Abstract
Coronavirus particles serve three fundamentally important functions in infection. The virion provides the means to deliver the viral genome across the plasma membrane of a host cell. The virion is also a means of escape for newly synthesized genomes. Lastly, the virion is a durable vessel that protects the genome on its journey between cells. This review summarizes the available X-ray crystallography, NMR, and cryoelectron microscopy structural data for coronavirus structural proteins, and looks at the role of each of the major structural proteins in virus entry and assembly. The potential wider conservation of the nucleoprotein fold identified in the Arteriviridae and Coronaviridae families and a speculative model for the evolution of corona-like virus architecture are discussed.
Keywords: Enveloped virus assembly, Virion structure, Structural proteins, Coronavirus evolution
1. Introduction
A virus particle is essentially a ruggedized viral genome that contains at least the minimal set of components necessary to propagate a virus infection. While virions from most viruses are considered to be metabolically inactive, some undergo internal structural changes after release, including protease-dependent retrovirus maturation (Konvalinka et al., 2015) and bicaudovirus elongation (Haring et al., 2005, Scheele et al., 2011). In addition to the viral genome and four conserved virally encoded structural proteins, coronavirus particles have been shown to contain a variety of packaged host-encoded proteins (Dent et al., 2015, Kong et al., 2010, Neuman et al., 2008, Nogales et al., 2012) including enzymes that may play important roles in promoting or preventing infection such as protein kinases (Neuman et al., 2008, Siddell et al., 1981), cyclophilin A (Neuman et al., 2008, Pfefferle et al., 2011), and APOBEC3G (Wang and Wang, 2009). Purified virions can also contain low levels of some virus-encoded replicase proteins (Neuman et al., 2008, Nogales et al., 2012), though packaged replicase proteins have not been shown to enhance infectivity. While the positive sense ssRNA genome is infectious for members of the genera Alphacoronavirus (Almazan et al., 2000, Donaldson et al., 2008b, Jengarn et al., 2015, Tekes et al., 2008, Thiel et al., 2001, Yount et al., 2000), Betacoronavirus (Donaldson et al., 2008a, Scobey et al., 2013, Yount et al., 2003), and Gammacoronavirus (Casais et al., 2001), it has been shown that expression of the viral nucleoprotein and nsp3 can together or separately promote infection (Hurst et al., 2010, Hurst et al., 2013, Pan et al., 2008, Schelle et al., 2005, Thiel et al., 2001). Taken together, this data demonstrate that packaged virion proteins are not essential for infection, but suggest that packaged proteins may confer a small replication advantage.
Coronaviruses encode three conserved membrane-associated proteins that are incorporated in virions: spike (S), envelope (E), membrane (M), and nucleoprotein (N; Fig. 1 ). These four proteins occur in the order S–E–M–N in every known coronavirus lineage (Woo et al., 2014). In between the S–E–M–N genes, coronaviruses encode species-specific accessory proteins, many of which appear to be incorporated in virions at low levels, ranging from one accessory in alphacoronaviruses including human coronavirus NL63 (Pyrc et al., 2004) to a predicted nine accessories in the gammacoronavirus HKU22 (Woo et al., 2014). The genomic position of these accessory genes varies, with accessories encoded before S in some betacoronaviruses, between S and E in most lineages, between M and N in most lineages, and after N rarely in alphacoronaviruses and gammacoronaviruses and commonly in deltacoronaviruses. Interestingly, the M gene appears to directly follow the E gene throughout the Coronaviridae, though there is not an obvious transcriptional or translational reason why this should necessarily be the case. Interestingly, in one study, deletion of E resulted in the evolution of spontaneously joined gene fragments of M encoded in the position normally occupied by E (Kuo and Masters, 2010).
Fig. 1.
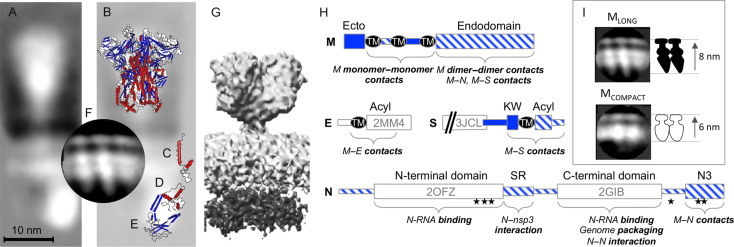
Structure and organization of proteins in the virion. Cryo-EM reconstruction of the SARS-CoV structural proteins from virions (A; Neuman et al., 2006) superimposed with solved structures of the MHV S ectodomain (B; 3JCL), SARS-CoV E (C; 2MM4), SARS-CoV C-terminal domain (D; 2GIB), SARS-CoV N-terminal domain (E; 2OFZ), and a cryo-EM reconstruction of the M proteins from MHV VLPs (F; Neuman et al., 2011). An alternative shorter, wider cryo-EM reconstruction of SARS-CoV virion proteins is shown for comparison (G; EMD-1423). Schematics are based on MHV proteins, following the annotation of Kuo et al. (2016). The orientation of N protein domains shown here is hypothetical, and is intended for illustrative purposes only. (H) Domain structure and annotation of MHV S, E, M, and N proteins showing domains outside the virion (solid blue), inside the virion (striped blue), and the position of solved structures. Transmembrane regions (TM), sites of palmitoylation (Acyl), a conserved sequence preceding the transmembrane of S (KW), a serine–arginine-rich unstructured region (SR), phosphorylation sites (stars), and the C-terminal M-interacting domain of N (N3) are marked. Comparison of the appearance of dimeric MLONG and MCOMPACT (I).
The coronavirus particle provides three important things for the genome: a durable transport vessel, a means of escape for newly synthesized genomes, and a means of entry into a cell. The following sections will examine each of the key functions of the coronavirus particle in durability, budding, and entry.
