Abstract
Protein (poly-)ubiquitination is a posttranslational modification that plays a key role in almost all cellular processes. It involves the installment of either single ubiquitin (Ub) moieties or one of eight different polyUb linkage types, each giving a distinct cellular outcome. Deubiquitinating enzymes (DUBs) reverse Ub signaling by disassembly of one or multiple poly-Ub chain types and their malfunction is often associated with human disease. The Ub system displays significant crosstalk with structurally homologous ubiquitin-like proteins (Ubls), including SUMO, Nedd8, and ISG15. This can be seen with the existence of heterogeneous chains made from Ub–Ubl mixtures as well as the proteolytic cross reactivity displayed by several DUBs toward other Ubl systems. In addition, numerous pathogens have been found to encode Ub(l)-ligases and deconjugating enzymes in order to facilitate infection and fight the host immune response. Studying the activity of DUBs and Ubl-specific proteases, both human as well as pathogen-derived, gives fundamental insights into their physiological roles. Activity-based probes (ABPs) have proven to be valuable tools to achieve this, as they report on enzyme activities by making a (often irreversible) covalent complex, rather than on their relative abundance. In this chapter, we explain the potential of ABPs to assess substrate preferences, structural features, and activity of Ub and Ubl deconjugating enzymes. We further demonstrate the practical use of ABPs to (1) characterize the activity of viral proteases toward Ub and Ubls and (2) to gain more insight in the structural determinants of substrate preference of DUBs.
Keywords: Deubiquitinating enzymes, Ubl-specific proteases, Activity-based probes, Ub linkage specificity, (di)ubiquitin-based probes, Viral DUBs, MERS-CoV PLpro, SARS-CoV PLpro, Ub–Ubl crosstalk
1. Introduction
Protein ubiquitination is a posttranslational modification that plays a major role in almost all cellular processes in eukaryotes (Hochstrasser, 2009; Komander & Rape, 2012). It involves the covalent attachment of ubiquitin (Ub) via its C-terminal glycine carboxylate to a primary amine of a target protein, generally to a lysine side chain resulting in an isopeptide bond. Ub itself can also be ubiquitinated and as such give rise to polyUb chains. This conjugation occurs at the side chain of one of the seven internal lysine residues (Lys-6, 11, 27, 29, 33, 48, 63), resulting in an isopeptide bond, or at the N-terminus (Met-1), resulting in a linear Ub chain, and it has been shown that all eight linkages coexist in cells (Xu et al., 2009).
Counteraction of the build-up of (poly-)ubiquitinated proteins is achieved by a group of deubiquitinating proteases (DUBs) that remove or trim the ubiquitin modification, liberating the substrate protein, recycling Ub and ending the Ub-induced signal. Nearly a hundred genes encoding DUBs have been identified in the human genome, which can be classified in seven distinct families. The subfamilies of ubiquitin-specific proteases (USP), ubiquitin C-terminal hydrolases (UCH), Ovarian TUmor domain proteases (OTU), Machado-Joseph disease proteases (MJD), Motif interacting with ubiquitin-containing novel DUB family (MINDY), and Zinc finger with UFM1-specific peptidase domain protein (ZUFSP) are cysteine proteases, whereas JAB1/MPN/MOV34 proteases (JAMMs) are zinc-dependent metalloproteases (Abdul Rehman et al., 2016; Komander, Clague, & Urbe, 2009; Kwasna et al., 2018; Nijman et al., 2005; Reyes-Turcu, Ventii, & Wilkinson, 2009). As distinct Ub linkages result in distinct biological signals (Komander & Rape, 2012; Yau & Rape, 2016), the determination of the linkage specificities of DUBs gives fundamental insights into the biological pathways they are involved in. It has been shown that some DUBs, mainly USPs, are able to process all isopeptide linked chains (Faesen et al., 2011) whereas others, especially OTUs, display a preference for one or a few Ub chain types (Mevissen et al., 2013).
Another level of complexity is based on the existence of Ub-like proteins (Ubls) (Kerscher, Felberbaum, & Hochstrasser, 2006). These posttranslational modifiers share structural homology to Ub as well as a highly similar system for conjugation and deconjugation. The most studied examples are the small ubiquitin-like modifiers (SUMO), the neural precursor cell-expressed developmentally downregulated 8 (Nedd8) and interferon-stimulated gene of 15 kDa (ISG15). SUMOylation plays a key role in genome stability, and many of its protein targets are involved in DNA-damage responses (e.g., PCNA and BRCA1) (Flotho & Melchior, 2013). Nedd8 plays an important role in cell cycle control and its main targets are Cullin proteins, which are Ub ligase subunits (Soucy, Smith, & Rolfe, 2009). ISG15 is strongly induced by Type-I interferons as part of the innate immune response to viral and bacterial infections (Zhang & Zhang, 2011). Similar to the Ub system, specific proteases deconjugate Ubls from their targets. These include SENPs acting on SUMO (Hickey, Wilson, & Hochstrasser, 2012), USP18 acting on ISG15 (Malakhov, Malakhova, Kim, Ritchie, & Zhang, 2002) and DENs acting on Nedd8 (Gan-Erdene et al., 2003) and we refer to this group of proteases as Ubl-specific proteases.
Despite their functions in their own respective modification systems, there is growing evidence of crosstalk between Ub and Ubls, increasing the complexity of cellular responses even further. Best studied is the crosstalk between Ub and SUMO signaling, which includes the identification of ubiquitinated SUMO and SUMOylated Ub (Hendriks et al., 2014; Hendriks & Vertegaal, 2016; Nie et al., 2016; Nie & Boddy, 2016). Furthermore, ubiquitinated Nedd8 and crosstalk between Ub and Nedd8 signaling pathways have also been reported (Leidecker, Matic, Mahata, Pion, & Xirodimas, 2012; Singh, Sundar, & Fushman, 2014), as the existence of ISGylated ubiquitin (Fan et al., 2015). However, these so-called heterogeneous chains have so far remained largely unstudied, and their functions remain unknown (Swatek & Komander, 2016). In addition, several DUBs have been found to act on Nedd8 or ISG15 as well (Catic et al., 2007; Gan-Erdene et al., 2003; Geurink, El Oualid, Jonker, Hameed, & Ovaa, 2012; Hjerpe et al., 2012).
Since both the Ub and ISG15 systems are crucial for the innate immune response, many prokaryotic and viral pathogens have evolved ways to hijack them in order to create a “window-of-opportunity” for efficient replication. Several viral and bacterial proteins have been found to directly target these systems via their deubiquitinating or deISGylating activity (Li, Chai, & Liu, 2016). For example, proteases derived from severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and Crimean-Congo hemorrhagic fever virus (CCHFV) have been shown to deconjugate both Ub and ISG15 in order to suppress innate immune responses (Barretto et al., 2005; Bekes et al., 2016; Frias-Staheli et al., 2007; Lindner et al., 2005; Mielech, Kilianski, Baez-Santos, Mesecar, & Baker, 2014). In contrast to eukaryotic DUBs, proteases encoded by pathogens (bacteria and viruses) often deconjugate more than one type of Ubl. Another example is the CE clan bacterial effector proteases from, for instance, Rickettsia bellii and Chlamydia trachomatis, which were shown to display both deubiquitinating and deNeddylating activities (Lin & Machner, 2017; Pruneda et al., 2016).
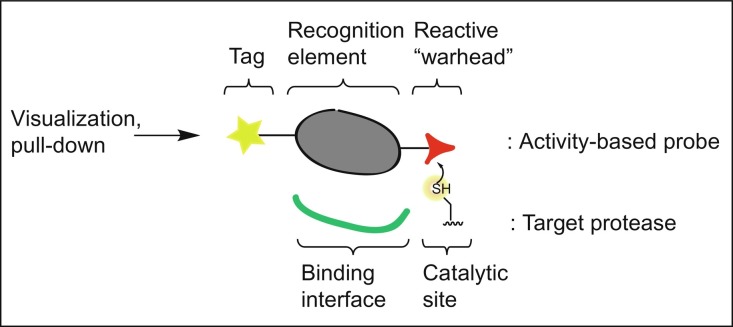
For many human DUBs and Ubl-specific proteases, it has been shown that their malfunction contributes to human disease, including cancer and neurodegenerative disorders (Harrigan, Jacq, Martin, & Jackson, 2018); therefore, tools to study them in detail and on a molecular level are of great interest. Often, proteases are translated as inactive proenzymes, requiring posttranslational activation by their natural regulators. In addition, their activity may be controlled by posttranslational modifications, such as acetylation, phosphorylation, ubiquitination, or methylation. In order to study the role of DUBs in biological processes, it is therefore insufficient to simply monitor the enzyme's abundance by antibody staining, proteomics, or mRNA quantification because this is not necessarily related to a protein's activity (Hewings, Flygare, Bogyo, & Wertz, 2017). A powerful method to visualize enzyme activities in a complex biological setting is the use of Activity-Based Probes (ABPs) (Ovaa, 2007; Verdoes & Verhelst, 2016). ABPs come in many flavors and their design is predominantly determined by their respective protein target(s) and the particular application of the ABP. Generally, ABPs comprise a recognition element, that directs the ABP toward its target, attached to a reactive group (or “warhead”) that reacts with the enzyme's active site to form a covalent adduct, either reversible or irreversible, depending on the type of enzyme and reactive group installed (see Fig. 1 ). The recognition element is designed to resemble structural and functional motifs of the natural substrate of the target, in the form of a short peptide, carbohydrate, nucleoside, or even a small protein. A variety of ABPs has been developed to study the activities of DUBs and these all share a common recognition element derived from full-length Ub. Typically, but not necessarily, an ABP is also equipped with a reporter group, such as a fluorophore, radioactive label or affinity tag, which is used for visualization, purification or identification of the ABP-bound target(s).
