Abstract
The ongoing battle against current and rising viral infectious threats has prompted increasing effort in the development of vaccine technology. A major thrust in vaccine research focuses on developing formulations with virus-like features towards enhancing antigen presentation and immune processing. Herein, a facile approach to formulate synthetic virus-like particles (sVLPs) is demonstrated by exploiting the phenomenon of protein corona formation induced by the high-energy surfaces of synthetic nanoparticles. Using an avian coronavirus spike protein as a model antigen, sVLPs were prepared by incubating 100 nm gold nanoparticles in a solution containing an optimized concentration of viral proteins. Following removal of free proteins, antigen-laden particles were recovered and showed morphological semblance to natural viral particles under nanoparticle tracking analysis and transmission electron microscopy. As compared to inoculation with free proteins, vaccination with the sVLPs showed enhanced lymphatic antigen delivery, stronger antibody titers, increased splenic T-cell response, and reduced infection-associated symptoms in an avian model of coronavirus infection. Comparison to a commercial whole inactivated virus vaccine also showed evidence of superior antiviral protection by the sVLPs. The study demonstrates a simple yet robust method in bridging viral antigens with synthetic nanoparticles for improved vaccine application; it has practical implications in the management of human viral infections as well as in animal agriculture.
Keywords: Protein corona, Virus-like particles, Gold nanoparticles, Coronavirus, Infectious bronchitis virus, Spike proteins
Graphical abstract
1. Introduction
Vaccine is historically the most effective countermeasure against infectious threats, as agents resembling pathogens are administered to mount an immune response against specific targets. Amidst continuing and emerging viral threats, vaccine technology continues to advance with the aim of effectively promoting antiviral immune responses, and a major development effort lies in retaining or integrating virus-like features in vaccine formulations for improved immune processing. Several morphological and antigenic characteristics of viral particles have been demonstrated to promote immune potentiation. For example, particles at the nanoscale have been shown to have better lymphatic transport as compared to smaller subunit antigens [1], [2]. In addition, the display of multiple antigens on a single particle facilitates more effective antigen presentation to immune cells [1]. As compared to traditional vaccine formulations, vaccines preserving virus-like features have shown superior capability in eliciting immune responses [3], [4], [5]. These results and observations have also prompted material scientists to apply synthetic nanomaterials towards mimicking viral features for vaccine development [6], [7], [8], [9].
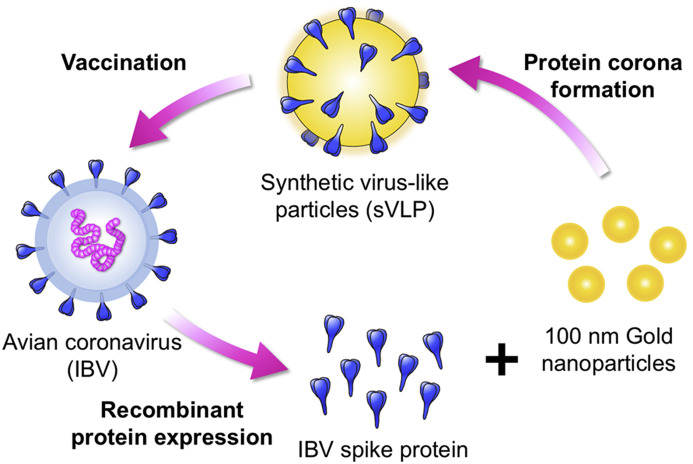
Given their high radii of curvature, synthetic nanoparticles frequently possess high surface energies that induce adsorption of biomolecules in a phenomenon known as protein corona formation. In protein-rich media, strong nanoparticle/protein association occurs spontaneously as a means to passivate surface energies, and the resulting particles are encased in a protein layer that dictates the particles' interactions with the environment [10], [11]. While protein corona formation is gaining increasing scientific interest owing to its implications in biomedical applications [10], [12], [13], we herein demonstrate harnessing this phenomenon can be beneficial towards mimicking viral features for vaccine applications. We show that synthetic virus-like particles (sVLPs) with close semblance to native virions in physicochemical properties and antigen display can be facilely prepared through spontaneous antigen-particle association in optimized incubation conditions. Using 100 nm gold nanoparticles (AuNP), a biologically inert material commonly used for biomedical research [14], [15], [16], and a spike glycoprotein derived from an avian infectious bronchitis virus (IBV), a single-stranded positive-sense RNA virus that belongs to the family Coronaviridae [17], we controlled the incubation condition to prepare spike glycoprotein-laden sVLPs (Fig. 1 ). The morphological features and antigen display by the sVLPs were compared to native IBV viral particles using nanoparticle tracking analysis and immunogold staining. In addition, vaccination potency between the sVLPs and free spike glycoproteins was compared in an avian model of coronavirus infection. A commercial whole inactivated virus (WIV) formulation that is the current standard vaccine for IBV management was examined in parallel.
Fig. 1.