2. Virion Structure and Durability
The genome likely comprises a relatively small part of the internal volume of a virion. MHV produces virions that are approximately 80 nm in diameter, which is typical for hydrated coronavirus particles in vitreous ice (Barcena et al., 2009, Neuman et al., 2006, Neuman et al., 2011). The average radius of MHV virions is about 42 nm (Neuman et al., 2011); subtracting 8 nm occupied by the viral membrane and M protein at each side (Neuman et al., 2011), and assuming a spherical virion, gives a predicted interior volume of 1.6 × 105 nm3 for a coronavirus particle. The partial specific volume of a coronavirus genome, calculated using a density of 0.57 cm3/g for ssRNA (Voss and Gerstein, 2005) and the molecular mass of the smallest coronavirus genome (porcine deltacoronavirus HKU15, 25.4 kb) and the largest (bottlenose dolphin coronavirus HKU22, 31.8 kb), gives a volume of 0.8–1.0 × 104 nm3 per genome, equivalent to 5–6% of the virion. From the estimated 0.7–2.2 × 103 N proteins per virion (Neuman et al., 2011) and the volume of about 60 nm3 per N protein (Neuman et al., 2006), we can calculate that N proteins should occupy 0.4–1.3 × 105 nm3 or 25–80% of the virion interior, and that each N is associated with 14–40 nt of genomic RNA (Neuman et al., 2011).
In their native state, virions are filled with water, and preimaging procedures that remove the water from the virion have led to the mistaken but strangely persistent ideas that both arteriviruses (Horzinek et al., 1971) and coronaviruses (Risco et al., 1996) contain an icosahedrally organized ribonucleoprotein core. The shape of hydrated coronavirus particles in water ice as observed by cryoelectron microscopy is roughly spherical, and shows no sign of icosahedral organization (Barcena et al., 2009, Beniac et al., 2006, Beniac et al., 2007, Neuman et al., 2006, Neuman et al., 2011). The same is true of Arteriviridae (Spilman et al., 2009), and probably also of Mesoniviridae (Nga et al., 2011). Other members of the Nidovirales produce bacillus-shaped particles, including Bafinivirus (Schutze et al., 2006) and Okavirus (Spann et al., 1997). In cell culture toroviruses produce a mixture of spherical and bacilliform particles (B. Neuman, unpublished data). Careful measurement of coronavirus particles in cryoelectron micrographs and tomograms reveals that most coronavirus particles are slightly prolate spheroids that differ from the shape of more spherical exosomal vesicles that appear in the same images (Neuman et al., 2011). While the rigidity of coronavirus particles remains to be investigated, the shape difference, along with the observation that coronavirus particles remain roughly spherical despite repeated prodding with a carbon nanotube during atomic force microscopy (Liu et al., 2012, Ng et al., 2004), suggests that coronavirus particles are relatively resistant to deformation, as recently reported for influenza A virus (Li et al., 2011).
Just because a virion is enveloped, does not mean it is necessarily fragile or quickly inactivated, as demonstrated by the enveloped pithovirus that was successfully recovered from 30,000-year-old permafrost (Legendre et al., 2014). Coronavirus particles are relatively robust compared to HIV-1, with SARS-CoV virions remaining infectious for 1–4 days on the relatively harsh environment of hard surfaces (reviewed in Sobsey and Meschke, 2003). MERS-CoV virions are somewhat less robust than SARS-CoV, with half lives on the order of an hour on hard surfaces and a maximum survival time of 2–3 days, but are considerably more durable than pandemic influenza A virus under the same conditions (van Doremalen et al., 2013). The persistent infectivity of coronaviruses outside the body has been used as evidence to suggest that direct contact with contaminated surfaces as well as respiratory droplets is a potential route of MERS-CoV spread (Assiri et al., 2013, Goh et al., 2013).
3. Viral Proteins in Assembly and Fusion
3.1. Membrane Protein
The M protein facilitates viral assembly by interacting with other M (Arndt et al., 2010, de Haan et al., 2000), E (Boscarino et al., 2008, Corse and Machamer, 2003, Lim and Liu, 2001), S (de Haan et al., 1999, Godeke et al., 2000), and N proteins (Escors et al., 2001, Hurst et al., 2005, Narayanan et al., 2000, Sturman et al., 1980). The MHV M protein may also interact with the RNA packaging signal that mediates incorporation of the viral genome into viral particles (Narayanan et al., 2003), but direct M-genome interactions are not efficient enough to rescue viruses in which the C-terminal region of N that interacts with M has been perturbed, suggesting that M-genome interactions are less important than M–N interactions for recovery of infectious virus (Kuo et al., 2016). Communication between the carboxyl termini of the M and N proteins has also been observed from mutagenesis and second-site reversion studies (Hurst et al., 2005, Kuo and Masters, 2002).
M proteins in SARS-CoV, FCoV, and MHV virions and virus-like particles (VLPs) form homodimers (Neuman et al., 2011), which appear to be functionally analogous to the M-GP5 heterodimers of Arteriviridae (de Vries et al., 1995, Faaberg et al., 1995, Snijder et al., 2003). Coronavirus M dimers resemble the shape of a Greek amphora, with the ectodomain forming the lip, transmembrane region forming narrow neck, and the endodomain forming the lower chamber (Fig. 1I). In virions it appears that the transmembrane region does not make contact between adjacent M dimers, suggesting that reported M–M interaction domains in the transmembrane region (de Haan et al., 2000) are between the two monomers that make up an M dimer. M protein deletion mutants were not assembly competent in the absence of wild-type M (de Haan et al., 2000). Thus M–M interactions are necessary, but not always sufficient for VLP assembly (de Haan et al., 2000).
In contrast, the endodomains of M dimers appeared to make close contact in the cryo-EM reconstructions (Neuman et al., 2011), suggesting that interactions between M dimers are likely to occur in the endodomain. The endodomain of MHV M is important for M–M interactions that could involve monomers or dimers. Purified SARS-CoV M endodomains can dimerize and can form M dimer–dimer interactions (Neuman et al., 2011), suggesting that the endodomain is the primary site of M dimer–dimer interaction.