Fig. 1.
General design of an activity-based probe (ABP).
These chemical tools are designed in such a way that they only bind to active enzymes covalently but do not react with their inactive counterparts. The application of ABPs is widespread. For example, these tools are commonly used in combination with mass spectrometry, to capture, isolate, and identify active enzymes from cells or cell extracts (Cravatt, Wright, & Kozarich, 2008). In addition, ABPs can be applied to determine the active fraction of a recombinantly expressed and purified enzyme or to study the effect of specific enzyme modifications or mutations with respect to the enzyme's activity and its substrate specificities (Mevissen et al., 2013, Mevissen et al., 2016). ABPs are also very useful tools for gaining insight into the structural characteristics of an enzyme, where an enzyme–ABP complex mimics a certain state of the reaction between the enzyme and its substrate (Basters et al., 2017; van Tilburg, Elhebieshy, & Ovaa, 2016). Also, by designing and testing different structural variants of an ABP (Flierman et al., 2016; Mulder, El Oualid, ter Beek, & Ovaa, 2014), one can identify preferences of a given enzyme for certain structural features (Bekes et al., 2015, Bekes et al., 2016; Mevissen et al., 2016). Finally, since only the active fraction of an enzyme is labeled by the ABP, it is possible to check the inhibitory potential of an inhibitor toward one or multiple enzymes in a cell or cell lysate, e.g., by means of an ABP competition assay (Altun et al., 2011; de Jong et al., 2012).
Here, we showcase the toolbox frequently used for the analysis of DUB activity and illustrate its application by profiling pathogen-derived proteases toward Ub and Ubls.
2. Activity-based probes
2.1. Probes based on a monoUb or Ubl recognition element
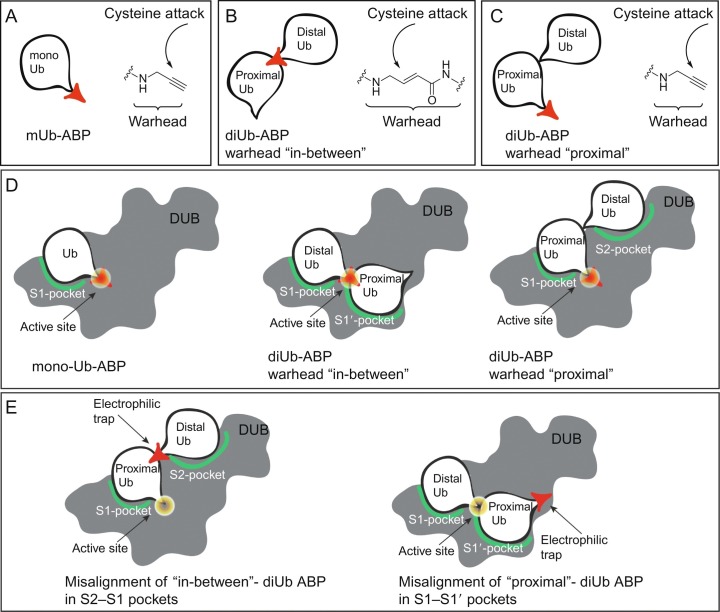
The first activity-based DUB probe was based on the replacement of the C-terminal Gly76 in Ub with an aldehyde moiety as reactive “warhead” (Ubal) (Pickart & Rose, 1986). Replacement of this reactive C-terminal element with a nitrile moiety (UbCN) (Lam, Xu, DeMartino, & Cohen, 1997), a glycine vinylsulfone (UbVS), or glycine vinylmethylester (UbVME) (Borodovsky et al., 2001, Borodovsky et al., 2002) led to the development of a larger panel of ABPs able to capture the active site cysteine of DUBs. The electron-poor vinyl motifs in UbVME and UbVS act as a Michael acceptor elements that trap the sulfur nucleophile of the active site cysteine under the formation of a covalent, irreversible bond. Later on, the total chemical synthesis of Ub (and mutants thereof) using solid phase peptide synthesis (SPPS) (El Oualid et al., 2010; Kumar, Haj-Yahya, Olschewski, Lashuel, & Brik, 2009; Pasunooti et al., 2009) opened the way to prepare Ub-ABPs carrying fluorescent labels or affinity handles on a large scale (de Jong et al., 2012). Unexpectedly, it was found that the C-terminal amide derivative of Ub1–75 with propargylamine (Ub-Prg or Ub-PA) is also able to covalently trap the active site cysteine of DUBs by formation of a stable vinyl thioether (Fig. 2A) (Ekkebus et al., 2013).
Fig. 2.
Activity-based probes to target DUB specificity. (A) monoUb-PA probe. (B) Details of “in-between” diUb probe. (C) Details of “proximal” diUb probe. (D) Different probes targeting S1, S1–S1′ or S1–S2 interactions. (E) Misalignment of probes on DUBS prevents formation of covalent complexes.
In general, specificity of such probes for DUBs originates from the interaction of the Ub-recognition element in the ABP with a Ub-binding interface in the DUB. This so-called S1 pocket in the protease holds the Ub molecule so that its C-terminus with the reactive element is positioned in close proximity to the active site cysteine of the protease. With this positioning, the two reacting partners are optimally aligned to allow formation of a covalent adduct. In a normal reaction with a ubiquitinated species, the DUB would be able to cleave the amide bond between substrate and Ub, resulting in free substrate and Ub, but due to the nature of the warhead, a covalent adduct is formed in the case of ABPs. Along this line, similar ABPs for Ubl-specific proteases have been developed by replacing the C-terminal residue in the respective Ubl for an electrophilic moiety, such as -Prg, -VME, or -VS and these include ABPs based on SUMO1,- 2, − 3, Nedd8, ISG15, and UFM1. Currently, many of these Ub and Ub-like ABPs are commercially available but are also readily obtained by chemical synthesis (Basters et al., 2017; Ekkebus et al., 2013; Mulder et al., 2018; Witting et al., 2018), or semisynthesis, such as the use of intein chemistry for Ub(l)ΔG-thioester formation, followed by reaction of this thioester with an amine nucleophile (Hemelaar et al., 2004).
2.2. Probes based on a diUb recognition element
While monoUb ABPs have greatly increased our understanding of DUB reactivity, these ABPs offer no information on poly-Ub chain recognition and processing. Classically, in order to study possible linkage preferences, a recombinantly expressed and purified DUB is incubated with each of the seven native isopeptide-linked diUb molecules. Cleavage of the diUb molecules is then monitored over time using gel-based analyses. A major limitation of this methodology is that the results are not readily extrapolatable to the substrate preference of a DUB in a more complex environment, such as cell-lysate, which might modulate DUB activity and preference due to other factors present in such samples. To overcome these issues, ABPs to investigate linkage specific proteolysis of DUBs have been developed, that can be utilized in complex biological systems such as cell lysates. These probes generally consist of two ubiquitin moieties equipped with a Michael acceptor element in the isopeptide linkage region in between the two Ub moieties (often referred to as “in-between” diUb probes). Initial reports show the two Ub regions to be linked together using nonnative connections such as a triazole (McGouran, Gaertner, Altun, Kramer, & Kessler, 2013) and a thiolether linkage (Li, Liang, Gong, Tencer, & Zhuang, 2014). Two types of probes, containing either a dehydroalanine (Dha) (Haj-Yahya et al., 2014) or VME-like electrophilic trap (Mulder et al., 2014) (see Fig. 2B), mimic the native lysine–glycine isopeptide linkage the closest.
A panel of all seven isopeptide linked diUb probes can be prepared and used to covalently capture the active site cysteine of DUBs, showing their reactivity and preference toward certain linkage types. When using such probes, the distal Ub molecule will be positioned in the so-called S1-pocket and the proximal Ub molecule in the so-called S1′-pocket, thereby placing the reactive element directly over the reactive cysteine. Due to the geometrical differences between all Lys-linked diUb probes, the DUB will only be able to position the probes mimicking its natural substrates in such a way that the active site cysteine is able to react with the reactive element. Although these covalent vinyl amide probes have allowed more detailed structural investigation of diubiquitin-specific DUB recognition (Mevissen et al., 2016), they do not allow investigation of additional Ub-binding sites.
