Abstract
Ebola virus (EBOV) causes a deadly hemorrhagic syndrome in humans with mortality rate up to 90%. First reported in Zaire in 1976, EBOV outbreaks showed a fluctuating trend during time and fora long period it was considered a tragic disease confined to the isolated regions of the African continent where the EBOV fear was perpetuated among the poor communities. The extreme severity of the recent 2014–16 EBOV outbreak in terms of fatality rate and rapid spread out of Africa led to the understanding that EBOV is a global health risk and highlights the necessity to find countermeasures against it. In the recent years, several small molecules have been shown to display in vitro and in vivo efficacy against EBOV and some of them have advanced into clinical trials. In addition, also existing drugs have been tested for their anti-EBOV activity and were shown to be promising candidates. However, despite the constant effort addressed to identify anti-EBOV therapeutics, no approved drugs are available against EBOV yet.
In this chapter, we describe the main EBOV life cycle steps, providing a detailed picture of the druggable viral and host targets that have been explored so far by different technologies. We then summarize the small molecules, nucleic acid oligomers, and antibody-based therapies reported to have an effect either in in silico, or in biochemical and cell-based assays or in animal models and clinical trials, listing them according to their demonstrated or putative mechanism of action.
Keywords: EBOV, Drugs, Small molecules, Inhibitors, Host targets, EBOV entry, VP35, VP24, Interferon, EBOV RNA polymerase
1. Introduction
Ebola virus (EBOV) is one of the deadliest pathogens faced by humankind being the etiological agent of a viral hemorrhagic fever named Ebola virus disease (EVD) affecting humans and nonhuman primates (NHPs) with high mortality rates. In less than 45 years, since its first discovery in 1976, EBOV claimed more than 11,000 deaths through several highly mortal outbreaks. In particular, across the years 2014 and 2016 a dreadful outbreak of EVD spread in West Africa, claiming more cases and deaths than all other previous epidemics combined.1 The ability of the infection to spread between countries, starting in Guinea and subsequently moving across land borders to Sierra Leone and Liberia, and also exporting cases in extra-African countries, turned the spotlight on EVD as a global health emergency to be strongly counteracted. EBOV belongs to the viral order Mononegavirales that gathers together related viruses with a nonsegmented, linear, and single-stranded negative-sense RNA genome (ssRNA −). Ebolavirus genera is one of the three genera along with Marburgvirus (MARV) and Cuevavirus, within the family Filoviridae and includes five distinct species: Bundibugyo ebolavirus, Reston ebolavirus, Sudan ebolavirus, Taï Forest ebolavirus, and the type species Zaire ebolavirus. The EBOV ssRNA(−) genome is approximately 19 kb in size and encodes for seven proteins: the nucleoprotein NP, the polymerase cofactor VP35, the matrix proteins VP40 and VP24, the surface glycoprotein GP, the transcription activator VP30, and the large (L) RNA-dependent RNA polymerase.
While EBOV is considered a zoonotic pathogen capable to affect humans and several animal species, its natural reservoir is not conclusively defined yet, even though many reports pointed the attention to the fruit bat from Pteropodidae family.2, 3 Animals carrying the virus can infect other animals and, after a spillover event introducing EBOV into the human population, the disease can spread among human communities. Viral transmission, including human-to-human, can occur through several mechanisms: direct contact of broken skin or mucous membranes with infected blood, secretions, organs, or other bodily fluids, in utero (during delivery from infected mother), and with surfaces and materials (e.g., bedding and clothing) contaminated with body fluids.4, 5
EVD is a severe acute viral illness (2–21 days of incubation). Fever onset is a typical sign of the disease which begins through a nonspecific symptom period (2–3 days of fatigue, malaise, muscular soreness, and gastrointestinal manifestations) followed by a fast deteriorating period (2–4 days of severe sore throat, chest and abdominal pain, maculopapular skin rash, gastrointestinal, circulatory and vascular function impairment manifested by diarrhea, vomiting, and in some cases both internal and external bleeding). In the highly frequent lethal cases, this clinical picture evolves in septic shock, multiorgan failure, and death (6–9 days from clinical disease onset). Importantly, EVD survivors show long-term consequences affecting different body districts (eyes, ears, brain, joints, muscles, uterus, and testis).6, 7 EVD laboratory findings include early leucopenia, lymphopenia, and subsequent neutrophilia, followed by the presence of atypical lymphocytes, thrombocytopenia, hyperproteinemia, and proteinuria.8
Although EBOV exerts a broad cell tropism, cells of the monocyte/macrophage lineage, and dendritic cells are early and preferred replication sites of the virus, followed by a variety of other cell targets.2 In fact, an innate immune response dysregulation is the first event in EBOV infection, mainly occurring through a robust inhibition of the type I interferon α/β (IFN-α/β) responses mediated by the viral proteins VP24 and VP35.9 Extensive infection of dendritic cells also determine a massive release of proinflammatory cytokines and chemokines, leading to the typical EVD cytokine storm additionally contributing to disease progression and blunting of the adaptive immune response.2
The lack of proven specific treatments, the challenge of early diagnosis and the high number of fatalities justify the consideration of EVD as a global threat.10 Primarily driven by concerns on the potential misuse of the virus as a bioweapon (the Centers of Diseases Control and Prevention classifies EBOV as Category A agent), the search for effective countermeasures to treat EBOV infections has been in progress for several decades. In the last 40 years, a number of antiviral compounds have shown some therapeutic promises in both in vitro and animal studies and some of these were administered to EVD patients, or to persons undergoing clinical trial evaluation, in particular during the 2014–16 West African epidemic. The unprecedented magnitude and scale of the West Africa outbreak, combined with the potential spread to other corners of the world, led to a recent renewed focus on medical interventions for EVD.10 However, despite the tremendous efforts spent in the discovery of therapeutics and in conducting some clinical trials during the biggest outbreak setting, no EBOV-specific therapy has been conclusively proven efficacious, nor has any therapy achieved regulatory approval for use in humans to date.10 A promising vaccine candidate, the rVSVΔG-ZEBOV-GP, has been granted Breakthrough Therapy Designation by the FDA and PRIority Medicines status from the European Medicines Agency (Merck Press Release, July 25, 2016) and is currently awaiting a license.11 The vaccine showed 100% protection during a ring vaccination trial in Guinea, Sierra Leone, and Conakry, and now it is still in use in the ongoing outbreak of Democratic Republic of Congo (May 2018).11
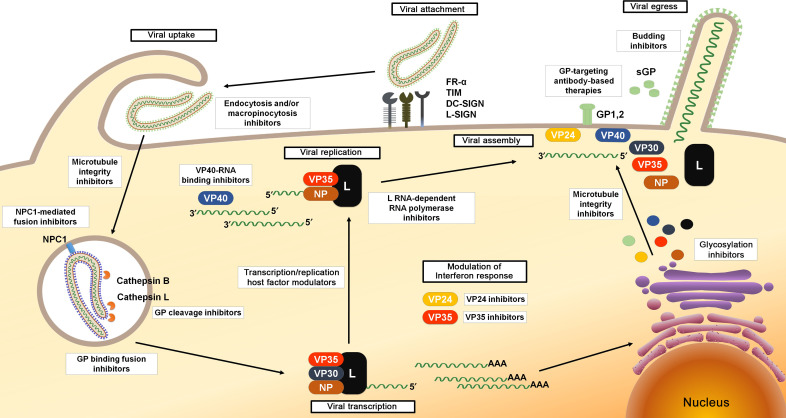
This chapter focuses on the different agents, including small molecules, antisense therapies, and immunotherapeutics, shown to be able to counteract EBOV in either in silico, in vitro, in vivo, or in clinical studies. The anti-EBOV agents reported in this chapter are categorized based on their proposed mechanism of action against specific steps of the viral life cycle, including entry, replication, packaging, and release of viral progeny from target cells (Fig. 1 ).
Fig. 1.
EBOV life cycle and viral steps targeted by antiviral agents.
2. EBOV Entry Inhibitors
Viral attachment on target cells occurs through the binding of the fusogenic virus envelope glycoprotein (GP1,2) produced by the cleavage of a precursor (GP0) obtained by the translation of the mRNA of the homonymous fourth gene on the EBOV genome. The glycoprotein GP1,2 is constituted by two subunits: GP1, responsible for the interaction of the viral particle with surface cell receptors, and GP2, containing a fusion loop critical for membrane fusion. Filoviruses have been shown to engage host cell attachment factors including the C-type lectin DC-SIGN (dendritic cell-specific ICAM3-grabbing nonintegrin), L-SIGN (liver and lymph node SIGN), and the folate receptor-α (FR-α).12 The receptor binding domain of the EBOV GP has been also shown to bind T-cell immunoglobulin mucin (TIM) domain-containing proteins 1 and 4.12 Moreover, different cell surface proteins such as integrins and tyrosine protein kinase receptor 3 (TYRO3) family components have been shown to enhance filovirus entry even if they do not act as physical viral receptors.13 Following binding to the cell surface, the uptake of EBOV virions into cells can involve different processes. In fact, EBOV entry mainly occurs through a macropinocytic pathway characterized by actin-dependent membrane ruffling. However, it is believed that also endocytic pathways can take place, even if studies characterizing the type of endocytosis, dependent on clathrin or caveolin, have shown conflicting results.14, 15, 16 From the early endosome, the virus is subsequently trafficked to a late endocytic compartment containing the cysteine proteases cathepsin B (CatB) and cathepsin L (CatL).12, 13 These proteases digest GP1,2 to a 19 kDa form, which is then triggered to initiate fusion between the viral and endosomal membranes.13
Preventing the entry of EBOV into host cells is an attractive antiviral strategy. A consistent number of small molecules have been found to be active against EBOV entry through multiple mechanisms, including some drugs approved by the Food and Drug Administration (FDA) for the treatment of malaria, cancer, Parkinson's disease, and other diseases. The first promising target for antiviral development is GP1,2, the only surface-expressed protein. Furthermore, since the process of viral entry involves the modulation of different cellular signaling pathways, therapeutic strategies were also aimed to identify compounds that act by targeting host cell components, such as G protein-coupled receptors (GPCRs) (histamine, serotonin, and estrogen receptors), cholesterol transporter, and multiple ion channels.
2.1. Endocytosis and/or Macropinocytosis Inhibitors
Through a large-scale compound screening (319,855 small molecules), Anantpadma and colleagues selected nine compounds that blocked filoviral infection (both EBOV and MARV) acting on different key steps of the infection process such as the macropinocytosis-mediated uptake and the endosomal trafficking. Among them, MLS000394177 and MLS000733230 exhibited a potent inhibition of EBOV infection in HeLa cells (EC50 = 1.9 and 6.7 μM, respectively) and a cell toxicity lower than the commonly accepted macropinocytosis inhibitor ethylisopropylamiloride.17 Importantly, both compounds inhibited EBOV infection in primary human macrophages.17
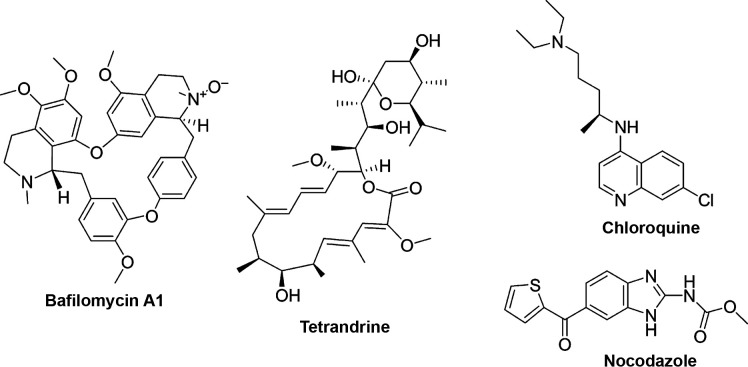
It has been demonstrated that low pH is required for the GP-mediated membrane fusion.12 Indeed, inhibitors of endosomal acidification bafilomycin A1 (Fig. 2 ) and clathrin-mediated endocytosis chlorpromazine have been shown to inhibit EBOV entry into the host cells in an in vitro replication-competent viral assay (inferred EC50 values around 10 nM and 13 μM, respectively), providing a promising scaffold for the development of novel EBOV therapeutics.18
Fig. 2.
Chemical structures of uptake and trafficking inhibitors.
