilization of the recombinant Env trimer ectodomain by structure guided mutagenesis (de Taeye et al., 2015, Joyce et al., 2017, Kulp et al., 2017, Torrents de la Peña et al., 2017). Furthermore, recent experiments immunizing rabbits with a SOSIP version of a transmission founder virus succeeded in eliciting antibodies neutralizing heterologous tier 2 viruses, but only with the engineered version that stabilized the closed form and did not expose the V3 loop (Saunders et al., 2017). These results highlight the importance of stabilization of the prefusion form in the closed conformation, limiting as much as possible its dynamic conformational behavior. These approaches remain very challenging, however, as all the identified the bNAbs exhibit very high somatic hypermutation (SHM) and recombinant antibodies with the inferred germline sequence do not bind the virus, except in the rare cases where the Env sequence of the founder virus is known. In this HIV-1 specific context, the available structures have also allowed the engineering of germline-targeting SOSIP trimer variants (Medina-Ramírez et al., 2017a) that could be used to test immunization of knock-in mice expressing antibodies with the inferred bNAb unmutated ancestor sequence. These experiments involved priming and boosting strategies conceived to shepherd the B cell response toward making the corresponding bNAb. Sequential boosting with stabilized SOSIP trimers having sequences closer and closer to wild-type has indeed led to induction of neutralizing antibodies in these mice with high levels of SHM that recapitulated many of the key mutations in the targeted bNAb, providing the proof of concept that this approach is viable, and can potentially be tried in future vaccine approaches (Escolano et al., 2016, Steichen et al., 2016).
Other Enveloped Viruses in Search of a Vaccine
The same principles outlined for HIV are being used in the development of an RSV vaccine where the prefusion form is also highly unstable. The difference is that this virus does not exhibit very high variability, and is therefore more tractable. Similar approaches are likely to be essential for developing efficient immunogens to use in a subunit vaccine against other enveloped viruses from the other classes. Important examples are the human pathogenic Epstein-Barr virus and the cytomegalovirus, belonging to the Herpesviridae family, for which a structure of the prefusion form of their class III fusion protein would be extremely valuable as a target for vaccine design, and where specific stabilization of the pre-fusion form is an absolute requirement.
The Dengue Viruses
As discussed above, for class II viruses the situation is different as the prefusion form of the fusion protein is involved in formation of a glycoprotein shell surrounding the viral membrane. Parallels still exist, as indicated by the flavivirus data, and in particular for the four dengue virus serotypes, where heterotypic secondary infection is the greatest risk factor for severe dengue disease (Katzelnick et al., 2017). The antibodies elicited during a primary infection normally protect from subsequent autologous infection; however, these antibodies also bind heterologous virus without neutralizing it. The resulting opsonized particles are efficiently internalized via Fcγ receptors into macrophages and other cells of the immune system, where the virus particle, which is not neutralized by the bound antibody, fuses with the endosomal membranes and infects the cell in an antibody-dependent enhancement (ADE) of the infection. As reviewed recently (Rey et al., 2018), the mechanism behind ADE appears to be related to particle heterogeneity introduced by partial maturation of the UGP as well as conformational dynamics of the fusion protein on the mature virions (illustrated in Figure 3B).
As illustrated in Figure 3A, flavivirus particles are different from the other class II virions because they lack the UGP cap in the mature form. Furthermore, although the UGP does cap the underlying DGP layer in immature flavivirus particles such that the fusion loop cannot insert into the endosomal membrane, the available structures show that the fusion loop is not buried, but is instead exposed on the sides of the (prM/E)3 spikes (reviewed in (Rey et al., 2018)) such that it is recognized by antibodies and also by B cell receptors. Because furin maturation is often incomplete, partially mature particles (Figure 3B) are released by infected cells and circulate in the host, stimulating B cells to produce antibodies against the laterally exposed fusion loop, and also against the highly immunogenic prM. Both of these classes of antibodies have been shown to be cross-reactive, poorly neutralizing and ADE prone. These antibodies can bind mature heterologous particles only when the conserved fusion loop is exposed, either by dynamic “breathing” of the E dimer, or in immature patches on the virion. The number of such antibodies coating the particle depends of the breathing kinetics of the E dimers, and is often insufficient to reach the neutralization threshold (Burton et al., 2001). In the absence of other antibodies targeting additional regions of the particle (which is the case during an heterologous infection, since the other exposed regions of the E dimer are variable across serotypes), the non-neutralized virion/antibody complex is internalized via Fcγ receptors, enhancing the overall infection by accessing to immune cells that are normally not infectable.