Schematics illustrating the preparation of an avian coronavirus sVLPs. sVLPs are prepared in optimized mixtures containing viral proteins and 100 nm gold nanoparticles via spontaneous protein corona formation.
Coronaviruses are a major viral family of which the most publicized examples include the pathogens behind severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) [18]. In animals, IBV is a prime example of coronavirus that infects the respiratory and urogenital tracts of chickens, posing a serious economic threat as one of the most important pathogens in the poultry industry. The IBV spike glycoprotein, which forms the large, pental-shaped spikes on the surface of the virion, is chosen as the antigen candidate as it is implicated as a determinant of virus pathogenicity. Among coronaviruses, spike glycoproteins possess a variety of biological functions, including triggering cell attachment, inducing cell-cell fusion, and binding to cellular receptors [19], [20]. As spike glycoproteins are the primary targets in ongoing vaccine development efforts for coronavirus vaccinations, the present study has broad implications across both human and animal disease management [21], [22].
2. Materials and methods
2.1. Cells and gold nanoparticles
S. frugiperda Sf9 (ATCC CRL-1711) insect cells were cultured in Grace's insect cell medium (Invitrogen, Carlsbad, CA) and supplemented with 10% FBS (Thermo Fisher, Rockford, IL) and 1% P/S/A antibiotics (Biological Industries, Beit-Haemek, Israel) at 27 °C. 100 nm gold nanoparticle (AuNP) solution was purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Propagation of IBV
Avian coronavirus IBV strain 2575/98 was propagated in 10-day-old specific-pathogen-free (SPF) chicken embryos via the allantoic route as previously described [23]. The virus titers of IBVs were determined with the method of Reed and Muench [24] in SPF chicken embryos and expressed as 50% embryo infectious dose (EID50) [25]. The virus-containing allantoic fluid was concentrated and purified using sucrose gradient solution as previously described to derive the native virions [23].
2.3. Preparation of recombinant IBV spike proteins
Full spike (S) protein of avian coronavirus IBV was cloned and expressed using the Bac-to-Bac baculovirus expression system (Invitrogen). Briefly, a recombinant plasmid was constructed by inserting full spike protein gene of IBV strain 2575/98 (accession no. DQ646405) [26] into the pFastBac-1 vector using the following primer set: IBV-S-BamHI-f: 5′- TTGGG ATCCG ATGTT GGTGA AGTCA C-3′; IBV-S-SalI-f: 5′-CTTGT CGACA TTAAA CAGAC TTTTT AGGT-3′. The recombinant pFastBac-1 shuttle vector was then transposed to the bacmid in E. coli strain DH10Bac, and recombinant bacmid was purified using the HiPure Plasmid Midiprep kit (Invitrogen). Sf9 cells were used for transfection with the recombinant bacmid, and recombinant baculoviruses were then harvested in the supernatant and designated rBac-2575S. Recombinant spike proteins (r2575S) were harvested from Sf9 cells infected with rBac-2575S (multiplicity of infection = 1). Sf9 cells were washed and lysed with the I-PER insect cell protein extraction reagent (Thermo Fisher). Recombinant proteins were purified using the Glycoprotein Isolation Kit, ConA (Thermo Fisher) according to the manufacturer's instructions. After purification, r2575S protein was stored in 10% sucrose at −20 °C.
2.4. Preparation of synthetic virus-like particles
Citrate-buffered 100 nm gold nanoparticles were washed repeatedly in water to remove the citrate stabilizer, and the resulting pellet was resuspended in 10% sucrose. Protein solutions ranging in concentrations between 100 μg/mL to 3 mg/mL of purified spike proteins were then mixed with 1 × 1011/mL of gold nanoparticles (determined by nanoparticle tracking analysis) in 10% sucrose. The mixtures were bath sonicated for 1 min followed by incubation in an ice bath for 30 min. The nanoparticles were then removed from unbound spike proteins via centrifugation at 1500g for 3 min. Following 3 centrifugal washes with 10% sucrose, pelleted nanoparticles were mixed with 1X PBS and sonicated in a bath sonicator for 30 s. Dispersible, stabilized sVLPs were retrieved and their protein content was quantified using a BCA protein assay (Thermo Fisher) with 25 μL of 1 × 1011 particles/mL following the manufacturer's protocol. Visualization of unstable nanoparticles and colloidally stable sVLPs was performed using a 200 kV high resolution transmission electron microscope (FEI Tecnai TF20). Particle stability was assessed by monitoring the size of sVLPs for 7 days. Particle size, polydispersity index (PDI), and concentrations were measured by nanoparticle tracking analysis using Nanosight NS-500 (Malvern, UK) at a concentration of 1 × 108 particles/mL based on the manufacturer's instructions. Particle size and zeta potential were also measured by dynamic light scattering using Zetasizer Nano ZS at a concentration of 1 × 1010 particles/mL (Malvern, UK) based on the manufacturer's instructions.
