Highlights
-
•
The heterogenous crtEBI operon was overexpressed in 10 LPS mutant strains of E. coli W3110.
-
•
ΔwaaC/pWSK29-crtEBI and ΔwaaF/pWSK29-crtEBI produced more lycopene than others.
-
•
Overexpressing dxr, dxr, ispA and idi in ΔwaaC/pWSK29-crtEBI and ΔwaaF/pWSK29-crtEBI enhanced lycopene production.
-
•
The maximum yield of 5.39 mg/g was produced in ΔwaaC/pWSK29-crtEBI-SRA.
Keywords: Lipopolysaccharide, Lycopene, Escherichia coli, waaC, Permeability
Abstract
Lipopolysaccharides, the major molecules in the outer membrane of Escherichia coli, affect the behavior of bacteria including outer membrane permeability, but its influence on lycopene production in E. coli has never been reported. In this study, the effects of lipopolysaccharides with different structures on lycopene biosynthesis were investigated. Firstly, the heterogenous crtEBI operon were overexpressed in 10 LPS mutant strains of E. coli W3110 (ΔwaaC, ΔwaaF, ΔwaaY, ΔwaaG, ΔwaaR, ΔwaaO, ΔwaaU, ΔwaaP, ΔwaaY and ΔwaaB), and their ability to produce lycopene were compared. ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI and ΔwaaY/pWSK29-crtEBI produced 4.19, 4.20, and 3.81 mg/g lycopene, respectively, while the control W3110/pWSK29-crtEBI produced 3.71 mg/g lycopene; the other strains produced less lycopene than the control. In order to enhance lycopene production, genes dxr, dxr, ispA, and idi were overexpressed in ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI individually or in combination, and the lycopene production in each strain was analyzed. The maximum yield of 5.39 mg/g was achieved in ΔwaaC/pWSK29-crtEBI-SRA, which is 142% higher than that in W3110/pWSK29-crtEBI. The results indicate that the length of lipopolysaccharide affects lycopene biosynthesis in E. coli, and the shorter lipopolysaccharide and higher outer membrane permeability might be beneficial to lycopene biosynthesis.
1. Introduction
Escherichia coli is an important platform for protein expressing and various product biosynthesis [1], however, the limited permeability of its outer membrane hinders the performance of E. coli as whole-cell biocatalyst [2]. The expression of SARS coronavirus small envelope protein [2] and deletion of lpp encoding lipoprotein in E. coli [3] were effective for increasing the outer membrane permeability. We have constructed E. coli mutants that could synthesize different length of lipopolysaccharides (LPS), the major molecules in the outer membrane, and found that the structure of LPS is closely relevant to the outer membrane permeability [4].
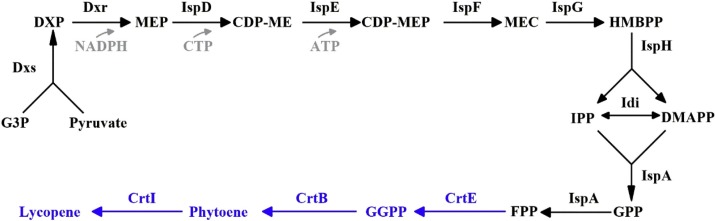
Lycopene is a bright red carotenoid pigment with 11 conjugated double bonds, and is an efficient singlet oxygen quencher. Recent advance in metabolic engineering has provided a very promising route for the heterologous production of lycopene within various microorganisms. E. coli has been widely utilized as microbial cell factory for the synthesis of various carotenoids [5]. E. coli can naturally synthesize (2E, 6E)-farnesyl diphosphate (FPP) from 3-phospho-d-glycerate and pyruvate through a series of catalytic reactions; expressing heterologous crtE, crtB and crtI in E. coli can convert FPP into lycopene (Fig. 1 ). Different types of plasmids have been used to carry the genes crtE, crtB and crtI to synthesize lycopene in E. coli [6]. Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), are synthesized via MEP pathway in E. coli [7]. In the MEP pathway, isopentenyl diphosphate isomerase encoded by idi, 1-deoxyxylulose-5-phosphate synthase encoded by dxs, 4-diphosphocytidyl-2C-methyl-d-erythritol synthase encoded by ispD, and 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthase encoded by ispF play important roles; therefore, over-expressing these genes has been used to elevate isoprenoid accumulation [5,8,9]. For example, overexpressing idi could significantly stimulate carotenoid synthesis [[10], [11], [12], [13]]. Enhancing the gene expression by chromosomal promoter replacement [14] or introduction of a heterologous mevalonate pathway to increase IPP and DMAPP supply [15] have also been used for carotenoid synthesis.
Fig. 1.
The biosynthetic pathway of lycopene in the recombinant E. coli. The last three reactions do not exist in E. coli but was introduced into E. coli in this study. Dxs, 1-deoxy-d-xylulose-5-phosphate synthase; Dxr, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; IspD, 4-diphosphocytidyl-2C-methyl-d-erythritol synthase; IspE, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase ; IspG, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; IspH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; IDI, isopentenyl-diphosphate Δ-isomerase; IspA, geranyl diphosphate farnesyl diphosphate synthase; CrtE, geranylgeranyl pyrophosphate synthase subunit; CrtB, phytoene synthase ; CrtI, phytoene dehydrogenase ; G3P, 3-phospho-d-glycerate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol; MEC, 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; DMAPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; GPP, geranyl diphosphate; FPP, (2E,6E)-farnesyl diphosphate; GGPP, geranylgeranyl diphosphate.
