Abstract
Cystatins are thiol proteinase inhibitors (TPI), present ubiquitously in animals, plants and micro-organisms. These are not merely inhibitors rather they are at heart of many pathological conditions ranging from diabetes to renal failure. These are essential for maintenance of protein balance of the cell; once this balance gets disturbed, it may lead to cell death. Thus, cystatins cannot be merely regarded as TPI’s as these have been found to play a pivotal role in tumorigenesis and neurodegenerative diseases. Many studies have reported the variation in cystatin level in incidences of different types of cancer; highlighting an important role played by these inhibitors in cancer development and progression. Cystatin C is increasingly replacing creatinine as a biomarker of glomerular filtration rate (GFR) thereby highlighting the importance of this important inhibitor. Some recent studies have also reported the interaction pattern of various anti-cancer drugs with cystatins in a bid to find how these drugs affect this important inhibitors and whether these drugs have any side effect on cystatins. Thus, in this growing disease era it can be said that cystatins are no more just inhibitors blocking the activity of thiol proteases rather they play a pivotal role in variety of pathological conditions.
Keywords: Cystatins, Cancer, Kidney
1. General
1.1. Proteases
Proteases are enzymes that irreversibly hydrolyze a peptide bond in an amino acid sequence by nucleophilic attack. Traditionally, proteases were thought of as digestive enzymes that cleave proteins into smaller peptides and amino acids and their role was thought of as merely being involved in turnover of cellular proteins or digestion of nutrient proteins. In recent times, this perception has evolved and proteases are now seen as vital signaling molecules that are involved in variety of processes. Protease signaling pathways are strictly regulated; any dysregulation of protease activity can lead to pathologies such as cardiovascular and inflammatory diseases, cancer, osteoporosis and neurological disorders [1]. Proteases are also known as proteolytic enzymes or proteinases. There are different ways in which these proteinases can be classified viz. on the basis of the pH range over which they are active (acid, neutral, or alkaline), on the basis of their ability to hydrolyze specific proteins (keratinase, elastase, collagenase etc.), or on the basis of their similarity to well-characterized proteinases such as pepsin, trypsin, chymotrypsin, or the mammalian cathepsins. The most acceptable classification (Table 1 ) is the one which classifies proteases on the basis of their catalytic mechanism [2]. Table 1 gives an overview of four different types of proteases: Aspartic proteinases, Metalloproteinases, Serine proteinases and Cysteine proteinases; based on their sensitivity to various inhibitors. Thus, we can say that proteases can be grouped according to the key catalytic group present at their active site: serine (Ser), threonine (Thr), cysteine (Cys), aspartate (Asp), glutamate (Glu), or zinc in metalloproteases. Ser, Cys and Thr can act directly as nucleophiles that attack an amide carbonyl C, whereas Asp, Glu and metalloproteases activate a water molecule which then acts as a nucleophile. These proteases can also be classified depending upon the position of the peptide bond cleaved by them: Endopeptidases- which cleaves an internal bond or Exopeptidases − which truncate one or several amino acids from either the N- or the C-terminus of a peptide.
Table 1.
Proteases classification: Classification of proteases on the basis of their catalytic mechanism (Hartley [2]).
| Type | Specific Inhibitors: characteristics of enzyme type | Other Inhibitors | Activators |
|---|---|---|---|
| Aspartic proteinases (EC 3.4.23) |
Pepstatin S-PI (acetyl pepstatin) Diazoacetyl norleucine methyl ester N— Diazoacetyl-N-‘ − 2,4- dinitrophenylethylenediamine Epoxy (p-nitrophenoxy) propane |
||
| Metalloproteinases (EC 3.4.24) |
Chelating agents EDTA, ethylene glycol-bis- (β- aminoethyl ether)- N, N- tetraacetic acid o-phenanthroline 8- hydoxyquinoline α,α- dipyridyl Phosphoramidon (not acid metalloproteinases |
||
| Serine proteinases (EC 3.4.21) |
PMSF DIFP |
TLCK TPCK Antipain Leupeptin Chymostatin |
|
| Cysteine proteinases (EC 3.4.22) |
Iodoacetamide, iodoacetate Heavy metals N-ethyl maleimide |
p-Chloromercurinenzoate (also inhibits some serine proteases) TLCK TPCK Antipain Leupeptin Chymostatin |
Reducing agents Cysteine DTT EDTA |
1.2. Cysteine proteases
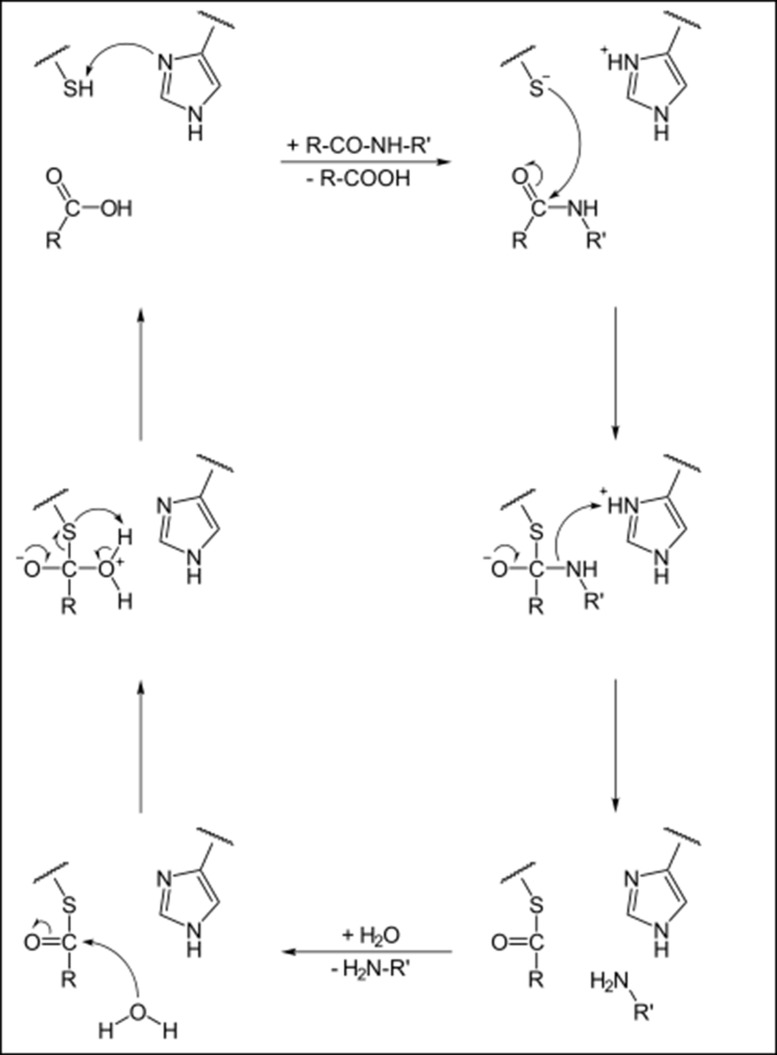
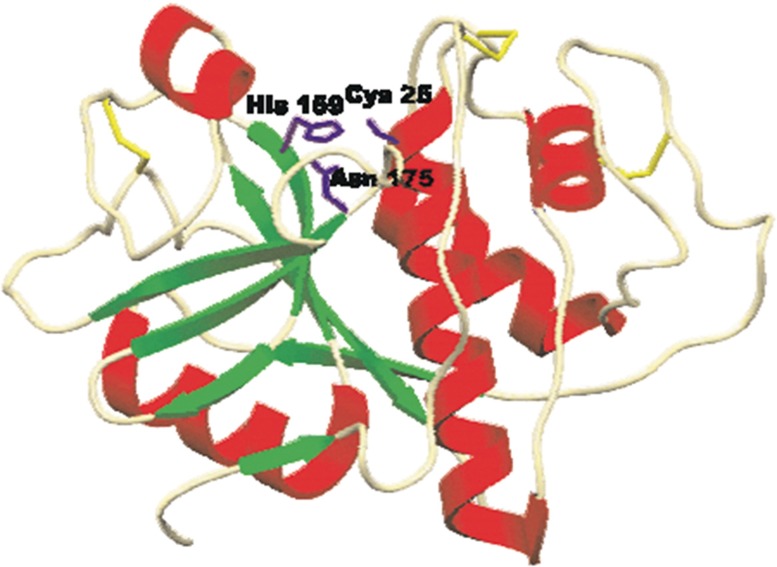
Cysteine proteases, also referred to as thiol proteases, are characterized by presence of cysteine residue at their active site for hydrolysis. Fig. 1 gives an overview of reaction mechanism of the cysteine protease mediated cleavage of a peptide bond. The catalytic site of CA-clan papain-like cysteine proteases consists of Cys, histidine (His) and Asp residues and is highly conserved among members of the enzyme family [3]. Till now, many families of cysteine proteases have been discovered [4] almost half of these in viruses. The best characterized family of cysteine proteases is that of papain. The papain family contains peptidases which are structurally related to papain, for example lysosomal cathepsins. Papain is characterized by a two-domain structure (Fig. 2 ). The active site (catalytic pocket), where the substrate is bound, is located between the domains. The catalytic residues of papain are Cys25 and His159, and they are evolutionarily preserved in all cysteine proteases.
Fig. 1.
Reaction Chemistry: Reaction mechanism of cysteine protease mediated cleavage of peptide bond.
Fig. 2.
Papain structure: Structure of papain (Kozak et al., 1996).
1.3. Mammalian cysteine proteases
In mammals, there are four main groups of cysteine proteases viz. lysosomal cathepsins (cathepsins: B, C, H, K, L, M, N, S, T, V, and W) [5], [6], cytosolic calpains (calpain type I, calpain type II), caspases and legumains.
1.3.1. Cathepsins
Cathepsins are a group of protease enzymes originally discovered in the cell lysosome, with several members ubiquitous in the human body. They are not catalytically conserved: cathepsins A, G are serine proteases; cathepsins D, E are aspartate proteases; and the remainder are lysosomal cysteine proteases, including the human isoforms B, C, F, H, K, L, O, S, V, X and W [7]. The optimum pH for cathepsins activity is slightly acidic, corresponding to the environment found in the lysosome. Although, they have been traditionally viewed as enzymes involved in terminal protein degradation, knockout (KO) mice have revealed major roles in cell regulation, i.e. of cell proliferation and adhesion, apoptosis, lipid metabolism and immune response [8], [9]. Cathepsin B is exclusive among the cathepsins in that it has an occluding loop, a peptide sequence which when closed can hinder access to the primed side of the substrate pocket. Thus, cathepsin B can function as an endopeptidase/exopeptidase depending upon pH [10]. The occluding loop has been targeted for the design of non-electrophilic cathepsin B inhibitors [11]. The lysosomal cathepsin K occurs in osteoclasts and is a major factor in bone resorption and a target for treating osteoporosis. Table 2 shows residue preference of cathepsin B in peptide substrates in each position [12], [13].
Table 2.
Residue preference: Residue preference of cathepsin B in peptide substrates in each position.
| Unprimeda | Preference | Primeda | Preference |
|---|---|---|---|
| P1 | Gly > Ala, Met, Gln | P1′ | Phe > Gly |
| P2 | Val > Phe, Tyr | P2′ | Val, Ile > Gly, Thr |
| P3 | Gly > Lys, Phe | P3′ | Gly |
1.3.2. Caplains
These are neutral, cytosolic cysteine proteases with 15 reported isoforms; 11 identified in humans [14], [15]. Calpain is an intracellular Ca2+-dependent cysteine protease. The first reports characterizing members of this family emerged in 1964, naming the enzyme calcium-activated neutral protease (CANP). The name calpain was introduced in 1981 which refers to the analogy of the cysteine protease with papain [16]. Calpains are found in almost all eukaryotes and some bacteria, but not in archaebacteria. Fifteen genes within the human genome encode a calpain-like protease domain. Interestingly, some human calpains, particularly those with non-classical domain structures, are very similar to calpain homologs identified in evolutionarily distant organisms. Three-dimensional structural analysis has helped to identify calpains unique mechanism of activation; the calpain protease domain comprises of two core domains that fuses to form a functional protease only when bound to Ca2+ via well-conserved amino acids.
1.3.3. Caspases
Caspases belong to a family of highly conserved cysteine-dependent aspartate-specific acid proteases that uses a cysteine residue as their catalytic nucleophile and share a stringent specificity for cleaving their substrates after aspartic acid residues in target proteins [17]. A member of this family was first identified as the protease responsible for the proteolytic maturation of pro-IL-1β (interleukin-1β-converting enzyme: ICE) [18], [19] and later it was purified and cloned as caspase-1 [20], [21]. The first three-dimensional model structure of caspase-1 was constructed in 1994 [22] along with the identification of first mammalian caspase homologue (caspase-2) [23], followed by the development of first caspase-1 knockout mice [24], [25]. All caspases exist within the cell as inactive latent pro-forms as precursor zymogen. X-ray crystal structure and peptide sequencing studies have confirmed that these nascent caspases are originally synthesized as catalytically-dormant tripartite proenzyme (a single polypeptide chain of 32–55 kDa representing 3 domains in common) with a 17–21 kDa central large internal domain (p20) containing a large catalytic subunit (active site), a 10–13 kDa small C-terminal domain (p10) also called small catalytic subunit, and a 3–24 kDa NH2-terminus prodomain called death domain (DD) [26].
1.3.4. Legumains
Legumain is the name that was given in 1993 to an endopeptidase that is present in many leguminous and other seeds, after it was isolated and characterized from Vigna aconitifolia (moth bean). Legumain is a cysteine endopeptidase showing strict specificity for hydrolysis of asparaginyl bonds. The enzyme belongs to peptidase family C13, and is unrelated to the known cysteine peptidases of the papain family, C1 [27].
2. Cystatins
Cystatins are virtually the most widely represented class of cysteine proteinase inhibitors, ubiquitously distributed in plants and animals, and devoted to regulating degradation of both intracellular and extracellular proteins. Cystatins function as competitive inhibitors which coordinate the biological activity of C1 class (papain-like) cysteine proteases that are involved in proteolytic processes [28]. Cystatins bind to the active site of cysteine proteases as pseudo-substrates thus making target cysteine proteases unable of cleaving peptide bonds. Ensuring proper equilibrium between free cysteine proteases and their complexes with inhibitors is critical for proper functioning of all living systems and in this regard cystatins are general regulators of harmful cysteine protease activities. The first cystatin to be isolated was a papain and ficin inhibitor characterized from chicken egg white [29]. The name ‘cystatin’ was proposed for the first time by [30] and was later used to describe homologous proteins in the same superfamily [31].
2.1. Cystatin superfamily
At the First International Symposium (Portoroz, Yugoslavia, September 1985) on cysteine proteinases and their Inhibitors, the subject and nomenclature of the cysteine proteinase inhibitors homologous with chicken egg white cystatin were discussed. In the symposium, it was decided that proteins that can be shown statistically to have an evolutionary relationship to chicken cystatin would comprise the cystatin superfamily. The cystatin superfamily encompasses proteins that contain multiple cystatin-like sequences. Some of the members are active cysteine protease inhibitors, while others have lost or perhaps never acquired this inhibitory activity. In recent years, several new members of the superfamily have been characterized, including proteins from insects and plants. Based on partial amino acid homology, new members, such as the invariant chain (Ii), and the transforming growth factor-β receptor type II (TGF-β receptor II) may, in fact, represent members of an emerging family within the superfamily that may have used some common building blocks to form functionally diverse proteins. Cystatin super-family members have been found throughout evolution and members of each family of the superfamily are present in mammals today. endogenous or external proteolysis [32]. The proteins which collectively form the cystatin superfamily have been subdivided into three families, namely the stefins (Family 1), the cystatins (Family 2) and the kininogens (Family 3); recently determined protein sequences indicate that the classification could be extended to include additional families. In present times, a fourth family has been included which is known as phytocystatins and it includes all cysteine proteinase inhibitors isolated from plant [33].
2.2. Stefin family (TYPE I CYSTATINS)
These are intracellular cystatins that are present in the cytosol of many cell types, but can also appear in body fluids at significant concentrations [34]. They are single-chain polypeptides of ∼100 amino acid residues which lack disulfide bonds and also devoid of carbohydrate content. Although, it was suggested that stefins are intracellular proteins, they have also been found in extracellular fluid [34]. The type 1 cystatins are called as ‘stefins’ to stress their difference from other cystatin superfamily members. However, they have a general structure very similar to the ‘cystatin fold’ of other cystatins and consequently a similar activity as cysteine protease inhibitors. Stefins are stable in the neutral and alkaline pH range; heat stability is another trait shown by stefins. Stefins are potent reversible and competitive inhibitors of cysteine proteinases. The inhibition constant for papain with human stefin A is 1.9 x l0−11 M. [32]. The stefins have been isolated and characterized from different mammalian tissues including humans. This family comprises mainly of stefin A (cystatin A or α) and stefin B (cystatin B or β) [35]. Additionally, stefin C was discovered in bovine thymus [36] and stefin D in pigs [37].
