Abstract
A new world of possibilities for “virus discovery” was opened up with high-throughput sequencing becoming available in the last decade. While scientifically metagenomic analysis was established before the start of the era of high-throughput sequencing, the availability of the first second-generation sequencers was the kick-off for diagnosticians to use sequencing for the detection of novel pathogens. Today, diagnostic metagenomics is becoming the standard procedure for the detection and genetic characterization of new viruses or novel virus variants. Here, we provide an overview about technical considerations of high-throughput sequencing-based diagnostic metagenomics together with selected examples of “virus discovery” for animal diseases or zoonoses and metagenomics for food safety or basic veterinary research.
Keywords: Virus discovery, Vriome, Diagnostic metagenomics, Second-generation sequencing, Technical considerations, Veterinary virology
1. Introduction
Since nucleic acids are the general genetic material of all organisms and viruses, shotgun sequencing is the most generic approach for the detection of any pathogen. Hence, it is logical to apply the different high-throughput sequencing technologies for the detection of novel or unexpected viruses. Therefore, already with the advent of the first commercially available second-generation sequencing platforms, diagnosticians and researchers started evaluating and optimizing the detection of pathogens by sequencing. Likewise, researchers interested in exploring the full diversity of the virome started applying shotgun high-throughput sequencing for the analysis of diverse samples to identify novel viruses and virus variants.
Expanding our knowledge of the virosphere is not only scientifically interesting but also important for disease detection and control. As shown later in selected examples, various diseases of unknown etiology are caused by pathogens that were recently detected by novel technologies, namely, by microarrays or high-throughput sequencing. Regardless of the platform or the specific sample selection and preprocessing, an unambiguous association of the detected pathogen with the observed disease requires the fulfillment of Koch's postulates as he formulated them in a talk (Redactions Comité, 1891). Due to the problems that may arise in fulfilling these, a number of amendments of Koch's postulates have been suggested (Mokili et al., 2012). Nevertheless, still the stringent criteria formulated in the late 19th century by Koch and coworkers are the golden standard. In some of the examples provided in a later section of the article and discussed in detail in the following chapters, Koch's postulates were fulfilled, showing that in veterinary science this important proof can be achieved.
Another advantage of metagenomics for diagnostics is the possibility to detect coinfections. This may be of rising importance as we progress with detecting new agents that may infect the host without causing clinical signs of illness in a certain host without a coinfecting agent but cause serious or varying symptoms when coming together with different other facultative pathogens, as for instance analyzed by Blomström et al. (2016), Zhang et al. (2014), or Hanke et al. (2017). In case of coinfections, Koch's postulates might be hard to fulfill if not even impossible. Nevertheless, selecting individuals with the same symptoms and from epidemiologically related vs unrelated cases may help gather evidence of the coinfecting agents to be the cause of the observed disease.
2. Technical Considerations of Diagnostic Metagenomics
2.1. The Right Sample
The choice of the samples is of utmost importance for successful virus detection. Even though a systematic assessment of the impact of the sample so far has not been conducted, published data imply this (Hoffmann et al., 2015, Pfaff et al., 2017a). Moreover, it is logical that due to differing organ tropism of different pathogens different organs will be positive for pathogen nucleic acids. Noteworthy to mention that also the time point of sampling is crucial. Looking at the studies of Wernike and colleagues (Bilk et al., 2012, Hoffmann et al., 2012, Wernike et al., 2012, Wernike et al., 2013) dealing with Schmallenberg virus (SBV), it is evident that owing to the short viremia sampling will yield genome copies only in a relatively narrow period and hence allow virus detection.
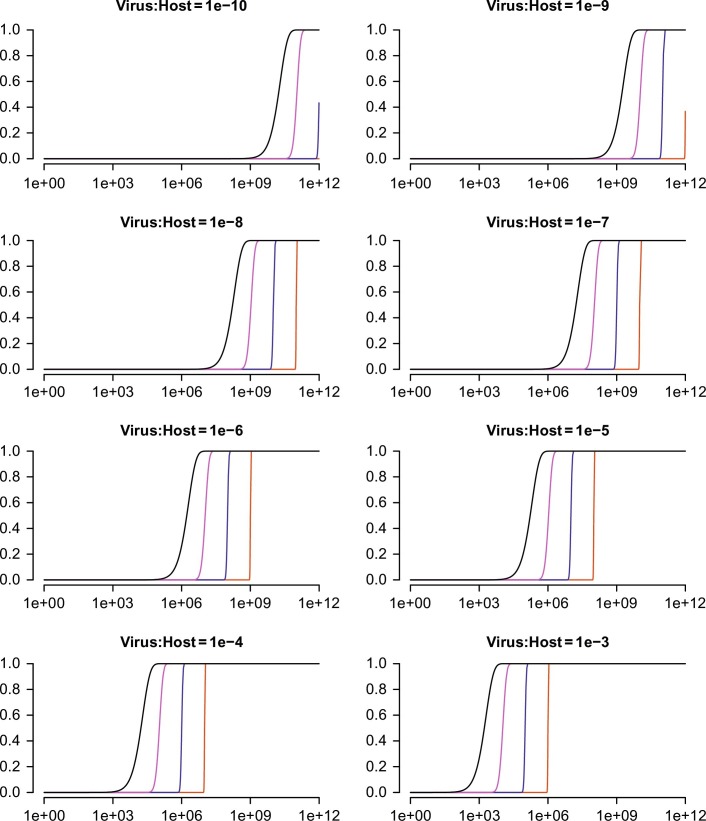
Fig. 1 displays the impact of the pathogen content on detection probability. The plots on the one hand illustrate that at a certain pathogen load it is nearly impossible to fail with detection. On the other hand, Fig. 1 also shows that if the pathogen content is too low, astronomic datasets may be necessary for the detection. However, it is logical that the larger and more complex the dataset gets, the longer the data analysis to find hints at the potential causative agent takes. Although substantial efforts have been put into optimizing sample preprocessing for pathogen detection, the intention of all these is to bias the sequencing (Briese et al., 2015, Conceicao-Neto et al., 2015, Kohl et al., 2015). However, this bias naturally comes together with information loss due to the selection of only a fraction of the nucleic acids, and this loss can render the analysis useless if the necessary information is lost.