Cryo-EM and cryoelectron tomography analysis also suggests that M can exist in two forms (Neuman et al., 2011). The main form found on virions and VLPs is an elongated conformation that makes contact with the ribonucleoprotein and imparts a spherical membrane curvature of about 5–6 degrees per M dimer. The minor form is more compact, has indistinct boundaries suggestive of a disordered aggregate, and does not appear to impart membrane curvature. The long conformation could be partially converted to the shorter conformation by transient acidification, weakening M–RNP interactions. This suggests a model in which formation of MLONG–RNP interactions drives the budding process, and membrane fusion is preceded by release of MLONG–RNP interactions. However, further work is needed to test the accuracy of this model.
The structure of the M protein is not known, but may be partially inferred from sequence comparison and secondary structure prediction algorithms. M proteins possess a short glycosylated ectodomain of variable sequence (Oostra et al., 2006), followed by three closely spaced hydrophobic transmembrane helix signatures, and a relatively long cytoplasmic tail region that may fold into a compact beta-dominated structure (Masters et al., 2006).
The weight of evidence suggests that coronavirus M and E proteins are the critical components required for assembly of coronavirus virions and VLPs (Bos et al., 1996, Corse and Machamer, 2000, Vennema et al., 1996). However the SARS-CoV M protein appears to readily form VLPs in the absence of E (Tseng et al., 2010), as does the M and S protein of IBV (Liu et al., 2013). M protein sequences from different coronaviruses are highly conserved, approaching the level of conservation of some of the viral enzymes and replicase accessory proteins from pp1a (Stadler et al., 2003). During the assembly process, N protein contributes to the formation of MLONG, narrowing the size range of resulting VLPs (Fig. 2 ; Neuman et al., 2011) and increasing the efficiency of VLP production (Boscarino et al., 2008, Siu et al., 2008). A study of VLP size and organization showed that MHV VLPs and virions with different components formed particles with a constrained minimum size, and that the size range of particles became narrower and approached the minimum size of VLPs as more structural components were incorporated (Fig. 2; Neuman et al., 2011). This suggests that incorporation of S and the genome, which are both essential for virion infectivity but dispensable for VLP production, affected the efficiency of budding in the sense that larger particles incorporate more M proteins.
Fig. 2.
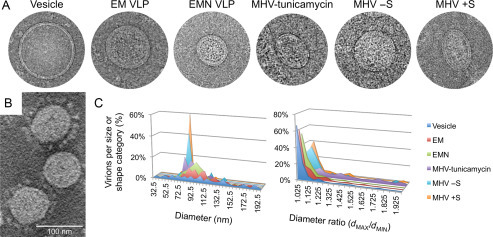
Appearance and characteristics of MHV virions, MHV VLPs, and copurified exosomal vesicles. Cryo-EM images show a vesicle, VLPs produced after coexpression of E and M (EM VLP) or E, M and N (EMN VLP), a virion from tunicamycin-treated cells, and virions from the same preparation that spontaneously formed lacking (− S) or displaying (+ S) visible spikes. A cryoelectron tomography image of MHV is shown to highlight the variation in spike incorporation in virions from the same preparation (B). Sizes (diameter) and shapes (dMAX/dMIN) of virions, VLPs, and vesicles were categorized to illustrate differences in the shape of particles that incorporate different combinations of structural proteins (C).
3.2. Nucleoprotein
Coronavirus nucleoproteins are phosphoproteins, and are encoded near the 3′ end of the genome. MHV N is phosphorylated at six sites (S162, S170, T177, S389, S424, and T428; White et al., 2007) by host kinases like cyclin-dependent kinase, glycogen synthase kinase, mitogen-activated protein kinase, and casein kinase II (Surjit et al., 2005). The protein is also sumoylated (at Lys62 in the SARS-CoV N protein), a posttranslational modification that enhances the protein's tendency to homooligomerize and affects typical N protein-mediated interference in host cell division (Li et al., 2005b). Several groups have successfully expressed soluble protein (both full-length and partial domains), but its unusually high positive charge, tendency to oligomerize, structural flexibility, and extremely low stability have hampered structural studies of the whole protein (Chang et al., 2006). N possesses two RNA-binding domains: an N-terminal domain with adjacent S/R-rich motif (He et al., 2004) and the C-terminal 209 amino acids (Ma et al., 2010, Surjit et al., 2004, Yu et al., 2005). The N protein also binds with nanomolar affinity to human cyclophilin A, though the physiological significance of this finding is still unknown (Luo et al., 2004, Pfefferle et al., 2011).
N protein supports coronavirus infection in several ways: the C-terminal domain (CTD) of N is important for binding the genomic RNA packaging signal leading to selective genome incorporation (Kuo et al., 2014, Molenkamp and Spaan, 1997), the N3 domain interacts with the endodomain of M to form virions (Kuo et al., 2016), and the serine–arginine repeat region of N (SR) interacts with the first ubiquitin-like domain of nsp3 in a critical early replication step (Hurst et al., 2010, Hurst et al., 2013). It has also been demonstrated that N can oligomerize through interactions in the CTD (Chang et al., 2013), bind viral RNA through the N-terminal domain (Fan et al., 2005), unwind double-stranded nucleic acid in the manner of an RNA chaperone (Neuman et al., 2008, Zuniga et al., 2007), and pack in a helix through the N-terminal domain (Saikatendu et al., 2007), though none of these other functions has yet been demonstrated to be important for infection. N protein is dynamically associated with sites of viral RNA replication, suggesting that N may also function to protect the genome or possibly mediate genome transport to the budding site (Verheije et al., 2010).
The nucleoprotein of coronavirus is not a good match for nucleoproteins of other members of the Nidovirales at the level of amino acid sequence. However, the small N protein of arteriviruses EAV (Deshpande et al., 2007) and PRRSV (Doan and Dokland, 2003) adopts a similar fold to the CTD of coronavirus N (Yu et al., 2006). Alignment of predicted protein secondary structures from N proteins from other nidoviruses with the structures of coronavirus and arenavirus N suggests that a helix–strand–strand–helix motif may form a conserved functional domain near the C-terminus of all nidovirus N proteins (Fig. 3 ).
Fig. 3.