2.2.1. Stefin A (Cystatin A or α)
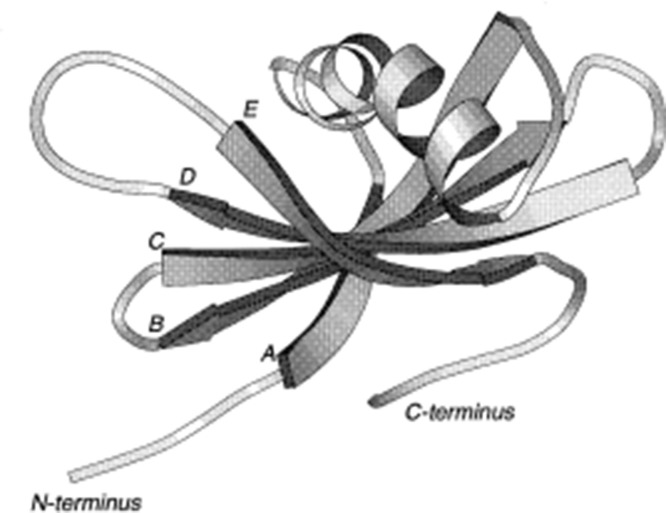
Human stefin A is a member of the cystatin superfamily of proteins that is found in high concentrations in various types of epithelial cells and in polymorphonuclear leucocytes. Human stefin A occurs in multiple isoelectric forms, with pI values in the range of 4.5–5.0 [38]. Immunohistochemical studies have shown the presence of stefin A in skin and epithelium, suggesting that the major function of stefin A is related to protection of these organs against over reactivity of cysteine proteases [39]. The gene coding for human cystatin A has been assigned to chromosome 3 [40]. It has been suggested that stefin A provides an important protective function as an inhibitor of cysteine proteinases that is being utilized as invasive tools by many infective agents. Stefin A is also implicated in number of diseased states viz. in several epimeroid carcinomas including squamous cell carcinomata of the lung, skin, vulva, cervix and oesophagus [41], [42] but was absent from a variety of other carcinomas [43]. Additionally, the serum level of stefin A has been shown to increase significantly in patients with cardiovascular disease [44] and the inhibitor has been found in the upper spinous layer of psoriatic cells. Stefin A has been found to be a sensitive marker during HIV-1infection and for regeneration of follicular lymphoid tissue [45]. Cystatin α is a species variant of cystatin A, found in rats and this variant was characterized by Jarvinen [46] as a specific inhibitor of cysteine proteases from rat skin having molecular weight of 13 kDa. Fig. 3 gives an illustration of the polypeptide fold of the average energy-minimized structure of stefin A.
Fig. 3.
Stefin A: Ribbon representation of the minimized average structure of stefin A, illustrating 5-stranded antiparallel β-sheet (with the strands marked A to E) wrapped around the central α-helix with the C-terminal loop running along the convex face of the sheet. This figure was created using the program MOLSCRIPT (Kraulis, 1991).
2.2.2. Stefin B (cystatin B)
Stefin B (cystatin B) is an endogenous cysteine cathepsin inhibitor localized in cytosol, mitochondria and nucleus. Its distribution throughout the tissues suggests that this inhibitor interacts with cathepsins liberated from lysosomes. The gene coding for human stefin B is assigned to chromosome 21 [47]. Unlike human stefin A, human stefin B is a more neutral protein with pI values in the range 5.9-6.5. It was detected as an inhibitor of cathepsin B and H in human tissues [48]. There is one cysteine residue in the stefin B sequence, and it can be converted into an intermolecular disulfide bridge (—Cys3 —Cys3′ —) resulting in the formation of inactive dimers; easily transformed back into active monomers under reducing conditions [49]. The appearance of dimers in small quantities has already been reported [50]. Cystatin β is a species variant of cystatin B; found in rat liver [51]. It has a pI value ranging from 5.04–5.6 [52]. It is more abundant than cystatin α in all tissues except skin. Cystatin B has been found to be a potential serum marker in hepatocellular carcinoma [53]. Mutations in the gene of stefin B are associated with the neurodegenerative disease known as Unverricht-Lundborg disease (EPM1) [54].
2.2.3. Stefin C
This protein consists of 101 amino acid residues, with an estimated molecular weight of 11,546 Da. This arises from the cleavage of Asn 5 − Leu 6 bond in the inhibitor [36]. The inhibitor was found to be acidic with pI values from 4.5 and 5.6. This inhibitor exhibits considerable sequence homology with all type B stefins and especially with bovine stefin B. Stefin C differs from bovine stefin B in a way that it has prolonged N-terminus, presence of a tryptophan residue, occurrence of one methionine on the N terminus unlike bovine stefin B which has two methionine and existence of leucine residue instead of cysteine residue [36].
2.3. Cystatin family (type 2 cystatins)
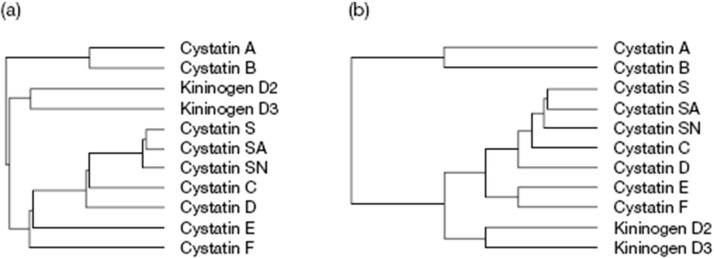
The name cystatin was first used to describe an inhibitor that had been discovered and partially characterized from chicken egg-white of papain, ficin and other related cysteine endopeptidases [29]. When other protein inhibitors of cysteine proteinases were characterized and their amino acid sequences determined, it became obvious that they are related to chicken cystatin, and thus they are members of the cystatin superfamily [31]. Simultaneously, it was decided that the second family within the cystatin superfamily may also be called the cystatin family. The main characteristic of the inhibitors of the cystatin family is the presence of two disulfide bonds located towards the carboxyl-terminus. They consist of 120–122 amino-acid residues and are synthesized as proproteins containing a signal peptide (20 residues), which suggests that cystatins display an extracellular activity [55]. They are broadly distributed and found in most body fluids [34]. The members of this family contain a conserved Gln-Xaa-Val-Xaa-Gly segment known as the ‘cystatin motif’ in the central part of their sequences. Mammalian and avian type 2 cystatins contain two conserved disulphide bridges in the C-terminal end of the sequence, with 10 and 20 residues in between the respective cysteines, and a characteristic Pro-Trp pair in their C-terminal segments. The members of this family may be phosphorylated (Laber et al., 1989). The cystatin family comprises of the following human cystatins: C (hCC), D (hCD), E (hCE), F (hCF), S (hCS), SA (hCSA) and SN (hCSN). Their homologues have also been found in other mammalian organisms and birds. Human cystatins are coded on chromosome 20 [56]. Cystatins S, SA, and SN (S-type cystatins) consisting of 121 amino-acid residues with molecular mass of 14.2–14.4 kDa display high sequence homology (90%). Post-translational phosphorylation of cystatins S and SA leads to formation of several isoforms. Expression of these cystatins is very restricted: cystatin SN is found only in saliva and tears, whereas variants S and SA are also present in seminal fluid [57]. Fig. 4 gives an overview of the relationship between various human cystatins. Some of the important members of this family are discussed in detail below:
Fig. 4.
Relationship Analysis: Relationships between human cystatins. Human cystatin sequences were analyzed using algorithm in the GCG package, to show their similarity (a) and evolutionary relationships (b). (Abrahamson et al., 2003).
2.3.1. Cystatin C
Human cystatin C (hCC) is a small protein belonging to the cystatin family of papain-like cysteine proteinase inhibitors. hCC is broadly distributed and found in most body fluids [58]. This small protein consists of 120 amino acids forming a single polypeptide chain. Cystatin C contains four conserved cysteine residues that can form two disulfide bonds but, unlike other family members, was neither shown to be glycosylated (cystatin E/M and cystatin F) nor phosphorylated (chicken cystatin) [59]. Interestingly, hCC can be found as monomer, dimer or oligomer also in fibril states [60]. Biologically, hCC is a target of proteolysis, and primarily functions as a protease inhibitor. It is degraded by cathepsin D and elastase [61]. The hCC concentration in blood correlates with the glomerular filtration rate (GFR) [62] an important marker of kidney health and the progression of diabetes, chronic kidney disease, etc. [63], [64]. Considering its biological importance, hCC concentrations have been linked to a number of other diseases (Table 3 ). The physiologically relevant oligomeric state of hCC is a monomer [65]. Fig. 5 depicts ribbon + representation of hCC dimer [66], [67].
Table 3.
Relation between altered hCC level and human diseases.
| Observation | hCC response |
|---|---|
| Hereditary cystatin C amyloid angiopathy (Icelandic form) |
Lower mutant hCC levels correspond to disease. hCC is deposited as amyloid plaques |
| Rheumatoid arthritis | Higher hCC levels corresponds to disease and inflammation |
| Cardiovascular disease | Higher hCC levels correspond to disease. Results uninfluenced by age, sex or body-mass index of patients |
| Sub clinical brain infarction | Higher hCC levels corresponds to disease |
| Stroke | Higher hCC levels corresponds to disease |
| Alzheimer’s disease | High hCC concentrations are toxic, optimal hCC concentrations protect neurons against amyloid deposition and degeneration |
| Multiple sclerosis | Cleavage of carboxy terminus of hCC |
| Diabetes | Higher hCC levels corresponds to disease |
| Neurodegenerative diseases | optimal hCC concentrations protect neurons against amyloid deposition and degeneration |
| Atherosclerosis, abdominal aortic aneurysm | Lower hCC levels cause an increased activity in cysteine protease |
| Shrunken pore syndrome | A specific pattern of the ratio of five glomerular filtration rate markers suggests that pore diameter in glomerular membrane is reduced |
| Dementia | Polymorphism of hCC gene is associated with higher or lower risk of disease |
| Age related macular degeneration | Recessive inheritance of the hCC A25 M mutant leads to a higher incidence of disease which is characterized by plaque formation. The A25T mutation is also related to this disease and the mutation occurs in an aggregation-prone region that also affects cystatin C signal cleavage. |
| Breast cancer | Cystatin C is inducible by p53 leading to suppressed protein concentrations in breast cancer. Low cystatin C concentrations correlate with poor breast cancer prognosis |
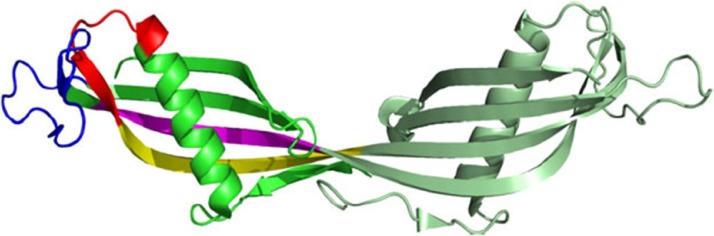
Fig. 5.
Ribbon representation of the hCC dimer: The interacting regions of β2 and β3 strands proposed by He (2013) are highlighted in yellow and magenta, respectively, whereas Shen’s (2012) fragments are marked in red (helix-β2) and blue (appendant structure). Figure prepared using PYMOL software using the hCC dimer. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3.2. Cystatin D
Cystatin D has been found mostly in saliva and tears. Fully active cystatin D, formed after removal of the 20-residue signal peptide, consists of 122 amino acid residues with molecular mass about 13.8 kDa. The protein exists in 2 polymorphic forms: [Cys26] hCD and [Arg26] hCD which have identical activity, stability, and distribution. The low homology with other cystatins (51–55%) suggests that, on a phylogenetic tree, cystatin D is located between cystatins S and C [68].Cystatin D has a more restricted pattern of tissue expression and a narrower inhibitory profile than other cystatins: it inhibits cathepsin S, H, and L but not cathepsin B [69]. In human colorectal tumors, expression of cystatin D is correlated with expression of VDR and E-cadherin, and loss of cystatin D is correlated with poor tumor differentiation. In recent times, it was reported that a proportion of cystatin D protein localizes within the cell nucleus at specific active chromatin sites and regulates gene transcription [70]. Cystatin D has been found to be a potent inhibitor of coronavirus replication [71].
2.3.3. Cystatin F
Cystatin F was discovered by three independent groups. Two of them identified the new inhibitor by cDNA cloning and named it leukocystatin and cystatin F [72], [73]. The third group found over expressed mRNA encoding cystatin F in liver metastatic tumors and identified it as CMAP (cystatin-like metastasis associated protein) [74]. Human cystatin F is synthesized as a 145 amino acids pre-protein with a putative 19 residues signal peptide. Although it is made with a signal sequence, only a small proportion is secreted and importantly, it is secreted as a disulphide- linked dimer [75] which is inactive until it is reduced to its monomeric form [76]. It is glycosylated and mannose-6-phosphate modification of its N-linked saccharides is used for targeting to the endosomes and lysosomes [77]. Glycosylation at Asn62 is proposed to protect the intermolecular disulphide from reduction, explaining unusually strong reducing conditions needed to monomerize dimeric cystatin F in vitro [78]. Cystatin F tightly inhibits cathepsins F, K, V, whereas cathepsins S and H are inhibited with lower affinities and cathepsin X is not inhibited at all [76]. It is quite evident that cystatin F plays a role in immune response-related processes through inhibition of specific enzyme target. C13 cysteine protease involved in antigen processing, mammalian legumain or asparaginyl endopeptidase (AEP), is also inhibited by cystatin F, although it shows reduced affinity for AEP compared with cystatins C and E/M [79]. The inhibitor is expressed selectively in immune cells such as cytotoxic T cells, natural killer cells (NK cells), monocytes, DCs (Fig. 6 ).
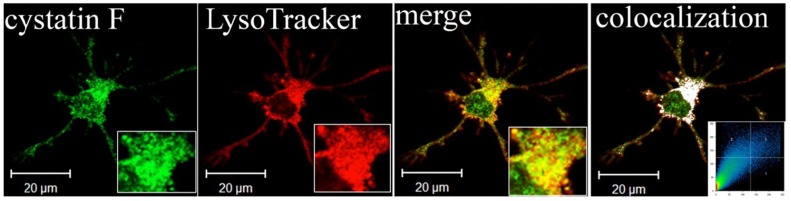
Fig. 6.
Lysosomal localization of cystatin F in adherent dendritic cells (DCs): Part of image is magnified in lower right corner. The white colour indicates colocalization of two labelled antigens, confirming the presence of cystatin F in lysosomes. The threshold value for colocalization was set to one half of the maximal brightness level. The mask of the pixels above the threshold in both channels (significant colocalization, blue colour) and the contour plot are shown (Magister et al., 2012). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3.4. Cystatin E/M
For cystatin M a down regulated mRNA in metastatic breast tumor cells was identified by differential display when compared to normal and primary breast tumor cells. 82 Independently, the same molecule was found by others in cDNA libraries derived from epithelial cells, and was designated cystatin E [80]. Cystatin E/M is a 14.5 kDa secreted protein that has an overall structure similar to other cystatin family members, such as a signal peptide and two intra-chain disulfide bonds. It possesses the unusual trait of being a glycoprotein, carrying an N-linked carbohydrate chain at position 108 [81]. This protein is only distantly related to the other known family members as reflected by the genomic position of the cystatin E/M gene on chromosome 11q13 [82], whereas all other family II cystatin genes are clustered in a narrow region on chromosome 20p11.2 [83]. Cystatin E/M is synthesized as a pre-protein with a putative 28 residues while mature cystatin E/M contains 121 amino acids [80]. High expression levels are largely confined to skin epithelia, which emphasize its prominent role in cutaneous biology [84]. Its expression was also recently reported in breast tissue [85] and in oligodendrocyte and astrocyte-like cells of human brain [86]. Cystatin E/M is a high-affinity inhibitor of cathepsins V, L [87] and asparaginyl endopeptidase/legumain, it binds to cathepsin B with lower affinity. In vivo mouse models have revealed that cystatin E/M is a key molecule in a biochemical pathway that controls skin barrier formation [88], [89].
