Abstract
Background
The leading cause of acute illnesses, respiratory viruses, typically cause self-limited diseases, although severe complications can occur in fragile patients. Rhinoviruses (RVs), respiratory enteroviruses (EVs), influenza virus, respiratory syncytial viruses (RSVs), and coronaviruses are highly prevalent respiratory pathogens, but because of the lack of reliable animal models, their differential pathogenesis remains poorly characterized.
Objective
We sought to compare infections by respiratory viruses isolated from clinical specimens using reconstituted human airway epithelia.
Methods
Tissues were infected with RV-A55, RV-A49, RV-B48, RV-C8, and RV-C15; respiratory EV-D68; influenza virus H3N2; RSV-B; and human coronavirus (HCoV)–OC43. Replication kinetics, cell tropism, effect on tissue integrity, and cytokine secretion were compared. Viral adaptation and tissue response were assessed through RNA sequencing.
Results
RVs, RSV-B, and HCoV-OC43 infected ciliated cells and caused no major cell death, whereas H3N2 and EV-D68 induced ciliated cell loss and tissue integrity disruption. H3N2 was also detected in rare goblet and basal cells. All viruses, except RV-B48 and HCoV-OC43, altered cilia beating and mucociliary clearance. H3N2 was the strongest cytokine inducer, and HCoV-OC43 was the weakest. Persistent infection was observed in all cases. RNA sequencing highlighted perturbation of tissue metabolism and induction of a transient but important immune response at 4 days after infection. No majority mutations emerged in the viral population.
Conclusion
Our results highlight the differential in vitro pathogenesis of respiratory viruses during the acute infection phase and their ability to persist under immune tolerance. These data help to appreciate the range of disease severity observed in vivo and the occurrence of chronic respiratory tract infections in immunocompromised hosts.
Key words: Respiratory virus, rhinovirus, cytotoxicity, pathogenesis, cytokines, mucociliary clearance, immune response, persistence
Abbreviations used: dpi, Days after infection; EV, Enterovirus; HCoV, Human coronavirus; HTS, High-throughput RNA sequencing; IP-10, Interferon-inducible protein 10; LDH, Lactate dehydrogenase; MCC, Mucociliary clearance; qPCR, Real-time quantitative PCR; RSV, Respiratory syncytial virus; RV, Rhinovirus; TEER, Transepithelial electrical resistance
Acute respiratory tract infections are the leading cause of both illness and death in postneonatal children at less than 5 years of age.1 Viruses represent the main cause of respiratory tract infections,2 among which the predominant causative agent is rhinovirus (RV) (40% of all viral respiratory tract infections),3 followed by influenza virus and respiratory syncytial virus (RSV). This latter infection has been reported as the main viral cause of hospitalization in infants, followed by RV and influenza virus.4 Unexpected outbreaks can also be caused by emerging or re-emerging pathogens, such as enterovirus (EV)–D68, which was responsible for the nationwide US outbreak of mild-to-severe respiratory illness in 2014 that especially affected the pediatric population.5 Even though respiratory viruses generally cause self-limited illnesses, the societal effect is significant because of work and school absenteeism, medical visits, and misuse of antibiotics.6 Furthermore, lower respiratory tract infections, chronic infections, and complications, such as bronchiolitis, pneumonia, and asthma exacerbations, are frequent in young children, immunocompromised subjects, or those with underlying conditions. Despite this considerable burden to individual well-being and the public health care system, supportive care is the only therapeutic option against most respiratory viruses.7 Therefore a better understanding of their pathogenesis is crucial to develop more effective antiviral drugs or vaccines.
Because cellular receptors are species specific for most human respiratory viruses, only a few animal models exist to study this prevalent and diverse group of pathogens. Selected mouse strains or transgenic mice expressing human receptors, as well as mouse-adapted viruses, are used8, 9, 10; however, they mimic poorly pathogenesis in a human host. Most in vitro studies are performed in immortalized cells and sometimes in primary cells derived from human airways, but these cell lines cannot fully reproduce the complexity of the human airway epithelium. In addition, some respiratory viruses, such as RVs from species C, grow poorly in standard cell lines.11 In this respect much effort has been dedicated to develop more representative models, such as airway epithelia reconstituted from primary cells isolated from nasal or bronchial surgeries. These air-liquid interface tissue-culture systems have been validated for the study of respiratory viruses, such as RSV,12 influenza,13 human coronaviruses (HCoVs), and RVs.14, 15, 16 They reproduce the pseudostratified architecture of the human airway epithelium, which is composed of basal, ciliated, and goblet cells, as well as its defense mechanisms (ie, mucociliary clearance [MCC] and epithelial cell immunity).17, 18, 19
Using standardized in vitro–reconstituted human airway epithelia and clinical viral strains, we could compare infection patterns by the most prevalent respiratory viruses, including 5 RVs, a respiratory EV, RSV, influenza, and HCoV. For each infection, we compared the replication kinetics, cellular tropism, and effect of the virus on tissue integrity or cytokine secretion. In addition, we performed high-throughput RNA sequencing (HTS) on viruses and host tissues collected at different times after infection to investigate changes in viral populations and tissue response. Our data highlight the differential in vitro pathogenesis of these representative respiratory pathogens and reveal the systematic viral persistence in the absence of a fully competent immune response.
Methods
Detailed procedures are provided in the Methods section in this article's Online Repository at www.jacionline.org.
Human airway epithelia reconstituted in vitro (MucilAir)
MucilAir tissues (Epithelix Sàrl, Geneva, Switzerland) were cultured at the air-liquid interface from a mixture of nasal polyp epithelial cells originating from 14 healthy donors, as previously described.16
Viruses
The apical surface was inoculated with 100 μL of infected sample. Sample collection was performed, as previously reported.16 To prepare viral stocks, apical samples collected 2 days after infection (dpi) were used to infect 5 new tissues. Samples were collected daily, and the 4 samples with highest viral loads were pooled. Isolated viruses were purified as described,20 and viral stocks were quantified by using real-time quantitative PCR (qPCR).
Viral load quantification
RNA extracted with NucliSens easyMAG (bioMérieux, Marcy L'Etoile, France) was quantified by using qPCR with the QuantiTect kit (#204443; Qiagen, Hilden, Germany) in a StepOne ABI Thermocycler.
Immunofluorescence
RV and EV were detected with anti–double-stranded RNA antibody (mAbJ2; SCICONS, Szirák, Hungary)16; influenza with anti–influenza A antibody (MAB5001; Millipore, Temecula, Calif), RSV with anti-RSV antibody (MAB5006; Millipore), and HCoV-OC43 with anti–HCoV-OC43 nucleoprotein antibody (MAB9013; Millipore). Ciliated, goblet, and basal cells were stained with the anti–beta IV tubulin antibody (179504; Abcam, Cambridge, United Kingdom), anti–mucin 5A antibody (bs1022R-Cy3; Bioss, Woburn, Mass), and anti–p63-α mAb (13109; Cell Signaling, Danvers, Mass), respectively. The Alexa Fluor 594–goat anti-mouse antibody (A11032; Life Technologies, Grand Island, NY) and the Alexa Fluor 488–goat anti-rabbit antibody (A11008; Life Technologies) were used as secondary antibodies. Images were acquired with the Zeiss LSM700 Meta Confocal Microscope (Zeiss, Oberkochen, Germany) and processed by Imaris (Bitplane, Zurich, Switzerland).16
Lactate dehydrogenase assay and transepithelial electrical resistance
Lactate dehydrogenase (LDH) release in basal medium was measured with the Cytotoxicity Detection Kit (04744926001; Roche, Mannheim, Germany), and transepithelial electrical resistance (TEER) was measured with an EVOM volt ohmmeter (World Precision Instruments, Sarasota, Fla).
ELISA
Cytokines were quantified in basal medium by means of ELISA, according to the manufacturer's instructions.
MCC and cilia beating
Displacement velocity of polystyrene microbeads (84135; Sigma, St Louis, Mo) at the tissue apical surface was measured with contrast phase microscopy and Image Pro-Plus software (Media Cybernetics, Rockville, Md). Cilia beating movies were created with a Sony XCD-U60CR microscope (Sony, Tokyo, Japan), Sony zcl software, and ImageJ software (National Institutes of Health, Bethesda, Md).