Some DUBs are able to recognize Ub chain topologies using other binding surfaces positioned further away from the active site, such as the S2-site. A set of probes targeting these S2-binding sites has been developed where a diUb molecule is constructed carrying a reactive element at the proximal C-terminus (Flierman et al., 2016) (see Fig. 2C). These probes are only able to react with DUBs that contain an S2-site that plays a determining role in positioning the diUb molecule in the S2- and S1-sites, thereby placing the reactive warhead directly over the active site cysteine (see Fig. 2D). Noteworthy is that the isopeptide linkage between proximal and distal Ub has been substituted for a protease-stable triazole linkage, prohibiting the protease of interest from degrading the probe during the assays.
If a DUB recognizes such a “proximal” diUb probe using its S1- and S1′-sites, the reactive warhead will not be in the vicinity of the active site cysteine and no covalent adduct will be formed. Conversely, if a “in-between” diUb probe will react with a DUB recognizing the diUb moiety using its S2- and S1-sites, no reaction will occur either since the reactive element will not be optimally aligned with the reactive cysteine (see Fig. 2E). Having access to both “proximal’ and “in-between” diUb probes offers an exciting combination to investigate the binding interfaces that play a role in determining binding preferences of DUBs and cast a light on their molecular mechanisms of action as showcased in the next section.
3. Characterization of coronavirus-encoded DUBs with activity-based probes
To study the activity of specific DUBs, investigators frequently take advantage of the ~ 10 kDa (monoUb or Ubl ABP) or ~ 20 kDa (DiUb ABP) increase in MW on probe labeling. SDS-PAGE analysis or blotting for an individual DUB, and comparing the intensities of the larger (labeled) band to the smaller (unlabeled) band, which allows the reactivity of the DUB toward the probe to be inferred. Here, we demonstrate how a typical experiment can be performed, by showing analysis of the MERS-CoV-encoded papain like cysteine protease (abbreviated PLpro), a viral DUB, upon incubation with our panel of ABPs. Additionally, having access to both “proximal” and “in-between” diUb probes we will demonstrate their use in the investigation of the binding interfaces that play a role in the specificity of the coronavirus-encoded DUBs MERS-CoV PLpro and SARS-CoV PLpro.
3.1. Probes based on a monoUb or Ubl recognition element
We first demonstrate how a typical ABP labeling experiment can be performed using a panel of Ub-Prg and Ubl-Prg probes in combination with their known proteases. The panel of ABPs consists of untagged constructs of human Ub, Nedd8, SUMO1, SUMO2, SUMO3, ISG15, and the C-terminal domain of ISG15 (see Table 1 ). The C-terminal glycine is replaced by propargylamine in all ABP reagents.
Table 1.
Overview of Ub and Ubl ABPs used in this study
| Protein | Abbreviation | UniProt ID | Residuesa | MW (kDa) |
|---|---|---|---|---|
| Ubiquitin | Ub | P0CG47b | 1-75 | 8.5 |
| Nedd8 | N8 | Q15843 | 1-75 | 8.5 |
| SUMO1 | S1 | P63165 | 1-96 | 11.1 |
| SUMO2 | S2 | P61956 | 1-92 | 10.6 |
| SUMO3 | S3 | P55854 | 1-91 | 10.5 |
| ISG15 C-domain | I15c | P05161 | 79–156 | 8.9 |
| ISG15 (C78S)c | I15 | P05161 | 1–156 | 17.1 |
The C-terminal Gly residue is not included in this list.
Ub is only listed as a polyubiquitin in UniProt; the UniProt ID refers to polyubiquitin-B (UBB).
The C78S-mutation was introduced to solubilize the ISG15-protein.
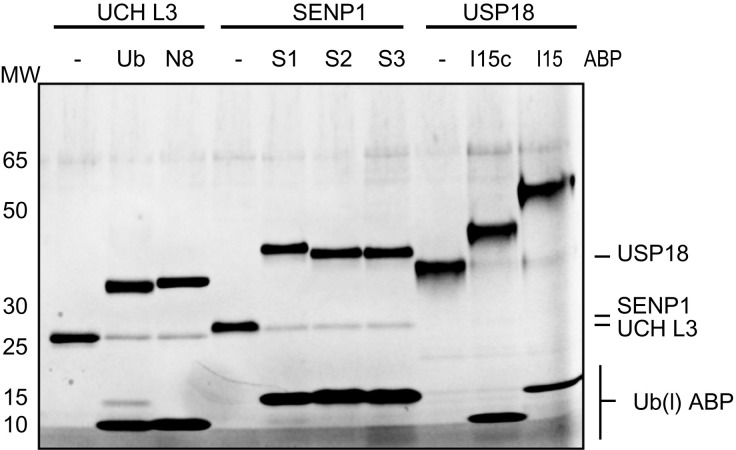
The reaction of a DUB or Ub-like protease with an ABP can be confirmed by incubation of the enzyme with the ABP followed by SDS-PAGE analysis. Fig. 3 shows the image of a typical ABP labeling experiment in which the ABPs were incubated with three proteases known to act on them: The DUB UCH-L3 is known to process Ub and Nedd8 (Gan-Erdene et al., 2003), SUMO protease SENP1 is active on all three SUMO proteins (Gong, Millas, Maul, & Yeh, 2000), and deISGylase USP18 targets ISG15 (Malakhov et al., 2002). The reaction between the enzyme and an ABP becomes apparent from the shift of a protein band to a higher molecular weight equal to the total mass of enzyme plus ABP. Here, the UCH-L3 band around 25 kDa shifts to a ~ 35 kDa band with either the Ub- or Nedd8 ABP. Similarly, the SENP1 band shifts from ~ 26 to ~ 40 kDa when incubated with either of the SUMO probes. It is to note here that SUMO proteins run somewhat higher than what would be expected from their mass. Finally, the USP18 corresponding band shifts from ~ 38 to ~ 43 kDa and ~ 50 kDa upon incubation with the truncated ISG15c ABP or the full-length ISG15, ABP respectively. From this result it follows that USP18 does not require full-length ISG15 for proper binding but that the C-terminal domain alone is enough, which corroborates earlier published results (Basters et al., 2017).
Fig. 3.
Profiling of proteases UCH-L3, SENP1, and USP18 against Ub(l)-Prg ABPs.
Upon closer examination of the gel image in Fig. 3, it can be seen that in most cases where the enzyme is incubated with the ABP, a small protein band remains at the molecular weight corresponding to the unbound enzyme. This indicates that not all enzyme reacted with the ABP and that most likely the enzyme is not 100% active. Quantification of the band intensities will give an estimate of the active fraction of the enzyme.
An experiment as shown in Fig. 3 can also be used to validate the properties of an ABP that was constructed and purified, by incubation of the ABP with its known protease target. An appropriately folded and active ABP will result in a proper reaction with its protease, which can be checked and quantified by SDS-PAGE analysis.
3.2. Profiling of MERS-CoV PLpro using monoUb and Ubl ABPs
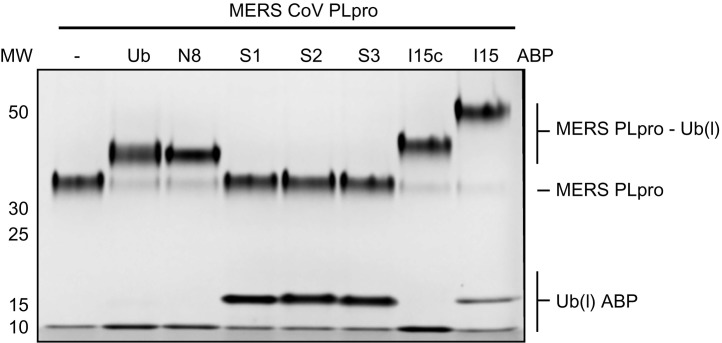
These Ub and Ubl ABPs are also particularly well suited to profile the activity of pathogen-derived proteases toward Ub and Ubls. We here present how a typical experiment can be performed, by showing analysis of a cysteine protease encoded by MERS-CoV, named MERS-CoV PLpro (Mielech et al., 2014), upon incubation with our panel of ABPs. The experiment is similar to the one described above and the result is shown in Fig. 4 . Unbound MERS-CoV PLpro gives a band around 32 kDa (outer left lane) and incubation with Ub, Nedd8 or either of the ISG15 ABPs shows a clear shift to a higher molecular weight band, whereas no change is observed with any of the three SUMO ABPs. It also becomes apparent that, like USP18, MERS PLpro only requires the C-terminal domain of ISG15 for proper binding, which is consistent with previously described results (Daczkowski, Goodwin, Dzimianski, Farhat, & Pegan, 2017). In all cases where a reaction takes place with the Ub(l)-ABP it shows almost full conversion, meaning that the enzyme preparation is close to 100% active.