Stabilizing the Dengue Virus Prefusion E Dimer
Structural studies have shown that highly neutralizing antibodies against the dengue viruses target readily accessible sites at the virus surface (reviewed in Lok, 2016) and that the poorly neutralizing antibodies target cryptic epitopes. Furthermore, although laboratory adapted strains appear to be highly dynamic, readily exposing the fusion loop (and therefore being easily neutralized by the fusion loop antibodies, similar to the autologous tier 1 HIV-1 strains by the V3 loop antibodies), clinical strains appear to be less dynamic, and only expose the fusion loop at few locations around the particle, such that the stoichiometry required for neutralization is never reached. It is important therefore to design immunogens that avoid eliciting antibodies against the fusion loop and to prM. In addition, a vulnerability site has been identified at the flavivirus surface, precisely in the region of the fusion loop, which is recognized in the context of the E dimer and not via the side chains that are buried at the dimer interface (Dejnirattisai et al., 2015). Antibodies targeting this site, termed “E dimer epitope” (EDE) are capable of potently neutralizing the four dengue viruses as well as Zika virus (Barba-Spaeth et al., 2016). The identification of the EDE site has opened the possibility of designing immunogens that are able to elicit antibodies broadly neutralizing all four dengue viruses as well as Zika virus. Designing such immunogens also requires avoiding exposure of cryptic epitopes, in particular the fusion loop. A step in this direction has been made recently, with cysteine residues introduced at the dimer interface (marked with stars in Figure 3C) that covalently cross-link the E dimer such that the fusion loop is not exposed (Rouvinski et al., 2017). Binding studies have shown that the double-disulfide cross-linked E dimers are efficiently recognized by the EDE bNAbs but not by antibodies targeting the fusion loop. It remains to test the type of immune response induced by these new immunogens, although ideally, the best approach would be to immunize with virion-like particles stabilized in their closed form, exposing exclusively locked E dimers incapable of breathing (such as the one illustrated in Figure 3A), analogous to the engineered HIV Env closed trimers.
Concluding Remarks
In summary, despite clear differences in their fusion proteins, the viruses within the individual classes discussed above have common properties. Although the virions of classes I and II have unrelated architectures, they display common features and sites of vulnerability that can be targeted for vaccine design. In both cases, stabilization of the prefusion form appears to be key to elicit only the type of antibodies required for protection. A similar approach is therefore likely to be valid for any other enveloped virus for which protective vaccines are not currently available.
A Broader Perspective
A further important insight comes from influenza virus. The structures of HA in complex with broadly neutralizing antibodies targeting the conserved stem region (Ekiert et al., 2009) (Figure 2), which neutralize by blocking the fusogenic conformational change, have shown us how to use the complementarity determining region of the bound antibodies to design small proteins targeting the same site (Fleishman et al., 2011). Further studies identified small peptides targeting this site, with the same potent inhibitory effect (Kadam et al., 2017). These results suggest that it may be possible to identify non-peptidic, orally bioavailable antiviral compounds using the same strategy to treat flu infections. These studies thus pave the way for deriving specific and potent drugs to treat infection by other pathogenic viruses. Structural biology of the viral membrane fusion proteins and their complexes with neutralizing antibodies is thus an exceptionally powerful approach to identify vulnerability sites and to extract the necessary information required to efficiently combat the emerging viral diseases threatening our planet.