EBOV requires calcium channels, termed two-pore channels (TPCs), in endosomal membranes for successful entry. The alkaloid tetrandrine (Fig. 2), a very well-known remedy in Traditional Chinese Medicine (TCM) for the treatment of a number of diseases such as tuberculosis, dysentery, malaria, cancer, and fever,19 isolated from different species of plants from the Menispermaceae family, was shown to inhibit EBOV entry into host cells and infection of human macrophages (EC50 = 55 nM) by binding to TPC. Tetrandrine also showed therapeutic efficacy in an EBOV mice model.20
Madrid et al. showed that the antimalarial chloroquine (CQ) (Fig. 2) inhibited EBOV entry in both viral pseudotype entry and replication assays with EC50 values of 4.7 and 16 μM, respectively.21 They also demonstrated that the treatment of infected mice with 90 mg/kg of CQ 4 h before infection was able to significantly reduce mortality, leading to a 90% survival benefit 13 days postinfection. CQ has been shown to act by multiple mechanisms. First, it was shown to directly perturb virus trafficking that results in aggregation of the viral particles, inhibiting their progression through the cytoplasm. Second, its antiviral activity appeared to be in part due to its inhibitory effects on the pH-dependent CatB and CatL, which have been shown to play a crucial role in EBOV GP1,2 processing events prior to fusion.21 However, more recent studies showed that CQ fails to protect against infection and disease in mice, hamsters, and guinea pig models as well as in patients.22, 23, 24
2.2. Microtubule Integrity Inhibitors
Entry of enveloped viruses requires the presence of an intact microtubule network to allow the correct trafficking from the cell surface to the appropriate acidified vesicular compartment.12 Agents that affect the integrity and/or function of actin filaments such as cytochalasin B, cytochalasin D, latrunculin A, and jasplakinolide, and the microtubule-disrupting agent nocodazole (Fig. 2), were investigated and showed to impair the microfilament function thus affecting the entry and fusion of EBOV GP pseudotype with EC50 values between 0.04 and 2 μM.16 An image-based high-throughput screening identified D011-2120, with EBOV inhibitory ability in the micromolar range (EC50 = 54 μM). It has been reported that D011-2120 acts, similarly to nocodazole, by depolymerizing microtubules and disrupting trafficking of virions from the Golgi to the cell membrane during viral egress.25 Among microtubule inhibitors also vinblastine (EC50 = 48 nM), vinorelbine (EC50 = 66 nM), vincristine (EC50 = 140 nM), colchicine (EC50 = 238 nM), mebendazole (EC50 = 3.4 μM), and albendazole (EC50 = 4.8 μM) displayed anti-EBOV entry activity.26
2.3. GP Cleavage Inhibitors
The digestion of EBOV GP1 is a crucial step necessary for membrane fusion. Blocking the GP cleavage with the use of the cysteine and serine protease inhibitor leupeptin led to a reduction of EBOV infection in macrophages (EC50 = 117 μM).27, 28 Similarly, other cysteine protease inhibitors, E-64 and E-64d, reduced viral antigen titer by 0.47 log10 and 1.5 log10, respectively, in a quantitative ELISA assay.29, 30 The treatment of infected cells with the CatL inhibitors also showed to reduce EBOV infection. In particular, a tetrahydroquinoline oxocarbazate (PubChem CID 23631927) blocked EBOV replication in HEK293T cells (EC50 = 193 nM)28 and the triazine derivatives 5705213 and 7402683 were reported to inhibit the entry of pseudotyped viruses carrying GP1,2 (EC50 = 15 and 10 μM, respectively).31 Other studies showed that the CatB inhibitors CA-074 and CA-074Me (EC50 = 20 μM and 4 μM, respectively), the CatL inhibitor Z-FY(t-Bu)-DMK (EC50 = 1 μM), and the CatB/CatL inhibitor FY-DMK (EC50 = 2 μM) also markedly reduced infectious EBOV progeny production.27, 30, 32
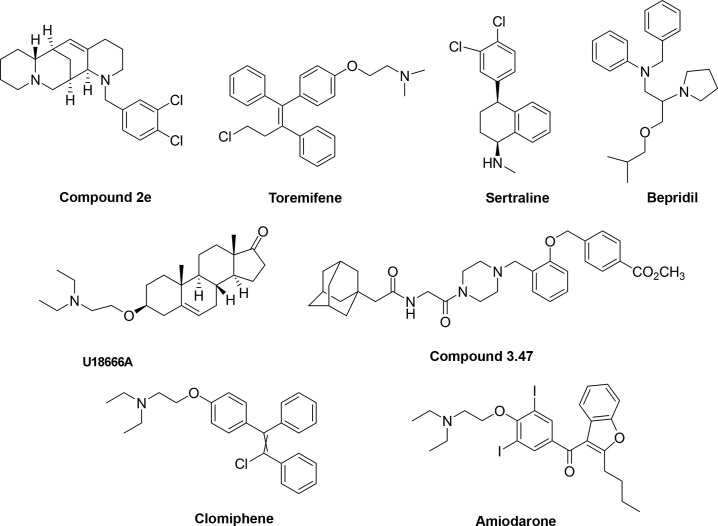
More recently, a series of derivatives of the Chinese natural product aloperine, substituted with a 12N-dichlorobenzyl group, were reported.33 Among the 23 derivatives synthesized, compound 2e (Fig. 3 ) exhibited the most potent activity in vitro (EC50 = 4.8 μM) and also in vivo (a dose of 1 mg/mL led to 58% of reduction in an anti-EBOV assay performed on BALB/c mice infected with human immunodeficiency virus [HIV] pseudotyped viruses with GP1,2 of EBOV), and a good pharmacokinetic and safety profile (low median lethal doses when administered intragastrically, intraperitoneally, and intravenously in mice). Mechanism of action studies showed that compound 2e could block a late stage of viral entry, mainly targeting the CatB activity.33
Fig. 3.
Chemical structures of fusion inhibitors.
2.4. GP Binding Fusion Inhibitors
In a recent study, Cheng et al. showed that known antagonists of different classes of GPCRs, including histamine receptors, 5-HT (serotonin) receptors, muscarinic acetylcholine receptors, and adrenergic receptors, were potent inhibitors of EBOV and MARV entry. In particular, benztropine, a drug clinically used for the treatment of Parkinson's disease, effectively blocked replication of both infectious viruses (EC50 = 3.7 and 13.2 μM, respectively), also suggesting a crucial role of GPCRs in filoviral entry mechanisms, and pointing out to the potential broad antiviral activity of GPCR antagonists.34 Interestingly, time-of-addition experiments and microscopic studies have provided evidence that GPCR antagonists block filoviral entry inhibiting a step following the initial attachment and prior to the viral/cell membrane fusion.34 It has been thus proposed either that filoviral GP can directly bind to one or multiple GPCRs and that such interaction could be critical for viral entry, or that EBOV can somehow trigger GPCR signaling, suggesting that GPCRs would be indirectly involved in filoviral entry.34
Several antagonists of histamine receptors and serotonin receptors have been shown to block the entry of EBOV virus-like particles (VLPs).26 Among them, the anticancer drug toremifene (Fig. 3), an estrogen receptor modulator, was capable to block VLPs entry into host cells mediated by EBOV surface GP at submicromolar concentrations (EC50 = 0.56 μM).26 Treatment of infected mice with toremifene resulted in statistically significant survival benefit (50%).35 The modulatory activity of toremifene was confirmed by Zhao et al. in 2016, showing that both the toremifene and the painkiller ibuprofen bind EBOV GP1,2 decreasing its thermal stability.36 Toremifene and ibuprofen bind in a cavity between the attachment (GP1) and fusion (GP2) subunit. This interaction triggers premature release of GP2 resulting in inhibition of viral fusion to the endosome membrane.36 Of note, crystal studies showed that the ibuprofen binding site is different to the one predicted by an in silico analysis37 suggesting the possible existence of two different binding sites for the compound on the EBOV GP.38 Recent results in cell culture assays (EC50 = 100 μM) seem to confirm its potential as a molecular template for the further drug development in the search for an anti-EBOV treatment.38 Following the same thread, another library screening (of about 2600 compounds among FDA-approved drugs and common mechanistic probes) identified 30 compounds with anti-EBOV activity in both Vero E6 and human HepG2 cell lines, including the selective serotonin reuptake inhibitors sertraline (Fig. 3) (EC50 = 3.1 and 1.4 μM, respectively) and paroxetine (EC50 = 7.4 and 1.4 μM, respectively) and the calcium channel blocker bepridil (Fig. 3) (EC50 = 5 and 3.2 μM, respectively).39 Sertraline and bepridil showed also a strong in vivo antiviral activity by affecting EBOV entry at a step after the internalization, inhibiting VLP-GP entry in a dose-dependent manner.39 More recently, crystallographic analysis and thermal shift assays demonstrated that sertraline, bepridil, paroxetine, and benztropine interact with the same toremifene EBOV GP pocket, thus affecting the thermal stability of the protein, destabilizing it and consequently leading to the block of virus/endosomal membrane fusion.40 However, it must be mentioned that high dosage of sertraline did not protect rhesus macaques in a subsequent challenge with a lethal EBOV dose (Makona strain), having no protective effect on viral load, morbidity, or survival.41
Finally, a benzodiazepine derivative (compound 7) has been identified to inhibit EBOV and MARV in cell-based entry assays with EC50 values of 10 and 12 μM, respectively.42 Compound 7 has been shown to act at an early stage of viral entry, probably by binding to a hydrophobic pocket (S2) in the prefusion conformation of EBOV GP. Its high selectivity sets the bases for further optimization toward novel filoviral inhibitors.42
2.5. NPC1-Mediated Fusion Inhibitors
During the fusion process, additional host factors in the endosome are required for viral/membrane fusion. One of them is the endo/lysosomal cholesterol transporter protein Niemann–Pick C1 (NPC1). Studies using primary fibroblasts derived from human NPC1 disease patients and Npc1-null Chinese hamster ovary (CHO) cells demonstrated that NPC1 function impairment led to total resistance to EBOV infections.43 Small molecules such as the androstenon derivative U18666A (Fig. 3) and the antidepressant imipramine caused a phenotype similar to NPC1 deficiency, probably by targeting the NPC1 pathway. A brief exposure of Vero cells with U18666A strongly inhibited EBOV infection (EC50 = 6 μM).43 In the same study, the treatment of HAP1 cells with imipramine (2 μM) led to a ~ 80% reduction in the number of cells infected by a replication-competent recombinant vesicular stomatitis virus (rVSV) bearing the EBOV GP.43
After cleavage of the glycosylated domains of GP1 by CatB, the aminoterminal domain is available for the binding to NPC1, thus promoting membrane fusion by the GP2 subunit. A benzylpiperazine adamantane diamide (compound 3.0) inhibited infection of Vero cells by VSV particles pseudotyped with EBOV GP (EC50 = 2.5 μM). The addition of a methoxycarbonyl-benzyl group at the ortho-position of the benzene ring, compound 3.47 (Fig. 3), significantly increased the potency of inhibition (EC50 = 30 nM). Importantly, it has been demonstrated that these molecules interfere with the binding of the cleaved GP to NPC1 in a concentration-dependent manner.44
Multiple cationic amphiphiles (AY 9944, clomiphene, Ro 48-8071, terconazole, triparanol) inhibited EBOV entry (EC50 = 1.6–8.0 μM) blocking EBOV fusion in an NPC1-dependent mode with a mechanism distinct from the one shown by compound 3.47, suggesting that there are at least two mechanisms of counteracting the NPC1-dependent pathway.45 In particular, clomiphene (Fig. 3), a drug approved for treating women anovulation, showed EC50 values of 11 and 3.8 μM against the two strains EBOV-95 and EBOV-76, respectively, and a 90% of survival benefit for infected mice.35
Compounds that are nonspecific ion channel inhibitors such as amiodarone (Fig. 3) and dronedarone, inhibitors of L-type calcium channels (verapamil) and sodium–potassium pump inhibitors (digoxin) were also investigated for the inhibition of EBOV infection. In vitro cell-based assays showed that all these agents were able to strongly inhibit EBOV entry (EC50 = 2, 3, 40, and 0.8 μM, respectively).26, 46
In particular, amiodarone is a pleiotropic drug that targets calcium, potassium, sodium channels, and α- and β-adrenergic receptors. In one efficacy study, treatment of mice with amiodarone initially resulted in a small, but significant, increase of survival, however in the same study such observation was not confirmed.47 It has been suggested that amiodarone might target different early phases of EBOV life cycle, affecting the entry of viral particles blocking the fusion of the EBOV GP with the endosomal membrane, and also modifying the cellular distribution of glycosphingolipids, sphingomyelin, cholesterol, and its transporter NPC-1.48, 49 Moreover, it has been demonstrated that amiodarone interferes with the progression of the viral particles along the endocytic pathway at multiple steps, decreasing the efficiency of the viral binding to target cells, and diminishing the acquisition of late endocytic markers to the compartment in which the virus is internalized.50 Noteworthy, the finding that molecules such as verapamil, targeting the L-type calcium channel, inhibited EBOV fusion also suggests a role for calcium currents in filoviral entry.46
2.6. Combination of Molecules With Different Entry Targets
