Abstract
Autophagy is a degradative transport route conserved among all eukaryotic organisms. During starvation, cytoplasmic components are randomly sequestered into large double‐membrane vesicles called autophagosomes and delivered into the lysosome/vacuole where they are destroyed. Cells are able to modulate autophagy in response to their needs, and under certain circumstances, cargoes, such as aberrant protein aggregates, organelles, and bacteria can be selectively and exclusively incorporated into autophagosomes. As a result, this pathway plays an active role in many physiological processes, and it is induced in numerous pathological situations because of its ability to rapidly eliminate unwanted structures. Despite the advances in understanding the functions of autophagy and the identification of several factors, named Atg proteins that mediate it, the mechanism that leads to autophagosome formation is still a mystery. A major challenge in unveiling this process arises from the fact that the origin and the transport mode of the lipids employed to compose these structures is unknown. This compendium will review and analyze the current data about the possible membrane source(s) with a particular emphasis on the yeast Saccharomyces cerevisiae, the leading model organism for the study of autophagosome biogenesis, and on mammalian cells. The information acquired investigating the pathogens that subvert autophagy in order to replicate in the host cells will also be discussed because it could provide important hints for solving this mystery.
I. Introduction
In eukaryotic cells, the principal locations where protein catabolism occurs are the proteasome and the lysosome. The proteasome mostly recognizes and degrades cytosolic factors that have been specifically marked with polyubiquitin chains (Roos‐Mattjus and Sistonen, 2004). The lysosome in contrast, requires active transport in order for the different substrates destined for elimination to reach its interior where the proteases are located. Four different pathways can deliver intracellular proteins into the lysosome lumen: endosomal transport routes, chaperone‐mediated autophagy (CMA), microautophagy, and macroautophagy, the latter generally referred to as autophagy (Dunn 2005, Katzmann 2002, Klionsky 2004, Majeski 2004). The endosomal transport routes and CMA are mostly devoted to the transport of polypeptides, whereas microautophagy and autophagy deliver other cellular constituents because these pathways are the only ones able to internalize entire organelles and bacteria. Eukaryotes, in particular fungi, can use microautophagy to eliminate peroxisomes and is the only cellular function that has indisputably been assigned to this pathway (Dunn et al., 2005). Autophagy, on the other hand, can deliver various cargoes to the lysosome interior and has multiple physiological roles.
A. Molecular Mechanism for Autophagy
The hallmark of this catabolic pathway is the sequestration of cargoes by large cytosolic double‐membrane vesicles called autophagosomes (Reggiori and Klionsky, 2005). The autophagosomes successively dock and fuse with mammalian lysosomes or the yeast and plant vacuoles releasing the inner vesicles into the lumen of these organelles (Reggiori and Klionsky, 2005). The biogenesis and consumption of these structures can be divided into six discrete steps: induction, expansion, vesicle completion, docking, fusion, and breakdown (Fig. 1 ).
Figure 1.
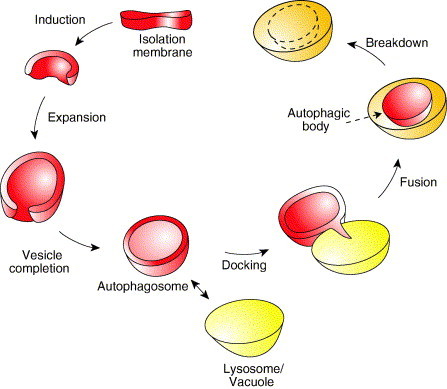
Conceptual model for autophagy. The basic mechanism of autophagy is the sequestration of the cargo material (bulk cytoplasm, protein aggregates, organelles, or pathogens) by a cytosolic double‐membrane vesicle named an autophagosome. Extracellular stimuli or the recognition of a specific intracellular cargo induce the expansion of the isolation membrane. Upon vesicle completion, the autophagosome docks with the lysosome/vacuole and successively fuses with it. In this way the inner vesicle is liberated inside the vacuole where it is finally consumed together with the cargo by resident hydrolases. This schematic represents nonspecific autophagy and does not show specific types of autophagy including the Cvt pathway.
1. Induction
Autophagosomes are generated by the elongation of a small template membrane, termed the isolation membrane or phagophore (Fengsrud 2004, Mizushima 2001, Noda 2002, Reggiori 2005). There are several of these structures per cell but it still remains unknown where they are derived from. The surface of this small compartment is decorated with Atg5 and Atg16, and its formation requires phosphatidylinositol (PtdIns)‐3‐kinase activity (Mizushima 2001, Mizushima 2003). There are two ways of triggering the expansion of the isolation membrane, and they differ depending if the process of autophagy is selective or nonselective (Section I.B) (Reggiori and Klionsky, 2005). When this pathway is selective, the binding to the isolation membrane of the cargo that has to be specifically eliminated (or in the case of resident hydrolases, activated) leads to the expansion of this structure (Ogawa 2005, Shintani 2004b). In contrast to selective autophagy, which is induced by intracellular components, the nonselective process is governed by extracellular stimuli such as nutrients or cytokines (Gutierrez 2004, Lum 2005, Shintani 2004a). In both cases, covalent conjugation of the ubiquitin‐like Atg12–Atg5 seems to be the step that initiates the expansion of the isolation membrane (Mizushima et al., 2001).
2. Expansion
The expansion of the isolation membrane is basically the simultaneous elongation and nucleation of this little cisterna (Fig. 1). It is not known how the Atg12–Atg5 complex recruits additional membranes, but the crescent autophagosome acquires more Atg12–Atg5 and Atg16 along with a second ubiquitin‐like molecule, Atg8/LC3, that is unconventionally linked to phosphatidylethanolamine (PE), and probably the rest of the Atg proteins (Mizushima 2001, Mizushima 2003). Two expansion mechanisms are possible, one that relies on delivery of lipid bilayer by vesicular traffic (vesicular expansion) and one based on the fusion of small compartments (cisternal expansion) (Reggiori and Klionsky, 2005). In addition, it has been suggested that retrograde traffic balances double‐membrane vesicle biogenesis by recycling some Atg proteins, such as Atg9, and also recovering from the forming autophagosome membrane components specific to the compartment(s) of origin (Meiling‐Wesse 2005, Nazarko 2005, Nice 2002, Reggiori 2003, Reggiori 2004a, Reggiori 2004b).
3. Vesicle Completion
When the two extremities of the forming autophagosomes reach each other, they fuse together sealing the vesicle (Fig. 1). This fusion event, at least in yeast, appears to be SNARE‐independent and triggers an uncoating reaction where the externally localized components dissociate from the vesicle surface (Ishihara 2001, Reggiori 2005, Reggiori 2004b). In particular, the ubiquitin‐like protein Atg8–PE is proteolytically released from its lipid moiety by the Atg4 protease, whereas the transmembrane protein Atg9 is completely retrieved (Kirisako 1999, Reggiori 2004a). It is still unknown which factor senses completion of the double‐membrane vesicle and initiates this disassembly.
4. Docking and Fusion
Once uncoated, the double‐membrane vesicle docks with the lysosomes/vacuoles (Fig. 1). In mammalian cells, this association is facilitated by microtubules and seems to require dynein whereas in yeast it is independent of these structures (Aplin 1992, Fengsrud 1995, Kirisako 1999, Punnonen 1990, Ravikumar 2005, Webb 2004). The fusion between the autophagosome and the lysosome/vacuole occurs as soon as these organelles dock and it is mediated by a set of proteins also used for other fusion reactions with the lysosome/vacuole (Section I.C). During this event, the external membrane of the autophagosome becomes part of the lysosome/vacuole surface whereas the inner autophagosomal vesicle is liberated in the interior of this organelle and now called an autophagic body (Fig. 1).
5. Breakdown
The limiting membrane of the autophagic body is immediately attacked and consumed by resident lysosomal/vacuolar hydrolases allowing these enzymes to gain access to the content of this vesicle. As a result, the cargoes are also degraded into their basic constituents (or in the case of certain resident hydrolases, processed to their active form; Fig. 1).
