Summary
Background
An epidemic of severe acute respiratory syndrome (SARS) has been associated with an outbreak of atypical pneumonia originating in Guangdong Province, People's Republic of China. We aimed to identify the causative agent in the Guangdong outbreak and describe the emergence and spread of the disease within the province.
Methods
We analysed epidemiological information and collected serum and nasopharyngeal aspirates from patients with SARS in Guangdong in mid-February, 2003. We did virus isolation, serological tests, and molecular assays to identify the causative agent.
Findings
SARS had been circulating in other cities of Guangdong Province for about 2 months before causing a major outbreak in Guangzhou, the province's capital. A novel coronavirus, SARS coronavirus (CoV), was isolated from specimens from three patients with SARS. Viral antigens were also directly detected in nasopharyngeal aspirates from these patients. 48 of 55 (87%) patients had antibodies to SARS CoV in their convalescent sera. Genetic analysis showed that the SARS CoV isolates from Guangzhou shared the same origin with those in other countries, and had a phylogenetic pathway that matched the spread of SARS to the other parts of the world.
Interpretation
SARS CoV is the infectious agent responsible for the epidemic outbreak of SARS in Guangdong. The virus isolated from patients in Guangdong is the prototype of the SARS CoV in other regions and countries.
Introduction
Since November, 2002, an infectious agent has caused outbreaks of an atypical pneumonia in Guangdong Province, southern China.1 The illness usually began with high fever and mild respiratory symptoms, but rapidly progressed to pneumonia within a few days.2 At the end of February, 2003, the disease had spread to neighbouring regions and countries.3, 4 The disease was severe, transmissible from person to person, and seemed to cause clusters of disease in health-care workers. It was named severe acute respiratory syndrome (SARS), and WHO issued a global alert about the disease on March 13, 2003.5, 6 SARS outbreaks occurred in South-East Asia, North America, and Europe, and gave rise to the first pandemic of the 21st century.
By July 11, 2003, WHO had recorded 8437 cases of SARS worldwide and attibuted 813 deaths to the disease. Most cases were in mainland China and Hong Kong.7 Although a novel coronavirus, at the end of March, 2003, SARS coronavirus (CoV), was identified as the infectious agent in the syndrome.8, 9, 10 The origin of this pandemic remains unclear. Here we report epidemiological data from the early phase of the SARS outbreak in Guangdong and the findings of virological investigation of 55 patients with SARS who were admitted to hospitals in Guangzhou, in mid-February, 2003.
Methods
Epidemiological investigation
To track the emergence of SARS, we gathered epidemiological data from clinical records of patients admitted with a diagnosis of atypical pneumonia, or we used reports issued by local health authorities and interviews done with doctors in charge of the treatment of SARS patients in Guangzhou. These data were used to assess how the disease spread, first within Guangdong Province and then to the rest of world. We used the WHO definition of SARS in our investigations.11
To identify the causative agent in SARS, we studied 55 patients admitted to the First Affiliated Hospital of Guangzhou Medical College and Guangzhou Chest Hospital between Jan 24, and Feb 18, 2003. 27 of these patients were health-care workers from hospitals in Guangzhou. All patients were diagnosed with atypical pneumonia, confirmed by chest radiographs, and had symptoms and signs that matched the WHO definition for SARS.11 Table 1 shows patients' characteristics. To isolate and identify the infectious agent, we took nasopharyngeal aspirates for virus isolation, and serum for antibody detection.
Table 1.
Characteristics of patients with SARS from Guangdong, China (Jan 24 to Feb 18, 2003)
| Patients (n=55) | |
|---|---|
| Geographic distribution | |
| Guangzhou | 37 |
| Foshan | 12 |
| Shenzhen | 2 |
| Heyuan | 1 |
| Quingyuan | 1 |
| Jiangmen | 1 |
| Zengcheng | 1 |
| Men | 28 (51%) |
| Age (years) | 38·4 (12·7) |
| Duration of fever (days) | 11·4 (6·8) |
| Contact history* | 41 (75%) |
| Shortness of breath | 42 (76%) |
| Health-care workers | 27 (49%) |
| White blood cell count (×109/L) | 6·8 (4·6) |
| Lymphocyte count (×109/L) | 0·9 (0·4) |
| Shadows on chest radiograph | 55 (100%) |
| Impaired liver-function test | 15 (27%) |
Data are mean (SD) or n (%) unless otherwise stated.