Since EBOV entry involves endocytosis, macropinocytosis, intracellular trafficking, and fusion, that require different host factors, in particular CatB and CatL,30 NPC1 protein,43, 44 acid sphingomyelinase (ASM),51 and TPCs,20 the synergistic effect of different compound combinations in counteracting EBOV entry was investigated. Zheng et al. screened 795 unique three-drug combinations in an EBOV entry assay.52 Among them, two sets of three-drug combinations, toremifene–mefloquine–posaconazole and toremifene–clarithromycin–posaconazole, effectively blocked EBOV entry via functional inhibition of NPC1, ASM, and NAADP-stimulated lysosomal calcium release showing ≥ 5-fold reduction in EC50 values with respect to the single drug EC50 values. This result was further validated in a mice model for EBOV infection in which the rate of survival for both combinations was 74% and > 90%, respectively. Importantly, the individual drug concentrations in the combinations were reduced to clinically relevant levels with no cytotoxic effect.52
2.7. GP-Targeting Antibody-Based Therapies
Among the different treatments developed to counteract EBOV entry into the host cells, therapeutic antibodies showed promising results. Human survivors to EVD tend to mount an early, vigorous, and long-lasting neutralizing antibody (NAb) response. NAbs can bind to EBOV structural envelope GP.53 However, since a single antibody may not be able to neutralize every single viral particle, a cocktail of Abs is typically required. Produced in vitro from convalescent blood, NAbs are in the form of cocktails of monoclonal antibodies (mAbs) against EBOV GP.54
Among them, MB-003 (Mapp), ZMAb (Defyrus/PHAC), and in particular ZMapp (a cocktail combining the best components of the two former treatments), have demonstrated in animal models some of the most significant therapeutic potential for treating EVD, showing to be highly effective in postexposure prophylaxis of NHPs against EBOV. NHPs treatment with 50 mg/kg cocktail MB-003, manufactured in the tobacco plant Nicotiana benthamiana, protected either 43%55 or 67%56 of animals challenged with EBOV, whereas controls with the same challenge stocks succumbed to infection.55, 56 Administration of 25 mg/kg murine mAb cocktail ZMab (mAbs: m1H3, m2G4, and m4G7) to NHPs 24 h after EBOV challenge showed 100% effectiveness (50% after 48 h), while 50 mg/kg ZMapp treatment showed 100% protection when administered 3 days postinfection.57 Furthermore, the coadministration of ZMab and an adenovirus-vectored IFN-α (Ad-IFN) as investigated by Qiu et al. demonstrated high effectiveness in seven out of eight NHPs when the treatment was provided 3 days postinfection.58 In a further experimental design, NHPs were treated with both Ad-IFN 1 day postinfection after challenge and ZMAb at 4 days postinfection after challenge, with two out of four cynomologus macaques surviving with robust specific immune responses.58 ZMab was also tested on rhesus macaques in combination with IFN-β showing to prolong the survival, but not impact on mortality.59 Thus, to extend the antibody half-life in humans and to facilitate the clinical acceptance, the ZMab cocktail was chimerized with human constant regions and produced in N. benthamiana, obtaining the Ab cocktail ZMapp (cocktail of three chimeric mAbs: c13C6, c2G4, and c4G7) that targets the GP base and glycan cap.60, 61 To solve the limitation of Abs manufacturing in N. benthamiana and its supplies in an outbreak context, the manufacturing of ZMapp in CHO cells was also attempted.62 However, recently, a third-generation anti-CD20 antibody, Obinutuzumab, was instead produced in N. benthamiana leaves with yield amounts comparable to the ones observed in CHO cells, opening a new perspective in this field.63 Finally, a more detailed structural analysis of ZMapp revealed a redundancy in mAbs c2G4 and c4G7 that recognize overlapping epitopes on the EBOV GP and compete with each other for binding. To solve this conflict, a chimeric MIL77E mAb cocktail was recently produced in engineered CHO cells on the basis of the mAbs c13C6 and c2G4 and showed to be fully protective in NHPs when administered at 50 mg/kg 3 days after challenge with a lethal dose of EBOV (variant Makona, responsible for the 2014–15 Western Africa outbreak).64
Indeed, ZMAb and ZMapp cocktails were used under emergency compassionate protocols in humans to treat EBOV infections during the 2014 EVD outbreak in Western Africa. Among them, seven patients were reported to be treated with ZMapp, with five surviving, while six patients were reported to be treated with ZMAb, with 100% surviving.60, 65, 66 In the PREVAIL II study during the 2014–16 outbreak, a randomized controlled trial performed in the countries of Guinea, Liberia, Sierra Leone, and the United States, ZMapp was administered to patients (often in combination with other therapies), however, due to low enrollment numbers, the study did not produce any statistically significant conclusion on the efficacy of ZMapp treatment.67
3. EBOV Transcription and Replication Inhibitors
After entry and membrane fusion, the ribonucleoprotein (RNP) complex is released into the cytoplasm and serves for transcription and replication of the viral genome (Fig. 1). In the RNP, the nucleoprotein NP, the polymerase cofactor VP35, the transcription activator VP30, and the L RNA-dependent RNA polymerase are tightly associated with the EBOV ssRNA(−). Since both the genomic ssRNA(−) and the antigenomic ssRNA(+) are present only in the RNP form during the entire virus replication cycle, the molecular interactions between these proteins within the RNP complex represent the driving force for transcription and replication. In particular, the L protein must interact with VP35 to form a functional replicase and transcriptase complex. Moreover, the binding of VP35 (N-terminal peptide) to NP (C-terminal domain) promotes the release of the RNA from the NP-RNA complex allowing RNA to be ready for transcription. The EBOV mRNA synthesis starts at 6–7 h postinfection. The leader region of each ssRNA(−) is targeted by the EBOV RNA-dependent RNA polymerase which slides along the RNA template, transcribing genes with a mechanism strongly dependent by VP30. Among the seven genes, NP is the most transcribed. When enough NP is present to encapsidate neosynthetized genomes, the replication process could start. During replication, a full-length, positive sense antigenomic RNA is synthetized and serves as a template for production of progeny negative sense genomes.12, 68
3.1. L RNA-Dependent RNA Polymerase Inhibitors
Most of the advanced therapeutics to counteract EVD are small molecule inhibiting the viral L protein. Coded by the homonymous, seventh EBOV gene, this large protein of about 2200 amino acids (250 kDa) constitutes the catalytic site of the EBOV RNA-dependent RNA polymerase complex.10 Nucleoside and nucleotide analogs have thus far been the most successful class of antivirals given their success in treating HIV, hepatitis B and C virus, and herpes viruses, and some of them have been reported to be also effective as anti-EBOV agents.
The pyrazinecarboxamide derivative T-705 (favipiravir) is licensed in Japan for the treatment of pandemic influenza virus infections not responding to conventional therapies, shows a broad-spectrum antiviral activity against RNA viruses of different virus families including EBOV (EC50 = 67 μM), and has been used in the European centers that treated EVD in the last outbreak.69 The drug is converted intracellularly into its active, phosphoribosylated form, which is recognized as a substrate by the viral RNA-dependent RNA polymerase. In rodent models, favipiravir induced rapid EBOV clearance when administered in infected mice lacking the type I IFN receptor as late as day 6 following EBOV inoculation,69 and led to 100% survival rate in infected A129 mice when first administered at 1 h postinfection.70 However, in a NHP model, oral dosing of favipiravir did not result in a survival benefit but only in reduced levels of viral RNA and in an extended time-to-death.10 In agreement with this, in a nonrandomized study conducted during the 2014–16 Western Africa outbreak, no reduction in viral load or in mortality rate was observed among 99 EVD patients (adults and adolescents), treated with favipiravir when compared to untreated historical control subjects (91% mortality rate in those treated with favipiravir vs 85% in historic controls).10, 71
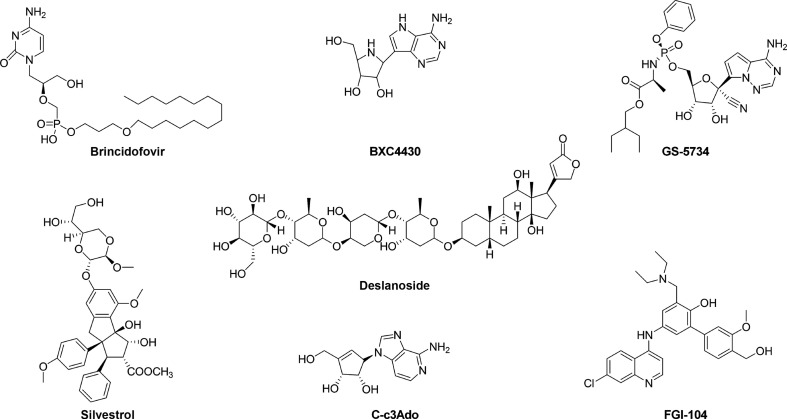
Brincidofovir (CMX001) (Fig. 4 ) is the 3-hexadecyloxy-1-propanol (HDP) lipid conjugate of the acyclic nucleoside phosphonate cidofovir (CDV), a drug originally developed as a treatment for double-stranded DNA virus infections and approved for the treatment of CMV retinitis. Even though the mechanism of action against RNA viruses is not well characterized, in vitro studies demonstrated that the anti-EBOV activity (EC50 = 120 nM to 1.3 μM) of brincidofovir was associated with the lipid nucleotide conjugate. In fact, the lipid alone was 29-fold less active than the conjugate while the nucleotide CDV alone had no detectable activity.72 Brincidofovir has been experimentally used in emergency situations to treat one of the first EBOV patients transferred in the United States, and underwent a phase II clinical trial that was however halted due to the limited number of enrolled patients.73, 74, 75
Fig. 4.
Chemical structures of transcription and replication inhibitors.
BCX4430 (Fig. 4) is an adenosine nucleoside analog currently under investigation as a treatment for a number of viral infections including EBOV.76 This compound showed efficacy in preexposure treatment of EBOV in vitro (EC50 = 11.8 μM) and in small animal models.73 When administered in a NHP MARV model 48 h after infection it demonstrated 100% protection, hence, further NHP studies were planned to verify its potential against EVD.10
As BCX4430, GS-5734 (Fig. 4) is another but newest RNA polymerase inhibitor being evaluated as a therapeutic for EBOV. It is a monophosphoramidate prodrug of an adenosine analog that is rapidly converted to its pharmacologically active triphosphate form (EC50 = 0.086–0.14 μM).77 In an NHP EBOV model, intravenous administration of 10 mg/kg for 12 days resulted in 100% survival. Importantly, since GS-5734 has been found to distribute into the testes, epididymis, eyes, and brain, its use has been envisaged also in post-EVD patients (in which viral replication is still ongoing) and for eradication of persistent viral replication in the genital tract.77 For this reason, the drug was used in the case of a nurse who suffered an EBOV relapse causing meningoencephalitis and in an infant in Guinea, and both individuals survived infection.78 In addition, given that GS-5734 rapidly distributes to cells of the blood mononuclear lineage maintaining drug levels sufficient for viral inhibition for approximately 24 h, it may also be valuable for postexposure prophylaxis of high-risk exposures.77
In the fall of 2014, it was reported that EVD patients in Liberia were successfully treated with the antiviral drug lamivudine, typically used to treat HIV and hepatitis B virus infections.79, 80 Thirteen patients out of 15 treated with the drug survived to the disease and were declared free from EVD. However, subsequent studies aimed at further investigating the anti-EBOV activity of lamivudine showed divergent results.81 To solve this contrast, the activity of lamivudine and another antiretroviral drug, zidovudine, was thoroughly evaluated in diverse cell lines (including Vero E6, Huh-7, HeLa, Hep G2, and 293T cells) and primary monocyte-derived macrophages, with different time courses of drug treatment and assay endpoints, and using varying multiplicities of infections against two variants of wild-type EBOV (Kikwit and Makona). As a result of this investigation, both compounds showed to be ineffective against EBOV replication, never achieving greater than 30% viral inhibition.81 Collectively, the results of this study found no evidence to support the therapeutic use of lamivudine for the treatment of EVD.81
Filone et al. identified an indoline alkaloid-type compound, CMLDBU3402, efficiently reducing EBOV infection in cell-based assays (EC50 = 20 μM). CMLDBU3402 exerted its antiviral effect blocking viral gene expression at an early stage of infection. While its mechanism of action has not been elucidated yet, CMLDBU3402 might inhibit the viral RNA polymerase activity.82
Nucleic acid-based inhibitors and small-interfering RNAs (siRNAs) had shown promise as EBOV therapeutics in nonclinical studies prior to the West Africa EBOV outbreak. In particular, siRNAs were shown to be effective in reducing EBOV mortality in NHPs.83, 84 Among them, TKM-Ebola (formerly Tekmira), a combination of siRNAs formulated in stable nucleic acid lipid particles targeting the EBOV L polymerase, VP24 and 35 protected all of six NHPs from EBOV infection, with good tolerance of the drug.83, 84 However, during phase I clinical trials, the FDA placed the drug on hold due to safety concerns consequent to induction of high cytokine levels.