2.5. Examination of antigen display and retention
Antigen display was examined using freshly prepared sVLPs. Antigen retention was examined by mixing sVLPs in protein-poor (PBS) or in protein-rich (10% BSA) conditions for varying periods of time. At 0, 3, 10, and 24 h marks, sVLPs were pelleted from their respective solutions. The particles were then processed using a previously published protocol with SDS-PAGE loading buffer for protein removal and quantification [27]. IBV spike proteins eluted from the sVLP were analyzed in 6% discontinuous SDS-PAGE under non-reducing condition. Protein gel was then transferred onto a 0.45 μm nitrocellulose membrane (Bio-Rad). After transfer, the membrane was soaked in blocking buffer (5% skim milk in PBS) at room temperature for 1 h and probed with anti-S monoclonal antibody (mAb) for another 1 h. After three washes, the membrane was incubated with peroxidase-conjugated goat anti-mouse IgG (H + L) (Jackson ImmunoResearch Laboratories, West Grove, PA) in blocking buffer at room temperature for 1 h. After three washes, the protein blots were detected with either TMB Membrane Peroxidase Substrate (KPL) or enhanced chemiluminescence (ECL) substrate (Pierce). Band intensities were analyzed via imaging analysis using ImageJ. Presence of IBV spike proteins on the sVLPs was further verified by immunogold staining, and purified IBV 2575/98 virions were used as a control. Briefly, 3 μl of sVLP or virion samples were deposited onto a glow-discharged carbon-coated grid for 2 min. The virion sample was fixed with 4% paraformaldehyde for 5 min. After 3 washes with PBS, the samples were blocked with 1% BSA for 15 min. The samples were then incubated with anti-S mAb for 1 h. After PBS washes, the samples were incubated with 6 nm gold-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) for another 1 h. After PBS washes, native virions were further stained with 1% uranyl acetate for 15 s. All experiments were performed at room temperate. Particles were visualized under a 200 kV high resolution transmission electron microscope (FEI Tecnai TF20).
2.6. Antigen delivery quantification
The care and use of animals were approved by the Institute Animal Care and Use Committee, National Taiwan University (approval no. NTU-102-EL-89). All animal experiments were carried out in accordance with the approved guidelines. 8-week old BALB/c mice were injected with 50 μL of PBS, free protein formulation, or sVLPs containing 2 μg of viral antigens via the intra-footpad route. After 24 h, the mice were sacrificed and the popliteal lymph nodes were harvested (n = 6). Cryosections (6 μm) were made and fixed for 10 min in acetone, followed by 8 min in 1% paraformaldehyde. Sections were blocked by 5% normal goat serum (Invitrogen) in PBS for 10 min and stained with anti-S mAb for 4 h at room temperature. After washes, sections were further incubated with FITC-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Nuclei were counterstained with DAPI (Invitrogen). Fluorescence signal was observed under a fluorescence microscope (Leica DMi8), and quantified via imaging analysis using ImageJ.
2.7. Animal immunization
8-week old BALB/c mice were injected intramuscularly in the thigh with 100 μL of formulations containing PBS, free protein, or sVLPs (10 μg of viral antigens) mixed with the complete Freund's adjuvant. Mice blood was collected on day 14 and 21 for antibody titer quantification (n = 4–5 per group). Three-week-old SPF chickens were obtained from JD-SPF Biotech (Miaoli, Taiwan). Chickens were randomly divided into four different experimental groups (n = 4–6 per group) receiving PBS, free protein (r2575S), whole inactivated virus (WIV) vaccine (Merial Laboratories, Lyon, France), or sVLPs. Briefly, free protein or sVLPs (10 μg of viral antigen in 100 μL) were emulsified with the complete Freund's adjuvant and administered via an intramuscular route. The commercially available WIV vaccine (oily-adjuvanted) was administered to chickens according to the manufacturer's recommendation (0.3 ml per chick). Chicken sera and tears were collected on day 0 (before immunization), 14, and 21 post-immunization. All chickens were intranasally challenged with IBV 2575/98 live virus (106 EID50) on day 21, and were observed for disease signs for 7 days. Chickens were sacrificed on day 28.
2.8. Antibody quantification
For serum IgA and IgG virus-specific ELISA, 100 ng of purified IBV 2575/98 virions was diluted with coating buffer (15 mM Na2CO3 and 35 mM NaHCO3, pH 9.6) and coated onto flat-bottomed microtiter plates (Nunc) at room temperature overnight. The wells were washed with PBST (0.1% Tween 80 in PBS) three times and blocked with blocking reagent (5% skim milk in PBST) at 37 °C for 1 h. After washes, 100 μl of chicken serum was added and incubated at room temperature for 1 h. Following three washes, 100 μl of peroxidase-conjugated goat anti-chicken IgY (H + L) or IgA (Jackson ImmunoResearch) in blocking buffer was added into each well and incubated at room temperature for 1 h. After three washes, 100 μl of SureBlue Reserve TMB Microwell Peroxidase Substrate (KPL) was added to each well and incubated in the dark at room temperature for 10 min. The reaction was stopped by adding 100 μl of TMB stop solution (KPL). The OD was measured at 450 nm using an automated plate reader (Thermo Fisher). For total tear IgA quantification, ELISA was performed with Chicken IgA ELISA Kit (ab157691, Abcam) according to the manufacturer's protocol.