Definitive history of contact with known SARS patients.
Virus isolation and identification
Between Feb 11, and Feb 18, 2003, we gathered 40 specimens of nasopharyngeal aspirate from patients with SARS. The specimens were stored in viral transport medium and inoculated into 9-to-10-day-old chicken embryos and cultured for conventional respiratory pathogens on MDCK, Hep-2, and A549 cell lines on the day of sample collection. Subsequently, these frozen and thawed samples were cultured on fetal rhesus kidney (FRhK-4) cells for SARS CoV in methods described elsewhere.8 All virus isolates were identified by serological tests and RT-PCR.
Serological tests and antibody detection
To confirm the infectious agent responsible for the outbreak of SARS, we collected sera from 55 patients and tested for antibodies. 22 paired sera were obtained, mainly from patients from Guangzhou in the acute stage of the disease. Another 33 single serum samples came from patients, some of whom had been transferred from neighbouring cities, who presented more than 2 weeks after the onset of symptoms. We used 60 serum samples from healthy adults from Guangzhou as controls. We tested all sera for the presence of antibodies against coronavirus using an immunofluorescence assay (IFA).8 For sera that were positive, we titrated to the endpoint using serial dilutions. We selected one positive paired set of samples for use in detection of viral infected cells in patient's nasopharyngeal aspirate samples.
Genetic analysis
We extracted viral RNA from the filtered supernatant of virus-infected FRhK-4 cells with the RNesay Mini Kit (Qiagen, Chatsworth, CA, USA). Based on the first fulllength genome sequence of a coronavirus isolated from a SARS patient in Canada,12 a primer (5′TTTTTTTTTTTTTTTGTGATT3′) was designed for reverse transcription of viral RNA. Subsequently, we did PCR using a series of primers prepared in our laboratory (primer sequences available on request from author).
After the purification of PCR products, cycling sequencing reactions were done to determine nucleotide sequence, by use of methods reported elsewhere.13 All sequence data were translated and analysed by Wisconsin Software Package, Version 10.1 (Genetics Computer Group, Madison, WI, USA). We constructed the phylogenetic relation of samples with the neighbourjoining method with bootstrap analysis (1000 replicates) using molecular evolutionary genetics analysis 2 (MEGA 2).14 The sequences generated by our study are in GenBank (accession numbers AY304490, AY304491, and AY304495).
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Epidemiology
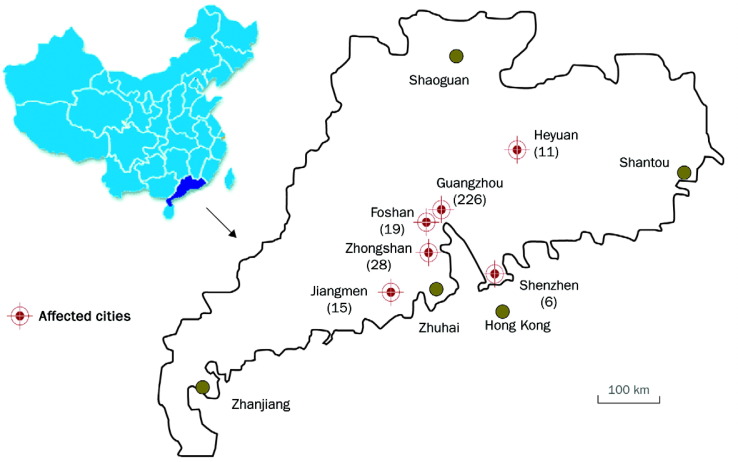
Before the major outbreak of SARS in February, 2003, smaller outbreaks compatible with the disease had been noted in five cities around Guangzhou in the preceding 2 months. The sequence of the SARS outbreaks in the Guangdong region is shown in figure 1 .
Figure 1.
SARS outbreaks in Guangdong Province, People's Republic of China
The geographic distribution of SARS outbreak in Guangdong Nov 16, 2002, to Feb 9, 2003. Number of cases are shown in brackets. Approximate dates of the onset of the outbreaks for each city were Foshan, Nov 16, 2002; Heyuan, Dec 17, 2002; Zhongshan, Dec 26, 2003; Guangzhou, Jan 31, 2003; Jiangmen, Jan 10, 2003; Shenzhen, Jan 15, 2003.