Fig. 1.
Theoretical probabilities for pathogen detection. The graphs display the probabilities (plotted on the ordinate) to detect at least one read (plotted black), at least 10 reads (magenta), at least 100 reads (blue), or at least 1000 reads (red) representing a pathogen present in a sample with a certain pathogen: host ratio (plotted as the title of each graph) in a dataset with a certain size (plotted on the abscissa).
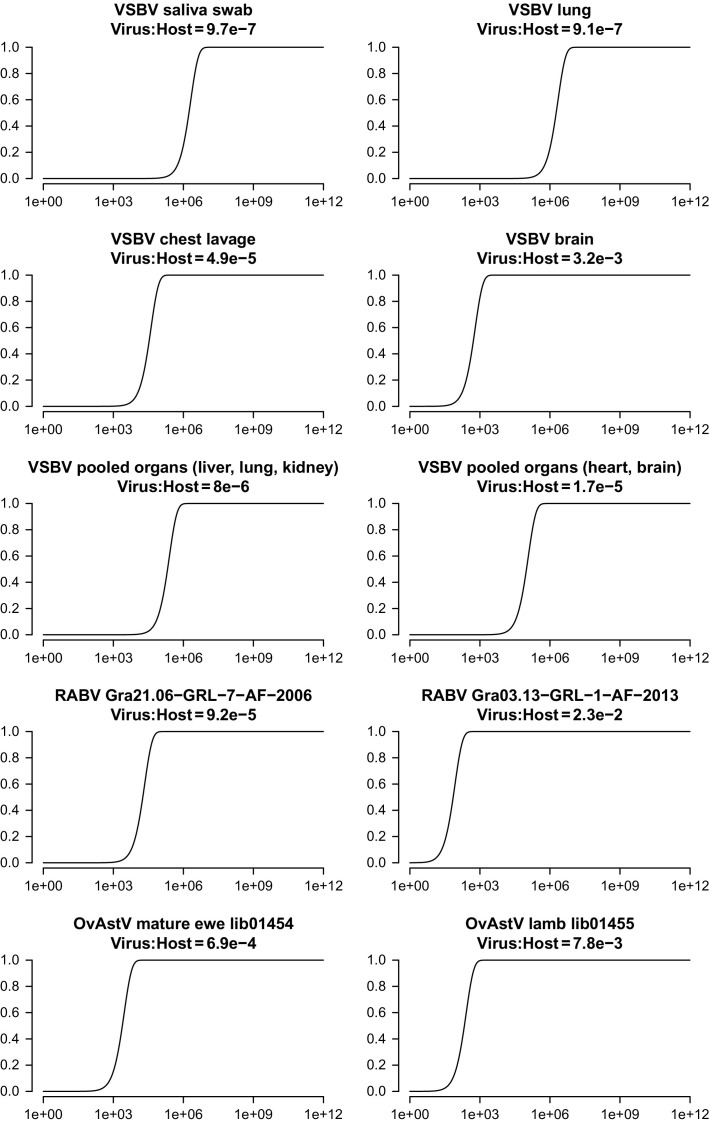
As a consequence, the applied preprocessing procedures often sacrifice the generic character of the shotgun sequencing approach. Instead, selection of a more suitable sample can solve the problem of the detection limits. As shown in Fig. 2 for the variegated squirrel bornavirus 1 (VSBV-1), the probability of detection of a certain number of viral reads in different samples from the same individual greatly varies. This variation clearly is a result of the organ tropism of the virus in quest. Therefore, analyzing a variety of sample materials might be necessary for success. As also shown in Fig. 2, samples from different animals infected with the same viral species may have a greatly varying pathogen content, as depicted for a pair of rabies virus (RABV) infected animals (data taken from Hanke et al., 2016). This is to a lesser extent also true for a pair of samples from cases of ovine astrovirus (OvAstV) infected animals (data taken from Pfaff et al., 2017a).
Fig. 2.
Example probabilities for pathogen detection. Observed virus: host ratios taken from published studies in which pathogens were detected by shotgun metagenomics. The graphs display the probabilities (plotted on the ordinate) to detect at least one read representing a pathogen present in a sample with a certain pathogen: host ratio (plotted as the title of each graph) in a dataset with a certain size (plotted on the abscissa). VSBV, variegated squirrel bornavirus, data taken from Hoffmann et al. (2015); RABV, Rabies virus, data taken from Hanke et al. (2016); OvAstV, ovine astrovirus, data taken from Pfaff et al. (2017a).
2.2. Pooling
In some cases, pooling of samples might appear useful to reduce the expenses per sample. However, calculating the probabilities for detection of VSBV-1 in pooled samples shows (Fig. 2) that the sequencing effort increases, at least when negative samples or samples with a low virus content are part of the pool. As shown, eventually the expenses can be reduced without significantly reducing the probability of detection if a sample with sufficiently high pathogen content is part of the sample pool. However, when working with pooled samples and aiming for complete genomes, individual positive samples have to be identified after detection of the virus, and individual libraries and sequencing have to be conducted afterward.
2.3. DNA or RNA?
Another issue that has to be considered carefully is the choice of the nucleic acids to sequence, DNA or RNA. Possible criteria to take into account for this decision are at first of course the expected type of virus, i.e., DNA or RNA virus. Next, if dealing with samples where virus replication can be expected, preferably RNA should be chosen since this will enable the detection of both DNA and RNA viruses. On the contrary, choosing DNA will make the detection of RNA viruses fundamentally impossible. In case of samples in which no viral replication can be expected, for instance feces or environmental samples, DNA and RNA must be analyzed and should preferably be processed separately (also see Section 2.2).
