Summary
Programmed –1 ribosomal frameshifting (−1PRF) is an mRNA recoding event utilized by cells to enhance the information content of the genome and to regulate gene expression. The mechanism of –1PRF and its timing during translation elongation are unclear. Here, we identified the steps that govern –1PRF by following the stepwise movement of the ribosome through the frameshifting site of a model mRNA derived from the IBV 1a/1b gene in a reconstituted in vitro translation system from Escherichia coli. Frameshifting occurs at a late stage of translocation when the two tRNAs are bound to adjacent slippery sequence codons of the mRNA. The downstream pseudoknot in the mRNA impairs the closing movement of the 30S subunit head, the dissociation of EF-G, and the release of tRNA from the ribosome. The slippage of the ribosome into the –1 frame accelerates the completion of translocation, thereby further favoring translation in the new reading frame.
Graphical Abstract

Highlights
-
•
Kinetics of –1 ribosomal frameshifting are monitored at single-codon resolution
-
•
Frameshifting occurs when the backward movement of the 30S subunit head is impeded
-
•
Two tRNAs at the XXXYYYZ mRNA sequence are stalled in chimeric POST states
-
•
The shift to the –1 reading frame occurs prior to EF-G release from the ribosome
Programmed ribosomal frameshifting is an mRNA recoding process that allows cells to maximize gene expression. Kinetic deconstruction of this process reveals that the binding of tRNAs to adjacent slippery sequence codons of the mRNA and the resulting pseudoknot impair movement of the 30S ribosome and slip it to the −1 position into a new translational frame.
Introduction
Programmed ribosomal frameshifting (PRF) is a recoding event that leads to a change of the translational reading frame and allows the translation of messenger RNAs (mRNAs) with overlapping open reading frames, often yielding two protein products from one mRNA. PRF is particularly frequent in the decoding of mRNAs from viral and mobile genes but is found also in some cellular genes in all domains of life. During PRF, the reading frame may shift in +1, –1, or even –2 direction. The mechanisms of +1 and –1PRF appear to be quite different and require a different set of stimulatory elements. The classic example of −1PRF is slippage triggered by two elements in the mRNA, a “slippery sequence” (Jacks et al., 1988) and a downstream secondary structure element (Brierley et al., 1989). The slippery site is made up by sequences such as X XXY YYZ, where XXY and YYZ are codons in the original frame (0 frame) that presumably favor tRNA sliding on the mRNA and anticodon misalignment, changing the codons to XXX and YYY in the –1 frame. The stimulatory secondary structure elements of the mRNA, which usually consist of a stem loop or a pseudoknot located at a particular distance downstream of the slippery sequence, may enhance –1PRF by inducing pausing of the ribosome or by perturbing normal decoding. In some cases, –1PRF is additionally stimulated by a Shine-Dalgarno (SD)-like sequence in the mRNA located at a particular distance upstream of the slippery sequence (Larsen et al., 1994). Sequence comparison and molecular genetic analysis of a variety of frameshifting sequences identified canonical structures that can cause –1PRF in both bacterial and eukaryotic systems, although details of how the pro- and eukaryotic ribosomes respond to frameshifting signals may be different (Brierley et al., 2010, Dinman, 2012, Farabaugh, 1997, Fayet and Prère, 2010, Gesteland and Atkins, 1996).
The mechanism of –1PRF is not clear, and there are at least four groups of models as to when and why it occurs (Figure 1 A). The slippage can take place during translocation as the ribosome enters the slippery site, when deacylated tRNA is bound at the codon (nnX) preceding the slippery sequence and peptidyl-tRNA at the XXY codon (pept-tRNAXXY) (Bekaert and Rousset, 2005, Léger et al., 2007) (Figure 1A, pathway I). Alternatively, the shift may occur during the accommodation of aminoacyl-tRNAYYZ on the ribosome after reading the second slippery codon YYZ by tRNAYYZ and/or following peptidyl transfer with two tRNAs at the slippery site (Harger et al., 2002, Jacks et al., 1988, Plant et al., 2003) (pathway II). Several models propose cotranslocational shifting of pept-tRNAYYZ with or without concomitant slippage of deacylated tRNAXXY as the ribosome exits the slippery site (Farabaugh, 1996, Horsfield et al., 1995, Namy et al., 2006, Weiss et al., 1989, Yelverton et al., 1994)—i.e., when the XXY YYZ mRNA sequence moves from the P and A to the E and P sites, respectively (pathway III). Finally, it has been suggested that frameshifting might occur during the accommodation of tRNA after the slippery sequence or as a result of competition between the tRNAs decoding the 0 and –1 frame codons (Léger et al., 2004, Yelverton et al., 1994) (pathway IV). Although there is evidence for and against each of these models, recent kinetic models predicted that –1PRF is simultaneously accessible through several pathways governed by the kinetic parameters of aa-tRNA binding, peptide bond formation, and translocation (Liao et al., 2011). However, these kinetic constants are not known for any –1PRF system. This prompted us to establish a fully reconstituted in vitro translation system to study real-time kinetics of –1PRF and to identify the steps that control frameshifting. Here, we report the results of the analysis, which include the kinetics of stepwise translation of the slippery sequence, the kinetics of amino acid incorporation into 0 and –1 frame translation products, and the detailed kinetics of the translocation reactions that govern –1PRF.
Figure 1.
Kinetics of Product Formation in 0 and –1 Frames
(A) Potential kinetic pathways leading to –1PRF (–1 frame). On pathway I, –1PRF occurs during translocation (TL) of tRNAnnX (gray) and tRNAXXY (green). Pathway II, –1PRF after correct reading the 0 frame codon and upon accommodation (Acc) of tRNAYYZ (magenta); the following codon is read in –1 frame. Pathway III, during translocation of tRNAXXY and tRNAYYZ that exposes –1 frame codon Zab in the A site. Pathway IV, upon decoding of the codon following the slippery sequence; 0 frame tRNA is shown in blue.
(B) Schematic of the frameshifting mRNA containing a modified IBV 1a/1b gene fragment. The encoded amino acids in 0 and –1 frame are indicated above the nucleotide sequence. As a result of –1PRF, Val is the first –1 frame amino acid incorporated into the peptide chain.
(C) Schematic of the quench-flow experiment. 70S initiation complexes were mixed with EF-G–GTP and EF-Tu–GTP–aa-tRNA complexes with a defined set of purified aa-tRNAs (Y, Tyr-tRNATyr; L, Leu-tRNALeu [anticodon AAU]; K, Lys-tRNALys; F, Phe-tRNAPhe; V, Val-tRNAVal; EF-Tu was omitted for clarity) and incubated for desired times before quenching the reaction with KOH.
(D) Time courses of synthesis of fMetTyr (MY), fMetTyrLeu (MYL), fMetTyrLeuLys (MYLK), fMetTyrLeuLysPhe (MYLKF), and fMetTyrLeuLysVal (MYLKV) upon translation of the mRNA without a slippery sequence or a pseudoknot (–/–). The sequence of the mRNA coding region is given above the graph with the fourth codon (Lys) indicated in bold for orientation.
(E) Same as (D), but mRNA contained a Val codon in the 0 frame. Symbols as in (D).
(F) Same as (D) but with mRNA containing a slippery sequence (+/−).
(G) Same as (D) but with mRNA containing a pseudoknot (–/+).
(H) Same as (D) but with mRNA containing both slippery sequence and pseudoknot (+/+). Global fits (continuous lines) were performed by numerical integration as described in Extended Experimental Procedures.
See also Figures S1 and S3 and Tables 1, 2, and S1.
Results
Kinetics of –1 Frameshifting In Vitro
To study the mechanism of –1PRF, we have chosen a minimal fragment of the infectious bronchitis virus (IBV) 1a/1b gene for which significant levels of –1 frameshifting were demonstrated in vivo in both eukaryotic and bacterial systems (Brierley et al., 1989, Brierley et al., 1997, Napthine et al., 2003). The construct used for in vitro translation contained codons for fMet and Tyr followed by the slippery site and a pseudoknot (Figure 1B). The native IBV 1a/1b slippery sequence, U UUA AAC, was replaced with U UUA AAG, which results in a higher –1PRF efficiency in E. coli (Brierley et al., 1997, Napthine et al., 2003). This slippery sequence is decoded by tRNALeu (anticodon 3 ′AAU5′) and tRNALys (anticodon 3 ′UUU5′), resulting in the synthesis of fMetTyrLeuLysPhe (MYLKF) in the 0 frame and fMetTyrLeuLysVal (MYLKV) in the –1 frame. In addition to the frameshifting mRNA construct that contained both slippery sequence and pseudoknot (designated as +/+), a set of model mRNAs was generated that had different combinations of stimulatory elements—i.e., had only the slippery sequence but lacked the pseudoknot (+/−), no slippery sequence but the pseudoknot (–/+), or lacked both stimulatory elements (–/–). To disrupt the slippery sequence, the UUA codon was replaced with UUG, which is decoded by the same tRNALeu (3 ′AAU5′). Additional mRNA constructs used as controls are described in Extended Experimental Procedures.