Acknowledgments
F.A.R. was funded by Institut Pasteur, the CNRS, the ERC Advanced grant “Celcelfus” (Grant UE 340371), the ANR-13-ISV8-0002-01 grant “Flavistem,” the European Infect-ERA “HantaHunt” Project (Grant ANR 15 IFEC), from the “Integrative Biology of Emerging Infectious Diseases” Labex (Laboratoire d’Excellence) grant ANR-10-LABX-62-IBEID. Part of the research was performed as part of the Zoonoses Anticipation and Preparedness Initiative (ZAPI project; IMI Grant Agreement n°115760). S.-M.L. acknowledges support by the Singapore Ministry of Education Tier 3 grant (MOE2012-T3-1-008), the National Research Foundation Investigatorship award (NRF-NRFI2016-01) and the National Research Foundation competitive Research Project grant (NRF2015NRF-CRP001-063). Given the focus of this Review, we have not attempted to be comprehensive and only aselection of recent primary literature has been cited.
Contributor Information
Felix A. Rey, Email: felix.rey@pasteur.fr.
Shee-Mei Lok, Email: sheemei.lok@duke-nus.edu.sg.
References
- Altamura L.A., Bertolotti-Ciarlet A., Teigler J., Paragas J., Schmaljohn C.S., Doms R.W. Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein. J. Virol. 2007;81:6632–6642. doi: 10.1128/JVI.02730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi R., Su C.Y., Liang C.H., Shivatare S.S., Briney B., Voss J.E., Nawazi S.K., Wu C.Y., Wong C.H., Burton D.R. Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development. Immunity. 2017;47:524–537 e523. doi: 10.1016/j.immuni.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero E., Albertini A.A., Gaudin Y. Recent mechanistic and structural insights on class III viral fusion glycoproteins. Curr. Opin. Struct. Biol. 2015;33:52–60. doi: 10.1016/j.sbi.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Barba-Spaeth G., Dejnirattisai W., Rouvinski A., Vaney M.C., Medits I., Sharma A., Simon-Lorière E., Sakuntabhai A., Cao-Lormeau V.M., Haouz A. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Binley J.M., Sanders R.W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D.J., Maddon P.J., Olson W.C., Moore J.P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S., Jardetzky T.S., Lamb R.A. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology. 2015;479-480:518–531. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri D.J., da Palma J.R., Kunz S., Pasquato A. Envelope glycoprotein of arenaviruses. Viruses. 2012;4:2162–2181. doi: 10.3390/v4102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Saphire E.O., Parren P.W. A model for neutralization of viruses based on antibody coating of the virion surface. Curr. Top. Microbiol. Immunol. 2001;260:109–143. doi: 10.1007/978-3-662-05783-4_7. [DOI] [PubMed] [Google Scholar]
- Chen L., Gorman J.J., McKimm-Breschkin J., Lawrence L.J., Tulloch P.A., Smith B.J., Colman P.M., Lawrence M.C. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., Jumnainsong A., Edwards C., Quyen N.T.H., Duangchinda T. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessau M., Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. USA. 2013;110:1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R.W. What Came First-the Virus or the Egg? Cell. 2017;168:755–757. doi: 10.1016/j.cell.2017.02.012. [DOI] [PubMed] [Google Scholar]
- DuBois R.M., Vaney M.C., Tortorici M.A., Kurdi R.A., Barba-Spaeth G., Krey T., Rey F.A. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Iourin O., Harlos K., Grimes J.M., Stuart D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R., Schmaljohn C. Sixth Edition. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. Bunyaviridae. [Google Scholar]
