Abstract
Effective antiviral agents are needed to treat severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infection. We assessed the antiviral potential of recombinant interferons against two clinical isolates of SARS-CoV—FFM-1, from Frankfurt patients, and Hong Kong—replicated in Vero and Caco2 cells. Interferon β was five to ten times more effective in Caco2 cells. Interferon α effectively inhibited SARS-CoV replication, but with a selectivity index 50–90 times lower than that for interferon β. Interferon γ was slightly better than interferon α in Vero cell cultures, but was completely ineffective in Caco2 cell cultures. Interferon β could be useful alone or in combination with other antiviral drugs for the treatment of SARS.
Several clinical isolates of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) have been isolated, including FFM-1 and FFM-2 from SARS patients admitted to the Clinical Centre of Frankfurt University.1 So far, no specific treatment has been identified for SARS, but attempts are being made to identify antiviral agents that could be effective in treating the SARS-CoV infection.2
We assessed the antiviral potential of recombinant interferons α, β, and γ against two clinical isolates (FFM-1 and Hong Kong: obtained from Dr Niedrig, Robert Koch Institute, Berlin) of SARS-CoV. We used commercially available interferons (Intron A [interferon α-2b], Essex Pharma, Munich, Germany; Betaferon [interferon β-1b], Schering, Berlin; and Imukin [interferon γ-1b], Boehringer, Ingelheim) that have been used for their antiviral, antitumour, and immunomodulating activities. We visually scored cytopathogenic effects induced by the virus 72 h after infection in 96-well microplates on confluent layers of Vero and Caco2 cells, the latter derived from human colonic tumour, treated with interferons 24 h before and immediately after virus infection (mutiplicity of infection 0–01). The selectivity index was defined as the ratio of the concentration of the interferon that reduced cell viability to 50% (CC50) to the concentration of the compound needed to inhibit the cytopathic effect to 50% of control value (EC50). We assessed the cytotoxic effects of the drugs with an MTT cell-proliferative Kit I (Roche, Mannheim, Germany). Viral titres in supernatants collected from infected cultures 72 h after infection were measured by the 50% tissue culture infective dose (TCID50) calculated from cytopathogenic effects induced in monolayers incubated with varying dilutions of the supernatants. To validate the interferon effects on SARS-CoV, we included vesicular stomatitis virus (strain Indiana), replication of which is extremely sensitive to interferon action.
Interferon β was the most potent inhibitor of SARS-CoV (table ). In Vero cells, selectivity index of interferon β activity for SARS-CoV strain FFM-1 was 50-fold and 25-fold higher than that of interferon α and interferon γ, respectively. Sensitivity of the Hong Kong isolate was similar to that for the FFM-1 strain. Interferon β (EC50 105 IU/mL) was more potent than interferons α or γ (EC50s 6500 IU/mL and 1700 IU/mL, respectively). Vesicular stomatitis virus strain Indiana was much more sensitive to interferon treatment than SARS-CoV; interferon β had the highest antiviral activity in Vero cell cultures (EC50 18 IU/mL).
Table.
Effect of Interferons on virus-induced cytopathogenic effects of SARS-CoV and vesicular stomatitis virus
| Mean (SD) EC50 (IU/mL)* | CC50 (IU/mL) | Selectivity index | |||||
|---|---|---|---|---|---|---|---|
| SARS-CoV FFM-1 | SARS-CoV Hong Kong | VSV Indiana | SARS-CoV FFM-1 | SARS-CoV Hong Kong | VSV Indiana | ||
| Vero† | |||||||
| Interferon α | 4950 (890) | 6500 (980) | 638 (98) | >10000 | >2 | >1·5 | >16 |
| Interferon β | 95 (17) | 105 (21) | 18·0 (3·7) | >10000 | >105 | >95 | >556 |
| Interferon γ | 2500 (340) | 1700 (290) | 508 (62) | >10000 | >4 | >5·9 | >20 |
| Caco2† | |||||||
| Interferon α | 1530 (220) | 880 (130) | 3700 (420) | >10000 | >6·5 | >11·4 | >2·7 |
| Interferon β | 21·0 (3·9) | 9·2 (2·1) | 1030 (150) | >10000 | >476 | >1087 | >9·7 |
| Interferon γ | >10000 | >10000 | >10000 | >10000 | NC | NC | NC |
VSV=vesicular stomatitis virus. NC=not calculable
Mean (SD) of eight assays.
Cells treated with interferons 24 h before and during virus infection.
We studied the action of interferons on SARS-CoV-induced cytopathogenic effects in Vero cells when the drugs were added after infection (1 h adsorption period). Interferon β was active against FFM-1 strain (EC50 560 IU/mL) but interferons α and β were not. Therefore, only interferon β can be used as an antiviral agent after infection, even though its EC50 value shows a six fold increase over the value obtained in pretreated cultures (table).