3.2. VP40-RNA Binding Inhibitors
The VP40 matrix protein has a role during the replication process, participating in viral and host cell RNA metabolism, virus assembly, and egress.85, 86 The X-ray structure of the complex VP40-RNA (PBD: 1H2C) suggested that the VP40 amino acid residues F125 and R134 mainly interact with RNA.87 These interactions play a crucial role in VP40 octamer formation necessary for promoting EBOV replication in the early stage of infection.88 Mutations of the amino acid residue F125 partially abolished the ability of VP40 to bind RNA, while mutations of the amino acid R134 completely disrupt VP40-RNA binding and octamer formation.89
In order to identify novel EBOV inhibitors, different in silico studies have been performed to characterize VP40-RNA interaction.90, 91, 92 In particular, the TCM Database (TCMD)93 was used to identify inhibitors of this interaction and two hits, emodin-8-beta-d-glucoside and tonkinochromane_G, were found to form a hydrogen bond with R134 and interact with other key residues, negatively influencing the VP40-RNA binding and being hence potentially able to inhibit EBOV replication process.90
Another in silico study by Shah et al. characterized the binding affinity to VP40 of a few compounds and reported that deslanoside (Fig. 4) (score = − 11.6), a drug approved for congestive heart failure and cardiac arrhythmia, and its analog ZINC67911768 (score = − 8.6), show a strong interaction with the VP40 amino acid residue N130.91
3.3. Targeting Host Factors Involved in Transcription and Replication
Silvestrol (Fig. 4) is a secondary metabolite, isolated from the plant Aglaia foveolata, that acts as a potent and highly specific inhibitor of the ATP-dependent DEAD-box RNA helicase eIF4A,94 implicated in disabling the recruitment of ribosomes to the 5′-UTR of mRNAs in eukaryotes.95 It has been proposed that silvestrol increases the affinity of eIF4A to mRNA, thereby stalling the helicase on its RNA substrate, which leads to a depletion of eIF4A from mRNA-bound eIF4F complexes.96 Biedenkopf et al. observed that silvestrol inhibited EBOV replication at low nanomolar concentration (EC50 = 0.8 nM).97 More recently, silvestrol has been also shown to inhibit positive sense RNA genome viruses of the genera Coronavirus, Picornavirus, and Flavivirus, suggesting its potential role as broad range viral replication inhibitor.98, 99 Phosphorylation and dephosphorylation of VP30, which involve N-terminal serine residues and T52, are functionally important to modulate EBOV transcription. In particular, nonphosphorylated VP30 is distributed throughout the cytoplasm where transcription is activated, while the phosphorylation of two N-terminal VP30 serines positively regulates its binding to NP-induced inclusions to form the nucleocapsid structure and negatively regulates the VP30 transcription activation function. Since the VP30 dephosphorylation is mediated by intracellular phosphatases (PP1, PP2A, and PP2C), treatment of EBOV-infected cells with okadaic acid, an inhibitor of PP1 and PP2A, led to an increase of VP30 phosphorylation, an accumulation of inactive VP30, a reduction in transcription activation, and a block of viral replication (EC50 = 130 nM).100
A number of structural adenosine analogs, for example, 3-deazaaristeromycin (C-c3Ado) (Fig. 4) and 3-deazaneplanocin A (c3-Npc A), were reported to inhibit EBOV replication in vitro by blocking S-adenosyl-l-homocystein (SAH) hydrolase (EC50 = 30 and 2 μM, respectively).101, 102 SAH hydrolases are enzymes involved in methylation reactions that depend on S-adenosylmethionine (SAM) as the methyl donor. They play a crucial role in the methylation of 5′-end guanine of viral mRNA, regulating the capping process. Adenosine analogs are potential SAH hydrolase inhibitors, blocking the cleavage of SAH into homocysteine and adenosine, which can be further metabolized into AMP, adenine, and inosine. SAH hydrolase inhibition leads to its accumulation into the cell, causing the inhibition of the SAM-dependent methylation processes including those necessary for the maturation (i.e., 5′′-capping) of viral mRNAs. As a consequence, the production of progeny virus particles is suppressed. When EBOV-infected mice were treated twice a day with C-c3Ado and c3-Npc A, a prolonged survival was observed. Treatment with C-c3Ado doses ≥ 0.7 mg/kg every 8 h on day 0 or 1 after challenge resulted in dose-dependent protection, with complete mortality prevention.101, 102, 103, 104 Interestingly, Bray et al. reported that the protective effect of c3-Npc A was due to massive increase of IFN-α production in EBOV-infected, but not uninfected, mice.105 The c3-Npc A analog 1″,6″-isoneplanocin A enantiomers (d-like and l-like), also showed antiviral activity against several viruses, including EBOV in the submicromolar range (d-like EC50 = 0.38 μM, l-like EC50 = 0.76 μM).106 An in vitro screening utilizing a variant of EBOV-GFP resulted in the identification of the compound FGI-103, that inhibited EBOV infection in a dose-dependent manner (EC50 = 100 nM), with > 90% EBOV inhibition with 0.8 μM of compound.107 A single intraperitoneal FGI-103 dose (10 mg/kg) delivered 24 h postinfection was sufficient to completely protect mice against a lethal EBOV challenge.107 The same dose of a FGI-103 derivative, FGI-104 (Fig. 4), delivered 2 h before infection resulted in a 100% of survival rate in the same EBOV mouse model.108 Moreover, a diazacrysene derivative, named FGI-106, and its analogs, FGI 106-a and FGI-106-b, also showed significant antiviral activity against EBOV.109, 110, 111 Interestingly, FGI-106 was able to reduce dose dependently the mortality rate in mice, leading a survival benefit of 100% with a dose of 5 mg/kg administered 1 h before infection.109 It has been demonstrated that FGI-106 inhibits replication of both ssRNA(−) and ssRNA(+) viruses, suggesting it may affect a cellular pathway, however the molecular mechanism of action of these derivatives is not understood yet.109
4. Glycosylation Inhibitors
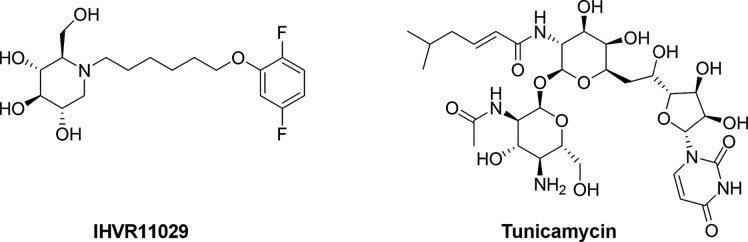
N-linked glycosylation is the most common glycosylation type and is needed for the correct folding, sorting, secretion, and function of glycoproteins including viral envelope glycoproteins.112 The process occurs in the ER lumen of eukaryotes and involves the sequential cleavage of the three glucose residues from the high-mannose N-linked glycans attached to nascent glycoproteins by the two ER glycoside cleaving enzymes, α-glucosidases I and II. This allows glycoproteins to properly fold and interact with ER chaperones, calnexin and calreticulin, to be moved to the Golgi complexes for further processing.113 Abrogation of glycoprotein trafficking and processing by inhibition of ER α-glucosidases I and II results in the production of misfolded glycoproteins, which may be retained in the ER and ultimately driven to degradation.114 Although the viral envelope is rich in glycoproteins, mammalian viruses do not codify for their own glucosidases, thus host cell glucosidases represent validated antiviral targets for multiple enveloped viruses including EBOV. The antiviral effects of glucosidase inhibitors, such as the iminosugar compound deoxynojirimycin (DNJ) and its derivatives, in which an oxygen has been substituted by a nitrogen atom that serves as competitive substrate and inhibit ER α-glucosidases I and II,115 have been observed for many types of viruses.116 Miglustat is a clinically approved glucosidase inhibitor used for treating lysosomal storage diseases (Gaucher's disease) that have also been evaluated in phase II trials for HIV infection showing limited efficacy due to its low potency and for the difficulties to achieve steady-state therapeutic concentrations.117, 118 Three Miglustat-derived iminosugars IHVR11029 (Fig. 5 ), IHVR17028, and IHVR19029 dose dependently blocked the assembly and secretion of EBOV envelope GP pseudotyped lentiviral particles in cell-based assays (EC50 = 30 μM) and were active against lethal infections of EBOV in mice (50%–80% of survival rate).119
Fig. 5.
Chemical structures of glycosylation inhibitors.
Another known N-glycosylation inhibitor, tunicamycin (Fig. 5), has been also shown to decrease > 90% EBOV infection in HeLa and HOP cells at a concentration of 3 and 15 μg/mL, respectively.120
A different possible strategy against N-glycosylated viruses including EBOV is the use of GP-targeting immunotherapeutics such as mannose-binding lectin (MBL). MBL is a C-type lectin that recognizes hexose sugars including mannose, glucose, fucose, and N-acetylglucosamine on the surface of many pathogens, preferentially glycosylated viruses. It has been shown that recombinant human MBL (rhMBL) bind EBOV (Zaire) and MARV (Musoke) envelope GPs, mediating complement-dependent virus neutralization.121 In vitro studies showed that 100 ng/mL of rhMBL was the minimum concentration needed to inhibit > 90% infectivity of HepG2 cells using EBOV GP pseudotyped lentiviral particles and to inhibit > 90% EBOV infectivity in Vero E6 cells using recombinant EBOV-eGFP. Treatment of infected mice with a higher single intraperitoneal dose of 350 μg rhMBL either 1 h before or 12 h after EBOV challenge led to an increased survival ≥ 40% and allowed the development of an effective adaptive immune response (higher B lymphocyte and CD11b+ granulocyte counts and downregulation of intrahepatic cytokines that may mitigate the detrimental effects of the characteristic cytokine storm following the infection).10, 122
5. Modulation of IFN Response
The severity of the disease associated with EBOV infection is determined by systemic virus replication, with the subsequent cellular disability to implement an efficacious defense. At the base of the abundant EBOV replication and spread is its ability to effectively counteract the cellular antiviral response, in particular the type I IFN system. This suppression, together with the excessive virus-induced expression of several inflammatory mediators such as cytokines, TNFα, and reactive oxygen and nitrogen species, results in an immunological imbalance that strongly contributes to the progression of EVD. EBOV-mediated IFN inhibition occurs through multiple mechanism123 and is carried out by the VP35 and VP24 proteins that block its production and signaling, respectively.124, 125, 126, 127 Since the IFN suppression is a key feature in filovirus pathogenesis, considerable efforts have been made to develop agents able to restore IFN response. This aim can be achieved by either targeting the viral proteins involved in this process, VP35 and VP24, or enhancing the IFN response acting on cellular host factors. Since, as already mentioned, VP35 and VP24 are not only involved in IFN response evasion, the following sections will list all VP35 and VP24 targeted inhibitors even if they affect other steps of the virus life cycle.
5.1. VP35 Inhibitors
The VP35 multifunctional protein potently inhibits the RIG-I-like receptors (RLR), RIG-I and MDA5 signaling,128, 129, 130 resulting in (i) an IFN production reduction, (ii) a downregulation of various antiviral genes subsets and, importantly, (iii) in the inhibition of dendritic cell maturation, likely contributing to the suppression of adaptive immune responses development.131 Located at the very C-terminal portion of the protein, the EBOV VP35 IFN inhibitory domain (IID) is the fundamental region important for the powerful suppression of the host innate immune response. It includes two basic patches with a high degree of sequence similarity among filoviruses: a first basic patch (FBP), important for replication complex formation through interactions with the viral NP, and central basic patch (CBP) important for VP35-dsRNA binding, IFN inhibition, and innate immune antagonism.128, 132 This latter feature led to term the IID also as RNA binding domain (RBD) and shows a unique fold that does not follow the α-β-β-β-α organization observed for canonical RBDs. Previous studies revealed that mutation of key basic residues within the VP35 IID results in significant EBOV attenuation, diminished, or abolished suppression of IFN-β induction, both in vitro and in vivo.132, 133, 134, 135, 136, 137 Therefore, VP35 is a validated key target for drug development and a potential avenue for therapeutic intervention would be to target it to restore the innate immune functions.
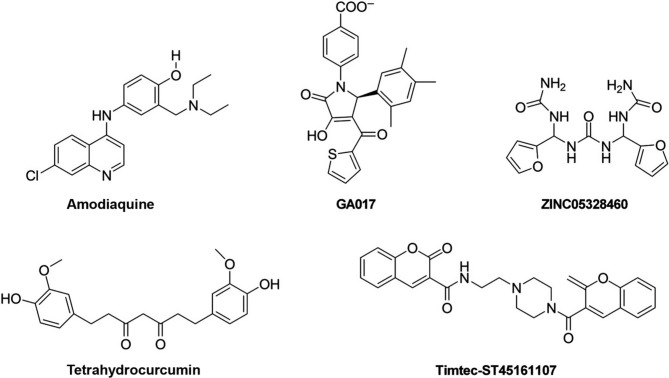
Structure-based in silico screening studies were performed and small molecules targeting a VP35 binding pocket were identified. In particular, different compounds with similar structure containing a pyrrolidinone scaffold have been shown to potentially counteract VP35 function. Docking studies highlighted the FDA-approved antimalarials CQ and amodiaquine (Fig. 6 ) as compounds possibly targeting VP35.21, 138 However, as already mentioned, a few studies showed that CQ was not able to increase the survival rate either in animals or in EVD patients.22, 23, 24 Differently, when 71 patients with confirmed EVD were treated with a combination of artesunate–amodiaquine, it was observed a lower risk of death compared to other 194 EVD-confirmed patients treated with artemether–lumefantrine, a first-line antimalarial combination drug.139
Fig. 6.
Chemical structures of VP35 inhibitors.
Other compounds with the pyrrolidinone scaffold were also reported. In particular, the pyrrole-based compound (GA017) (Fig. 6) was shown to bind the VP35 IID FBP (PDB-ID: 3FKE), disrupting the VP35-NP interaction by in vitro pull down assay and inhibit EBOV minigenome replication (EC50 = 50 μM).140 Another study identified five GA017 derivatives that inhibited VP35 with IC50 values < 200 μM, with one of them, ZINC05328460 (Fig. 6), that showed an IC50 value of 4 μM.141
A virtual screening study combining a pharmacophore model (HypoA of 4-acetyl-3-hydroxy-1-phenyl-1H-pyrrol-2(5H)-one derivatives), with 3D QSAR and docking models, was performed identifying seven compounds (named cpd 1–7), as new VP35 potential inhibitors with novel scaffolds.142
In another in silico study, more than 45,000 compounds were docked to the EBOV IID FBP and 13 compounds were selected for their potential to inhibit VP35, showing binding affinities between − 9.2 and − 7.0 kcal/mol.143 Among them, the dicoumarin carboxamide derivative, Timtec-ST45161107 (Fig. 6) displayed the best binding energy.