Statistics
Values are expressed as means ± SEMs. The ANOVA F-test was run and, on significance (P < .05), completed by using multiple comparisons with the max-t test.21 ANOVA was replaced by a Kruskal-Wallis nonparametric case, if needed. Analyses were made with R software (https://www.R-project.org/). For each biological replicate, infections were done in duplicates, and N represents the number of infected tissues.
High-throughput sequencing
RNA extracted from apically collected specimens or from tissues by using TRIzol (5596018; Ambion, Austin, Tex) was used for library preparation with the TruSeq Stranded Total RNA Kit (Illumina, San Diego, Calif). For transcriptome analysis, library preparation was preceded by using ribodepletion Ribo-Zero Gold. Sequencing was performed with TruSeq SBS HS v3 chemistry on an Illumina HiSeq2500 sequencer. Differential expression analyses were performed with R/Bioconductor package DESeq2. Viral genomes were assembled de novo by using IDBA-UD.22 Variant calling was performed with Lofreq2,23 SNVer,24 and SAMtools 1.3.25 Only the variants agreed upon by at least 2 of the 3 callers were considered for downstream analysis.
Results
Respiratory viruses have varied growth kinetics
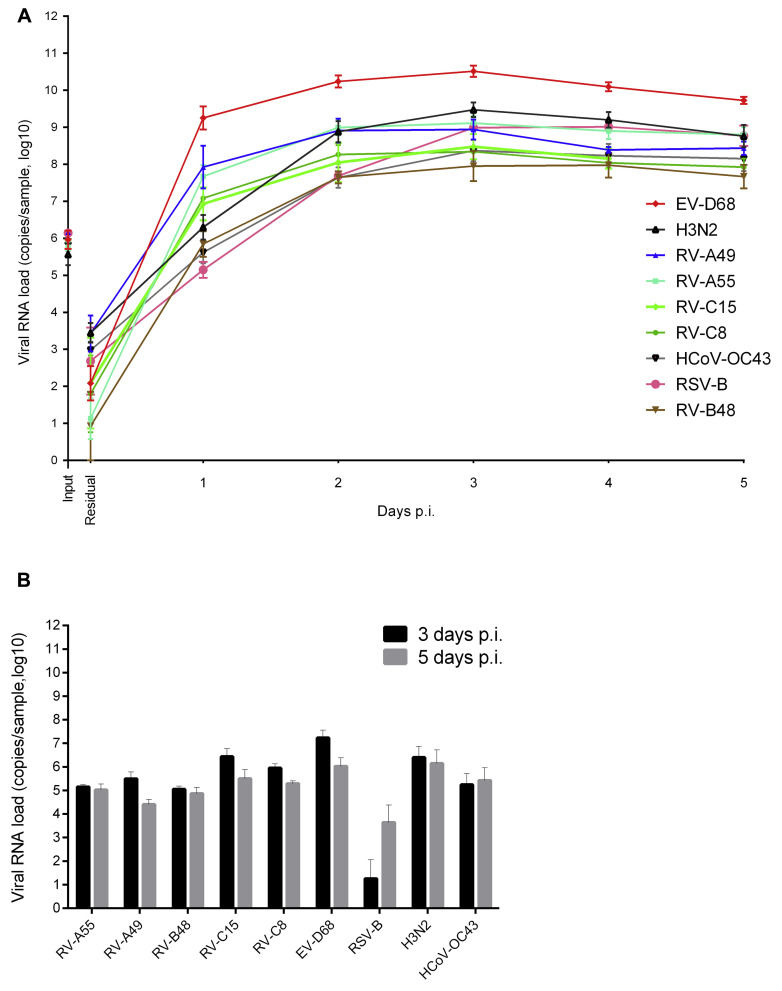
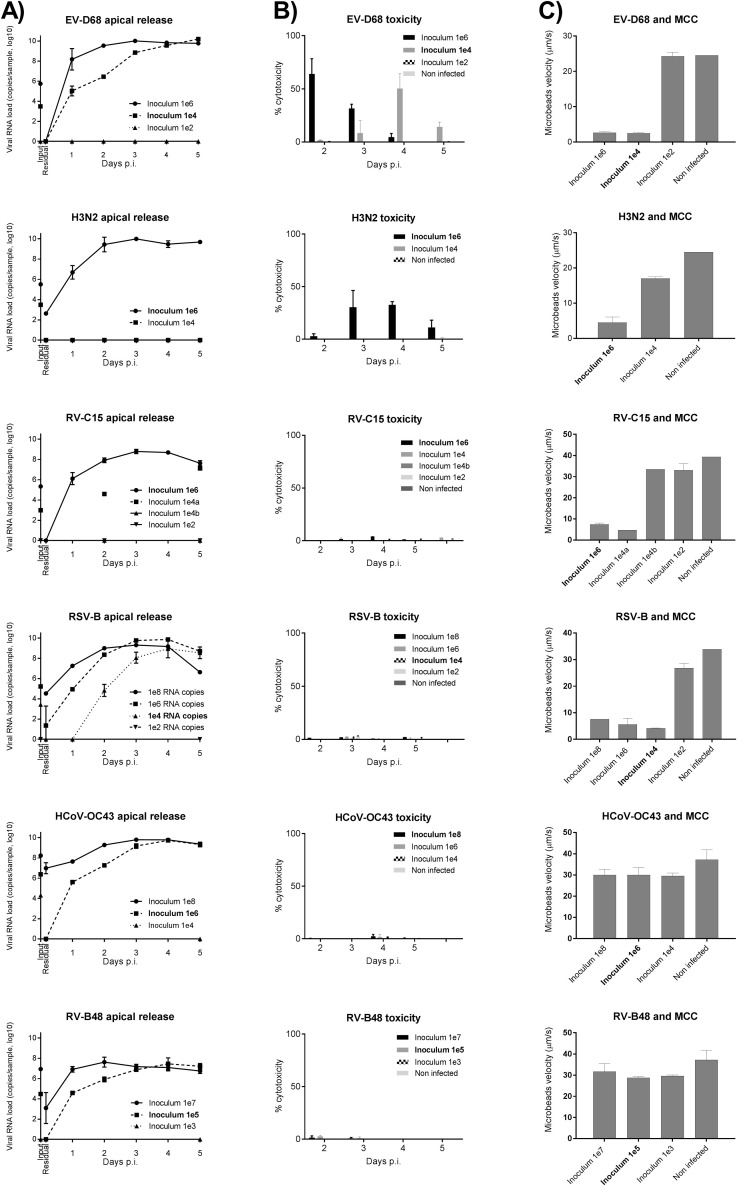
Viral stocks were prepared for RV-A55 (RV from species A, major group), RV-A49 (RV from species A, minor group), RV-B48 (RV from species B), RV-C15 (RV from species C), RV-C8 (RV from species C, isolated from a patient's serum with disseminated infection),26 EV-D68 (respiratory EV from species D), influenza A H3N2 Victoria like, RSV subgroup B, and HCoV-OC43 (Table I )16, 26, 27, 28, 29 directly in reconstituted airway epithelia to prevent acquisition of cell adaptation mutations. Viral RNA copies (106) corresponding to the average load found in clinical samples30, 31, 32 were used in each round of infection. Viruses on the apical surfaces of infected tissues were quantified by using qPCR to assess residual viral loads (remaining after 3 wash steps, 4 hours after inoculation) and subsequent virion replication (collected at 24-hour intervals thereafter). There was no reproducibility in the amount of residual virus for each virus type, probably because of tissue properties, washing efficiency, or both rather than the physicochemical characteristics of the virus. In contrast, apical viral release from day 1 onward was reproducible among replicates (Fig 1 , A), peaking at days 2 to 4 after inoculation and ranging from 107 to 1011 viral RNA copies for RV-B48 and EV-D68, respectively. Overall, replication rates were significantly greater for EV-D68 versus all other viruses, for H3N2 and RV-A49 versus RV-B48 and HCoV-OC43, and for RV-A55 versus RV-B48 (all P < .01). The amount of viral RNA released in the basal chamber was 2- to 4-log lower than virus released apically (Fig 1, B).
Table I.