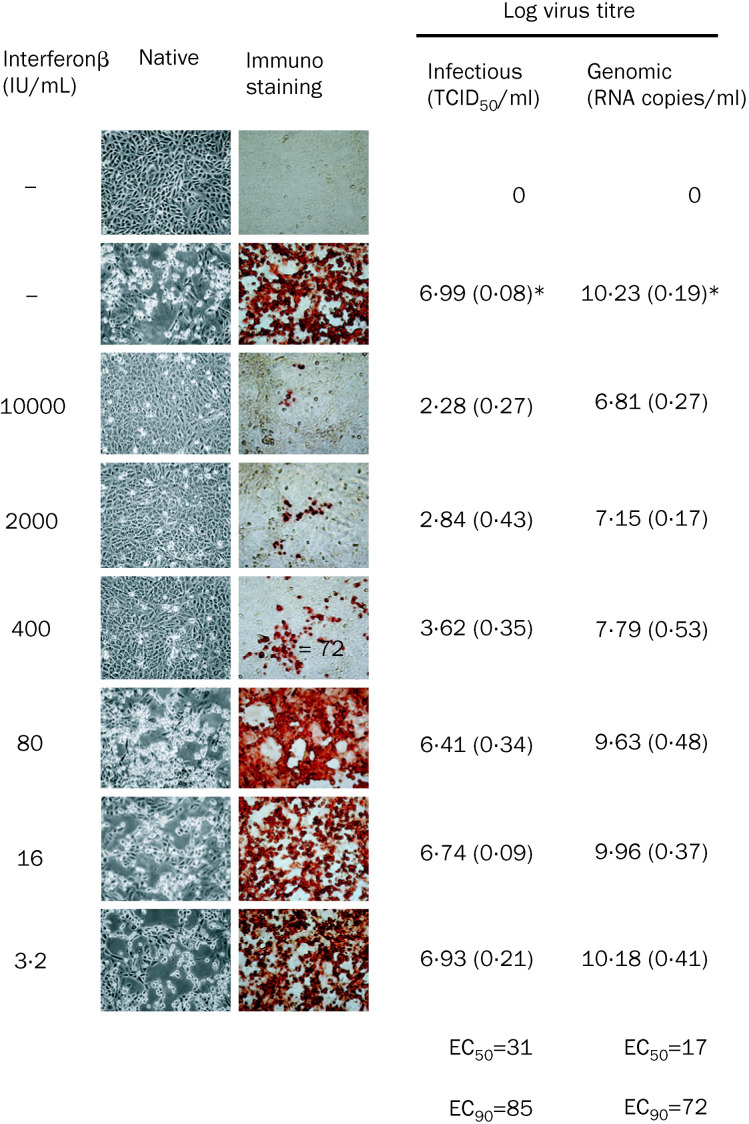
We assessed also the relevance of inhibition of virus replication for suppression of virus-induced cytopathogenic effects in cultures treated with interferon β 24 h before and immediately after virus infection. We infected Vero cells with FFM-1 strain at multipicity of infection 0·01 and assessed viral replication by peroxidase staining with immune serum after treatment with different concentrations of interferon (figure ). Native cells, or cells fixed with 60 parts methanol to 40 parts acetone were assessed 72 h after infections. We also quantified the genomic titre (RNA copies/mL) with real-time PCR. Interferon β showed a dose-dependent inhibition of the production of infectious virus in culture. In the absence of virus induced cytopathogenic effects, however, interferon β treatment did not completely reduce virus titre and antigen production. In cultures treated with 10 000 IU/mL, a reduction of about 5×104-fold was noted compared with the untreated control (1·9×102 vs 9·7×106 TCID50 /mL). The number of gene copies are reduced by interferon β in a dose-dependent way; however, the magnitude is around 3·3–4·5 log higher than the infectious virus titre measured in cell culture. This difference may be due to the presence of non-infectious viral RNA or the overexpression of the sequence amplified in our real-time PCR system. Similar observations have been made by Garcia and colleagues3 on the quantitative detection of Rift valley fever virus gene copies to assess the antiviral effect of drugs.
Figure.
Effect of interferon β on replication of SARS-CoV strain: FFM-1 in Vero cells 72 h after infection
Virus detected in serum with peroxidase staining. *Values represent mean (SD) from three experiments, each done in triplicate.
Interferons α and β share components of the same receptor and are type I interferons. Interferon γ uses a different receptor system and is a type II interferon. We noted that interferon β was by far more effective than interferon α against SARS-CoV and vesicular stomatitis virus. Although interferons α and β primarily induce the same proteins, some proteins may be induced by one better than the other. By use of oligonucleotide arrays, Der and colleagues4 identified the genes differentially regulated by interferons. They reported more than a 21-fold increase of MxA protein by interferon α, more than a 31-fold increase by interferon β, and none by interferon γ. Gene-array analysis of Caco2 cells infected with FFM-1 in our laboratory has shown a nine-fold increase of MxA gene expression in cultures 24 h after infection. This increase could be one of the reasons for amplification of interferon β effect that yields the higher antiviral potential.
Interferons α and γ, alone or in combination, are partly effective against animal coronaviruses, mouse hepatitis virus, and transmissible gastroenteritis virus; a few data on human studies report a limited response of interferon α by the intranasal route.5 We showed that interferons inhibit SARS-CoV replication in vitro. Interferon β was most potent, showing prophylactic protection and antiviral potential after infection. Interferon β could be the drug of choice, alone or in combination with other antiviral drugs,2 in the treatment of SARS.
Acknowledgments
Acknowledgments
This study was supported by the Ministry of Science and Culture of the State of Hessen, Germany. We thank Marhild Kortenbusch and Gesa Meincke for expert technical assistance.
References
- 1.Drosten C, Gunther S, Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia S, Crance JM, Billecocq A. Quantitative real-time PCR detection of Rift Valley fever virus and its application to evaluation of antiviral compounds. J Clin Microbiol. 2001;39:4456–4461. doi: 10.1128/JCM.39.12.4456-4461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitkaranta A, Nokso-Koivisto J, Jantti V, Takala A, Kilpi T, Hovi T. Lowered yields of virus-induced interferon production in leukocyte cultures and risk of recurrent respiratory infections in children. J Clin Virol. 1999;14:199–205. doi: 10.1016/S1386-6532(99)00056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]