2.4. Considerations for Sequencing Platform Selection
Despite the fact that some HTS platforms are no longer available, there is still a considerable range of technologies and instruments from which researchers and diagnosticians can select. All of these have their specific pros and cons and all proved to be more or less suitable for pathogen detection. We will not evaluate the different platforms here nor will we argue for the use of a specific platform, rather we will only briefly discuss a number of characteristics to consider for platform selection. Inter alia, characteristics influencing the choice of the most suitable sequencing platform are the read length, which is a major determinant of the reliable taxonomic classification, the runtime to complete an analysis, the possible size of the datasets, or the overall cost for an analysis.
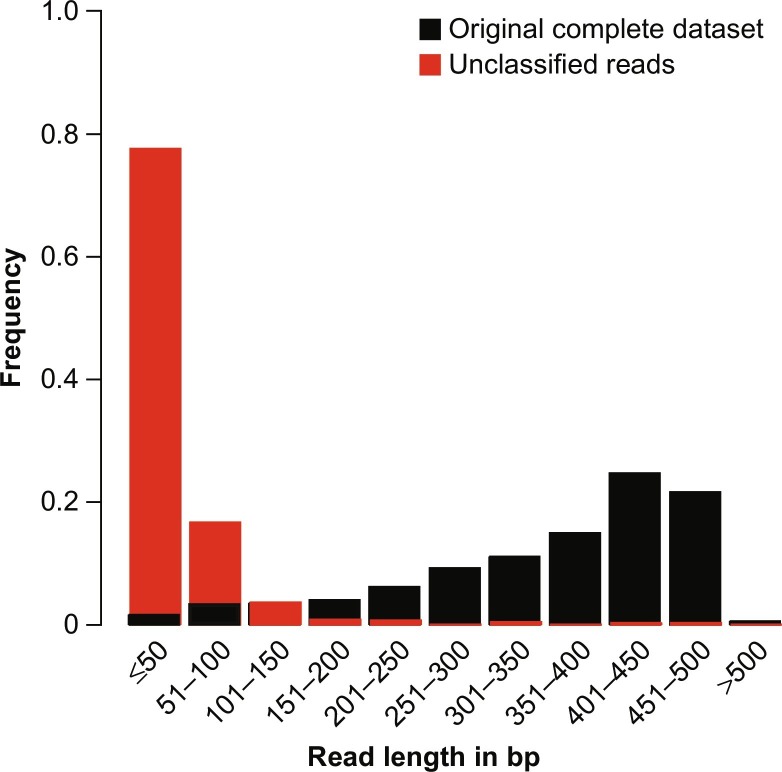
Fig. 3 and Table 1 show an example for the impact of read length on the sensitivity of detection. The original dataset (from Hoffmann et al., 2012) and the same dataset with shortened reads were analyzed using the software pipeline RIEMS (Scheuch et al., 2015) with identical settings and databases. It is clearly visible that longer reads are more readily classified than shorter ones. On the contrary, to a certain extent errors can be tolerated and do not influence a sensitive reliable classification (Scheuch et al., 2015) since in case of diagnostic metagenomics not exact genotyping but classification at higher taxonomic ranks is the aim. This tolerance depends on the software workflow in use (Scheuch et al., 2015). Of course, if the error rate rises too high, no sufficient sequence identity will be detectable and hence no classification possible. Finally, it has to be concluded that there is a trade-off between read length and error rates that can vary depending on the availability of reference sequences with sufficient similarity enabling recognition of the pathogen.
Fig. 3.
Read length dependency of sensitive classification. The figure displays two overlaid histograms of the length distributions of the complete Schmallenberg virus dataset (black; Hoffmann et al., 2012) and those reads for which no sequence with significant identity could be detected (red) using an early version of the RIEMS analysis pipeline (Scheuch et al., 2015).
Table 1.
Main Figures of the Classification Efficiency Depending on Read Length Using the Dataset From the Schmallenberg Virus Identification (Hoffmann et al., 2012)
| Original Read Length | Shortened Reads | |
|---|---|---|
| Mean read length | 315 | 96 |
| Total no. reads | 27,420 | 27,420 |
| Classified reads | 26,128 | 25,310 |
| Orthobunyavirus hits | 7 | 1 |
| Identity of the orthobunyavirus reads with subject sequence | 68.4%–95.6% | 98.7% |
| Unclassified reads | 1292 | 2110 |
Data were analyzed using an early version of the RIEMS analysis pipeline (Scheuch et al., 2015) with identical settings and databases.
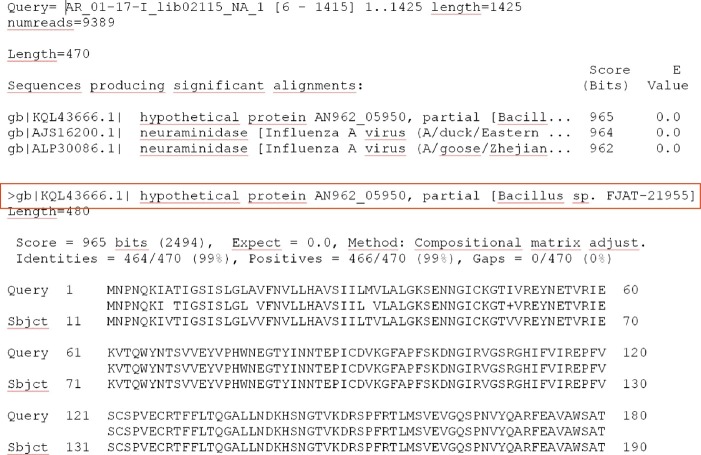
Of utmost importance are the potential inherent sources of contamination like carryover between runs or missorting of/mislabeling with molecular barcodes due to impurity of the barcoded adapters (Sigma Aldrich, 2017) used for deconvolution of the datasets. Such missorting is the likely reason for errors seen in the INSDC databases. For instance, Mukherjee et al. (2015) reported on integration of the PhiX phage into bacterial genomes of diverse families, even those that had never before been associated with PhiX. The authors report that 10% of the contaminated genomes had even been published in literature. By thorough analysis, they found out that the integration of the phage genome into the bacterial genomes was by missorting of the raw data, which included the used sequencing control sample into all datasets. Fig. 4 depicts another example of probable missorting and its result in the generated sequence. In this case, the Influenza A neuraminidase gene was assembled into a genome sequence of a bacterium taxonomically classified as Bacillus sp. In the respective genome assembly, also the Influenza A virus Hemagglutinin gene was included (not shown). These two examples of the incorporation of foreign sequences are most likely also a result of improper data deconvolution. Moreover, these examples show that care must be taken when analyzing datasets, since such errors will always occur and need manual inspection and correction of the database content by trained data curators.