The first wave—the first case of SARS that fulfilled the WHO definition was reported in Foshan, a city about 20 km from Guangzhou, on Nov 16, 2002. However, there were no clinical samples from this patient available for virological investigation. On Dec 17, 2002, a chef from Heyuan who worked at a restaurant in Shenzhen was reported to have atypical pneumonia, and became the second identified case of SARS. He felt unwell in Shenzhen but sought medical treatment in Heyuan. He had high fever and mild respiratory symptoms; radiographic examination showed shadows in both lungs. His wife, two sisters, and seven medical staff were infected, and all had the same clinical manifestations. This patient, as a chef, came into regular contact with several types of live caged animals used as exotic game food.
Soon after the Heyuen outbreak, similar cases were also noted on Dec 26, 2002, in Zhongshan, a city about 90 km from Guangzhou. Between Dec 26, 2002, and Jan 20, 2003, 28 cases (10 men, 18 women, aged 1–53 years) were clinically recognised as atypical pneumonia with an unknown pathogen (figure 2 ). 13 of the 28 patients were health-care workers, but none of their family members was infected. Four patients required mechanical ventilation.2
Figure 2.
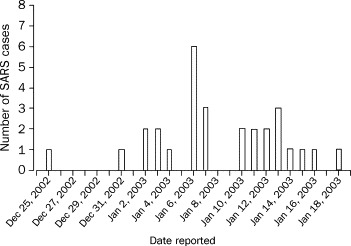
The outbreak of SARS in Zhongshan, People's Republic of China
28 cases were reported from Dec 26, 2002 to Jan 19, 2003. 13 were health-care workers.
The second wave—since Jan 12, 2003, some complicated SARS cases from the neighbouring cities were transferred to the major hospitals in Guangzhou for better medical care. The index community SARS case of Guangzhou was a 46-year-old male seafood merchant who was admitted to the Second Affiliated Hospital of Zhongshan University on Jan 31, 2003. He had travelled to Zhongshan in mid-January. The patient stayed in the hospital for only 18 h, but caused more than 30 hospital staff to become infected. On Feb 1, 2003, the Guangzhou index patient was transferred to the Third Affiliated Hospital of Zhongshan University. During the transfer, the ambulance driver, two doctors, and two nurses were infected. Within the next 8 days, 20 more medical staff who had been in contact with the patient became infected. At the same time, 19 family members or relatives of the patient fell ill with similar clinical manifestations after close personal contact with the patient. This patient generated a large cluster of secondary infections that further spread and gave rise to the outbreak in Guangzhou. In mid-February, one doctor working at the Second Affiliated Hospital of Zhongshan travelled to Hong Kong, and became the major source of the SARS outbreak in Hong Kong, which led on to the pandemic.4, 8, 9, 10
From Nov 16, 2002 to Feb 9, 2003, 305 cases of SARS were reported, 105 of which were in health-care workers (figure 1). The outbreak was characterised by infection within hospitals and family clusters, suggesting that transmission arises through close contact with infected patients. By April 20, 2003, 1317 SARS cases and 48 deaths had been reported.1
Clinical features
Characteristics of 55 patients with SARS who acquired the disease between Jan 24, and Feb 18, 2003, are shown in table 1. 18 cases were transferred from neighbouring cities of Guangzhou during outbreaks from December, 2002, to January, 2003. About half (49%) of infected people in our study group were health-care workers, mainly from two hospitals, and the group who were infected during the transfer of the first index SARS case of Guangzhou. Other patients were from the community of Guangzhou.
All patients had high fever (>38°C) lasting for a mean of 11 days (SD 6·8), and developed pneumonia within 1–4 days of admission. Most had a contact history with a SARS patient 1–5 days before the onset of symptoms. Many patients presented with shortness of breath; some had myalgia and fatigue. A few had other mild respiratory infection symptoms, such as cough and sore throat. Radiography confirmed the diagnosis of pneumonia in all patients. White blood-cell count was slightly decreased, and lymphocyte count was strikingly reduced (table 1).