Fig. 4.
Profiling of MERS PLpro against Ub(l)-Prg ABPs.
3.3. DiUb ABPs to characterize MERS CoV PLpro and SARS CoV PLpro activity
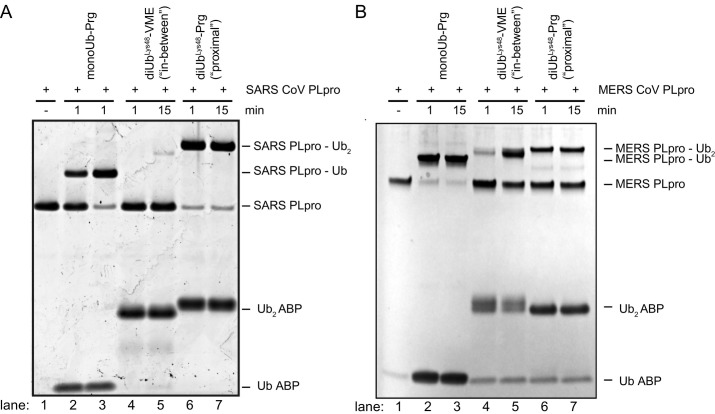
Virus-encoded DUBs such as MERS-CoV PLpro and SARS-CoV PLpro (both papain-like proteases) have been shown to counteract the host cell's ubiquitination machinery. The SARS-CoV and MERS-CoV PLpro enzymes both can bind and cleave K48-linked Ub chains, however they do this in distinctive manners. It was shown that MERS-CoV PLpro has a monodistributive mechanism, meaning it nibbles one Ub-molecule off the K48-polyUb chain at a time (Bekes et al., 2015). The SARS-CoV PLpro on the other hand was shown to have a di-distributive mechanism, as it recognizes a diUb molecule and cleaves this off the polyUb chain as one unit (Bekes et al., 2016). ABPs with a diUb recognition element (Flierman et al., 2016; Mulder et al., 2014) were used to study this interesting difference between SARS-CoV- and MERS-CoV PLpro as demonstrated in Fig. 5 .
Fig. 5.
SARS PLpro and MERS PLpro profiling using monoUb-, K48 linked diUb “in-between”-, and K48-linked diUb “proximal”-ABPs. (A) SDS-PAGE analysis of SARS PLpro reactivity toward the three types of probes. (B) SDS-PAGE analysis of MERS PLpro reactivity toward the three types of probes.
Panel A: Adapted from Bekes, M., van der Heden van Noort, G. J., Ekkebus, R., Ovaa, H., Huang, T. T., & Lima, C. D. (2016). Recognition of Lys48-linked Di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Molecular Cell, 62(4), 572–585. doi:10.1016/j.molcel.2016.04.016.
SARS-CoV PLpro has a strong preference for the K48-linked “proximal”-probe as full complex formation is observed within 1 min of reaction time (see Fig. 5A, lanes 6–7). In contrast, the monoUb-probe does react with the protease but more slowly, showing a similar extend of labeling only after 15 min of incubation time (see Fig. 5A, lanes 2–3). The protease shows little reactivity toward the K48-linked “in-between”-probe, showing only marginal complex formation after 15 min (see Fig. 5A, lanes 4–5). This experiment shows the strong dependency of this DUB on an S2-site to bind diUb-molecules or longer Ub chains (Bekes et al., 2016). These results are in line with previous findings showing SARS-CoV PLpro to have a di-distributive mode of action (Bekes et al., 2015). A crystal structure of the K48-diUb “proximal”-probe in complex with the enzyme has allowed a detailed look at this S2-site and pin-pointed important interactions contributing to this strong S2 dependency in the DUB's proteolytic profile (Bekes et al., 2016).
Reactivity of MERS-CoV PLpro toward monoUb-, diUb “in-between”- and diUb “proximal”-probes shows a distinct profile compared to SARS-CoV PLpro (see Fig. 5A and B). The MERS-CoV DUB reacts fast with the monoUb-probe showing complete labeling within 1 min (Fig. 5B, lanes 2–3). Both diUb-probes are also processed by the DUB, but more slowly as roughly 50% of the protease is labeled after 15 min (Fig. 5B, lanes 5 and 7). When looking at the “in-between” probe a notable increase in labeling is observed when comparing the two time points (Fig. 5B, lanes 4 and 5), whereas for the “proximal” probe no increase in labeling is observed after 1 min (Fig. 5B, lanes 6 and 7). These results might indicate that recognition of the Ub-substrate by MERS-CoV PLpro primarily occurs using the S1-site and additional substrate context either on the proximal or distal site of this recognized Ub-moiety is tolerated, however slowing down the proteolysis event.
When using ABPs to study DUB substrate preferences, finding the correct reaction time window is crucial because measurements at different times may result in different outcomes. The SARS-CoV PLpro for instance (Fig. 5A) shows a preference for the K48-linked diUb “proximal” probe (lane 6) over the monoUb-Prg probe (lane 2) at short incubation times, whereas at longer incubation times the amount of labeling for both seems to be identical (lanes 3 and 7). The same holds true for MERS PLpro (Fig. 5B), where at 15 min incubation the difference between “in-between”-probe and “proximal”-probe (lanes 5 and 7) is less pronounced then at short incubation times (lanes 4 and 6). One way to overcome making assumptions based on such time-dependent snapshots of enzyme activity is to measure full kinetic parameters of the DUBs. Michaelis–Menten kinetics can be measured based on fluorogenic (di)Ub substrates that start to emit a fluorescent signal upon DUB activity. Rather than forming a covalent complex with the DUB, these substrates are processed by the DUB in a normal way, liberating a fluorescent molecule. As such, the appearance of the fluorescent signal is a direct measure of DUB activity, that can be followed in real-time. Both “proximal” diUb and monoUb substrates have been generated and compliment the toolbox of ABPs to study DUBs (Dang, Melandri, & Stein, 1998; Flierman et al., 2016).
4. Methods
4.1. Preparation of Ub-like-PRG probes using intein chemistry
4.1.1. Equipment
-
•
Incubator
-
•
Vortex mixer
-
•
Centrifuge
-
•
Sonicator (we used Fisher Scientific FB120, 120 W, 20 kHz with a CL-18 tip)
-
•
Sealable column
-
•
RP-HPLC system (we used a Waters HPLC system equipped with a Waters XBridge Prep C18 5-μm OBD column (30 × 150 mm)
-
•
LC–MS system (we used a Waters 2795 Separation Module (Alliance HT) using a Phenomenex Kinetex C18-column (2.1 × 50, 2.6 μm), Waters 2996 Photodiode Array Detector (190–750 nm) and LCT™ ESI-Mass Spectrometer
-
•
Spinfilter (3000-Da cutoff)
-
•
Gel filtration system (FPLC) (we used a BioRAD NGC)
-
•
Hi-Load Superdex75 16/600 (GE-Healthcare) size-exclusion chromatography column
-
•
Nanodrop
-
•
Cold room (4°C)
-
•
50 mL Falcon tubes
-
•
Lyophilizer
4.1.2. Buffers and reagents
-
•
Lysis buffer: 50 mM HEPES, 100 mM NaOAc, pH 6.5
-
•
SEC buffer: 50 mM MES, pH 6.5, 100 mM NaCl
-
•
Chitin resin, stored in EtOH (New England BioLabs, catalog number S6651S)
-
•
Protease inhibitor cocktail (Complete, Roche)
-
•
β-mercaptoethanesulfonic acid sodium salt (MesNa)
-
•
Propargylamine (Sigma Aldrich, catalog number P50900)
-
•
DMSO
-
•
Acetic acid
-
•
Deionized water
4.1.3. Procedure
Expression of Ubl-intein-chitin-binding domain fusion proteins can be performed in Escherichia coli BL21 cells as reported elsewhere (Basters et al., 2017; Hemelaar et al., 2004). The Ubl-PRG probes can be prepared from the bacterial cell pellet as follows:
-
1.
Resuspend the bacterial cell pellet from a 2.5 L culture in 80 mL lysis buffer (+ protease-inhibitor cocktail) by vigorous vortexing.
-
2.
Lyse the cells by sonication: 6 × (30 s ON, 45 s OFF, amplitude 50%).
-
3.
Centrifuge for 10 min at 3500 rpm at 4°C.
-
4.
Collect the supernatant by decantation.
-
5.
Prepare a 30 mL chitin-bead column, remove the EtOH and flush the column with 120 mL lysis buffer.
-
6.
Load the supernatant onto the chitin-bead column at a flow rate of 0.5 mL/min.
-
7.
Wash the column with 120 mL lysis buffer, followed by 60 mL lysis buffer containing 50 mM MesNa.
-
8.