2.3.5. Cystatin S
Cystatins S, SN and SA were first isolated from human saliva and later identified as a cysteine protease inhibitors and their role in controlling cysteine proteases derived from bacteria has been suggested [90], [91], [92], [93]. These three proteins with similar sequence (88% identity at the protein level) are distantly related to cystatin D (less than 60% identity at the protein level with cystatins S, SN and SA). Although they possess inhibitory activity, they are all poorer inhibitors of cysteine cathepsins than cystatin C [94]. Inhibition of bacterial cysteine proteases has been tested by cystatins C, S N, S and chicken cystatin and they are generally inactive against bacterial cysteine proteases [95], [96], [97]. Cystatins S, SA, and SN (S-type cystatins) consisting of 121 amino-acid residues with molecular mass of 14.2–14.4 kDa. Post-translational phosphorylation of cystatins S and SA leads to formation of several isoforms. Expression of these cystatins is very restricted: cystatin SN has been found only in saliva and tears, whereas variants S and SA are also present in seminal fluid [98]. In contrast to bacterial cysteine proteases, they are more efficient against cysteine proteases from parasites; for example, cystatins SA and SN are inhibitors of trypanosome cruzain [99]. Cystatin SN has been found also in dendritic cells exposed to Toxoplasma gondii [100] and it could thus modulate antigen presentation. Cystatins S, SA and SN can suppress the infectivity of adenovirus [101] and herpes simplex virus 1 [102].
2.4. Kininogens family (type 3 cystatins)
The third family consists of 3 members: human high molecular mass kininogen (hHK), about 120 kDa; human low molecular mass kininogen (hLK), about 68 kDa; and kininogen T, discovered so far only in rats. These proteins are high-molecular-mass cystatins representing three tandemly repeated type 2-like cystatin domains, with a total of eight disulphide bridges (six conserved and two additional at the beginning of cystatin domains D2 and D3). They are glycosylated proteins, but the carbohydrate attachments sites are not present in the cystatin domains. In these cystatins, only second and third cystatin domains (D2 and D3) present cysteine peptidase inhibitory activity [103]. Kininogens are intravascular in localization; they are found in blood plasma and, as a result of diffusion, in synovial and amniotic fluids. Like type 2 cystatins, cystatin domains D2 and D3 with inhibitory activity in human kininogens are grouped in Subfamily B of MEROPS Family I25. The first chemically characterized kinin was the nonapeptide bradykinin (Arg1-Pro2-Pro3-Gly4-Phe5-Ser6-Pro7-Phe8-Arg9). Following a preliminary note by Abelous & Bardier in 1909 [104] between 1926 and 1939, Werle and colleagues described in their pioneering studies the nature and functional relationships of the basic components of the kallikrein-kinin-system (KKS): (tissue) kallikreins, kinins, kininogens and kininases and particularly numerous pharmacological effects of peptide hormone kinins [105]. Kininogens of low molecular weight have a heavy chain with four domains (D1, D2, D3 and D4, which contains the bradykinin moiety) and a light chain with the D5 domain and carboxy terminal. Kininogens of high molecular weight have the same structure as kininogens of low molecular weight except that the light chain has two domains (D5 and D6). T-kininogen (also called as thiostatin) is found only in rats. It is an exception to the general rule for kininogens, since it is not susceptible to kallikrein hydrolysis. However, cathepsins D can liberate T-kinin from T-Kininogen [106]. T-kininogen has been reported to have strong inhibitory capacity against papain and cathepsin L [107]. A recent study shows that kallikrein-kinin system (KKS) is involved in cardiac function changes and myocardial inflammatory infiltration in response to local irradiation.
2.5. Phytocystatin family (type 4 cystatins)
Peptidase inhibitory proteins are a complex group of molecules involved in the regulation of the protein degradation caused by peptidases. The MEROPS database (http://merops.sanger.ac.uk) is an integrated source of information that classifies peptidases and their inhibitors into families and clans [108]. Members of 21 of the 78 families recognized in the current MEROPS 10.0 version have been identified in plants [109]. From that, some of the most abundant peptidase inhibitors are the phytocystatins, which belong to the ubiquitous family of the cystatins (MEROPS identifier I25) [110]. Unique structural features and phylogenetic inferences suggest a specific evolution for phytocystatins and support their inclusion in a specific plant cystatin family referred to as phytocystatin family. Most phytocystatins are small proteins with a molecular mass in the 12–16 kDa range and are inhibitors of the cysteine proteases from the C1A papain-like family. The unique feature of this superfamily is a highly conserved region of the G58 residue, the QVVAG motif and a PW motif. Studies on oryzacystatin and its various truncated forms have identified the conserved QVVAG motif as a primary region of interaction between the inhibitor and its cognate enzyme. The PW motif is believed to act like a cofactor [111]. In land plants, some members with a molecular weight of approximately 23 kDa have a carboxy-terminal extension involved in the inhibition of a second family of cysteine proteases, the C13 legumains [112]. In addition, several 85–87 kDa multi cystatins, with eight cystatin domains, have been described in dicots [113]. The inhibitory properties of phytocystatins are a consequence of a tight and reversible interaction with their target enzymes. It involves a conserved tripartite wedge formed by the partially flexible N-terminus containing one or two glycine residues and two hairpin loops carrying a conserved QxVxG motif and a tryptophan residue, respectively [114].
2.6. Othet cystatins and cystatin-like proteases
A number of proteins have been described which, in spite of high sequence homology, show distinct differences in structure and biological activity in comparison with cystatins. Histidine-rich glycoproteins (HRG) and fetuins are examples of such cystatin-like proteins. Histidine-rich glycoprotein (HRG) contains two cystatin-like domains and the conserved disulfide bond but is not inhibitory in protease assays [115]. The physiological function of HRG is not known. On the other hand, a related protein known as fetuin-A [116] has been shown to be involved in a number of physiological functions including some that it shares with the other cystatin-family members. Like HRP, bovine fetuin-A and its human homologue alpha 2HS glycoprotein (ahsg) contain two cystatin like domains but lack the inhibitory activity. The subject of cystatins and cystatin-like proteins has been reviewed [115]. On the other hand, there are also proteins, like the intensely sweet plant protein monellin which, in spite of very low sequence homology and lack of inhibitory function, have a cystatin-like three-dimensional structure [117]. Other proteins that contain cystatin like domains but lack the inhibitory activities are the cystatin-related protein (CRP), an androgen-regulated protein expressed in prostate [118], [119] and Spp24, a bone phosphoprotein. These duos contain disulfide bonds, but only a single cystatin domain which lack inhibitory activity and may have evolved from a family-2 precursor. Celostatin is a cystatin from crested cock’s comb (Celosiacristata) has been cloned and characterized [120]. Crestatin was another one that was found in mouse epididymis, showing substantial homology with those of well-established protein inhibitors, cystatins [121].
3. Cysteine protease-cystatin interaction
Numerous spectroscopic, kinetic, and crystallographic studies have been carried out to explain the mechanism of cysteine protease inhibition by cystatins. The results have shown that the inhibitor binds in a one-step process that is simple, reversible, and second-order type. In addition, those studies have revealed that enzymes with a blocked active centre could still bind cystatins, albeit with lower affinity. This indicates that cysteine protease–cystatin interactions are not based on a simple reaction with the catalytic cysteine residue of the enzyme, as is typical of substrates, but that they consist of hydrophobic contacts between the binding regions of cystatins and the corresponding residues forming the binding pockets of the enzyme. Despite their structural homology and similar mode of inhibition, cystatins display quite different enzyme affinity (Table 4 )
Table 4.
DISSOCIATION CONSTANT (Ki) values of different cystatins.
| Cystatin | Papain | Cathepsin B | Cathepsin H | Cathepsin L | Cruzipain |
|---|---|---|---|---|---|
| Stefin A | 0.019 | 8.2 | 0.31 | 1.3 | 0.0072 |
| Stefin B | 0.12 | 73 | 0.58 | 0.23 | 0.060 |
| Cystatin C | 0.00001 | 0.27 | 0.28 | <0.005 | 0.014 |
| Cystatin D | 1.2 | >1000 | 7.5 | 18 | n. d. |
| Cystatin E/M | 0.39 | 32 | n. d. | n. d. | n. d. |
| Cystatin F | 1.1 | >1000 | n. d. | 0.31 | n. d. |
| Cystatin S | 108 | n. d. | n. d. | n. d. | n. d. |
| Cystatin SA | 0.32 | n. d. | n. d. | n. d. | n. d. |
| Cystatin SN | 0.016 | 19 | n. d. | n. d. | n. d. |
| Chicken cystatin | 0.005 | 1.7 | 0.06 | 0.019 | 0.001 |
| L- kininogen | 0.015 | 600 | 0.72 | 0.017 | 0.041 |
| H-kininogen | 0.02 | 400 | 1.1 | 0.109 | n. d. |
4. Role of cystatins in various diseases
4.1. Cystatins in cancer and tumor immune response
There are an accumulating number of studies showing that cystatins are implicated in the progression of various types of cancer. In general, their localization and regulation of tumor associated proteolytic activity can be recognized by two distinct mechanisms. Type I cystatins, stefins A and B are up-regulated in tumor tissue and, up to a certain level could counter-balance the over-expressed tumor- associated proteolytic activity. Their function as tumor suppressors is supported by survival analysis, which associated their high levels with better outcome of cancer patients. For example, in brain tumors, increase of cathepsin B expression and lower levels of its inhibitor stefin A were associated with tumor malignancy [122].
On the other hand, type II cystatins, at least cystatins C and E/M are generally down-regulated in tumors. Although their role remains protective, their lower levels could allow a surplus of harmful tumor associated proteolytic activity. Outside the cells, higher levels of type II cystatins may impair extracellular activity of cysteine proteases, associated directly or indirectly with the degradation of ECM and resulting in tumor cell invasion and metastasis. However, higher levels of cystatins in body fluids have been associated with poor prognosis in cancer patients supporting their role in regulation of proteases involved in the regression of tumors. In melanoma and colorectal cancer, increased extracellular levels of cystatin C as well as stefins correlated significantly with high risk of adverse outcome in cancer patients [123]. Cathepsin B/cystatin C complex was also found to be less abundant in sera of patients with tumors suggesting an imbalance between the enzyme and its inhibitor in cancer patients [123]. Animals with excluded expression of type I or type II cystatins experience better outcome with regard to tumor growth and metastasis as the wild type ones. These contradictive results can be explained by the fact that type II cystatins are involved also in processes resulting in tumor regression such as anti- tumor immune response, apoptosis, cell migration and seeding [124]. In particular, the role of cysteine proteases is very important for proper maturation of antigen presenting cells, antigen processing and the presentation to T cells; therefore, the enhanced inhibition may affect the activation of naive T cells by tumor associated antigens and impair T cell dependent anti-tumor immune response, known to be effective for eradication of tumor cells [125], [126]. On the other hand, patients with higher local levels of stefins A and B in non-small cell lung tissue than in control lung tissue exhibited a better survival probability implying on their ability to counteract harmful tumor-associated proteolytic activity [127]. Also, the over-expression of cystatin C in tumor cells inhibited melanoma metastasis formation [128].
4.2. Type II cystatins in pathological processes
Changes in cystatin C expression and localization have been associated with various neurodegenerative pathologies. For example, a point mutation inthe cystatin C gene, resulting in the substitution of Leu to Gln, is responsible for the dominantly inherited icelandic type of amyloidosis, hereditary cystatin C amyloid angiopathy (HCCAA). Patients with this disease suffer from successive brain hemorrhages concluding in death at the age of 30. In first line the formation of cystatin C aggregates are responsible for the outcome and progression of the disease, however, the loss of inhibitory function due to aggregation and lower concentration in brain cannot be excluded [129].The levels of cystatin C were highly reduced in cerebrospinal fluid of individuals suffering from multiple sclerosis (MS) compared with healthy individuals. Its low concentration suggests that the inhibition of cysteine proteinases is impaired in this disease; hence higher activity of cysteine proteinases could initiate or increase the breakdown of myelin [130]. Cystatin C was shown to be implicated in Alzheimer's disease (AD). It co-deposits with amyloid-beta (Aβ) in amyloid plaques of AD patients [131], associates with Aβ and inhibits Aβ oligomerization in vitro and in vivo [132]. Thus, cystatin C could protect the brain from amyloid-induced toxicity and may have therapeutic implications for AD. However, there are opposing results demonstrating neuronal cell death upon injecting cystatin C into the brain of AD mouse model, showing that cystatin C triggers Aβ accumulation by inhibiting cathepsin B-induced Aβ degradation [133]. Cystatin C may have an important role in brain injuries as its enhanced expression has been observed in response to different types of insults to the brain, such as ischemia and epilepsy [134], [135]. Regarding constant serum concentrations of cystatin C it is a suitable marker for glomerular filtration rate (GFR) and kidney function and thus has a possible application as a replacement for creatinine [136], [137]. Studies on the sensitivity and specificity of cystatin C for detecting impaired GFR strongly suggest it to be superior diagnostic marker to creatinine for detecting impaired GFR [138]. It has been suggested that disturbance of the cystatin E/M–cathepsin pathway could contribute to dysregulated skin barrier function as observed in the inflammatory dermatoses. Atopic dermatitis and psoriasis are two common chronic inflammatory skin diseases in which the expression of many genes and the formation of the epidermal barrier are altered. Recent genetic studies have revealed that abnormalities in epithelium-expressed genes are an important etiological factor [139], [140], [141].
4.3. Role of cystatins in neurodegenerative diseases
Cerebrovascular amyloid deposition (CAA) is a disease characterized by amyloid protein deposition in the blood vessels of the brain. Severe forms of the disease may cause cerebrovascular disorders such as lobar cerebral hemorrhage and leukoencephalopathy and may present with dementia such as that observed in Alzheimer’s disease (AD) [142]. Several cerebrovascular amyloid proteins have been characterized and include amyloid-β-protein type (Aβ), cystatin C, prion protein, variant transthyretins (ATTR) in meningovascular amyloidoses, mutated gelsolin (AGE l) in familial amyloidosis of Finnish type, disease associated prion protein (PrP(Sc)) in a variant of the Gerstmann-Straussler-Scheinker syndrome [143]. Among the several types of CAA, that of the Aβ type is the most commonly found in elderly individuals and in patients with AD and mutations found in the genes encoding amyloid precursor proteins are associated with hereditary CAA. Studies have shown cystatin C to be immunohistochemically co-localized with Aβ in sporadic CAA [144], [145] and to co-immunoprecipitate with amyloid-beta precursor protein [146]. However, in such cases examined to date, cystatin C has not been shown to be an intrinsic component of the amyloid fibrils and more definitive studies must be conducted to elucidate the significance of the immunohistochemical co-localization of cystatin C. The association of cystatin C with Aβ in AD patients may have a hitherto unexplored role. Apart from simply being co-localized with Aβ in the brain, there are studies showing that cystatin C can also polymerize and give rise to amyloid bodies. In hereditary cerebral hemorrhage with amyloidosis icelandic-type (HCHWA-1), an autosomal dominant disorder in icelanders, cystatin C is directly involved in the pathogenesis of CAA. This disorder is associated with a (68Leu—Gln) mutation and a loss of 10 amino acids in the N-terminus of the cystatin C [147]. Concerning the formation of Aβ fibrils in the brain, it is generally believed that globular amyloidogenic proteins partially unfold and then switch into alternatively folded conformations to self assemble into fibrils (amyloid deposits) [148]. More specifically, cystatins and other amyloidogenic proteins have been shown to participate in domain swapping, which leads to formation of stable dimers and eventually stable fibrils in amyloidogenic deposits [149], [150]. Other than amyloidogenesis, cystatin C has been shown to be associated with multiple sclerosis (MS), another neurodegenerative disease. Studies have demonstrated that the level of cystatin C is highly reduced in the cerebrospinal fluid of individuals suffering from MS compared with healthy controls [151]. In other studies of patients with inflammatory neurological disorders, such as Guillain-Barre syndrome (GBS) and chronic inflammatory demyelinating polyneuropathy, levels of cystatin C were shown to be consistently lower in the CSF compared with control subjects. In the neurodegenerative disorder known as progressive myoclonus epilepsy of Unverricht-Lundborg type (EPMI), a significant reduction in cystatin B activity was recorded in lymphoblastoid cells of EPMI [152]. Concomitant to the decreased level of cystatin B, levels of cathepsin B, L, and S shot up significantly, as expected. Furthermore, mice with disruptions in the cystatin-B gene (loss of function mutation) display myoclonic seizures, progressive ataxia and cerebellar pathology, which closely parallels EPMI in humans [153].