In this study, a series of E. coli LPS mutants strains [4] were used to synthesize lycopene by overexpressing the genes crtE, crtB, crtI, dxr, dxs and ispA, and the effects of different structures of LPS on lycopene biosynthesis were investigated.
2. Material and methods
2.1. Strains, media and growth conditions
All E. coli strains used in this study were listed in Table 1 . E. coli strains were grown in LB medium (10 g/L trypton, 5 g/L yeast extract, 10 g/L NaCl) at 37 °C or 30 °C at 200 rpm. LB medium or 2 × YT + G medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, 1 g/L glycerol), was used for shaking flask. When necessary, the medium was supplemented with ampicillin (100 μg/mL), kanamycin (30 μg/mL) or chloramphenicol (30 μg/mL) for plasmid maintenance or strain selection. Isopropyl-β-dthiogalactoside (IPTG) or arabinose were used as the inducer.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| W3110 | Wild type E. coli, F−, λ− | Novagen |
| ATCC14067 | Wild type Corynebacterium glutamicum | ATCC |
| ΔwaaC | W3110 ΔwaaC | [4] |
| ΔwaaF | W3110 ΔwaaF | [4] |
| ΔwaaG | W3110 ΔwaaG | [4] |
| ΔwaaO | W3110 ΔwaaO | [4] |
| ΔwaaR | W3110 ΔwaaR | [4] |
| ΔwaaU | W3110 ΔwaaU | [4] |
| ΔwaaP | W3110 ΔwaaP | [4] |
| ΔwaaQ | W3110 ΔwaaQ | [4] |
| ΔwaaY | W3110 ΔwaaY | [4] |
| ΔwaaB | W3110 ΔwaaB | [4] |
| ΔwaaC/pWSK29-crtEBI | ΔwaaC harboring pWSK29-crtEBI | This work |
| ΔwaaF/pWSK29-crtEBI | ΔwaaF harboring pWSK29-crtEBI | This work |
| ΔwaaG/pWSK29-crtEBI | ΔwaaG harboring WSK29-crtEBI | This work |
| ΔwaaO/pWSK29-crtEBI | ΔwaaO harboring pWSK29-crtEBI | This work |
| ΔwaaR/pWSK29-crtEBI | ΔwaaR harboring pWSK29-crtEBI | This work |
| ΔwaaU/pWSK29-crtEBI | ΔwaaU harboring pWSK29-crtEBI | This work |
| ΔwaaP/pWSK29-crtEBI | ΔwaaP harboring pWSK29-crtEBI | This work |
| ΔwaaQ/pWSK29-crtEBI | ΔwaaQ harboring pWSK29-crtEBI | This work |
| ΔwaaY/pWSK29-crtEBI | ΔwaaY harboring pWSK29-crtEBI | This work |
| ΔwaaB/pWSK29-crtEBI | ΔwaaB harboring pWSK29-crtEBI | This work |
| ΔwaaC/pWSK29-crtEBI-ispA | ΔwaaC harboring pWSK29-crtEBI-ispA | This work |
| ΔwaaF/pWSK29-crtEBI-ispA | ΔwaaF harboring pWSK29-crtEBI-ispA | This work |
| ΔwaaC/pWSK29-crtEBI-dxs | ΔwaaC harboring pWSK29-crtEBI-dxs | This work |
| ΔwaaF/pWSK29-crtEBI-dxs | ΔwaaF harboring pWSK29-crtEBI-dxs | This work |
| ΔwaaC/pWSK29-crtEBI-dxr | ΔwaaC harboring pWSK29-crtEBI-dxr | This work |
| ΔwaaF/pWSK29-crtEBI-dxr | ΔwaaF harboring pWSK29-crtEBI-dxr | This work |
| ΔwaaC/pWSK29-crtEBI-SR | ΔwaaC harboring pWSK29-crtEBI-SR | This work |
| ΔwaaF/pWSK29-crtEBI-SR | ΔwaaF harboring pWSK29-crtEBI-SR | This work |
| ΔwaaC/pWSK29-crtEBI-SRA | ΔwaaC harboring pWSK29-crtEBI- SRA | This work |