Add 30 mL lysis buffer containing 50 mM MesNa to the chitin beads, seal the column tube and incubate for 15 h at 37°C.
-
9.
Collect the 30 mL elution (this contains the protein-MesNa thioester) and wash the beads with another 25 mL lysis buffer containing 50 mM MesNa and collect this as well.
-
10.
Pool the fractions and concentrate them to a concentration of ~ 5 mg/mL by ultrafiltration using 3000 Da cutoff centrifugal filter units.
-
11.
Prepare a solution of 2 M propargylamine in lysis buffer and add this to the protein-MesNa thioester such that the final concentration of propargylamine becomes 225 mM.
-
12.
Check the pH. It should be around pH 8.5, otherwise adjust the pH accordingly by addition of 1 M HCl or 1 M NaOH.
-
13.
Incubate the mixture at room temperature and follow the reaction by LC–MS analysis. A typical reaction time is 90 min to achieve complete conversion.
-
14.
Acidify the mixture to pH 4.5 by addition of acetic acid.
-
15.
Purify the Ub-like-PRG protein by RP-HPLC purification: 20%–60% CH3CN in MQ with 0.1% TFA over 15 min at a flow rate of 37.5 mL/min.
-
16.
Combine and lyophilize the fractions containing pure Ub-like-PRG protein.
-
17.
Dissolve the dried protein in DMSO to a concentration of 10 mM.
-
18.
Slowly dilute this into SEC buffer to a concentration of 1 mM.
-
19.
Load the protein solution onto a size-exclusion chromatography Superdex 75 (16/600) column equilibrated with SEC buffer and elute in the same buffer.
-
20.
Combine pure fractions and concentrate them where necessary by ultrafiltration.
-
21.
The pure Ub-like PRG protein solution can be stored at − 80°C and remain stable for more than a year.
4.1.4. Notes
-
1.
Unless noted otherwise, keep everything on ice or in a cold room at 4°C.
-
2.
It is important to predissolve propargylamine in the lysis buffer and adjust the pH before adding it to the Ubl-MesNa thioester solution to prevent local pH increases.
4.2. Preparation of synthetic ubiquitin probes
4.2.1. Equipment
-
•
Rotation film evaporator
-
•
Round bottom flasks
-
•
Fritted syringes 5 mL
-
•
Magnetic stirrer
-
•
Multitech Syro II peptide synthesizer
-
•
LC–MS system for analysis
-
•
RP-HPLC system with C18 column
-
•
Lyophilizer
-
•
Eppendorf tubes 1.5 mL
-
•
Gel filtration system (FPLC) equipped with a Superdex 75 16/600 size-exclusion chromatography column
4.2.2. Reagents
-
•
Preloaded trityl resin TentaGel® R TRT-Gly Fmoc (Rapp Polymere GmbH; RA1213)
-
•
1,1,1,3,3,3-hexafluoro-2-propanol
-
•
1,2-Dichloroethane
-
•
Dichloromethane
-
•
TFA cleavage cocktail: 90% trifluoroacetic acid, 5% H2O, 2.5% triisopropylsilane, 2.5% phenol (v/v/v/v)
-
•
Diethylether
-
•
n-Pentane
-
•
PyBOP: (Benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate
-
•
Triethylamine
-
•
Propargylamine
-
•
Sodium ascorbate
-
•
TBTA analog (prepared according to (Zhou & Fahrni, 2004))
-
•
Cu(II)SO4
-
•
EDTA: ethylenediaminetetraacetic acid
-
•
MPAA: 4-mercaptophenylacetic acid
-
•
TCEP: tris(2-carboxyethyl)phosphine
-
•
Chaotropic buffers: 8 M Urea containing 100 mM sodium phosphate pH 7 or 6 M Guanidinium·HCl containing 150 mM sodium phosphate pH 7
-
•
2,5-dibromohexandiamide
4.2.3. Procedure
Ub-mutants were synthesized as reported elsewhere (El Oualid et al., 2010) on an automated solid phase peptide synthesizer from Multitech Syro II on 25 μm scale. Preloaded trityl resin TentaGel® R TRT-Gly Fmoc (Rapp Polymere GmbH; RA1213) was used to allow mild acidic release of the final peptide from the resin without removing all side chain functionality protective groups. After automated synthesis the crude Ub-mutants were processed as follows:
4.2.3.1. Ub-Prg probe synthesis
-
1.
React Ub1–75 resin with 4 mL 20% v/v hexafluoro-2-propanol in dichloromethane for 20 min in a fritted syringe while gently shaking at room temperature.
-
2.
Collect the filtrate in a 25 mL round bottom flask and concentrate using a rotation film evaporator.
-
3.
Repeat the treatment of the resin with 4 mL 20% v/v hexafluoro-2-propanol in dichloromethane for 20 min and concentrate the combined filtrates. Coevaporate with 1,2-dichloroethane three times to remove all traces of hexafluoro-2-propanol.
-
4.
Dissolve the partially protected peptide in a round bottom flask in 5 mL dichloromethane and add 5 eq. PyBOP, 5 eq. triethylamine and 10 eq. propargylamine. React for 16 h at room temperature while stirring with a magnetic stirrer.
-
5.
Concentrate the reaction mixture using a rotation film evaporator and redissolve in 5 mL TFA cleavage cocktail and react for 2.5 h at room temperature while stirring with a magnetic stirrer.
-
6.
Add the reaction mixture to chilled (− 20°C) 3:1 v/v diethylether:pentane and centrifuge for 10 min at 3500 rpm.
-
7.
Collect the precipitate and remove traces of diethylether:pentane using a N2 flow for 5 min.
-
8.
Dissolve the crude peptide in 3 mL warm DMSO and add this solution to 27 mL warm MilliQ, by pipetting the DMSO stock up and down quickly when adding it to the MilliQ, to avoid precipitation.
-
9.
Filter the sample over a 0.2 μm filter and purify the product on a RP-HPLC system. (We used a Waters XBridge OBD (150 × 30) C18 column with a linear gradient between 20% and 45% B over 25 min (A = 95/5/0.05 v/v/v H2O/acetonitrile/trifluoroacetic acid; B = 5/95/0.05 v/v/v H2O/acetonitrile/trifluoroacetic acid).
-
10.
Check purity of fractions using LC–MS analysis and pool and lyophilize pure fractions (> 95%).
Mutants containing azido-ornithine or alloc-diaminopropionic acid at respective lysine positions were prepared using SPPS as reported elsewhere (Flierman et al., 2016; Mulder et al., 2014) and used to construct diUb probes as described below.
4.2.3.2. Proximal diUb-Prg probe synthesis
-
1.
Dissolve Ub1–75Prg and K48 Azido-ornithine Ub1–75-thioester in DMSO (50 mg/mL final concentration) and dilute in 8 M Urea chaotropic buffer to reach a final concentration of 5 mg/mL in an Eppendorf tube.
-
2.
Add 150 μL catalyst solution containing 25 mg/mL Cu(II)SO4 in MilliQ, 120 mg/mL sodium ascorbate in MilliQ and 52 mg/mL TBTA-analog (Chan, Hilgraf, Sharpless, & Fokin, 2004) in acetonitrile (1:1:1, v/v/v) to the Ub mutants solutions followed by a short vortex, repeat this addition two times in total in 15 min intervals.
-
3.
Perform LC–MS analysis by diluting 1 μL of reaction mixture in 48 μL of MilliQ and 1 μL of 0.5 M EDTA (to quench the copper source).
-
4.
After the reaction is finished, quench the reaction by the addition of 100 μL of 0.5 M EDTA, pH 7.0 and purify by RP-HPLC.
-
5.
Check the purity of fractions using LC–MS analysis and pool and lyophilize pure fractions (> 95%). Dissolve the lyophilized fractions in 10% final concentration DMSO and add to 150 mM NaCl, 20 mM Tris–HCl pH 7.6, and purify further over a Superdex 75 pg 16/600 column (GE).
-
6.
Collect appropriate fractions and concentrate using Amicon (10 MWCO) spinfilters. Snap freeze in liquid N2 and store at − 80°C until used in labeling experiments.
4.2.3.3. In-between diUb probe synthesis
-
1.
Dissolve Ub1–75SEt and Ub1–76 K48 mutant in 10% final concentration of DMSO and dilute into 6 M Guanidinium chaotropic buffer containing 250 mM MPAA (50 mg/mL final concentration) in an Eppendorf tube and shake the mixture overnight at 37°C.
-
2.
Follow the reaction progress by LC–MS analysis by diluting 1 μL of reaction mixture in 48 μL of MilliQ and 1 μL of 1 M TCEP (to reduce the MPAA disulfide).
-
3.
After the reaction is finished, add TCEP and the dilute the guanidinium concentration to a maximum of 2 M with MilliQ.
-
4.
Purify the obtained mixture by RP-HPLC.
-
5.
Pool and lyophilize pure fractions (> 95%), as judged by LC–MS analysis.