5. Structure of cystatins
Clues as to which parts of the cystatin molecule are involved in binding to C1 peptidases were first obtained from functional studies. Controlled enzymatic cleavage, site-directed mutagenesis and fluorescence analysis of human cystatin C, CEW cystatin and kininogens [154], [155], [156] indicated three main peptidase-interacting segments. The first is the N-terminal region, where the well conserved Gly11 residue (human cystatin C numbering) is positioned. The second segment is located in the central part of the sequence, displaying the Gln-Xaa-Val-Xaa-Gly ‘cystatin motif’ at positions 55–59. The third segment includes the conserved Pro105-Trp106 pair in type 2 and 3 cystatins (type 1 cystatins present Pro-His/Gly in the corresponding positions). The three-dimensional structures of types 1 and 2 cystatins and of the plant cystatin, oryzacystatin confirmed the biochemical results and gave us an insight into how cystatins inhibit C1 peptidases. The structure models showed a very conserved tertiary structure, the so-called ‘cystatin fold’ (Fig. 7 ) which comprises a five-stranded anti-parallel β pleated sheet wrapped around a 5-turn α helix. The major difference between type 1 and 2 cystatins is the nearly 20-residue-long ‘appendix loop’ between strands 3 and 4 of the β-sheet in type 2 cystatins, which is missing from type 1 cystatins.
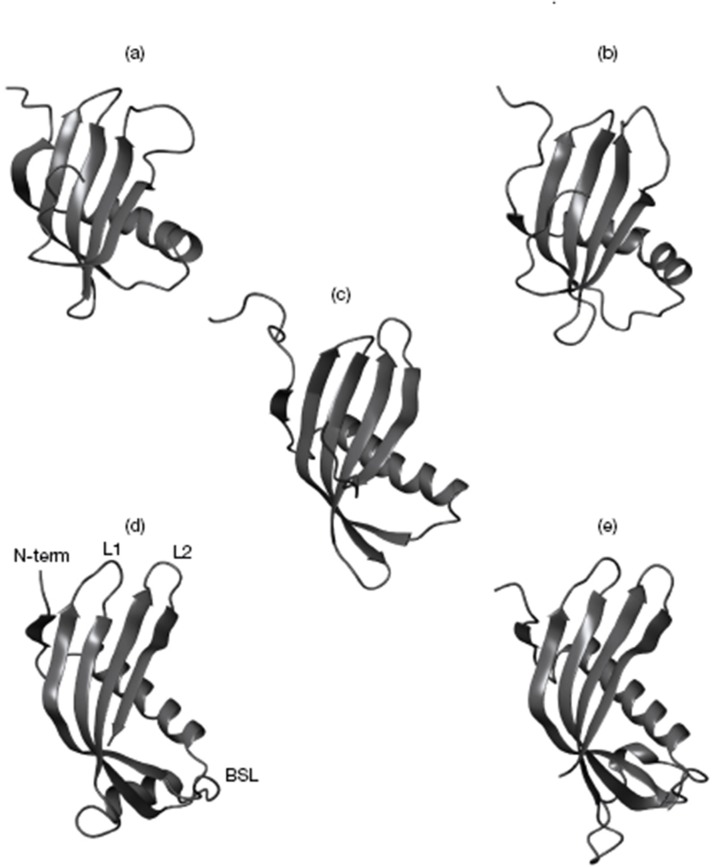
Fig. 7.
Cystatin structures: The cystatin structures known from X-ray crystallography or NMR spectroscopy studies are illustrated. (a) Human cystatin A (PDB code 1dvd) (Ni et al., 1998); (b) human cystatin B (1STF) (Stubbs et al. [159]); (c) oryzacystatin from rice (1EQG) (Nagata et al., 2000); (d) CEW cystatin (1CEW) (Bode et al. [162]); (e) theoretical cystatin C monomer, obtained by cutting the structure of the human cystatin C dimer (1G96) (Janowski et al., 2001) in half and modelling the L1 loop. The cystatin parts responsible for C1 peptidase binding and inhibition are indicated: the N-terminal segment (Nterm), the first (L1) and second (L2) hairpin loops. Also indicated is the BSL involved in the inhibition of leguman-like (family C13) cysteine peptidases (Abrahamson et al., 2003).
5.1. Primary structure
Few distinctive features can be owed to members of cystatin superfamily in terms of amino acid composition. Stefins are devoid of disulphide linkages (human cystatin A and rat cystatin α lack cysteine residues while human cystatin B and rat cystatin β have 1 and 2 cysteine residues, respectively) and tryptophan. However the presence of tryptophan in stefin C has also been reported. A unique feature of stefin B is the conserved QVVAG region in the stefins of mammalian origin, with Val 54 replaced by Leu 54. Grubb [157] identified the disulphide linked peptides from human cystatin C and chicken cystatin between Cys 71- Cys 81and Cys 95- Cys 115. Cystatin E apart from other usual conserved sequences characteristic of family II cystatins has a five residue insertion between amino acid 76 and 77 and a deletion of residue 91 (cystatin C numbering). Cystatin F is reported to have two more cysteine residues apart from the usual four cysteine residues, which is believed to stabilize the N-terminal part of the molecule in addition to the presence of second tryptophan residue, along with the conserved Trp 106 which is a characteristic of type II cystatins. The alignment of sequences of cystatins reveals common features, significant to the structure and activity of the proteins. Four residues are common to all the sequences of cystatins and inhibitory kininogen segments: Gly9, Gln53, Val55 and Gly57. These residues are considered to be of functional importance since they are absent from the non-inhibitory segment D1 of kininogens. Other six-conserved residues are Val47, Val55, Ala56, Tyr60, Cys71 and Tyr100. The segment Gln53 to Gly57 is the most highly conserved region.
5.2. Secondary structure
Based on sequence analysis of amino acids and overall structure of cystatin, the distribution of hydrophilic and hydrophobic residues were predicted and it was found that the hydrophilic residues are on the surface of proteins whereas hydrophobic regions are buried inside the core of protein, similar to other compact globular proteins. Each sequence possesses a large N-terminal hydrophilic region (15–42 residues) and a prominent central hydrophobic region (55–70 residues) followed by another large hydrophilic region at C-terminus. The structures of chicken cystatin and human cystatin C have been explored in depth. The secondary structure of hCC is very close to the structure of chicken cystatin [158]. The crystalline form of chicken cystatin exposed a new fold, a five-stranded anti-parallel β- sheet wrapped around the central N-terminal helix. This fold has been shown to exist in human cystatin C (hCC) (Fig. 8 a), chicken cystatin, cystatin D (Fig. 8b) as well as family I cystatin A and B [159], [160]. An appending segment of partial α-helix geometry is present in chicken cystatin [161], but absent in hCC [162]. Tryptophan was found only in second hairpin loops of cystatin. A unique feature was observed in crystals structure of cystatin F in a dimeric “off” state. The two monomers interacted in a fashion not seen before for cystatins or cystatin like proteins. This interaction was found to be crucially dependent on an unusual intermolecular disulphide bridge. The core sugars for one of the two N-linked glycosylation sites of cystatin F are well ordered and probably their conformation and interactions with the protein modulates its inhibitory properties in particular its reduced affinity towards asparaginyl endopeptidase compared with other cystatins [78]. Human cystatin C can undergo dimerization under mild denaturing conditions [150] significantly affecting the cysteine proteinase binding site [65] and account for the activity loss.
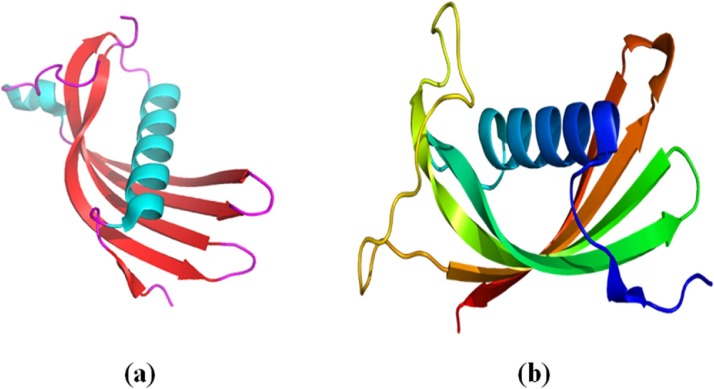
Fig. 8.
Crystal structure of: (a) human cystatin C monomer and (b) Human Cystatin D (Alvarez-Fernandez, 2005).
6. Emerging properties of cystatins
6.1. Stabilization of matrix metalloproteinases by cystatins
Matrix metalloproteinases (MMPs) are homologous Zn2+−dependent proteinases that participate in the physiological and pathological processes, which require tissue remodeling [163]. Their expression profiles are highly regulated and defects in their regulatory pathways are likely to give rise to diseases ranging from cancer, arthritis, and cardiovascular disorders to periodontal diseases [164], [165], [166], [167], [168]. Matrix metalloproteinases, like other proteinases, are secreted as zymogens and assume their activities in the extracellular milieu after activation processes [169]. For some MMPs such as MMP-9, the activation process is mediated by other MMPs [170]. Once activated, several proteinases including matrix metalloproteinases have the capacity for autolytic inactivation when they are in their active state [171], [172]. There are key mechanisms that have evolved in vivo to regulate the activities of these enzymes and prevent their autolytic inactivation. Tissue inhibitors of matrix metalloproteinases (TIMPs), the natural inhibitors of MMPs, interact with the enzymes in vivo and inhibit their activities. In the absence of these inhibitors, the activated enzymes if left unchecked, may turn against them and undergo autolysis. Therefore, under physiological conditions, which favor the active state of the enzymes and limited expression of the TIMPs, matrix metalloproteinases are unstable and can easily undergo autolysis unless some other protective mechanisms are in place.
6.2. Role of cystatin C as a new marker of glomerular filtration rate
For many years, serum creatinine has been the benchmark marker for the renal function assay also known as glomerular filtration rate (GFR) [173], [174]. The ideal marker for GFR should be produced endogenously and at a constant rate, regardless of age, sex, weight, and disease state. It should be filtered and excreted by the kidney only without renal tubular secretion and reabsorption, and once in urine it should be stable to allow for later analysis. Although creatinine meets some of these conditions, it has a number of limitations. Creatinine is produced in muscle, and the amount produced depends on muscle mass. It does not bind plasma proteins, and is freely filtered by the kidney although it is also secreted by the renal tubules. One major drawback for creatinine as a marker for GFR is that it is not a sensitive marker of early GFR, necessitating the collection of timed urinary samples and using a formula that requires measurement of serum and urinary forms of creatinine. Rhabdomyolisis and eating uncooked meats can dramatically raise serum creatinine, which can skew the readings of urinary creatinine. Because of these drawbacks related to creatinine, other endogenous markers for GFR have been evaluated. So far, the most promising marker which can potentially replace creatinine is cystatin C. Cyst C has proven to be a suitable marker because it satisfies most of the above established criteria. Simonsen [175] first noted the excellent correlation between Cyst C secretion and GFR when compared with the gold standard exogenous markers of GFR, such as Cr-EDTA. Cyst C is present in high concentrations in serum, saliva, and seminal, synovial, and cerebrospinal fluids. It is produced and secreted at a constant rate by most nucleated cells and is freely filtered by the glomerular because of its small size. Unlike creatinine, serum Cyst C is not secreted by renal tubular epithelial cells, although they reabsorb and catabolyze it so that Cyst C does not return to the bloodstream [176].
6.3. Immunomodulatory properties of cystatins
Fetuin-A and its human homologue (ahsg) are defined as negative acute-phase proteins. Normal circulating level of these proteins which is ∼0.5 mg/ml fall significantly during injury and infection [177], [178]. These cystatins have been shown to be immunomodulators that can mediate bacterial phagocytosis by neutrophils [179] and promotion of endocytosis by mouse macrophages [180], [181]. Apart from the fetuin-A, family-2 cystatin members have also been shown to be important immunomodulatory proteins. For example, during inflammatory processes, cystatin C release is down-regulated, contributing to increased cysteine protease activities in the macrophage microenvironment [182]. Cyst C is a powerful inhibitor of cathepsins S and L. High levels of Cyst C are detectable in class II-positive lysosomes of immature dendritic cells (DCs) and Langerhans cells. The maturation process of DCs leads to reduced levels of cyst C, and allows the up-regulation of cysteine proteases cathepsins L and S. For this reason, it has been suggested that Cyst C plays a role in the intracellular control of invariant chain (li) degradation and antigen presentation [183]. Members of the three cystatin families have been shown to up-regulate the release of nitric oxide (NO) from IFN-gamma activated macrophages [184], [185]. This feature applies only to natural cysteine protease inhibitors, as synthetic inhibitors are unable to up-regulate NO productions [186]. The inducible NOS (iNOS) is induced by IFN-gamma and accounts for rapid production of large quantities of NO [187]. The increased NO production by chicken cystatin was demonstrated to be potent enough to cure mice from potentially lethal visceral Leishmaniasis [188].
6.4. Kinetic analysis
Cystatins are the first group of cysteine protein inhibitors of CPs for which the kinetics of inhibition was investigated. All the cystatins are non-covalent, competitive, reversible, tight binding inhibitors which inhibits the target enzymes in micromolar to picomolar range. Their affinities for some of the target enzymes such as papain are so high that it is difficult to demonstrate reversibility or competition with substrate directly. They form tight equimolar complexes with CPs [189]. Dissociation constants (Ki) of enzyme-cystatin complexes have already been depicted in Table 4. The affinity differences as evident from these Ki values can be owed to differences in the active site regions of endopeptidase/exopeptidase.
7. Kidney
7.1. Kidney structure
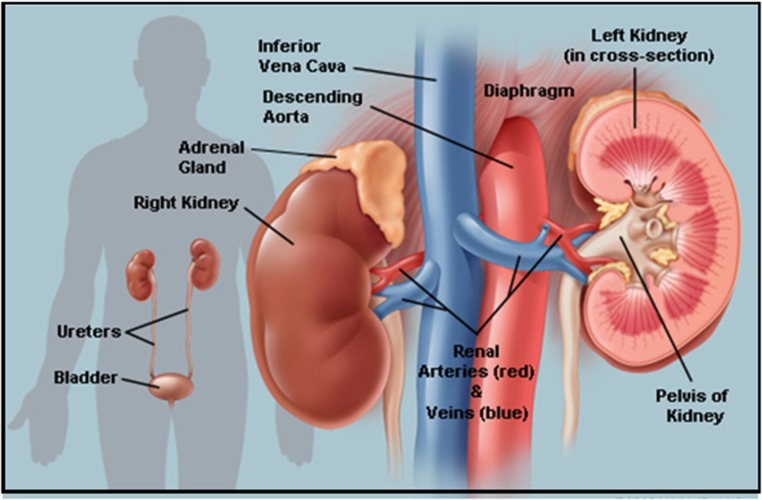
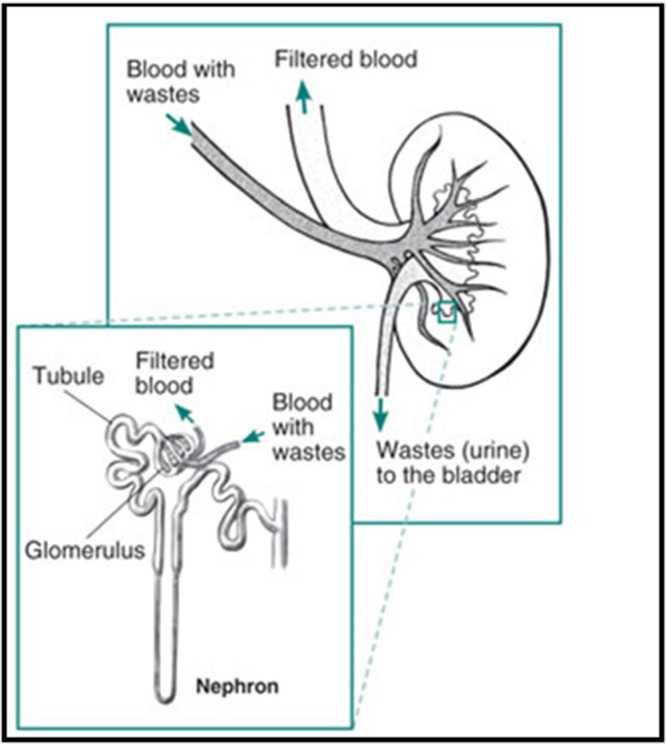
The kidneys are two bean-shaped organs (Fig. 9 ), each about the size of a fist. They are located just below the rib cage, one on each side of the spine. Every day, the two kidneys filter about 120–150 quarts of blood to produce about 1–2 quarts of urine, composed of wastes and extra fluid. The urine flows from the kidneys to the bladder through two thin tubes of muscle called ureters, one on each side of the bladder. The bladder stores urine. The muscles of the bladder wall remain relaxed while the bladder fills with urine. As the bladder fills to capacity, signals sent to the brain tell a person to find a toilet soon. When the bladder empties, urine flows out of the body through a tube called the urethra, located at the bottom of the bladder. In men the urethra is long, while in women it is short. The kidneys are important because they keep the composition, or makeup, of the blood stable, which lets the body function. They prevent the buildup of wastes and extra fluid in the body, keep levels of electrolytes stable, such as sodium, potassium, and phosphate and make hormones that help regulate blood pressure, make red blood cells and also help bones stay strong. The kidney is not one large filter. Each kidney is made up of about a million filtering units called nephrons (Fig. 10 ). Each nephron filters a small amount of blood. The nephron includes a filter, called the glomerulus, and a tubule. The nephrons work through a two-step process. The glomerulus lets fluid and waste products pass through it; however, it prevents blood cells and large molecules, mostly proteins, from passing. The filtered fluid then passes through the tubule, which sends needed minerals back to the bloodstream and removes wastes. The final product becomes urine.