Extended Experimental Procedures.
Ribosomes, mRNAs, tRNAs, and Translation Factors
Chemicals were from Roche, Sigma, or Merck. Radioactive compounds were from Hartmann Analytic. Ribosomes from E. coli MRE 600, fMet-tRNAfMet, Phe-tRNAPhe, EF-Tu, EF-G, and initiation factors were prepared as described (Cunha et al., 2013, Rodnina et al., 1997, Rodnina et al., 1999, Savelsbergh et al., 2003). Fluorescein-labeled tRNALeu was prepared according to published protocols (Milon et al., 2007, Wintermeyer et al., 1979). tRNATyr, tRNALeu (reading the UUA/UUG codon), tRNALys, and tRNAVal were prepared from bulk E. coli tRNA by consecutive column chromatographies on Sepharose 4B (GE Healthcare), Phenyl Sepharose (GE Healthcare), and DEAE Toyopearl (Tosoh Bioscience) (Holmes et al., 1975). Aminoacylation of purified tRNAs with 14C-labeled and unlabeled amino acids was performed as described (Kothe et al., 2006), and aa-tRNAs were purified by reversed-phase HPLC on a WP 300 RP-18 HPLC column (250 mm x 10.5 mm, Merck) equilibrated with buffer (20 mM ammonium acetate, pH 5.0, 10 mM magnesium acetate, 400 mM NaCl) using a gradient of 0%–15% ethanol. Aa-tRNAs were precipitated with ethanol, dissolved in water, and the concentrations were determined photometrically.
We used a modified variant of the original IBV pseudoknot, with loop 2 shortened from 35 nucleotides to 8 nucleotides, and placed it six nucleotides downstream of the slippery site, which ensured efficient –1PRF (Brierley et al., 1992, Napthine et al., 1999). The 5′ leader of the mRNA had an SD sequence seven nucleotides upstream of the AUG start codon, which was necessary to achieve high initiation efficiency. The following mRNA constructs were used (slippery sequence/pseudoknot are indicated by + or –, start codon underlined):
+/+ mRNA: 5’GGGAAUUCAAAAAUUGUUAAGAAUUAAGGAGAUAUACAUAUGUAUUUAAAGUUUCGCGGGGUAUCAGUCAGGCUCGGCUGGUACCCCUUGCAAAGCGAGCCGUAAUAA 3′;
+/− mRNA: 5′GGGAAUUCAAAAAUUGUUAAGAAUUAAGGAGAUAUACAUAUGUAUUUAAAGUUUCGCUAAUAA 3′;
–/– mRNA, Phe in 0-frame: 5′GGGAAUUCAAAAAUUGUUAAGAAUUAAGGAGAUAUACAUAUGUAUUUGAAGUUUCGCA 3′;
–/– mRNA, Val in 0-frame, AAG codon for Lys: 5’GGGAAUUCAAAAAUUGUUAAGAAUUAAGGAGAUAUACAUAUGUAUUUGAAGGUUUCGCA 3′;
–/+ mRNA: 5′GGGAAUUCAAAAAUUGUUAAGAAUCAAGGAGAUAUACAUAUGUA
UUUGAAGUUUCGCGGGGUAUCAGUCAGGCUCGGCUGGUACCCCUUGCAAAGCGAGCCGUAAUAA 3′;
mRNA with AAA codon for Lys: 5′GGGAAUUCAAAAAUUGUUAAGAAUUAAGGAGA UAUACAUAUGUAUUUGAAAGUUUCGCA 3′.
The mRNAs were prepared by T7 RNA-polymerase transcription and purified on a RNeasy Maxi Prep column (QIAGEN) according to the manufacturer’s instructions. BOF-Lys-tRNALys was prepared as described (Mittelstaet et al., 2013). Expression, purification, and refolding of S13 were carried out as described (Hickerson et al., 2005). Labeling of S13 with Atto540Q and reconstitution of 30S subunits were performed according to published protocols (Cunha et al., 2013). The 30S subunits were labeled to more than 90%, as determined photometrically. 70S ribosomes carrying L12 labeled at position 118 with Alx488 were prepared as described (Mohr et al., 2000, Savelsbergh et al., 2005). Labeling of EF-G with QSY9 was performed by incubating EF-G containing a His6-tag and a single engineered Cys at position 209 (Wilson and Noller, 1998) with a five-fold excess of QSY9-maleimide in labeling buffer (50 mM Tris-HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 300 mM NaCl) in the dark at 4°C overnight. Affinity purification of labeled EF-G by the C-terminal His-tag was carried out using a Protino gravity-flow column (Macherey-Nagel) (Cunha et al., 2013). The protein was concentrated and stored in buffer A with 50% glycerol. The concentration of EF-G(QSY9) was calculated from the absorbance at 280 nm.
Rapid Kinetics
Quench-flow experiments were performed in buffer A at 37°C using a KinTek RQF3 apparatus. Translation experiments were performed by rapidly mixing initiation complexes (0.2 μM after mixing) with the respective ternary complexes as indicated (1.5 μM) and EF-G (2 μM). Reactions were quenched by the addition of KOH (0.5 M), and peptides were released by hydrolysis for 30 min at 37°C. After neutralization with acetic acid, samples were analyzed by HPLC (LiChroSpher100 RP-8 HPLC column, Merck) using an adapted gradient of acetonitrile in 0.1% TFA. The elution positions of the reaction products were determined using a set of peptides synthesized in vitro (f[3H]Met[14C]Tyr, f[3H]MetTyr[14C]Leu, f[3H]MetTyrLeu[14C]Lys, f[3H]MetTyrLeuLys[14C]Phe, or f[3H]MetTyrLeuLys[14C]Val). The extent of product formation was determined from the ratio of f[3H]Met in the respective peaks to the total 3H-radioactivity in the eluate.
Time courses were evaluated by numerical integration using the Micromath Scientist software according to the following model:
Where k Tyr, k Leu, k Lys, k Phe and k Val are rate constants of incorporation of the respective amino acids into the peptide and k Tl MY is the rate of the 1st translocation event. The fraction of ribosomes that did not react was taken into account by the respective concentration correction parameters. Standard deviations of the values determined by global fitting are indicated in Tables 2, S1, S2, and S3.
The Pmn assay was carried out in buffer A at 37°C. Initiation complexes (0.2 μM after mixing) were first incubated with ternary complexes as indicated (1.5 μM; with 14C-labeled Lys) and EF-G (2 μM) for different times and then rapidly mixed with Pmn (1 mM), incubated for another 0.5 s, and finally quenched with KOH (0.5 M) in the collection tube. After hydrolysis and neutralization, samples were analyzed by HPLC.
Fluorescence experiments were carried out in buffer A at 37°C using a stopped-flow apparatus (SX-20MV; Applied Photophysics). Labeled ribosomes, tRNAs, and EF-G were used as indicated. Alx488 and Flu fluorescence was excited at 470 nm and detected after passing a KV500 cut-off filter (Schott). Equal volumes of post-translocation complexes carrying MYL-tRNALeu in the P site were rapidly mixed with ternary complexes as indicated (0.25 μM) and EF-G (1 μM). At least seven individual traces were averaged and evaluated by numerical integration using the Micromath Scientist software. Evaluation of individual time courses was carried out by numerical integration as indicated in the caption of Table S3. Because such fitting requires that some values are fixed during fitting (see caption of Table S3), we also performed global fitting of all time courses together, which does not require that any particular value is fixed and validates the overall consistency of the calculated values. For the global fitting of translocation, the following time courses were combined: FRET change upon interaction of EF-G(QSY9) with L12(Alx488) (Figure 2C); fluorescence intensity change upon translocation of tRNALeu(Flu) (Figure 4C); and FRET change upon movement of tRNALeu(Flu) relative to S13(AttoQ) upon addition of Lys-tRNALys alone or together with Phe-tRNAPhe and Val-tRNAVal (Figure 4E). The following kinetic model was used:
where k Lys, k Phe, and k Val are the incorporation rates of the respective amino acid; k EFG is the rate of EF-G binding; k Tl1 is the rate of movement into the chimeric state where MYLK-tRNALys becomes Pmn-reactive; k Tl2 for Phe and Val are the rates of rearrangements reported by the label in tRNALeu; k Tl3 for Phe and Val are the rates leading to completion of translocation; PRE and POST states refer to the respective pept-tRNA in the A site (PRE) or P site (POST) (Table S4). The relative fluorescence of each intermediate was expressed by the respective F value.