| ΔwaaF/pWSK29-crtEBI- SRA | ΔwaaF harboring pWSK29-crtEBI- SRA | This work |
| ΔwaaC/pWSK29-crtEBI/pACYC184-SRA | ΔwaaC harboring pWSK29-crtEBI and pACYC184-SRA | This work |
| ΔwaaF/pWSK29-crtEBI/pACYC184-SRA | ΔwaaF harbor the pWSK29-crtEBI and pACYC184-SRA | This work |
| ΔwaaC/pWSK29-crtEBI/pACYC184-SRAI | ΔwaaC harbor the pWSK29-crtEBI and pACYC184-SRAI | This work |
| ΔwaaF/pWSK29-crtEBI/pACYC184-SRAI | ΔwaaF harbor the pWSK29-crtEBI and pACYC184-SRAI | This work |
| Plasmids | ||
| pWSK29 | Expression vector | [26] |
| pACYC184 | Expression vector | [27] |
| pBlueScript II SK+ | Cloning vector, ColE1, lacZ, AmpR | Stratagene |
| pWSK29-crtEBI | pWSK29 harboring the crtEBI operon from C. glutamicum ATCC14067 | This work |
| pWSK29-crtEBI-ispA | pWSK29 harboring the crtEBI operon from C. glutamicum ATCC14067, and ispA from E. coli W3110 | This work |
| pWSK29-crtEBI-dxs | pWSK29 harboring the crtEBI operon from ATCC14067 and dxs from W3110 | This work |
| pWSK29-crtEBI-dxr | pWSK29 harboring the crtEBI operon from ATCC14067 and dxr from W3110 | This work |
| pWSK29-crtEBI-SR | pWSK29 harboring the crtEBI operon from ATCC14067, and dxs and dxr from W3110 | This work |
| pWSK29-crtEBI-SRA | pWSK29 harboring the crtEBI operon from ATCC14067, and ispA, dxs and dxr from W3110 | This work |
| pACYC184-SRA | pACYC184 harboring the genes ispA, dxs and dxr from W3110 | This work |
| pACYC184-SRAI | pACYC184 harboring the genes ispA, dxs, dxr and idi from W3110 | This work |
2.2. Construction of expression plasmids and recombinant strains
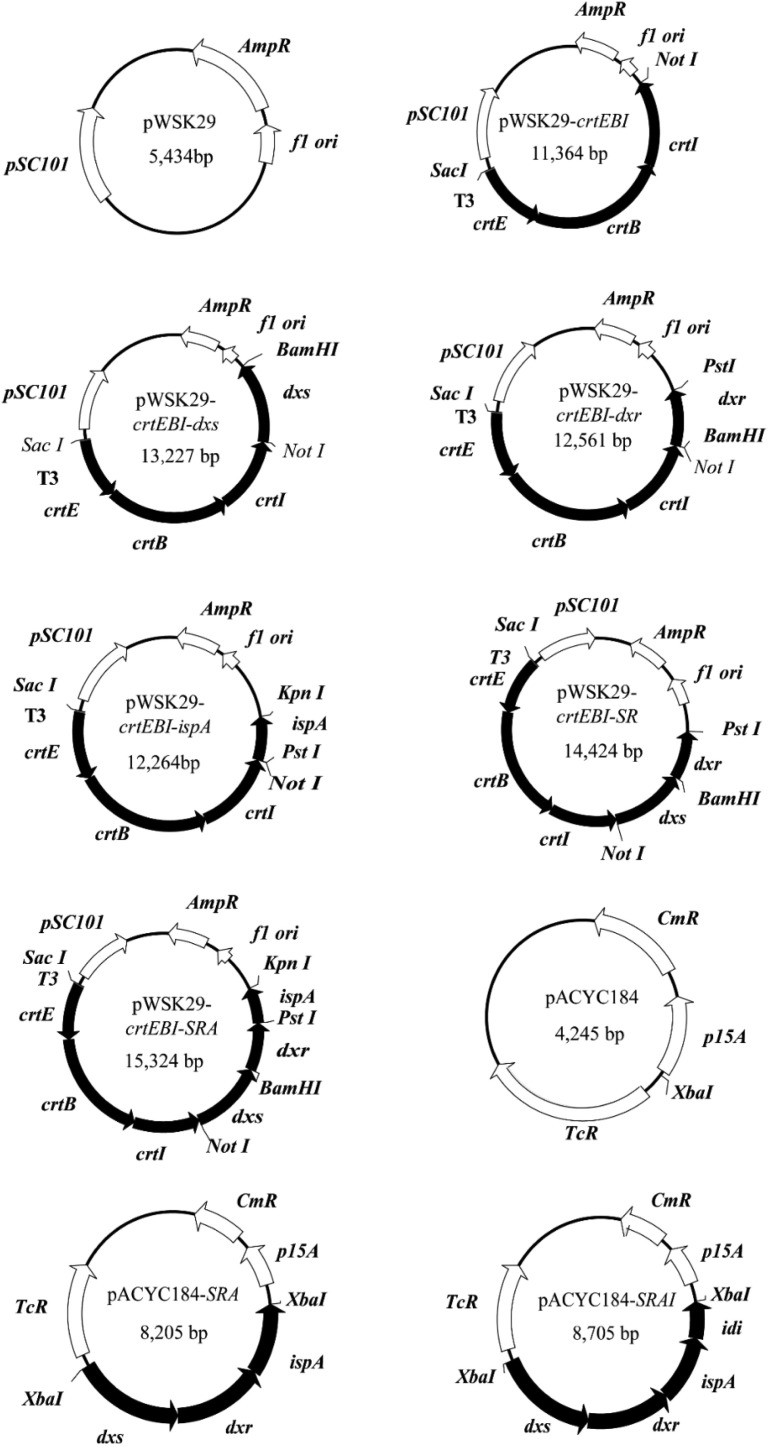
The maps of different plasmids carrying the genes crtE, crtB, crtI, dxr, dxs or ispA individually or in combination are shown in Fig. 2 . Restriction enzymes, shrimp alkaline phosphatase, T4 DNA ligase and DNA ladder were purchased from Sangon (Shanghai, China). Plasmid DNA was prepared by using the EZ-10 spin column plasmid mini-preps kit from Bio Basic Inc. (Markham, Canada). PCR reaction mixtures (50 μL) contained: 5 μL 10 × Ex Taq buffer, 4 μL dNTP mixture (2.5 mM each), 1 μL plasmid template (100 ng/μL), 1 μL forward primer (20 μM), 1 μL reverse primer (20 μM) and 0.5 μL TaKaRa Ex Taq DNA polymerase. The PCR reaction was first heated to 94 °C and maintained for 10 min, followed by 35 cycles of denaturation (94 °C for 30 s), annealing (30 s at 55 °C or 58 °C) and extension (72 °C for 3 min). At the end, an additional 10 min incubation at 72 °C was used. PCR products were purified by using the TIANgel midi purification kit from Tiangen (Beijing, China). Primer synthesis and DNA sequencing were performed by Sangon. The sequences of primers used in this study are listed in Table 2 .