B. A Multitask Pathway
Autophagy has been known for a long time as an adaptation response to starvation and as the major factor in the turnover of long‐lived proteins. But in recent years, it has become evident that autophagy plays an active role in several other physiological tasks highlighting its versatility and adaptability. We now know that this catabolic pathway participates in cellular processes such as development, cellular differentiation and rearrangement, elimination of aberrant structures, lifespan extension, MHC class II presentation of cytoplasmic antigens, and type II programmed cell death, as well as protecting against pathogens (both viruses and bacteria) and tumors (Cuervo 2005, Debnath 2005, Deretic 2005, Edinger 2003, Edinger 2004, Kirkegaard 2004, Komatsu 2005, Kondo 2005, Levine 2004, Paludan 2005, Rubinsztein 2005, Shintani 2004a). As a result, this degradative transport route plays a relevant role in the pathophysiology of neurodegenerative, cardiovascular, muscular, and autoimmune diseases, and some malignancies (Edinger 2003, Edinger 2004, Kondo 2005, Rubinsztein 2005, Shintani 2004a, Towns 2005).
Autophagy provides one effective way to adjust and cope with these various situations by rapidly delivering large fractions of the cytoplasm, aberrant protein aggregates, superfluous or damaged organelles, and invading pathogens into the lysosome/vacuole interior where they are destroyed by resident hydrolases (Reggiori and Klionsky, 2005).
The adaptability of this pathway is due to its ability to select specific cargoes when forced by circumstances. It has been believed for a long time that autophagy was a nonspecific process because when induced by starvation, cytoplasmic components and organelles were randomly sequestered into autophagosomes; however, this pathway can also be selective (Table I ) (Reggiori and Klionsky, 2005). In the yeast Saccharomyces cerevisiae, for example, aminopeptidase I (Ape1) and α‐mannosidase (Ams1) form a large oligomer that is unconventionally delivered from the cytoplasm directly to the vacuole interior through a process known as the cytoplasm to vacuole targeting (Cvt) pathway (Kim 1997, Shintani 2002). This transport route is specific and biosynthetic. Precursor Ape1 (prApe1) is packed into double‐membrane vesicles called Cvt vesicles, which are four to eight times smaller in surface area than autophagosomes (Baba 1997, Scott 1997). In the same organism, dysfunctional mitochondria are preferentially eliminated by autophagy (mitophagy) as well as superfluous peroxisomes (pexophagy) (Table I) (Hutchins 1999, Priault 2005). The specific sequestration of peroxisomes into double‐membrane vesicles and their subsequent degradation has also been very well described in other fungi such as Pichia pastoris, Hansenula polymorpha, and Yarrowia lipolytica (Dunn et al., 2005). Mammalian cells on the other hand, seem not to possess a transport route similar to the Cvt pathway, but there are indications that mitophagy could occur (Bota 2001, Elmore 2001, Rodriguez‐Enriquez 2006). Pexophagy has also been reported (Luiken 1992, Yokota 1993, Yokota 1994). It has lately been shown that autophagy can be selective in mammalian cells as well, as evidenced by the specific recognition and disposal of invading bacteria and potentially also of intracellular viruses (Table I) (Deretic 2005, Kirkegaard 2004, Levine 2005). In addition, a study analyzing conditional knock‐out mice defective for autophagy has revealed that the mutant animal accumulates numerous ubiquitinated aggregates in the cytosol, suggesting that this covalent protein modification could serve to specifically target to autophagosomes large structures that have to be eliminated (Komatsu et al., 2005).
Table I.
Types of Selective Autophagy
| Name | Cargo | Organism |
|---|---|---|
| Cvt pathway | prApe1, prAms1 | S. cerevisiae, P. pastoris |
| Pexophagy | Peroxisomes | S. cerevisiae, P. pastoris, H. polymorpha, Y. lipolytica, and mammals |
| Mitophagy | Mitochondria | S. cerevisiae and mammals |
| Xenophagy | Bacteria and virus | Plants and mammals |
The different types of selective autophagy, their specific cargoes, and the organisms that have been described are indicated.
C. Autophagy‐Related Genes
The process of autophagy has been known for at least 40 years, but because none of the specific components involved in this pathway were known, the studies about this degradative transport route were limited to morphological and phenomenological observations. In the last 15 years, genetic screens, mostly in the yeast S. cerevisiae and fungi such as P. pastoris and H. polymorpha, have lead to the isolation of 18 genes termed AuTophaGy‐related (ATG) genes whose products are specifically involved in this catabolic pathway (Table II ) (Klionsky 2004, Klionsky 2003).
Table II.
Yeast S. cerevisiae Genes Specifically Involved in Autophagy, Cvt Pathway, and Pexophagy
| Protein | Step | Role | Interactions | Orthologs |
|---|---|---|---|---|
| Atg1 | Formation/ expansion | Serine/ threonine kinase | Atg13, Atg11, Atg17 | D.d., C.e., P.p., H.p. |
| Atg2 | Formation/ expansion | Atg9 recycling | Atg9, Atg18 | P.p. |
| Atg3 | Formation/ expansion | Atg8 conjugation system (E2) | Atg7, Atg8, Atg12 | H.s., D.m., P.p. |
| Atg4 | Formation/ expansion | Cysteine protease | Atg8 | H.s., M.m., D.m., P.p. |
| Atg5 | Formation/ expansion | Atg12 conjugation system | Atg12, Atg16 | H.s., M.m., D.d. |
| Atg6a | Formation/ expansion | PtsIns‐3‐P synthesis | Atg14, Vps15, Vps34 | H.s., M.m., D.d., C.e. |
| Atg7 | Formation/ expansion | Atg8 and Atg12 conjugation systems (E1) | Atg3, Atg8, Atg12 | H.s., M.m., D.d., C.e., A.t., P.p. |
| Atg8 | Formation/ expansion | Ubiquitin‐like protein | Atg3, Atg4, Atg7, Atg19 | H.s., M.m., R.n., D.d., C.e., A.t., P.p., H.p. |
| Atg9 | Formation/ expansion | Transmembrane protein | Atg2, Atg18, Atg23 | H.s., M.m., A.t., P.p. |
| Atg10 | Formation/ expansion | Atg12 conjugation system (E2) | Atg12 | H.s., M.m. |
| Atg12 | Formation/ expansion | Ubiquitin‐like protein | Atg3, Atg5, Atg7, Atg10, Atg16, Atg17 | H.s., M.m., D.d. |
| Atg13 | Formation/ expansion | Modulates Atg1 activity | Atg1, Atg17, Vac8 | – |
| Atg14 | Formation/ expansion | PtsIns‐3‐P synthesis | Atg6, Vps15, Vps34 | – |
| Atg16 | Formation/ expansion | Associates with the Atg12–Atg5 complex | Atg5, Atg12, Atg16 | H.s., M.m., P.p. |
| Atg17b | Formation/ expansion | Modulates Atg1 activity | Atg1, Atg13, Atg11, Atg12, Atg24 | – |
| Atg18 | Formation/ expansion | PtsIns‐3‐P binding protein | Atg2, Atg9 | H.s., A.t., P.p. |
| Atg22c | Vesicle breakdown | Transmembrane protein | – | – |
| Atg23 | Formation/ expansion | Cycling factor | Atg9 | – |
H.s. = Homo sapiens; M.m. = Mus musculus; R.n. = Rattus norvegicus; D.m. = Drosophila melanogaster; D.d. = Dictyostelium discoideum; C.e. = Caenorhabditis elegans; A.t. = Arabidopsis thaliana; P.p. = Pichia pastoris; H.p. = Hansenula polymorpha; Y.l. = Yarrowia lipolytica.
In yeast, Atg6 plays an important role in endosomal trafficking.
Atg17 is required for autophagy and pexophagy but not Cvt pathway.
Atg22 is not necessary for both pexophagy and the Cvt pathway.
The extent of the conservation of this pathway between eukaryotes was first revealed by comparing the genomes once various sequencing projects were completed (Reggiori and Klionsky, 2002). It became immediately evident that most of the ATG genes had one or more homologs in higher eukaryotic organisms. The cellular role of some of them has now been explored and in all the analyzed cases, it has been demonstrated that the homologs function as orthologs (Table II) (Levine 2004, Reggiori 2002).
The same genetic approaches have also led to the discovery of nine ATG genes dispensable for bulk autophagy but essential for the Cvt pathway and/or pexophagy (Table III ). Their products are mostly involved in cargo selection and the final sealing of the double‐membrane vesicle, indicating that additional components are required for the autophagosomes to be able to enwrap specific cargoes. It is important to note that these genes involved in specific types of autophagy do not have clear homologs in higher eukaryotes sustaining the idea that the Cvt pathway and pexophagy are probably only present in fungi (Reggiori and Klionsky, 2002).