As already stated, plants metabolites are potential antiviral molecules. As an example molecular dynamics simulations showed that curcumin, curcuminoids, and the metabolite tetrahydrocurcumin (Fig. 6) could bind to simulated structures of EBOV proteins. In particular, etrahydrocurcumin showed the best binding energy against EBOV VP35.144
Recently, it was also found that the extract from the leaves of Asphodelus microcarpus Salzm et Vivi (Asphodelaceae) was highly effective in counteracting EBOV VP35-mediated inhibition of IFN-β production in a luciferase reporter assay.145, 146
5.2. VP24 Inhibitors
EBOV VP24 is also an excellent target for drug development because of its role exerted in suppressing the IFN signaling cascade that contributes to EBOV virulence and pathogenesis.147, 148 VP24 inhibition of both IFN type I and II signaling occurs through the block of the interaction between phosphorylated STAT1 (P-STAT1) and karyopherin-α (KPNA) which is essential for P-STAT1 nuclear translocation and the consequent expression of the IFN stimulating genes (ISGs).125, 126, 127 Recently, it has been showed that VP24 could also directly bind P-STAT1.149 The crucial role of VP24 determined the development of novel drug discovery techniques and the identification of a number of inhibitors.150, 151, 152, 153
AVI-7537, a phosphorodiamidate morpholino oligomer containing five positively charged linkages (PMOplus) was designed based on the EBOV VP24 sequence.150 AVI-7537 acts as steric inhibitor of gene expression by physically blocking the binding of translation complex elements to the VP24 transcript RNA. The suppression of VP24 translation resulted in mice and guinea pigs protection against EBOV lethal challenge.150 AVI-6002, a combination of AVI-7539, targeting VP35, and AVI-7537 for VP24, led to a survival of 61% in NHPs, while the survival rate for AVI-6003, a combination of AVI-7287 (VP24) and AVI-7288 (NP), was 100%.150 In a subsequent study to determine the minimum PMO elements required for protection against EBOV in NHPs, the efficacy of AVI-7537 or AVI-7539 alone compared to the one of AVI-6002 was analyzed revealing that the PMOplus targeting VP24 alone was sufficient to confer protection from lethal EBOV infection but no protection was achieved with PMOplus targeting VP35 alone.151
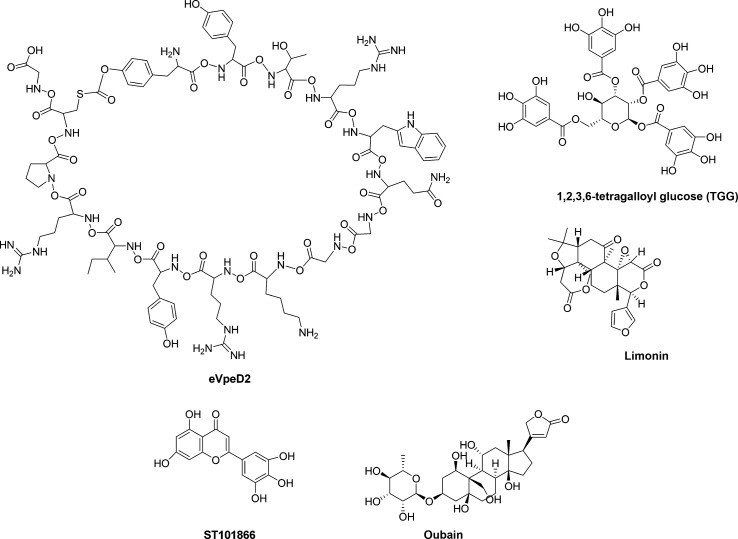
These studies confirmed the importance of VP24 as key antiviral target for EBOV. However, there is currently a lack in molecules that specifically counteract the VP24 function in suppressing the IFN signaling. Only a recent work provided the evidence that the binding between VP24 and KPNA may be druggable. Indeed, a macrocyclic peptide, eVpeD2 (Fig. 7 ), has been identified that blocks the VP24-KPNA5 interaction at low micromolar range in vitro (EC50 = 9 μM).152 Moreover, the recent resolution of the structure of the VP24-KPNA5 complex (PDB-ID: 4U2X) allowed to perform a number of in silico studies to identify possible inhibitors.154, 155, 156 Docking studies showed a certain VP24 binding affinity to some plant polyphenols such as epigallocatechin gallate, 1,2,3,6-tetragalloyl glucose (1,2,3,6-TGG) (Fig. 7), and theaflavin-3,3′-digallate.157 Three small molecules extracted from Indian medicinal plants, limonin (Fig. 7), samarcandin and gummosin were reported to have high docked energy in complex with VP24.158 Moreover, a screening of 4500 compounds resulted in high docking score (− 8.252) and glide energy (− 34.633 kcal/mol) for the interaction between VP24 (PDB-ID: 4M0Q) and ST059622, also known as gossypetin, a flavonol compound isolated from Hibiscus sabdariffa and Alchemilla mollis, already known from their antiviral activity on other viruses including influenza virus A/WSN/33.159, 160, 161 In the same in silico screening, a strong interaction was identified between VP24 and ST060285 ortaxifolin, a flavonoid commonly present in different conifer species including Taxus chinensis and Larix sibirica, whose antiviral properties have been known for a long time.161, 162, 163 While gossypetin and taxifolin have been found to interact also with other protein targets, VP40, VP35, and VP30, the flavonoid ST101866 (Fig. 7) was specific for VP24.161 In another pharmacophore-based virtual screening and molecular docking study, deslanoside and digoxin with their respective analogs (ZINC85911633 and ZINC77291634) have been identified, among 56 FDA-approved drugs, to possess high binding affinity for VP24.91 Using a structural system pharmacology approach, Zhao et al. identified indinavir, a HIV protease inhibitor, as a possible ligand for VP24. Comparing the binding pattern of indinavir with its primary target, HIV protease (PDB-ID: 2AVO), it has been shown that the same indinavir atoms (O2, N4, and O4) forming hydrogen bonds with HIV protease also interact with VP24. Hence, the predicted binding mode of indinavir to VP24 suggests that the compound may function as VP24 inhibitor.164
Fig. 7.
Chemical structures of VP24 inhibitors.
VP24 is not only involved in IFN signaling antagonism but it plays several functions during the EBOV life cycle. A study using affinity tagging coupled to label-free quantitative mass spectrometry was aimed to identify cellular proteins that could potentially interact with VP24.165 Among approximately 50 proteins that were found to bind VP24, a high binding ratio was reported for an ATPase involved in establishing and maintaining Na+ and K+ electrochemical gradients, ATP1A1. Treatment of EBOV-infected MRC-5 cells at 24 and 48 h postinfection with 20 nM of ouabain (Fig. 7), a small molecule inhibitor of ATP1A1, resulted in a 30% and 50% decrease in progeny virus, respectively, suggesting that this protein may have a functional role in EBOV replication, even if it has not been demonstrated that ouabain acts counteracting the binding between ATP1A1 and VP24.165
5.3. IFN Response Enhancers
A very recent high-throughput screening assay identified the anthracycline antibiotics doxorubicin, daunorubicin, epirubicin, and aklavine hydrochloride, together with the topical antiseptic aminacrine (9-aminoacridine), as IFN response inducers. Anthracycline antibiotics are a class of compounds that include commonly used cancer chemotherapy drugs that inhibit type II topoisomerase intercalating DNA, hence triggering the DNA damage response. These compounds induced IFN responses in an ATM kinase-dependent manner, triggered the DNA-sensing cGAS-STING pathway of IFN induction and suppressed EBOV replication in vitro, also inducing IFN in the presence of IFN-antagonist proteins from multiple negative sense RNA viruses.166
6. EBOV Budding Inhibitors
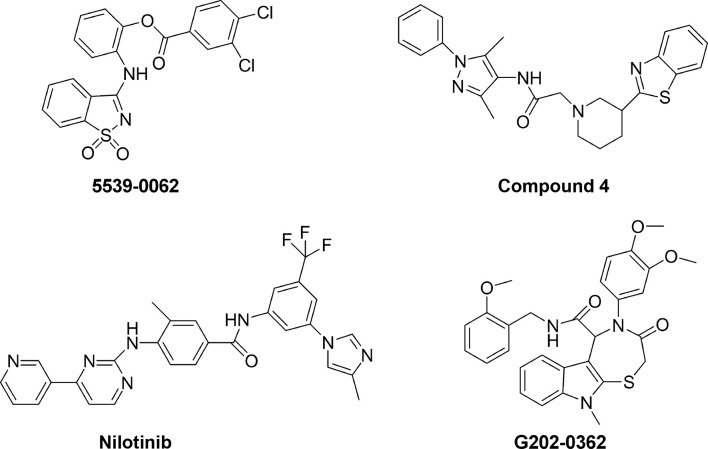
Drugs that counteract filovirus budding are attractive therapeutics since they would be expected to potently reduce the spread of the viral particles allowing the immune system to gain time for contrasting the infection. Among the seven proteins encoded by EBOV, NP, VP35, and VP24 are requested for NC assembly, and expression of the matrix protein VP40 allows the budding of filamentous virions. The interaction of NP with VP40 is demonstrated to be crucial for NC transport to the cell surface and for its incorporation into virions, leading to the formation of mature virus particles.167 EBOV VP40 contains two late-(L-) domain motifs (PTAP and PPEY) that are critical for efficient egress of virions, as they specifically bind cellular proteins involved in vacuolar protein sorting pathways to facilitate the final step of virus–cell separation.168 The PTAP L-domain recruits host Tsg101, involved in the protein endosomal sorting pathway, while the PPxY motif interacts with the host Nedd4 ubiquitin ligase leading to ubiquitination of VP40 and enhancing viral egress. Small molecules that inhibit these interactions would be predicted to reduce virus budding and spread. In an in silico screening on 2.4 million compounds investigated for inhibiting the binding between PTAP L-domain and Tsg101, the top six scoring molecules identified were tested for their ability to disrupt the VP40-Tsg101 interaction. Of them, 5539-0062 (Fig. 8 ) exhibited PTAP-specific inhibition of the VP40-Tsg101 complex formation and the VP40 VLP egress (40% at 50 μM and > 90% at 100 μM). The compound showed specific antiviral activity also in cells infected with a recombinant VSV (VSV-M40) expressing the PTAP L-domain of EBOV VP40.168 Another in silico study combined with functional VLP and virus budding assays identified compounds 4 (Fig. 8) and 5 that have broad-spectrum antibudding activity, including antifiloviruses, by competitively disrupting viral PPxY-host Nedd4 interaction.169 Compound 4 inhibited egress of EBOV VP40 VLPs by 3- and 10-folds at concentrations of 0.5 and 1.0 μM, respectively, while compound 5 inhibited it by 3.3- and 7-folds, respectively, at the same concentrations.169
Fig. 8.
Chemical structures of budding inhibitors.
Tyrosine kinase c-Abl1 is involved in phosphorylation of tyrosine 13 of VP40 which modulates the release of EBOV particles. This step of the virus life cycle therefore may represent a target for antiviral therapy. The Abl-family kinase inhibitors imatinib (for the treatment of chronic myelogenous leukemia) and nilotinib (Fig. 8) (for the treatment of chronic myelogenous leukemia) blocked the egress of VLPs in a cell culture cotransfection system (EC50 = 15 and 10 μM, respectively) through the mechanism involving phosphorylation of VP40 and significantly inhibited the productive replication of the highly pathogenic EBOV Zaire strain.170
Another compound affecting viral egress is the oxo-thiazepino-indole-carboxamide derivative, G202-0362 (Fig. 8) (EC50 = 16.1 μM) that has been demonstrated to block viral budding from the trans Golgi.74 The finding that it does not alter either the Golgi structure or the cytoskeleton organization, cell, or nucleus size makes it a promising hit for drug development.25
7. Conclusions
EBOV is an extremely virulent pathogen responsible of a severe hemorrhagic fever in humans and NHPs. The high mortality rate and the rapid spread of the recent outbreak highlight the need to identify efficacious anti-EBOV therapeutics. To date, several antiviral agents showed inhibitory potency in in vitro studies as well as in animal models and patients. Among them, different drugs already approved have been proposed to be repurposed with the aim of accelerating the process of FDA approval. Analysis of the so far identified anti-EBOV agents based on their mode of action against specific viral proteins or host factors involved in the viral life cycle, shows that the majority of these agents target the viral entry or proteins involved in viral in transcription and genome replication. In addition, a number of compounds targeting cell signaling pathways have also been reported. An example is the modulation of the IFN response, a crucial step at the base of the pathogenesis of EBOV infection. Human clinical data have been reported for different agents such as BCX4430, GS-5734, AVI-6002, AVI-7537 which completed phase I trials, while compounds such as brincidofovir, amiodarone were evaluated in phase II clinical trials that, however, were early terminated. ZMapp is currently considered the best option to treat EBOV patients, in particular in combination with other antiviral agents, such as favipiravir or GS-5734, that likely contributed to the improved outcome. In conclusion, we provide a description of the state-of-the-art in EBOV drug development, focusing on the molecular mechanisms of EBOV infection and reporting the possible strategies that could be exploited by anti-EBOV agents to subvert its virulence.