2.9. Antigen-specific cytokine expression analysis
On day 28 post immunization, chicken spleens were minced and passed through a 70-μm cell strainer (Corning) to obtain single-cell suspensions. Red blood cells (RBCs) were lysed using an RBC lysis buffer (eBiosciences), and cells were resuspended in RPMI 1640 medium (Gibco, Grand Island, NY) containing 10% FBS. Viable cells were determined by trypan blue staining. 106 splenocytes were plated in 96-well U-bottom plates (Corning), and were stimulated with 1 μg of purified IBV 2575/98 virions in the presence of brefeldin A (GolgiPlug, BD Biosciences) for 6 h at 37 °C. For the quantification of cytokine expression, the stimulated splenocytes were lysed, and total RNA was isolated by TRIzol (Invitrogen) according to the manufacturer's manual. Real-time RT-PCR was performed using iScript (Bio-Rad) and iQ SYBR Green Supermix Kit (Bio-Rad) with previously described primers for chicken IFN-γ and GAPDH [28]. Melting curve analysis following real-time PCR was conducted to verify the specificity for each primer set. All obtained Ct values were normalized to GAPDH. The relative expression of chicken IFN-γ (fold change of naive control) was determined by a 2−ΔΔCt method [29].
2.10. Clinicopathological assessment
Disease signs of chickens were recorded on a daily basis after virus challenge. The clinical score index of IBV infection was interpreted according to a previously described method [30]. The clinical signs were evaluated as: 0 = no clinical signs; 1 = lacrimation, slight shaking, watering feces or tracheal rales; 2 = lacrimation, presence of nasal exudate, depression, water feces, apparent sneezing or cough; 3 = high degree of lacrimation, nasal exudate, and severe watery feces; 4 = death. After necropsy, gross lesions at the tracheas and kidneys were recorded. Chicken kidneys were further harvested and homogenized in tryptose phosphate broth (BD Biosciences). Viral load in kidneys was assessed by quantitative RT-PCR described below.
2.11. Viral RNA quantification
RNA in chicken kidneys was extracted using TRIzol (Invitrogen) according to the manufacturer's manual. For viral load assessment, Quantitative RT-PCR was performed with iScript (Bio-Rad) and iQ SYBR Green Supermix Kit (Bio-Rad) using previously described primer sets that target the S protein gene of IBV (rC2U and rC3L) [31] and chicken 28S rRNA [32]. Quantitative RT-PCR experiments were performed in duplicates. Data was expressed as arbitrary units.
2.12. Statistical analysis
Data was analyzed by ANOVA followed by Dunnett's multiple comparison tests using GraphPad Prism (GraphPad Software, San Diego, CA). p values smaller than 0.05 were considered significant.
3. Results and discussion
Following AuNP incubation in solutions of different protein concentrations, the resulting nanoparticles were pelleted from free proteins and re-dispersed through sonication in PBS. Consistent with previous studies on nanoparticle/protein interactions [33], it was observed that higher protein concentrations yielded particles with increased colloidal stability as evidenced by the disappearance of a discernable pellet and a purple solution characteristic of AuNP suspensions (Fig. 2 A). sVLPs prepared from the 3 mg/mL protein suspension were readily dispersible and manifest as distinct, non-clustered nanoparticles under transmission electron microscopy (Fig. 2B), indicating passivation of the high particle surface energy upon sufficient protein coating. In contrast, particle preparations with lower protein content (1000 μg/mL) yielded clustered AuNPs. To further characterize sVLPs, we assessed AuNPs, sVLPs, and native IBV virions (Fig. 2C) using nanoparticle tracking analysis, which examines particle samples on a particle-by-particle basis via tracking of scattered laser light from individual particles [34]. Between AuNPs and sVLPs, we observed an overall reduction in the light scattering intensity. Given that AuNPs are known to scatter light at an extraordinary efficiency, the intensity reduction in sVLPs can be attributed to successful protein coating, which restricts light passage to the AuNP surfaces. Likewise, native virions have the lowest light scattering under the analysis as they are comprised entirely of organic materials. The result demonstrates the feasibility of studying the evolution of nanoparticle protein corona formation using nanoparticle tracking analysis, which reveals changes in light scattering and size simultaneously.
Fig. 2.