Clinical specimens used to prepare viral stocks directly in tissues
| Virus | Specimen | Patient | Collection year | Study |
|---|---|---|---|---|
| RSV-B | NPS | Newborn (CH) | 2014 | Essaidi-Laziosi et al28 |
| H3N2 | NPS | Adult (CH) | 2013 | This study |
| HCoV-OC43 | NPS | Elderly (CH) | 2014 | This study |
| EV-D68 | NPS or OPS | Child (Tanzania) | 2008 | L'Huillier et al29 |
| RV-A55 (M) | NPS | Elderly (CH) | 2012 | This study |
| RV-A49 (m) | NPA | Child (CH) | 2009 | Tapparel et al27 |
| RV-B48 | NPA | Infant (CH) | 2012 | This study |
| RV-C8 | Serum | Child (Fr) | 2012 | Lupo et al26 |
| RV-C15 | NPA | Child (CH) | 2009 | Tapparel et al16 |
CH, Switzerland; Fr, France; NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab; OPS, oropharyngeal swab.
Fig 1.
Virus production at the apical (A) and basal (B) sides of air-liquid interface culture of reconstituted human airway epithelia. Viral RNA loads measured by using qPCR from infected samples collected at the apical (Fig 1, A) or basal (Fig 1, B) side of tissues infected with 9 different respiratory viruses (n ≥ 4) are shown. Each tissue was inoculated apically with 106 RNA copies of the indicated virus and washed 3 times after 4 hours. Samples were then collected from the apical or basal face at the indicated time point. Input, Viral RNA load present in the inoculum for each viral stock; p.i., post-inoculation; Residual, residual bound virus after 3 washes.
Respiratory viruses have varied cell tropisms
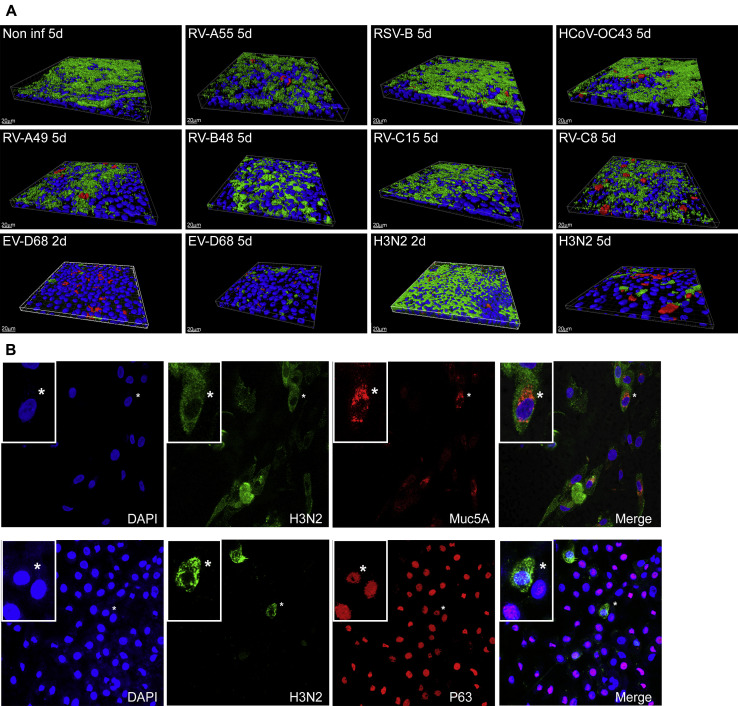
All viruses tested were localized in ciliated cells, but only infections with EV-D68 and H3N2 resulted in large-scale ciliated cell destruction (Fig 2 , A). H3N2 was the only tested virus to be detected rarely in goblet and basal cells (Fig 2, B). EV-D68 was found in nonciliated cells without goblet or basal cell markers (data not shown), likely being undifferentiated cells.
Fig 2.
Immunofluorescence of epithelia infected with the indicated respiratory virus. A, Three-dimensional view of noninfected and infected tissues, with ciliated cells stained in green, viruses in red, and cell nuclei in blue. For all viruses, immunofluorescence was performed at day 5 after infection, whereas for H3N2 and EV-D68, immunofluorescence was also done at day 2, as indicated. B, Two-dimensional view of tissues infected with H3N2 at day 5 after infection (virus stained in green, mucus cells in red [Muc5A], basal cells in red [P63], and cell nuclei in blue), with magnification insets of selected regions (*) in each panel. DAPI, 4′-6-Diamidino-2-phenylindole dihydrochloride.
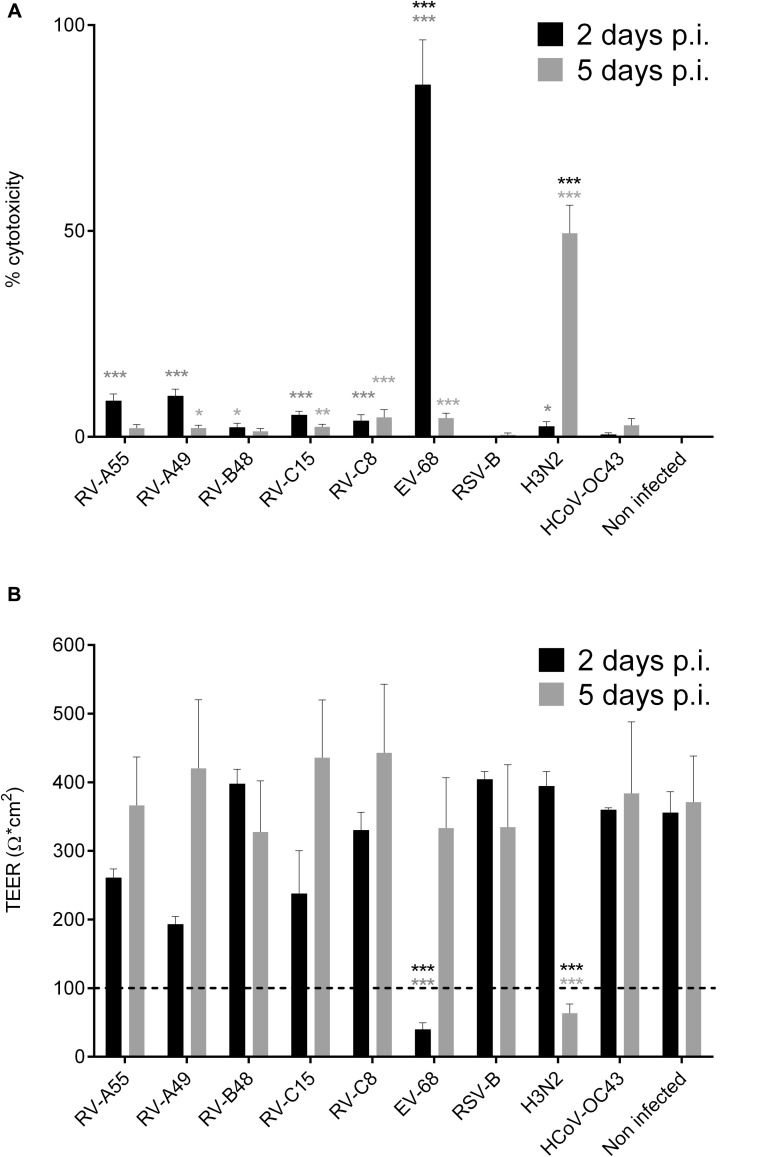
EV-D68 and H3N2 are the most cytotoxic viruses
All viruses showed cytotoxicity, as measured based on LDH release in the basal medium, but EV-D68 and H3N2 were significantly more cytotoxic (Fig 3 , A) and were the only viruses to cause loss of tissue integrity evidenced by decreased TEER (at later time points for H3N2; Fig 3, B). This observation is coherent with immunofluorescence results (Fig 2, A). TEER and LDH values returned to normal at 5 dpi for EV-D68, despite a low number of ciliated cells and at 11 dpi for 8 of 12 H3N2-infected tissues (data not shown), which suggests a rapid proliferation of undifferentiated cells, contributing to tissue recovery.
Fig 3.
Effect of infections on epithelia. Viral toxicities calculated from the amount of LDH released by damaged cells in the basal medium (n = 8; A) and TEER (n ≥ 2; B) were measured at days 2 and 5 post infection (p.i.). The 48-hour time point represents LDH accumulated during 48 hours; for later time points, the medium was replaced every day, and data correspond to LDH secretions within the preceding 24 hours. Dotted lines in Fig 3, B, correspond to the established threshold of tissue integrity. *P < .05, **P < .01, and ***P < .001. Asterisks in light gray show statistical significance relative to noninfected tissue, whereas black asterisks show statistical significance compared with all other respiratory viruses at each time point.