- Escolano A., Steichen J.M., Dosenovic P., Kulp D.W., Golijanin J., Sok D., Freund N.T., Gitlin A.D., Oliveira T., Araki T. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458 e1412. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedry J., Liu Y., Pehau-Arnaudet G., Pei J., Li W., Tortorici M.A., Traincard F., Meola A., Bricogne G., Grishin N.V. The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell. 2017;168:904–915 e910. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fleishman S.J., Whitehead T.A., Ekiert D.C., Dreyfus C., Corn J.E., Strauch E.M., Wilson I.A., Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Long F., Edeling M.A., Lin H., van Duijl-Richter M.K.S., Fong R.H., Kahle K.M., Smit J.M., Jin J., Simmons G. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell. 2015;163:1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado-Calvo P., Bignon E.A., Stettner E., Jeffers S.A., Pérez-Vargas J., Pehau-Arnaudet G., Tortorici M.A., Jestin J.L., England P., Tischler N.D., Rey F.A. Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc. PLoS Pathog. 2016;12:e1005813. doi: 10.1371/journal.ppat.1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado-Calvo P., Atkovska K., Jeffers S.A., Grau N., Backovic M., Pérez-Vargas J., de Boer S.M., Tortorici M.A., Pehau-Arnaudet G., Lepault J. A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion. Science. 2017;358:663–667. doi: 10.1126/science.aal2712. [DOI] [PubMed] [Google Scholar]
- Halldorsson S., Behrens A.J., Harlos K., Huiskonen J.T., Elliott R.M., Crispin M., Brennan B., Bowden T.A. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc. Natl. Acad. Sci. USA. 2016;113:7154–7159. doi: 10.1073/pnas.1603827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldorsson S., Li S., Li M., Harlos K., Bowden T.A., Huiskonen J.T. Shielding and activation of a viral membrane fusion protein. Nat. Commun. 2018;9:349. doi: 10.1038/s41467-017-02789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.S., Sevvana M., Kuhn R.J., Rossmann M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018;25:13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- Hastie K.M., Zandonatti M.A., Kleinfelter L.M., Heinrich M.L., Rowland M.M., Chandran K., Branco L.M., Robinson J.E., Garry R.F., Saphire E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E.E., Lou H., Bender F.C., Cohen G.H., Eisenberg R.J., Harrison S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Tappe D., Höper D., Herden C., Boldt A., Mawrin C., Niederstraßer O., Müller T., Jenckel M., van der Grinten E. A Variegated Squirrel Bornavirus Associated with Fatal Human Encephalitis. N. Engl. J. Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- Jae L.T., Brummelkamp T.R. Emerging intracellular receptors for hemorrhagic fever viruses. Trends Microbiol. 2015;23:392–400. doi: 10.1016/j.tim.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Joyce M.G., Georgiev I.S., Yang Y., Druz A., Geng H., Chuang G.Y., Kwon Y.D., Pancera M., Rawi R., Sastry M. Soluble Prefusion Closed DS-SOSIP.664-Env Trimers of Diverse HIV-1 Strains. Cell Rep. 2017;21:2992–3002. doi: 10.1016/j.celrep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- Julien J.P., Lee J.H., Cupo A., Murin C.D., Derking R., Hoffenberg S., Caulfield M.J., King C.R., Marozsan A.J., Klasse P.J. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. USA. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam R.U., Juraszek J., Brandenburg B., Buyck C., Schepens W.B.G., Kesteleyn B., Stoops B., Vreeken R.J., Vermond J., Goutier W. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J., Loureiro S., Abrescia N.G., Stuart D.I., Jones I.M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key T., Sarker M., de Antueno R., Rainey J.K., Duncan R. The p10 FAST protein fusion peptide functions as a cystine noose to induce cholesterol-dependent liposome fusion without liposome tubulation. Biochim. Biophys. Acta. 2015;1848:408–416. doi: 10.1016/j.bbamem.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Khan A.G., Whidby J., Miller M.T., Scarborough H., Zatorski A.V., Cygan A., Price A.A., Yost S.A., Bohannon C.D., Jacob J. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wasserman H., Amarasinghe G.K., Saphire E.O. Filovirus Structural Biology: The Molecules in the Machine. Curr. Top. Microbiol. Immunol. 2017;411:381–417. doi: 10.1007/82_2017_16. [DOI] [PubMed] [Google Scholar]