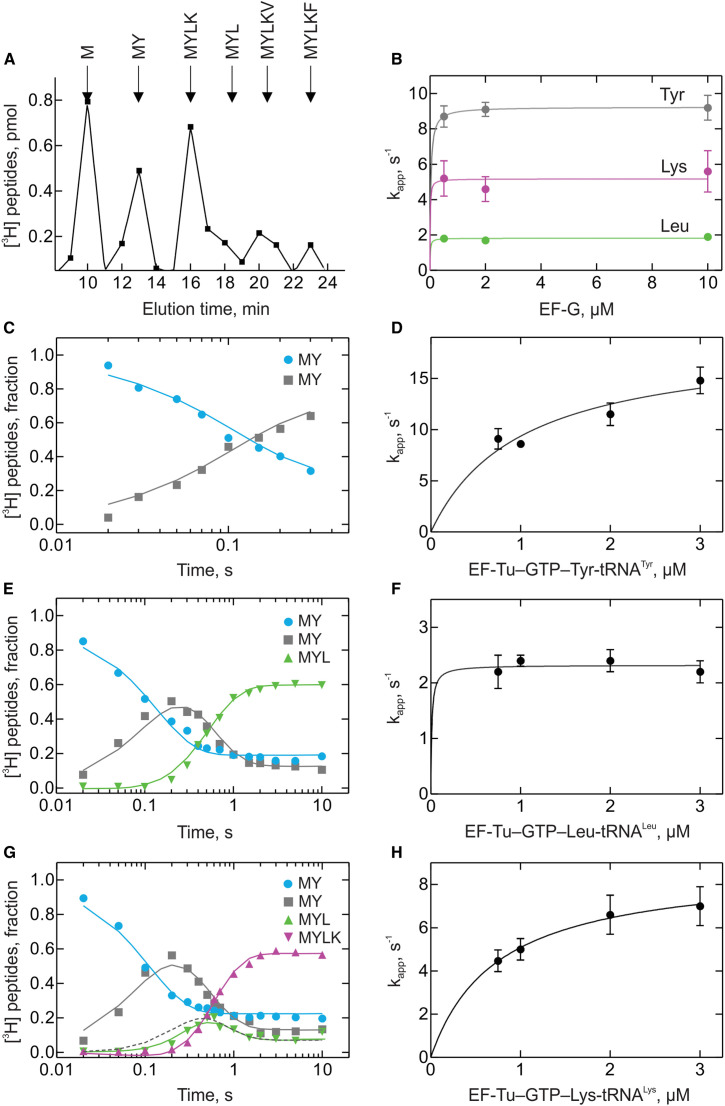
To study –1PRF in vitro, we utilized a reconstituted translation system consisting of purified components from E. coli. Translation of consecutive codons was measured after mixing 70S initiation complexes programmed with one of the model mRNAs and containing f[3H]Met-tRNAfMet in the P site with excess of ternary complexes, EF-Tu–GTP–aa-tRNA, formed with purified individual Tyr-, Leu-, [14C]Lys-, Phe-, and Val-tRNA, and EF-G–GTP (Figure 1C). The formation of translation products in 0 frame or –1 frame was monitored using the quench-flow technique. After the desired incubation time, the reaction was quenched, and the products were analyzed by high-performance liquid chromatography (HPLC) (Figure S1 A available online). Concentrations of aa-tRNAs and Elongation Factor G (EF-G) were optimized to achieve the maximum translation rate (Figures S1B–S1H). With the control –/– mRNA, which lacked slippery sequence and pseudoknot, the major end product was MYLKF, which is consistent with the fifth codon of the 0 frame specifying Phe (Figure 1D). Likewise, when the fifth codon in the 0 frame specified Val, mostly MYLKV peptide was formed (Figure 1E). In contrast, with mRNA constructs containing only the slippery sequence (+/−), only the pseudoknot (–/+), or both elements together (+/+), both 0-frame (MYLKF) and –1 frame (MYLKV) peptides were produced (Figures 1F–1H), with –1PRF taking place on up to about 75% of ribosomes on +/+ mRNA (Table 1 ).
Figure S1.
Optimization of the In Vitro Assay, Related to Figure 1 and Tables 1 and 2
(A) Peptide separation by HPLC. The composition of peptides is indicated.
(B) Dependence on EF-G concentration. Time courses of sequential Tyr, Leu and Lys incorporation on –/– mRNA were obtained as in Figure 1C and the rates (kapp) were evaluated by numerical integration (Extended Experimental Procedures).
(C) A representative experiment showing time courses of fMet-tRNAfMet consumption (M) and fMetTyr formation (MY).
(D) Concentration dependence of the observed kTyr rate (kapp) of MY formation.
(E) Same as (C) for fMetTyrLeu (MYL).
(F and G) (F) Concentration dependence of the rate (kapp) of MYL formation (G) Same as (C) for fMetTyrLeuLys (MYLK) fitted to a model . An alternative fit which does not take into account Tl MY as a separate step is shown for MYL for comparison (dashed line). Fitting the incorporation kinetics of Lys, Phe and Val did not require that the respective preceding translocation steps were taken into account.
(H) Concentration dependence of the rate (kapp) of MYLK formation.
Error bars in (B), (D), (F), and (G) are SDs of rate constants obtained by global fitting.
Table 1.
Efficiency of –1PRF In Vivo and In Vitro
| Stimulatory Elements | –1PRF Efficiency (%a) | |
|---|---|---|
| SS/PKb | in vitroc | in vivod |
| +/+ | 75 ± 10 | 78 ± 9 |
| +/− | 28 ± 4 | 23 ± 10 |
| –/+ | 60 ± 10 | 36 ± 9 |
See also Figure S2.
–1PRF efficiency was calculated as the product of –1 frame translation divided by the sum of the products of –1 and 0 frame translation.
SS, slippery sequence; PK, pseudoknot.
Calculated from the end levels (after 10–15 s) of –1 frame Val and 0 frame Phe incorporation shown in Figures 1F–1H. SDs were calculated from six to ten experiments.
The efficiency of –1PRF was calculated from the ratio of Rluc/Fluc activities in the –1 and 0 frames as described in the Extended Experimental Procedures. SDs were calculated from three different cultures with at least five independent extracts from each culture.
Validation In Vivo
To validate the efficiency of the –1PRF construct in vivo, we used a reporter assay in which the minimal IBV 1a/1b gene fragment was cloned into a dual luciferase reporter plasmid coding for firefly (Fluc) and renilla (Rluc) luciferases in such a way that translation of Rluc required –1PRF in the inserted 1a/1b fragment (Figure S2 ). The efficiency of –1PRF was calculated as the ratio of relative Rluc/Fluc synthesis in –1 and 0 frames. The Rluc/Fluc ratio for –1 frame translation in the absence of the two –1PRF stimulatory elements was considered as background and subtracted from all other –1 frame values obtained in the presence of stimulatory elements; thus, by definition, the efficiency of –1PRF in the –/– construct is set to 0, and the reported values for the +/+, +/−, and –/+ constructs represent lower-limit estimates for –1PRF. When both slippery sequence and pseudoknot were present, the relative efficiency of Rluc synthesis in the –1-frame was 78% (Table 1). This value is somewhat higher than the previously reported frameshifting efficiency of 40% on a similar mRNA construct (Brierley et al., 1997) but is consistent with our results obtained in vitro. For –1PRF in E. coli, the requirement for a pseudoknot structure in IBV 1a/1b is not strict (Brierley et al., 1997). In our construct, the removal of either the pseudoknot or the slippery sequence reduced –1PRF to 23% and 36%, respectively, which is in reasonable agreement with the in vitro results (Table 1).
Figure S2.
Dual Luciferase Construct, Related to Table 1
The genes coding for firefly (Fluc) and renilla (Rluc) luciferases were linked by a fragment of the frameshifting site of the IBV 1a/1b gene. The complete Fluc-Rluc fusion protein was produced when the linker region was translated, which required the –1PRF. Frameshifting efficiency of –1PRF in vivo was calculated in the same way as in the in vitro experiment, FS% = –1-frame product/(−1-frame + 0-frame products) × 100%.
Stepwise Kinetics of –1PRF
To obtain the rate constants that govern –1PRF, we evaluated time courses of stepwise translation (Figures 1D–1H) by numerical integration, using the combined data for the synthesis and consumption of each peptide. The initial simplest kinetic model included individual sequential steps for the incorporation of Tyr, Leu, and Lys and a branch allowing for the alternative incorporation of Phe (0 frame) or Val (–1 frame) (Extended Experimental Procedures). The MY peptide was formed rapidly in a one-step fashion at a rate of about 10 s−1 (Table 2 ). Fitting the time courses of MYL formation to a simple model with only two steps (for the incorporation of Tyr and Leu, respectively) did not yield a satisfactory solution and required including a delay between the formation of the MY and the MYL peptides (Figures 1D–1H and S1G). This additional step presumably reflects the first translocation event, which displaces tRNAfMet from the P site and moves MY-tRNATyr from the A to the P site; the rate of that translocation event was around 2 s−1 independent of the mRNA construct (Table 2), which is consistent with earlier reports on the rate of translocation involving initiator tRNAfMet (Dorner et al., 2006). We expect that, after the first round of translocation, the SD-aSD interaction is weakened or resolved, as it is destabilized after the formation of the first peptide bond (Uemura et al., 2007). The subsequent incorporation of Leu was rapid, about 15 s−1. The next step, incorporation of Lys, followed a one-step mechanism MYL→MYLK with a rate of around 5 s−1.
Table 2.