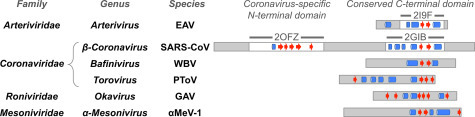
Evidence for conservation of the N protein C-terminal domain fold across the Nidovirales. Proteins are depicted as rectangles with resolved regions of solved protein structures (white regions) and unsolved or unresolved regions (gray regions) indicated. Alpha helix (blue cylinders) and beta strand (red arrows) regions of solved protein secondary structures or JPRED secondary structure predictions are shown manually aligned by secondary structure to facilitate comparison.
3.3. Envelope Protein
E proteins are encoded by all known coronavirus genomes and are found at low levels in the virion (Godet et al., 1992, Liu and Inglis, 1991). As pointed out by Kuo and Masters (Kuo et al., 2016), E appears to have three distinct functions that contribute to infection: regulating aggregation-prone M–M interactions (Boscarino et al., 2008), disrupting Golgi organization in a way that produces larger vesicles capable of transporting virions (Machamer and Youn, 2006, Ruch and Machamer, 2011, Ruch and Machamer, 2012), and interacting with host factors in a way that affects pathogenesis (DeDiego et al., 2007, DeDiego et al., 2011, Dediego et al., 2008, Nieto-Torres et al., 2015, Regla-Nava et al., 2015, Teoh et al., 2010). E proteins of several coronaviruses have been reported to have ion channel activity (Liao et al., 2004, Madan et al., 2005, Wilson et al., 2004), which appears to enhance viral growth (Wilson et al., 2006, Ye and Hogue, 2007). In addition to these three roles, E proteins have been speculated to be involved in scission of the membrane to free newly budded virions based on evidence from MHV VLP formation in the presence of E (Vennema et al., 1996a) and aberrant virion formation in E mutant viruses (Fischer et al., 1998), analogous to the function of M2 in influenza A virus (Rossman et al., 2010). However, it is not clear whether the role of E in virion release is distinct from its role in limiting M aggregation (Boscarino et al., 2008) and SARS-CoV M does not appear to require the presence of E to generate VLPs (Tseng et al., 2010), suggesting that the role of E differs, or differs in importance, in some coronaviruses.
E is probably best viewed as a multifunctional accessory gene that contributes to both virus growth and pathogenesis. Expression of SARS-CoV E protein is dispensable for coronavirus growth, but deletion results in severe defects in virus growth, presumably related to inefficient assembly (DeDiego et al., 2007, Kuo and Masters, 2003). Interestingly, deletion of the SARS-CoV E gene has less effect on replication than the corresponding E gene deletion in MHV, suggesting that the function of E may be duplicated elsewhere in the genome. A candidate for the compensating factor could be the SARS-CoV 3a protein, which similarly displays ion channel activity (Lu et al., 2006) and serves to increase the viral growth rate when present (Yount et al., 2005), or possibly the SARS-CoV ORF6-encoded protein, which is predicted to adopt a similar membrane topology to E and induces intracellular membrane rearrangement (Zhou et al., 2010). In MHV, duplication of the amino-terminal part of M can partially compensate for the deletion of E, suggesting that the functions of MHV M and E proteins may overlap (Kuo and Masters, 2010). The 229E-4a accessory protein also has ion channel activity, but it is not known whether this can compensate for deletion of E (Zhang et al., 2014). In contrast, deletion of E in TGEV prevented the formation of infectious virions (Ortego et al., 2007).
A number of general structural features can be identified in all coronavirus E proteins, including a short hydrophilic N-terminal region, followed by a hydrophobic putative transmembrane region, and a relatively long hydrophilic C-terminal tail (Liu et al., 2007). SARS-CoV E protein forms pentamers (Pervushin et al., 2009) that conduct cations (Pervushin et al., 2009), which is likely a conserved feature of E proteins. There is evidence that IBV E protein has different functions that correlate to different oligomerization states in mutant E proteins: monomeric E is sufficient to disrupt the Golgi, while oligomerization-competent E supports VLP release (Westerbeck and Machamer, 2015). The target of ion channel activity may be disruption of intracellular processes by releasing calcium ions stored in the endoplasmic reticulum, ultimately driving an inflammatory immune response (Nieto-Torres et al., 2015). The E protein is palmitoylated in at least some coronaviruses (Corse and Machamer, 2002, Liao et al., 2006), and palmitylation is important for the role of MHV E protein in limiting M–M aggregation (Boscarino et al., 2008). The transmembrane and palmitoylated domains of E are each sufficient to colocalize with M protein (Nieto-Torres et al., 2011).
3.4. Spike Protein
The first open reading frame downstream of the SARS-CoV replicase encodes the S glycoprotein which is conserved in all coronaviruses. The ~ 180-kDa spike protein plays a central role in the host cell attachment and entry processes. The spike is organized into an amino-terminal S1 domain that contains receptor-binding determinants, and a carboxyl-terminal S2 domain that contains the membrane anchor and fusion motor domains (Belouzard et al., 2012, Heald-Sargent and Gallagher, 2012). Coronavirus S proteins contain short amino-terminal hydrophobic signal sequence motifs (von Heijne, 1984). Although some coronavirus spike proteins are cleaved between the S1 and S2 regions as part of the activation process, the SARS-CoV spike is not appreciably cleaved before it is internalized in a host cell (Xiao et al., 2003). The more variable amino-terminal region of the spike protein (S1) has been demonstrated to contain the receptor-binding activity (Wong et al., 2004). The more conserved S2 region contains the transmembrane anchor, palmitic acid acylation site (Thorp et al., 2006) that is important for membrane fusion (McBride and Machamer, 2010, Shulla and Gallagher, 2009), and the coiled-coil fusion motor domain (Bosch et al., 2003, Duquerroy et al., 2005, Liu et al., 2004, Tripet et al., 2004, Xu et al., 2004a, Xu et al., 2004b). One or more protease cleavage events are necessary to prime S for membrane fusion. An apparent fusion peptide of coronaviruses resides in the S2 region (Bosch et al., 2003, Sainz et al., 2005), where it is exposed by cleavage that takes place on tetraspanin-enriched membranes where host proteases including TMPRSS2 and HAT localize for some coronaviruses including MERS-CoV, while other spikes can be primed for entry by cathepsins (Bertram et al., 2013, Earnest et al., 2015, Glowacka et al., 2011, Heurich et al., 2014, Huang et al., 2006, Shulla et al., 2011, Simmons et al., 2005). This is consistent with host gene knockdown experiments that show that coronavirus entry is dependent on several elements that are important in the endosomal and lysosomal trafficking (Burkard et al., 2014, Wong et al., 2015). Expression of IFITM proteins inhibits entry driven by several coronavirus spike proteins (Huang et al., 2011, Wrensch et al., 2014), but paradoxically appears to promote infection by HCoV-OC43 (Zhao et al., 2014).