Preparation and characterizations of sVLPs. (A) Visualization of nanoparticle solutions following incubation with and isolation from different concentrations of IBV spike proteins. (B) TEM visualization of nanoparticles prepared with a low protein concentration (1000 μg/mL; left) and sVLPs prepared with a high protein concentration (3000 μg/mL; right). Scale bars = 1 μm. (C) Particle-by-particle examination of AuNPs, sVLPs, and native IBV virions under nanoparticle tracking analysis. (D) Size and zeta potential of AuNPs, sVLPs, and native IBV virions as analyzed by nanoparticle tracking analysis. Bars represent means ± s.d. (n = 3). (E) sVLP stability in PBS observed over 7 days. Bars represent means ± s.d. (n = 3). (D) Western blotting analysis confirms the presence of IBV spike proteins on sVLPs. (E) Transmission electron microscopy of sVLPs (left) and native IBV virions (right) following immunogold staining against IBV spike proteins. Scale bars = 50 nm. (H) Western blotting analysis of IBV spike protein retention on sVLPs following different incubation periods in PBS or in 10% BSA.
Upon examining the size distributions of the different particles, sVLPs showed a broader distribution as compared to the sharply distributed 100 nm AuNPs. Protein corona formation increased the nanoparticle size from 100.6 nm (PDI = 0.012) to 139.2 nm (PDI = 0.073) and increased the zeta potential from −23.2 mV to −16.7 mV (Fig. 2C,D). In comparison to native IBV virions, which have an average diameter of 147.3 nm (PDI = 0.081) and a zeta potential of −16.6 mV, the sVLPs are similar in overall physicochemical properties. Examination of particle stability showed that the sVLPs remained stable in PBS over a 7-day period with its size ranging from 136.7 nm (PDI = 0.071) to 140.2 nm (PDI = 0.091) (Fig. 2E). Analysis of antigen display with freshly prepared sVLPs showed that 1 × 1011 AuNPs retained 23.5 ± 2.2 μg of spike proteins, corresponding to approximately 900 IBV spike proteins per particle. Western blotting using analysis revealed a sharp protein band of approximately 160 kDa (Fig. 2F), which is characteristic of the viral antigen [17]. Transmission electron microscopy and immunogold staining further highlight the similarity between sVLPs and native IBV virions. It was observed that immunogold clustered around the sVLPs, mirroring the staining pattern on the native virions (Fig. 2G). These observations demonstrate the close semblance between the sVLPs and native virions regarding their physicochemical properties and antigen display.
Examination of antigen retention in protein-poor (1X PBS) and protein-rich (10% BSA in 1X PBS) conditions also shed light on the characteristics of the protein corona around the sVLPs. In PBS, particle-bound antigen level remained steady over a span of 24 h, yielding similar IBV spike protein band intensities across the different incubation samples (Fig. 2H). A rapid drop-off in particle-bound spike protein was observed upon incubation in 10% BSA. Immediate retrieval of sVLPs from the BSA solution resulted in ∼65% reduction in the spike protein level, and at the 24 h mark, ∼25% of the initial antigen remained on the sVLPs. This observation suggests the formation of two distinctive corona layers distinguishable by their interaction dynamics with surrounding biomolecules, reflecting the presence of both a reversible “soft corona” and an irreversible “hard corona” that have been frequently observed in prior nanoparticle studies [35], [36], [37]. The results indicate that approximately 200–250 IBV spike proteins are stably bound to each sVLPs. These proteins are expected to remain in the particulate form in complex biological environments upon in vivo administration.
To examine antigen delivery and lymphatic transport by the sVLPs as compared to free spike proteins, sVLP formulation was administered to mice through a footpad injection. Popliteal lymph nodes, which are the draining lymph nodes by the footpads, were subsequently collected and sectioned for immunofluorescence assay. IBV spike protein-specific immunofluorescence staining showed a significantly enhanced antigen delivery by the sVLPs as compared to the free protein formulation, resulting in an increased number of fluorescent punctates (green) in the lymph node sections (Fig. 3 A). Imaging analysis on multiple lymph node sections showed that the sVLP formulation increased lymphatic delivery by approximately 6 fold (Fig. 3B). The observation of increased delivery attests to the strong protein/particle binding in the “hard corona” layer as the particle carrier is capable of facilitating antigen transport in vivo. The enhanced lymph node localization of the sVLPs is consistent with prior observations on nanoparticles and virus-like particles [2]. Owing to their nanoscale morphology and physicochemical properties, these nanoparticles are known to facilitate free lymphatic drainage via convective transport [38], [39] as well as cell-mediated lymphatic delivery via increased cellular uptake [2].
Fig. 3.
Antigen delivery and immunogenicity of sVLPs. (A) Sections of popliteal lymph nodes were examined under bright field (top panel) and using immunofluorescence assay (bottom panel). Lymph node sections were stained with DAPI (blue) and FITC-conjugated anti-IBV spike protein antibodies (green) to examine antigen content in the lymph node 24 h following footpad injections with free proteins or sVLPs. Scale bars = 100 μm. (B) Quantification of antigen-specific fluorescence signals in the lymph node. Bars represent means ± s.d. (n = 6). (C) Quantification of anti-IBV spike protein IgG titers 14 and 21 days following vaccination. Lines and boxes represent upper extreme, 25th, 50th, 75th percentile, and lower extreme (n = 4–5). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Immunogenicity of the sVLPs was also examined following intramuscular inoculation in mice. Anti-IBV IgG serum titers were compared between mice vaccinated with sVLPs and with free IBV spike proteins (Fig. 3C), and it was observed that the sVLPs elicited significantly higher IgG levels, demonstrating improved vaccination potency over the free protein formulation. The improved immunogenicity can be explained in part by the enhanced antigen delivery to the lymph node, where a high number of antigen presenting cells reside. In addition, the particulate nature of the sVLPs likely also favors other immune activation mechanisms, such as improved cellular uptake, enhanced complement activation [38] and presentation by follicular dendritic cells [40]. These nanoparticle-specific immunological features make the sVLPs a promising vaccine candidate for disease management.