Table III.
Yeast Genes Specifically Involved in the Cvt Pathway and/or Pexophagy
| Protein | Cvt | Pexophagy | Step | Role | Organism |
|---|---|---|---|---|---|
| Atg11 | + | + | Formation/ expansion | Cargo receptor/ adaptor | S.c., P.p., H.p. |
| Atg19 | + | − | Formation/ expansion | Cargo receptor | S.c. |
| Atg20a | + | + | Formation/ expansion | PtdIns‐3‐P binding protein | S.c. |
| Atg21b | + | ? | Formation/ expansion | PtdIns‐3‐P binding protein | S.c., H.p. |
| Atg24a | + | + | Formation/ expansion | PtdIns‐3‐P binding protein | S.c., P.p. |
| Atg25c | − | + | Fusion | Coiled‐coil protein | H.p. |
| Atg26c | − | + | Vesicle completion | UDP‐glucose:sterol glucosyltransferase | P.p. |
| Atg27 | + | N.D. | Formation/ expansion | PtdIns‐3‐P binding protein | S.c. |
| Atg28c | − | + | Vesiculation | Coiled‐coil protein | P.p. |
| Tlg1a | + | N.D. | Formation/ expansion | vSNARE | S.c. |
| Tlg2a | + | N.D. | Formation/ expansion | tSNARE | S.c. |
| Vps45a | + | N.D. | Formation/ expansion | Sec1 homolog | S.c. |
N.D. = not determined.
A plus or a minus mark indicates whether the protein is required for a pathway. S.c. = Saccharomyces cerevisiae; P.p. = Pichia pastoris; H.p. = Hansenula polymorpha.
?One report has indicated that Atg21 is essential for pexophagy, another affirms that Atg21 is not required for this process.
In S. cerevisiae, these proteins also catalyze the retrieval transport from early endosomes.
Atg21 is not required for pexophagy in S. cerevisiae but is essential for the same process in H. polymorpha.
These factors have no counterparts in S. cerevisiae or the homologs do not have a role in pexophagy.
In addition to the Atg proteins, the genetic screens in yeast have also permitted the identification of additional components required for the normal progression of autophagy that are shared with other intracellular transport routes (Table IV ). The function of several of these factors in the other pathways was already known and that has helped in clarifying the mechanism of autophagy. For example, yeast vacuoles can fuse with late endosomes [multivesicular bodies (MVB) pathway] or possibly with vesicles derived from the endosome [carboxypeptidase Y (CPY) pathway], Golgi‐derived vesicles [alkaline phosphatase (ALP) pathway], and with themselves (homotypic fusion). In all these cases, cells use an identical fusion machinery, which consists of SNARE proteins, Sec18 (NSF), Sec17 (α‐SNAP), a Rab‐GTPase, and the class C Vps protein complex also known as the HOPS complex. The same components have also been found to be exploited for the fusion of double‐membrane vesicles (Table IV) (Reggiori 2002, Wang 2003). Similarly, it is also now evident that the dissolution of autophagic bodies is mediated by the same hydrolases that degrades the MVB internal vesicles once these are released into the vacuole lumen (Table IV) (Epple 2003, Reggiori 2002).
Table IV.
Yeast S. cerevisiae Genes Involved in Autophagy, Cvt Pathway, and Pexophagy but Also in Other Endosomal Transport Routes
| Protein/complex | Step | Role |
|---|---|---|
| Atg15 | Vesicle breakdown | Lipase |
| Ccz1–Mon1 complex (Ccz1, Mon1) | Docking/fusion | Tethering/docking factor |
| HOPS complex/class C Vps protein complex | ||
| (Vps11, Vps16, Vps18, Vps33, Vps39, Vps41) | Docking/fusion | Tethering factor/Rab effector |
| Pep4 | Vesicle breakdown | Vacuolar protease |
| Prb1 | Vesicle breakdown | Vacuolar protease |
| PtsIns‐3‐kinase complex (Vps15, Vps34) | Formation/expansion | PtsIns‐3‐P synthesis |
| Trs85 | Formation/expansion | Tethering factor |
| Vac8 | Formation/expansion | Vacuole landmark |
| Vam3 | Docking/fusion | tSNARE |
| Vam7 | Docking/fusion | vSNARE |
| VFT complex (Vps51, Vps52, Vps53, Vps54) | Formation/expansion | Tethering factor |
| Ykt6 | Docking/fusion | vSNARE |
| Vti1 | Docking/fusion | vSNARE |
| Ypt7 | Docking/fusion | Rab‐GTPase |
II. Membrane Source in the Yeast S. cerevisiae
A. Pre‐autophagosomal Structure
Most of the Atg components are peripheral membrane proteins that transiently associate with the nascent autophagosomes. In contrast to mammalian cells where several isolation membranes can be simultaneously activated, a single perivacuolar site of organization for double‐membrane vesicle formation (named the pre‐autophagosomal structure, PAS) is observed in the yeast S. cerevisiae (Kim 2002, Suzuki 2001). The PAS is believed to be the yeast counterpart of a mammalian isolation membrane and in this unicellular eukaryote, most of the Atg proteins appear to be primarily restricted to this location. This unique site seems also to be present in H. polymorpha (Monastyrska 2005a, Monastyrska 2005b). In P. pastoris, however, several Atg components are distributed to more than one punctate structure (Ano 2005, Chang 2005, Kim 2001b, Mukaiyama 2004, Stromhaug 2001). It is unclear if this represents a difference between organisms or is due to different growth conditions. P. pastoris is mostly used for the study of pexophagy and therefore grown in special media containing carbon sources that induce peroxisome proliferation.
It is unclear where the PAS is derived from and at which point it becomes membranous. The study of the Cvt pathway has provided insights into how this structure is generated. After synthesis, prApe1 forms a large oligomer that first associates with the Atg19 cargo receptor and then with the Atg11 adaptor to form the Cvt complex (Shintani et al., 2002). This large cytosolic protein aggregate then moves in close proximity to the vacuole surface, where it induces the recruitment of the rest of the Atg factors, triggering the formation of the Cvt vesicle (Shintani 2004b, Yorimitsu 2005). Neither the PAS nor the vesicles are efficiently formed in the absence of any of the Cvt complex components, indicating that the cargo stimulates the biogenesis of these structures (Shintani and Klionsky, 2004b). This requirement is overcome when cells are nitrogen‐starved (Kim 2001b, Shintani 2004b).
Because of its dynamic properties, the PAS should not be seen as a static or defined organelle but more as a structure in constant remodeling. It remains unclear at which stage and how membranes are transported at the PAS, but because of their association with lipid bilayers, two proteins, Atg8 and Atg9, could be important for dissecting this event.
B. Atg8
Atg8 is a soluble ubiquitin‐like protein and its carboxy‐terminal arginine is removed by the Atg4 cysteine protease leaving a glycine residue at the new carboxy terminus (Kim 2001a, Kirisako 2000). Atg8 is activated by the E1 enzyme Atg7 through a thioester bond between its carboxy‐terminal glycine and cysteine 507 of Atg7 (Kim 1999, Kirisako 2000, Komatsu 2001). Atg8 is subsequently transferred to the E2 enzyme Atg3 via a new thioester bond between these two proteins (Ichimura 2000, Kim 2001a). Atg8 is finally covalently conjugated to a PE molecule, becoming tightly membrane associated (Ichimura et al., 2000). This linkage is reversible because Atg8 can be proteolitically released from its lipid moiety by Atg4, an event that takes place once the double‐membrane vesicles are completed (Section I.A.3) (Kirisako 1999, Reggiori 2004a).
It is unclear where the Atg8 conjugation to PE occurs. This protein is normally lipidated in mutants unable to form the PAS indicating that this modification takes place at a different subcellular location (Suzuki et al., 2001). This is supported by the fact that Atg8–PE localizes to the PAS but also to tiny cytosolic vesicles (Kirisako et al., 1999). These data, however, do not exclude the possibility that Atg8–PE conjugates are formed at the PAS as well. The association of Atg8–PE with membranes prior to getting concentrated at the PAS suggests that the Atg8–PE structures could be at least in part the source of autophagosome lipid bilayers. This idea is supported by the observation that in the absence of Atg8, membranes fail to be delivered to the PAS and therefore the size of autophagosomes is strongly reduced (Abeliovich 2000, Kirisako 1999, Lang 1998).