Near-atomic resolution cryo-EM structures have been published for the ectodomains of trimeric MHV (Walls et al., 2016) and HKU1 (Kirchdoerfer et al., 2016) up to the second heptad repeat region. The high-resolution spike ectodomain structures are similar to the profile to the upper part of SARS-CoV S in one study (Neuman et al., 2006) and taller and less square than the spikes reconstructed by another group (Beniac et al., 2006, Beniac et al., 2007). High-resolution X-ray crystallography structures have also been obtained for two domains of the coronavirus spike protein. The structure of the minimal receptor-binding domain from SARS-CoV was solved first in conjunction with angiotensin-I converting enzyme 2 (ACE2; Towler et al., 2004), the primary cellular receptor for SARS-CoV (Li et al., 2003) and human coronavirus NL63 (Hofmann et al., 2005). More recently, S1 receptor structures have been solved for betacoronaviruses MHV (Peng et al., 2011) and MERS-CoV (Wang et al., 2013) and alphacoronaviruses NL63 (Wu et al., 2009), TGEV (Reguera et al., 2012), and PRCoV (Reguera et al., 2012). The receptor-binding domain structure of SARS-CoV consists of a core subdomain containing a five-stranded antiparallel β sheet with three short connecting α helices, and an extended loop subdomain that contacts ACE2 (Li et al., 2005a), and the domains that mediate S1 receptor contact in other betacoronaviruses also seem to involve curved β sheets at the center with stabilizing loop interactions at the side. In contrast, the receptor-binding domain of alphacoronaviruses makes contact via a series of loops positioned at the edge of a β sheet. Image analysis of cryoelectron micrographs of SARS-CoV (Beniac et al., 2006, Neuman et al., 2006) and other coronaviruses (Neuman et al., 2006) confirms that spikes exist as a homotrimer in the native prefusion state on virions. However, some biochemical characterizations have revealed that S1 interacts with the receptor protein as a dimer, even within the context of the trimeric spike (Lewicki and Gallagher, 2002, Xiao et al., 2004).
In the model of coronavirus spike protein-mediated fusion, receptor binding triggers conformational changes including disulfide reshuffling (Gallagher, 1996, Lavillette et al., 2006) that release HR1 and HR2 to form the coiled-coil fusion motor structure, thereby driving fusion of the viral and host cell membranes and release of the viral ribonucleoprotein into the cytosol (Bosch et al., 2003). The fusion motor complex of S2 consisting of two hydrophobic amino acid 4-3 heptad repeat regions (HR1 and HR2) which form amphipathic helices of a coiled-coil structure has also been solved in several forms (Bosch et al., 2003, Duquerroy et al., 2005, Liu et al., 2004, Tripet et al., 2004, Xu et al., 2004a, Xu et al., 2004b). In the structure representing the largest region of S2 structure, HR1 forms a trimeric 120 Å coiled coil (Duquerroy et al., 2005). Coordinated chloride ions in the structure are instrumental in the formation of hydrogen bonds that stretch both ends of the HR2 region into an extended conformation surrounding the central alpha helix (Duquerroy et al., 2005). After insertion of the fusion peptide into the target membrane, a single-particle fusion study revealed that it takes about 15 s to go from membrane insertion to hemifusion, 15–60 more seconds until a pore is formed, then another 30 s for complete lipid mixing between the virion and cell (Costello et al., 2013).
Spike protein is incorporated into virions through interactions with the membrane protein M (Godeke et al., 2000). It can be generally accepted that all the determinants for virion incorporation reside in the transmembrane and carboxyl-terminal regions of the spike protein (Godeke et al., 2000, Kuo et al., 2000). However, a recent study found that a chimeric MHV with SARS-CoV M and S transmembrane and endodomain was severely deficient in incorporating S into virions, suggesting that cellular localization signals or more complex interactions among the structural proteins may help support S incorporation (Kuo et al., 2016).
4. Evolution of the Structural Proteins
Working on nidoviruses is a mixed blessing—on one hand, there is sufficient evolutionary divergence and complexity with a few common threads to make it seem possible to reconstruct an evolutionary path from a simple coronavirus-like progenitor to the present array of nidoviruses. However, the extreme divergence between homologous proteins means that common ancestry is sometimes only evident at the level of protein fold and conserved function, meaning that homologs cannot necessarily be recognized by amino acid alignment.
Two structural proteins stand out as being conserved: coronavirus M and N. One or more M-like three-pass transmembrane proteins with endodomains rich in predicted β structure are found in coronaviruses, toroviruses, bafiniviruses, arteriviruses, the arteri-like possum nidovirus (Dunowska et al., 2012), and the newly discovered toro-like ball python nidovirus (Stenglein et al., 2014). Roniviridae also encode a structural polyprotein of GP116 and GP64 that includes a superficially similar three-pass transmembrane protein known as 3N at its amino terminus, though 3N has not yet been detected in infected cells or virions to date. Mesoniviridae lack a three-pass M protein, but two single-pass transmembrane proteins with predicted β-rich endodomains known as M and 3b may serve the same function as coronavirus M in the virion. Every member of the Nidovirales also encodes a positively charged N-like protein. Although the proteins differ in size, common predicted structure elements near the C-terminus suggest that nidovirus N proteins may in fact be homologous, as shown in Fig. 3. In comparison, proposed S-like proteins are highly divergent, and E-like proteins are absent in several nidovirus lineages, suggesting S and E are later refinements to nidovirus virion architecture.