Fig. 9.
Kidneys: Picture of kidneys depicting the entire morphology.
Fig. 10.
Nephrons: Building blocks of kidney.
7.2. Different kidney conditions
7.2.1. Pyelonephritis (infection of kidney pelvis)
Bacteria may infect the kidney, usually causing back pain and fever. A spread of bacteria from an untreated bladder infection is the most common cause of pyelonephritis.
7.2.2. Glomerulonephritis
An overactive immune system may attack the kidney, causing inflammation and some damage. Blood and protein in the urine are common problems that occur with glomerulonephritis. It can also result in kidney failure.
7.2.3. Kidney stones (nephrolithiasis)
Minerals in urine form crystals (stones), which may grow large enough to block urine flow. It's considered one of the most painful conditions. Most kidney stones pass on their own but some are too large and need to be treated.
7.2.4. Nephrotic syndrome
Damage to the kidneys causes them to spill large amounts of protein into the urine. Leg swelling (edema) may be a symptom.
7.2.5. Polycystic kidney disease
A genetic condition resulting in large cysts in both kidneys that impair their function.
7.2.6. Acute renal failure (kidney failure)
A sudden worsening in kidney function. Dehydration, a blockage in the urinary tract, or kidney damage can cause acute renal failure, which may be reversible.
7.2.7. Chronic renal failure
A permanent partial loss of kidney function. Diabetes and high blood pressure are the most common causes.
7.2.8. End stage renal disease (ESRD)
Complete loss of kidney function, usually due to progressive chronic kidney disease. People with ESRD require regular dialysis for survival.
7.2.9. Papillary necrosis
Severe damage to the kidneys can cause chunks of kidney tissue to break off internally and clog the kidneys. If untreated, the resulting damage can lead to total kidney failure
7.2.10. Diabetic nephropathy
High blood sugar from diabetes progressively damages the kidneys, eventually causing chronic kidney disease. Protein in the urine (nephrotic syndrome) may also result.
7.2.11. Hypertensive nephropathy
Kidney damage caused by high blood pressure. Chronic renal failure may eventually result.
7.2.12. Kidney cancer
Renal cell carcinoma is the most common cancer affecting the kidney. Smoking is the most common cause of kidney cancer.
7.2.13. Interstitial nephritis
Inflammation of the connective tissue inside the kidney, often causing acute renal failure. Allergic reactions and drug side effects are the usual causes.
7.2.14. Minimal change disease
A form of nephrotic syndrome in which kidney cells look almost normal under the microscope. The disease can cause significant leg swelling (edema). Steroids are used to treat minimal change disease.
7.2.15. Nephrogenic diabetes insipidus
The kidneys lose the ability to concentrate the urine, usually due to a drug reaction. Although it's rarely dangerous, diabetes insipidus causes constant thirst and frequent urination.
7.2.16. Renal cyst
A benign hollowed-out space in the kidney. Isolated kidney cysts occur in many normal people and almost never impair kidney function.
7.3. Different kidney tests
7.3.1. Urinalysis
A routine test of the urine by a machine and often by a person looking through a microscope. Urinalysis can help detect infections, inflammation, microscopic bleeding, and kidney damage.
7.3.2. Kidney ultrasound
A probe placed on the skin reflects sound waves off the kidneys, creating images on a screen. Ultrasound can reveal blockages in urine flow, stones, cysts, or suspicious masses in the kidneys.
7.3.3. Computed tomography (CT scan)
A CT scanner takes a series of X-rays and a computer creates detailed images of the kidneys.
7.3.4. Magnetic resonance imaging (MRI scan)
A scanner uses radio waves in a magnetic field to make high-resolution images of the kidneys.
7.3.5. Urine and blood cultures
If an infection is suspected, cultures of the blood and urine may identify the bacteria responsible. This can help target antibiotic therapy.
7.3.6. Ureteroscopy
An endoscope (flexible tube with a camera on its end) is passed through the urethra into the bladder and ureters. Ureteroscopy generally cannot reach the kidneys themselves, but can help treat conditions that also affect the ureters.
7.3.7. Kidney biopsy
Using a needle inserted into the back, a small piece of kidney tissue is removed. Examining the kidney tissue under a microscope may help diagnose a kidney problem.
7.4. Different kidney treatments
7.4.1. Antibiotics
Kidney infections caused by bacteria are treated with antibiotics. Often, cultures of the blood or urine can help guide the choice of antibiotic therapy.
7.4.2. Nephrostomy
A tube (catheter) is placed through the skin into the kidney. Urine then drains directly from the kidney, bypassing any blockages in urine flow.
7.4.3. Lithotripsy
Some kidney stones may be shattered into small pieces that can pass in the urine. Most often, lithotripsy is done by a machine that projects ultrasound shock waves through the body.
7.4.4. Nephrectomy
Surgery to remove a kidney. Nephrectomy is performed for kidney cancer or severe kidney damage.
7.4.5. Dialysis
Artificial filtering of the blood to replace the lost function of damaged kidneys. Hemodialysis is the most common method of dialysis in the U.S.
7.4.6. Hemodialysis
A person with complete kidney failure is connected to a dialysis machine, which filters the blood and returns it to the body. Hemodialysis is typically done three days per week in people with ESRD.
7.4.7. Peritoneal dialysis
Placing large amounts of a special fluid in the abdomen through a catheter, allows the body to filter the blood using the natural membrane lining the abdomen. After a while the fluid with the waste is drained and discarded.
7.4.8. Kidney transplant
Transplanting a kidney into a person with ESRD can restore kidney function. A kidney may be transplanted from a living donor, or a recently deceased organ donor.
8. Fates of protein during folding process: overview of protein folding study
Protein folding is a spontaneous process under suitable physiological conditions which is determined mainly by amino acid sequence. The tendency of a polypeptide chain with proper primary structure to fold into its native form, without external help, completes the vital link in the series leading to the expression of genetic information. Once the native form is achieved, the intrinsically flexible, irregular polymer chain folds into a more compact form with the specific structure required for its biological activity. Folding of protein into their condensed three dimensional form is the most basic and universal example of self-assembly. Native and functionally active protein is formed by the folding of newly synthesized polypeptide chain. It results in convey of information encoded into amino acid sequence to a well defined 3-D structure [190]. Although, it has long been known that the amino acid sequence in one way or the other dictates the biological active conformation of protein, but the tools required to detect the intermediate states along the folding pathway are accessible in the past two decades. These tools are revealing a tightly regulated pathway where multiple factors guide nascent polypeptide to select the correct shape from a large number of possibilities. Revealing the process through which protein folding takes place is one of the grand challenge of modern science. There are strict quality-control checks that come into play if the folding process fails, ensuring that the misfolded products are targeted for destruction before they cause any harm. Those that escape this cellular scrutiny are prone to forming aggregates that can damage or kill cells through various mechanisms.
8.1. Transition/intermediate states in protein folding
To find out different equilibrium intermediates, different studies on globular protein were reported [191], [192], these were later confirmed to be associated to some specific folding intermediates. Nevertheless, these equilibrium intermediates, reported in different proteins, shows prominent similarity among themselves. They all possess native-like structure and lack prominent tertiary structure. Later, Ohgushi and Wada [193] introduced the concept of “MG”. According to them, MGs are considered to be the equilibrium intermediates that belong to a common physical state of globular protein. It has been observed that under appropriate conditions, many proteins can form a specific, compact and denatured conformation, a pre-MG state [194]. Characteristics of this state includes presence of native like secondary structure, but in a much lesser quantity than that of MG state, a considerably less compact structure than MG state but much more than corresponding random coil. It can also react with hydrophobic dyes like ANS, but the bond is much weaker compared to that of its counterpart MG.
8.2. Protein misfolding and aggregation
The outcome of polypeptides as to whether they fold correctly into native functionally active molecules or form aggregates is determined by antagonism between protein folding, degradation and aggregation [195]. Concentration of misfolded proteins and molecular crowding inside the cell leads to the formation of aggregates. Evidences have shown that genetic mutation including gene mutations, gene dose and promoter polymorphisms; environmental factors like oxidative or metabolic stress and/or changes in the intracellular conditions (aging) can instigate protein misfolding and aggregation, but the exact mechanisms underlying protein aggregation in different neurodegenerative disorders are still not explored completely. Recent evidences have also indicated that the aggregated proteins may spread from one cell or brain area to another and function as seeds to prompt protein misfolding and aggregation in these previously unaffected cells/areas [196]. Environmental factors and genetic mutations can affect a protein's structure, or 3D shape, causing it to misfold. Misfolded proteins often clump together resulting in aggregates. The aggregates are toxic to some cells such as neurons and lead to diseases such as Alzheimer's, Parkinson’s and Huntington’s. Cell death occurs as a consequence of a rise of misfolded/unfolded polypeptides and their toxic early aggregates overpowering the chaperone–ubiquitin–proteasome clearing efficiency. Generally, clumping and deposition of proteins in the human body, in the form of well organized, fibrillar aggregates termed as Amyloid Fibrils (AF), is the main cause of a major part of protein misfolding diseases. During the past decade, numerous etiologically distinct diseases have been linked by the possibility that they result from the progressive misfolding of specific proteins into aggregates that can injure and kill cells. Protein aggregates can elicit immune response in body or sometimes they may affect vital body functions. Together, these disorders inflict enormous personal and societal burdens, making it crucial to understand their origins and to learn how to prevent them.
8.3. Different types of aggregates
Depending upon solubility and their mode of interaction, aggregates can be of different types. On the basis of solubility, these are of two types: soluble aggregates and insoluble aggregates. Soluble aggregates are invisible particles that cannot be viewed by naked eye and cannot be removed by filtration. Insoluble aggregates are large enough to be removed by filtration and are often visible to human eye. Aggregates can also be covalently linked by disulfide bond formation between free thiol groups [197]. Oxidation of Tyr residues can lead to formation of bityrosine which often results in aggregation. Aggregates may be linked via weaker interactions like those in reversible protein aggregates [198].
8.4. Amyloid states results from protein abnormalities
Proteins responsible for neurodegenerative diseases are natively unfolded and thus can interact with different partners to form several multimeric complexes. It can result in their potential propensity, if not kept under check, to form abnormal multimeric complexes. Physiologic and pathologic data suggests the formation of at least three different types of protein complexes namely; Physiologic Protein Mosaics (PM), Protein Aggregates and Pathologic PM [199]. Furthermore, the β-sheet conformation present in aggregates may have a physiologic role and even the amorphous aggregates may function to shield the potentially toxic diffusible oligomers. In fact, fibril formation (amyloidogenesis) may be an evolutionary conserved mechanism for creating biologically active quaternary structures. Although aggregation does not rule out detrimental effects of amyloid deposits in particular contexts (eg. obstructive vascular amyloid), they clearly show that amyloid formation can be beneficial. Several studies suggest that the formation of tightly packed Huntingtin deposits is beneficial for cell survival.
8.5. Toxic species in aggregation
Even though substantial evidences suggests that aggregates are the toxic end states achieved in aberrant protein folding but the actual culprits are the transient, pre-fibrillar species preceding aggregates that are most toxic in nature [200], [201]. The toxicity of immature protein aggregate “preamyloid” seems to be a quite general phenomenon. For example, the prefibrillar forms of non-disease-related HypF-N from E. coli, the SH3 domain from bovine phosphatidylinositol 3-kinase, lysozyme from horse, and apomyoglobin from sperm whale are all extremely toxic to cultured fibroblasts and neurons, whereas their monomeric native states and amyloid-like fibrils, formed in vitro, displayed very little toxicity [202].
9. Conclusion
Cystatins are of immense significance as these are considered to play vital roles in many regulatory processes. Owing to their physiological importance these have been isolated from variety of sources ranging from buffalo heart to buffalo kidney. In recent times a new family of cystatins called as phytocystatins has also evolved and is being given utmost importance. Phytocystatins play an important role during biotic and abiotic stress and seed germination and also regulate endogenous cysteine proteinases of insects and other pathogens. Thus cystatins can no more be considered as just the inhibitors of thiol proteases, rather they are being explored for more and more important roles played by these inhibitors.
Conflict of interest
The authors declare that they have no competing financial interests.