Fig. 2.
Maps of different plasmids used in this study. The genes crtE, crtI, and crtB were amplified from the genome of Corynebacterium glutamicum ATCC14067.
Table 2.
Primers used in this study.
| Primer | Sequences (5′→3′) | Restriction site |
|---|---|---|
| crtEBI-29-F | ATGGGAGCTCAGGAGGTATATACCATGGACAATGGCATGACAAT | SacI |
| crtEBI-29-R | GGAGCGGCCGCTTAATGATCGTATGAGGTC | NotI |
| dxs-29-F | GAGGCGGCCGCAGGAGGTATATACCATGAGTTTTGATATTGCCAA | NotI |
| dxs-29-R | AACGGATCCTTATGCCAGCCAGGCCTTGAT | BamHI |
| dxr-29-F | AACGGATCCAGGAGGTATATACCATGAAGCAACTCACCATTCT | BamHI |
| dxr-29-R | AAGCTGCAGTCAGCTTGCGAGACGCATCAC | PstI |
| ispA-29-F | AACCTGCAGAGGAGGATTATAGGATGGACTTTCCGCAGCAACT | PstI |
| ispA-29-R | GCCGGTACCTTATTTATTACGCTGGATGATG | KpnI |
| idi-29-F | AACGGTACCAGGAGGATTATAGGATGCAAACGGAACACGTCATT | KpnI |
| idi-29-R | GGACTCGAGTTATTTAAGCTGGGTAAATGC | XhoI |
| 184-srA-F | GGCTCTAGTAGGAGGTATATACCATGAGTTTTGATATTGCCAA | XbaI |
| 184-srA-F | GCGTCTAGT TTATTTATTACGCTGGATGATG | XbaI |
| 184-srAi-F | GGCTCTAGTAGGAGGTATATACCATGAGTTTTGATATTGCCAA | XbaI |
| 184-srAi-R | GCGTCTAGTTTATTTAAGCTGGGTAAATGC | XbaI |
Fragments crtE, crtB and crtI were amplified from the genomic DNA of Corynebacterium glutamicum ATCC14067. Fragments ispA, dxs, dxr and idi were amplified from the genomic DNA of E. coli W3110, using different primer pairs listed in Table 2. PCR products were digested with the corresponding restriction enzymes and ligated with the vector pWSK29 or pACYC184 similarly digested, resulting in the plasmids pWSK29-crtEBI, pWSK29-crtEBI-ispA, pWSK29-crtEBI-dxs, pWSK29-crtEBI-dxr, pWSK29-crtEBI-SR, pWSK29-crtEBI-SRA, pACYC184-SRA, and pACYC184-SRAI, respectively.
The plasmid pWSK29-crtEBI was transformed into E. coli W3110 and its mutant strains ΔwaaC, ΔwaaF, ΔwaaG, ΔwaaO, ΔwaaR, ΔwaaU, ΔwaaP, ΔwaaQ, ΔwaaY, and ΔwaaB, resulting in the strains ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI, ΔwaaG/pWSK29-crtEBI, ΔwaaO/pWSK29-crtEBI, ΔwaaR/pWSK29-crtEBI, ΔwaaU/pWSK29-crtEBI, ΔwaaP/pWSK29-crtEBI, ΔwaaQ/pWSK29-crtEBI, ΔwaaY/pWSK29-crtEBI, and ΔwaaB/pWSK29-crtEBI. The plasmids pWSK29-crtEBI, pWSK29-crtEBI-ispA, pWSK29-crtEBI-dxs, pWSK29-crtEBI-dxr, pWSK29-crtEBI-SR and pWSK29-crtEBI-SRA was transformed into ΔwaaC and ΔwaaF, respectively, resulting in the strains ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI, ΔwaaC/pWSK29-crtEBI-ispA, ΔwaaF/pWSK29-crtEBI-ispA, ΔwaaC/pWSK29-crtEBI-dxs, ΔwaaF/pWSK29-crtEBI-dxs, ΔwaaC/pWSK29-crtEBI-dxr, ΔwaaF/pWSK29-crtEBI-dxr, ΔwaaC/pWSK29-crtEBI-SR, ΔwaaF/pWSK29-crtEBI-SR, ΔwaaC/pWSK29-crtEBI-SRA, and ΔwaaF/pWSK29-crtEBI-SRA. The plasmids pACYC184-SRA, pACYC184-SRAI was transformed into ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI, resulting in the strains ΔwaaC/pWSK29-crtEBI/pACYC184-SRA, ΔwaaF/pWSK29-crtEBI/pACYC184-SRA, ΔwaaC/pWSK29-crtEBI/pACYC184-SRAI, and ΔwaaF/pWSK29-crtEBI/pACYC184-SRAI. Transformation of E. coli was performed according to the published protocol [16].
2.3. Analytical methods
Cell growth during the cultivations was monitored by measuring the optical density at 600 nm (OD600). For dry cell weight (DCW) determination, a known volume of fermentation broth was centrifuged for 10 min in pre-weighed test tubes at 4 °C and 4000 rpm, washed once with water, and dried for 24 h at 90 °C to a constant weight.
To extract lycopene, the E. coli cells were grown in a 500-ml shaking flask at 37 °C and 200 rpm, harvested by centrifugation at 4000 rpm and 4 °C for 10 min, and rinsed twice with deionized water. Then the cells were suspended in 1 ml of acetone and incubated at 55 °C for 15 min in the dark. The samples were centrifuged at 4000 rpm for 10 min, and the acetone supernatant containing lycopene was transferred to a clean tube. The lycopene content of the extracts was measured according to the previous reported method [8]. Lycopene (purchased from Sigma) dissolved in acetone was used as the standard. The results were the mean from three independent determinations, and the standard deviations were in the range of ± 10% of the means.