-
6.
Dissolve the K48-linked precursor in 10% final concentration of DMSO and dilute into 50 mM sodium phosphate buffer (pH 8, 0.5–1 mg/mL final concentration).
-
7.
Add 2,5-dibromohexandiamide (100 eq.) and react the mixture while shaking overnight at 37°C.
-
8.
Spin down the reaction to remove insoluble dibromide and perform LC–MS analysis.
-
9.
After the reaction is finished, purify the reaction using RP-HPLC.
-
10.
Pool and lyophilize pure fractions (> 95%), as judged by LC–MS analysis.
-
11.
Dissolve the lyophilized fractions in 10% final concentration DMSO and add to 150 mM NaCl, 20 mM Tris–HCl pH 7.6, and purify further over a Superdex 75 pg 16/600 column (GE).
-
12.
Collect appropriate fractions and concentrated using Amicon (10 MWCO) spinfilters. Snap freeze in liquid N2 and store at − 80°C until used in labeling experiments.
4.3. Batch-purification of His-tagged MERS-CoV PLpro
4.3.1. Equipment
-
•
Eppendorf centrifuge
-
•
Table centrifuge for 50 mL Falcon tubes
-
•
Falcon tubes (50 mL), Eppendorf tubes (1.5 mL), screw-cap tubes (2 mL)
-
•
Erlenmeyer flasks (for E. coli growth)
-
•
Sonicator (we used MSE Soniprep 150)
-
•
SDS-PAGE gel electrophoresis equipment (we used Bio-Rad Mini-PROTEANR gel kit)
-
•
Roller bench
-
•
End-over-end rotator
4.3.2. Buffers and reagents
-
•
E. coli (strain C2523/pCG1; expressing ubiquitin-specific protease Ubp1) transformed with pASK3 plasmid encoding His-tagged MERS-CoV PLpro (Bailey-Elkin et al., 2014)
-
•
Standard LB E. coli growth medium, ampicillin, chloramphenicol
-
•
Talon beads (GE Healthcare)
-
•
Lysis buffer: 20 mM HEPES, pH 7.0, 200 mM NaCl, 10% glycerol (vol/vol), 0.1 mg/mL lysozyme
-
•
Anhydrotetracycline
-
•
Imidazole
-
•
Dialysis buffer: 20 mM HEPES, pH 7.0, 100 mM NaCl, 50% glycerol (vol/vol), 2 mM DTT
4.3.3. Procedure
4.3.3.1. Preparation of Talon beads
-
1.
Take 400 μL resuspended Talon beads (stored in ethanol), add 10 mL of water for washing, centrifuge at 1000 × g for 2 min, wash beads two times in lysis buffer (centrifugation at 1000 × g for 2 min).
4.3.3.2. E. coli culture
-
1.
Inoculate 50 mL LB + ampicillin (pASK resistance) + chloramphenicol (pCG1 resistance) with transformed E. coli colony from fresh plate or sample from glycerol stock.
-
2.
Grow bacteria at 37°C, 210 rpm until OD600 ~ 0.7.
-
3.
Cool the culture to RT.
-
4.
Induce protein expression with anhydrotetracycline (200 μg/mL final).
-
5.
Incubate for protein expression at 20°C, 190 rpm, overnight.
-
6.
Harvest cells by centrifugation at 3000 × g, 20 min, 4°C.
4.3.3.3. Lysis
-
1.
Freeze–thaw cell pellet at 20°C once to ease lysis.
-
2.
Resuspend pellet in 5 mL lysis buffer (1 mL per 10 mL culture) + 0.1 mg/mL lysozyme.
-
3.
Perform enzymatic lysis in 50 mL Falcon tube for 1 h at 4°C.
-
4.
Perform subsequent mechanical lysis by sonication (10 times 10 s with cooling on ice in between).
-
5.
Clarify suspension by centrifugation at 20,000 × g, 20 min, 4°C.
-
6.
Take a sample of the supernatant and the pellet for SDS-PAGE analysis.
4.3.3.4. Purification
-
1.
Incubate the clarified supernatant with Talon beads for 1–2 h at 4°C on the roller bench.
-
2.
Centrifuge at 1000 × g, 2 min, 4°C.
-
3.
Discard supernatant (take a sample of this supernatant for SDS-PAGE analysis).
-
4.
Add 10 mL lysis buffer + 20 mM imidazole to beads.
-
5.
Incubate 15 min at 4°C on the roller bench.
-
6.
Repeat this washing step 3 more times (with the same buffer).
-
7.
Transfer beads to 2 mL tube after last wash step.
-
8.
Elute protein from beads with 300 μL lysis buffer + 250 mM imidazole for 15 min at 4°C with end-over-end rotation.
-
9.
Collect supernatant (take sample for SDS-PAGE gel analysis).
-
10.
Repeat elution with another 300 μL buffer (take sample for SDS-PAGE gel analysis).
-
11.
Pool both supernatants and dialyze overnight at 4°C against dialysis buffer.
4.4. Procedure for labeling of enzymes with Ub-Prg and Ubl-Prg probes
4.4.1. Equipment
-
•
Micropipettes (2, 20, 200 μL) with appropriate tips
-
•
Vortex mixer
-
•
0.5 mL and 1.5 mL Eppendorf tubes
-
•
15 mL Falcon tubes
-
•
Table top mini centrifuge (6 × 1.5 mL tubes)
-
•
ThermoMixer with thermoblock (we used: Eppendorf Thermomixer C equipped with a Smartblock 24 × 1.5 mL) or incubator at 37°C
-
•
Heating block
-
•
SDS-PAGE gel tank (we used: Invitrogen Mini Gel Tank, catalog number A25977, coupled to a Biorad PowerPac™ Basic Power Supply)
-
•
Precast gels (we used: Invitrogen NuPAGE™ 4%–12% Bis-Tris Gel, 1.0 mm, 10 and 12 wells)
-
•
Plastic container to store the gel
-
•
Reciprocating shaker (we used GFL reciprocating shaker 3018)
-
•
Gel scanner (we used: GE Amersham Imager 600 RGB)
4.4.2. Buffers and reagents
-
•
Reaction buffer: 50 mM Tris–HCl, pH 7.6, 100 mM NaCl, 5 mM dithiothreitol (DTT), 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)
-
•
Sample buffer: 900 μL Invitrogen NuPage® LDS Sample Buffer (4 ×) (cat. number NP0007), 210 μL Milli-Q® deionized water, 90 μL β-mercaptoethanol
-
•
Protein ladder (we used: ThermoFisher PageRuler™ Plus Prestained Protein Ladder, 10 to 250 kDa, cat. number 26619)
-
•
SDS-PAGE running buffer: Novex NuPage® MOPS SDS Running Buffer (cat. number NP0001)
-
•
Expedeon InstantBlue™ protein staining solution or other Coomassie-based protein staining solution
-
•
Deionized water
-
•
monoUb(l) ABPs (see Table 1): Ub-Prg (Ekkebus et al., 2013) and ISG15c-PRG (Basters et al., 2017) were prepared by SPPS as reported. Nedd8-Prg, SUMO1-Prg, SUMO2-Prg, SUMO3-Prg and ISG15-Prg (Basters et al., 2017) were prepared with intein chemistry using a method similar to a reported procedure (Hemelaar et al., 2004)
-
•
diUb ABPs. K48-linked diUb “in-between”-probe was prepared as reported by (Mulder et al., 2014); K48 linked diUb “proximal”-probe was prepared as reported by Flierman et al. (2016)
-
•
Enzymes: human UCH-L3, expressed and purified according to Larsen, Price, and Wilkinson (1996), SENP1, expressed and purified according to Mikolajczyk et al. (2007) and USP18, expressed and purified according to Basters et al. (2017), viral MERS-CoV PLpro (see protocol under Section 4.3) and SARS-CoV PLpro, expressed and purified according to Bekes et al. (2015)
4.4.3. Procedure for labeling of enzymes with Ub-Prg and Ubl-Prg probes
-
1.
Dilute all enzymes to a concentration of 2 μM in reaction buffer and incubate them for 10 min on ice.
-
2.
Dilute all ABPs to a concentration of 10 μM in reaction buffer.
-
3.
Transfer 8 μL of the enzyme solutions to an empty 1.5 mL tube.
-
4.
Add 8 μL of the ABP solutions to the appropriate enzyme solutions and pipette the solutions up and down a few times for proper mixing.
-
5.
Spin down the samples for a few seconds.
-
6.
Incubate the samples at 37°C for 45 min.
-
7.
Add 8 μL sample buffer to each sample.
-
8.
Vortex and spin down the samples a few seconds.
-
9.
Put the samples in a preheated heating block at 95°C for 10 min.
-
10.
Prepare the gel tank with the gel and SDS-PAGE running buffer.
-
11.
Load the samples (whole sample) onto the gel.
-
12.