References
- 1.Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 2006;5(9):785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 2.Hartley B.S. Proteolytic enzymes. Annu. Rev. Biochem. 1960;29:45–72. doi: 10.1146/annurev.bi.29.070160.000401. [DOI] [PubMed] [Google Scholar]
- 3.Lecaille F., Kaleta J., Brömme D. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chem. Rev. 2002;102(12):4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 4.Otto H.H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97(1):133–172. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- 5.Rawlings N.D., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27(1):325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGrath M.E. The lysosomal cysteine proteases. Annu. Rev. Biophys. Biomol. Struct. 1999;28(1):181–204. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 7.Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng B., Liu X., Gong H., Huang L., Chen H., Zhang X., Li C., Yang M., Ma B., Jiao L., Zheng L. Coffee components inhibit amyloid formation of human islet amyloid polypeptide in vitro: possible link between coffee consumption and diabetes mellitus. J. Agric. Food Chem. 2011;59(24):13147–13155. doi: 10.1021/jf201702h. [DOI] [PubMed] [Google Scholar]
- 9.Turk V., Turk B., Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20(17):4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illy C., Quraishi O., Wang J., Purisima E., Vernet T., Mort J.S. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 1997;272(2):1197–1202. doi: 10.1074/jbc.272.2.1197. [DOI] [PubMed] [Google Scholar]
- 11.Schenker P., Alfarano P., Kolb P., Caflisch A., Baici A. A double-headed cathepsin B inhibitor devoid of warhead. Protein Sci. 2008;17(12):2145–2155. doi: 10.1110/ps.037341.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotrin S.S., Puzer L., de Souza Judice W.A., Juliano L., Carmona A.K., Juliano M.A. Positional-scanning combinatorial libraries of fluorescence resonance energy transfer peptides to define substrate specificity of carboxydipeptidases: assays with human cathepsin B. Anal. Biochem. 2004;335(2):244–252. doi: 10.1016/j.ab.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Biniossek M.L., Nagler D.K., Becker-Pauly C., Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B: L, and S. J. Proteome Res. 2011;10(12):5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y., Wang K.K. The calpain family and human disease. Trends Mol. Med. 2001;7(8):355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- 15.Sorimachi H., Hata S., Ono Y. Impact of genetic insights into calpain biology. J. Biochem. 2011;150(1):23–37. doi: 10.1093/jb/mvr070. [DOI] [PubMed] [Google Scholar]
- 16.Murachi T., Tanaka K., Hatanaka M., Murakami T. Intracellular Ca2+-dependent protease (calpain) and its high-molecular-weight endogenous inhibitor (calpastatin) Adv. Enzyme Regul. 1981;19:407–424. doi: 10.1016/0065-2571(81)90026-1. [DOI] [PubMed] [Google Scholar]
- 17.Alnemri E.S., Livingston D.J., Nicholson D.W., Salvesen G., Thornberry N.A., Wong W.W., Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87(2):171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 18.Black R.A., Kronheim S.R., Sleath P.R. Activation of interleukin-1β by a co-induced protease. FEBS Lett. 1989;247(2):386–390. doi: 10.1016/0014-5793(89)81376-6. [DOI] [PubMed] [Google Scholar]
- 19.Kostura M.J., Tocci M.J., Limjuco G., Chin J., Cameron P., Hillman A.G., Chartrain N.A., Schmidt J.A. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc. Natl. Acad. Sci. 2016;86(14):5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerretti D.P., Kozlosky C.J. Molecular cloning of the interleukin-1beta converting enzyme. Science. 1992;256(5053):97. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 21.Thornberry N.A., Bull H.G., Calaycay J.R., Chapman K.T., Howard A.D., Kostura M.J., Miller D.K., Molineaux S.M., Weidner J.R., Aunins J., Elliston K.O. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature. 1992;356(6372):768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 22.Walker N.P., Talanian R.V., Brady K.D., Dang L.C., Bump N.J., Ferenza C.R., Franklin S., Ghayur T., Hackett M.C., Hammill L.D., Herzog L. Crystal structure of the cysteine protease interleukin-1β-converting enzyme: a (p20/p10) 2 homodimer. Cell. 1994;78(2):343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang L., Miura M., Bergeron L., Zhu H., Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 2016;78(5):739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 24.Kuida K., Lippke J.A., Ku G., Harding M.W. Altered cytokine export and apoptosis in mice deficient in interleukin-1beta converting enzyme. Science. 2000;1995(5206):267. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 25.Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., Salfeld J., Towne E. Mice deficient in IL-1β-converting enzyme are defective in production of mature IL-1β and resistant to endotoxic shock. Cell. 1995;80(3):401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury I., Tharakan B., Bhat G.K. Caspases—an update. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2008;151(1):10–27. doi: 10.1016/j.cbpb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Rawlings N.D., Barrett A.J. [32] Families of cysteine peptidases. Methods Enzymol. 1994;244:461–486. doi: 10.1016/0076-6879(94)44034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wyk S.G., Kunert K.J., Cullis C.A., Pillay P., Makgopa M.E., Schluter U., Vorster B.J. Review The future of cystatin engineering. Plant Sci. 2016;246:119–127. doi: 10.1016/j.plantsci.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Sen L.C., Whitaker J.R. Some properties of a ficin-papain inhibitor from avion egg white. Arch. Biochem. Biophys. 1973;158(2):623–632. doi: 10.1016/0003-9861(73)90554-7. [DOI] [PubMed] [Google Scholar]
- 30.Barrett A.J. [57] Cystatin, the egg white inhibitor of cysteine proteinases. Methods Enzymol. 1981;80:771–778. [Google Scholar]
- 31.Barrett A.J. Cysteine proteinase inhibitors of the cystatin superfamily. Proteinase Inhib. 1986:515–569. [Google Scholar]
- 32.Turk V., Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285(2):213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 33.Shamsi A., Ahmed A., Bano B. Biochemical, immunological and kinetic characterization and partial sequence analysis of a thiol proteinase inhibitor from Bubalus bubalis kidney: an attempt targeting kidney disorders. Int. J. Biol. Macromol. 2016;2(January) doi: 10.1016/j.ijbiomac.2015.12.084. [DOI] [PubMed] [Google Scholar]
- 34.Abrahamson M., Barrett A., Salvesen G., Grubb A. Isolation of six cysteine proteinase inhibitors from human urine: their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem. 1986;261(24):11282–11289. [PubMed] [Google Scholar]
- 35.Turk B., Turk V., Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol. Chem. 1996;378(3–4):141–150. [PubMed] [Google Scholar]
- 36.Turk B., Krizaj I., Kralj B., Dolenc I., Popovic T., Bieth J.G., Turk V. Bovine stefin C, a new member of the stefin family. J. Biol. Chem. 1993;268(10):7323–7329. [PubMed] [Google Scholar]
- 37.Lenarcic B., Ritonja A., Dolenc I., Stoka V., Berbic S., Pungercar J., Strukelj B., Turk V. Pig leukocyte cysteine proteinase inhibitor (PLCPI): a new member of the stefin family. FEBS Lett. 1993;336(2):289–292. doi: 10.1016/0014-5793(93)80822-c. [DOI] [PubMed] [Google Scholar]
- 38.Brzin J., KOPITAR M., Turk V., MACHLEIDT W. Protein inhibitors of cysteine proteinases I. Isolation and characterization of stefin, a cytosolic protein inhibitor of cysteine proteinases from human polymorphonuclear granulocytes. Hoppe-Seyleŕ s Zeitschrift für physiologische Chemie. 1983;364(2):1475–1480. doi: 10.1515/bchm2.1983.364.2.1475. [DOI] [PubMed] [Google Scholar]
- 39.Lenarcic B., Ritonja A., Sali A., Kotnik M., Turk V., Machleidt W. Properties and structure of human spleen stefin B—A low molecular weight protein inhibitor of cysteine proteinases. Cysteine Proteinases Their Inhib. 1986;47:3–87. [Google Scholar]
- 40.Abrahamson M. Cystatins-protein inhibitors of papain-like cysteine proteinases. Ciencia e Cultura. 1993;45(5):299-. [Google Scholar]
- 41.Rinne A. University of Oulu; 1979. Epidermal SH-Protease Inhibitor: Occurrence in Human and Rat Tissues and in Human Neoplasms. [Google Scholar]
- 42.Rinne A., Jarvinen M., Rasanen Dorn O.A. Demonstration of an epidermal SH-protease inhibitor in normal epithelium and in some human neoplasms–an immunological study (author's transl) Acta Histochem. Suppl. 1979;22:325–329. [PubMed] [Google Scholar]
- 43.Rinne A., Rasanen O., Jarvinen M., Dammert K., Kallioinen M., Hopsu-Havu V.K. Occurrence of acid and neutral cysteine proteinase inhibitors in epidermal malignancies: immunohistochemical study. Acta Histochem. 1984;74(1):75IN1–79IN2. doi: 10.1016/S0065-1281(84)80030-6. [DOI] [PubMed] [Google Scholar]
- 44.Hopsu-Havu V.K., Jarvinen M., Rinne A. Separation of cysteine proteinase inhibitors from psoriatic scale. Br. J. Dermatol. 1983;109:77–85. [PubMed] [Google Scholar]
- 45.Voltersvik P., Bostad L., Dyrhol-Riise A.M., Eide G.E., Rosok B.I., Olofsson J., Asjo B., Cystatin A. HIV-1 p24 antigen expression in tonsillar lymphoid follicles during HIV-1 infection and during highly active antiretroviral therapy. JAIDS J. Acq. Immune Defic. Syndr. 2006;41(3):277–284. doi: 10.1097/01.qai.0000199234.77081.a2. [DOI] [PubMed] [Google Scholar]
- 46.Jarvinen M. Purification and properties of two protease inhibitors from rat skin inhibiting papain and other SH-proteases. Acta Chem. Scand. B. 1976;30(10) doi: 10.3891/acta.chem.scand.30b-0933. [DOI] [PubMed] [Google Scholar]
- 47.Pennacchio L.A., Lehesjoki A.E., Stone N.E., Willour V.L. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1) Science. 1996;271(5256):1731. doi: 10.1126/science.271.5256.1731. [DOI] [PubMed] [Google Scholar]
- 48.Lenney J.F., Tolan J.R., Sugai W.J., Lee A.G. Thermostable endogenous inhibitors of cathepsins B and H. Eur. J. Biochem. 1979;101(1):153–161. doi: 10.1111/j.1432-1033.1979.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 49.Lenarcic B., Krizaj I., Zunec P., Turk V. Differences in specificity for the interactions of stefins A, B and D with cysteine proteinases. FEBS Lett. 1996;395(October (2–3)):113–118. doi: 10.1016/0014-5793(96)00984-2. [DOI] [PubMed] [Google Scholar]
- 50.Green G.D., Kembhavi A.A., Davies M.E., Barrett A.J. Cystatin-like cysteine proteinase inhibitors from human liver. Biochem. J. 1984;218(3):939–946. doi: 10.1042/bj2180939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finkenstaedt J.T. Intracellular distribution of proteolytic enzymes in rat liver tissue. Exp. Biol. Med. 1957;95(2):302–304. doi: 10.3181/00379727-95-23201. [DOI] [PubMed] [Google Scholar]
- 52.Kominami E., Wakamatsu N., Katunuma N. Endogenous thiol protease inhibitor from rat liver. Biochem. Biophys. Res. Commun. 1981;99(2):568–575. doi: 10.1016/0006-291x(81)91783-6. [DOI] [PubMed] [Google Scholar]
- 53.Lee M.J., Yu G.R., Park S.H., Cho B.H., Ahn J.S., Park H.J., Song E.Y., Kim D.G. Identification of cystatin B as a potential serum marker in hepatocellular carcinoma. Clin. Cancer Res. 2008;14(4):1080–1089. doi: 10.1158/1078-0432.CCR-07-1615. [DOI] [PubMed] [Google Scholar]
- 54.Kopitar-Jerala N. The role of stefin B in neuro-inflammation. Front. Cell. Neurosci. 2015:9. doi: 10.3389/fncel.2015.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freije J.P., Abrahamson M., Olafsson I., Velasco G., Grubb A., Lopez-Otin C. Structure and expression of the gene encoding cystatin D: a novel human cysteine proteinase inhibitor. J. Biol. Chem. 1991;266(30):20538–20543. [PubMed] [Google Scholar]
- 56.Saitoh E., Sabatini L.M., Eddy R.L., Shows T.B., Azen E.A., Isemura S., Sanada K. The human cystatin C gene (CST3) is a member of the cystatin gene family which is localized on chromosome 20. Biochem. Biophys. Res. Commun. 1989;162(3):1324–1331. doi: 10.1016/0006-291x(89)90818-8. [DOI] [PubMed] [Google Scholar]
- 57.Isemura S., Saitoh E., Sanada K., Minakata K. Identification of full-sized forms of salivary (type) cystatins (cystatin SN, cystatin SA, cystatin S, and two phosphorylated forms of cystatin S) in human whole saliva and determination of phosphorylation sites of cystatin S. J. Biochem. 1991;110(October (4)):648–654. doi: 10.1093/oxfordjournals.jbchem.a123634. [DOI] [PubMed] [Google Scholar]
- 58.Grubb A.O. Cystatin C-properties and use as diagnostic marker. Adv. Clin. Chem. 2001;35:63–99. doi: 10.1016/S0065-2423(01)35015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turk V., Stoka V., Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front. Biosci. 2008;13(5):5780–5786. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- 60.Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., Jaskolski M. Human cystatin C: an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Mol. Biol. 2001;8(4):316–320. doi: 10.1038/86188. [DOI] [PubMed] [Google Scholar]
- 61.Lenarcic B., Krasovec M., Ritonja A., Olafsson I., Turk V. Inactivation of human cystatin C and kininogen by human cathepsin D. FEBS Lett. 1991;280(2):211–215. doi: 10.1016/0014-5793(91)80295-e. [DOI] [PubMed] [Google Scholar]
- 62.Grubb A., Horio M., Hansson L.O., Bjork J., Nyman U., Flodin M., Larsson A., Bokenkamp A., Yasuda Y., Blufpand H., Lindstrom V. Generation of a new cystatin C–based estimating equation for glomerular filtration rate by use of 7 assays standardized to the international calibrator. Clin. Chem. 2014;60(7):974–986. doi: 10.1373/clinchem.2013.220707. [DOI] [PubMed] [Google Scholar]
- 63.Roos J.F., Doust J., Tett S.E., Kirkpatrick C.M. Diagnostic accuracy of cystatin C compared to serum creatinine for the estimation of renal dysfunction in adults and children—a meta-analysis. Clin. Biochem. 2007;40(5):383–391. doi: 10.1016/j.clinbiochem.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 64.Jung Y.J., Lee H.R., Kwon O.J. Comparison of serum cystatin C and creatinine as a marker for early detection of decreasing glomerular filtration rate in renal transplants. J. Korean Surg. Soc. 2012;83(2):69–74. doi: 10.4174/jkss.2012.83.2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ekiel I., Abrahamson M. Folding-related dimerization of human cystatin C. J. Biol. Chem. 1996;271(3):1314–1321. doi: 10.1074/jbc.271.3.1314. [DOI] [PubMed] [Google Scholar]
- 66.He J., Xu L., Zhang S., Guan J., Shen M., Li H., Song Y. Steered molecular dynamics simulation of the binding of the β2 and β3 regions in domain-swapped human cystatin C dimer. J. Mol. Model. 2013;19(2):825–832. doi: 10.1007/s00894-012-1609-7. [DOI] [PubMed] [Google Scholar]
- 67.Shen M., Guan J., Xu L., Yu Y., He J., Jones G.W., Song Y. Steered molecular dynamics simulations on the binding of the appendant structure and helix-β2 in domain-swapped human cystatin C dimer. J. Biomol. Struct. Dyn. 2012;30(6):652–661. doi: 10.1080/07391102.2012.689698. [DOI] [PubMed] [Google Scholar]
- 68.Balbin M., Freije J.P., Abrahamson M., Velasco G., Grubb A., López-Otín C. A sequence variation in the human cystatin D gene resulting in an amino acid (Cys/Arg) polymorphism at the protein level. Hum. Genet. 1993;90(6):668–669. doi: 10.1007/BF00202491. [DOI] [PubMed] [Google Scholar]
- 69.Balbin M., Hall A., Grubb A., Mason R.W., Lopez-Otin C., Abrahamson M. Structural and functional characterization of two allelic variants of human cystatin D sharing a characteristic inhibition spectrum against mammalian cysteine proteinases. J. Biol. Chem. 1994;269(37):23156–23162. [PubMed] [Google Scholar]