Summary of Rate Constants of Elemental Translation Steps upon –1PRF on the IBV 1a/1b Construct
| SS/PK | Rates (s−1) | |||||
|---|---|---|---|---|---|---|
| Tyra | TlMYb | Leua | Lysa | Phea | Vala | |
| 0 frame | –1 frame | |||||
| +/+ | 10.1 ± 0.5 | 1.9 ± 0.1 | 11.1 ± 3.5 | 4.3 ± 0.5 | 0.09 ± 0.01 | 0.3 ± 0.05 |
| +/– | 9.8 ± 0.4 | 1.4 ± 0.1 | 14.9 ± 6.3 | 6.0 ± 0.9 | 5.9 ± 1.1 | 2.4 ± 0.5 |
| –/+ | 8.8 ± 0.4 | 3.1 ± 0.2 | 17.8 ± 5.6 | 4.2 ± 0.4 | 0.04 ± 0.02 | 0.06 ± 0.02 |
| –/– | 9.9 ± 0.5 | 2.7 ± 0.1 | 13.6 ± 3.2 | 5.1 ± 0.4 | 5.3 ± 0.6 | 0.2 ± 0.05 |
| 0 frame | ||||||
| –/– | 11.1 ± 0.8 | 2.2 ± 0.1 | 14.4 ± 6 | 6.3 ± 1.2 | NA | 4.0 ± 0.4 |
See also Figures S1 and S3 and Table S1.
Rates of incorporation of the respective amino acid into peptide, as determined by global fitting of data shown in Figures 1D–1H.
Rates of the first translocation event (TlMY; translocation of MY-tRNATyr), which is manifested by a delay of Leu incorporation (Figures 1D–1H).
If –1PRF occurred through pathway I, one would expect that translocation of tRNATyr and MYL-tRNALeu—and, consequently, Lys incorporation—are affected by the frameshifting signals. Similarly, if pathway II were operational, then frameshifting signals would change the kinetic characteristics of tRNALys accommodation, resulting in altered rates of Lys incorporation. The kinetic analysis shows that the rates of Tyr, Leu, or Lys incorporation and the rate of the first translocation were independent of the presence of –1PRF-stimulatory elements in the mRNA (Table 2). Thus, it is unlikely that –1PRF takes place through pathways I and II. In contrast, the incorporation of Phe (0 frame) and Val (–1 frame) was strongly affected by the presence of the slippery sequence and the pseudoknot (Table 2), which is predicted by the pathway III scenario in which slippage occurs during translocation over the XXY YYZ sequence. In the absence of –1PRF stimulatory elements (–/– mRNA), the rate of Phe or Val incorporation in the 0 frame was 4–5 s−1, compared to 0.2 s−1 (Val) or 0.09 s−1 (Phe in the –1 frame), i.e., translation in 0 frame was strongly favored kinetically. The pseudoknot inhibited translation in both –1 and 0 frames, whereas the slippery sequence dramatically increased the rate of Val incorporation in the –1 frame. Together, the two stimulatory elements led to a faster incorporation of Val in the –1 frame than of Phe in 0 frame (Table 2). These results suggest that –1PRF is kinetically governed by the reactions that follow Lys incorporation into the nascent peptide but precede the incorporation of Val or Phe.
Commitment to the Alternative Reading Frame
To address more directly whether pathway II was operational, we followed the accommodation of the 3′ end of Lys-tRNALys on the 50S subunit using a fluorescent derivative of Lys-tRNALys, BOP-Lys-tRNALys. The rates of tRNA accommodation or peptide bond formation are similar with the modified and unmodified tRNALys (Mittelstaet et al., 2013). To study the A site binding of Lys-tRNA, rather than the preceding steps, we prepared ribosome complexes with MYL-tRNALeu in the P site, and the reaction was started by adding EF-Tu–GTP–BOP-Lys-tRNALys and EF-G–GTP in a stopped-flow apparatus (Figure S3 ). Accommodation of BOP-Lys-tRNALys was observed as a fluorescence decrease with a rate of about 2 s−1, independent of the frameshifting signals (Figure S3). These results provide further support to the notion that –1PRF via pathway II, involving accommodation, is unlikely.
Figure S3.
Accommodation of BOP-Lys-tRNALys, Related to Table 2
(A) Ribosome complexes with MYL-tRNALeu (0.05 μM) were mixed with fluorescent BOP-Lys-tRNALys (2 μM) in the complex with EF-Tu–GTP (10 μM).
(B) Time courses of BOP-Lys-tRNALys accommodation on ribosome complexes prepared with –/– mRNA (blue) or +/+ mRNA (red). Fitting was done by numerical integration using a two-step sequential model, where the first step includes all decoding events prior to accommodation followed to the accommodation step. The first step led to a small fluorescence decrease (3%–5%); the estimated rate was about 20 ± 10 s−1. The major change (20%) was due to the accommodation; the rates of accommodation were 2.0 ± 0.1 and 2.2 ± 0.1 for –/– and +/+ mRNAs, respectively.
Next, we asked the question whether –1PRF took place after the translocation over the XXY YYZ sequence, e.g., when Phe or Val codons are decoded (pathway IV, Figure 1A). If slippage occurred at that step, the presence and relative concentrations of Val-tRNAVal and Phe-tRNAPhe should influence the efficiency of –1PRF. This was tested by adding Val-tRNAVal and Phe-tRNAPhe in different ratios (4:1 or 1:4) or omitting one of the two during the translation of +/+ mRNA (Table S1). The data indicate that the efficiency of amino acid incorporation in the 0 or –1 frames was not affected by the presence or absence of the aa-tRNA decoding the alternative reading frame. This suggests that, at the time of Phe or Val decoding, the fraction of ribosomes that have switched to the –1 frame was already fixed. The slippage must have occurred after decoding the Lys codon in the 0 frame but before the Phe or Val codons are read. The most likely explanation for this observation is that –1PRF occurs during the translocation of tRNALeu and MYLK-tRNALys which are both bound to their codons in the slippery sequence, i.e., through pathway III.
Delayed Dissociation of EF-G
One potential reason for inaccurate translocation during –1PRF may be an altered interaction of EF-G with the ribosome, which could lead to prolonged pausing at the pseudoknot. To examine this possibility, we measured the rate of EF-G binding to and dissociation from the ribosome using a fluorescence resonance energy transfer (FRET) reporter assay with a FRET donor (Alexa 488, Alx) placed on ribosomal protein L12, which is known to recruit translation factors to the ribosome, and a nonfluorescent FRET acceptor (QSY9) in the G domain of EF-G (Figure 2 A). The fluorescence labels did not affect the translation rates or the efficiency of –1PRF (Table S2). To study specifically the translocation of tRNAs bound to the slippery sequence, we prepared L12-labeled ribosome complexes with MYL-tRNALeu in the P site. The reaction was initiated by adding EF-Tu–GTP–Lys-tRNALys and QSY-labeled EF-G–GTP in a stopped-flow apparatus, and the recruitment of EF-G(QSY) was monitored by a decrease of L12(Alx) fluorescence due to quenching upon complex formation. The interaction is maintained until the end of translocation when EF-G is released, which leads to fluorescence recovery (Figure 2B). The rate constants of EF-G binding and dissociation estimated by numerical integration, which took into account the Lys incorporation step preceding translocation (Table S2), suggested that EF-G binding to the ribosomes in the absence of the pseudoknot (–/– and +/− mRNA constructs) was rapid (>10 s−1) and limited by the preceding Lys decoding step; from experiments with simplified model systems, the rate of EF-G binding is expected to be about 60 s−1 (C. da Cunha, A. Lehwess-Litzmann, F.P., and M.V.R., unpublished data). In the presence of the pseudoknot, EF-G binding was somewhat slower, 4.8 s−1, regardless of the presence of the slippery sequence but clearly not rate limiting for the following translocation steps. In contrast, EF-G dissociation was much slower when the pseudoknot was present (Figure 2C); the dissociation rate (3 s−1 from ribosomes translating –/– or +/− mRNAs) was reduced >15-fold to 0.2 s−1 when both –1PRF stimulatory elements were present (Table S3). Furthermore, the latter rate was very similar to the rate of Val incorporation in the –1 frame (0.3 s−1), suggesting that decoding of the next codon in the –1 frame is rate limited by EF-G dissociation. Dissociation of EF-G from ribosomes translating the –/+ mRNA was even slower (<0.1 s−1; Figure 2C) and comparable to slow decoding of the next codon both in –1 and 0 frames (Table 2). These data indicate that (1) the pseudoknot causes delayed EF-G release and (2) –1PRF is induced while EF-G is still bound to the ribosome. In comparison, EF-G binding and dissociation upon translocation of tRNATyr and MYL-tRNALeu were very similar on –/– and +/− mRNAs (Figure 2B) and too fast to be resolved kinetically.
Figure 2.