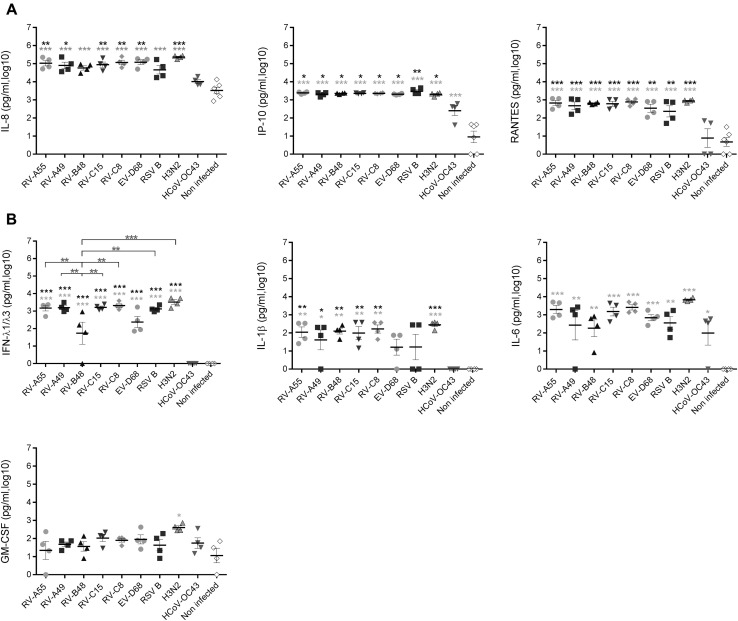
Respiratory viruses induce varied patterns of cytokine secretion, ranging from strong induction by H3N2 to weak induction by HCoV-OC43
Release of chemokines (IL-8, interferon-inducible protein 10 [IP-10], and RANTES) and cytokines (IFN-λ1/λ3, IL-1β, IL-6, and GM-CSF) induced by viral infections was evaluated at 4 dpi (Fig 4 ). H3N2 was the only virus to significantly induce all cytokines. RV-As and RV-Cs induced an intermediate response, whereas RV-B48 induced significantly less IFN-λ. Overall, the response to RSV-B and EV-D68 was similar to the response to RV-As and RV-Cs, except for IL-1β, which was not significantly induced by either virus. Finally, HCoV-OC43 induced significantly less IL-8, IP-10, RANTES, IFN-λ, and IL-1β than the other viruses.
Fig 4.
Effect of respiratory viruses on cytokine production. Chemokine (A) and cytokine (B) levels were measured in basal medium 96 hours after infection with the indicated respiratory virus (n = 4). Data correspond to cytokine secretions within the preceding 24 hours. *P < .05, **P < .01, and ***P < .001. Asterisks in light gray show statistical significance relative to noninfected tissue, whereas black asterisks show statistical significance compared with HCoV-OC43. Differences between other viruses are shown with bars. Additional cytokines (IL-33, TGF-β, IL-25, IFN-β, and thymic stromal lymphopoietin [TSLP]) were also tested but either were not significantly induced by viral infections or were not detected in basal medium (Fig E1).
Other cytokines involved in the pathophysiology of allergic diseases (IL-33, IL-25, thymic stromal lymphopoietin, and IFN-β) and anti-inflammatory cytokines (TGF-β) were either not significantly induced compared with control values (IL-33 and TGF-β) or not detected (IL-25, thymic stromal lymphopoietin, and IFN-β; see Fig E1 in this article's Online Repository at www.jacionline.org).
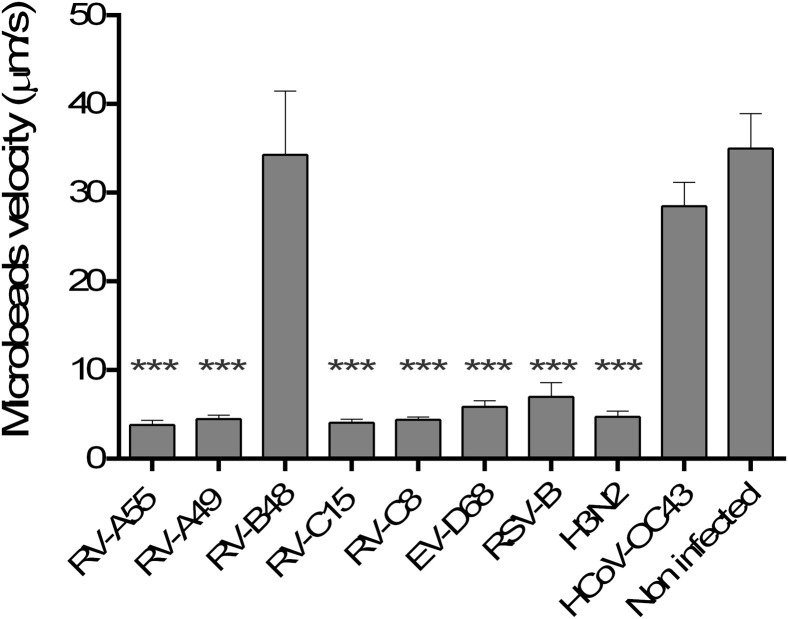
All but RV-B48 and HCoV-OC43 affect cilia beating and MCC independent of viral input
MCC was affected by all viruses except RV-B48 and HCoV-OC43 (Fig 5 ). This was confirmed by an observed decrease in cilia beating frequency at 2 dpi for EV-D68 and 4 dpi for H3N2, RV-A55, RV-A49, RV-C8, RV-C15, and RSV (data not shown).
Fig 5.
Effect of respiratory viruses on MCC. Displacement velocity of polystyrene microbeads applied at the apical side of the tissue at 5 dpi with indicated virus is shown (n ≥ 5). ***P < .001 (vs noninfected).
Infections with 100-fold serial dilutions of the inoculum (starting with higher concentrations for less cytotoxic viruses) were compared for the 2 most cytotoxic viruses (H3N2 and EV-D68), the 2 least cytotoxic viruses (HCoV-OC43 and RV-B48), and 2 viruses with intermediate toxicity (RV-C8 and RSV-B) to assess whether these observed differences were linked to different amounts of infectious particles in viral stocks. As shown in Fig E2 (in this article's Online Repository at www.jacionline.org), EV-D68 exhibits a similar but delayed toxicity, even after inoculation of 104 viral RNA copies, whereas RV-B48 and HCoV-OC43 do not affect MCC, even after inoculation of 107 and 108 viral RNA copies, respectively. Of note, the low cytokine induction by HCoV-OC43 was also confirmed at these higher inoculum doses (data not shown).