To evaluate the sVLPs' effectiveness against viral infections, we vaccinated SPF chickens with free IBV spike proteins or sVLPs (10 μg of total viral antigens) via the intramuscular route. As an additional reference, a commercial WIV vaccine for IBV was administered based on the manufacturer's suggested dosage. Following vaccination, blood and tear were collected for analysis and a live IBV challenge was performed (Fig. 4 A). ELISA analysis showed that the sVLPs were superior in generating both IgG and IgA titers as compared to the free protein formulation and the WIV vaccine (Fig. 4B,C). The total IgA in the tears of the vaccinated chickens were also quantified. Despite that intramuscular vaccination is generally known to be non-ideal for promoting mucosal immunity [41], elevation of tear IgA level was observed for all three vaccine formulations (Fig. 4D). It is expected that mucosal vaccination in future studies may further increase tear IgA levels and better highlight the differences among the formulations in eliciting mucosal immunity. Besides humoral immunity, cellular immunity, a major component of effective antiviral immune responses [42], was analyzed using splenocytes extracted on day 28. The sVLP sample showed a significant increase in the IFN-γ mRNA level as compared to the control, free protein, and the WIV vaccine samples (Fig. 4E), demonstrating superior promotion of antigen-specific cellular immunity.
Fig. 4.
Immunopotentiation following vaccinations with sVLPs. (A) Vaccination, tissue sample collection, and virus challenge schedule in an avian model of coronavirus infection. (B) Virus-specific serum IgG titers observed in animals vaccinated with free proteins, a commercial whole inactivated virus (WIV) vaccine, and sVLPs. Lines and boxes represent upper extreme, 25th, 50th, 75th percentile, and lower extreme (n = 6). (C) Virus-specific serum IgA titers in animals vaccinated with the different formulations. Lines and boxes represent upper extreme, 25th, 50th, 75th percentile, and lower extreme (n = 6). (D) Virus-specific tear IgA titers in animals vaccinated with the different formulations. Bars represent means ± s.e.m (n = 6). (E) Relative IFN-γ mRNA levels observed from the splenocytes of the different vaccinated groups following a viral antigen challenge. Bars represent means ± s.e.m (n = 4). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
We further examined the effect of the different vaccinations in protecting against a viral challenge. Clinical scores evaluated based on stamina, posture, and voice show that the sVLP group had the lowest overall symptoms, on par with animals vaccinated with the WIV formulation (Fig. 5 A,B). In comparison, vaccination with the free protein formulation was less effective and highly variable in moderating the disease symptoms. On day 28, necropsies were performed to examine the tracheas and kidneys, which are characteristic sites for infections by IBV [43]. As indicated in the gross lesion photos, the best antiviral protection was observed in the sVLP-immunized group, whereas organs from the free protein group and the WIV vaccine group showed observable mucus secretion and petechiae in tracheas (Fig. 5D, upper panel, arrowed) and swollen lesions and hemorrhages in kidneys (Fig. 5D, lower panel, arrowed). The prophylactic effect of the sVLP vaccination was further demonstrated by examining the viral load in kidneys. Analysis by quantitative RT-PCR showed that immunization with sVLPs more consistently reduced the viral content, resulting in the lowest relative viral mRNA expression across the animal samples (Fig. 5C). The results further corroborate the protective effect by the sVLP vaccination, which enhanced both humoral and cellular immunity for increased protection against the viral challenge.
Fig. 5.
Protection against a viral challenge following sVLP vaccination. (A) Daily clinical scores of the different vaccinated groups as evaluated by the subjects' stamina, posture, and voice following IBV viral challenge. (B) Averaged clinical scores over a 6-day observation period. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (C) Viral RNA levels in the kidneys of the different vaccinated samples as measured by quantitative RT-PCR. Bars represent means ± s.e.m (n = 4). (D) Necropsies of vaccinated chicken samples following viral challenge by IBV. Examinations of the trachea (top panel) and kidneys (bottom panel) show different damages reflective of the viral infection. Petechiae in tracheas and swollen lesions/hemorrhages in kidneys are indicated by arrows.