2.4. Outer membrane permeability assay
To determined outer membrane permeability of E. coli, the fluorescent probe 1-N-phenyl-1-naphthylamine (NPN) assay was used [17]. E. coli strains were cultivated in LB broth at 37 °C, harvested by centrifugation at 4000 rpm for 10 min, washed and resuspended in phosphate buffer (10 mM, pH = 7.4), The value of OD600 was adjusted to 0.5 in the final cell suspension. Then 1.92 mL of cell suspension was mixed with 80 μL NPN (1 mM) into quartz cuvette, immediately. Fluorescence was measured using a Fluorescence Spectrophotometer (650-60, Hitachi, Japan), using a slit width of 5 nm, an excitation wavelength of 350 nm and an emission wavelength of 420 nm.
3. Results
3.1. Lycopene biosynthesis in E. coli W3110 overexpressing the genes crtE, crtB, and crtI from C. glutamicum is affected by LPS structure
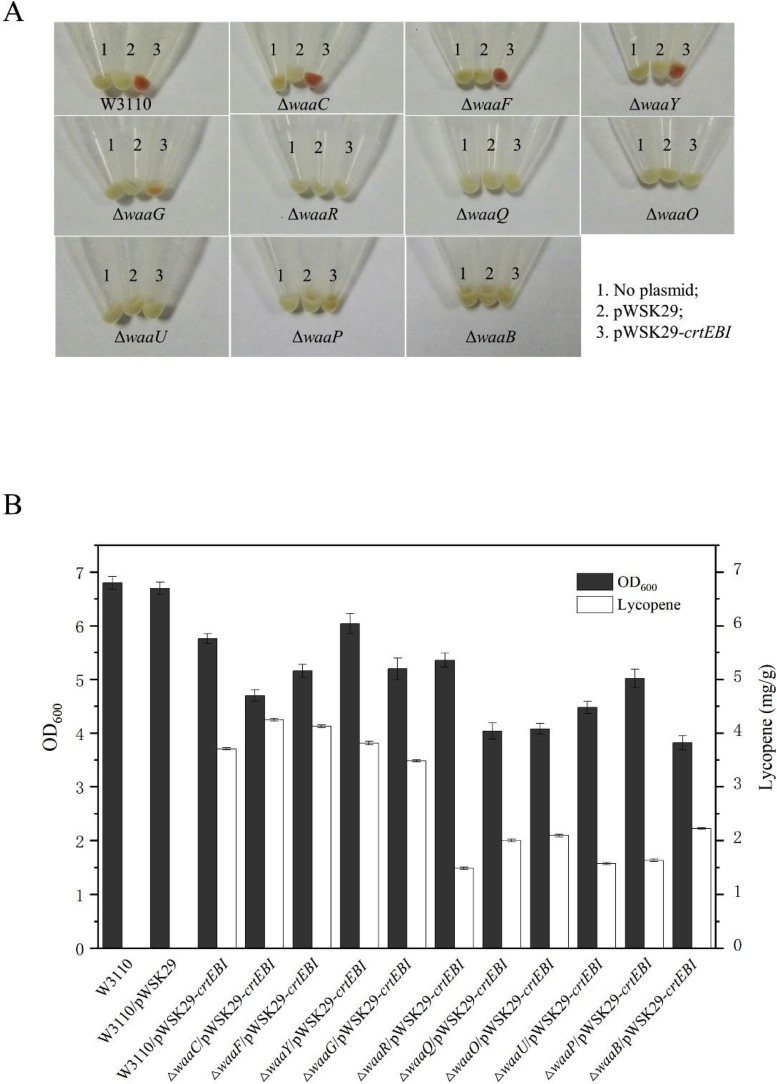
LPS mutants ΔwaaC, ΔwaaF, ΔwaaG, ΔwaaO, ΔwaaR, ΔwaaU, ΔwaaP, ΔwaaQ, ΔwaaY and ΔwaaB from E. coli W3110 synthesized different length of LPS and showed different outer membrane permeability [4]. In order to effect of LPS structure on lycopene biosynthesis, pWSK29-crtEBI and pWSK29 were transformed into these strains and their lycopene productions were investigated [19].
As shown in Fig. 3 A, the color of the bacterial cells can be used to determine the amount of lycopene. W3110/pWSK29-crtEBI cells were red, but not W3110 and W3110/pWSK29 cells, suggesting that lycopene was produced only when the genes crtE, crtB, and crtI from C. glutamicum were overexpressed. ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI and ΔwaaY/pWSK29-crtEBI cells were also red, ΔwaaG/pWSK29-crtEBI cells were light red, suggesting lycopene was synthesized in these cells. These results indicate that LPS structure affects lycopene synthesis in E. coli. These cells were broken and lycopene was extracted to quantify. As shown in Fig. 3B, all the strains harboring pWSK29-crtEBI can synthesize lycopene, but more lycopene was produced in ΔwaaC/pWSK29-crtEBI (4.19 mg/g), ΔwaaF/pWSK29-crtEBI (4.20 mg/g), ΔwaaY/pWSK29-crtEBI (3.81 mg/g). This quantification is consistent with the red shades of the cells (Fig. 3A vs B). Based on their OD600 value, the cell growth was retarded when pWSK29-crtEBI was introduced in E. coli. By comparison, ΔwaaC/pWSK29-crtEBI and ΔwaaF/pWSK29-crtEBI grew better and produced more lycopene than other strains, therefore, they were used in further study.