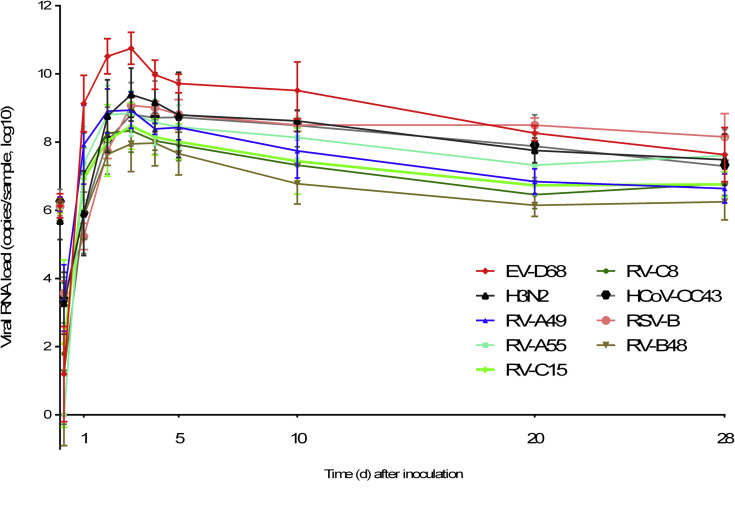
All respiratory viruses cause persistent infection, which is not linked to majority mutations in the viral population but rather a contained innate immune response and tissue recovery
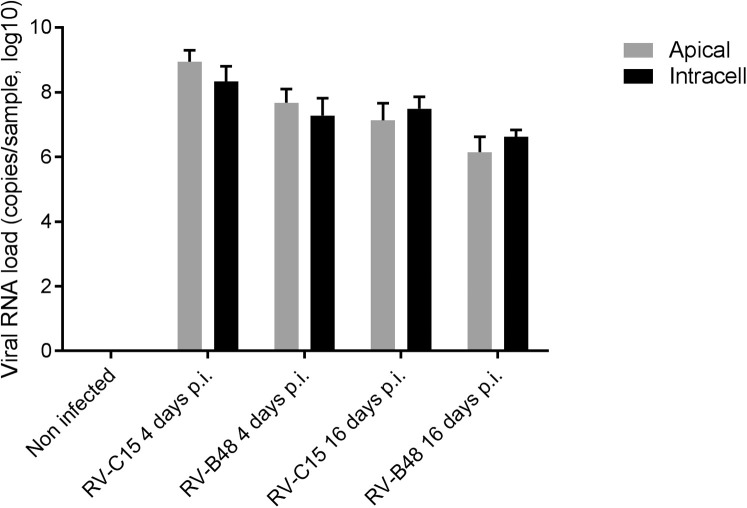
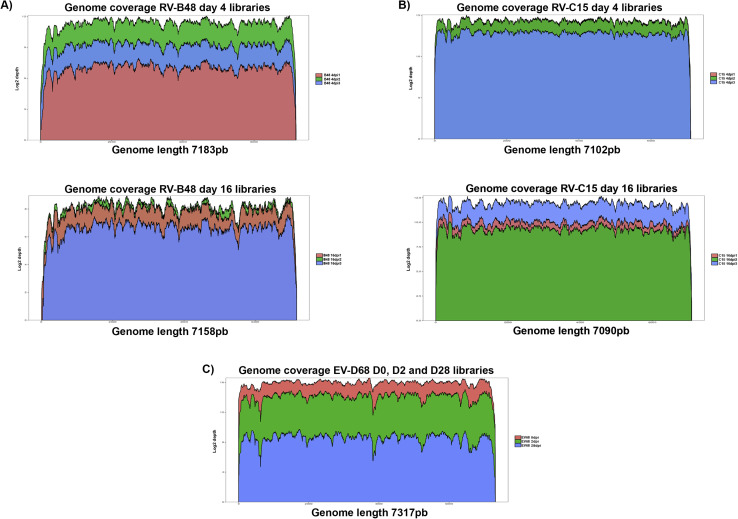
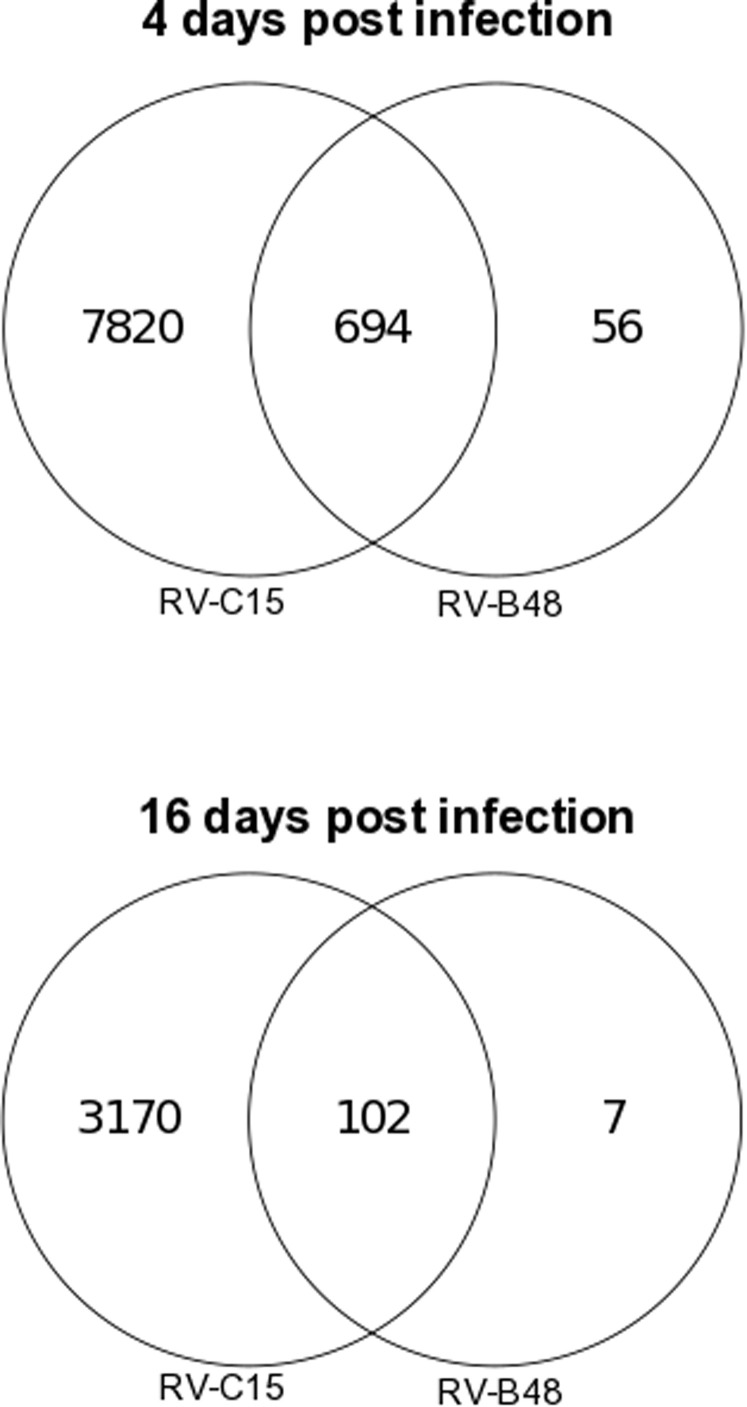
Throughout the 28-day culture period, viral genomic RNA was produced at the tissue apical surface despite repeated epithelial washes (Fig 6 ). The infectivity of viruses collected at 28 dpi was confirmed in tissue culture. To find out whether this persistence was linked to changes in the composition of the viral population, viruses produced in the tissue (RV-B48 and RV-C15, see Fig E3 in this article's Online Repository at www.jacionline.org) or released apically (EV-D68, Fig 6) were sequenced by means of HTS during the acute (4 dpi for RV-B48 and RV-C15 and 2 dpi for EV-D68) and persistent (16 dpi for RV-B48 and RV-C15 and 28 dpi for EV-D68) phases of the infection. The EV-D68 inoculum was also analyzed by using HTS. Changes were observed in the composition of the viral population at different time points, indicating evolution of the viral quasispecies (see Table E1 in this article's Online Repository at www.jacionline.org). However, the proportion of variants in the population stayed less than 50% at all time points. Mutations highlighted in the coding sequence were silent except for 1 substitution (Leu to Pro) in the VP2 protein of RV-C15 present in the population at 4 dpi (30.6%) and 16 dpi (24.2, see Table E1). Of note, 3 changes observed in the RV-B48 5′ untranslated region with low frequency at 4 dpi (23.8%, 30.7%, and 25%) were no longer detected at 16 dpi (see Table E1). Comparison of genome coverage at the different time points did not highlight the presence of defective interfering particles (see Fig E4 in this article's Online Repository at www.jacionline.org).
Fig 6.
Virus production at the apical site of air-liquid interface cultures of reconstituted human airway epithelia over a prolonged time period. Viral RNA loads measured by using qPCR from infected samples collected at the apical side of tissues infected with 9 different respiratory viruses are shown. Each tissue was inoculated with 106 RNA copies of the indicated virus, and tissues were washed 3 times after 4 hours; samples were then collected from the apical surface at the indicated time point (n = 4).