Fig. 4.
Example for a result of improper data deconvolution and review. An Influenza A virus neuraminidase gene included in the whole-genome shotgun assembly of a Bacillus sp. An Influenza A H5N8 neuraminidase amino acid sequence was searched using blastp (Camacho et al., 2009) with default settings in the NCBI nr database.
2.5. Read Classification
After sample selection, processing, and sequencing, with data analysis another important part of the detection process starts. Since the huge datasets that are generated by shotgun metagenomics may comprise mostly host sequences masking the relevant information, sensitive and specific workflows for classification of the obtained reads are urgently needed. A major pitfall of data analysis is the fact that if dealing with a novel pathogen, no suitable reference sequence allowing its easy recognition may be available. Hence, although generally seen as a method that works without a priori knowledge because the starting point of the complete procedure can be true shotgun sequencing, regardless of the chosen type of nucleic acid, DNA or RNA, metagenomics heavily relies on a priori knowledge. Until we overcome the necessity that initial identification of the pathogen is based on similarity of the sequencing reads with known sequences available in the INSDC databases, we will not have a system that is independent of a priori knowledge and hence truly unbiased. This stresses the need for algorithms that can determine the source of a sequence with sufficient reliability solely based on low-level characteristics without calculating sequence similarities. On the one hand, novel optimized procedures like for instance Kraken (Wood and Salzberg, 2014) or Diamond (Buchfink et al., 2015) have the potential to speed up the classification. On the other hand, if it is possible to determine characteristics for classes of genomes (classified by higher orders like for instance a superkingdom), e.g., composition rather than sequence-based characteristics, this may help increase the sensitivity of detection. Approaches for classification of sequences that do not rely on calculation of alignments but are based on oligonucleotide frequencies have been proposed and also shown to be suitable for classification (A. Belka et al., unpublished; Diaz et al., 2009, Gregor et al., 2016, Leung et al., 2011, McHardy et al., 2007). The mentioned approaches let it seem likely that it will in the near future be possible to improve the detection of novel viruses without a suitable known reference. Furthermore, the increasing number of sequencing projects results in a fast growing number of sequences from novel virus species, genera, or even families. This new data sets continuously improve the analysis and detection rate for new pathogens.
2.6. Data Quality
Another important issue that is frequently stressed (Byrd et al., 2014, Wood and Salzberg, 2014) is the quality of the sequences used for building the reference datasets for classification. As pointed out in Section 2.4, this is at the time of writing a problem. This problem may even be increasing due to the increasing democratization of sequencing and the consequent increase of sequencing efforts. Therefore, it is urgently necessary to have (more) trained personnel for database curation to ensure the quality and reliability of the nucleotide sequence database content.
2.7. Sensitivity and Specificity of Diagnostic Metagenomics
Even after some years of continuous use, it is still not reliably possible to establish the sensitivity and specificity of metagenomic pathogen detection. With regard to the specificity, it is noteworthy to mention that with a truly unbiased sample preparation and sequencing protocol, only the final step of the identification procedure, i.e., the algorithms and databases used for taxonomic sequence classification, determines the specificity. On the contrary, all protocols including sample preprocessing will strongly influence the specificity of the overall process. All these procedures in the worst case will render detection of pathogens that are not in the focus of the preprocessing procedure nearly impossible. Nevertheless, due to common and divergent characteristics of groups of pathogens, the specificity of the preprocessing protocols cannot be assessed.
At the same time, sample processing and data analysis have a strong impact on the sensitivity. As an example, searching for an unknown pathogen but only using a viral sequence database for data analysis will cause the sensitivity for the detection of bacterial pathogens drop to zero, and vice versa. The same applies for novel pathogens for which no suitable reference sequence is available. In these cases, the sensitivity may also drop to zero when no suitable alternative criteria can be applied to determine the pathogen. Likewise, even in case of the combination of unbiased sequencing with unbiased data analysis, the algorithm at least in part determines the sensitivity (see for instance (Scheuch et al., 2015) for a comparison of the sensitivities of different software). In summary, a general assessment of the sensitivity and specificity of high-throughput sequencing-based pathogen detection is not possible.
Taken together, in approximately 10 years of high-throughput sequencing-based diagnostic metagenomics, mostly driven by second-generation sequencing, a substantial number of viruses has been detected and genomically characterized, thereby proving the suitability of sequencing approaches for diagnostics.
3. Recent Examples in “Virus Discovery” for Animal Diseases and Zoonoses
In former times, the discovery of new viruses or novel virus variants happened mainly as the consequence of isolation on cell culture, embryonated chicken eggs, or in animal models. Virus growth was detected due to disease development, cytopathic effects, and the use of broad diagnostics like electron microscopy or antisera for neutralization or staining. Detection of completely new viruses was often by accident only, depending, e.g., on the possibility to grow the virus. Today, “virus discovery” is driven by the new developments in molecular diagnostics, mainly broad PCRs, microarrays, and new sequencing technologies. Especially for infectious animal diseases or zoonoses, and in the field of virus reservoirs, like viruses transmitted from bats or voles, tremendous progress in the detection and characterization of new viruses was achieved.
A growing number of examples exists now where viruses were detected in samples from diseased animals or in animal reservoirs. Table 2 provides a short overview of some prominent examples. It is important to mention that in several of the listed cases, the experimental confirmation of the causation was possible, and the conclusions were not based exclusively on the sequence information. Various important examples are presented later in much more detail in this issue.
Table 2.