Cause
Culture of 40 nasopharyngeal specimens in cell lines used for respiratory diseases yielded four influenza A (H3N2) viruses and one adenovirus. However, serological investigation suggested that these conventional viruses were not the primary infectious agent in other patients with SARS for whom we had paired sera samples (data not shown). The H3N2 virus isolates were partially sequenced and genotyped—we noted typical human H3N2 viruses in all gene segments without evidence of reassortment with avian influenza viruses. So, all samples of nasopharyngeal aspirate were recultured on FRhK-4 cells. Positive cytopathic effect was noted in three samples after the inoculation. All three isolates were confirmed to be SARS CoV by RTPCR and sequencing PCR products with specific primers.12
The three viruses were isolated from specimens from a 36-year-old female health-care worker (GZ43), a 49-year-old female clerk (GZ50), and a 39-year-old male health-care worker (GZ60). All were from Guangzhou and had had contact with a patient with SARS before the onset of their disease. Their specimens were all collected on Feb 18, 2003 at Guangzhou Chest Hospital.
To determine whether these SARS CoV isolates were responsible for the Guangdong outbreak, paired sera from 22 patients and single sera from 33 patients were tested for antibodies to SARS CoV. For paired sera, the first samples were obtained from 18 patients at 3–5 days from disease onset, and four patients at days 7–10. Samples from days 3–5 had no detectable specific antibodies at 1:10 dilution of sera. The titre of specific antibodies was 10–160 in samples taken on days 7–10, but the antibody titres were at least four-fold higher in the second serum sample, taken 15 days after the onset, for these four patients. SARS CoV antibodies were also detected in 28 of 33 single sera from SARS patients. All 18 patients who had been transferred from neighbouring cities and the ambulance driver had SARS CoV antibodies (table 2 ). The four patients from whom H3N2 influenza was isolated had no detectable SARS CoV antibody in their serum during convalescence. No specific antibodies against SARS CoV were detected in sera taken from 60 healthy donors during the same period of the study. Our findings suggest that most patients from Guangdong were recently infected by SARS CoV.
Table 2.
Antibodies against coronavirus in patients and controls
| Positive rate | Mean titre in positive sera (SD) | |
|---|---|---|
| Patients with SARS | ||
| Paired (n=22) | 20 (91%) | 1312 (790) |
| Single (n=33) | 28 (85%) | 860 (583) |
| Controls | ||
| Single (n=60) | 0 | NA |
NA=not applicable.
Paired sera—one sample taken during the acute stage of the disease, and one during convalescence—were used to detect viral antigen by IFA in cells from the nasopharyngeal samples from 23 patients in whom residual specimens were available. 14 samples from the convalesent stage showed viral antigen, but none was present in the acute serum (figure 3 ). Most SARS CoVinfected cells were bronchial columnar cells and a few positive cells were squamous cells.
Figure 3.
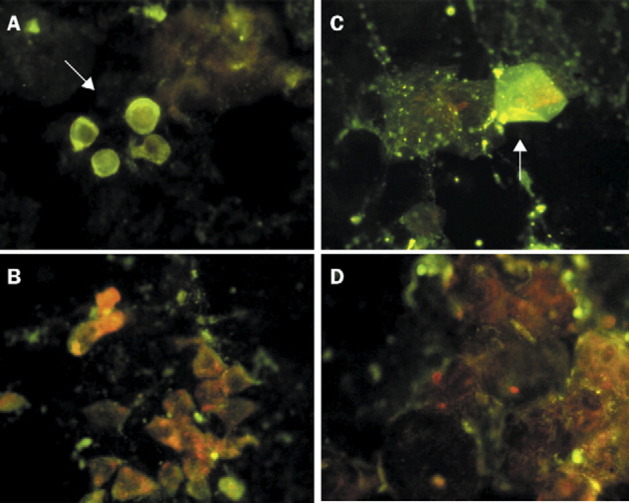
Immunofluorescent identification of coronavirus-infected cells in nasopharyngeal samples
Arrows show respiratory epithelia cells: bronchial columnar cells (A) and squamous cells (B), which were reactive with the convalescent serum, but not with the acute serum (C and D) (×400). Paired sera used in the immunofluorescent tests were obtained from a patient with confirmed coronavirus infection. Titre of coronavirus-specific antibodies was 1:1280 in the convalescent serum and less than 1:10 in acute serum.