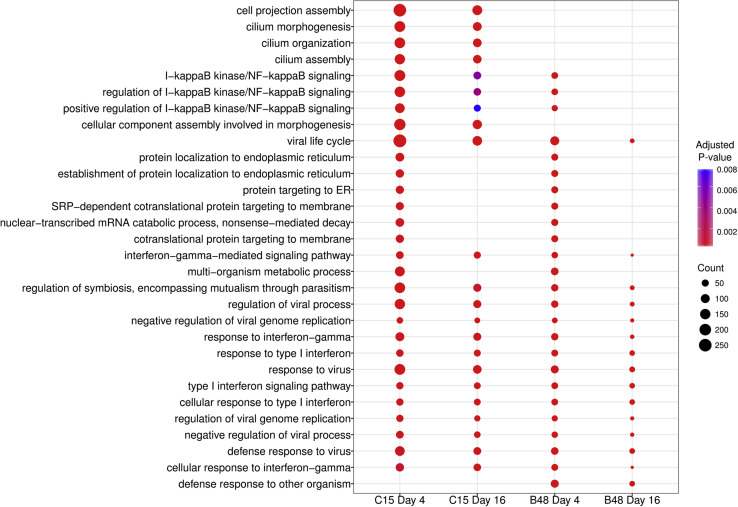
Tissue response was then addressed through transcriptomic analysis of tissues infected with RV-C15 (moderate cytotoxicity) and RV-B48 (low cytotoxicity) at 4 and 16 dpi. For RV-C15, 8514 and 3272 genes showed altered expression at 4 and 16 dpi, respectively, whereas 750 (4 dpi) and 109 (16 dpi) genes were affected by RV-B48 (see Fig E5 and Table E2 in this article's Online Repository at www.jacionline.org). Among those, only 56 (4 dpi) and 7 (16 dpi) genes were affected specifically by RV-B48. Biological processes (Fig 7 and see Table E3 in this article's Online Repository at www.jacionline.org) and reactome pathway enrichment analysis (see Table E4 in this article's Online Repository at www.jacionline.org) both highlighted modifications of metabolic processes at 4 dpi but not 16 dpi. Defense and immunity pathways were also strongly activated at 4 dpi (particularly for RV-C15) and decreased between 4 and 16 dpi. Importantly, pathways linked to cilia morphogenesis, organization, and assembly were affected only in RV-C15–infected tissues.
Fig 7.
Comparison between the top 30 most significantly enriched biological processes in tissues infected with RV-C15 and RV-B48 at the indicated time. Circle size represents the number of differentially expressed genes found on that pathway, whereas the color represents the adjusted P value. The top 100 genes showing significant changes in each condition is available in Table E2, as well as the enrichment analysis for gene ontology biological processes (see Table E3) and the reactome pathway (see Table E4). SRP, Signal recognition particle.
Together, these findings indicate that after important modifications of tissue metabolism and activation of innate immunity at 4 dpi, tissues recover and develop tolerance to the infection at 16 dpi without incurring major changes in the viral population.
Discussion
Using reconstituted human airway epithelia, we analyzed the first steps of pathogenesis of the most prevalent human respiratory pathogens. Viruses were amplified in human tissues directly from clinical specimens to best reflect in vivo conditions. Moreover, identical batches of tissues were inoculated in parallel to ensure optimal comparisons of infections by the different respiratory viruses. Our data revealed similarities but also marked differences among the tested viruses.
Of the 9 clinical isolates tested (RV-A55, RV-A49, RV-B48, RV-C8, RV-C15, EV-D68, RSV-B, H3N2, and HCoV-OC43), replication patterns were generally similar, with an increase in apical release during the first 48 hours, followed by a peak between 48 and 96 hours and a slight decrease thereafter. Surprisingly, all respiratory viruses tested were detected constantly over the 28-day culture period. Viral persistence was reported in similar tissue culture systems for a recombinant RSV–green fluorescent protein,33 as well as in calf tracheal organ cultures for bovine RV and influenza A.34 Here we extend these observations and show that 9 single-stranded RNA respiratory viruses with highly diverse biological characteristics systematically produced persistent infections in cultured human respiratory tissues. Persistence could not be explained by major viral changes because HTS of RV-C15, RV-B48, and EV-D68 collected at 16 dpi or later were not significantly different from those isolated in acute infection, either in viral population composition or in the appearance of defective interfering viral particles. On the other hand, transcriptome analysis of host tissues infected with RV-C15 and RV-B48 showed that innate immunity is strongly induced during the acute phase of infection but decreases thereafter and is insufficient to clear the infection. Although all these viruses usually cause self-limited diseases, chronic infections have been documented in immunocompromised populations,27 supporting our observation and confirming that an effective immune response is required for viral clearance.
All 9 viruses exhibited preferential apical replication with specific tropism for ciliated cells. Only H3N2 was also detected in rare goblet and basal cells. These results are consistent with previous studies on RSV, RV-A, RV-C, HCoV-OC43, and influenza.33, 35, 36, 37, 38 In a previous study on sinus mucosal organ culture, the RV genome was present in both ciliated and nonciliated cells; however, this study included mechanical tissue injury before infection, which might support viral spread.14 Similarly, it has been demonstrated that mechanical tissue injury allows RSV to extend to basal cells.39 However, cell tropism of EV-D68 has not yet been described. Here we detected EV-D68 RNA in ciliated and nonciliated cells. Interestingly, these nonciliated cells had no goblet or basal cell markers and might represent a population of differentiating cells involved in tissue repair. Of note, influenza, EV-D68, and HCoV-OC43 use sialic acid as a receptor and showed differential cell tropism in the tissues. Therefore specific coreceptors might be required for infection or other viral functions and could play a role in this distinct tropism.
After inoculation with the different viruses, marked differences in tissue toxicity were observed. First, all viruses, except HCoV-OC43 and RV-B48, affected cilia beating and MCC. RV-B has already been shown to be less cytotoxic and induce less cytokine production (including interferons) in vitro than RV-A and RV-C.20 In our study it also induced less IFN-λ than the other viruses. In addition, HTS of tissues infected with RV-B48 showed significantly fewer changes in gene expression profiles than tissues infected with RV-C15. In particular, processes related to cilia morphogenesis were not affected by RV-B48, supporting the observed absence of perturbation of MCC and cilia beating by this virus. Concerning HCoV-OC43, the absence of effect on tissue was associated with an even weaker induction of cytokines. Among all tested cytokines, only IP-10 and IL-6 were induced, although at significantly lower levels than by all other respiratory viruses. The ability of coronaviruses to remain undetected by innate immune sensors has been unraveled recently.40 Interestingly, for most viruses that affected cilia beating and MCC, a limited number of cells was infected, and the ciliated cell layer was preserved. This observation suggests a paracrine effect, propagated disorganization of cilia beating, or both. It has been shown that infection by HCoV-E229 does not affect tissue integrity in vitro and cilia beating in vivo.41 Cilia dyskinesia was observed, and Wang et al41 suggested that it would affect MCC; however, HCoV-E229 might have a more expanded cell tropism compared with other HCoV species and might thus be more virulent than HCoV-OC43.37
Regarding the other respiratory viruses, RVs from species A and C, as well as RSV-B, affected MCC but without causing significant cell death or loss in tissue integrity. These observations were confirmed with RSV-A (data not shown). Previous studies with RSV-A in air-liquid interface cultures showed no change in cilia beating frequencies at 72 hours after infection, but ciliary dyskinesia was increased.42 However, this finding does not contradict our data because in our case inhibition of cilia beating for this virus started at only 4 dpi. In addition, our RSV-B was produced in tissue directly from a clinical sample, whereas in the study by Smith et al,42 the RSV-A stock was produced in BSC-1 monkey kidney cells and was potentially cell adapted. For RVs, the low number of infected cells and a weak effect on mucosal integrity has already been demonstrated.43, 44, 45 Similarly, in vitro and in vivo RSV infections were also associated with little tissue damage,12 which is an unexpected finding for a virus that is highly pathogenic in vivo. However, it has been previously proposed that RSV pathogenicity in vivo is mainly immune mediated.12 Surprisingly, the RV-C8 strain that was associated with a disseminated human infection26 was not particularly virulent in our model. This result suggests that the in vivo dissemination and virulence is linked to host rather than viral factors.
We also investigated the ability of respiratory viruses to induce cytokines involved in asthma exacerbation and atopy. However, no correlation was found, even for viruses known to exacerbate allergic diseases, such as RV-Cs. Similar studies performed in tissues originating from asthmatic donors will certainly bring more insight into these associations and the role of host factors.