Overview of Recent Examples in Virus Discovery for Animal and Zoonotic Diseases Detected Using High-Throughput Sequencing Technologies
| Year | Newly Detected Virus | References |
|---|---|---|
| 2009 | Lujo virus/hemorrhagic fever-associated arenavirus | Briese et al. (2009) |
| 2010 | Shaking mink syndrome astovirus | Blomström et al. (2010) |
| 2011 | Hepacivirus from a dog | Kapoor et al. (2011) |
| 2012 | Middle East respiratory syndrome (MERS) | Zaki et al. (2012) |
| 2012 | Schmallenberg orthobunyavirus | Hoffmann et al. (2012) |
| 2012 | Bat Influenza A virus | Tong et al. (2012) |
| 2013 | Swine Influenza C virus | Hause et al. (2013) |
| 2013 | Rodent hepaciviruses | Drexler et al. (2013) |
| 2013 | Primate pegi- and hepaciviruses | Quan et al. (2013) |
| 2013/2017 | Ovine astroviruses | Li et al. (2013) and Pfaff et al. (2017a) |
| 2014 | Bokeloh bat lyssavirus | Nolden et al. (2014) |
| 2014/2016 | Bovine astroviruses | Bouzalas et al. (2014) and Schlottau et al. (2016) |
| 2015 | Bovine hepacivirus | Baechlein et al. (2015) |
| 2015 | Novel tick-borne flavivirus subtype (louping-ill-related) in goats | Mansfield et al. (2015) |
| 2015 | Bluetongue virus serotype 27 | Jenckel et al. (2015) |
| 2015 | Variegated squirrel bornavirus 1 | Hoffmann et al. (2015) |
| 2015 | Atypical porcine pestivirus | Hause et al. (2015) |
| 2016 | Porcine pegivirus | Baechlein et al. (2016) |
| 2017 | Penguin alphaherpesvirus | Pfaff et al. (2017b) |
| 2017 | Lateral-shaking inducing neuro degenerative agent (LINDA) | Lamp et al. (2017) |
One of the most prominent examples is the detection of SBV in serum samples from cattle showing rather unspecific signs of illness in 2011 in Germany. The impact of SBV was huge since it spread all over Europe within only a few years due to the naïve population of ruminants. Metagenomics allowed the detection and whole-genome characterization of virus from acutely infected animals before the further spread and also before malformed offspring was born in 2012. Furthermore, the sequence information from sequencing enabled the fast development of specific molecular diagnostics. SBV and the current situation is reviewed in a later chapter of this issue.
Further examples include also the detection of diverse astroviruses from various ruminant species, especially sheep and cattle. Most importantly, these cases were all associated with neurological disease. Before, astrovirus-induced encephalitis was only reported for very few human cases. This additional and new knowledge about astroviruses and their impact on differential diagnostics of encephalitis cases is also presented in detail in the chapter “The expanding field of mammalian astroviruses: opportunities and challenges in clinical virology” by Boujon et al.
Another field in veterinary virology where metagenomics provided completely new insights and expanded our knowledge on animal diseases was the first detection of the so-called atypical porcine pestiviruses (APPV). Although initially not associated with clear symptoms and identified during the NGS-based screening of porcine samples in the United States, several groups could link APPV infection with the congenital tremor of newborn piglets. More details on the novel pestiviruses can be found in the chapter “New leaves in the growing tree of pestiviruses” by Blome et al.
With the same NGS-based analyses, the Orbivirus family was substantially expanded by a number of new serotypes of bluetongue viruses. The metagenomics-driven growth of the species BTV is shown in a dedicated chapter summarizing not only the detection but also the further characterization of isolated BTV in vitro and in vivo. A very similar “growth” happened in the genus Lyssavirus of the Rhabdoviridae family (see chapter about “Novel lyssaviruses” by Eggerbauer et al.).
Examples which are not further elaborated in this issue but which are of importance comprise, e.g., a novel Herpesvirus from Penguins (SpAHV). This report is a good example for the use of novel sequences to complete the phylogeny of a virus family and to provide data for a better understanding of the relationship of similar viruses from different animal species (Pfaff et al., 2017b). The same is true for all the novel members of the genus Hepacivirus with new complete genomes of viruses detected in dogs, horses, voles, rats, and cattle (Baechlein et al., 2015, Drexler et al., 2013, Kapoor et al., 2011, Quan et al., 2013). These sequences changed the picture of Hepaciviruses as a virus restricted to only humans into that of a huge virus group present in many animal species including humans. This is one of the most impressive examples how NGS-based metagenomics can change our view on important viruses within years or even a few months. The same is true for pegiviruses (Baechlein et al., 2016, Quan et al., 2013) and influenza A viruses.
Especially the novel bat influenza viruses H17N10 and H18N11 expanded even the well-studied influenza A viruses (Tong et al., 2012) in a new reservoir host. Bat influenza virus is in addition a perfect example for the de novo generation of a replicating virus by using only the sequence information for reverse genetics and recovery of infectious virus particles without the need of virus isolation from positive sample material (Moreira et al., 2016). High-quality whole genomes of viruses are therefore in many cases the basis of further studies like gene expression, construction of chimeric viruses, or generation of recombinant clones.
4. Metagenomics for Food Safety
Beside veterinary virology, food safety, which is closely linked to veterinary virology by, for instance, zoonotic pathogens, is another field for which diagnostic metagenomics can improve pathogen detection (Stasiewicz et al., 2015). Foodborne pathogenic bacteria like Salmonella, Listeria, or toxin-producing Escherichia coli strains, but also norovirus and Hepatitis A virus and parasites (Trichinella, Giardia) can cause disease outbreaks accompanied by vomiting and more or less severe diarrhea. For instance, a severe outbreak of gastroenteritis and the hemolytic–uremic syndrome caused by Shiga toxin-producing E. coli in Germany in 2011 mostly affecting adult women was likely caused be the consumption of sprouts (Frank et al., 2011). In another case, frozen strawberries caused a norovirus gastroenteritis outbreak in Germany in 2012. Samples from this outbreak analyzed by real-time RT-PCR assay revealed a combination of three different genotypes that had not been reported in Germany so far resulting in the suggestion that the strawberries were polluted from sewage rather than from a single infected food handler (Mäde et al., 2013).