Interaction of EF-G with the Ribosome
(A) Experimental design. POST complexes formed with L12(Alx)-labeled ribosomes containing MYL-tRNALeu in the P site were rapidly mixed with EF-Tu–GTP–Lys-tRNALys and EF-G(QSY) with GTP in a stopped-flow apparatus. EF-G binding to the ribosome is manifested in the decrease of Alx fluorescence due to FRET and the dissociation of EF-G by the recovery of fluorescence.
(B) Time courses of EF-G binding and dissociation upon translocation of MYLK-tRNALys on ribosomes programmed with –/– mRNA (blue) together with the fit obtained by numerical integration (Extended Experimental Procedures) (black smooth line). For comparison, an analogous experiment is shown for translocation of MYL-tRNALeu on –/– (black trace) or +/+ (gray trace) mRNA; the respective fits could not be obtained due to low amplitudes of fluorescence change.
(C) Time courses of EF-G binding and dissociation upon translocation of MYLK-tRNALys on ribosomes programmed with –/– (same trace as in B), +/−; –/+; or +/+ mRNA. The data were fitted by numerical integration, as described in Extended Experimental Procedures; fits are shown in black. The results for the –/– and +/+ mRNAs are given in Table S3. For –/+ mRNA, the rates of the fluorescence decrease and increase, respectively, were 4.8 s−1 and < 0.1 s−1 (the latter value cannot be determined with precision because the end level was not reached within the time of experiment). For +/− mRNA, the fit was ambiguous due to a low amplitude of the fluorescence change.
Movements of tRNALys and tRNALeu
Translocation is a dynamic process that entails movements of two tRNAs together with the mRNA from the A to P to E site coupled to the rotation of the 30S and 50S ribosomal subunits relative to one another and to a movement of the 30S head. To understand when exactly the change in the reading frame takes place, we measured the rates of several individual motions of tRNAs during translocation. We first examined whether MYLK-tRNALys moved from the pretranslocation state (PRE) to the posttranslocation state (POST) using a time-resolved puromycin (Pmn) assay. High Pmn reactivity is indicative of the positioning of pept-tRNA in the POST state on the 50S subunit, as, in comparison, the Pmn reactivity of pept-tRNA in both classical or hybrid PRE states is very low (Semenkov et al., 1992, Sharma et al., 2004). Translation was initiated in the quench-flow apparatus as described above (Figure 3 A). After different incubation periods, Pmn was added in saturating concentration, and the reaction was quenched after 0.5 s. This fixed time is sufficient for the completion of the Pmn reaction of pept-tRNA in the POST state; therefore, the increase of the Pmn reactivity over time reflects the movement of MYLK-tRNALys from the PRE to the POST state on the 50S subunit. The apparent reaction rate, as estimated by numerical integration, was about 7 s−1, independent of –1PRF elements (Figure 3B), suggesting that the movement of the pept-tRNA from the A to P site on the 50S subunit is not altered by frameshifting, and therefore, the slippage is likely to occur later than the movement of MYLK-tRNALys into the Pmn-reactive state. The value of 7 s−1 is the lower limit for the rate constant of A to P site translocation because the rate of the Pmn reaction itself contributed to the observed rate.
Figure 3.
Translocation on the 50S Subunit as Monitored by Pmn Assay
(A) Experimental design. 70S initiation complexes were mixed in a quench-flow apparatus with EF-G–GTP and EF-Tu–GTP–aa-tRNA complexes with Tyr-tRNATyr, Leu-tRNALeu, and Lys-tRNALys, incubated for desired times (1), mixed with Pmn (2), and reacted for another 0.5 s before stopping the reaction (3).
(B) Time courses of Pmn reaction with MYLK-tRNALys upon translocation on ribosomes programmed with different mRNAs. The average rate of the Pmn reaction for all mRNA constructs was 7 ± 4 s−1 with a delay of 4 ± 1 s−1; fit is shown in black.
Movement of MYLK-tRNALys to the POST state on the 50S subunit implied that tRNALeu should have vacated the P site on the 50S subunit and moved toward the E site. To test this directly, we prepared ribosome complexes containing MYL-tRNALeu labeled with fluorescein (Flu) at position 8 (Figure 4 A). Introducing the fluorescence label did not affect the rates of translation (Table S2). By adding EF-Tu–GTP–Lys-tRNALys and EF-G–GTP, one round of Lys incorporation and translocation was induced, which resulted in the displacement of tRNALeu(Flu) from the P site, as manifested in a decrease of fluorescence (Figure 4B). After the completion of this step, tRNALeu(Flu) may reside in the E site or dissociate into solution (Chen et al., 2013, Uemura et al., 2010). In comparison, when Val-tRNAVal and Phe-tRNAPhe were added together with Lys-tRNALys, decoding of two consecutive codons (regardless in which frame) and two translocation events should completely displace tRNALeu from the ribosome, thereby providing a measure for the maximum fluorescence change associated with tRNALeu(Flu) movement from the P site to the solution. The apparent rates of elemental reactions were estimated by numerical integration, taking into account the rates of Lys incorporation (Table S2) and EF-G binding steps (Figure 2 and Table S3), which as such do not lead to fluorescence changes of tRNALeu(Flu). In the absence of –1PRF stimulatory elements, the fluorescence of tRNALeu(Flu) changed rapidly, with a predominant rate of about 9 s−1 (Figures 4B, 4D, and 4H and Table S3), reflecting rapid tRNA movement toward the POST state. The additional step, at 3 s−1, contributed relatively little to the fluorescence change (Figure 4H) but was required to obtain satisfactory fits. The dissociation of tRNALeu(Flu) from ribosomes translating –/– mRNA was not monitored as a separate step, as the fluorescence changes observed after one or two translocation rounds were not significantly different (Figures 4B and 4H).
Figure 4.
Movement of tRNALeu
(A) Experimental design. 70S POST complexes (with protein S13 unlabeled or labeled with AttoQ) with MYL-tRNALeu(Flu) in the P site. The complexes were mixed with EF-G–GTP, EF-Tu–GTP, and either Lys-tRNALys alone, leading to one round of translocation (Lys; blue traces in B, C, E, and F), or Lys-tRNALys, Val-tRNAVal, and Phe-tRNAPhe, leading to two consecutive rounds of translocation (Lys/Val/Phe; red in B, C, E, and F).
(B) Time courses of movement upon translation of –/– mRNA monitored by changes in tRNALeu(Flu) fluorescence (unlabeled S13). The fluorescence decrease is indicative of tRNALeu(Flu) moving out of the P site.
(C) Same as (B) with +/+ mRNA.
(D) Comparison of time courses with –/– and +/+ mRNAs upon two rounds of translocation (Lys/Val/Phe, from B and C). For better comparison, time courses were normalized with the maximum value in each curve set to 1 and the minimum set to 0. Apparent rate constants of predominant reactions are indicated.
(E) Time courses of the detachment of tRNALeu(Flu) from the 30S subunit upon translation of –/– mRNA monitored by FRET between S13(AttoQ) and tRNALeu (Flu).
(F) Same as (E) with +/+ mRNA.
(G) Comparison of time courses with –/– and +/+ mRNAs upon two rounds of translocation (Lys/Val/Phe, from E and F); data were normalized as in (D). Apparent rate constants of predominant reactions are indicated.
(H and I) Summary of the minimum set of apparent rate constants and fluorescence changes required to evaluate translocation time courses without (H) and with (I) frameshifting signals (tRNALys binding was included as the first step preceding translocation in all fittings; see also Table S3).
(J) Global fit of translocation kinetics on +/+ mRNA monitored by FRET between protein L12 and EF-G; fluorescence changes of tRNALeu and FRET between protein S13 and tRNALeu during one round or two rounds of translocation.
In the presence of both –1PRF stimulatory elements (+/+ mRNA), the fluorescence change observed with tRNALeu(Flu) was multiphasic (Figure 4C). Numerical integration (Table S3) suggested that, following EF-G binding, there was a very fast step (>10 s−1), followed by a slower step (0.9 s−1) that contributed most of the fluorescence change (Figure 4I). The rapid step likely reflects the movement of tRNALeu(Flu) concomitantly with the movement of MYLK-tRNALys into the Pmn-reactive POST state on the 50S subunit, which, in the following, we refer to as POST1. The step with the rate of 0.9 s−1 was significantly slower than the lower limit for the rate of movement into the Pmn-reactive POST1 step (7 s−1) and therefore constitutes a separate, second step during which tRNALeu moves further to a POST2 state and the tRNALeu(Flu) fluorescence decreases. The rate of tRNALeu dissociation from the ribosome was about 0.2 s−1, as estimated from the kinetic differences between one and two rounds of translocation (Figures 4C, 4D, and 4I; see also below).