Fig. 3.
A. Color comparison of different E. coli cells. B. Comparison of cell growth and lycopene yield in different E. coli cells. Data represent the average of three experiments and the error bars represent the standard deviation.
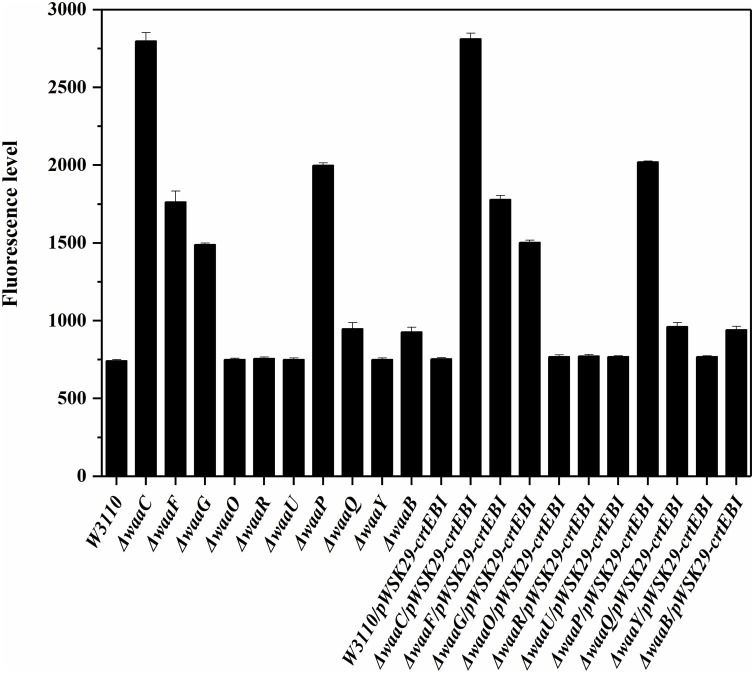
The outer membrane permeability of the 10 E. coli LPS mutants ΔwaaC, ΔwaaF, ΔwaaG, ΔwaaO, ΔwaaR, ΔwaaU, ΔwaaP, ΔwaaQ, ΔwaaY and ΔwaaB with or without pWSK29-crtEBI were analyzed, using the wild type W3110 as the control (Fig. 4 ). Similar outer membrane permeabilities were observe for the same strain with or without pWSK29-crtEBI, suggesting that the lycopene produced in the strains ΔwaaC/pWSK29-crtEBI, ΔwaaF/pWSK29-crtEBI, ΔwaaG/pWSK29-crtEBI, ΔwaaO/pWSK29-crtEBI, ΔwaaR/pWSK29-crtEBI, ΔwaaU/pWSK29-crtEBI, ΔwaaP/pWSK29-crtEBI, ΔwaaQ/pWSK29-crtEBI, ΔwaaY/pWSK29-crtEBI and ΔwaaB/pWSK29-crtEBI does not affect the outer membrane permeability and might locate in the inner membranes. ΔwaaC/pWSK29-crtEBI showed high outer membrane permeability and produced a large amount of lycopene, but the outer membrane permeability and the lycopene production were not proportionally increased for other strains such as ΔwaaY/pWSK29-crtEBI and ΔwaaP/pWSK29-crtEBI. The results suggested that outer membrane permeability is not the only factor affecting lycopene production in E. coli.
Fig. 4.
Comparison of cell outer membrane permeability of different E. coli cells. Data represent the average of three experiments and the error bars represent the standard deviation.
3.2. More lycopene was produced in ΔwaaC/pWSK29-crtEBI and ΔwaaF/pWSK29-crtEBI after enhancing the MEP pathway
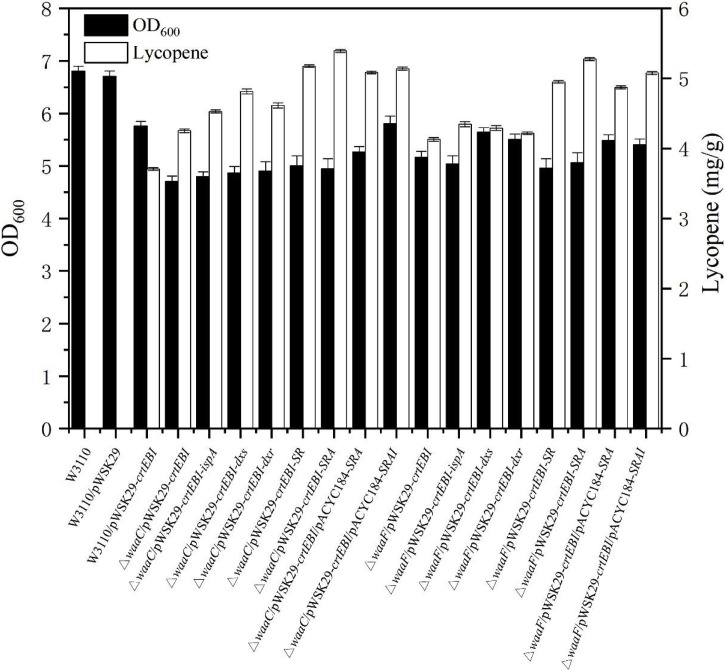
To further improve lycopene yield, the key genes dxs, dxr, ispA and idi in the MEP pathway were overexpressed, individually or in combination, in ΔwaaC/pWSK29-crtEBI and ΔwaaF/pWSK29-crtEBI; the cell growth and lycopene production in these recombinant strains were investigated (Fig. 5 ).