Genomes of foodborne pathogens have been often sequenced for genomic characterization of strains of special interest and for comparative genomic studies, but food products were rather seldom handled and evaluated for metagenomics. To systematically evaluate the suitability of shotgun metagenomics for the assessment of the quality of animal-derived foods and foods in general, we conducted a pilot study in which we investigated conventional food samples of animal or plant origin. We analyzed various untreated and highly processed food sample using our in-house metagenomics sample processing workflow. The investigated sample matrices included rocket, mushrooms, ham, salmon, meat loaf, cheese, oat flakes pizza, chocolate, oysters, strawberries, tap water, and parasite-containing wild boar meat. The used workflow starts with RNA instead of DNA extraction. In general, we could obtain RNA from all tested food products. The resulting concentration and especially the quality of the RNA obtained from processed food samples were clearly lower since their RNA content and integrity is inherently very low. Special matrices like fatty (e.g., cheese) and fiber-containing foods (e.g., oat flakes) were challenging and possibly need sample-dependent pretreatments, like defatting. Nucleic acids of samples with a low pH value (e.g., frozen berries suspicious to be contaminated by norovirus) are difficult to isolate and should be pretreated with a neutralization step.
Nevertheless, we were able to detect viruses in different data sets resulting from both untreated and processed foods. In a rocket sample, we found a novel yellows virus (0.3% of reads) that was only about 91% identical (at nucleotide level) to yellows viruses known from turnip and brassica. Meat loaf is an example of processed food in which we were able to detect a well-known pepper mild mottle virus (0.1%) that might cause different symptoms like fever or pruritus (Colson et al., 2010). Interestingly, we discovered a novel mycovirus in untreated (mushrooms) and processed samples (pizza with mushrooms). This virus is only 35% similar (at amino acid level) to known mycoviruses; however, mycovirus sequences found in both mushroom samples are nearly identical to each other. In the examples mentioned here, the detected viruses are RNA viruses, which prove our RNA-based strategy to be very successful in shedding light on the virosphere. In addition, parasite-specific sequences matching the expected pathogens could be detected in wild boar meat containing parasites (trichina 0.02% and liver fluke 0.000002%). Thus, the used protocols seem to be applicable over a wide range of foodborne pathogens and food categories. Routinely applied for foodborne pathogens, resulting metagenomics data can be helpful for surveillance as well as foodborne outbreak investigation and will improve the hazard identification by increased specificity and potentially by a fundamental change in the definition of the hazard being rather a specific virulent strain, subtype, or gene instead of a not well-specified species.
5. Metagenomics for Basic Research in (Veterinary) Virology
While the aforementioned examples illustrate the impact of high-throughput sequencing on veterinary diagnostics and the closely connected field of food safety, there are of course diverse applications beyond. Whereas anthropologists already successfully analyzed ancient samples immediately after the early second-generation sequencers became available (Green et al., 2006, Green et al., 2010, Maricic and Pääbo, 2009, Noonan et al., 2006), researchers only recently started using shotgun high-throughput sequencing to systematically assess the pathogen content of historic samples available in museums or other collections and ancient DNA from archaeologic specimens. Systematic investigation of historic samples will not only help discover the diversity of the virome but will essentially contribute to the elucidation of viral evolution and the potential impact of vaccination on the evolving virome, for instance. At the moment, however, no reports on the analysis of historic animal samples have been published, but the studies of historic DNA samples are focused to human samples. For instance, in a recently published paper (Feldman et al., 2016), the authors describe the analysis of an ancient Yersinia pestis genome. The DNA was recovered from a 6th-century skeleton, a putative victim of the Justinianic plague, found in a southern German grave. The authors were able to assemble a complete genome with high coverage. Their sequence analysis revealed a number of unique variants and some structural differences compared to the previously available Y. pestis sequences. In another report, Duggan et al. (2016) sequenced a complete variola virus genome from a 17th century child mummy from Lithuania. The authors were able to reconstruct the complete viral genome, which was found to be basal to all strains from the 20th century. The authors concluded that much of variola virus evolution and diversification occurred recently driven by the impact of vaccination. Another study published by Pajer et al. (2017) shed further light on the long-term evolution of variola virus. In their study, they report on the sequencing and analysis of two complete variola virus genomes from historic human specimens from a museum in Prague. Together these two studies provide a new level of insight into long-term virus evolution.
This impressively demonstrates the power of high-throughput sequencing for the investigation of the evolutionary history of viruses. Another example for the possibilities that high-throughput sequencing opens in this field is the work published by Toppinen et al. (2015). They analyzed DNA obtained from bones of putative Finnish casualties from World War II for parvovirus B19 DNA. Interestingly, they found only viral genotypes that disappeared from Northern Europe in the 1970s or had never previously been reported to be found there. Moreover, using molecular clock analysis the authors were able to date the most recent common ancestors for all sequenced viruses back to the early 19th century. Noteworthy to mention that in addition to virus analysis, the obtained sequences also enabled tracing the origin of the casualties. The authors were able to show that one of the casualties was most likely of Russian origin. This highlights an additional facet of genome analysis using shotgun high-throughput sequencing. A similar approach was used for in-depth spatio-temporal investigation of arctic rabies viruses and their reservoir hosts in Greenland (Hanke et al., 2016). The study explored virus dynamics over a period of roughly 10 years. The authors did not find a link between viral genotypes and the reservoir host population. All these studies together demonstrate the utility of shotgun high-throughput sequencing for investigations into virus dynamics and evolution.
6. Conclusions and Future Directions
For sensitive, reliable, and in the optimal case comparable results, a certain degree of standardization is necessary. This encompasses the procedures from sample to sequence and the subsequent analyses of the generated datasets for the detection of the viruses. An important determinant of success is the selection of suitable samples and the sampling and sample handling and storage conditions. To this end, awareness must be raised as to the preservation of samples that are intended for metagenomic analysis. A common preservation is formalin fixation followed by paraffin embedding. However, this minimizes the prospects of success for sequencing-based pathogen detection. Moreover, since the complete characterization of viruses and their unambiguous connection with the disease imperatively requires virus isolates, native material must be available. Therefore, it is necessary to preserve samples by other means than fixation and embedding. Possible procedures have to be evaluated for their suitability and subsequently need to be validated on a range of samples. Finally, this will help improve the complete procedure of diagnostic metagenomics.