References
- 1.World Health Organization Situat. Rep. 2016;(June):1–2. [Google Scholar]
- 2.Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. Annu. Rev. Pathol. Mech. Dis. 2017;12(1):387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- 3.Paweska J.T., Storm N., Grobbelaar A.A., Markotter W., Kemp A., Van Vuren P.J. Viruses. 2016;8(2):1–11. doi: 10.3390/v8020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen C.F., Kidd S., Sillah A.R.M., Davis E., Mermin J., Kilmarx P.H. MMWR Morb. Mortal. Wkly Rep. 2015;64(1):20–27. [PMC free article] [PubMed] [Google Scholar]
- 5.Bebell L.M., Oduyebo T., Riley L.E. Birth Defects Res. 2017;109(5):353–362. doi: 10.1002/bdra.23558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deen G.F., McDonald S.L.R., Marrinan J.E., Sesay F.R., Ervin E., Thorson A.E., Xu W., Ströher U., Ongpin P., Abad N., Ariyarajah A., Malik T., Liu H., Ross C., Durski K.N., Gaillard P., Morgan O., Formenty P., Knust B., Broutet N., Sahr F. PLoS Negl. Trop. Dis. 2017;11(9) doi: 10.1371/journal.pntd.0005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez L.L., De Roo A., Guimard Y., Trappier S.G., Sanchez A., Bressler D., Williams A.J., Rowe A.K., Bertolli J., Khan A.S., Ksiazek T.G., Peters C.J., Nichol S.T. J. Infect. Dis. 1999;179(s1):S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- 8.Feldmann H., Geisbert T.W. Lancet. 2011;377(9768):849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misasi J., Sullivan N.J. Cell. 2014;159(3):477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bixler S.L., Duplantier A.J., Bavari S. Curr. Treat. Options Infect. Dis. 2017;9(3):299–317. doi: 10.1007/s40506-017-0130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medaglini D., Santoro F., Siegristb C.A. Semin. Immunol. 2018 doi: 10.1016/j.smim.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Yu D.S., Weng T.H., Wu X.X., Wang F.X.C., Lu X.Y., Wu H.B., Wu N.P., Li L.J., Yao H.P. Oncotarget. 2017;8(33):55750–55759. doi: 10.18632/oncotarget.18498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White J.M., Schornberg K.L. Nat. Rev. Microbiol. 2012;10:317–322. doi: 10.1038/nrmicro2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Empig C.J., Goldsmith M.A. J. Virol. 2002;76(10):5266–5270. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmons G., Rennekamp A.J., Chai N., Vandenberghe L.H., Riley J.L., Bates P. J. Virol. 2003;77(24):13433–13438. doi: 10.1128/JVI.77.24.13433-13438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonezawa A., Cavrois M., Greene W.C. J. Virol. 2005;79(2):918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anantpadma M., Kouznetsova J., Wang H., Huang R., Kolokoltsov A., Guha R., Lindstrom A.R., Shtanko O., Simeonov A., Maloney D.J., Maury W., Lacount D.J., Jadhav A., Davey A. Antimicrob. Agents Chemother. 2016;60(8):4471–4481. doi: 10.1128/AAC.00543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya S., Warfield K.L., Ruthel G., Bavari S., Aman M.J., Hope T.J. Virology. 2010;401(1):18–28. doi: 10.1016/j.virol.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhagya N., Chandrashekar K.R. Phytochemistry. 2016:5–13. doi: 10.1016/j.phytochem.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai Y., Kolokoltsov A.A., Chen C.-C., Tidwell M.W., Bauta W.E., Klugbauer N., Grimm C., Wahl-Schott C., Biel M., Davey R.A. Science. 2015;347(6225):995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madrid P.B., Chopra S., Manger I.D., Gilfillan L., Keepers T.R., Shurtleff A.C., Green C.E., Iyer L.V., Dilks H.H., Davey R.A., Kolokoltsov A.A., Carrion R., Patterson J.L., Bavari S., Panchal R.G., Warren T.K., Wells J.B., Moos W.H., Burke R.L.L., Tanga M.J. PLoS One. 2013;8(4):e60579. doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop B.M. Ann. Pharmacother. 2015;49(2):196–206. doi: 10.1177/1060028014561227. [DOI] [PubMed] [Google Scholar]
- 23.Dowall S.D., Bosworth A., Watson R., Bewley K., Taylor I., Rayner E., Hunter L., Pearson G., Easterbrook L., Pitman J., Hewson R., Carroll M.W. J. Gen. Virol. 2015;96(12):3484–3492. doi: 10.1099/jgv.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falzarano D., Safronetz D., Prescott J., Marzi A., Feldmann F., Feldmann H. Emerg. Infect. Dis. 2015;21(6):1065–1067. doi: 10.3201/eid2106.150176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mudhasani R., Kota K.P., Retterer C., Tran J.P., Whitehouse C.A., Bavari S. PLoS Negl. Trop. Dis. 2014;8(8):e3095. doi: 10.1371/journal.pntd.0003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouznetsova J., Sun W., Martinez-Romero C., Tawa G., Shinn P., Chen C.Z., Schimmer A., Sanderson P., McKew J.C., Zheng W., Garcia-Sastre A. Emerg. Microbes Infect. 2014;3(12) doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnirß K., Kühl A., Karsten C., Glowacka I., Bertram S., Kaup F., Hofmann H., Pöhlmann S. Virology. 2012;424(1):3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah P.P., Wang T., Kaletsky R.L., Myers M.C., Purvis J.E., Jing H., Huryn D.M., Greenbaum D.C., Smith A.B., Bates P., Diamond S.L. Mol. Pharmacol. 2010;78(2):319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrientos L.G., Rollin P.E. Virology. 2007;358(1):1–9. doi: 10.1016/j.virol.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Science. 2005;308(5728):1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elshabrawy H.A., Fan J., Haddad C.S., Ratia K., Broder C.C., Caffrey M., Prabhakar B.S. J. Virol. 2014;88(8):4353–4365. doi: 10.1128/JVI.03050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. J. Virol. 2006;80(8):4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Liu Q., Zhang N., Li Q.Q., Liu Z.D., Li Y.H., Gao L.M., Wang Y.C., Deng H.B., Song D.Q. Eur. J. Med. Chem. 2018;149:45–55. doi: 10.1016/j.ejmech.2018.02.061. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H., Lear-Rooney C.M., Johansen L., Varhegyi E., Chen Z.W., Olinger G.G., Rong L. J. Virol. 2015;89(19):9932–9938. doi: 10.1128/JVI.01337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., DeWald L.E., Schornberg K.L., Scully C., Lehar J., Hensley L.E., White J.M., Olinger G.G. Sci. Transl. Med. 2013;5(190):190ra79. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Nature. 2016;535(7610):169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veljkovic V., Goeijenbier M., Glisic S., Veljkovic N., Perovic V.R., Sencanski M., Branch D.R., Paessler S. F1000Research. 2015;4(104):1–10. doi: 10.12688/f1000research.6436.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paessler S., Huang C., Sencanski M., Veljkovic N., Perovic V., Glisic S., Veljkovic V. Front. Biosci. (Landmark Ed.) 2018;23:947–953. doi: 10.2741/4627. [DOI] [PubMed] [Google Scholar]
- 39.Johansen L.M., DeWald L.E., Shoemaker C.J., Hoffstrom B.G., Lear-Rooney C.M., Stossel A., Nelson E., Delos S.E., Simmons J.A., Grenier J.M., Pierce L.T., Pajouhesh H., Lehar J., Hensley E., Glass P.J., White J.M., Olinger G.G. Sci. Transl. Med. 2015;7(290):290ra89. doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- 40.Ren J., Zhao Y., Fry E.E., Stuart D. J. Med. Chem. 2018;61:724–733. doi: 10.1021/acs.jmedchem.7b01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honko A.N., Johnson J.C., Marchand J.S., Huzella L., Ricky D., Oberlander N., Torzewski L.M., Bennett R.S., Hensley L.E., Jahrling P.B., Olinger G.G. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-06179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basu A., Li B., Mills D.M., Panchal R.G., Cardinale S.C., Butler M.M., Peet N.P., Majgier-Baranowska H., Williams J.D., Patel I., Moir D.T., Bavari S., Ray R., Farzan M.R., Rong L., Bowlin T.L. J. Virol. 2011;85(7):3106–3119. doi: 10.1128/JVI.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Cin P.D., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Nature. 2011;477(7364):344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoemaker C.J., Schornberg K.L., Delos S.E., Scully C., Pajouhesh H., Olinger G.G., Johansen L.M., White J.M. PLoS One. 2013;8(2):e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gehring G., Rohrmann K., Atenchong N., Mittler E., Becker S., Dahlmann F., Pöhlmann S., Vondran F.W.R., David S., Manns M.P., Ciesek S., von Hahn T.J. Antimicrob. Chemother. 2014;69(8):2123–2131. doi: 10.1093/jac/dku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. ACS Infect. Dis. 2016;1(7):317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 48.Salata C., Baritussio A., Munegato D., Calistri A., Ha H.R., Bigler L., Fabris F., Parolin C., Palù G., Mirazimi A. Pathog. Dis. 2015;73(5):1–9. doi: 10.1093/femspd/ftv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salata C., Calistri A., Parolin C., Baritussio A., Palù G. Expert Rev. Anti. Infect. Ther. 2017;15(5):483–492. doi: 10.1080/14787210.2017.1305888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salata C., Munegato D., Martelli F., Parolin C., Calistri A., Baritussio A., Palù G. New Microbiol. 2018;2(41):1. [PubMed] [Google Scholar]
- 51.Miller M.E., Adhikary S., Kolokoltsov A.A., Davey R.A. J. Virol. 2012;86(14):7473–7483. doi: 10.1128/JVI.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun W., He S., Martínez-Romero C., Kouznetsova J., Tawa G., Xu M., Shinn P., Fisher E.G., Long Y., Motabar O., Yang S., Sanderson P.E., Williamson P.R., García-Sastre A., Qiu X., Zheng W. Antivir. Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shedlock D.J., Bailey M.A., Popernack P.M., Cunningham J.M., Burton D.R., Sullivan N.J. Virology. 2010;401(2):228–235. doi: 10.1016/j.virol.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trad M.A., Naughton W., Yeung A., Mazlin L., O’sullivan M., Gilroy N., Fisher D.A., Stuart R.L. J. Clin. Virol. 2017;86:5–13. doi: 10.1016/j.jcv.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Pettitt J., Zeitlin L., Kim D.H., Working C., Johnson J.C., Bohorov O., Bratcher B., Hiatt E., Hume S.D., Johnson A.K., Morton J., Pauly M.H., Whaley K.J., Ingram M.F., Zovanyi A., Heinrich M., Piper A., Zelko J., Olinger G.G. Sci. Transl. Med. 2013;5(199):199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 56.Olinger G.G., Pettitt J., Kim D., Working C., Bohorov O., Bratcher B., Hiatt E., Hume S.D., Johnson A.K., Morton J., Pauly M., Whaley K.J., Lear C.M., Biggins J.E., Scully C., Hensley L., Zeitlin L. Proc. Natl. Acad. Sci. U. S. A. 2012;109(44):18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu X., Audet J., Wong G., Pillet S., Bello A., Cabral T., Strong J.E., Plummer F., Corbett C.R., Alimonti J.B., Kobinger G.P. Sci. Transl. Med. 2012;4(138):138ra81. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 58.Qiu X., Wong G., Fernando L., Audet J., Bello A., Strong J., Alimonti J.B., Kobinger G.P. Sci. Transl. Med. 2013;5(207):207ra143. doi: 10.1126/scitranslmed.3006605. [DOI] [PubMed] [Google Scholar]