We can therefore imagine two evolutionary paths that gradually built up to the current complexity of nidovirus structural proteins (Fig. 4 ). The first model involves an ancestral enveloped virus that encodes a hypothetical protein similar to coronavirus M (but capable of RNA packaging and attachment to host cells) served as the original structural protein in a progenitor of the last common ancestor of nidoviruses. In this model, first proto-N, then S-like and E-like proteins would be added. M protein equivalents of coronavirus, arterivirus, and torovirus are similar in appearance, and are consistent with the size of two protein chains (B. Neuman, unpublished data) suggesting that networks formed of dimers may be an ancestral trait that originated in the hypothetical proto-M. M has many of the characteristics that would be expected of a primitive movement factor. M is generally the most abundant protein in virions, is essential for VLP formation, and for S and N incorporation into VLPs. MHV M protein has been reported to mediate incorporation of RNA containing the genomic packaging signal into VLPs in the absence of N inefficiently (Narayanan et al., 2003), and the structurally similar 3a accessory protein of SARS-CoV may also bind RNA (Sharma et al., 2007), suggesting that RNA packaging could be an ancestral feature of M-like proteins. The M-like GP5 protein of arteriviruses may also be involved in attachment to host cells via its glycosylated ectodomain (Tian et al., 2012), though studies with chimeric PRRSV proteins demonstrated that M and GP5 are not solely responsible for differences in tropism (Lu et al., 2012). These observations, together with the previously mentioned instance of M protein duplication compensating for deletion of E (Kuo and Masters, 2010), suggest that M-like proteins as a group have the potential to carry out some of the essential functions of E, N, and S.
Fig. 4.
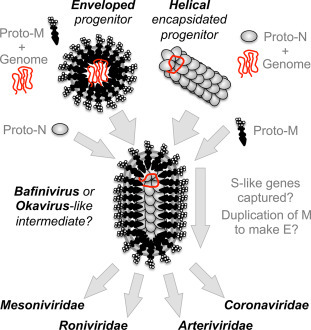
Models of coronavirus virion evolution by gradual accumulation of structural proteins. Evolution of potential progenitors with enveloped pleomorphic or helical encapsidated virion architecture is shown leading to a filamentous enveloped intermediate stage, superficially resembling virions of the genus Bafinivirus or the family Roniviridae. Structural diversification by capture of attachment and fusion proteins from an unknown source, and partial duplication of M to make E in some lineages then leads to modern nidovirus lineages.
The other potential evolutionary path we will consider is an encapsidated helical progenitor virus that encoded a proto-N related to the CTD of coronavirus N. Evidence for this is the potential conservation of fold in the CTD, suggesting a common origin for nidovirus N proteins. As described earlier, N selectively binds the genomic RNA and interacts with other N proteins to form a helical ribonucleoprotein (Barcena et al., 2009) reminiscent of the encapsidated forms of helical + RNA viruses of the Tymovirales, Virgaviridae, or Closteroviridae. In this model, a primitive virus with a helical capsid would gain an advantage in attaching to host cells by capturing a gene encoding a membrane-spanning protein like M.
Considered individually, or as a group, viral particles formed by nidoviruses are pleomorphic. Whatever their origin, coronavirus structural proteins demonstrate a remarkable plasticity to accommodate gene deletion, gene duplication, and genetic divergence, while still facilitating the entry, egress, and protection of the genome. It seems fitting, therefore, that pleomorphic nidovirus particles should be formed from a set of structural components that could themselves collectively also be described as pleomorphic.
References
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 2000;97(10):5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84(21):11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena M., Oostergetel G.T., Bartelink W., Faas F.G., Verkleij A., Rottier P.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U.S.A. 2009;106(2):582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13(8):751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., deVarennes S.L., Andonov A., He R., Booth T.F. Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion. PLoS One. 2007;2(10):e1082. doi: 10.1371/journal.pone.0001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87(11):6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218(1):52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82(6):2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10(11):e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75(24):12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Chen C.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein—implication for virus ribonucleoprotein packaging. PLoS One. 2013;8(5):e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74(9):4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 2002;76(3):1273–1284. doi: 10.1128/JVI.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello D.A., Millet J.K., Hsia C.Y., Whittaker G.R., Daniel S. Single particle assay of coronavirus membrane fusion with proteinaceous receptor-embedded supported bilayers. Biomaterials. 2013;34(32):7895–7904. doi: 10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Smeets M., Vernooij F., Vennema H., Rottier P.J. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 1999;73(9):7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A., Post S.M., Raamsman M.J., Horzinek M.C., Rottier P.J. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 1995;69(8):4668–4674. doi: 10.1128/jvi.69.8.4668-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Alvarez E., Almazan F., Rejas M.T., Lamirande E., Roberts A. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81(4):1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376(2):379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Alvarez E., Oliveros J.C. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7(10):e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent S.D., Xia D., Wastling J.M., Neuman B.W., Britton P., Maier H.J. The proteome of the infectious bronchitis virus Beau-R virion. J. Gen. Virol. 2015;96:3499–3506. doi: 10.1099/jgv.0.000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A., Wang S., Walsh M.A., Dokland T. Structure of the equine arteritis virus nucleocapsid protein reveals a dimer-dimer arrangement. Acta Crystallogr. Sect. D. 2007;63(Pt. 5):581–586. doi: 10.1107/S0907444907008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan D.N., Dokland T. Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Structure. 2003;11(11):1445–1451. doi: 10.1016/j.str.2003.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Sims A.C., Baric R.S. Systematic assembly and genetic manipulation of the mouse hepatitis virus A59 genome. Methods Mol. Biol. 2008;454:293–315. doi: 10.1007/978-1-59745-181-9_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82(23):11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunowska M., Biggs P.J., Zheng T., Perrott M.R. Identification of a novel nidovirus associated with a neurological disease of the Australian brushtail possum (Trichosurus vulpecula) Vet. Microbiol. 2012;156(3–4):418–424. doi: 10.1016/j.vetmic.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquerroy S., Vigouroux A., Rottier P.J., Rey F.A., Bosch B.J. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335(2):276–285. doi: 10.1016/j.virol.2005.02.022. S0042-6822(05)00120-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Hantak M.P., Park J.E., Gallagher T. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J. Virol. 2015;89(11):6093–6104. doi: 10.1128/JVI.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75(3):1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faaberg K.S., Even C., Palmer G.A., Plagemann P.G. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J. Virol. 1995;69(1):613–617. doi: 10.1128/jvi.69.1.613-617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13(12):1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72(10):7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J. Virol. 