It remains a mystery where the tiny Atg8–PE containing vesicles are derived from, but one possibility is that that they originate from early compartments of the secretory pathway, for example, the endoplasmic reticulum (ER) and/or the Golgi apparatus. This hypothesis is based on two experimental findings. First, Atg8 binds two vSNAREs required for both anterograde and retrograde transport between the ER and Golgi apparatus (Legesse‐Miller et al., 2000). Second, this ubiquitin homolog has been detected on autophagosome‐like structures derived from the Golgi complex and/or ER (Section II.D.2) (Reggiori et al., 2004b).
C. Atg9
Atg9 is the only integral membrane protein essential for double‐membrane vesicle formation (Noda et al., 2000). This protein is probably transported to the PAS with at least part of the lipids or lipid bilayers required to create this structure. This notion is corroborated by the fact that the totality of Atg9 is associated with membranes (Noda 2000, Reggiori 2005b). Atg9 cycles between the PAS and several unknown punctate structures dispersed in the cytosol supporting the idea that it could partially supply the forming autophagosomes with membranes (Reggiori et al., 2004a). A fraction of these punctate structures are Atg9 aggregates residing on the mitochondria surface (Reggiori et al., 2005b). This suggests that this organelle could provide the nascent autophagosomes with at least part of its lipid bilayers.
However, it cannot be excluded that Atg9 trafficking carries out other functions. Under certain conditions, autophagy becomes one of the principal sources of energy for the cell (Kuma 2004, Lum 2005). Because the mitochondria provide the other primary supply of energy, one could imagine that Atg9 is used to coordinate the two sources.
The sorting mechanism for Atg9 transit from mitochondria to the PAS is unknown, but under growing conditions this event is induced by Cvt complex assembly and requires actin (Reggiori 2005a, Shintani 2004b). In contrast, the retrieval transport of this transmembrane protein from the PAS has been characterized in more detail and shown to be regulated by the Atg1–Atg13 signaling complex and requires Atg2, Atg18, and the PtdIns‐3‐phosphate generated by the Atg14‐containing PtdIns 3‐kinase complex (Reggiori et al., 2004a). This recycling event, however, seems to be to some extent differently organized in P. pastoris during micropexophagy, possibly because other membranous structures are used and assembled in a different way during this invagination process (Chang et al., 2005).
D. Yeast Organelles and Autophagy
1. Endoplasmic Reticulum
An initial analysis concerning the role of yeast early secretion (sec) mutants in autophagy has revealed that several of them are essential for autophagosome formation (Ishihara et al., 2001). This class of genes is involved in transport out of the ER (Kaiser and Schekman, 1990). Successive studies, however, have shown that these mutants have an indirect negative effect on both the Cvt pathway and autophagy (Hamasaki 2003, Reggiori 2004b). One possible explanation of their phenotype is that they alter the ER morphology and consequently impair several functions of this organelle, including the putative one to supply membranes for double‐membrane vesicle formation. For example, the ER is structurally connected with the mitochondria and the disruption of the ER organization in the early sec mutants causes the fragmentation of the mitochondrial reticulum (Prinz et al., 2000). As mentioned, Atg9 partially localizes to mitochondria, and in this class of mutants its trafficking out of this compartment is severely impaired (Reggiori and Klionsky, submitted).
2. Golgi Apparatus
Atg20, Atg24, Tlg1, Tlg2, Trs85, Vps45, and the subunits of the Vps‐fifty‐three (VFT) complex are part of retrieval transport routes from endosomal compartments back to the Golgi apparatus, and consequently they are important in maintaining certain functions of this organelle (Hettema 2003, Holthuis 1998, Sacher 2000, Sacher 2001, Siniossoglou 2001). These proteins have also been shown to be required for the Cvt pathway and some of them also play an important role in double‐membrane vesicle biogenesis during pexophagy and autophagy (Table III, Table IV) (Abeliovich 1999, Meiling‐Wesse 2005, Nazarko 2005, Nice 2002, Reggiori 2003). It is unclear, however, why these pathways are impaired in the absence of these factors. One possibility is that retrograde traffic from the forming double‐membrane vesicles is essential for the expansion and/or completion of these structures (Meiling‐Wesse 2005, Reggiori 2004a, Reggiori 2004b). A second hypothesis is that similarly to what was predicted for early sec mutants, an alteration of the Golgi apparatus functions could interfere with the lipid bilayer delivery essential for the creation of these large vesicles.
The major difficulty in investigating the contribution to autophagy of both the ER and the Golgi apparatus is that these two organelles depend on each other for their proper function. Mutations that affect one of these two compartments indirectly perturb the other one. Along these lines, the interpretation of the block of both the Cvt pathway and autophagy in the sec7 mutant is not simple (Reggiori et al., 2004b). Nevertheless, the analysis of this strain has led to important information. Sec7 is a GDP/GTP exchange factor required for trafficking through the Golgi complex (Franzusoff 1989, Jackson 2000). The inactivation of this protein provokes the accumulation of unsealed, autophagosome‐like structures that are decorated with Atg8 (Reggiori et al., 2004b). These membranous arrangements enwrap ribosomes and cytosol and have been previously named Berkeley bodies (Esmon 1981, Novick 1980). This surprising result indicates that potentially, double‐membrane vesicles can be created in large part by altering the activity of a single enzyme; however, it cannot be excluded that this is an indirect phenomenon.
3. Endosomes
Vps4 is a protein essential for the invagination of the late endosome limiting membranes and therefore MVB biogenesis (Babst 1998, Katzmann 2002). A unique VPS4 allele was isolated in a screen for mutations that result in autophagy induction even in the presence of nutrients (Shirahama et al., 1997). This led to an initial interpretation that endosomes play a relevant role in autophagosomes biogenesis. However, reports where the functions of these compartments have been severely impaired by specific gene deletions have revealed that the integrity of the endosomal system is not essential for either the Cvt pathway or autophagy (Epple 2003, Reggiori 2004b).
III. Lipid Bilayer Origin in Mammalian Cells
A. Uncertain Origin of the Isolation Membrane
In contrast to the late stages of the autophagosome biogenesis where lipid bilayers are derived from endosomal compartments, the origin of the mammalian isolation membrane or phagophore remains uncertain. It is still unknown if this small sequestering cisterna is formed de novo or derived from a preexisting organelle (Fengsrud et al., 2004). A major problem in trying to investigate its origin is that these structures and autophagosomes are mostly composed of lipids and depleted in transmembrane proteins making particularly difficult the detection of specific organelle markers (Fengsrud 2000, Hirsimaki 1982, Punnonen 1989, Reunanen 1985, Stromhaug 1998). This unique characteristic is one line of evidence that the isolation membranes and autophagosomes differ structurally from the other subcellular organelles. This observation also implies that whatever the origin of the lipid bilayers used to form autophagosomes, integral membrane components are segregated away from them.
Two models could explain how protein‐depleted membranes are obtained. In the first, isolation membranes are derived from a specialized organelle subdomain where autochthonous proteins are gradually excluded. A similar process has been shown to occur during peroxisome biogenesis from the ER (Geuze 2003, Tabak 2003, Tam 2005). In the second model, the same cisterna is progressively emptied of integral membrane factors by retrieval transport— a phenomenon hypothesized to occur during double‐membrane vesicle formation in yeast (Reggiori 2004a, Reggiori 2004b). It is also possible that both mechanisms coexist.
Numerous studies have been published investigating the source for autophagosome lipid bilayers in mammalian cells but their conclusions often contrast. Thus various organelles, such as the ER, the Golgi complex, and the plasma membrane, have been suggested to be the origin of double‐membrane vesicles. Because of the heterogeneity in the results, no unanimous agreement in the field has been reached. For example, several studies have reported the presence of ER‐marker proteins in the isolation membranes and autophagosomes but others have shown that these structures lack ER‐resident factors (Arstila 1968, Dunn 1990a, Furuno 1990, Reunanen 1985, Stromhaug 1998, Yamamoto 1990, Yokota 1994).
As with the ER, the role of the Golgi complex as a lipid donor for the early autophagosome intermediates remains ambiguous. Some studies have shown the presence of Golgi protein markers in these structures whereas others have failed to detect them (Arstila 1968, Dunn 1990a, Frank 1968, Locke 1975, Yang 1997, Yokota 1994). The membranes of the cis‐Golgi network have been shown to possess the same compositional characteristics of the isolation membrane (Fengsrud 2004, Locke 1975, Reunanen 1988, Yamamoto 1990).