Run the gel for 40 min at 200 V.
-
13.
Remove the wet gel slab from the cassette and put it into the plastic container that is half-filled with water.
-
14.
Decant the water and wash the gel three times with water.
-
15.
Add InstantBlue™ staining solution and stain the gel under gently shaking until clear bands appear.
-
16.
Decant the InstantBlue™ solution and wash the gel three times with water.
-
17.
Add water to the gel and let it shake gently until proper background destaining has been achieved.
-
18.
Transfer the wet gel slab to the gel scanner and capture the gel image using the colorimetric settings.
4.4.4. Procedure for labeling of viral enzymes with diUb probes
-
1.
Dilute all enzymes to a concentration of 2 μM in reaction buffer and incubate them for 10 min on ice.
-
2.
Dilute all ABPs to a concentration of 10 μM in reaction buffer.
-
3.
Transfer 12.5 μL of the enzyme solutions to an empty 0.5 mL tube.
-
4.
Add 12.5 μL of the ABP solutions to the appropriate enzyme solutions and pipette the solutions up and down a few times for proper mixing.
-
5.
Spin down the samples for a few seconds.
-
6.
Incubate the samples at 37°C while gently shaking (~ 500 rpm.) for indicated amount of time.
-
7.
Take 10 μL sample and add it to 5 μL sample buffer and 5 μL distilled water.
-
8.
Vortex and spin down the samples a few seconds.
-
9.
Prepare the gel tank with the gel and SDS-PAGE running buffer.
-
10.
Load the samples (17.5 μL) onto the gel.
-
11.
Run the gel for 45 min at 190 V.
-
12.
Remove the wet gel slab from the cassette and put it into the plastic container that is half-filled with water.
-
13.
Decant the water and wash the gel three times with water.
-
14.
Add InstantBlue™ staining solution and stain the gel under gently shaking until clear bands appear.
-
15.
Decant the InstantBlue™ solution and wash the gel three times with water.
-
16.
Add water to the gel and let it gently shake until proper background destaining has been achieved.
-
17.
Transfer the wet gel slab to the gel scanner and capture the gel image using the colorimetric settings.
4.4.5. Notes
-
1.
Keep all enzyme and ABP solutions on ice until needed.
-
2.
Addition of CHAPS detergent to the reaction buffer is not always required but in some cases (here in the case of USP18) gives more pronounced protein bands.
-
3.
The 10 min incubation of the enzymes with reaction buffer helps to reduce the possibly oxidized active site cysteine of the enzymes by DTT.
-
4.
The type of gel, type of running buffer and running time is determined by the type of proteins used and must be chosen as such to achieve maximum separation between the unbound enzyme and ABP-bound enzyme.
-
5.
To achieve optimal protein staining the InstantBlue™ staining and destaining were performed for 12 h each.
5. Conclusions and outlook
The advent of numerous ABPs has aided greatly in unraveling the complex and highly sophisticated ubiquitination system. Since the first ABP targeting DUBs, the field has brought forth an assortment of tools enabling profound insights into the structural, biochemical, and biological role of these enzymes. Although these advancements have helped gain insights into the functions of DUBs, it is becoming increasingly clear that these ABPs require innovation to address outstanding questions. The generation of tools specifically designed for dissecting the proteolytic processing of ubiquitin chains have revealed profound differences among these proteases in their specificity. Adding to this complexity, the discovery of heterotypic and hybrid Ub chains warrants the development of customized tools in order to understand the regulatory roles of DUBs in this context. Other outstanding questions include the development of ABPs capable of capturing metalloprotease DUBs, ABPs targeting a single DUB-type specifically and optimization of cell delivery methodologies for ABPs to enable in-cell enzymology.
The introduction of ABPs into living cells permit visualization and in-cell enzymology in a spatial, temporal, and substrate context, allowing study of the intrinsic regulation by cellular signaling events such as phosphorylation of DUBs to enhance their proteolytic activity as highlighted by the necessity of serine phosphorylation of OTUD5/DUBA (Huang et al., 2012). Most ABP profiling experiments are currently performed using either recombinant enzymes or cell lysates, although several methods allowing their biochemical study in a functional cellular environment are emerging, such as electroporation (Mulder et al., 2016) or the use of cell-penetrating peptides (Gui et al., 2018; Shahul Hameed, Sapmaz, Gjonaj, Merkx, & Ovaa, 2018).
Given the intrinsic role of Ub in the pathogenesis of a variety of diseases, enzymes involved in this system are emerging drug targets. Without a doubt the next generation of Ub-based tools will help increase our knowledge, ultimately leading to new diagnostic tools or therapeutics making it to the clinic.
References
- Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Molecular Cell. 2016;63(1):146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chemistry & Biology. 2011;18(11):1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., Van Kasteren P.B. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. Journal of Biological Chemistry. 2014;289(50):34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. Journal of Virology. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basters A., Geurink P.P., Rocker A., Witting K.F., Tadayon R., Hess S. Structural basis of the specificity of USP18 toward ISG15. Nature Structural & Molecular Biology. 2017;24(3):270–278. doi: 10.1038/nsmb.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes M., Rut W., Kasperkiewicz P., Mulder M.P.C., Ovaa H., Drag M. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochemical Journal. 2015;468(2):215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-linked Di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Molecular Cell. 2016;62(4):572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A., Kessler B.M., Casagrande R., Overkleeft H.S., Wilkinson K.D., Ploegh H.L. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO Journal. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chemistry & Biology. 2002;9(10):1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Catic A., Fiebiger E., Korbel G.A., Blom D., Galardy P.J., Ploegh H.L. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2(7) doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.R., Hilgraf R., Sharpless K.B., Fokin V.V. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Organic Letters. 2004;6(17):2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Wright A.T., Kozarich J.W. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annual Review of Biochemistry. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Daczkowski C.M., Goodwin O.Y., Dzimianski J.V., Farhat J.J., Pegan S.D. Structurally guided removal of DeISGylase biochemical activity from papain-like protease originating from Middle East respiratory syndrome coronavirus. Journal of Virology. 2017;91(23) doi: 10.1128/JVI.01067-17. e01067-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L.C., Melandri F.D., Stein R.L. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37(7):1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- de Jong A., Merkx R., Berlin I., Rodenko B., Wijdeven R.H., El Atmioui D. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. Chembiochem. 2012;13(15):2251–2258. doi: 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkebus R., van Kasteren S.I., Kulathu Y., Scholten A., Berlin I., Geurink P.P. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. Journal of the American Chemical Society. 2013;135(8):2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oualid F., Merkx R., Ekkebus R., Hameed D.S., Smit J.J., de Jong A. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angewandte Chemie, International Edition. 2010;49(52):10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen A.C., Luna-Vargas M.P.A., Geurink P.P., Clerici M., Merkx R., van Dijk W.J. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chemistry & Biology. 2011;18(12):1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Fan J.B., Arimoto K., Motamedchaboki K., Yan M., Wolf D.A., Zhang D.E. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Science Reports. 2015;5 doi: 10.1038/srep12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierman D., van der Heden van Noort G.J., Ekkebus R., Geurink P.P., Mevissen T.E., Hospenthal M.K. Non-hydrolyzable diubiquitin probes reveal linkage-specific reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell Chemical Biology. 2016;23(4):472–482. doi: 10.1016/j.chembiol.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho A., Melchior F. Sumoylation: A regulatory protein modification in health and disease. In: Kornberg R.D., editor. Annual Review of Biochemistry. Vol. 82. 2013. pp. 357–385. Annual Reviews, CA, USA. [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host & Microbe. 2007;2(6):404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan-Erdene T., Nagamalleswari K., Yin L., Wu K., Pan Z.Q., Wilkinson K.D. Identification and characterization of DEN1, a deneddylase of the ULP family. Journal of Biological Chemistry. 2003;278(31):28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- Geurink P.P., El Oualid F., Jonker A., Hameed D.S., Ovaa H. A general chemical ligation approach towards isopeptide-linked ubiquitin and ubiquitin-like assay reagents. Chembiochem. 2012;13(2):293–297. doi: 10.1002/cbic.201100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Millas S., Maul G.G., Yeh E.T. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. Journal of Biological Chemistry. 2000;275(5):3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Gui W., Ott C.A., Yang K., Chung J.S., Shen S., Zhuang Z. Cell-permeable activity-based ubiquitin probes enable intracellular profiling of human deubiquitinases. Journal of the American Chemical Society. 2018;140(39):12424–12433. doi: 10.1021/jacs.8b05147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj-Yahya N., Hemantha H.P., Meledin R., Bondalapati S., Seenaiah M., Brik A. Dehydroalanine-based diubiquitin activity probes. Organic Letters. 2014;16(2):540–543. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- Harrigan J.A., Jacq X., Martin N.M., Jackson S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nature Reviews Drug Discovery. 