- 59.Smith L.M., Hensley L.E., Geisbert T.W., Johnson J., Stossel A., Honko A., Yen J.Y., Geisbert J., Paragas J., Fritz E., Olinger G., Young H.A., Rubins K.H., Karp C.L. J. Infect. Dis. 2013;208(2):310–318. doi: 10.1093/infdis/jis921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson E., Bryan C., Fong R.H., Barnes T., Pfaff J.M., Mabila M., Rucker J.B., Doranz B.J. J. Virol. 2015;89(21):10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., Goodman C., Kim D., Pauly M.H., Velasco J., Pettitt J., Olinger G.G., Whaley K., Xu B., Strong J.E., Zeitlin L., Kobinger G.P. Nature. 2014;514(7520):47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettit D.K., Rogers R.S., Arthur K., Brodsky Y., Clark R.H., Crowell C., Ennis J., Gillespie A., Gillespie R., Livingston B., Nalbandian E., Pace D., Smidt P., Pauly M., Timmons K., Trentalange M., Whaley K.J., Zeitlin L., Thomas J.N. mAbs. 2016;8(2):347–357. doi: 10.1080/19420862.2015.1127492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.W., Heo W., Lee J., Jin N., Yoon S.M., Park K.Y., Kim E.Y., Kim W.T., Kim J.Y. PLoS One. 2018;13(1):1–16. doi: 10.1371/journal.pone.0191075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu X., Audet J., Lv M., He S., Wong G., Wei H., Luo L., Fernando L., Kroeker A., Bovendo H.F., Bello A., Li F., Ye P., Jacobs M., Ippolito G., Saphire E.O., Bi S., Shen B., Gao G.F., Zeitlin L., Feng J., Zhang B., Kobinger G.P. Sci. Transl. Med. 2016;8(329):1–19. doi: 10.1126/scitranslmed.aad9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mora-Rillo M., Arsuaga M., Ramirez-Olivencia G., de la Calle F., Borobia A.M., Sanchez-Seco P., Lago M., Figueira J.C., Fernandez-Puntero B., Viejo A., Negredo A., Nunez C., Flores E., Carcas A.J., Jimenez-Yuste V., Lasala F., Garcia-de-Lorenzo A., Arnalich F., Arribas J.R. Lancet Respir. Med. 2015;3(7):554–562. doi: 10.1016/S2213-2600(15)00180-0. [DOI] [PubMed] [Google Scholar]
- 66.Schibler M., Vetter P., Cherpillod P., Petty T.J., Cordey S., Vieille G., Yerly S., Siegrist C.-A., Samii K., Dayer J.-A., Docquier M., Zdobnov E.M., Simpson A.J.H., Rees P.S.C., Sarria F.B., Gasche Y., Chappuis F., Iten A., Pittet D., Pugin J., Kaiser L. Lancet Infect. Dis. 2015;15(9):1034–1040. doi: 10.1016/S1473-3099(15)00229-7. [DOI] [PubMed] [Google Scholar]
- 67.Davey R.T.J., Dodd L., Proschan M.A., Neaton J., Neuhaus Nordwall J., Koopmeiners J.S., Beigel J., Tierney J., Lane H.C., Fauci A.S., Massaquoi M.B.F., Sahr F., Malvy D.N. Engl. J. Med. 2016;375(15):1448–1456. doi: 10.1056/NEJMoa1604330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong S., Yang P., Li G., Liu B., Wang W., Liu X., Xia B., Yang C., Lou Z., Guo Y., Rao Z. Protein Cell. 2015;6(5):351–362. doi: 10.1007/s13238-015-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oestereich L., Lüdtke A., Wurr S., Rieger T., Muñoz-Fontela C., Günther S. Antivir. Res. 2014;105(1):17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 70.Smither S.J., Eastaugh L.S., Steward J.A., Nelson M., Lenk R.P., Lever M.S. Antivir. Res. 2014;104(1):153–155. doi: 10.1016/j.antiviral.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 71.Sissoko D., Laouenan C., Folkesson E., M’Lebing A.-B., Beavogui A.-H., Baize S., Camara A.-M., Maes P., Shepherd S., Danel C., Carazo S., Conde M.N., Gala J.-L., Colin G., Savini H., Bore J.A., Le Marcis F., Koundouno F.R., Petitjean F., Lamah M.-C., Diederich S., Tounkara A., Poelart G., Berbain E., Dindart J.-M., Duraffour S., Lefevre A., Leno T., Peyrouset O., Irenge L., Bangoura N., Palich R., Hinzmann J., Kraus A., Barry T.S., Berette S., Bongono A., Camara M.S., Chanfreau Munoz V., Doumbouya L., Harouna S., Kighoma P.M., Koundouno F.R., Lolamou R., Loua C.M., Massala V., Moumouni K., Provost C., Samake N., Sekou C., Soumah A., Arnould I., Komano M.S., Gustin L., Berutto C., Camara D., Camara F.S., Colpaert J., Delamou L., Jansson L., Kourouma E., Loua M., Malme K., Manfrin E., Maomou A., Milinouno A., Ombelet S., Sidiboun A.Y., Verreckt I., Yombouno P., Bocquin A., Carbonnelle C., Carmoi T., Frange P., Mely S., Nguyen V.K., Pannetier D., Taburet A.-M., Treluyer J.-M., Kolie J., Moh R., Gonzalez M.C., Kuisma E., Liedigk B., Ngabo D., Rudolf M., Thom R., Kerber R., Gabriel M., Di Caro A., Wölfel R., Badir J., Bentahir M., Deccache Y., Dumont C., Durant J.-F., El Bakkouri K., Gasasira Uwamahoro M., Smits B., Toufik N., Van Cauwenberghe S., Ezzedine K., Dortenzio E., Pizarro L., Etienne A., Guedj J., Fizet A., Barte de Sainte Fare E., Murgue B., Tran-Minh T., Rapp C., Piguet P., Poncin M., Draguez B., Allaford Duverger T., Barbe S., Baret G., Defourny I., Carroll M., Raoul H., Augier A., Eholie S.P., Yazdanpanah Y., Levy-Marchal C., Antierrens A., Van Herp M., Günther S., de Lamballerie X., Keïta S., Mentre F., Anglaret X., Malvy D., Group J.S. PLoS Med. 2016;13(3):e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McMullan L.K., Flint M., Dyall J., Albariño C., Olinger G.G., Foster S., Sethna P., Hensley L.E., Nichol S.T., Lanier E.R., Spiropoulou C.F. Antivir. Res. 2016;125(Suppl. C):71–78. doi: 10.1016/j.antiviral.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 73.Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Nature. 2014;508:402. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Florescu D.F., Kalil A.C., Hewlett A.L., Schuh A.J., Stroher U., Uyeki T.M., Smith P.W. Clin. Infect. Dis. 2015;61(6):969–973. doi: 10.1093/cid/civ395. [DOI] [PubMed] [Google Scholar]
- 75.Dunning J., Kennedy S.B., Antierens A., Whitehead J., Ciglenecki I., Carson G., Kanapathipillai R., Castle L., Howell-Jones R., Pardinaz-Solis R., Grove J., Scott J., Lang T., Olliaro P., Horby P.W., Boyd M., Das D., Dawson H., Fardolo A.F., Gozalbes J.G., Horace M.T.C., Jackson M., Kerrie J., Leligdowicz A., Longuere K.S., Mambo A., McSwiggan S., Merson L., Moore M., Mulbah F.M., Odam M., Rojek A., Scott P., Welsley E., Wemmah B. PLoS One. 2016;11(9):e0162199. doi: 10.1371/journal.pone.0162199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor R., Kotian P., Warren T., Panchal R., Bavari S., Julander J., Dobo S., Rose A., El-Kattan Y., Taubenheim B., Babu Y., Sheridan W.P. J. Infect. Public Health. 2017;9(3):220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.-S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Nature. 2016;531:381. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacobs M., Rodger A., Bell D.J., Bhagani S., Cropley I., Filipe A., Gifford R.J., Hopkins S., Hughes J., Jabeen F., Johannessen I., Karageorgopoulos D., Lackenby A., Lester R., Liu R.S.N., MacConnachie A., Mahungu T., Martin D., Marshall N., Mepham S., Orton R., Palmarini M., Patel M., Perry C., Peters S.E., Porter D., Ritchie D., Ritchie N.D., Seaton R.A., Sreenu V.B., Templeton K., Warren S., Wilkie G.S., Zambon M., Gopal R., Thomson E.C. Lancet London, England. 2016;388(10043):498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hart G.J., Orr D.C., Penn C.R., Figueiredo H.T., Gray N.M., Boehme R.E., Cameron J.M. Antimicrob. Agents Chemother. 1992;36(8):1688–1694. doi: 10.1128/aac.36.8.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai C.Y., Tseng T.C., Wong G.L.H., Huang J.F., Wong V.W.S., Liu C.J., Yu M.L., Chuang W.L., Kao J.H., Yuen Chan H.L., Chen D.S. J. Antimicrob. Chemother. 2013;68(10):2332–2338. doi: 10.1093/jac/dkt193. [DOI] [PubMed] [Google Scholar]
- 81.Cong Y., Dyall J., Hart B.J., DeWald L.E., Johnson J.C., Postnikova E., Zhou H., Gross R., Rojas O., Alexander I., Josleyn N., Zhang T., Michelotti J., Janosko K., Glass P.J., Flint M., McMullan L.K., Spiropoulou C.F., Mierzwa T., Guha R., Shinn P., Michael S., Klumpp-Thomas C., McKnight C., Thomas C., Eakin A.E., O’Loughlin K.G., Green C.E., Catz P., Mirsalis J.C., Honko A.N., Olinger G.G., Bennett R.S., Holbrook M.R., Hensley L.E., Jahrling P.B. PLoS One. 2016;11(11):e0166318. doi: 10.1371/journal.pone.0166318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filone C.M., Hodges E.N., Honeyman B., Bushkin G.G., Boyd K., Platt A., Ni F., Strom K., Hensley L., Snyder J.K., Connor J.H. Chem. Biol. 2013;20(3):424–433. doi: 10.1016/j.chembiol.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geisbert T.W., Lee A.C., Robbins M., Geisbert J.B., Honko A.N., Sood V., Johnson J.C., de Jong S., Tavakoli I., Judge A., Hensley L.E., MacLachlan I. Lancet. 2010;375(9729):1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thi E.P., Mire C.E., Lee A.C.H., Geisbert J.B., Zhou J.Z., Agans K.N., Snead N.M., Deer D.J., Barnard T.R., Fenton K.A., Maclachlan I., Geisbert T.W. Nature. 2015;521(7552):362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stahelin R.V. Front. Microbiol. 2014;5:300. doi: 10.3389/fmicb.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruigrok R.W., Schoehn G., Dessen A., Forest E., Volchkov V., Dolnik O., Weissenhorn W. Klenk HD. J. Mol. Biol. 2000;300(1):103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 87.Gomis-Rüth F.X., Dessen A., Timmins J., Bracher A., Kolesnikowa L., Becker S., Klenk H.D., Weissenhorn W. Structure. 2003;11(4):423–433. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bornholdt Z.A., Noda T., Abelson D.M., Halfmann P., Wood M.R., Kawaoka Y., Saphire E.O. Cell. 2013;154(4):763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoenen T., Volchkov V., Kolesnikova L., Mittler E., Timmins J., Ottmann M., Reynard O.S.B., Weissenhorn W. J. Virol. 2005;79(3):1898–1905. doi: 10.1128/JVI.79.3.1898-1905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karthick V., Nagasundaram N., Doss C.G.P., Chakraborty C., Siva R., Lu A., Zhang G., Zhu H. Infect. Dis. Poverty. 2016;5(1):1–10. doi: 10.1186/s40249-016-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shah R., Panda P.K., Patel P., Panchal H. World J. Pharm. Pharm. Sci. 2015;4(5):1268–1282. [Google Scholar]
- 92.Palamthodi S., Patil D., Sankpal A., Zarekar S., Patil Y. J. Comput. Methods Mol. Des. 2012;2(2):76–84. [Google Scholar]
- 93.Chen C.Y.C. PLoS One. 2011;6(1):1–5. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bordeleau M.E., Robert F., Gerard B., Lindqvist L., Chen S.M.H., Wendel H.G., Brem B., Greger H., Lowe S.W., Porco J.A., Pelletier J. J. Clin. Invest. 2008;118(7):2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pelletier J., Graff J., Ruggero D., Sonenberg N. Cancer Res. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sadlish H., Galicia-Vazquez G., Paris C.G., Aust T., Bhullar B., Chang L., Helliwell S.B., Hoepfner D., Knapp B., Riedl R., Roggo S., Schuierer S., Studer C., Porco J.A., Pelletier J., Movva N.R. ACS Chem. Biol. 2013;8(7):1519–1527. doi: 10.1021/cb400158t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Biedenkopf N., Lange-Grünweller K., Schulte F.W., Weißer A., Müller C., Becker D., Becker S., Hartmann R.K., Grünweller A. Antivir. Res. 2017;137:76–81. doi: 10.1016/j.antiviral.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 98.Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elgner F., Sabino C., Basic M., Ploen D., Grünweller A., Hildt E. Viruses. 2018;10(4):149. doi: 10.3390/v10040149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Modrof J., Mühlberger E., Klenk H.D., Becker S. J. Biol. Chem. 2002;277(36):33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- 101.Huggins J.W., Zhang Z.X., Monath T.P. Antivir. Res. 1991;15:122. [Google Scholar]
- 102.Huggins J., Zhang Z.X., Bray M. J. Infect. Dis. 1999;179(Suppl. 1):S240–S247. doi: 10.1086/514316. [DOI] [PubMed] [Google Scholar]
- 103.Huggins J., Zhang Z.X., Davis K., Coulombe R.A. Antivir. Res. 1995;26(3):A301. [Google Scholar]
- 104.Bray M., Driscoll J., Huggins J.W. Antivir. Res. 2000;45(2):135–147. doi: 10.1016/s0166-3542(00)00066-8. [DOI] [PubMed] [Google Scholar]
- 105.Bray M., Raymond J.L., Geisbert T., Baker R.O. Antivir. Res. 2002;55(1):151–159. doi: 10.1016/s0166-3542(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 106.Ye W., Schneller S.W. Bioorg. Med. Chem. 2014;22(19):5315–5319. doi: 10.1016/j.bmc.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 107.Warren T.K., Warfield K.L., Wells J., Enterlein S., Smith M., Ruthel G., Yunus A.S., Kinch M.S., Goldblatt M., Aman M.J., Bavari S. Antimicrob. Agents Chemother. 2010;54(5):2152–2159. doi: 10.1128/AAC.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kinch M.S., Yunus A.S., Lear C., Mao H., Chen H., Fesseha Z., Luo G., Nelson E.A., Li L., Huang Z., Murray M., Ellis W.Y., Hensley L., Christopher-Hennings J., Olinger G.G., Goldblatt M. Am. J. Transl. Res. 2009;1(1):87–98. [PMC free article] [PubMed] [Google Scholar]