Only a few reports have indicated that the autophagic cisternae are derived from the plasma membrane and their conclusions have been challenged when other investigators have failed to detect plasma membrane protein markers in these structures (Araki 1995, Arstila 1968, Bosabalidis 1994, Ericsson 1969, Fengsrud 2004, Oledzka‐Slotwinska 1969, Reunanen 1988). Importantly, autophagosomes have a low cholesterol content validating the idea that their membranes are not derived from the plasma membrane (Reunanen et al., 1985).
The discrepancy between all these analyses could be due, in part, to different experimental approaches and techniques used in the various laboratories. But one possibility that should not be discarded a priori is that autophagosomes could be a mosaic of membranes derived from more than one organelle. For example, the isolation membrane could originate from one compartment and the additional lipid bilayers required for its expansion be acquired from other sources. In addition, the different contributions could vary depending on the tissues with cells able to derive the membranes from the most suitable reservoirs.
B. Atg8
The Atg8 conjugation system is highly conserved in higher eukaryotic cells (Table II) (Ohsumi 2004, Tanida 2004). In mammals, there are at least three Atg8 homologs: the microtubule‐associated protein 1 (MAP1) light chain 3 (LC3), the Golgi‐associated ATPase enhancer of 16 kDa (GATE‐16), and the γ‐aminobutyric acid (GABA)A‐receptor‐associated protein (GABARAP) (Mann 1994, Sagiv 2000, Wang 1999). It should be noted that these three proteins were first isolated because of their involvement in other trafficking pathways. The mammalian counterparts of Atg4 process these three Atg8 homologs by exposing their conserved C‐terminal glycine which then interacts with mammalian Atg7 and Atg3 homologs before being covalently linked to a lipid (Hemelaar 2003, Scherz‐Shouval 2003, Tanida 2001, Tanida 2002). The target phospholipid has not yet been unequivocally identified, but strong evidence suggests that it is PE (Kabeya 2004, Tanida 2004).
Of the three homologs, LC3 has been best characterized as an autophagosomal marker in mammalian autophagy. The newly synthesized LC3 precursor is processed cotranslationally to generate a soluble LC3 form (LC3‐I) that, upon starvation, becomes membrane‐bound and has greater mobility than LC3‐I when resolved by SDS‐PAGE (Kabeya et al., 2000). The lipidated protein, called LC3‐II, localizes on both autophagosomes and autolysosomes (Kabeya et al., 2000). These in vitro results have been confirmed using transgenic mice expressing GFP‐LC3 (Mizushima et al., 2004). Unfortunately, the small amount of LC3‐II generated prior to induction of autophagy is already associated with the double‐membrane vesicles formed by the basal activity of this pathway and LC3‐I is not clearly associated with a distinct membranous structure (Kabeya et al., 2004). Therefore, the subcellular localization of these molecules has not furnished insights about the lipid bilayer source.
Both GATE‐16 and GABARAP possess a form II and localize to LC3‐positive autophagosomes that are induced by starvation (Kabeya et al., 2004). Thus, it remains a possibility that they participate in autophagy in addition to, or instead of, their originally described functions. Because the three mammalian Atg8 homologs are differently expressed in various tissues (Tanida et al., 2004), another intriguing option is that these proteins are involved in supplying the autophagosome with membranes derived from different compartments depending on the cell type; for example, GATE‐16 from the Golgi complex and GABARAP from the same organelle as well as the synaptic cisternae (Kittler 2001, Kneussel 2000, Sagiv 2000).
C. Atg9
A report has demonstrated that the two human proteins with high homology to Atg9, HsAtg9L1 and HsAtg9L2, are its orthologs (Yamada et al., 2005). In human adult tissues, HsATG9L1 is ubiquitously expressed, whereas HsATG9L2 is highly expressed in placenta and pituitary gland. Importantly, the authors have also shown that these two factors are not distributed on mitochondria. Instead they localize to a perinuclear region, suggesting that in higher eukaryotes Atg9 could supply autophagy with membranes by deriving lipid bilayers from a different reservoir. This observation could also explain why HsAtg9L1 and HsAtg9L2 cannot substitute for the yeast Atg9 (Reggiori 2005b, Yamada 2005). However, HsAtg9L2 possesses a nonfunctional mitochondrial targeting sequence that is also present in its closest higher eukaryote homologs (Yamada et al., 2005). This characteristic raises the possibility that this is an ancient localization signal. Because the subcellular distribution of HsAtg9L1 and HsAtg9L2 have not been carefully examined and the preliminary localization analysis was performed with overexpressed proteins, the identification of the precise localization of these two proteins could provide insights into membrane dynamics during autophagosome biogenesis in mammals.
D. Autophagosome Maturation
In mammalian cells, autophagosomes, also called initial autophagic vacuoles (AVi), undergo a stepwise maturation process that can be followed ultrastructurally by monitoring the disintegration status of their internal lipid bilayer and cargoes (Fig. 2 ) (Dunn 1990b, Eskelinen 2005, Fengsrud 2004, Rabouille 1993). These morphological changes correlate with the increasing acquisition of lysosomal makers (Berg 1998, Dunn 1990b, Liou 1997, Tanaka 2000). Autophagosomes, which contain intact cytosol and organelles, fuse first with endosomal vesicles and MVB turning into early degradative autophagic vacuoles (AVd) or amphisomes. These structures successively fuse together or with lysosomes becoming late AVd or autolysosomes. The degradation of the internal material starts in the early AVd and continues in the late AVd until completion.
Figure 2.
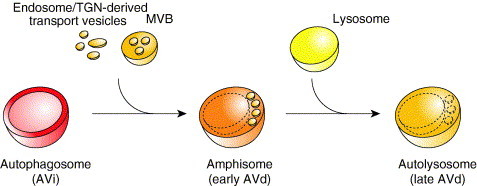
Autophagosome maturation in mammalian cells. Once sealed, the autophagosome (or AVi) fuses with endosome‐ and/or TGN‐derived transport vesicles and the MVB becoming an amphisome (or early AVd). This event leads to the acquisition of hydrolytic enzymes that initiate the consumption of the autophagosome cargo. The amphisome then fuses with a lysosome generating a new organelle termed autolysosome (or late AVd) where the degradation of the content of the initial autophagosome is completed.
In contrast to yeast, the endosomal system plays an essential role in mammalian autophagy (see Section II.D.3). This divergence between species has been highlighted by the discovery that SKD1 is necessary for autophagosome maturation in mouse cells (Nara et al., 2002). SKD1 is the mouse homolog of yeast Vps4 and, as its counterpart, it is also essential to maintain endosome morphology and endosomal transport (Yoshimori et al., 2000). As mentioned, Vps4 is not required for autophagy in yeast (Section II.D.3) (Reggiori 2004b, Shirahama 1997).
It is unclear why mammalian autophagosomes need the additional maturation step characterized by their fusion with endosome‐ and/or trans‐Golgi network (TGN)‐derived transport vesicles and MVB. In yeast, double‐membrane vesicles fuse with a much larger vacuole one after the other. Therefore, their cargoes do not influence the hydrolytic capacity of this compartment by altering, for example, the pH because the volume of their contents is just a fraction of that of the entire vacuole. Lysosomes, in contrast, are much smaller than vacuoles and their size is comparable to that of autophagosomes. Consequently, if these two structures would immediately fuse together, an important dilution of the lysosome content could occur impairing its internal enzymatic activity. Addition of extra hydrolytic enzymes prior to autolysosome formation could help to compensate for this dilution phenomenon.
E. Pathogens
Autophagy provides a cellular defense against invading pathogens but unfortunately, some of them have developed systems to avoid the sequestration and elimination by double‐membrane vesicles (Deretic 2005, Kirkegaard 2004, Levine 2005). In addition, there are virus and bacteria that exploit this transport route to enter and replicate inside the host cell (Kirkegaard et al., 2004). The study of this latter class of pathogens has furnished some indications about the possible origin of autophagosome membranes even if it should be kept in mind that these invading microorganisms are also altering other cellular pathways, and therefore autophagy could progress in part differently in the infected cells.