1996;70(7):4683–4690. doi: 10.1128/jvi.70.7.4683-4690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke G.J., de Haan C.A., Rossen J.W., Vennema H., Rottier P.J. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74(3):1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., L'Haridon R., Vautherot J.F., Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188(2):666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G.K., Dunker A.K., Uversky V. Prediction of intrinsic disorder in MERS-CoV/HCoV-EMC supports a high oral-fecal transmission. PLoS Curr. 2013;5 doi: 10.1371/currents.outbreaks.22254b58675cdebc256dbe3c5aa6498b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M., Vestergaard G., Rachel R., Chen L., Garrett R.A., Prangishvili D. Virology: independent virus development outside a host. Nature. 2005;436(7054):1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316(2):476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4(4):557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. 0409465102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzinek M., Maess J., Laufs R. Studies on the substructure of togaviruses. II. Analysis of equine arteritis, rubella, bovine viral diarrhea, and hog cholera viruses. Arch. Gesamte Virusforschung. 1971;33(3):306–318. [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281(6):3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79(21):13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. 79/21/13285 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84(19):10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87(16):9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96(8):2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Xue C., Ren X., Zhang C., Li L., Shu D. Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteome Sci. 2010;8:29. doi: 10.1186/1477-5956-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konvalinka J., Krausslich H.G., Muller B. Retroviral proteases and their roles in virion maturation. Virology. 2015;479–480:403–417. doi: 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76(10):4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77(8):4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Evolved variants of the membrane protein can partially replace the envelope protein in murine coronavirus assembly. J. Virol. 2010;84(24):12872–12885. doi: 10.1128/JVI.01850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74(3):1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Koetzner C.A., Hurst K.R., Masters P.S. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88(8):4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst-Hess K.R., Koetzner C.A., Masters P.S. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J. Virol. 2016;90:4357–4368. doi: 10.1128/JVI.03212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette D., Barbouche R., Yao Y., Boson B., Cosset F.L., Jones I.M., Fenouillet E. Significant redox insensitivity of the functions of the SARS-CoV spike glycoprotein: comparison with HIV envelope. J. Biol. Chem. 2006;281(14):9200–9204. doi: 10.1074/jbc.M512529200. M512529200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M., Bartoli J., Shmakova L., Jeudy S., Labadie K., Adrait A. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 2014;111(11):4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki D.N., Gallagher T.M. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 2002;277(22):19727–19734. doi: 10.1074/jbc.M201837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. 309/5742/1864 [pii] [DOI] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579(11):2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Eghiaian F., Sieben C., Herrmann A., Schaap I.A. Bending and puncturing the influenza lipid envelope. Biophys. J. 2011;100(3):637–645. doi: 10.1016/j.bpj.2010.12.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lescar J., Tam J.P., Liu D.X. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 2004;325(1):374–380. doi: 10.1016/j.bbrc.2004.10.050. S0006-291X(04)02295-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349(2):264–275. doi: 10.1016/j.virol.2006.01.028. S0042-6822(06)00056-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276(20):17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185(2):911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: a small membrane protein with multiple functions. Cell. Mol. Life Sci. 2007;64(16):2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Liu M., Cao B., Tan H., Wang J., Li X. Responses of three different ecotypes of reed (Phragmites communis Trin.) to their natural habitats: leaf surface micro-morphology, anatomy, chloroplast ultrastructure and physio-chemical characteristics. Plant Physiol. Biochem. 2012;51:159–167. doi: 10.1016/j.plaphy.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Liu G., Lv L., Yin L., Li X., Luo D., Liu K. Assembly and immunogenicity of coronavirus-like particles carrying infectious bronchitis virus M and S proteins. Vaccine. 2013;31(47):5524–5530. doi: 10.1016/j.vaccine.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U.S.A. 2006;103(33):12540–12545. doi: 10.1073/pnas.0605402103. 0605402103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Zhang J., Huang C.M., Go Y.Y., Faaberg K.S., Rowland R.R. Chimeric viruses containing the N-terminal ectodomains of GP5 and M proteins of porcine reproductive and respiratory syndrome virus do not change the cellular tropism of equine arteritis virus. Virology. 2012;432(1):99–109. doi: 10.1016/j.virol.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321(3):557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Tong X., Xu X., Li X., Lou Z., Rao Z. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell. 2010;1(7):688–697. doi: 10.1007/s13238-010-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C.E., Youn S. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. Adv. Exp. Med. Biol. 2006;581:193–198. doi: 10.1007/978-0-387-33012-9_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Garcia Mde J., Sanz M.A., Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005;579(17):3607–3612. doi: 10.1016/j.febslet.2005.05.046. S0014-5793(05)00654-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Kuo L., Ye R., Hurst K.R., Koetzner C.A., Hsue B. Genetic and molecular biological analysis of protein-protein interactions in coronavirus assembly. Adv. Exp. Med. Biol. 2006;581:163–173. doi: 10.1007/978-0-387-33012-9_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Machamer C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology. 2010;405(1):139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenkamp R., Spaan W.J. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74(17):8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77(5):2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80(16):7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82(11):5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Lee J.W., Leong M.L., Ling A.E., Tan H.C., Ooi E.E. Topographic changes in SARS coronavirus-infected cells at late stages of infection. Emerg. Infect. Dis. 2004;10(11):1907–1914. doi: 10.3201/eid1011.040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Dediego M.L., Alvarez E., Jimenez-Guardeno J.M., Regla-Nava J.A., Llorente M. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Castano-Rodriguez C., Fernandez-Delgado R. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A., Marquez-Jurado S., Galan C., Enjuanes L., Almazan F. Transmissible gastroenteritis coronavirus RNA-dependent RNA polymerase and nonstructural proteins 2, 3, and 8 are incorporated into viral particles. J. Virol. 2012;86(2):1261–1266. doi: 10.1128/JVI.06428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., de Groot R.J., Rottier P.J. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80(5):2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. 