The constantly growing number of available viral genome sequences will improve the chances of recognizing new pathogens. Moreover, the ongoing efforts for the implementation of algorithms to identify sequences of viral origin without sequences comparison will enable the identification of viruses even stronger deviating from those known.
Despite all these successes, further improvements and standardization are urgently necessary for routine use of shotgun metagenomics in diagnostics. With the ongoing dissemination of high-throughput sequencing in diagnostic laboratories, the need for quality control will rise. Therefore, in the near future it will be of utmost importance to establish platform-independent quality measures and ring trials for diagnostic laboratories that use high-throughput sequencing for pathogen detection in general and for virus detection in particular. To make high-throughput sequencing-based diagnostics as generic as its foundation is, the necessary procedures must be designed in a way to abolish the necessity of differentiation between viral, bacterial, and parasitic pathogens for reliable detection.
References
- Baechlein C., Fischer N., Grundhoff A., Alawi M., Indenbirken D., Postel A. Identification of a novel hepacivirus in domestic cattle from Germany. J. Virol. 2015;89(14):7007–7015. doi: 10.1128/JVI.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechlein C., Grundhoff A., Fischer N., Alawi M., Hoeltig D., Waldmann K.-H., Becher P. Pegivirus infection in domestic pigs, Germany. Emerg. Infect. Dis. 2016;22(7):1312–1314. doi: 10.3201/eid2207.160024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilk S., Schulze C., Fischer M., Beer M., Hlinak A., Hoffmann B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet. Microbiol. 2012;159(1–2):236–238. doi: 10.1016/j.vetmic.2012.03.035. [DOI] [PubMed] [Google Scholar]
- Blomström A.L., Fossum C., Wallgren P., Berg M. Viral metagenomic analysis displays the co-infection situation in healthy and pmws affected pigs. PLoS One. 2016;11(12):e0166863. doi: 10.1371/journal.pone.0166863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström A.-L., Widén F., Hammer A.-S., Belák S., Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010;48(12):4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzalas I.G., Wüthrich D., Walland J., Drögemüller C., Zurbriggen A., Vandevelde M. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J. Clin. Microbiol. 2014;52(9):3318–3324. doi: 10.1128/JCM.01195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Paweska J.T., McMullan L.K., Hutchison S.K., Street C., Palacios G. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., Lipkin W.I. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6(5):e01491–15. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Byrd A.L., Perez-Rogers J.F., Manimaran S., Castro-Nallar E., Toma I., McCaffrey T. Clinical PathoScope. Rapid alignment and filtration for accurate pathogen identification in clinical samples using unassembled sequencing data. BMC Bioinformatics. 2014;15:262. doi: 10.1186/1471-2105-15-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST +. Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Richet H., Desnues C., Balique F., Moal V., Grob J.-J. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao-Neto N., Zeller M., Lefrere H., de Bruyn P., Beller L., Deboutte W. Modular approach to customise sample preparation procedures for viral metagenomics. A reproducible protocol for virome analysis. Sci. Rep. 2015;5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz N.N., Krause L., Goesmann A., Niehaus K., Nattkemper T.W. TACOA. Taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Müller M.A., Lukashev A.N., Gmyl A., Coutard B. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9(6) doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A.T., Perdomo M.F., Piombino-Mascali D., Marciniak S., Poinar D., Emery M.V. 17(th) century variola virus reveals the recent history of smallpox. Curr. Biol. 2016;26(24):3407–3412. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Harbeck M., Keller M., Spyrou M.A., Rott A., Trautmann B. A high-coverage Yersinia pestis genome from a sixth-century justinianic plague victim. Mol. Biol. Evol. 2016;33(11):2911–2923. doi: 10.1093/molbev/msw170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C., Werber D., Cramer J.P., Askar M., Faber M., der Heiden M.a. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011;365(19):1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- Green R.E., Krause J., Ptak S.E., Briggs A.W., Ronan M.T., Simons J.F. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444(7117):330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor I., Dröge J., Schirmer M., Quince C., McHardy A.C. PhyloPythiaS +. A self-training method for the rapid reconstruction of low-ranking taxonomic bins from metagenomes. PeerJ. 2016;4 doi: 10.7717/peerj.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Freuling C.M., Fischer S., Hueffer K., Hundertmark K., Nadin-Davis S. Spatio-temporal analysis of the genetic diversity of arctic rabies viruses and their reservoir hosts in Greenland. PLoS Negl. Trop. Dis. 2016;10(7) doi: 10.1371/journal.pntd.0004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Pohlmann A., Sauter-Louis C., Höper D., Stadler J., Ritzmann M. Porcine epidemic diarrhea in Europe: In-detail analyses of disease dynamics and molecular epidemiology. Viruses. 2017;9(7):177. doi: 10.3390/v9070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Ducatez M., Collin E.A., Ran Z., Liu R., Sheng Z. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9(2) doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Peddireddi L., Yuan F., Chen Z., Hesse R.A. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J. Gen. Virol. 2015;96(10):2994–2998. doi: 10.1099/jgv.0.000251. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Scheuch M., Höper D., Jungblut R., Holsteg M., Schirrmeier H. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 2012;18(3):469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Tappe D., Höper D., Herden C., Boldt A., Mawrin C. A variegated squirrel bornavirus associated with fatal human encephalitis. N. Engl. J. Med. 2015;373(2):154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- Jenckel M., Breard E., Schulz C., Sailleau C., Viarouge C., Hoffmann B. Complete coding genome sequence of putative novel bluetongue virus serotype 27. Genome Announc. 