2018;17(1):57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J., Borodovsky A., Kessler B.M., Reverter D., Cook J., Kolli N. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Molecular and Cellular Biology. 2004;24(1):84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I.A., D'Souza R.C.J., Yang B., Verlaan-de Vries M., Mann M., Vertegaal A.C.O. Uncovering global SUMOylation signaling networks in a site-specific manner. Nature Structural & Molecular Biology. 2014;21(10):927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I.A., Vertegaal A.C.O. A comprehensive compilation of SUMO proteomics. Nature Reviews. Molecular Cell Biology. 2016;17(9):581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- Hewings D.S., Flygare J.A., Bogyo M., Wertz I.E. Activity-based probes for the ubiquitin conjugation-deconjugation machinery: New chemistries, new tools, and new insights. FEBS Journal. 2017;284(10):1555–1576. doi: 10.1111/febs.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey C.M., Wilson N.R., Hochstrasser M. Function and regulation of SUMO proteases. Nature Reviews. Molecular Cell Biology. 2012;13(12):755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochemical Journal. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang O.W., Ma X., Yin J., Flinders J., Maurer T., Kayagaki N. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nature Structural & Molecular Biology. 2012;19(2):171–175. doi: 10.1038/nsmb.2206. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annual Review of Cell and Developmental Biology. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbe S. Breaking the chains: Structure and function of the deubiquitinases. Nature Reviews. Molecular Cell Biology. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annual Review of Biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kumar K.S.A., Haj-Yahya M., Olschewski D., Lashuel H.A., Brik A. Highly efficient and chemoselective peptide ubiquitylation. Angewandte Chemie, International Edition. 2009;48(43):8090–8094. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]
- Kwasna D., Abdul Rehman S.A., Natarajan J., Matthews S., Madden R., De Cesare V. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Molecular Cell. 2018;70(1):150–164. doi: 10.1016/j.molcel.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y.A., Xu W., DeMartino G.N., Cohen R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385(6618):737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Larsen C.N., Price J.S., Wilkinson K.D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry. 1996;35(21):6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- Leidecker O., Matic I., Mahata B., Pion E., Xirodimas D.P. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle. 2012;11(6):1142–1150. doi: 10.4161/cc.11.6.19559. [DOI] [PubMed] [Google Scholar]
- Li J., Chai Q.Y., Liu C.H. The ubiquitin system: A critical regulator of innate immunity and pathogen-host interactions. Cellular & Molecular Immunology. 2016;13(5):560–576. doi: 10.1038/cmi.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liang Q., Gong P., Tencer A.H., Zhuang Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chemical Communications. 2014;50(2):216–218. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Machner M.P. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. Journal of Cell Science. 2017;130(12):1985–1996. doi: 10.1242/jcs.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. Journal of Virology. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhov M.P., Malakhova O.A., Kim K.I., Ritchie K.J., Zhang D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. Journal of Biological Chemistry. 2002;277(12):9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- McGouran J.F., Gaertner S.R., Altun M., Kramer H.B., Kessler B.M. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chemistry & Biology. 2013;20(12):1447–1455. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E.T., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154(1):169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E.T., Kulathu Y., Mulder M.P.C., Geurink P.P., Maslen S.L., Gersch M. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature. 2016;538(7625):402–405. doi: 10.1038/nature19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk J., Drag M., Bekes M., Cao J.T., Ronai Z., Salvesen G.S. Small ubiquitin-related modifier (SUMO)-specific proteases: Profiling the specificities and activities of human SENPs. Journal of Biological Chemistry. 2007;282(36):26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- Mulder M.P.C., El Oualid F., ter Beek J., Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: Diubiquitin as a case study. Chembiochem. 2014;15(7):946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M.P.C., Merkx R., Witting K.F., Hameed D.S., El Atmioui D., Lelieveld L. Total chemical synthesis of SUMO and SUMO-based probes for profiling the activity of SUMO-specific proteases. Angewandte Chemie (International ed. in English) 2018;57(29):8958–8962. doi: 10.1002/anie.201803483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M.P.C., Witting K., Berlin I., Pruneda J.N., Wu K.P., Chang J.G. A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes. Nature Chemical Biology. 2016;12(7):523–530. doi: 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M., Arner E., Prudden J., Schaffer L., Head S., Boddy M.N. Functional crosstalk between the PP2A and SUMO pathways revealed by analysis of STUbL suppressor, razor 1-1. PLoS Genetics. 2016;12(7) doi: 10.1371/journal.pgen.1006165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M., Boddy M.N. Cooperativity of the SUMO and ubiquitin pathways in genome stability. Biomolecules. 2016;6(1):14. doi: 10.3390/biom6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M.B., Luna-Vargas M.P.A., Velds A., Brummelkamp T.R., Dirac A.M.G., Sixma T.K. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ovaa H. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nature Reviews. Cancer. 2007;7:613. doi: 10.1038/nrc2128. [DOI] [PubMed] [Google Scholar]
- Pasunooti K.K., Yang R.L., Vedachalam S., Gorityala B.K., Liu C.F., Liu X.W. Synthesis of 4-mercapto-l-lysine derivatives: Potential building blocks for sequential native chemical ligation. Bioorganic & Medicinal Chemistry Letters. 2009;19(22):6268–6271. doi: 10.1016/j.bmcl.2009.09.107. [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Rose I.A. Mechanism of ubiquitin carboxyl-terminal hydrolase. Borohydride and hydroxylamine inactivate in the presence of ubiquitin. Journal of Biological Chemistry. 1986;261(22):10210–10217. [PubMed] [Google Scholar]
- Pruneda J.N., Durkin C.H., Geurink P.P., Ovaa H., Santhanam B., Holden D.W. The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Molecular Cell. 2016;63(2):261–276. doi: 10.1016/j.molcel.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual Review of Biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahul Hameed D.A., Sapmaz A., Gjonaj L., Merkx R., Ovaa H. Enhanced delivery of synthetic ubiquitin into live cells using NextGen Ub-TAT conjugates. Chembiochem. 2018;19:2553–2557. doi: 10.1002/cbic.201800649. [DOI] [PubMed] [Google Scholar]
- Singh R.K., Sundar A., Fushman D. Nonenzymatic rubylation and ubiquitination of proteins for structural and functional studies. Angewandte Chemie (International ed. in English) 2014;53(24):6120–6125. doi: 10.1002/anie.201402642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy T.A., Smith P.G., Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clinical Cancer Research. 2009;15(12):3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- Swatek K.N., Komander D. Ubiquitin modifications. Cell Research. 2016;26(4):399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilburg G.B., Elhebieshy A.F., Ovaa H. Synthetic and semi-synthetic strategies to study ubiquitin signaling. Current Opinion in Structural Biology. 2016;38:92–101. doi: 10.1016/j.sbi.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoes M., Verhelst S.H.L. Detection of protease activity in cells and animals. Biochimica et Biophysica Acta. Proteins and Proteomics. 2016;1864(1):130–142. doi: 10.1016/j.bbapap.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Witting K., van der Heden van Noort G., Kofoed C., Talavera Ormeno C., El Atmioui D., Mulder M.P.C. Generation of the UFM1 toolkit for profiling UFM1-specific proteases and ligases. Angewandte Chemie (International ed. in English) 2018;57:14164–14168. doi: 10.1002/anie.201809232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R., Rape M. The increasing complexity of the ubiquitin code. Nature Cell Biology. 2016;18(6):579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- Zhang D.X., Zhang D.E. Interferon-stimulated gene 15 and the protein ISGylation system. Journal of Interferon and Cytokine Research. 2011;31(1):119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Fahrni C.J. A fluorogenic probe for the copper(I)-catalyzed azide-alkyne ligation reaction: Modulation of the fluorescence emission via 3(n,π*)−1(π,π*) inversion. Journal of the American Chemical Society. 2004;126(29):8862–8863. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
Further reading
- Balakirev M.Y., Tcherniuk S.O., Jaquinod M., Chroboczek J. Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Reports. 2003;4(5):517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe G.J., Zhang Y., Przydacz M., Rolhion N., Yang Y., Pruneda J.N. The salmonella effector SpvD Is a cysteine hydrolase with a serovar-specific polymorphism influencing catalytic activity, suppression of immune responses, and bacterial virulence. Journal of Biological Chemistry. 2016;291(50):25853–25863. doi: 10.1074/jbc.M116.752782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E.T., Komander D. Mechanisms of deubiquitinase specificity and regulation. In: Kornberg R.D., editor. Annual Review of Biochemistry. Vol. 86. 2017. pp. 159–192. [DOI] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & Biology. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Scholte F.E.M., Zivcec M., Dzimianski J.V., Deaton M.K., Spengler J.R., Welch S.R. Crimean-Congo hemorrhagic fever virus suppresses innate immune responses via a ubiquitin and ISG15 specific protease. Cell Reports. 2017;20(10):2396–2407. doi: 10.1016/j.celrep.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]