EV-D68 and H3N2 were the most cytotoxic viruses in interfering both with MCC and tissue integrity, with marked loss of ciliated cells at days 2 and 5, respectively. For both viruses, tissues recovered in most cases, which is presumably linked to proliferation of undifferentiated cells, as previously described.46 To our knowledge, this study presents the first successful attempt to grow EV-D68 in human respiratory epithelia, and our data shed some light on the capacity of this virus to replicate and kill ciliated cells. In vitro virulence of EV-D68 is particularly striking when compared with that of RVs belonging to the same genus. This difference might account for the frequent complicated diseases observed during EV-D68 infections.5 Different strains of influenza (H3N2, H1N1, or influenza B) are also known to affect tissue integrity, cilia beating, and MCC in mice, rats, pigs, and human subjects.47, 48, 49, 50 In addition, we observed similar induction of LDH release and blockage of MCC with an H1N1 clinical isolate (data not shown). The mechanisms used by EV-D68 and influenza to destroy ciliated cells might differ. In our study EV-D68 toxicity was associated with higher replication but intermediate cytokine induction, whereas for H3N2, it was associated with strong cytokine induction but intermediate replication levels.
By providing insight into the tissue response to viral infections, this study reveals possible broad-spectrum host-targeted strategies to combat respiratory viruses. Based on cytokine (Fig 4) and transcriptome (Fig 7) profiles of infected tissues and on published data, several pathways have shown promise as targets for antiviral therapies. For example, inhibitors of the inflammatory IκB kinase,51 nuclear factor κB signaling pathways,52 and biological processes related to protein localization and targeting,53, 54 as well as type III interferon–based therapies (reviewed by Syedbasha and Egli55), have all been reported to exhibit a direct antiviral activity or to counteract virally activated pathways. Based on our data, these molecules represent valuable targets to fight the infection, as well as its subsequent biological consequences and associated symptoms. Other recently characterized broad-spectrum antiviral molecules known to be effective against enveloped and nonenveloped viruses could also be further validated in this system, such as oxysterols, which interfere with the formation of viral replication organelles56 or direct-acting antiviral drugs active in vitro against several types of viruses, such as sialidase-derived compounds57 or polymerase inhibitors.58 Thus this clinically relevant and unique tissue-culture model could be used to test promising compounds for their tissue toxicity and broad-spectrum antiviral activity (either as prevention or treatment). Because the model is applicable to diverse respiratory viruses circulating in the population (including poorly cultivable ones, such as RV-C), these studies will provide valuable and wide-ranging information that might better guide later in vivo models.
Taken together, our data provide a comprehensive overview of the differential pathogenesis of some of the most prevalent respiratory viruses in human subjects. We demonstrate that among the tested viruses, influenza H3N2 and EV-D68 are the most virulent, followed by RSV-B, RV-A, and RV-C, whereas HCoV-OC43 and RV-B are almost noncytotoxic. These significant differences in vitro might contribute to the diversity of disease severity associated with each of these pathogens in vivo.
Furthermore, we highlight viral persistence in these respiratory tissues and indicate that this persistence results from a contained epithelial cell response in addition to absence of immune cells rather than significant changes in viral population composition. To conclude, our results shed light on the complex interplay between viruses and the host tissue response and contribute to the understanding of the mechanism behind chronic respiratory tract infections in immunocompromised patients.
Key messages.
-
•
RV-B48 and HCoV-OC43 induce low toxicity in respiratory tissues because they affect neither MCC nor tissue integrity. RV-A55, RV-A49, RV-C8, RV-C15, and RSV exhibit low cytotoxicity but affect MCC. H3N2 and EV-D68 are the most pathogenic, affecting both MCC and tissue integrity.
-
•
In tissues deficient of immune cells, respiratory viruses are able to cause persistent infection. This persistence is not linked to the emergence of majority mutations in the viral population but rather involves a contained innate immune response and recovery of normal tissue metabolism.
-
•
This model system provides some clues to improve our understanding of the pathogenesis of these frequent pathogens and helps to appreciate the range of disease severity observed in vivo, as well as the frequent occurrence of chronic respiratory tract infections in immunocompromised hosts.
Acknowledgments
We thank Sophie Clément (Department of Pathology and Immunology, Faculty of Medicine, University of Geneva), Ludovic Wiszniewsk (Epithelix Sàrl), Bernadett Boda (Epithelix Sàrl), Rosy Bonfante (Epithelix Sàrl), and Patricia Boquete-Suter (Division of Genetic and Laboratory Medicine, Geneva University Hospital, Switzerland), for technical help, as well as the iGE3 genomics platform of the University of Geneva, for HTS and transcriptome analysis and Annie Hartley for editorial assistance.
Footnotes
Supported by the Swiss National Science Foundation (grant 310030_166218 to C.T.), the Marie-Heim Vögtlin Foundation (grant PMPDP3-158269 to M.E.-L.), the Leenaards Foundation (grant 4390 to C.T.), and the Sandoz Foundation (grant 9975 to C.T.).
Disclosure of potential conflict of interest: S. Huang and S. Constant serve as board members for, are employees of, and hold stock in Epithelix Sarl. The rest of the authors declare that they have no relevant conflicts of interest.
Supplementary data
Fig E1.
Fig E2.
Fig E3.
Fig E4.
Fig E5.
References
- 1.World Health Organization. Global Health Observatory (GHO) data—child mortality and causes of death 2015. Available at: http://www.who.int/gho/child_health/mortality/causes/en/. Accessed December 2016.
- 2.Denny F.W., Jr. The clinical impact of human respiratory virus infections. Am J Respir Crit Care Med. 1995;152(suppl):S4–S12. doi: 10.1164/ajrccm/152.4_Pt_2.S4. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosioni J., Bridevaux P.O., Wagner G., Mamin A., Kaiser L. Epidemiology of viral respiratory infections in a tertiary care centre in the era of molecular diagnosis, Geneva, Switzerland, 2011-2012. Clin Microbiol Infect. 2014;20:O578–O584. doi: 10.1111/1469-0691.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tregoning J.S., Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan F. Enterovirus D68: acute respiratory illness and the 2014 outbreak. Emerg Med Clin North Am. 2015;33:e19–32. doi: 10.1016/j.emc.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Bertino J.S. Cost burden of viral respiratory infections: issues for formulary decision makers. Am J Med. 2002;112(suppl 6A):42S–49S. doi: 10.1016/s0002-9343(01)01063-4. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen C., Kaku S., Tutera D., Kuschner W.G., Barr J. Viral respiratory infections of adults in the intensive care unit. J Intensive Care Med. 2016;31:427–441. doi: 10.1177/0885066615585944. [DOI] [PubMed] [Google Scholar]
- 8.Stokes K.L., Chi M.H., Sakamoto K., Newcomb D.C., Currier M.G., Huckabee M.M. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol. 2011;85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett N.W., Walton R.P., Edwards M.R., Aniscenko J., Caramori G., Zhu J. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat Med. 2008;14:199–204. doi: 10.1038/nm1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margine I., Krammer F. Animal models for influenza viruses: implications for universal vaccine development. Pathogens. 2014;3:845–874. doi: 10.3390/pathogens3040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkov Y.A., Watters K., Basnet S., Sijapati S., Hill M., Palmenberg A.C. Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa-E8 cells. Virology. 2016;499:350–360. doi: 10.1016/j.virol.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villenave R., Shields M.D., Power U.F. Respiratory syncytial virus interaction with human airway epithelium. Trends Microbiol. 2013;21:238–244. doi: 10.1016/j.tim.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell H., Levin D., Forrest S., Beauchemin C.A., Tipper J., Knight J. Higher level of replication efficiency of 2009 (H1N1) pandemic influenza virus than those of seasonal and avian strains: kinetics from epithelial cell culture and computational modeling. J Virol. 2011;85:1125–1135. doi: 10.1128/JVI.01722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochkov Y.A., Palmenberg A.C., Lee W.M., Rathe J.A., Amineva S.P., Sun X. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao W., Bernard K., Patel N., Ulbrandt N., Feng H., Svabek C. Infection and propagation of human rhinovirus C in human airway epithelial cells. J Virol. 2012;86:13524–13532. doi: 10.1128/JVI.02094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapparel C., Sobo K., Constant S., Huang S., Van Belle S., Kaiser L. Growth and characterization of different human rhinovirus C types in three-dimensional human airway epithelia reconstituted in vitro. Virology. 2013;446:1–8. doi: 10.1016/j.virol.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Knight D.A., Holgate S.T. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8:432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 18.Roche W.R., Montefort S., Baker J., Holgate S.T. Cell adhesion molecules and the bronchial epithelium. Am Rev Respir Dis. 1993;148(suppl):S79–S82. doi: 10.1164/ajrccm/148.6_Pt_2.S79. [DOI] [PubMed] [Google Scholar]