Genetic analysis
Sequence comparison—all three virus isolates were partially sequenced to cover the polyprotein (P) gene and the S gene. Blast search showed that all Guangzhou SARS CoV had the highest homology (>99%) with the sequences of the SARS CoV that were isolated from SARS cases in Hong Kong, Beijing, Singapore, Canada and the USA, suggesting that all of these SARS CoV are closely related. All of these sequences have very low homologies with known coronaviruses that arise in human beings and animals.12, 15
When the S gene sequences of three Guangzhou isolates were compared with that of the other 11 SARS CoV viruses available in GenBank, 11 asynonymous and six synonymous point mutations were recognised in the whole coding region (table 3 ). Five unique aminoacid residues, including 224R, 716L, 931R, 2332D, and 3536P (numbering starting from the coding region of S gene), were recognised only in those SARS CoV isolated from patients in the early phase of SARS outbreak of Guangzhou (table 3). One aminoacid deletion was identified in GZ43 and GZ60 (residue 214 to 216 nucleotide).
Table 3.
Nucleotide sequence variation of the S gene of SARS CoV viruses
| Nucleotide position numbering start on coding region* | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 146† | 224† | 230† | 654 | 716† | 731† | 931† | 1026 | 1031 | 1502 | 1729† | 1994† | 2332† | 2380† | 3075 | 3381 | 3536† | |
| Virus | |||||||||||||||||
| GZ01 | C (S) | C (T) | A (D) | C (P) | T (L) | C (T) | A (R) | G (R) | G (K) | A (F) | T (S) | T (L) | G (D) | T (S) | C (C) | T (L) | T (L) |
| GZ43 | C (S) | G (R) | A (D) | T (P) | T (L) | C (T) | A (R) | G (R) | G (K) | T (F) | T (S) | C (S) | G (D) | C (P) | C (C) | T (L) | C (P) |
| GZ60 | C (S) | G (R) | A (D) | T (P) | C (S) | C (T) | A (R) | G (R) | G (K) | T (F) | T (S) | C (S) | G (D) | C (P) | C (C) | T (L) | C (P) |
| GZ50 | T (L) | C (T) | A (D) | T (P) | T (L) | C (T) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | G (D) | C (P) | T (C) | T (L) | T (L) |
| HKU-36871 | C (S) | C (T) | A (D) | T (P) | C (S) | C (T) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| CUHK-W1 | C (S) | C (T) | A (D) | T (P) | C (S) | C (T) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| BJ01 | C (S) | C (T) | A (D) | T (P) | C (S) | C (T) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| HKU-39849 | C (S) | C (T) | G (G) | T (P) | C (S) | T (I) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| TOR2 | C (S) | C (T) | G (G) | T (P) | C (S) | T (I) | G (G) | A (R) | A (K) | T (F) | G (A) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| Urbani | C (S) | C (T) | G (G) | T (P) | C (S) | T (I) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | C (L) | T (L) |
| CUHK-Su10 | C (S) | C (T) | G (G) | T (P) | C (S) | T (I) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
| SIN 2677 | C (S) | C (T) | G (G) | T (P) | C (S) | T (I) | G (G) | A (R) | A (K) | T (F) | T (S) | T (L) | T (Y) | C (P) | T (C) | T (L) | T (L) |
For GZ43 and GZ60, there is a three-nucleotide deletion located at 214–216 (AAT) which encode isoleucine (I).
Asynonymous point mutations.
Phylogenetic analysis—investigations of the complete S gene nucleotide sequences of SARS CoV shows that these novel coronaviruses clustered into two distinct sublineages. The first one only contains viruses isolated from Guangzhou (GZ01, GZ43, and GZ60). The second includes SARS CoV isolates from other regions after SARS spread. Of note is that GZ50 also joined the second sublineage, but still kept an outgroup relation with all other SARS CoV from other regions (figure 4 ), suggesting that GZ50 might be the prototype of SARS CoV noted in other regions. The phylogenetic relation matches with the disease spread pathway—ie, from Guangzhou to Hong Kong, and then other regions.
Figure 4.