- 70.Ferrer-Mayorga G., Alvarez-Diaz S., Valle N., De LasRivas J., Mendes M., Barderas R., Canals F., Tapia O., Casal J.I., Lafarga M., Munoz A. Cystatin D locates in the nucleus at sites of active transcription and modulates gene and protein expression. J. Biol. Chem. 2015;290(44):26533–26548. doi: 10.1074/jbc.M115.660175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collins A.R., Grubb A., Cystatin D. A natural salivary cysteine protease inhibitor, inhibits coronavirus replication at its physiologic concentration. Oral Microbiol. Immunol. 1998;13(1):59–61. doi: 10.1111/j.1399-302X.1998.tb00753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halfon S., Ford J., Foster J., Dowling L., Lucian L., Sterling M., Xu Y., Weiss M., Ikeda M., Liggett D., Helms A. Leukocystatin, a new class II cystatin expressed selectively by hematopoietic cells. J. Biol. Chem. 1998;273(26):16400–16408. doi: 10.1074/jbc.273.26.16400. [DOI] [PubMed] [Google Scholar]
- 73.Ni J., Fernandez M.A., Danielsson L., Chillakuru R.A., Zhang J., Grubb A., Su J., Gentz R., Abrahamson M. Cystatin F is a glycosylated human low molecular weight cysteine proteinase inhibitor. J. Biol. Chem. 1998;273(38):24797–24804. doi: 10.1074/jbc.273.38.24797. [DOI] [PubMed] [Google Scholar]
- 74.Morita M., Hara Y., Tamai Y., Arakawa H., Nishimura S. Genomic construct and mapping of the gene for CMAP (leukocystatin/cystatin F: CST7) and identification of a proximal novel gene, BSCv (C20orf3) Genomics. 2000;67(1):87–91. doi: 10.1006/geno.2000.6237. [DOI] [PubMed] [Google Scholar]
- 75.Cappello F., Gatti E., Camossetto V., David A., Lelouard H., Pierres P. Cystatin F is secreted, but artificial modification of its C-terminus can induce its endocytic targeting. Exp. Cell Res. 2004;297(2):607–618. doi: 10.1016/j.yexcr.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 76.Langerholc T., Zavasnik-Bergant V., Turk B., Turk V., Abrahamson M., Kos J. Inhibitory properties of cystatin F and its localization in U937 promonocyte cells. FEBS J. 2005;272(6):1535–1545. doi: 10.1111/j.1742-4658.2005.04594.x. [DOI] [PubMed] [Google Scholar]
- 77.Colbert J.D., Plechanovova A., Watts C. Glycosylation directs targeting and activation of cystatin f from intracellular and extracellular sources. Traffic. 2009;10(4):425–437. doi: 10.1111/j.1600-0854.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schuttelkopf A.W., Hamilton G., Watts C., van Aalten D.M. Structural basis of reduction-dependent activation of human cystatin F. J. Biol. Chem. 2006;281(24):16570–16575. doi: 10.1074/jbc.M601033200. [DOI] [PubMed] [Google Scholar]
- 79.Alvarez-Fernandez M., Barrett A.J., Gerhartz B., Dando P.M., Ni J., Abrahamson M. Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J. Biol. Chem. 1999;274(27):19195–19203. doi: 10.1074/jbc.274.27.19195. [DOI] [PubMed] [Google Scholar]
- 80.Ni J., Abrahamson M., Zhang M., Fernandez M.A., Grubb A., Su J., Yu G.L., Li Y., Parmelee D., Xing L., Coleman T.A. Cystatin E is a novel human cysteine proteinase inhibitor with structural resemblance to family 2 cystatins. J. Biol. Chem. 1997;272(16):10853–10858. doi: 10.1074/jbc.272.16.10853. [DOI] [PubMed] [Google Scholar]
- 81.Sotiropoulou G., Anisowicz A., Sager R. Identification: cloning, and characterization of cystatin M, a novel cysteine proteinase inhibitor, down-regulated in breast cancer. J. Biol. Chem. 1997;272(2):903–910. doi: 10.1074/jbc.272.2.903. [DOI] [PubMed] [Google Scholar]
- 82.Stenman G., Aastroem A.K., Roijer E., Sotiropoulou G., Zhang M., Sager R. Assignment of a novel cysteine proteinase inhibitor (CST6) to 11q13 by fluorescence in situ hybridization. Cytogenet. Genome Res. 1997;76(1-2):45–46. doi: 10.1159/000134512. [DOI] [PubMed] [Google Scholar]
- 83.Schnittger S., Rao V.G., Abrahamson M., Hansmann I. Cystatin C. (CST3), the candidate gene for hereditary cystatin C amyloid angiopathy (HCCAA), and other members of the cystatin gene family are clustered on chromosome 20p11. 2. Genomics. 1993;16(1):50–55. doi: 10.1006/geno.1993.1139. [DOI] [PubMed] [Google Scholar]
- 84.Cheng T., Tjabringa G.S., Vlijmen-Willems V., Hitomi K., Van Erp P.E., Schalkwijk J., Zeeuwen P.L. The cystatin M/E-controlled pathway of skin barrier formation: expression of its key components in psoriasis and atopic dermatitis. Br. J. Dermatol. 2009;161(2):253–264. doi: 10.1111/j.1365-2133.2009.09156.x. [DOI] [PubMed] [Google Scholar]
- 85.Rivenbark A.G., Livasy C.A., Boyd C.E., Keppler D., Coleman W.B. Methylation-dependent silencing of CST6 in primary human breast tumors and metastatic lesions. Exp. Mol. Pathol. 2007;83(2):188–197. doi: 10.1016/j.yexmp.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qiu J., Ai L., Ramachandran C., Yao B., Gopalakrishnan S., Fields C.R., Delmas A.L., Dyer L.M., Melnick S.J., Yachnis A.T., Schwartz P.H. Invasion suppressor cystatin E/M (CST6): high-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab. Invest. 2008;88(9):910–925. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng T., Hitomi K., van Vlijmen-Willems I.M., de Jongh G.J., Yamamoto K., Nishi K., Watts C., Reinheckel T., Schalkwijk J., Zeeuwen P.L. Cystatin M/E is a high affinity inhibitor of cathepsin V and cathepsin l by a reactive site that is distinct from the legumain-binding site a novel clue for the role of cystatin m/e in epidermal cornification. J. Biol. Chem. 2006;281(23):15893–15899. doi: 10.1074/jbc.M600694200. [DOI] [PubMed] [Google Scholar]
- 88.Sundberg J.P., Boggess D., Hogan M.E., Sundberg B.A., Rourk M.H., Harris B., Johnson K., Dunstan R.W., Davisson M.T. Harlequin ichthyosis (ichq): a juvenile lethal mouse mutation with ichthyosiform dermatitis. Am. J. Pathol. 1997;151(1):293. [PMC free article] [PubMed] [Google Scholar]
- 89.Zeeuwen P.L., van Vlijmen-Willems I.M., Hendriks W., Merkx G.F., Schalkwijk J. A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet. 2002;11(23):2867–2875. doi: 10.1093/hmg/11.23.2867. [DOI] [PubMed] [Google Scholar]
- 90.Juriaanse A.C., Booij M. Isolation and characterisation of the main neutral protein from human submandibular saliva. Arch. Oral Biol. 1979;24(10–11):825–828. doi: 10.1016/0003-9969(79)90045-1. [DOI] [PubMed] [Google Scholar]
- 91.Juriaanse A.C., Booij M. Isolation and partial characterisation of three acidic proteins from human submandibular saliva. Arch. Oral Biol. 1979;24(8):621–625. doi: 10.1016/0003-9969(79)90024-4. [DOI] [PubMed] [Google Scholar]
- 92.Minakata K., Asano M. New protein inhibitors of cysteine proteinases in human saliva and salivary glands. Hoppe-Seyleŕ s Zeitschrift für physiologische Chemie. 1984;365(1):399–404. doi: 10.1515/bchm2.1984.365.1.399. [DOI] [PubMed] [Google Scholar]
- 93.Obenauf S.D., Fisher R.H., Cowman R.A., Fitzgerald R.J. Immunological relationship between anionic antimicrobial proteins from caries-free individuals and known salivary antimicrobial factors. Infect. Immun. 1984;46(3):797–801. doi: 10.1128/iai.46.3.797-801.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dickinson D.P. Salivary (SD-type) cystatins: over one billion years in the making—but to what purpose? Crit. Rev. Oral Biol. Med. 2002;13(6):485–508. doi: 10.1177/154411130201300606. [DOI] [PubMed] [Google Scholar]
- 95.Potempa J., Dubin A., Korzus G., Travis J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J. Biol. Chem. 1988;263(6):2664–2667. [PubMed] [Google Scholar]
- 96.Hiltkel T.R., Lee T.C., Bobekl L.A. Structure/function analysis of human cystatin SN and comparison of the cysteine proteinase inhibitory profiles of human cystatins C and SN. J. Dent. Res. 1999;78(8):1401–1409. doi: 10.1177/00220345990780080501. [DOI] [PubMed] [Google Scholar]
- 97.Abe N., Kadowaki T., Okamoto K., Nakayama K., Ohishi M., Yamamoto K. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J. Biochem. 1998;123(2):305–312. doi: 10.1093/oxfordjournals.jbchem.a021937. [DOI] [PubMed] [Google Scholar]
- 98.Isemura S., Saitoh E., Sanada K., Minakata K. Identification of full-sized forms of salivary (type) cystatins (cystatin SN: cystatin SA, cystatin S, and two phosphorylated forms of cystatin S) in human whole saliva and determination of phosphorylation sites of cystatin S. J. Biochem. 1991;110(4):648–654. doi: 10.1093/oxfordjournals.jbchem.a123634. [DOI] [PubMed] [Google Scholar]
- 99.Baron A.C., DeCarlo A.A., Featherstone J.D. Functional aspects of the human salivary cystatins in the oral environment. Oral Dis. 1999;5(3):234–240. doi: 10.1111/j.1601-0825.1999.tb00307.x. [DOI] [PubMed] [Google Scholar]
- 100.N. Kopitar-Jerala, Cystatins in cells of the immune systems: a current view. 2006. [DOI] [PubMed]
- 101.Ruzindana-Umunyana A., Weber J.M. Interactions of human lacrimal and salivary cystatins with adenovirus endopeptidase. Antiviral Res. 2001;51(3):203–214. doi: 10.1016/s0166-3542(01)00154-1. [DOI] [PubMed] [Google Scholar]
- 102.Gu M., Haraszthy G.G., Collins A.R., Bergey E.J. Identification of salivary proteins inhibiting herpes simplex virus 1 replication. Oral Microbiol. Immunol. 1995;10(1):54–59. doi: 10.1111/j.1399-302x.1995.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 103.Salvesen G., Parkes C., Abrahamson M., Grubb A., Barrett A.J. Human low-Mr kininogen contains three copies of a cystatin sequence that are divergent in structure and in inhibitory activity for cysteine proteinases. Biochem. J. 1986;234(2):429–434. doi: 10.1042/bj2340429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abelous J.E., Bardier E. Les substances hypotensives de l’urine humaine normale. CR Soc. Biol. 1909;66:511–520. [Google Scholar]
- 105.Kraut H., Frey E.K., Werle E. Uber die Inaktivierung des Kallikreins. (VI Mitteilung über dieses Kreislaufhormon.) Hoppe-Seyleŕ s Zeitschrift für physiologische Chemie. 1930;192(1–3):1–21. [Google Scholar]
- 106.Okamoto H., Greenbaum L.M. Isolation and structure of T-kinin. Biochem. Biophys. Res. Commun. 1983;112(2):701–708. doi: 10.1016/0006-291x(83)91519-x. [DOI] [PubMed] [Google Scholar]
- 107.Moreau T., Gutman N., Faucher D., Gauthier F. Limited proteolysis of T-kininogen (thiostatin): Release of comparable fragments by different endopeptidases. J. Biol. Chem. 1989;264(8):4298–4303. [PubMed] [Google Scholar]
- 108.Rawlings N.D., Barrett A.J., Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44(D1):D343–50. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Santamaría M.E., Diaz-Mendoza M., Diaz I., Martinez M. Plant protein peptidase inhibitors: an evolutionary overview based on comparative genomics. BMC Genomics. 2014;15(1):812. doi: 10.1186/1471-2164-15-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kordis D., Turk V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 2009;9(1):1. doi: 10.1186/1471-2148-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arai S., Watanabe H., Kondo H., Emori Y., Abe K. Papain-inhibitory activity of oryzacystatin: a rice seed cysteine proteinase inhibitor, depends on the central Gln-Val-Val-Ala-Gly region conserved among cystatin superfamily members. J. Biochem. 1991;109(2):294–298. [PubMed] [Google Scholar]
- 112.Martinez M., Diaz I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 2008;8(1):1. doi: 10.1186/1471-2148-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Green A.R., Nissen M.S., Kumar G.M., Knowles N.R., Kang C. Characterization of Solanum tuberosum multicystatin and the significance of core domains. Plant Cell. 2013;25(12):5043–5052. doi: 10.1105/tpc.113.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benchabane M., Schluter U., Vorster J., Goulet M.C., Michaud D. Plant cystatins. Biochimie. 2010;92(11):1657–1666. doi: 10.1016/j.biochi.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 115.Brown W.M., Dziegielewska K.M. Friends and relations of the cystatin superfamily—new members and their evolution. Protein Sci. 1997;6(1):5–12. doi: 10.1002/pro.5560060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brown W.M., Dziegielewska K.M., Saunders N.R., Møsllgard K. Fetuin-an old friend revisited. Bioessays. 1992;14(11):749–755. doi: 10.1002/bies.950141105. [DOI] [PubMed] [Google Scholar]
- 117.Murzin A.G. Sweet-tasting protein monellin is related to the cystatin family of thiol proteinase inhibitors. J. Mol. Biol. 1993;230(2):689–694. doi: 10.1006/jmbi.1993.1186. [DOI] [PubMed] [Google Scholar]
- 118.Winderickx J., Hemschoote K., De Clercq N., Van Dijck P., Peeters B., Rombauts W., Verhoeven G., Heyns W. Tissue-specific expression and androgen regulation of different genes encoding rat prostatic 22-kilodalton glycoproteins homologous to human and rat cystatin. Mol. Endocrinol. 1990;4(4):657–667. doi: 10.1210/mend-4-4-657. [DOI] [PubMed] [Google Scholar]
- 119.Devos A., De Clercq N., Vercaeren I., Heyns W., Rombauts W., Peeters B. Structure of rat genes encoding androgen-regulated cystatin-related proteins (CRPs): a new member of the cystatin superfamily. Gene. 1993;125(2):159–167. doi: 10.1016/0378-1119(93)90323-u. [DOI] [PubMed] [Google Scholar]
- 120.Gholizadeh A., Santha I.M., Kohnehrouz B.B., Lodha M.L., Kapoor H.C. Cystatins may confer viral resistance in plants by inhibition of a virus-induced cell death phenomenon in which cysteine proteinases are active: cloning and molecular characterization of a cDNA encoding cysteine-proteinase inhibitor (celostatin) from Celosia cristata (crested cock's comb) Biotechnol. Appl. Biochem. 2005;42(3):197–204. doi: 10.1042/BA20050029. [DOI] [PubMed] [Google Scholar]
- 121.Cornwall G.A., Orgebin-Crist M.C., Hann S.R. The CRES gene: a unique testis-regulated gene related to the cystatin family is highly restricted in its expression to the proximal region of the mouse epididymis. Mol. Endocrinol. 1992;6(10):1653–1664. doi: 10.1210/mend.6.10.1280328. [DOI] [PubMed] [Google Scholar]
- 122.Strojnik T., Zajc I., Bervar A., Zidanik B., Golouh R., Kos J., Dolenc V., Lah T. Cathepsin B and its inhibitor stefin A in brain tumors. Pflügers Archiv. 2000;439(1):r122–3. [PubMed] [Google Scholar]
- 123.Kos J., Krasovec M., Cimerman N., Nielsen H.J., Christensen I.J., Brunner N. Cysteine proteinase inhibitors stefin A, stefin B, and cystatin C in sera from patients with colorectal cancer: relation to prognosis. Clin. Cancer Res. 2000;6(2):505–511. [PubMed] [Google Scholar]
- 124.Kos J., Lah T.T. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review) Oncol. Rep. 1998;5(6):1349–1410. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- 125.Hamilton G., Colbert J.D., Schuettelkopf A.W., Watts C. Cystatin F is a cathepsin C-directed protease inhibitor regulated by proteolysis. EMBO J. 2008;27(3):499–508. doi: 10.1038/sj.emboj.7601979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pham C.T., Ley T.J. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc. Natl. Acad. Sci. 1999;96(15):8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Werle B., Schanzenbacher U., Lah T.T., Ebert E., Julke B., Ebert W., Fiehn W., Kayser K., Spiess E., Abrahamson M., Kos J. Cystatins in non-small cell lung cancer: tissue levels, localization and relation to prognosis. Oncol. Rep. 2006;16(4):647–656. [PubMed] [Google Scholar]
- 128.Cox J.L., Sexton P.S., Green T.J., Darmani N.A. Inhibition of B16 melanoma metastasis by overexpression of the cysteine proteinase inhibitor cystatin C. Melanoma Res. 1999;9(4):369–374. doi: 10.1097/00008390-199908000-00005. [DOI] [PubMed] [Google Scholar]
- 129.Olafsson I., Grubb A. Hereditary cystatin C amyloid angiopathy. Amyloid. 2000;7(1):70–79. doi: 10.3109/13506120009146827. [DOI] [PubMed] [Google Scholar]
- 130.Bollengier F., Cystatin C. Alias post-γ-globulin. a marker for multiple sclerosis? Clin. Chem. Lab. Med. 1987;25(9):589–594. doi: 10.1515/cclm.1987.25.9.589. [DOI] [PubMed] [Google Scholar]
- 131.Levy E., Sastre M., Kumar A., Gallo G., Piccardo P., Ghetti B., Tagliavini F. Codeposition of cystatin C with amyloid-β protein in the brain of Alzheimer disease patients. J. Neuropathol. Exp. Neurol. 2001;60(1):94–104. doi: 10.1093/jnen/60.1.94. [DOI] [PubMed] [Google Scholar]
- 132.Sastre M., Calero M., Pawlik M., Mathews P.M., Kumar A., Danilov V., Schmidt S.D., Nixon R.A., Frangione B., Levy E. Binding of cystatin C to Alzheimer’s amyloid β inhibits in vitro amyloid fibril formation. Neurobiol. Aging. 2004;25(8):1033–1043. doi: 10.1016/j.neurobiolaging.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 133.Sun B., Zhou Y., Halabisky B., Lo I., Cho S.H., Mueller-Steiner S., Devidze N., Wang X., Grubb A., Gan L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron. 2008;60(2):247–257. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palm D.E., Knuckey N.W., Primiano M.J., Spangenberger A.G., Johanson C.E. Cystatin C, a protease inhibitor: in degenerating rat hippocampal neurons following transient forebrain ischemia. Brain Res. 1995;691(1):1–8. doi: 10.1016/0006-8993(95)00520-z. [DOI] [PubMed] [Google Scholar]