2015;3(2) doi: 10.1128/genomeA.00016-15. e00016-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J.A. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108(28):11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl C., Brinkmann A., Dabrowski P.W., Radonić A., Nitsche A., Kurth A. Protocol for metagenomic virus detection in clinical specimens. Emerg. Infect. Dis. 2015;21(1):48–57. doi: 10.3201/eid2101.140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamp B., Schwarz L., Högler S., Riedel C., Sinn L., Rebel-Bauder B. Novel pestivirus species in pigs, Austria, 2015. Emerg. Infect. Dis. 2017;23(7):1176–1179. doi: 10.3201/eid2307.170163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H.C.M., Yiu S.M., Yang B., Peng Y., Wang Y., Liu Z. A robust and accurate binning algorithm for metagenomic sequences with arbitrary species abundance ratio. Bioinformatics. 2011;27(11):1489–1495. doi: 10.1093/bioinformatics/btr186. [DOI] [PubMed] [Google Scholar]
- Li L., Diab S., McGraw S., Barr B., Traslavina R., Higgins R. Divergent astrovirus associated with neurologic disease in cattle. Emerg. Infect. Dis. 2013;19(9):1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäde D., Trübner K., Neubert E., Höhne M., Johne R. Detection and typing of norovirus from frozen strawberries involved in a large-scale gastroenteritis outbreak in Germany. Food Environ. Virol. 2013;5:162–168. doi: 10.1007/s12560-013-9118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield K.L., Morales A.B., Johnson N., Ayllón N., Höfle U., Alberdi P. Identification and characterization of a novel tick-borne flavivirus subtype in goats (Capra hircus) in Spain. J. Gen. Virol. 2015;96(Pt. 7):1676–1681. doi: 10.1099/vir.0.000096. [DOI] [PubMed] [Google Scholar]
- Maricic T., Pääbo S. Optimization of 454 sequencing library preparation from small amounts of DNA permits sequence determination of both DNA strands. Biotechniques. 2009;46(1):51–57. doi: 10.2144/000113042. [DOI] [PubMed] [Google Scholar]
- McHardy A.C., Martin H.G., Tsirigos A., Hugenholtz P., Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007;4(1):63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2(1):63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira É.A., Locher S., Kolesnikova L., Bolte H., Aydillo T., García-Sastre A. Synthetically derived bat influenza A-like viruses reveal a cell type-but not species-specific tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113(45):12797–12802. doi: 10.1073/pnas.1608821113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Huntemann M., Ivanova N., Kyrpides N.C., Pati A. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand Genomic. Sci. 2015;10:18. doi: 10.1186/1944-3277-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden T., Banyard A.C., Finke S., Fooks A.R., Hanke D., Hoper D. Comparative studies on the genetic, antigenic and pathogenic characteristics of Bokeloh bat lyssavirus. J. Gen. Virol. 2014;95(Pt. 8):1647–1653. doi: 10.1099/vir.0.065953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J.P., Coop G., Kudaravalli S., Smith D., Krause J., Alessi J. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314(5802):1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajer P., Dresler J., Kabíckova H., Písa L., Aganov P., Fucik K. Characterization of two historic smallpox specimens from a Czech museum. Viruses. 2017;9(8):200. doi: 10.3390/v9080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff F., Schlottau K., Scholes S., Courtenay A., Hoffmann B., Höper D., Beer M. A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound. Emerg. Dis. 2017;64(3):677–682. doi: 10.1111/tbed.12623. [DOI] [PubMed] [Google Scholar]
- Pfaff F., Schulze C., König P., Franzke K., Bock S., Hlinak A. A novel alphaherpesvirus associated with fatal diseases in banded penguins. J. Gen. Virol. 2017;98:89–95. doi: 10.1099/jgv.0.000698. [DOI] [PubMed] [Google Scholar]
- Quan P.-L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 2013;110(20):8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redactions Comité Ueber bakteriologische Forschung. X. internationaler medicinischer Congress. Berlin, 1891; Berlin: Verlag von August Hirschwald; 1891. [Google Scholar]
- Scheuch M., Höper D., Beer M. RIEMS. a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinformatics. 2015;16(1):69. doi: 10.1186/s12859-015-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K., Schulze C., Bilk S., Hanke D., Höper D., Beer M., Hoffmann B. Detection of a novel bovine astrovirus in a cow with encephalitis. Transbound. Emerg. Dis. 2016;63(3):253–259. doi: 10.1111/tbed.12493. [DOI] [PubMed] [Google Scholar]
- Sigma Aldrich . Sigma Aldrich; Saint Louis, MO: 2017. Traditional HPLC is Incapable of Reducing Cross Contamination of Custom Next-Generation Sequencing Adapters to Acceptable Levels.http://www.sigmaaldrich.com/technical-documents/articles/biology/traditional-hplc-is-incapable.html Available online at. checked on 8/1/2017. [Google Scholar]
- Stasiewicz M.J., den Bakker H.C., Wiedmann M. Genomics tools in microbial food safety. Curr. Opin. Food Sci. 2015;4:105–110. doi: 10.1016/j.cofs.2015.06.002. [DOI] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A.A., Chen L.-M. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 2012;109(11):4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppinen M., Perdomo M.F., Palo J.U., Simmonds P., Lycett S.J., Söderlund-Venermo M. Bones hold the key to DNA virus history and epidemiology. Sci. Rep. 2015;5:17226. doi: 10.1038/srep17226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernike K., Eschbaumer M., Breithaupt A., Hoffmann B., Beer M. Schmallenberg virus challenge models in cattle. Infectious serum or culture-grown virus? Vet. Res. 2012;43:84. doi: 10.1186/1297-9716-43-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernike K., Hoffmann B., Bréard E., Bøtner A., Ponsart C., Zientara S. Schmallenberg virus experimental infection of sheep. Vet. Microbiol. 2013;166(3–4):461–466. doi: 10.1016/j.vetmic.2013.06.030. [DOI] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang B., Tang C., Yue H., Ren Y., Song Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J. Gen. Virol. 2014;95(Pt 7):1603–1611. doi: 10.1099/vir.0.063743-0. [DOI] [PubMed] [Google Scholar]