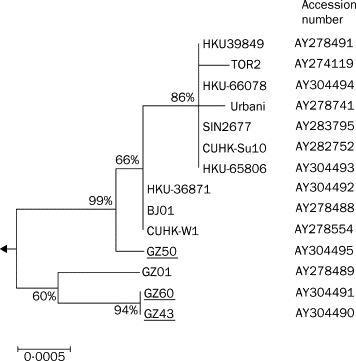
Phylogenetic analysis of nucleotide acid sequence of spike gene of SARS CoV viruses
Bootstrap values are shown as a percentage. The scale bar shows genetic distance estimated using Kimura's two parameter substitution model.14 The nucleotide sequences of representative SARS CoV S genes (S gene coding region residue, 3765 bp) were analysed. Viruses sequenced in this study are underlined, and the other sequences used in the analysis can be accessed in GenBank with accession numbers as shown.
Discussion
Virological surveillance in Guangdong Province in mid-February, 2003, showed that the outbreak of a disease fitting the WHO case definition of SARS started in mid-November, 2002. Epidemiological investigations suggest that SARS probably first emerged in satellite cities of Guangzhou, circulating for at least 2 months before causing a major outbreak in Guangzhou itself. Serological tests and virus isolation confirmed that SARS CoV was the primary infectious agent of these early cases of SARS. Genetic analysis of these SARS CoV isolates showed that they belonged to two different subgroups, one closely related to that noted in Hong Kong and other countries, and one that seemingly had not spread to people in other regions. Phylogenetic analysis also suggests that all SARS CoV share the same origin and most probably first originated in Guangdong, southern China.
In this report, our investigation covered 55 patients with SARS from Guangzhou and its neighbouring cities in the early phase of the SARS outbreak. The findings of our study show that most of the patients had serological evidence of infection with the novel SARS CoV, while healthy controls did not. Information generated here suggests that SARS CoV was also the primary infectious agent in the outbreaks in Guangdong Province. Because most people who had SARS in other countries were directly or indirectly linked to Hong Kong or Guangdong Province, we believe that the coronavirus might have originated from this region. Thus, SARS, a new emerging infectious disease that has become the first pandemic of the 21st century, probably originated in Guangdong.7, 8, 9, 10
Our epidemiological findings suggest that SARS had been spreading in an unusual pattern. The pandemic began with just a few index cases clustered mainly in families and hospital health-care workers in cities around Guangzhou. That specific antibodies were not present in any serum samples from healthy controls suggests that this coronavirus has not previously been present in human beings in Guangzhou.
Our analyses of genome function segment excludes the possibility that SARS CoV was generated by a recombination between human and animal, or animal and animal, coronavirus.12 SARS virus might be zoonotic and could have been introduced to humans by interspecies transmission quite recently. Since SARS CoV had very low homologies with all known coronaviruses,12 our postulation is that the new virus might have originally been resident in animals that have infrequent contact with humans, rather than in domestic animals.
The strains GZ43 and GZ60 from two health-care workers affected within the same hospital seem phylogenetically similar; wherease GZ50, which is an independent case from the community, is a distinct lineage. This finding suggests that there were multiple lineages of viruses circulating in Guangdong. One of these lineages (GZ50) maintains an outgroup relation to the major cluster of cases in Hong Kong and could represent the precursor virus of the Hong Kong outbreak. Whether some lineages are more transmissible than others is not clear.1 An analysis of more strains from Guangdong are needed to address this issue. The spread of SARS CoV in Foshan and Zhongshan in November and December, 2002, did not lead to an outbreak of the same scale as that in Guangzhou in January and February, 2003. Foshan and Zhongshan, like Guandong, are large urban conurbations of more than 1 million people. One possibility is that the virus was adapting to human beings—ie, human transmission with time. However, spread of the disease might have been promoted by the movement of people during the Chinese New Year holiday season that, in 2003, occurred at the end of January. Furthermore, Guangzhou, being a premier medical centre, received transfers of several patients with respiratory complications associated with SARS.
Viruses that undergo interspecies transmission tend to undergo more rapid genetic change as they adapt to a new host.16, 17 In fact, we noted that the genetic variation in the S gene was associated with mutations that were mainly asynonymous. The SCoV isolated in our study are some of the earliest isolates in the global outbreak. Further genetic analysis of these isolates might enable us to understand the origins of this virus and how it became established in human beings.