Fig. 5.
Comparison of cell growth and lycopene yield in different E. coli cells. Data represent the average of three experiments and the error bars represent the standard deviation.
ΔwaaC/pWSK29-crtEBI-ispA, ΔwaaC/pWSK29-crtEBI-dxs, and ΔwaaC/pWSK29-crtEBI-dxr produced 4.53, 4.81 and 4.61 mg/g lycopene, respectively; ΔwaaC/pWSK29-crtEBI-SR, ΔwaaC/pWSK29-crtEBI-SRA, ΔwaaC/pWSK29-crtEBI/pACYC184-SRA and ΔwaaC/pWSK29-crtEBI/pACYC184-SRAI produced 5.17, 5.39, 5.08 and 5.14 mg/g lycopene, respectively (Fig. 5). Compared to ΔwaaC/pWSK29-crtEBI, these strains grew better and produced more lycopene, suggesting that enhancing the MEP pathway facilitates lycopene synthesis in E. coli.
ΔwaaF/pWSK29-crtEBI-ispA, ΔwaaF/pWSK29-crtEBI-dxs, and ΔwaaF/pWSK29-crtEBI-dxr produced 4.35, 4.29 and 4.22 mg/g lycopene, respectively; ΔwaaF/pWSK29-crtEBI-SR, ΔwaaF/pWSK29-crtEBI-SRA, ΔwaaF/pWSK29-crtEBI/pACYC184-SRA, and ΔwaaF/pWSK29-crtEBI/pACYC184-SRAI produced 4.95, 5.28, 4.87 and 5.08 mg/g lycopene, respectively. Compared to ΔwaaF/pWSK29-crtEBI, these strains produced more lycopene, suggesting again that enhancing the MEP pathway facilitates lycopene synthesis in E. coli.
4. Discussion
Biocatalysis employing the whole cell had been increasingly developed as a green technology in the synthesis of various valuable products, while low permeability of cell membrane always lead to the low productivities [3]. The outer membrane of gram-negative bacteria provided the cell with an effective permeability barrier against external noxious agents [18], and at the same time to allow the influx of nutrient molecules [19]. In the previous publications, lpp deletion was developed as a general permeabilization method. The lpp mutant had higher permeability of outer membrane and higher ability to synthesize l-carnitine [3]. Besides, expression of SCVE viroporins in E. coli also improved the diffusivity of small molecules across outer membrane through introducing additional pores within the outer membrane. As expected, the biocatalysts efficiency of E. coli was enhanced [2].
LPS, as the major molecule in outer membrane, played important roles on membrane behavior. In E. coli, LPS typically consists of a hydrophobic domain known as lipid A, a nonrepeating core oligosaccharide, and a distal polysaccharide known as O-antigen repeats. The core oligosaccharide is assembled on lipid A via sequential glycosyl transfer from nucleotide sugar precursors. In E. coli, the chromosomal waa locus encodes enzymes required for biosynthesis of the core oligosaccharide [20]. Mutations in LPS could alter outer membrane stability, giving rise to pleiotropic phenotype [21,22]. Among 10 W3110 Δwaa mutant strains, W3110 ΔwaaC has the simplest LPS structure. In compared to W3110, W3110 ΔwaaC had four-fold higher membrane permeability, and this likely benefited for lycopene accumulation. Comparative transcriptome showed that mRNA levels of dxr and ispA in ΔwaaC and ΔwaaF were up-regulated, compared to W3110 (data not shown). The over-expression of the dxr gene provided more IPP precursors in the MEP pathway in tobacco, which consequently stimulated synthesis efficiency of isoprenoid downstream [23]. IPP and DMAPP supply likely limited lycopene biosynthesis [9]. Besides, the accumulation of metabolites in the MEP pathway stimulated the transcription of dxs and dxr in Arabidopsis cell culture [24]. In the current study, therefore, the genes dxs, dxr and idi were overexpressed individually or in combination, to increase these precursors supply. As expected, the overexpression of genes in MEP pathway stimulated lycopene synthesis [25].
A recombinant E. coli strain overexpressing the genes crtE, crtB, and crtI from Deinococcus radiodurans R12 and optimizing the Shine Dalgarno regions and aligned spacing sequence could produce 88 mg/g lycopene after 40 h fermentation [28]. E. coli K12f could efficiently uptake fructose; overexpressing the crtEBI operon from Pantoea ananatis. in K12f could produce 192 mg/g lycopene when grown on LB medium containing 10 g/L fructose [29]. Therefore, overexpressing the crtEBI operon from other bacteria in ΔwaaC or ΔwaaF might increase lycopene production
Acknowledgments
This study was supported by the National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-10) and the Collaborative Innovation Center of Jiangsu Modern Industrial Fermentation.
Contributor Information
Xiaoqing Hu, Email: hu.x.q@hotmail.com.
Xiaoyuan Wang, Email: xwang@jiangnan.edu.cn.