Impeded Rearrangement of the 30S Subunit
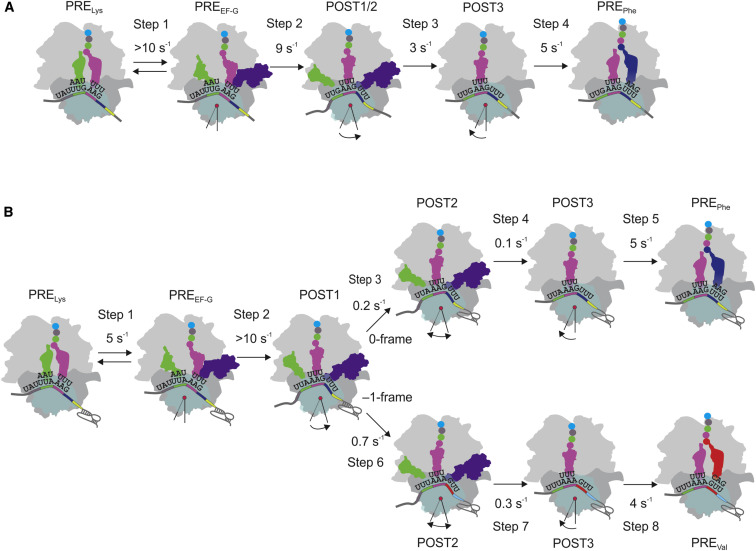
In addition to recording the fluorescence change of tRNALeu upon movement through the ribosome, we employed a FRET assay monitoring the proximity between tRNALeu(Flu) and a nonfluorescent acceptor (Atto540Q, denoted as AttoQ in the following) attached to protein S13. The FRET assay measures the timing of tRNALeu release from its P site position on the 30S subunit. Introducing the quencher did not affect the kinetics of translation (Table S2). As in the experiments described above, we prepared ribosome complexes carrying S13(AttoQ) (Cunha et al., 2013) and MYL-tRNALeu(Flu) in the P site and mixed them with Lys-, Val-, and Phe-tRNA bound to EF-Tu–GTP and EF-G–GTP (Figure 4A). Translocation on the –/– mRNA resulted in a fluorescence increase (reduced FRET) due to the movement of tRNALeu away from S13 (Figure 4E). After Lys-tRNALys and EF-G recruitment, there was a rapid small FRET change, taking place at a rate of about 9 s−1, which coincided with the rate of the transition reported by tRNALeu(Flu), and a major FRET change that took place at 3 s−1, concomitantly with EF-G dissociation (Figure 4H and Table S3). This suggests the following mechanism of translocation when there is no –1PRF: following EF-G binding to the PRE complex, tRNALeu and MYLK-tRNALys moved at a rate of 9 s−1 through POST1 to POST2 states; those two states cannot be distinguished in the experiments with –/– mRNA. The extent of FRET between tRNALeu(Flu) and S13(AttoQ) did not change at this stage, suggesting that the head of the subunit moved together with the tRNA. The interaction between tRNALeu and the 30S head was released at a later step, which proceeded at a rate of 3 s−1 and likely represented the backward rotation of the 30S head. This movement coincided with the dissociation of EF-G (Table S3) and resulted in the formation of the POST3 state (Figure 5 A).
Figure 5.
Kinetic Model of –1PRF
(A) Model of translocation in the absence of –1PRF. In the absence of –1PRF stimulatory elements (–/– mRNA), EF-G binds rapidly to the PRE complex (step 1). Subsequently, the tRNAs move into a chimeric state in which both deacylated tRNA and pept-tRNA move relative to the 50S subunit into a Pmn-reactive state (POST1/2), whereas their contacts with the 30S subunit are not disrupted (step 2). In step 3, tRNALeu detaches from the 30S head, probably during the backward 30S head rotation, and EF-G is released (POST3). Step 4, EF-Tu–GTP–Phe-tRNAPhe binds to the A site, and Phe is incorporated into the peptide.
(B) Kinetic partitioning during –1PRF. The slippage occurs during translocation of the two tRNAs bound to the slippery sequence (tRNALeu and MYLK-tRNALys). Recruitment of EF-G (step 1) to the PRE complex facilitates rapid tRNA movement (step 2) into a chimeric state (POST1); however, the following steps are inhibited by the presence of the pseudoknot. Further movement of tRNALeu proceeds in two steps. First, tRNALeu moves on the 50S subunit into a POST2 state while the distance to the 30S subunit is not changed (steps 3 and 6 in 0 frame and –1 frame, respectively). Second, tRNALeu and the 30S subunit move apart (steps 4 and 7) into a POST3 state. Steps 3 and 4 are particularly slow for the tRNA that remains in 0 frame, which limits the rate of the following Phe-tRNAPhe binding (step 5). In contrast, tRNALeu movement on those ribosomes which switched to the –1 frame, is faster (step 6), followed by dissociation of tRNALeu from the 30S subunit, 30S head rotation, and dissociation of EF-G (step 7) and binding of Val-tRNAVal (step 8).
Translation of the +/+ mRNA led to a much slower change of FRET between tRNALeu and S13 (Figures 4F and 4G). The reaction rates estimated by numerical integration suggested that, after Lys incorporation (about 5 s−1; Table S3) and EF-G binding (about 5 s−1; Figure 2 and Table S3), the FRET efficiency changed in two steps with rates of 0.9 s−1 (to POST2) and 0.2 s−1 (to POST3), which coincided with the rates reported by the fluorescence change of tRNALeu(Flu) and EF-G dissociation, respectively (Figure 4I and Table S3). Thus, on +/+ mRNA, the movement of the 30S head away from tRNALeu is delayed, and the rates of the respective reactions are >10-fold slower than in the absence of –1PRF stimulatory elements.
Taken together, the kinetic data lead us to the following model of –1PRF (Figure 5B). The ribosomes are committed to either 0 or –1 frame during translocation of the two tRNAs bound to the slippery sequence. Based on the assignment of the kinetic steps (Figures 3 and 4), we assume the existence of the following different intermediate positions of tRNALeu: POST1, which forms by the movement of MYLK-tRNALys from PRE to POST on the 50S subunit (Pmn reaction); POST2, which leads to the major change in tRNALeu(Flu) fluorescence; and POST3 corresponding to the movement of protein S13 away from tRNALeu. Formation of POST1 is rapid and takes place independent of frameshifting signals (Figure 3). The rates of the following reactions are different on the –/– and +/+ mRNAs, suggesting that partitioning between the reading frames is likely to happen at this step, with 75% of ribosomes continuing translation in the –1 frame (Table 1). For the partitioning model, the observed value of 0.9 s−1 obtained by model-free fitting of individual time courses (Table S3) represents the sum of the rate constants of ribosomes moving in the 0 and –1 frames, whereas the ratio between these two rate constants defines the efficiency of –1PRF. This yields a rate constant of 0.7 s−1 for the ribosomes, which shifted into the –1 frame, compared to 0.2 s−1 for those that remained in the 0 frame (Figure 5B). The following movement of the 30S head away from tRNALeu (transition from POST2 to POST3) was also slow, 0.2–0.3 s−1 in the –1 frame. This transition coincided with the release of EF-G (0.2 s−1) (Table S3) and limited the rate of Val-tRNAVal incorporation (0.3 s−1, cf. Table 2). The rate of the POST2 to POST3 transition in the 0 frame could not be measured directly because the fraction of ribosomes remaining in the 0 frame was too small. However, assuming that also Phe-tRNAPhe incorporation was limited by the transition from POST2 to POST3, the rate of that transition should be about 0.1 s−1 (cf. Table 2). To validate the rates of each elemental step and to ensure that all data collectively are consistent with the model, we combined the time courses reflecting the kinetics of EF-G binding and dissociation (Figure 2 and Table S3), as well as tRNALeu movement (Figure 4 and Table S3) with the kinetics of Lys, Val, and Phe incorporation, and performed global fitting of the data on the basis of the model depicted in Figure 5B using a unifying set of rate constants. The results of global fitting yielded a solution that was in perfect agreement with all measured kinetic data (Figure 4J) and provided the values for the rate constants that determine the efficiency of –1PRF (Figure 5B).
Discussion
Kinetic Model of Programmed –1 Frameshifting
The present kinetic analysis of –1PRF indicates that the –1 slippage occurs at a late step of tRNA translocation when two tRNAs are still bound to the slippery sequence of the mRNA (Figure 5). EF-G binding to the PRE complex with tRNALeu and MYLK-tRNALys drives a rapid movement of both tRNAs into a POST state on the 50S subunit, rendering MYLK-tRNALys reactive with Pmn. This notion is consistent with the results of recent single-molecule fluorescence studies that suggest that the translocation of the A site tRNA, as monitored by FRET to ribosomal protein L11, is not affected by the presence of a downstream pseudoknot (Chen et al., 2013). Our data further suggest that the head of the 30S subunit moves together with the tRNAs because the distance between FRET labels in tRNALeu and protein S13 does not change at this stage. Thus, the conformation of the ribosome in the state where –1PRF occurs may resemble a chimeric state with tRNAs in pe/E and ap/P states (Ramrath et al., 2013), although the degree of intersubunit rotation and 30S head swiveling/tilting may be different. A recent single-molecule study suggested that, in the presence of the frameshift-inducing dnaX hairpin, the ribosomal subunits are driven into a hyperrotated state with the L1 stalk in a predominantly open conformation (Qin et al., 2014). In our system, EF-G remains bound to the ribosome, engaged in an attempt to complete translocation; however, the backward rotation of the 30S head is inhibited by the pseudoknot, which could also explain why the E site tRNA is not released (this paper and Chen et al. [2013]). The action of EF-G and possibly the chimeric state of the ribosome appear to destabilize codon-anticodon interactions (Fredrick and Noller, 2002), which can then repair in either 0 or –1 frame. Apparently, slippage to the –1 frame is favorable, as those ribosomes that switched to the –1 frame can complete translocation and release EF-G about three times faster than those remaining in the 0 frame, which determines partitioning between the –1 and 0 frames and efficiency of –1PRF (Figure 5).