- 135.Lukasiuk K., Pirttila T.J., Pitkanen A. Upregulation of cystatin C expression in the rat hippocampus during epileptogenesis in the amygdala stimulation model of temporal lobe epilepsy. Epilepsia. 2002;43(s5):137–145. doi: 10.1046/j.1528-1157.43.s.5.20.x. [DOI] [PubMed] [Google Scholar]
- 136.Inker L.A., Okparavero A. Cystatin C as a marker of glomerular filtration rate: prospects and limitations. Curr. Opin. Nephrol. Hypertens. 2011;20(6):631–639. doi: 10.1097/MNH.0b013e32834b8850. [DOI] [PubMed] [Google Scholar]
- 137.Dworkin L.D. Serum cystatin C as a marker of glomerular filtration rate. Curr. Opin. Nephrol. Hypertens. 2001;10(5):551–553. doi: 10.1097/00041552-200109000-00001. [DOI] [PubMed] [Google Scholar]
- 138.Laterza O.F., Price C.P., Scott M.G. Cystatin C: an improved estimator of glomerular filtration rate? Clin. Chem. 2002;48(5):699–707. [PubMed] [Google Scholar]
- 139.Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., Goudie D.R., Sandilands A., Campbell L.E., Smith F.J., O'Regan G.M. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38(4):441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 140.Hollox E.J., Huffmeier U., Zeeuwen P.L., Palla R., Lascorz J., Rodijk-Olthuis D., van de Kerkhof P.C., Traupe H., de Jongh G., den Heijer M., Reis A. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40(1):23. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Cid R., Riveira-Munoz E., Zeeuwen P.L., Robarge J., Liao W., Dannhauser E.N., Giardina E., Stuart P.E., Nair R., Helms C., Escaramís G. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 2009;41(2):211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yamada M. Cerebral amyloid angiopathy: an overview. Neuropathology. 2000;20(1):8–22. doi: 10.1046/j.1440-1789.2000.00268.x. [DOI] [PubMed] [Google Scholar]
- 143.Revesz T., Ghiso J., Lashley T., Plant G., Rostagno A., Frangione B., Holton J.L. Cerebral amyloid angiopathies: a pathologic, biochemical, and genetic view. J. Neuropathol. Exp. Neurol. 2003;62(9):885–898. doi: 10.1093/jnen/62.9.885. [DOI] [PubMed] [Google Scholar]
- 144.Fujihara S., Shimode K., Nakamura M., Kobayashi S., Tsunematsu T. Cerebral amyloid angiopathy with the deposition of cystatin C (gamma-trace) and beta-protein. Prog. Clin. Biol. Res. 1989;317:939. [PubMed] [Google Scholar]
- 145.Maruyama K., Ikeda S.I., Ishihara T., Allsop D., Yanagisawa N. Immunohistochemical characterization of cerebrovascular amyloid in 46 autopsied cases using antibodies to beta protein and cystatin C. Stroke. 1990;21(3):397–403. doi: 10.1161/01.str.21.3.397. [DOI] [PubMed] [Google Scholar]
- 146.Vattemi G., King Engel W., McFerrin J., Askanas V. Cystatin C colocalizes with amyloid-β and coimmunoprecipitates with amyloid-β precursor protein in sporadic inclusion-body myositis muscles. J. Neurochem. 2003;85(6):1539–1546. doi: 10.1046/j.1471-4159.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 147.Jensson O., Gudmundsson G., Arnason A., Blondal H., Petursdottir I., Thorsteinsson L., Grubb A., Lofberg H., Cohen D., Frangione B. Hereditary cystatin C (γ-trace) amyloid angiopathy of the CNS causing cerebral hemorrhage. Acta Neurol. Scand. 1987;76(2):102–114. doi: 10.1111/j.1600-0404.1987.tb03553.x. [DOI] [PubMed] [Google Scholar]
- 148.Carrell R.W., Gooptu B. Conformational changes and disease—serpins: prions and Alzheimer's. Curr. Opin. Struct. Biol. 1998;8(6):799–809. doi: 10.1016/s0959-440x(98)80101-2. [DOI] [PubMed] [Google Scholar]
- 149.Staniforth R.A., Giannini S., Higgins L.D., Conroy M.J., Hounslow A.M., Jerala R., Craven C.J., Waltho J.P. Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily. EMBO J. 2001;20(17):4774–4781. doi: 10.1093/emboj/20.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., Jaskolski M. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Mol. Biol. 2001;8(4):316–320. doi: 10.1038/86188. [DOI] [PubMed] [Google Scholar]
- 151.Bollengier F. C. cystatin, alias post-γ-globulin. a marker for multiple sclerosis? Clin. Chem. Lab. Med. 1987;25(9):589–594. doi: 10.1515/cclm.1987.25.9.589. [DOI] [PubMed] [Google Scholar]
- 152.Rinne R., Saukko P., Jarvinen M., Lehesjoki A.E. Reduced cystatin B activity correlates with enhanced cathepsin activity in progressive myoclonus epilepsy. Ann. Med. 2002;34(5):380–385. doi: 10.1080/078538902320772124. [DOI] [PubMed] [Google Scholar]
- 153.Shannon P., Pennacchio L.A., Houseweart M.K., Minassian B.A., Myers R.M. Neuropathological changes in a mouse model of progressive myoclonus epilepsy: cystatin B deficiency and Unverricht-Lundborg disease. J. Neuropathol. Exp. Neurol. 2002;61(12):1085–1091. doi: 10.1093/jnen/61.12.1085. [DOI] [PubMed] [Google Scholar]
- 154.Auerswald E.A., Genenger G., Assfalg-Machleidt I., Machleidt W., Engh R.A., Fritz H. Recombinant chicken egg white cystatin variants of the QLVSG region. European journal of biochemistry. 1992;209(3):837–845. doi: 10.1111/j.1432-1033.1992.tb17355.x. [DOI] [PubMed] [Google Scholar]
- 155.Hall A., Dalbøge H., Grubb A., Abrahamson M. Importance of the evolutionarily conserved glycine residue in the N-terminal region of human cystatin C (Gly-11) for cysteine endopeptidase inhibition. Biochem. J. 1993;291(1):123–129. doi: 10.1042/bj2910123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hall A., Hakansson K., Mason R.W., Grubb A., Abrahamson M. Structural basis for the biological specificity of cystatin C identification of leucine 9 in the N-terminal binding region as a selectivity-conferring residue in the inhibition of mammalian cysteine peptidases. J. Biol. Chem. 1995;270(10):5115–5121. doi: 10.1074/jbc.270.10.5115. [DOI] [PubMed] [Google Scholar]
- 157.Grubb A., Lofberg H., Barrett A.J. The disulphide bridges of human cystatin C (γ-trace) and chicken cystatin. FEBS Lett. 1984;170(2):370–374. [Google Scholar]
- 158.Schwabe C., Anastasi A., Crow H., McDonald J.K., Barrett A.J. Cystatin Amino acid sequence and possible secondary structure. Biochem. J. 1984;217(3):813–817. doi: 10.1042/bj2170813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990;9(6):1939. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Martin J.R., Jerala R., Kroon-Zitko L., Zerovnik E., Turk V., Waltho J.P. Structural characterisation of human stefin A in solution and implications for binding to cysteine proteinases. Eur. J. Biochem. 1994;225(3):1181–1194. doi: 10.1111/j.1432-1033.1994.1181b.x. [DOI] [PubMed] [Google Scholar]
- 161.Saxena I., Tayyab S. Protein proteinase inhibitors from avian egg whites. Cell. Mol. Life Sci. 1997;53(1):13–23. doi: 10.1007/PL00000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bode W., Engh R., Musil D.J., Thiele U., Huber R., Karshikov A., Brzin J., Kos J., Turk V. The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 1988;7(8):2593. doi: 10.1002/j.1460-2075.1988.tb03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Matrisian L.M. Metalloproteinases and their inhibitors in matrix remodeling. Trends Genet. 1990;6:121–125. doi: 10.1016/0168-9525(90)90126-q. [DOI] [PubMed] [Google Scholar]
- 164.Coussens L.M., Werb Z. Matrix metal loproteinases and the development of cancer. Chem. Biol. 1996;3(11):895–904. doi: 10.1016/s1074-5521(96)90178-7. [DOI] [PubMed] [Google Scholar]
- 165.Rudolph-Owen L.A., Chan R., Muller W.J., Matrisian L.M. The matrix metalloproteinase matrilysin influences early-stage mammary tumorigenesis. Cancer Res. 1998;58(23):5500–5506. [PubMed] [Google Scholar]
- 166.Walakovits L.A., Moore V.L., Bhardwaj N., Gallick G.S., Lark M.W. Detection of stromelysin and collagenase in synovial fluid from patients with rheumatoid arthritis and posttraumatic knee injury. Arthritis Rheum. 1992;35(1):35–42. doi: 10.1002/art.1780350106. [DOI] [PubMed] [Google Scholar]
- 167.Salivary (SD-type) cystatins: over one billion years in the making–but to what purpose? Crit. Rev. Oral Biol. Med. 2002;13(6):485–508. doi: 10.1177/154411130201300606. [DOI] [PubMed] [Google Scholar]
- 168.Dollery C.M., McEwan J.R., Henney A.M. Matrix metalloproteinases and cardiovascular disease. Circ. Res. 1995;77(5):863–868. doi: 10.1161/01.res.77.5.863. [DOI] [PubMed] [Google Scholar]
- 169.Grant G.A., Goldberg G.I., Wilhelm S.M., He C., Eisen A.Z. Activation of extracellular matrix metalloproteases by proteases and organomercurials. Matrix (Stuttgart, Germany) 1991;1(Supplement):217–223. [PubMed] [Google Scholar]
- 170.Fridman R., Toth M., Pena D., Mobashery S. Activation of progelatinase B (MMP-9) by gelatinase a (MMP-2) Cancer Res. 1995;55(12):2548–2555. [PubMed] [Google Scholar]
- 171.Ellerbroek S.M., Wu Y.I., Stack M.S. Type I collagen stabilization of matrix metalloproteinase-2. Arch. Biochem. Biophys. 2001;390(1):51–56. doi: 10.1006/abbi.2001.2345. [DOI] [PubMed] [Google Scholar]
- 172.Ray S., Lukyanov P., Ochieng J. Members of the cystatin superfamily interact with MMP-9 and protect it from autolytic degradation without affecting its gelatinolytic activities. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2003;1652(2):91–102. doi: 10.1016/j.bbapap.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 173.Massey D. Commentary: clinical diagnostic use of cystatin C. J. Clin. Lab. Anal. 2004;18(1):55–60. doi: 10.1002/jcla.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Perrone R.D., Madias N.E., Levey A.S. Serum creatinine as an index of renal function: new insights into old concepts. Clin. Chem. 1992;38(10):1933–1953. [PubMed] [Google Scholar]
- 175.Simonsen O., Grubb A., Thysell H. The blood serum concentration of cystatin C (γ-trace) as a measure of the glomerular filtration rate. Scand. J. Clin. Lab. Investig. 1985;45(2):97–101. doi: 10.3109/00365518509160980. [DOI] [PubMed] [Google Scholar]
- 176.Reed C.H. Diagnostic applications of cystatin C. Br. J. Biomed. Sci. 2000;57(4):323. [PubMed] [Google Scholar]
- 177.Van Oss C.J., Gillman C.F., Bronson P.M., Border J.R. Opsonic properties of human serum alpha-2 hs glycoprotein. Immunol. Commun. 1974;3(4):329–335. doi: 10.3109/08820137409061113. [DOI] [PubMed] [Google Scholar]
- 178.Lebreton J.P., Joisel F., Raoult J.P., Lannuzel B., Rogez J.P., Humbert G. Serum concentration of human alpha 2 HS glycoprotein during the inflammatory process: evidence that alpha 2 HS glycoprotein is a negative acute-phase reactant. J. Clin. Invest. 1979;64(4):1118. doi: 10.1172/JCI109551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Oss C.J., Gillman C.F., Bronson P.M., Border J.R. Opsonic properties of human serum alpha-2 hs glycoprotein. Immunol. Commun. 1974;3(4):329–335. doi: 10.3109/08820137409061113. [DOI] [PubMed] [Google Scholar]
- 180.Lewis J.G., Andre C.M. Effect of human alpha 2HS glycoprotein on mouse macrophage function. Immunology. 1980;39(3):317. [PMC free article] [PubMed] [Google Scholar]
- 181.Lewis J.G., Andre C.M. Enhancement of human monocyte phagocytic function by alpha 2HS glycoprotein. Immunology. 1981;42(3):481. [PMC free article] [PubMed] [Google Scholar]
- 182.Chapman H.A., Jr, Reilly J.J., Jr, Yee R., Grubb A. Macrophages in vitro1-3. Am. Rev. Respir. Dis. 1990;141:698–705. doi: 10.1164/ajrccm/141.3.698. [DOI] [PubMed] [Google Scholar]
- 183.Pierre P., Mellman I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell. 1998;93(7):1135–1145. doi: 10.1016/s0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- 184.Verdot L., Lalmanach G., Vercruysse V., Hartmann S., Lucius R., Hoebeke J., Gauthier F., Vray B. Cystatins up-regulate nitric oxide release from interferon-γ-activated mouse peritoneal macrophages. J. Biol. Chem. 1996;271(45):28077–28081. doi: 10.1074/jbc.271.45.28077. [DOI] [PubMed] [Google Scholar]
- 185.Hartmann S., Schonemeyer A., Sonnenburg B., Vray B., Lucius R. Cystatins of filarial nematodes up-regulate the nitric oxide production of interferon-γ-activated murine macrophages. Parasite Immunol. 2002;24(5):253–262. doi: 10.1046/j.1365-3024.2002.00459.x. [DOI] [PubMed] [Google Scholar]
- 186.Marletta M.A. Nitric oxide synthase structure and mechanism. Am. Soc. Biochem. Mol. Biol. 1993;(June 15) [PubMed] [Google Scholar]
- 187.Kolb H., Kolb-Bachofen V. Nitric oxide: a pathogenetic factor in autoimmunity. Immunol. Today. 1992;13(5):157–160. doi: 10.1016/0167-5699(92)90118-Q. [DOI] [PubMed] [Google Scholar]
- 188.Das L., Datta N., Bandyopadhyay S., Das P.K. Successful therapy of lethal murine visceral leishmaniasis with cystatin involves up-regulation of nitric oxide and a favorable T cell response. J. Immunol. 2001;166(6):4020–4028. doi: 10.4049/jimmunol.166.6.4020. [DOI] [PubMed] [Google Scholar]
- 189.Anastasi A., Brown M.A., Kembhavi A.A., Nicklin M.J., Sayers C.A., Sunter D.C., Barrett A.J. Cystatin, a protein inhibitor of cysteine proteinases. Improved purification from egg white, characterization, and detection in chicken serum. Biochem. J. 1983;211(1):129–138. doi: 10.1042/bj2110129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Anfinsen C.B. Principles that govern folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 191.Kuznetsova I.M., Stepanenko O.V., Turoverov K.K., Zhu L., Zhou J.M., Fink A.L., Uversky V.N. Unraveling multistate unfolding of rabbit muscle creatine kinase. Biochimica et Biophysica Acta (BBA)-Protein Struct. Mol. Enzymol. 2002;1596(1):138–155. doi: 10.1016/s0167-4838(02)00212-1. [DOI] [PubMed] [Google Scholar]
- 192.Cardinale A., Filesi I., Biocca S. Aggresome formation by anti-Ras intracellular scFv fragments. Eur. J. Biochem. 2001;268(2):268–277. doi: 10.1046/j.1432-1033.2001.01876.x. [DOI] [PubMed] [Google Scholar]
- 193.Ohgushi M., Wada A. ‘Molten-globule state’: a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;164(1):21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 194.Uversky V.N., Karnoup A.S., Segel D.J., Seshadri S., Doniach S., Fink A.L. Anion-induced folding of Staphylococcal nuclease: characterization of multiple equilibrium partially folded intermediates. J. Mol. Biol. 1998;278(4):879–894. doi: 10.1006/jmbi.1998.1741. [DOI] [PubMed] [Google Scholar]
- 195.Bushmarina N.A., Kuznetsova I.M., Biktashev A.G., Turoverov K.K., Uversky V.N. Partially folded conformations in the folding pathway of bovine carbonic anhydrase II: a fluorescence spectroscopic analysis. ChemBioChem. 2001;2(11):813–821. doi: 10.1002/1439-7633(20011105)2:11<813::AID-CBIC813>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 196.Walker L.C., LeVine H. Corruption and spread of pathogenic proteins in neurodegenerative diseases. J. Biol. Chem. 2012;287(40):33109–33115. doi: 10.1074/jbc.R112.399378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Krebs M.R., Domike K.R., Donald A.M. Protein aggregation: more than just fibrils. Biochem. Soc. Trans. 2009;37(4):682–686. doi: 10.1042/BST0370682. [DOI] [PubMed] [Google Scholar]
- 198.Cromwell M.E., Hilario E., Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8(3):E572–9. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Agnati L.F., Leo G., Genedani S., Piron L., Rivera A., Guidolin D., Fuxe K. Common key-signals in learning and neurodegeneration: focus on excito-amino acids, β-amyloid peptides and α-synuclein. J. Neural Transm. 2009;116(8):953–974. doi: 10.1007/s00702-008-0150-4. [DOI] [PubMed] [Google Scholar]
- 200.Zhu Y.J., Lin H.A., Lal R. Fresh and nonfibrillar amyloid β protein (1–40) induces rapid cellular degeneration in aged human fibroblasts: evidence for AβP-channel-mediated cellular toxicity. FASEB J. 2000;14(9):1244–1254. doi: 10.1096/fasebj.14.9.1244. [DOI] [PubMed] [Google Scholar]
- 201.Aguzzi A., Calella A.M. Prions: protein aggregation and infectious diseases. Physiological reviews. 2009;89(4):1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 202.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]