References
- 1.Adams B.L. The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth. Biol. 2016;5(12):1328–1330. doi: 10.1021/acssynbio.6b00256. [DOI] [PubMed] [Google Scholar]
- 2.Patel T.N., Park A.H., Banta S. Genetic manipulation of outer membrane permeability: generating porous heterogeneous catalyst analogs in Escherichia coli. ACS Synth. Biol. 2014;3(12):848–854. doi: 10.1021/sb400202s. [DOI] [PubMed] [Google Scholar]
- 3.Ni Y., Reye J., Chen R.R. lppdeletion as a permeabilization method. Biotechnol. Bioeng. 2007;97(6):1347–1356. doi: 10.1002/bit.21375. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Wang J., Ren G., Li Y., Wang X. Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar. Drugs. 2015;13(6):3325–3339. doi: 10.3390/md13063325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews P.D., Wurtzel E.T. Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl. Microbiol. Biotechnol. 2000;53(4):396–400. doi: 10.1007/s002530051632. [DOI] [PubMed] [Google Scholar]
- 6.Xu J., Xu X., Xu Q., Zhang Z., Jiang L., Huang H. Efficient production of lycopene by engineered E. coli strains harboring different types of plasmids. Bioprocess Biosyst. Eng. 2018;41(4):489–499. doi: 10.1007/s00449-017-1883-y. [DOI] [PubMed] [Google Scholar]
- 7.Farmer W.R., Liao J.C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 2000;18(5):533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 8.Kim S.W., Keasling J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001;72(4):408–415. doi: 10.1002/1097-0290(20000220)72:4<408::aid-bit1003>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 9.Lv X.M., Xu H.M., Yu H.W. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl. Microbiol. Biotechnol. 2013;97(6):2357–2365. doi: 10.1007/s00253-012-4485-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang J., Nie Q., Liu H., Xian M. A novel MVA-mediated pathway for isoprene production in engineered E. coli. BMC Biotechnol. 2016;16:5. doi: 10.1186/s12896-016-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C., Gao X., Jiang Y., Sun B.B., Gao F., Yang S. Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli. Metab. Eng. 2016;37:79–91. doi: 10.1016/j.ymben.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y., Yang J., Qin B., Li Y., Sun Y., Su S., Xian M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl. Microbiol. Biotechnol. 2011;90(6):1915–1922. doi: 10.1007/s00253-011-3199-1. [DOI] [PubMed] [Google Scholar]
- 13.Choi H.S., Lee S.Y., Kim T.Y., Woo H.M. In silico identification of gene amplification targets for improvement of lycopene production. Appl. Environ. Microbiol. 2010;76(10):3097–3105. doi: 10.1128/AEM.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng F., Xu Y., Tao Y., Liu W., Lin B. Improving isoprene production by engineered heterologous mevalonate pathway in Escherichia coli. Chin. J. Biotechnol. 2015;31(7):1073–1081. [PubMed] [Google Scholar]
- 15.Haney J.T., Jr, Phillips T., Sielken R.L., Jr, Valdez-Flores C. Development of an inhalation unit risk factor for isoprene. Regul. Toxicol. Pharmacol. 2015;73(3):712–725. doi: 10.1016/j.yrtph.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Russell D.W. Vol. 1–3. Cold Spring Harbor Laboratory Press; New York: 2001. (Molecular Cloning: a Laboratory Manual). [Google Scholar]
- 17.Helander I.M., Mattila-Sandholm T. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 2010;88(2):213–219. doi: 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- 18.Richard S.B., Lillo A.M., Tetzlaff C.N., Bowman M.E., Noel J.P., Cane D.E. Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry. 2004;43(38):12189–12197. doi: 10.1021/bi0487241. [DOI] [PubMed] [Google Scholar]
- 19.Tao L., Yao H., Cheng Q. Genes from a Dietzia sp. For synthesis of C40 and C50 beta-cyclic carotenoids. Gene. 2007;386(1-2):90–97. doi: 10.1016/j.gene.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56(3):395. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grawert T., Rohdich F., Span I., Bacher A., Eisenreich W., Eppinger J., Groll M. Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction. Angew. Chem. Int. Ed. Engl. 2009;48(31):5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]
- 22.Kemp L.E., Bond C.S., Hunter W.N. Structure of 2C-methyl-d-erythritol 2,4- cyclodiphosphate synthase: an essential enzyme for isoprenoid biosynthesis and target for antimicrobial drug development. Proc. Natl. Acad. Sci. U. S. A. 2002;99(10):6591–6596. doi: 10.1073/pnas.102679799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W., Oldfield E. Bioorganometallic chemistry with IspG and IspH: structure, function and inhibition of the Fe4S4 proteins involved in Isoprenoid Biosynthe. Angew. Chem. Int. Ed. Engl. 2014;53(17):4294–4310. doi: 10.1002/anie.201306712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter M.H., Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 25.Zepeck F., Gräwert T., Kaiser J., Schramek N., Eisenreich W., Bacher A., Rohdich F. Biosynthesis of isoprenoids. Purification and properties of IspG protein from Escherichia coli. J. Org. Chem. 2005;70(23):9168–9174. doi: 10.1021/jo0510787. [DOI] [PubMed] [Google Scholar]
- 26.Rong F.W., Kushner S.R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100(100):195–199. [PubMed] [Google Scholar]
- 27.Martinez E., Bartolomé B., De l.C.F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68(1):159. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 28.Jin W., Xu X., Jiang L., Zhang Z., Li S., Huang H. Putative carotenoid genes expressed under the regulation of Shine-Dalgarno regions in Escherichia coli for efficient lycopene production. Biotechnol. Lett. 2015;37(11):2303–2310. doi: 10.1007/s10529-015-1922-1. [DOI] [PubMed] [Google Scholar]
- 29.Du W., Song Y., Liu M., Yang H., Zhang Y., Fan Y., Luo X., Li Z., Wang N., He H., Zhou H., Ma W., Zhang T. Gene expression pattern analysis of a recombinant Escherichia coli strain possessing high growth and lycopene production capability when using fructose as carbon source. Biotechnol. Lett. 2016;38(9):1571–1577. doi: 10.1007/s10529-016-2133-0. [DOI] [PubMed] [Google Scholar]