- 109.Aman M.J., Kinch M.S., Warfield K., Warren T., Yunus A., Enterlein S., Stavale E., Wang P., Chang S., Tang Q., Porter K., Goldblatt M., Bavari S. Antivir. Res. 2009;83(3):245–251. doi: 10.1016/j.antiviral.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 110.Opsenica I., Burnett J.C., Gussio R., Opsenica D., Todorović N., Lanteri C.A., Sciotti R.J., Gettayacamin M., Basilico N., Taramelli D., Nuss J.E., Wanner L., Panchal R.G., Šolaja B.A., Bavari S. J. Med. Chem. 2011;54(5):1157–1169. doi: 10.1021/jm100938u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selaković Ž., Opsenica D., Eaton B., Retterer C., Bavari S., Burnett J.C., Šolaja B.A., Panchal R.G. Viruses. 2012;4(8):1279–1288. doi: 10.3390/v4081279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ari H., Markus A. Annu. Rev. Biochem. 1995;73:1019–1049. [Google Scholar]
- 113.Hebert D.N., Foellmer B., Helenius A. Cell. 1995;81(3):425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 114.Alonzi D.S., Kukushkin N.V., Allman S.A., Hakki Z., Williams S.J., Pierce L., Dwek R.A., Butters T.D. Cell. Mol. Life Sci. 2013;70(15):2799–2814. doi: 10.1007/s00018-013-1304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dwek R.A., Butters T.D., Platt Frances M.Z.N. Nat. Rev. Drug Discov. 2002;1:65–75. doi: 10.1038/nrd708. [DOI] [PubMed] [Google Scholar]
- 116.Chang J., Block T.M., Guo J.T. Antivir. Res. 2013;99(3):251–260. doi: 10.1016/j.antiviral.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fischl M., Resnick L., Coomb R., Kremer A., Pottage J.J., Fass R., KH, F., Powderly W., Collier A., Aspinall R. J. Acquir. Immune Defic. Syndr. 1994;7(2):139–147. [PubMed] [Google Scholar]
- 118.Mehta A., Zitzmann N., Rudd P.M., Block T.M., Dwek R.A. FEBS Lett. 1998;430:17–22. doi: 10.1016/s0014-5793(98)00525-0. [DOI] [PubMed] [Google Scholar]
- 119.Chang J., Warren T.K., Zhao X., Gill T., Guo F., Wang L., Comunale M.A., Du Y., Alonzi D.S., Yu W., Ye H., Liu F., Guo J., Mehta A., Cuconati A., Butters T.D., Bavari S., Block T.M. Antivir. Res. 2013;98(3):432–440. doi: 10.1016/j.antiviral.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chan S.Y., Speck R.F., Ma M.C., Goldsmith M.A. J. Virol. 2000;74(10):4933–4937. doi: 10.1128/jvi.74.10.4933-4937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ji X., Olinger G.G., Aris S., Chen Y., Gewurz H., Spear G.T. J. Gen. Virol. 2005;86(9):2535–2542. doi: 10.1099/vir.0.81199-0. [DOI] [PubMed] [Google Scholar]
- 122.Michelow I.C., Lear C., Scully C., Prugar L.I., Longley C.B., Yantosca L.M., Ji X., Karpel M., Brudner M., Takahashi K., Spear G.T., Ezekowitz R.A.B., Schmidt E.V., Olinger G.G. J. Infect. Dis. 2011;203(2):175–179. doi: 10.1093/infdis/jiq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Versteeg G.A., García-Sastre A. Curr. Opin. Microbiol. 2010:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zinzula L., Tramontano E. Antivir. Res. 2013;100(3):615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reid S.P., Leung L.W., Hartman A.L., Martinez O., Shaw M.L., Carbonnelle C., Volchkov V.E., Nichol S.T., Basler C.F.D. J. Virol. 2006;80(11):5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reid S.P., Valmas C., Martinez O., Mauricio Sanchez F., Basler C.F. J. Virol. 2007;81(24):13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mateo M., Reid S.P., Leung L.W., Basler C.F., Volchkov V.E. J. Virol. 2010;84(2):1169–1175. doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leung D.W., Ginder N.D., Fulton D.B., Nix J., Basler C.F., Honzatko R.B., Amarasinghe G.K. Proc. Natl. Acad. Sci. U. S. A. 2009;106(2):411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cárdenas W.B., Loo Y.M., Gale M., Hartman A.L., Kimberlin C.R., Martínez-Sobrido L., Saphire E.O., Basler C.F. J. Virol. 2006;80(11):5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. J. Exp. Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yen B., Mulder L.C.F., Martinez O., Basler C.F. J. Virol. 2014;88(21):12500–12510. doi: 10.1128/JVI.02163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Prins K.C., Delpeut S., Leung D.W., Reynard O., Volchkova V.A., Reid S.P., Ramanan P., Cárdenas W.B., Amarasinghe G.K., Volchkov V.E., Basler C.F., Mahanty S., Bray M., Kash J.C., Mühlberger E., Carter V., Grosch M., Perwitasari O., Proll S.C., Thomas M.J., Weber F., Klenk H.-D., Katze M.G. J. Virol. 2010;84(6):3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cardenas W.B., Loo Y.M., Gale M., Hartman A.L., Kimberlin C.R., Martinez-Sobrido L., Saphire E.O., Basler C.F. J. Virol. 2006;80(11):5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hartman A.L., Ling L., Nichol S.T., Hibberd M.L. J. Virol. 2008;82(11):5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hartman A.L., Bird B.H., Towner J.S., Antoniadou Z.-A., Zaki S.R., Nichol S.T. J. Virol. 2008;82(159):2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leung D.W., Prins K.C., Borek D.M., Farahbakhsh M., Tufariello J.M., Ramanan P., Nix J.C., Helgeson L.A., Otwinowski Z., Honzatko R.B., Basler C.F., Amarasinghe G.K. Nat. Struct. Mol. Biol. 2010;17(2):165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zinzula L., Tramontano E. Antivir. Res. 2013;100(3):615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ekins S., Freundlich J.S., Coffee M.F. F1000Research. 2014;3(277):1–15. doi: 10.12688/f1000research.5741.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gignoux E., Azman A.S., de Smet M., Azuma P., Massaquoi M., Job D., Tiffany A., Petrucci R., Sterk E., Potet J., Suzuki M., Kurth A., Cannas A., Bocquin A., Strecker T., Logue C., Pottage T., Yue C., Cabrol J.-C., Serafini M., Ciglenecki I. N. Engl. J. Med. 2016;374(1):23–32. doi: 10.1056/NEJMoa1504605. [DOI] [PubMed] [Google Scholar]
- 140.Brown C.S., Lee M.S., Leung D.W., Wang T., Xu W., Luthra P., Anantpadma M., Shabman R.S., Melito L.M., Macmillan K.S., Borek D.M., Otwinowski Z., Ramanan P., Stubbs A.J., Peterson D.S., Binning J.M., Tonelli M., Olson M.A., Davey R.A., Ready J.M., Basler C.F., Amarasinghe G.K. J. Mol. Biol. 2014;426(10):2045–2058. doi: 10.1016/j.jmb.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Glanzer J.G., Byrne B.M., McCoy A.M., James B.J., Frank J.D., Oakley G.G. Bioorg. Med. Chem. 2016;24(21):5388–5392. doi: 10.1016/j.bmc.2016.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ren J.X., Zhang R.T., Zhang H., Cao X.S., Liu L.K., Xie Y. Biomed. Pharmacother. 2016;84:199–207. doi: 10.1016/j.biopha.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 143.Mirza M.U., Ikram N. Int. J. Mol. Sci. 2016;17(11) doi: 10.3390/ijms17111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baikerikar S. Pharm. Res. 2017;9(5):15. [Google Scholar]
- 145.Cannas V., Daino G.L., Corona A., Esposito F., Tramontano E. J. Infect. Dis. 2015;212(Suppl. 2):S277–S281. doi: 10.1093/infdis/jiv214. [DOI] [PubMed] [Google Scholar]
- 146.Di Petrillo A., Fais A., Pintus F., Santos-Buelga C., González-Paramás A.M., Piras V., Orrù G., Mameli A., Tramontano E., Frau A. BMC Microbiol. 2017;17(159):1–9. doi: 10.1186/s12866-017-1068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mateo M., Carbonnelle C., Reynard O., Kolesnikova L., Nemirov K., Page A., Volchkova V.A., Volchkov V.E. J. Infect. Dis. 2011;204(Suppl. 3):1011–1020. doi: 10.1093/infdis/jir338. [DOI] [PubMed] [Google Scholar]
- 148.Ebihara H., Takada A., Kobasa D., Jones S., Neumann G., Theriault S. PLoS Pathog. 2006;2(7):1–7. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang A.P.P., Bornholdt Z.A., Liu T., Abelson D.M., Lee D.E., Li S., Woods V.L., Saphire E.O. PLoS Pathog. 2012;8(2):e1002550. doi: 10.1371/journal.ppat.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Iversen P.L., Warren T.K., Wells J.B., Garza N.L., Mourich D.V., Welch L.S., Panchal R.G., Sina B. Viruses. 2012;4:2806–2830. doi: 10.3390/v4112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Warren T.K., Whitehouse C.A., Wells J., Welch L., Heald A.E., Charleston J.S., Sazani P., Reid P., Iversen P.L. mBio. 2015;6(1):1–4. doi: 10.1128/mBio.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Song X., Lu L., Passioura T., Suga H. Org. Biomol. Chem. 2017;15(24):5155–5160. doi: 10.1039/c7ob00012j. [DOI] [PubMed] [Google Scholar]
- 153.Fanunza E., Frau A., Sgarbanti M., Orsatti R., Corona A., Tramontano E. Viruses. 2018;10(2):98. doi: 10.3390/v10020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu W., Edwards M.R., Borek D.M., Feagins A.R., Mittal A., Alinger J.B., Berry K.N., Yen B., Hamilton J., Brett T.J., Pappu R.V., Leung D.W., Basler C.F., Amarasinghe G.K. Cell Host Microbe. 2015;16(2):187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Darapaneni V. AJC Microbiol. 2014;3(1):14–22. [Google Scholar]
- 156.Chakraborty S., Rao B.J., Asgeirsson B., Dandekar A.M. F1000Research. 2015;3(251):1–17. doi: 10.12688/f1000research.5573.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Plesko S., Volk H., Luksic M., Podlipnik C. Acta Chim. Slov. 2015;62(3):555–564. [PubMed] [Google Scholar]
- 158.Setlur A.S., Naik S.Y., Skariyachan S. Interdiscip. Sci. Comput. Life Sci. 2016;9(2):254–277. doi: 10.1007/s12539-016-0149-8. [DOI] [PubMed] [Google Scholar]
- 159.Todorov D., Hinkov A., Shishkova K., Shishkov S. Phytochem. Rev. 2014;13(2):525–538. [Google Scholar]
- 160.Makau J.N., Watanabe K., Kobayashi N. Drug Discov. Ther. 2013;7(5):189–195. [PubMed] [Google Scholar]
- 161.Raj U., Varadwaj P.K. Interdiscip. Sci. Comput. Life Sci. 2016;8(2):132–141. doi: 10.1007/s12539-015-0109-8. [DOI] [PubMed] [Google Scholar]
- 162.Willför S., Ali M., Karonen M., Reunanen M., Arfan M., Harlamow R. Holzforschung. 2009;63(5):551–558. [Google Scholar]
- 163.Kaul T.N., Middleton E., Ogra P.L. J. Med. Virol. 1985;15(1):71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 164.Zhao Z., Martin C., Fan R., Bourne P.E., Xie L. BMC Bioinformatics. 2016;17(1):1–12. doi: 10.1186/s12859-016-0941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Garcia-Dorival I., Wu W., Dowall S., Armstrong S., Touzelet O., Wastling J., Barr J.N., Matthews D., Carroll M., Hewson R., Hiscox J.A. J. Proteome Res. 2014;13(11):5120–5135. doi: 10.1021/pr500556d. [DOI] [PubMed] [Google Scholar]
- 166.Luthra P., Aguirre S., Yen B.C., Pietzsch C.A., Sanchez-Aparicio M.T., Tigabu B., Morlock L.K., García-Sastre A., Leung D.W., Williams N.S., Fernandez-Sesma A., Bukreyev A., Basler C.F. mBio. 2017;8(2) doi: 10.1128/mBio.00368-17. e00368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noda T., Ebihara H., Muramoto Y., Fujii K., Takada A., Sagara H., Jin H.K., Kida H., Feldmann H., Kawaoka Y. PLoS Pathog. 2006;2(9):0864–0872. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Y., Lee M.S., Olson M.A., Harty R.N. Adv. Virol. 2011;2011:1–10. doi: 10.1155/2011/341816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Han Z., Lu J., Liu Y., Davis B., Lee M.S., Olson M.A., Ruthel G., Freedman B.D., Schnell M.J., Wrobel J.E., Reitz A.B., Harty R.N. J. Virol. 2014;88(13):7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.García M., Coope A., Shi W., Bornmann W., Carrion R., Kalman D.J., Nabel G. 2012;4(123) doi: 10.1126/scitranslmed.3003500. 123a24. [DOI] [PMC free article] [PubMed] [Google Scholar]