80/5/2326 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Ceriani J.E., Patino C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3(10):e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108(26):10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7):e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7(10):e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89(7):3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordono D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8(8):e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70(7):4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman J.S., Jing X., Leser G.P., Lamb R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142(6):902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011;85(2):675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog. 2012;8(5):e1002674. doi: 10.1371/journal.ppat.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81(8):3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol. 2005;79(11):7195–7206. doi: 10.1128/JVI.79.11.7195-7206.2005. 79/11/7195 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele U., Erdmann S., Ungewickell E.J., Felisberto-Rodrigues C., Ortiz-Lombardia M., Garrett R.A. Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus. J. Virol. 2011;85(10):4812–4821. doi: 10.1128/JVI.00072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79(11):6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze H., Ulferts R., Schelle B., Bayer S., Granzow H., Hoffmann B. Characterization of White bream virus reveals a novel genetic cluster of nidoviruses. J. Virol. 2006;80(23):11598–11609. doi: 10.1128/JVI.01758-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Surjit M., Satija N., Liu B., Chow V.T., Lal S.K. The 3a accessory protein of SARS coronavirus specifically interacts with the 5'UTR of its genomic RNA, using a unique 75 amino acid interaction domain. Biochemistry. 2007;46(22):6488–6499. doi: 10.1021/bi062057p. [DOI] [PubMed] [Google Scholar]
- Shulla A., Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009;284(47):32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G., Barthel A., ter Meulen V. Coronavirus JHM: a virion-associated protein kinase. J. Gen. Virol. 1981;52(Pt. 2):235–243. doi: 10.1099/0022-1317-52-2-235. [DOI] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Dobbe J.C., Spaan W.J. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 2003;77(1):97–104. doi: 10.1128/JVI.77.1.97-104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey M.D., Meschke J.S. WHO; Geneva, Switzerland: 2003. WHO Virus Survival Report: Survival in the Environment with Special Attention to Survival in Sewage Droplets and Other Media of Fecal or Respiratory Origin.https://http://www.unc.edu/courses/2008spring/envr/421/001/WHO_VirusSurvivalReport_21Aug2003.pdf [Google Scholar]
- Spann K.M., Cowley J.A., Walker P.J., Lester R.J.G. A yellow-head-like virus from Penaeus monodon cultured in Australia. Dis. Aquat. Org. 1997;31:169–179. [Google Scholar]
- Spilman M.S., Welbon C., Nelson E., Dokland T. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J. Gen. Virol. 2009;90(Pt. 3):527–535. doi: 10.1099/vir.0.007674-0. [DOI] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F., Kincaid A., Gordon M. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in Python regius. MBio. 2014;5(5):e01484–e01514. doi: 10.1128/mBio.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33(1):449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317(4):1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79(17):11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2008;82(4):1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh K.T., Siu Y.L., Chan W.L., Schluter M.A., Liu C.J., Peiris J.S. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21(22):3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82(Pt. 6):1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80(3):1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. 80/3/1280 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wei Z., Zevenhoven-Dobbe J.C., Liu R., Tong G., Snijder E.J., Yuan S. Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture. J. Virol. 2012;86(7):3701–3712. doi: 10.1128/JVI.06836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. M311191200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279(20):20836–20849. doi: 10.1074/jbc.M400759200. M400759200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.T., Wang S.M., Huang K.J., Lee A.I., Chiang C.C., Wang C.T. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem. 2010;285(17):12862–12872. doi: 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Munster V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18(38) doi: 10.2807/1560-7917.es2013.18.38.20590. pii: 20590. [DOI] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S., de Haan C.A. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84(21):11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J. 1984;3(10):2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss N.R., Gerstein M. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. J. Mol. Biol. 2005;346(2):477–492. doi: 10.1016/j.jmb.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., Wang C.T. APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein. Virology. 2009;388(1):112–120. doi: 10.1016/j.virol.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J. Virol. 2015;89(18):9313–9323. doi: 10.1128/JVI.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.C., Yi Z., Hogue B.G. Identification of mouse hepatitis coronavirus A59 nucleocapsid protein phosphorylation sites. Virus Res. 2007;126(1–2):139–148. doi: 10.1016/j.virusres.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. doi: 10.1016/j.virol.2004.09.033. S0042-6822(04)00644-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. S0042-6822(06)00359-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. C300520200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Kumar P., Tay F.P., Moreau D., Liu D.X., Bard F. Genome-wide screen reveals valosin-containing protein requirement for coronavirus exit from endosomes. J. Virol. 2015;89(21):11116–11128. doi: 10.1128/JVI.01360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Tsang A.K., Hui S.W., Fan R.Y. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J. Virol. 2014;88(2):1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wrensch F., Winkler M., Pohlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6(9):3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 2009;106(47):19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. S0006291X03024136 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Feng Y., Chakraborti S., Dimitrov D.S. Oligomerization of the SARS-CoV S glycoprotein: dimerization of the N-terminus and trimerization of the ectodomain. Biochem. Biophys. Res. Commun. 2004;322(1):93–99. doi: 10.1016/j.bbrc.2004.07.084. S0006-291X(04)01554-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Liu Y., Lou Z., Qin L., Li X., Bai Z. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004;279(29):30514–30522. doi: 10.1074/jbc.M403760200. M403760200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279(47):49414–49419. doi: 10.1074/jbc.M408782200. M408782200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81(7):3597–3607. doi: 10.1128/JVI.01472-06. JVI.01472-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74(22):10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2003;100(22):12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79(23):14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. 79/23/14909 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Gustafson C.L., Diao J., Burgner J.W., 2nd, Li Z., Zhang J., Chen J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280(24):23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281(25):17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838(4):1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U.S.A. 2014;111(18):6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ferraro D., Zhao J., Hussain S., Shao J., Trujillo J. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J. Virol. 2010;84(7):3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357(2):215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]