1. Virus
Upon infection, positive‐strand viruses disassemble and release their genomic RNA into the cytoplasm of the host cell. The genomic RNA is subsequently translated to produce the replicase proteins that induce the formation of the RNA replication complexes. These complexes are assembled and anchored on membrane surfaces and this is an essential requisite for their virulence. Some of the positive‐strand viruses, such as the poliovirus, the mouse hepatitis virus (MHV), the equine arterivirus (EAV), and the severe acute respiratory syndrome (SARS) coronavirus, seen to use autophagosomes as a membrane platform (Kirkegaard et al., 2004).
Factors of the poliovirus RNA‐replication complex localize to double‐membrane vesicles that are derived from the ER by the action of viral proteins 2BC and 3A by a mechanism that excludes resident host proteins (Schlegel 1996, Suhy 2000). Importantly, these structures contain LC3/Atg8 and are highlighted with the fluorophore monodansylcadaverine, a dye that specifically stains autophagosomes (Jackson et al., 2005). The idea that poliovirus subverts components of the cellular autophagy machinery to promote its replication is also supported by the fact that inhibition of this pathway by 3‐methyladenine or by RNA interference against mRNAs that encode two different Atg proteins (LC3/Atg8 and Atg12) decrease the poliovirus yield (Jackson et al., 2005).
Coronaviruses (MHV and SARS) and arteriviruses (EAV) are the two families within the order Nidovirales. Cells infected by these viruses accumulate double‐membrane vesicles and the viral RNA‐replication complexes are associated with them (Goldsmith 2004, Gosert 2002, Pedersen 1999, Shi 1999, van der Meer 1998). In the case of the MHV and SARS coronaviruses, it has also been shown that these structures are decorated with LC3/Atg8, revealing that they are autophagosomes (Prentice 2004a, Prentice 2004b). For the MHV in addition, it has been demonstrated that the autophagy machinery is required to generate these compartments and in its absence the virus replication is severely blocked (Prentice et al., 2004a). Importantly, studies about the origin of these double‐membrane vesicles generated in cells infected by nidoviruses indicate that they are derived from the ER (Pedersen 1999, Prentice 2004a, Shi 1999, van der Meer 1998).
2. Bacteria
After endosomal uptake of Porphyromonas gingivalis and Brucella abortus, by the host cell, the endosomes that contain these bacteria immediately fuse with structures resembling autophagosomes (Dorn 2001, Pizarro‐Cerda 1998a, Pizarro‐Cerda 1998b, Progulske‐Fox 1999). This event prevents their delivery to the lysosome where they would be eliminated. In addition containing endosomal factors, the double membranes surrounding these two pathogens are decorated with ER protein markers and their formation is blocked by autophagy inhibitors such as 3‐methyladenine and wortmannin (Rich et al., 2003).
Legionella pneumophila is a Gram‐negative bacterium that can replicate within human macrophages. After being taken up by phagosomes, this pathogen becomes enwrapped within a double‐membrane compartment that contains the ER resident chaperone BiP through an unknown mechanism, and starts to replicate (Coers 2000, Joshi 2001, Sturgill‐Koszycki 2000, Swanson 1995). It has been shown that this compartment also progressively becomes decorated with typical autophagosome markers such as Atg7 and Atg8 (Amer and Swanson, 2005). However, it remains unclear if these structures are autophagosomes or similar conformations derived from the ER that at successive stage acquire autophagosomal membranes or subvert the autophagy machinery to complete their biogenesis (Kagan 2002, Tilney 2001). In Dictyostelium discoideum, a natural host for L. pneumophila, deletion of ATG genes leads to a defect in autophagy without affecting the formation of the double‐membrane compartment and therefore the replication of this invading microorganism is unaffected (Otto et al., 2004). But this could just reflect host‐specific differences.
Listeria monocytogenes is another Gram‐negative bacterium that after entering into host cells destroys the phagosome membrane using hemolysin to gain access to the cytoplasm where it starts to multiply. However, when infected cells are treated with chloramphenicol, an inhibitor of bacterial protein synthesis, or lack the actA gene, the bacteria become trapped into double‐membrane compartments shortly after phagosome lysis (Rich et al., 2003). These structures are autophagosomes because L. monocytogenes sequestration is enhanced by autophagic induction through serum withdrawal and blocked by autophagy inhibitors such as 3‐methyladenine and wortmannin (Rich et al., 2003). The formation of these autophagosomes seems to be mediated by the assembly of small vesicles and cisternae with variable morphology, which contain the ER protein marker protein disulfide isomerase (PDI). Importantly, PDI‐positive vesicular structures are accumulated around the cytoplasmic bacteria during the early stages of autophagosome biogenesis but not at later stages when these structures begin to acquire endosomal makers (Rich et al., 2003).
IV. Perspectives
Our knowledge about the physiological roles of autophagy has enormously increased and we have realized how important this pathway is for cell survival in several extreme situations. Despite the identification and partial characterization of the Atg proteins, however, the molecular mechanism of this catabolic transport route remains largely unknown. A major challenge in studying this process arises from the fact that the origin and the transport mode of the lipids employed to compose these structures is unknown. Investigations on this topic seem to indicate that the ER and possibly the Golgi complex are involved in supplying the nascent autophagosomes with membranes. Endosomal compartments, in contrast, play a relevant role only in mammalian cells and at a later stage during autophagosome maturation.
The large majority of the morphological characterization of autophagosome formation was done 10–15 years ago, when specific autophagy markers were unavailable. Atg proteins provide now the researchers with the long‐awaited markers that could be used to at least dissect this transport route at an ultrastructural level, thus solving some of the mysteries that surround the double‐membrane vesicle origin and biogenesis. Analysis of pathogens and their gene products has helped in the past to unveil and analyze numerous cellular pathways. The discovery of the existence of viruses and bacteria subverting autophagy will probably have a similar impact. The study of these microorganisms will help us to understand how lipid bilayers are derived from the membrane source(s) but will also potentially lead to the isolation of agents that will allow investigators to manipulate this process.
Acknowledgments
The author thanks Daniel Klionsky, Judith Klumperman, Catherine Rabouille, and Ger Strous for critically reading the chapter. The author also wishes to thank Marc van Peski and René Scriwanek for Figure 1, Figure 2.