- Kong L., Giang E., Nieusma T., Kadam R.U., Cogburn K.E., Hua Y., Dai X., Stanfield R.L., Burton D.R., Ward A.B. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krarup A., Truan D., Furmanova-Hollenstein P., Bogaert L., Bouchier P., Bisschop I.J., Widjojoatmodjo M.N., Zahn R., Schuitemaker H., McLellan J.S., Langedijk J.P. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. Sixth Edition. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. Togaviridae. [Google Scholar]
- Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D.W., Steichen J.M., Pauthner M., Hu X., Schiffner T., Liguori A., Cottrell C.A., Havenar-Daughton C., Ozorowski G., Georgeson E. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais E., Huang X., Havenar-Daughton C., Murrell B., Price M.A., Wickramasinghe L., Ramos A., Bian C.B., Simek M., Allen S. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 2016;12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Seitz S., Mattei S., Suh A., Beck J., Herstein J., Börold J., Salzburger W., Kaderali L., Briggs J.A.G., Bartenschlager R. Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses. Cell Host Microbe. 2017;22:387–399.e6. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ozorowski G., Ward A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikin S., Parsegian V.A., Rau D.C., Rand R.P. Hydration forces. Annu. Rev. Phys. Chem. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- Li K., Zhang S., Kronqvist M., Wallin M., Ekström M., Derse D., Garoff H. Intersubunit disulfide isomerization controls membrane fusion of human T-cell leukemia virus Env. J. Virol. 2008;82:7135–7143. doi: 10.1128/JVI.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Lok S.M., Yu I.M., Zhang Y., Kuhn R.J., Chen J., Rossmann M.G. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Li L., Jose J., Xiang Y., Kuhn R.J., Rossmann M.G. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang J., Kanai R., Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. USA. 2013;110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rissanen I., Zeltina A., Hepojoki J., Raghwani J., Harlos K., Pybus O.G., Huiskonen J.T., Bowden T.A. A Molecular-Level Account of the Antigenic Hantaviral Surface. Cell Rep. 2016;16:278. doi: 10.1016/j.celrep.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sun Z., Pryce R., Parsy M.L., Fehling S.K., Schlie K., Siebert C.A., Garten W., Bowden T.A., Strecker T., Huiskonen J.T. Acidic pH-Induced Conformations and LAMP1 Binding of the Lassa Virus Glycoprotein Spike. PLoS Pathog. 2016;12:e1005418. doi: 10.1371/journal.ppat.1005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libersou S., Albertini A.A., Ouldali M., Maury V., Maheu C., Raux H., de Haas F., Roche S., Gaudin Y., Lepault J. Distinct structural rearrangements of the VSV glycoprotein drive membrane fusion. J. Cell Biol. 2010;191:199–210. doi: 10.1083/jcb.201006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B., Murray C., Thiel H., Rice C. Sixth Edition. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. Flaviviridae. [Google Scholar]
- Lok S.M. The Interplay of Dengue Virus Morphological Diversity and Human Antibodies. Trends Microbiol. 2016;24:284–293. doi: 10.1016/j.tim.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Long F., Fong R.H., Austin S.K., Chen Z., Klose T., Fokine A., Liu Y., Porta J., Sapparapu G., Akahata W. Cryo-EM structures elucidate neutralizing mechanisms of anti-chikungunya human monoclonal antibodies with therapeutic activity. Proc. Natl. Acad. Sci. USA. 2015;112:13898–13903. doi: 10.1073/pnas.1515558112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangala Prasad V., Klose T., Rossmann M.G. Assembly, maturation and three-dimensional helical structure of the teratogenic rubella virus. PLoS Pathog. 2017;13:e1006377. doi: 10.1371/journal.ppat.1006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Yang Y., Graham B.S., Kwong P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Leung S., Graepel K.W., Du X., Yang Y., Zhou T., Baxa U., Yasuda E., Beaumont T. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramírez M., Garces F., Escolano A., Skog P., de Taeye S.W., Del Moral-Sanchez I., McGuire A.T., Yasmeen A., Behrens A.J., Ozorowski G. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 2017;214:2573–2590. doi: 10.1084/jem.20161160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramírez M., Sanders R.W., Sattentau Q.J. Stabilized HIV-1 envelope glycoprotein trimers for vaccine use. Curr. Opin. HIV AIDS. 2017;12:241–249. doi: 10.1097/COH.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Membrane fusion during poxvirus entry. Semin. Cell Dev. Biol. 2016;60:89–96. doi: 10.1016/j.semcdb.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J.B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D.R., Koff W.C., Courter J.R., Smith A.B., 3rd, Kwong P.D. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwuta J.O., Chen M., Modjarrad K., Joyce M.G., Kanekiyo M., Kumar A., Yassine H.M., Moin S.M., Killikelly A.M., Chuang G.Y. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci. Transl. Med. 2015;7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaus J., Warner J.M., O’Shaughnessy B., Herrmann A. The pathway to membrane fusion through hemifusion. Curr. Top. Membr. 2011;68:1–32. doi: 10.1016/B978-0-12-385891-7.00001-5. [DOI] [PubMed] [Google Scholar]
- Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C.A., Torres J.L., Copps J., Stanfield R.L., Cupo A., Pugach P. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Changela A., Kwong P.D. How HIV-1 entry mechanism and broadly neutralizing antibodies guide structure-based vaccine design. Curr. Opin. HIV AIDS. 2017;12:229–240. doi: 10.1097/COH.0000000000000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., Zhang S., Cui Y., Shi Y., Gao G.F., Qi J. Structures of human-infectingThogotovirusfusogens support a common ancestor with insect baculovirus. Proc. Natl. Acad. Sci. USA. 2017;114:E8905–E8912. doi: 10.1073/pnas.1706125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vargas J., Krey T., Valansi C., Avinoam O., Haouz A., Jamin M., Raveh-Barak H., Podbilewicz B., Rey F.A. Structural basis of eukaryotic cell-cell fusion. Cell. 2014;157:407–419. doi: 10.1016/j.cell.2014.02.020. [DOI] [PubMed] [Google Scholar]
- Qiao H., Pelletier S.L., Hoffman L., Hacker J., Armstrong R.T., White J.M. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J. Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Stiasny K., Vaney M.C., Dellarole M., Heinz F.X. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 2018;19:206–224. doi: 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe R.P., Sanders R.W., Yasmeen A., Kim H.J., Lee J.H., Cupo A., Korzun J., Derking R., van Montfort T., Julien J.P. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl. Acad. Sci. USA. 2013;110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Rouvinski A., Dejnirattisai W., Guardado-Calvo P., Vaney M.C., Sharma A., Duquerroy S., Supasa P., Wongwiwat W., Haouz A., Barba-Spaeth G. Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat. Commun. 2017;8:15411. doi: 10.1038/ncomms15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva I.J., Novoa R.R., Cabezas P., López-Iglesias C., Carrascosa J.L., Elliott R.M., Risco C. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 2003;77:1368–1381. doi: 10.1128/JVI.77.2.1368-1381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Vesanen M., Schuelke N., Master A., Schiffner L., Kalyanaraman R., Paluch M., Berkhout B., Maddon P.J., Olson W.C. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Verkoczy L.K., Jiang C., Zhang J., Parks R., Chen H., Housman M., Bouton-Verville H., Shen X., Trama A.M. Vaccine Induction of Heterologous Tier 2 HIV-1 Neutralizing Antibodies in Animal Models. Cell Rep. 2017;21:3681–3690. doi: 10.1016/j.celrep.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz S., Iancu C., Volz T., Mier W., Dandri M., Urban S., Bartenschlager R. A Slow Maturation Process Renders Hepatitis B Virus Infectious. Cell Host Microbe. 