Coronavirus spike proteins are the primary antigenic signatures on coronaviruses as they contribute to the characteristic crown-like morphology underlining this virus family. As these proteins comprise the outermost layer of coronaviruses, the spike proteins have a pivotal role in viral pathogenesis and are recognized as the primary target for vaccine preparations [44]. Present vaccination strategies for coronaviruses include recombinant viruses and virus-like particles, and there is a continuing effort in developing new strategies for improving vaccine potency and safety [22]. To the best of our knowledge, incorporating coronavirus spike protein with synthetic nanoparticles has not been previously explored. By exploiting the high surface energies of synthetic nanoparticles, spontaneous assembly of sVLPs covered with IBV spike proteins were demonstrated. The strong particle/antigen association resulted in virus-sized particulates displaying IBV spike proteins, and the sVLPs elicited strong immune protection against a live IBV challenge. The enhanced immunopotentiation by the particle carrier is consistent with previous studies and echoes the curious observation that gold nanoparticles not only promote humoral but also cellular immune responses upon association with antigens [14], [15]. As the increased cellular immune response suggests that the nanoparticles may play a role beyond a passive antigen carrier, future studies examining the impact of nanomaterials and nanoparticle surface energies on immunological interactions are warranted.
It should be noted that the phenomenon of protein corona formation is an evolving field of study in which scientists continue to examine nanomaterials in biological medium with increasing complexity [45], [46], [47]. Subtle changes on the environment and on nanoparticle properties can have dramatic and unpredictable impact on the overall corona identity with significant biological implications. To demonstrate a practical utility for the protein corona phenomenon, the present study adopts a reductionist approach in examining protein-particle interactions. AuNPs are incubated in a highly controlled condition with proteins of a singular species to form sVLPs with virus-mimetic features, and the dynamics of such association are expected to vary with different biomolecules and nanomaterials [48]. In general, inorganic nanoparticles promote stronger protein adsorption as compared to organic nanoparticles as inorganic nanoparticles tend to have higher surface energies. Decreasing particle size also tends to increase biomolecule interactions as it increases radii of curvature of nanoparticle surfaces. Other forces, such as electrostatic interactions, van der Waals forces and covalent interactions all play intertwining roles in governing the nano-bio interface, and factors including nanoparticle functionalizations, buffer conditions, and biomolecule species have significant impact on the corona formation [48]. Nonetheless, in a controlled and optimized condition, the phenomenon may be exploited to facilely prepare formulations with defined characteristics and favorable biological performance. The present work takes advantage of this spontaneous interaction between nanomaterials and biomolecules towards improving vaccine development. This strategy may find practical applications in disease management against coronaviruses as well as other infectious threats.
4. Conclusions
In summary, we demonstrate by incubating viral antigens with synthetic nanoparticles in optimized conditions, spontaneous formation of protein corona induces the assembly of virus-like nanostructures with viral antigens encasing the particulate core. Results from the present study validate the successful preparation of sVLPs via nanoparticles' innate tendency to induce protein coating. In comparison to typical virus-like particle preparations, the present strategy offers practical advantages owing to its simple and facile process. Amidst the growing health threats of coronavirus infections as well as the ongoing economic impact of IBV infections, virus-like particles are garnering increasing scientific interest as vaccine candidates owing to their improved efficacy in comparison to subunit antigens [49], [50]. In the present study, vaccination with the sVLPs resulted in enhanced humoral and cellular immune responses, improving protection against an avian model of coronavirus infection as compared to free protein antigens and a commercial WIV vaccine. Strong immunity against the viral challenge following sVLP vaccination was evidenced by multiple criteria, including improved physical symptoms, reduced organ lesions, and decreased overall viral load. The enhanced immunopotentiation by the sVLPs is attributable at least in part to increased lymphatic delivery and multivalent antigen display. Given the robustness and versatility of the approach, it can be envisioned the technique can be broadly applied for different vaccine development.
Acknowledgments
The study was supported by the Ministry of Science and Technology (103-2321-B-002-066, 104-2321-B-002-023, 105-2321-B-001-055) and National Taiwan University (104R7320).