The present model in principle is consistent with the notion that –1PRF is achieved by simultaneous slippage of two tRNAs bound to the codons of the slippery sequence (Jacks et al., 1988). However, in contrast to the original simultaneous-slippage model, which proposed that –1PRF occurs prior to peptide bond formation, our data suggest that the slippage takes place during translocation, as originally suggested by Atkins and Gesteland (Weiss et al., 1989), with EF-G bound to the ribosome during pausing at the pseudoknot (Namy et al., 2006). We note that, although simultaneous slippage during translocation appears to be predominant in our model system, other –1PRF pathways may be favored depending on the length of the slippery sequence and the structure of the mRNA secondary structure element. Furthermore, mutations of the ribosome, translation factors, or tRNAs that alter the kinetics of translation reactions or impairing ribosome functions with antibiotics may also alter the rate-determining step of frameshifting, thereby introducing alternative routes for –1PRF (for reviews see Brierley et al., 2010, Dinman, 2012, Farabaugh, 1997, Fayet and Prère, 2010, Gesteland and Atkins, 1996). Further kinetic analysis will be necessary to show which pathway for –1PRF is predominant in other systems, such as dnaX, HIV-1, or SARS-CoV.
Our findings suggest that the pseudoknot stalls the ribosome in a chimeric state with the 3′ end of the pept-tRNA in the POST state on the 50S subunit and an intermediate state on the 30S subunit. This provides a biochemical interpretation for the cryo-EM reconstruction of the eukaryotic frameshifting complex stalled at the pseudoknot in the absence of the slippery sequence. In that complex, a single tRNA apparently occupied the P site (a chimeric position of the small ribosomal subunit head could not be seen due to insufficient resolution), the tRNA appeared to be deformed, and the translocase (eEF2, the eukaryotic homolog of EF-G) interacted with the tRNA (Namy et al., 2006). In contrast to that structure, we find that the E site tRNA remains bound to the ribosome during –1PRF, which is consistent with the recent single-molecule (Chen et al., 2013) and previous biochemical studies (Horsfield et al., 1995). In the POST2 state, the codon-anticodon interaction in the E site may be weakened or dissolved (Lill and Wintermeyer, 1987), which may contribute to the high efficiency of –1PRF and explain why the E site tRNA appears to be bound to a near-cognate codon after shifting into the –1 frame (this paper and Brierley et al. [1997]) and dissociate upon sample dilution in cryo-EM experiments (Namy et al., 2006). Our data also predict that mutations of the E site should affect –1PRF (Devaraj et al., 2009, Léger et al., 2007, McGarry et al., 2005, Sergiev et al., 2005) by altering the kinetic parameters of slippage, e.g., by changing the stability of pe/E site tRNA binding or affecting the dynamics of the ribosome subunits in the chimeric state.
Role of the Slippery Sequence and the Pseudoknot
The present data shed light on the roles of the slippery sequence and the pseudoknot in promoting –1PRF. The slippery sequence alone can induce –1 frame decoding without significant ribosome pausing, increasing the rate of Val incorporation in the –1 frame about 100-fold. EF-G binding and the resulting destabilization of codon-anticodon interactions may allow a portion of the ribosomes to re-pair in the alternative frame within the short EF-G residence time on the ribosome, thus resulting in a rapid translation in either 0 or –1 frame. In the presence of the pseudoknot, the slippery sequence specifically enhances the –1 frame translation due to faster (about 5-fold) completion of the translocation step as monitored by EF-G release. These data suggest that slippery sequences in genomes may provide a high, as-yet-unappreciated potential for recoding and misreading.
The pseudoknot alone has a moderate inhibitory effect on EF-G binding, but its main effect is to inhibit the backward rotation of the 30S subunit head, which results in the retention of the E site tRNA and EF-G, independent of the slippery sequence. In the absence of the slippery sequence, the progression of the ribosome is stalled dramatically; EF-G release is extremely slow, which limits the rate of further decoding steps. The low rate of spontaneous passage through the pseudoknot in the absence of slippage suggests that passive unwinding, which is one of the two mechanisms employed by the ribosome to handle mRNA secondary structures (Qu et al., 2011), is too slow to promote further movement.
With both stimulatory elements present, the slippery sequence provides the necessary freedom for the ribosome to change its position with respect to the pseudoknot, allowing for the completion of translocation and continuation of translation in the new frame. The position of the pseudoknot relative to the slippery sequence appears to play a central role in –1PRF (Lin et al., 2012). We note that realignment of the ribosome in the –1 reading frame would place the pseudoknot at the ribosomal helicase active site formed by ribosomal proteins S3, S4, and S5 at position +11 of an mRNA bound to the ribosome (counting from the first nucleotide of the P site codon) (Takyar et al., 2005). One attractive hypothesis is that the precise ribosome positioning allows the helicase to act on the mRNA roadblock. This mechanism may work in both pro- and eukaryotic systems, as helicase residues in S3 and S5 are evolutionary conserved. In line with this hypothesis, frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome (Mouzakis et al., 2013). Our experiments suggest that –1 slippage allows for a 3-fold faster movement of the ribosome through the pseudoknot base. This suggests that the exact position of the secondary structure element relative to the translocating ribosome has a kinetic effect on unwinding, underscoring the important role of an active helicase mechanism in mRNA unwinding (Qu et al., 2011).
Implications for the Mechanism of Translocation
Translocation is a highly dynamic process that entails movements of tRNAs, mRNA, and EF-G, rotation of the ribosomal subunits relative to one another, 30S head swiveling and tilting, and motions of ribosomal elements, such as proteins L1 and L12. The present results provide insights into the nature of the rate-limiting steps that govern the stepwise motion of the tRNA-mRNA complex through the ribosome (Figure 5). In the PRE state, the anticodons of the two tRNAs occupy the P and A sites of the 30S subunit, whereas the tRNA 3′ ends on the 50S subunit are dynamic, allowing for different tRNA positions in classical, hybrid, or intermediate states. Binding of EF-G accelerates the rate-limiting ribosome unlocking step, which is followed by rapid translocation both on the 50S and 30S subunit to a state that is a Pmn-reactive POST state with respect to pept-tRNA (analogous to POST2 in Figure 5) (Cunha et al., 2013, Holtkamp et al., 2014, Savelsbergh et al., 2003). The next kinetically distinct step of translocation is the backward movement of the 30S head (Cunha et al., 2013, Guo and Noller, 2012), which takes place concomitantly with the dissociation of EF-G and the E site tRNA and results in the movement of mRNA by one codon. mRNA secondary structures are usually resolved by the unwinding activity of the ribosome (Takyar et al., 2005, Wen et al., 2008). If the ribosome mechanically pulls apart the mRNA strands of the closed junction (Qu et al., 2011), it is likely to do so at a late state of translocation when the 30S subunit head rotates backward.
Experimental Procedures
Ribosome Complexes
Ribosomes, translation factors, and tRNAs were from E. coli. The experiments were carried out in buffer A (50 mM Tris-HCl [pH 7.5], 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2) supplemented with GTP (1 mM) at 37°C. To prepare initiation complexes, 70S ribosomes (1–1.5 μM) or AttoQ-labeled 30S (0.5 μM) and 50S subunits (1 μM) were incubated with a 3-fold excess of mRNA and a 1.5-fold excess each of IF1, IF2, IF3, and f[3H]Met-tRNAfMet in buffer A for 30 min at 37°C. Initiation complexes were purified by centrifugation through a 1.1 M sucrose cushion in buffer A. Ternary complexes were prepared by incubating EF-Tu (2-fold excess over aa-tRNAs) with GTP (1 mM), phosphoenolpyruvate (3 mM), and pyruvate kinase (0.1 mg/ml) for 15 min at 37°C and then with the mixture of Tyr-tRNATyr, Leu-tRNALeu, Lys-tRNALys, Phe-tRNAPhe, and Val-tRNAVal (1 μM) for 1 min. Posttranslocation complexes with MYL-tRNALeu in the P site were prepared by incubating purified 70S initiation complexes (1.5 μM) with ternary complexes EF-Tu–GTP–Tyr-tRNATyr and –Leu-tRNALeu (3.5 μM each) and EF-G (3.5 μM) for 30 s at 37°C. Complexes were purified on a Sephacryl-300 column in buffer A. Ternary complexes with BOP were prepared at a 5-fold excess of EF-Tu–GTP over BOF-Lys-tRNALys to compensate for the decreased affinity of the modified tRNA for the factor as described (Mittelstaet et al., 2013).