References
- Abeliovich H., Darsow T., Emr S.D. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t‐SNARE‐Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H., Dunn W.A., Jr., Kim J., Klionsky D.J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A.O., Swanson M.S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Y., Hattori T., Oku M., Mukaiyama H., Baba M., Ohsumi Y., Kato N., Sakai Y. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol‐3‐phosphate. Mol. Biol. Cell. 2005;16:446–457. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin A., Jasionowski T., Tuttle D.L., Lenk S.E., Dunn W.A., Jr. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell. Physiol. 1992;152:458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- Araki N., Takashima Y., Makita T. Redistribution and fate of colchicine‐induced alkaline phosphatase in rat hepatocytes: Possible formation of autophagosomes whose membrane is derived from excess plasma membrane. Histochem. Cell. Biol. 1995;104:257–265. doi: 10.1007/BF01464321. [DOI] [PubMed] [Google Scholar]
- Arstila A.U., Trump B.F. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am. J. Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S.V., Klionsky D.J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., Emr S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T.O., Fengsrud M., Stromhaug P.E., Berg T., Seglen P.O. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Bosabalidis A.M. Developmental features of autophagy in aging secretory cells of Tamarix aphylla salt glands. J. Submicrosc. Cytol. Pathol. 1994;26:473–479. [Google Scholar]
- Bota D.A., Davies K.J. Protein degradation in mitochondria: Implications for oxidative stress, aging and disease: A novel etiological classification of mitochondrial proteolytic disorders. Mitochondrion. 2001;1:33–49. doi: 10.1016/s1567-7249(01)00005-8. [DOI] [PubMed] [Google Scholar]
- Chang T., Schroder L.A., Thomson J.M., Klocman A.S., Tomasini A.J., Stromhaug P.E., Dunn W.A., Jr. PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell. 2005;16:4941–4953. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J., Kagan J.C., Matthews M., Nagai H., Zuckman D.M., Roy C.R. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Bergamini E., Brunk U.T., Dröge W., Ffrench M., Terman A. Autophagy and aging: The importance of maintaining. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Debnath J., Baehrecke E.H., Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Dorn B.R., Dunn W.A., Jr., Progulske‐Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 2001;69:5698–5708. doi: 10.1128/IAI.69.9.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr. Studies on the mechanisms of autophagy: Formation of the autophagic vacuole. J. Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr. Studies on the mechanisms of autophagy: Maturation of the autophagic vacuole. J. Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr., Cregg J.M., Kiel J.A.K.W., van der Klei I.J., Oku M., Sakai Y., Sibirny A.A., Stasyk O.V., Veenhuis M. Pexophagy: The selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Thompson C.B. Defective autophagy leads to cancer. Cancer Cell. 2003;4:422–424. doi: 10.1016/s1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Thompson C.B. Death by design: Apoptosis, necrosis and autophagy. Curr. Opin. Cell. Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Elmore S.P., Qian T., Grissom S.F., Lemasters J.J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Epple U.D., Eskelinen E.‐L., Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Ericsson J.L. Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp. Cell Res. 1969;55:95–106. doi: 10.1016/0014-4827(69)90462-5. [DOI] [PubMed] [Google Scholar]
- Eskelinen E.‐L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Esmon B., Novick P., Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981;25:451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Roos N., Berg T., Liou W., Slot J.W., Seglen P.O. Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: Effects of vinblastine and asparagine on vacuole distributions. Exp. Cell Res. 1995;221:504–519. doi: 10.1006/excr.1995.1402. [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Erichsen E.S., Berg T.O., Raiborg C., Seglen P.O. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze‐fracture electron microscopy. Eur. J. Cell Biol. 2000;79:871–882. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Lunde Sneve M., Overbye A., Seglen P.O. Structural aspects of mammalian autophagy. In: Klionsky D.J., editor. “Autophagy”. Landes Bioscience; Georgetown, TX: 2004. pp. 11–25. [Google Scholar]
- Frank A.L., Christensen A.K. Localization of acid phosphatase in lipofuscin granules and possible autophagic vacuoles in interstitial cells of the guinea pig testis. J. Cell Biol. 1968;36:1–13. [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno K., Ishikawa T., Akasaki K., Lee S., Nishimura Y., Tsuji H., Himeno M., Kato K. Immunocytochemical study of the surrounding envelope of autophagic vacuoles in cultured rat hepatocytes. Exp. Cell Res. 1990;189:261–268. doi: 10.1016/0014-4827(90)90245-6. [DOI] [PubMed] [Google Scholar]
- Geuze H.J., Murk J.L., Stroobants A.K., Griffith J.M., Kleijmeer M.J., Koster A.J., Verkleij A.J., Distel B., Tabak H.F. Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double‐membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- Hemelaar J., Lelyveld V.S., Kessler B.M., Ploegh H.L. A single protease, Apg4B, is specific for the autophagy‐related ubiquitin‐like proteins GATE‐16, MAP1‐LC3, GABARAP, and Apg8L. J. Biol. Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Lewis M.J., Black M.W., Pelham H.R.B. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsimaki Y., Hirsimaki P., Lounatmaa K. Vinblastine‐induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze‐fracture electron microscopy. Eur. J. Cell Biol. 1982;27:298–301. [PubMed] [Google Scholar]
- Holthuis J.C., Nichols B.J., Dhruvakumar S., Pelham H.R.B. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M.U., Veenhuis M., Klionsky D.J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin‐like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Casanova J.E. Turning on ARF: The Sec7 family of guanine‐nucleotide‐exchange factors. Trends Cell. Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.D., Sturgill‐Koszycki S., Swanson M.S. Evidence that Dot‐dependent and ‐independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell. Microbiol. 2001;3:99–114. doi: 10.1046/j.1462-5822.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani‐Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form‐II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kagan J.C., Roy C.R. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- Kaiser C.A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Odorizzi G., Emr S.D. Receptor downregulation and multivesicular‐body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kim J., Scott S.V., Oda M.N., Klionsky D.J. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Dalton V.M., Eggerton K.P., Scott S.V., Klionsky D.J. Apg7p/Cvt2p is required for the cytoplasm‐to‐vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.‐P., Klionsky D.J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P.E., Guan J., Hefner‐Gravink A., Baba M., Scott S.V., Ohsumi Y., Dunn W.A., Jr., Klionsky D.J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.‐P., Stromhaug P.E., Klionsky D.J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. The reversible modification regulates the membrane‐binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M.P., Jackson W.T. Cellular autophagy: Surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler J.T., Rostaing P., Schiavo G., Fritschy J.M., Olsen R., Triller A., Moss S.J. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABAA receptors. Mol. Cell. Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. “Autophagy.”. Landes Bioscience; Georgetown, TX: 2004. [Google Scholar]
- Klionsky D.J., Cregg J.M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. A unified nomenclature for yeast autophagy‐related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Kneussel M., Haverkamp S., Fuhrmann J.C., Wang H., Wassle H., Olsen R.W., Betz H. The g‐aminobutyric acid type A receptor (GABAAR)‐associated protein GABARAP interacts with gephyrin but is not involved in receptor anchoring at the synapse. Proc. Natl. Acad. Sci. USA. 2000;97:8594–8599. doi: 10.1073/pnas.97.15.8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Tanida I., Ueno T., Ohsumi M., Ohsumi Y., Kominami E. The C‐terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1‐E2 complex formation. J. Biol. Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K. Impairment of starvation‐induced and constitutive autophagy in Atg7‐deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A., Kanzawa T., Sawaya R., Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Kuma Y., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lang T., Schaeffeler E., Bernreuther D., Bredschneider M., Wolf D.H., Thumm M. Aut2p and Aut7p, two novel microtubule‐associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse‐Miller A., Sagiv Y., Glozman R., Elazar Z. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J. Biol. Chem. 2000;275:32966–32973. doi: 10.1074/jbc.M000917200. [DOI] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: Autophagy‐related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. Development by self‐digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liou W., Geuze H.J., Geelen M.J., Slot J.W. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M., Sykes A.K. The role of the Golgi complex in the isolation and digestion of organelles. Tissue Cell. 1975;7:143–158. doi: 10.1016/s0040-8166(75)80012-7. [DOI] [PubMed] [Google Scholar]
- Luiken J.J., van den Berg M., Heikoop J.C., Meijer A.J. Autophagic degradation of peroxisomes in isolated rat hepatocytes. FEBS Lett. 1992;304:93–97. doi: 10.1016/0014-5793(92)80596-9. [DOI] [PubMed] [Google Scholar]
- Lum J.J., Bauer D.E., Kong M., Harris M.H., Li C., Lindsten T., Thompson C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Majeski A.E., Dice J.F. Mechanisms of chaperone‐mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Mann S.S., Hammarback J.A. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- Meiling‐Wesse K., Epple U.D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E.‐L., Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5‐deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD‐repeat protein, targets to the autophagic isolation membrane with the Apg12‐Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., Kiel J.A.K.W., Krikken A.M., Komduur J.A., Veenhuis M., van der Klei I.J. The Hansenula polymorpha ATG25 gene encodes a novel coiled‐coil protein that is required for macropexophagy. Autophagy. 2005;1:92–100. doi: 10.4161/auto.1.2.1832. [DOI] [PubMed] [Google Scholar]