- 19.Smith D.J., Gaffney E.A., Blake J.R. Modelling mucociliary clearance. Respir Physiol Neurobiol. 2008;163:178–188. doi: 10.1016/j.resp.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Nakagome K., Bochkov Y.A., Ashraf S., Brockman-Schneider R.A., Evans M.D., Pasic T.R. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134:332–341. doi: 10.1016/j.jaci.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 22.Peng Y., Leung H.C., Yiu S.M., Chin F.Y. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 23.Wilm A., Aw P.P., Bertrand D., Yeo G.H., Ong S.H., Wong C.H. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Z., Wang W., Hu P., Lyon G.J., Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39:e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupo J., Schuffenecker I., Morel-Baccard C., Bardet J., Payen V., Kaiser L. Disseminated rhinovirus C8 infection with infectious virus in blood and fatal outcome in a child with repeated episodes of bronchiolitis. J Clin Microbiol. 2015;53:1775–1777. doi: 10.1128/JCM.03484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapparel C., Cordey S., Junier T., Farinelli L., Van Belle S., Soccal P.M. Rhinovirus genome variation during chronic upper and lower respiratory tract infections. PLoS One. 2011;6:e21163. doi: 10.1371/journal.pone.0021163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essaidi-Laziosi M., Lyon M., Mamin A. Fernandes Rocha M, Kaiser L, Tapparel C. A new real-time RT-qPCR assay for the detection, subtyping and quantification of human respiratory syncytial viruses positive- and negative-sense RNAs. J Virol Methods. 2016;235:9–14. doi: 10.1016/j.jviromet.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 29.L'Huillier A.G., Kaiser L., Petty T.J., Kilowoko M., Kyungu E., Hongoa P. Molecular epidemiology of human rhinoviruses and enteroviruses highlights their diversity in Sub-Saharan Africa. Viruses. 2015;7:6412–6423. doi: 10.3390/v7122948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borg I., Rohde G., Loseke S., Bittscheidt J., Schultze-Werninghaus G., Stephan V. Evaluation of a quantitative real-time PCR for the detection of respiratory syncytial virus in pulmonary diseases. Eur Respir J. 2003;21:944–951. doi: 10.1183/09031936.03.00088102. [DOI] [PubMed] [Google Scholar]
- 31.Esposito S., Daleno C., Scala A., Castellazzi L., Terranova L., Sferrazza Papa S. Impact of rhinovirus nasopharyngeal viral load and viremia on severity of respiratory infections in children. Eur J Clin Microbiol Infect Dis. 2014;33:41–48. doi: 10.1007/s10096-013-1926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ngaosuwankul N., Noisumdaeng P., Komolsiri P., Pooruk P., Chokephaibulkit K., Chotpitayasunondh T. Influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses. Virol J. 2010;7:75. doi: 10.1186/1743-422X-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Peeples M.E., Boucher R.C., Collins P.L., Pickles R.J. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed S.E. Persistent respiratory virus infection in tracheal organ cultures. Br J Exp Pathol. 1969;50:378–388. [PMC free article] [PubMed] [Google Scholar]
- 35.Jakiela B., Gielicz A., Plutecka H., Hubalewska-Mazgaj M., Mastalerz L., Bochenek G. Th2-type cytokine-induced mucus metaplasia decreases susceptibility of human bronchial epithelium to rhinovirus infection. Am J Respir Cell Mol Biol. 2014;51:229–241. doi: 10.1165/rcmb.2013-0395OC. [DOI] [PubMed] [Google Scholar]
- 36.Griggs T.F., Bochkov Y.A., Basnet S., Pasic T.R., Brockman-Schneider R.A., Palmenberg A.C. Rhinovirus C targets ciliated airway epithelial cells. Respir Res. 2017;18:84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dijkman R., Jebbink M.F., Koekkoek S.M., Deijs M., Jonsdottir H.R., Molenkamp R. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibricevic A., Pekosz A., Walter M.J., Newby C., Battaile J.T., Brown E.G. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson B.D., Jaffe A.B., Fearns R., Danahay H. Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS One. 2014;9:e102368. doi: 10.1371/journal.pone.0102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G., Deering C., Macke M., Shao J., Burns R., Blau D.M. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J Virol. 2000;74:9234–9239. doi: 10.1128/jvi.74.19.9234-9239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith C.M., Kulkarni H., Radhakrishnan P., Rutman A., Bankart M.J., Williams G. Ciliary dyskinesia is an early feature of respiratory syncytial virus infection. Eur Respir J. 2014;43:485–496. doi: 10.1183/09031936.00205312. [DOI] [PubMed] [Google Scholar]
- 43.Arruda E., Boyle T.R., Winther B., Pevear D.C., Gwaltney J.M., Jr., Hayden F.G. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J Infect Dis. 1995;171:1329–1333. doi: 10.1093/infdis/171.5.1329. [DOI] [PubMed] [Google Scholar]
- 44.Winther B. Effects on the nasal mucosa of upper respiratory viruses (common cold) Dan Med Bull. 1994;41:193–204. [PubMed] [Google Scholar]
- 45.Hendley J.O. Clinical virology of rhinoviruses. Adv Virus Res. 1999;54:453–466. doi: 10.1016/S0065-3527(08)60375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tadokoro T., Wang Y., Barak L.S., Bai Y., Randell S.H., Hogan B.L. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc Natl Acad Sci U S A. 2014;111:E3641–E3649. doi: 10.1073/pnas.1409781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu N.H., Yang W., Beineke A., Dijkman R., Matrosovich M., Baumgartner W. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci Rep. 2016;6:39668. doi: 10.1038/srep39668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pittet L.A., Hall-Stoodley L., Rutkowski M.R., Harmsen A.G. Influenza virus infection decreases tracheal mucociliary velocity and clearance of Streptococcus pneumoniae. Am J Respir Cell Mol Biol. 2010;42:450–460. doi: 10.1165/rcmb.2007-0417OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iravani J., Melville G.N., Horstmann G. Tracheobronchial clearance in health and disease: with special reference to interciliary fluid. Ciba Found Symp. 1978;54:235–252. doi: 10.1002/9780470720356.ch12. [DOI] [PubMed] [Google Scholar]
- 50.Wilson R., Alton E., Rutman A., Higgins P., Al Nakib W., Geddes D.M. Upper respiratory tract viral infection and mucociliary clearance. Eur J Respir Dis. 1987;70:272–279. [PubMed] [Google Scholar]
- 51.Catley M.C., Sukkar M.B., Chung K.F., Jaffee B., Liao S.M., Coyle A.J. Validation of the anti-inflammatory properties of small-molecule IkappaB Kinase (IKK)-2 inhibitors by comparison with adenoviral-mediated delivery of dominant-negative IKK1 and IKK2 in human airways smooth muscle. Mol Pharmacol. 2006;70:697–705. doi: 10.1124/mol.106.023150. [DOI] [PubMed] [Google Scholar]
- 52.Mazur I., Wurzer W.J., Ehrhardt C., Pleschka S., Puthavathana P., Silberzahn T. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell Microbiol. 2007;9:1683–1694. doi: 10.1111/j.1462-5822.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 53.Kalies K.U., Romisch K. Inhibitors of protein translocation across the ER membrane. Traffic. 2015;16:1027–1038. doi: 10.1111/tra.12308. [DOI] [PubMed] [Google Scholar]
- 54.Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity. 2016;44:46–58. doi: 10.1016/j.immuni.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Syedbasha M., Egli A. Interferon lambda: modulating immunity in infectious diseases. Front Immunol. 2017;8:119. doi: 10.3389/fimmu.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lembo D., Cagno V., Civra A., Poli G. Oxysterols: An emerging class of broad spectrum antiviral effectors. Mol Aspects Med. 2016;49:23–30. doi: 10.1016/j.mam.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Koszalka P., Tilmanis D., Hurt A.C. Influenza antivirals currently in late-phase clinical trial. Influenza Other Respir Viruses. 2017;11:240–246. doi: 10.1111/irv.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdelnabi R., Morais A.T.S., Leyssen P., Imbert I., Beaucourt S., Blanc H. Understanding the mechanism of the broad-spectrum antiviral activity of favipiravir (T-705): key role of the F1 motif of the viral polymerase. J Virol. 2017;91(12) doi: 10.1128/JVI.00487-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.