Rapid Translation Experiments
Translation experiments were performed by rapidly mixing initiation complexes (0.2 μM) with the respective ternary complexes as indicated (1.5 μM) and EF-G (2 μM) in a quench-flow apparatus. After neutralization with acetic acid, the products were analyzed by HPLC (LiChroSpher100 RP-8 HPLC column, Merck). Fluorescence experiments were carried out using a stopped-flow apparatus after mixing equal volumes of posttranslocation complexes carrying MYL-tRNALeu in the P site (0.05 μM) with ternary complexes as indicated (0.25 μM) and EF-G (1 μM). Alx488 and Flu fluorescence was excited at 470 nm and detected after passing a KV500 cut-off filter (Schott). The time courses were evaluated by numerical integration using the Micromath Scientist software as described in Extended Experimental Procedures.
Frameshift Assay In Vivo
Vectors used for in vivo experiments contained Fluc and Rluc genes amplified by PCR from vectors pGEM-luc and pRL (Promega), respectively, and ligated into pET24a(+) (Novagen) (Figure S2). A synthetic fragment of the IBV 1a/1b gene was cloned into pTZ18 and then used as a template for further PCR amplification. All other vectors were generated by point mutation or deletion using PCR.
Dual luciferase constructs were transformed into E. coli Tuner (DE3) cells (Novagen), and cells were plated on LB with kanamycin (30 μg/ml). LBkan cultures were inoculated from single colonies and grown at 37°C to OD600 = 0.5. Expression was induced with IPTG (70 μM) and conducted for 30 min at 37°C. Cells were harvested by centrifugation (2 min at 10,000 g). Cells were resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 5 mg/ml lysozyme; 1 ml buffer per OD600 unit) and lysed on ice for 10 min. Cell debris was removed by centrifugation for 5 min at 10,000 g and 4°C. Fluc and Rluc activities were measured separately in a luminometer (Sirius Single, Berthold) with a delay time of 2 s and an integration time of 5 s. To measure Fluc activity, 5 μl of supernatant were mixed with 100 μl of Beetle Juice (PJK GmbH) and incubated at RT for 5 min prior to the measurement. Rluc activity was measured by mixing 5 μl of cell extract with 100 μl of Renilla Glow Juice (PJK GmbH) following an incubation of 10 min at RT. Frameshifting efficiencies were calculated as follows: –1PRF = Rluc/Fluc–1 frame/(Rluc/Fluc–1 frame + Rluc/Fluc0 frame). The Rluc/Fluc–1 frame value obtained with the –/– construct was considered as background and subtracted from the Rluc/Fluc–1 frame values obtained with all other mRNA constructs.
Author Contributions
N.C. performed most of the experiments; N.C., V.I.K., and R.B. prepared materials; all authors conceived the research, designed experiments, and analyzed the data; and N.C., F.P., R.B., and M.V.R. wrote the paper.
Acknowledgments
This paper is dedicated to the memory of Vladimir Katunin, who sadly passed away before submission of the manuscript. We thank Anja Lehwess-Litzmann and Carlos da Cunha for sharing their L12–EF-G FRET reporter system prior to publication. We thank Anna Bursy, Olaf Geintzer, Sandra Kappler, Christina Kothe, Theresia Uhlendorf, Tanja Wiles, and Michael Zimmermann for expert technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (RO 2004/9-1 and SFB860 to M.V.R.).
Published: June 19, 2014
Footnotes
Supplemental Information includes Extended Experimental Procedures, three figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.04.041.
Supplemental Information
References
- Bekaert M., Rousset J.P. An extended signal involved in eukaryotic -1 frameshifting operates through modification of the E site tRNA. Mol. Cell. 2005;17:61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Meredith M.R., Bloys A.J., Hagervall T.G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Gilbert R., Pennell S. Pseudoknot-dependent programmed –1 ribosomal frameshifting: Structures, mechanisms and models. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York: 2010. pp. 149–174. [Google Scholar]
- Chen C., Zhang H., Broitman S.L., Reiche M., Farrell I., Cooperman B.S., Goldman Y.E. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013;20:582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C.E., Belardinelli R., Peske F., Holtkamp W., Wintermeyer W., Rodnina M.V. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013;1:e24315. doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj A., Shoji S., Holbrook E.D., Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–265. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S., Brunelle J.L., Sharma D., Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Landes Bioscience; Austin, TX: 1997. Programmed Alternative Reading of the Genetic Code. [Google Scholar]
- Fayet O., Prère M.-F. Programmed Ribosomal −1 Frameshifting as a Tradition: The Bacterial Transposable Elements of the IS3 Family. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York: 2010. pp. 259–280. [Google Scholar]
- Fredrick K., Noller H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Guo Z., Noller H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J.W., Meskauskas A., Dinman J.D. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- Holtkamp W., Cunha C.E., Peske F., Konevega A.L., Wintermeyer W., Rodnina M.V. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J.A., Wilson D.N., Mannering S.A., Adamski F.M., Tate W.P. Prokaryotic ribosomes recode the HIV-1 gag-pol-1 frameshift sequence by an E/P site post-translocation simultaneous slippage mechanism. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Wills N.M., Gesteland R.F., Atkins J.F. rRNA-mRNA base pairing stimulates a programmed -1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger M., Sidani S., Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger M., Dulude D., Steinberg S.V., Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed -1 ribosomal frameshift. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P.Y., Choi Y.S., Dinman J.D., Lee K.H. The many paths to frameshifting: kinetic modelling and analysis of the effects of different elongation steps on programmed -1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:300–312. doi: 10.1093/nar/gkq761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- Lin Z., Gilbert R.J., Brierley I. Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 2012;40:8674–8689. doi: 10.1093/nar/gks629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry K.G., Walker S.E., Wang H., Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstaet J., Konevega A.L., Rodnina M.V. A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J. Am. Chem. Soc. 2013;135:17031–17038. doi: 10.1021/ja407511q. [DOI] [PubMed] [Google Scholar]
- Mouzakis K.D., Lang A.L., Vander Meulen K.A., Easterday P.D., Butcher S.E. HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome. Nucleic Acids Res. 2013;41:1901–1913. doi: 10.1093/nar/gks1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S., Vidakovic M., Girnary R., Namy O., Brierley I. Prokaryotic-style frameshifting in a plant translation system: conservation of an unusual single-tRNA slippage event. EMBO J. 2003;22:3941–3950. doi: 10.1093/emboj/cdg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Jacobs K.L., Harger J.W., Meskauskas A., Jacobs J.L., Baxter J.L., Petrov A.N., Dinman J.D. The 9-A solution: how mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Yu D., Zuo X., Cornish P.V. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014;15:185–190. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Wen J.D., Lancaster L., Noller H.F., Bustamante C., Tinoco I., Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath D.J., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H.F., Spahn C.M. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Semenkov Yu., Shapkina T., Makhno V., Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- Sergiev P.V., Lesnyak D.V., Kiparisov S.V., Burakovsky D.E., Leonov A.A., Bogdanov A.A., Brimacombe R., Dontsova O.A. Function of the ribosomal E-site: a mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Southworth D.R., Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Uemura S., Dorywalska M., Lee T.H., Kim H.D., Puglisi J.D., Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- Uemura S., Aitken C.E., Korlach J., Flusberg B.A., Turner S.W., Puglisi J.D. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Shuh M., Atkins J.F., Gesteland R.F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- Wen J.D., Lancaster L., Hodges C., Zeri A.C., Yoshimura S.H., Noller H.F., Bustamante C., Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelverton E., Lindsley D., Yamauchi P., Gallant J.A. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol. Microbiol. 1994;11:303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson R., Majumdar Z.K., Baucom A., Clegg R.M., Noller H.F. Measurement of internal movements within the 30 S ribosomal subunit using Förster resonance energy transfer. J. Mol. Biol. 2005;354:459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Holmes W.M., Hurd R.E., Reid B.R., Rimerman R.A., Hatfield G.W. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc. Natl. Acad. Sci. USA. 1975;72:1068–1071. doi: 10.1073/pnas.72.3.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U., Paleskava A., Konevega A.L., Rodnina M.V. Single-step purification of specific tRNAs by hydrophobic tagging. Anal. Biochem. 2006;356:148–150. doi: 10.1016/j.ab.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Milon P., Konevega A.L., Peske F., Fabbretti A., Gualerzi C.O., Rodnina M.V. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- Mohr D., Wintermeyer W., Rodnina M.V. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 2000;19:3458–3464. doi: 10.1093/emboj/19.13.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S., Liphardt J., Bloys A., Routledge S., Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient -1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Matassova N.B., Katunin V.I., Semenkov Y.P., Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. USA. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr D., Kothe U., Wintermeyer W., Rodnina M.V. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.S., Noller H.F. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W., Schleich H.G., Zachau H.G. Incorporation of amines or hydrazines into tRNA replacing wybutine or dihydrouracil. Methods Enzymol. 1979;59:110–121. doi: 10.1016/0076-6879(79)59073-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.