2016;20:25–35. doi: 10.1016/j.chom.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Steichen J.M., Kulp D.W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R.L., McCoy L.E., Ozorowski G., Hu X., Kalyuzhniy O. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J., Corper A.L., Basler C.F., Taubenberger J.K., Palese P., Wilson I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones G.B., Thomas P.V., Chen M., Druz A., Joyce M.G., Kong W.P., Sastry M., Soto C., Yang Y., Zhang B. A Cysteine Zipper Stabilizes a Pre-Fusion F Glycoprotein Vaccine for Respiratory Syncytial Virus. PLoS ONE. 2015;10:e0128779. doi: 10.1371/journal.pone.0128779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Xiang Y., Akahata W., Holdaway H., Pal P., Zhang X., Diamond M.S., Nabel G.J., Rossmann M.G. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.A., Settembre E.C., Shaw C.A., Dey A.K., Rappuoli R., Mandl C.W., Dormitzer P.R., Carfi A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. USA. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Jose J., Chipman P., Zhang W., Kuhn R.J., Baker T.S. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J. Mol. Biol. 2011;414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Peña A., Julien J.P., de Taeye S.W., Garces F., Guttman M., Ozorowski G., Pritchard L.K., Behrens A.J., Go E.P., Burger J.A. Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Chen R., Leal G., Forrester N., Higgs S., Huang J., Weaver S.C. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc. Natl. Acad. Sci. USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney M.C., Rey F.A. Class II enveloped viruses. Cell. Microbiol. 2011;13:1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- Vaney M.C., Duquerroy S., Rey F.A. Alphavirus structure: activation for entry at the target cell surface. Curr. Opin. Virol. 2013;3:151–158. doi: 10.1016/j.coviro.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Voss J.E., Vaney M.C., Duquerroy S., Vonrhein C., Girard-Blanc C., Crublet E., Thompson A., Bricogne G., Rey F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.B., Wilson I.A. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev. 2017;275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of Enveloped Viruses in Endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willensky S., Bar-Rogovsky H., Bignon E.A., Tischler N.D., Modis Y., Dessau M. Crystal Structure of Glycoprotein C from a Hantavirus in the Post-fusion Conformation. PLoS Pathog. 2016;12:e1005948. doi: 10.1371/journal.ppat.1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhu Y., Gao F., Jiao Y., Oladejo B.O., Chai Y., Bi Y., Lu S., Dong M., Zhang C. Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. USA. 2017;114:E7564–E7573. doi: 10.1073/pnas.1705176114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Helenius A. Virus entry at a glance. J. Cell Sci. 2013;126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- Yang Z.N., Mueser T.C., Kaufman J., Stahl S.J., Wingfield P.T., Hyde C.C. The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution. J. Struct. Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- Yin H.S., Paterson R.G., Wen X., Lamb R.A., Jardetzky T.S. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.S., Wen X., Paterson R.G., Lamb R.A., Jardetzky T.S. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Zhang W., Holdaway H.A., Li L., Kostyuchenko V.A., Chipman P.R., Kuhn R.J., Rossmann M.G., Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Hryc C.F., Cong Y., Liu X., Jakana J., Gorchakov R., Baker M.L., Weaver S.C., Chiu W. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., Zhou Z.H. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wu Y., Chai Y., Qi J., Peng R., Feng W.H., Gao G.F. The Postfusion Structure of the Heartland Virus Gc Glycoprotein Supports Taxonomic Separation of the Bunyaviral Families Phenuiviridae and Hantaviridae. J. Virol. 2017;92:92. doi: 10.1128/JVI.01558-17. [DOI] [PMC free article] [PubMed] [Google Scholar]