The index case of Guangzhou outbreak actually functioned as the trigger for the pandemic that generated a large number of secondary infections. Retrospective analysis of the pathway of the SARS spread from satellite cities to Guangzhou, then Hong Kong and other countries, showed that the major outbreak around the world could be linked to just a small number of so called super-spreading incidents. This finding implies that effective human-to-human transmission is firmly established and suggests that SARS CoV has become well adapted to human beings.16
One important question that has arisen from the SARS outbreak is why the new coronavirus emerged in Guangdong, southern China. There might be a link to the culinary habits of some southern Chinese who consume wild game meat as a delicacy. To satisfy this demand, different kinds of wild animals have been hunted for consumption, or raised in captivity. These animals are a likely source of new emergent infectious disease to human beings. In fact, the emergence of the highly pathogenic H5N1 influenza virus in humans in 199718, 19 and in early 2003 (unpublished data) is also zoonotic. The SARS outbreak provides evidence to support the hypothesis that southern China could be a site for emerging pandemic infectious disease in the future.20
The investigation of the infectious agent in SARS emphasises the importance of international collaboration as an effective strategy to control emerging diseases in people, especially for those diseases with the potential to become pandemic. The lesson from SARS outbreaks associated with coronavirus is that we cannot ignore or underestimate the effect of a new emerging infectious disease. The best way to control a new emerging infection is to intervene at an early stage in the outbreak.
Acknowledgments
Acknowledgments
We thank Jeffrey Huang, Vice-Minister of Ministry of Health in the Chinese Government; the Guangdong Government; the Chinese Centre for Disease Control and Prevention, and the Centre for Disease Control and Prevention of Guangdong Province; staff of the First Affiliated Hospital of Guangzhou Medical College, Guangzhou Pulmonary Hospital, and the Department of Microbiology, University of Hong Kong; and S W Luo, C L Cheung, Y J Guan, L J Zhang, and C Butt for technical assistance. This project was supported by research funding from Public Health Research (Grant A195357), the National Institute of Allergy and Infectious Diseases, USA.
Contributors
N S Zhong and B J Zheng contributed equally to this article. Y Guan and N S Zhong are coprincipal investigators who supervised the virological and clinical components of the study. B J Zheng did the serological studies and jointly wrote the paper. P H Li did the virus isolation. All other researchers were involved in collection and analysis of epidemiological and clinical data.
Conflict of interest statement
None declared.
References
- 1.Peng GW, He JF, Lin JY. Epidemiological study on severe acute respiratory syndrome in Guangdong province. Chin J Epidemiol. 2003;24:350–352. [PubMed] [Google Scholar]
- 2.Guangdong Public Health Office. 2003: Document No 2. Summary report of investigating an atypical pneumonia outbreak in Zhongshan (January 21, 2003).
- 3.Tsang KW, Ho PL, Ooi GC. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 4.Lee N, Hui D, Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 5.Update: outbreak of severe acute respiratory syndrome-Worldwide, 2003. MMWR Morb Mort Wkl Rep. 2003;52:241–248. [PubMed] [Google Scholar]
- 6.Anon Severe acute respiratory syndrome (SARS) Wkly Epidemiol Rec. 2003;78:81–83. [PubMed] [Google Scholar]
- 7.WHO. Cumulative number of reported probable cases of severe acute respiratory syndrome (SARS) http://www.who.int/csr/sarscountry/2003_07_11/en (accessed July 16, 2003).
- 8.Peiris J, Lai S, Poon L. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poutanen SM, Low DE, Henry B. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek TG, Erdman D, Goldsmith C. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.WHO. Case definitions for surveillance of severe acute respiratory syndrome (SARS) http://www.who.int/csr/sars/casedefinition/en/ (accessed May 1, 2003).
- 12.Marra MA, Jones SJ, Astell CR. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 13.Guan Y, Peiris JM, Lipatov AS. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci USA. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Tarnura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 15.Holmes KV. Coronaviruses. In: Knipe DM, Howley PM, editors. Field Virology. 4th edn. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1187–1203. [Google Scholar]
- 16.Holland JJ, de la Torre JC, Clarke DK. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:1960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 18.de Jong JC, Claas EC, Osterhaus AD. A pandemic warning? Nature. 1997;389:544. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbarao K, Klimov A, Katz J. Characterisation of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 20.Shortridge KF, Stuart-Harris CH. An influenza epicentre? Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]