- Monastyrska I., van der Heide M., Krikken A.M., Kiel J.A., van der Klei I.J., Veenhuis M. Atg8 is essential for macropexophagy in Hansenula polymorpha. Traffic. 2005;6:66–74. doi: 10.1111/j.1600-0854.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Baba M., Osumi M., Aoyagi S., Kato N., Ohsumi Y., Sakai Y. Modification of a ubiquitin‐like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell. 2004;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara A., Mizushima N., Yamamoto A., Kabeya Y., Ohsumi Y., Yoshimori T. SKD1 AAA ATPase‐dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 2002;27:29–37. doi: 10.1247/csf.27.29. [DOI] [PubMed] [Google Scholar]
- Nazarko T.Y., Huang J., Nicaud J.M., Klionsky D.J., Sibirny A.A. Early secretory pathway gene TRS85 is required for selective macroautophagy of peroxisomes in Yarrowia lipolytica. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice D.C., Sato T.K., Stromhaug P.E., Emr S.D., Klionsky D.J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to PtdIns(3)P at the pre‐autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.‐P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D.J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Suzuki K., Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post‐translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y., Mizushima N. Two ubiquitin‐like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Oledzka‐Slotwinska H., Desmet V. Participation of the cell membrane in the formation of “autophagic vacuoles.”. Virchows Arch. 1969;2:47–61. [PubMed] [Google Scholar]
- Otto G.P., Wu M.Y., Clarke M., Lu H., Anderson O.R., Hilbi H., Shuman H.A., Kessin R.H. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol. Microbiol. 2004;51:63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a‐encoded subunits of the arterivirus replicase induce endoplasmic reticulum‐derived double‐membrane vesicles which carry the viral replication complex. J. Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro‐Cerda J., Meresse S., Parton R.G., van der Goot G., Sola‐Landa A., Lopez‐Goni I., Moreno E., Gorvel J.P. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro‐Cerda J., Moreno E., Sanguedolce V., Mege J.L., Gorvel J.P. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome‐like compartments. Infect. Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M., Salin B., Schaeffer J., Vallette F.M., di Rago J.P., Martinou J.C. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;17:415–422. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., Rapoport T.A. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progulske‐Fox A., Kozarov E., Dorn B., Dunn W., Jr., Burks J., Wu Y. Porphyromonas gingivalis virulence factors and invasion of cells of the cardiovascular system. J. Periodontal Res. 1999;34:393–399. doi: 10.1111/j.1600-0765.1999.tb02272.x. [DOI] [PubMed] [Google Scholar]
- Punnonen E.‐L., Reunanen H. Effects of vinblastine, leucine, and histidine, and 3‐methyladenine on autophagy in Ehrlich ascites cells. Exp. Mol. Pathol. 1990;52:87–97. doi: 10.1016/0014-4800(90)90061-h. [DOI] [PubMed] [Google Scholar]
- Punnonen E.‐L., Pihakaski K., Mattila K., Lounatmaa K., Hirsimaki P. Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver. Cell Tissue Res. 1989;258:269–276. doi: 10.1007/BF00239447. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Strous G.J., Crapo J.D., Geuze H.J., Slot J.W. The differential degradation of two cytosolic proteins as a tool to monitor autophagy in hepatocytes by immunocytochemistry. J. Cell Biol. 1993;120:897–908. doi: 10.1083/jcb.120.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Acevedo‐Arozena A., Imarisio S., Berger Z., Vacher C., O'Kane C.J., Brown S.D., Rubinsztein D.C. Dynein mutations impair autophagic clearance of aggregate‐prone proteins. Nat. Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C.‐W., Stromhaug P.E., Shintani T., Klionsky D.J. Vps51 is part of the yeast Vps fifty‐three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K.A., Stromhaug P.E., Klionsky D.J. The Atg1‐Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre‐autophagosomal structure. Dev. Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.‐W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. Autophagosomes: Biogenesis from scratch? Curr. Opin. Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D.J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D.J. Atg9 cycles between mitochondria and the pre‐autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reunanen H., Punnonen E.‐L., Hirsimaki P. Studies on vinblastine‐induced autophagocytosis in mouse liver. V. A cytochemical study on the origin of membranes. Histochemistry. 1985;83:513–517. doi: 10.1007/BF00492453. [DOI] [PubMed] [Google Scholar]
- Reunanen H., Hirsimaki P., Punnonen E.‐L. Cytochemical studies on induced autophagocytosis in mouse exocrine pancreas. Comp. Biochem. Physiol. A. 1988;90:321–327. doi: 10.1016/0300-9629(88)91123-1. [DOI] [PubMed] [Google Scholar]
- Rich K.A., Burkett C., Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 2003;6:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Enriquez S., Kim I., Currin R.T., Lemasters J.J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos‐Mattjus P., Sistonen L. The ubiquitin‐proteasome pathway. Ann. Med. 2004;36:285–295. doi: 10.1080/07853890310016324. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., DiFiglia M., Heintz N., Nixon R.A., Qin Z.‐H., Ravikumar B., Stefanis L., Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Schieltz D., Yates J.R., III, Ferro‐Novick S. Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., Pypaert M., Ferro‐Novick S. TRAPP I implicated in the specificity of tethering in ER‐to‐Golgi transport. Mol. Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- Sagiv Y., Legesse‐Miller A., Porat A., Elazar Z. GATE‐16, a membrane transport modulator, interacts with NSF and the Golgi v‐SNARE GOS‐28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz‐Shouval R., Sagiv Y., Shorer H., Elazar Z. The COOH terminus of GATE‐16, an intra‐Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J. Biol. Chem. 2003;278:14053–14058. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Baba M., Ohsumi Y., Klionsky D.J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell. Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Schiller J.J., Kanjanahaluethai A., Baker S.C., Oh J.W., Lai M.M. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo‐synthesized viral RNA in infected cells. J. Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. Autophagy in health and disease: A double‐edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.‐P., Stromhaug P.E., Klionsky D.J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama K., Noda T., Ohsumi Y. Mutational analysis of Csc1/Vps4p: Involvement of endosome in regulation of autophagy in yeast. Cell. Struct. Funct. 1997;22:501–509. doi: 10.1247/csf.22.501. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., Pelham H.R.B. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P.E., Berg T.O., Fengsrud M., Seglen P.O. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P.E., Bevan A., Dunn W.A., Jr. GSA11 encodes a unique 208‐kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 2001;276:42422–42435. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- Sturgill‐Koszycki S., Swanson M.S. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 2000;192:1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: An autophagy‐like origin for virus‐induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre‐autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S., Isberg R.R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H.F., Murk J.L., Braakman I., Geuze H.J. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 2003;4:512–518. doi: 10.1034/j.1600-0854.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- Tam Y.Y., Fagarasanu A., Fagarasanu M., Rachubinski R.A. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E.‐L., Hartmann D., Lullmann‐Rauch R., Janssen P.M., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP‐2‐deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida‐Miyake E., Ueno T., Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein‐activating enzyme for multiple substrates including human Apg12p, GATE‐16, GABARAP, and MAP‐LC3. J. Biol. Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Tanida‐Miyake E., Komatsu M., Ueno T., Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE‐16, GABARAP, and MAP‐LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tilney L.G., Harb O.S., Connelly P.S., Robinson C.G., Roy C.R. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: Implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 2001;114:4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- Towns R., Kabeya Y., Yoshimori T., Guo C., Shangguan Y., Hong S., Kaplan M., Klionsky D.J., Wiley J.W. Sera from patients with type 2 diabetes and neuropathy induce autophagy and colocalization with mitochondria in SY5Y cells. Autophagy. 2005;1:163–170. doi: 10.4161/auto.1.3.2068. [DOI] [PubMed] [Google Scholar]
- van der Meer Y., van Tol H., Locker J.K., Snijder E.J. ORF1a‐encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.‐W., Stromhaug P.E., Kauffman E.J., Weisman L.S., Klionsky D.J. Yeast homotypic vacuole fusion requires the Ccz1‐Mon1 complex during the tethering/docking stage. J. Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Bedford F.K., Brandon N.J., Moss S.J., Olsen R.W. GABAA‐receptor‐associated protein links GABAA receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- Webb J.L., Ravikumar B., Rubinsztein D.C. Microtubule disruption inhibits autophagosome‐lysosome fusion: Implications for studying the roles of aggresomes in polyglutamine diseases. Int. J. Biochem. Cell Biol. 2004;36:2541–2550. doi: 10.1016/j.biocel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Yamada T., Carson A.R., Caniggia I., Umebayashi K., Yoshimori T., Nakabayashi K., Scherer S.W. Endothelial nitric‐oxide synthase antisense (NOS3AS) gene encodes an autophagy‐related protein (APG9‐like2) highly expressed in trophoblast. J. Biol. Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Masaki R., Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J. Histochem. Cytochem. 1990;38:573–580. doi: 10.1177/38.4.2319125. [DOI] [PubMed] [Google Scholar]
- Yang D.M., Chiang A.S. Formation of a whorl‐like autophagosome by Golgi apparatus engulfing a ribosome‐containing vacuole in corpora allata of the cockroach Diploptera punctata. Cell Tissue Res. 1997;287:385–391. doi: 10.1007/s004410050764. [DOI] [PubMed] [Google Scholar]
- Yokota S. Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate. Eur. J. Cell Biol. 1993;61:67–80. [PubMed] [Google Scholar]
- Yokota S., Himeno M., Kato K. Degradation of excess peroxisomes by cellular autophagy: Immuno‐cytochemical and biochemical analysis. Acta. Histochem. Cytoc. 1994;27:573–579. [Google Scholar]
- Yorimitsu T., Klionsky D.J. Atg11 links cargo to the vesicle‐forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;4:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T., Yamagata F., Yamamoto A., Mizushima N., Kabeya Y., Nara A., Miwako I., Ohashi M., Ohsumi M., Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]