References
- 1.Moon J.J., Suh H., Li A.V., Ockenhouse C.F., Yadava A., Irvine D.J. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand T-fh cells and promote germinal center induction. P Natl. Acad. Sci. U. S. A. 2012;109:1080–1085. doi: 10.1073/pnas.1112648109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann M.F., Jennings G.T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 3.Amanna I.J., Raue H.P., Slifka M.K. Development of a new hydrogen peroxide-based vaccine platform. Nat. Med. 2012;18:974–979. doi: 10.1038/nm.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noad R., Roy P. Virus-like particles as immunogens. Trends Microbiol. 2003;11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 5.Kanekiyo M., Wei C.J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R.R. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moon J.J., Suh H., Bershteyn A., Stephan M.T., Liu H.P., Huang B. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat. Mater. 2011;10:243–251. doi: 10.1038/nmat2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson M.C., Crespo M.P., Abraham W., Moynihan K.D., Szeto G.L., Chen S.H. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J. Clin. Investig. 2015;125:2532–2546. doi: 10.1172/JCI79915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuhn L., Vanparijs N., De Beuckelaer A., Lybaert L., Verstraete G., Deswarte K. pH-degradable imidazoquinoline-ligated nanogels for lymph node-focused immune activation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:8098–8103. doi: 10.1073/pnas.1600816113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang R.H., Hu C.M., Luk B.T., Gao W., Copp J.A., Tai Y. Cancer cell membrane-coated nanoparticles for anticancer vaccination and drug delivery. Nano Lett. 2014;14:2181–2188. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenzer S., Docter D., Kuharev J., Musyanovych A., Fetz V., Hecht R. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 11.Albanese A., Tang P.S., Chan W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 12.Schottler S., Becker G., Winzen S., Steinbach T., Mohr K., Landfester K. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016;11:372–377. doi: 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Fierens K., Zhang Z., Vanparijs N., Schuijs M.J., Van Steendam K. Spontaneous protein adsorption on graphene oxide nanosheets allowing efficient intracellular vaccine protein delivery. ACS Appl. Mater. Interfaces. 2016;8:1147–1155. doi: 10.1021/acsami.5b08963. [DOI] [PubMed] [Google Scholar]
- 14.Gao W., Fang R.H., Thamphiwatana S., Luk B.T., Li J., Angsantikul P. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett. 2015;15:1403–1409. doi: 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niikura K., Matsunaga T., Suzuki T., Kobayashi S., Yamaguchi H., Orba Y. Gold nanoparticles as a vaccine platform: influence of size and shape on immunological responses in vitro and in vivo. ACS Nano. 2013;7:3926–3938. doi: 10.1021/nn3057005. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh P., Han G., De M., Kim C.K., Rotello V.M. Gold nanoparticles in delivery applications. Adv. Drug Deliv. Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J. Gen. Virol. 1983;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- 18.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PloS One. 2013;8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32:3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H.W., Wang C.H., Cheng I.C. A type-specific blocking ELISA for the detection of infectious bronchitis virus antibody. J. Virol. Methods. 2011;173:7–12. doi: 10.1016/j.jviromet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- 25.Villegas P. Titration of biological suspensions. In: Swayne D.E., Glisson J.R., Jackwood M.W., Pearson J.E., Reed W.M., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. The American Association of Avian Pathologists; Pennsylvania: 1998. pp. 248–254. [Google Scholar]
- 26.Chen H.W., Huang Y.P., Wang C.H. Identification of Taiwan and China-like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res. 2009;140:121–129. doi: 10.1016/j.virusres.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Docter D., Distler U., Storck W., Kuharev J., Wunsch D., Hahlbrock A. Quantitative profiling of the protein coronas that form around nanoparticles. Nat. Protoc. 2014;9:2030–2044. doi: 10.1038/nprot.2014.139. [DOI] [PubMed] [Google Scholar]
- 28.Liu H., Zhang M., Han H., Yuan J., Li Z. Comparison of the expression of cytokine genes in the bursal tissues of the chickens following challenge with infectious bursal disease viruses of varying virulence. Virol. J. 2010;7:364. doi: 10.1186/1743-422X-7-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Avellaneda G.E., Villegas P., Jackwood M.W., King D.J. In vivo evaluation of the pathogenicity of field isolates of infectious bronchitis virus. Avian Dis. 1994;38:589–597. [PubMed] [Google Scholar]
- 31.Huang Y.P., Lee H.C., Cheng M.C., Wang C.H. S1 and N gene analysis of avian infectious bronchitis viruses in Taiwan. Avian Dis. 2004;48:581–589. doi: 10.1637/7186-033004R. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H., Yang H., Kapczynski D.R. Chicken interferon alpha pretreatment reduces virus replication of pandemic H1N1 and H5N9 avian influenza viruses in lung cell cultures from different avian species. Virol. J. 2011;8:447–458. doi: 10.1186/1743-422X-8-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moerz S.T., Kraegeloh A., Chanana M., Kraus T. Formation mechanism for stable hybrid clusters of proteins and nanoparticles. ACS Nano. 2015;9:6696–6705. doi: 10.1021/acsnano.5b01043. [DOI] [PubMed] [Google Scholar]
- 34.Filipe V., Hawe A., Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monopoli M.P., Aberg C., Salvati A., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 36.Milani S., Bombelli F.B., Pitek A.S., Dawson K.A., Radler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 37.Lundqvist M., Stigler J., Cedervall T., Berggard T., Flanagan M.B., Lynch I. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 38.Reddy S.T., van der Vlies A.J., Simeoni E., Angeli V., Randolph G.J., O'Neil C.P. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 39.Reddy S.T., Berk D.A., Jain R.K., Swartz M.A. A sensitive in vivo model for quantifying interstitial convective transport of injected macromolecules and nanoparticles. J. Appl. Physiol. 2006;101:1162–1169. doi: 10.1152/japplphysiol.00389.2006. [DOI] [PubMed] [Google Scholar]
- 40.Heesters B.A., Myers R.C., Carroll M.C. Follicular dendritic cells: dynamic antigen libraries. Nat. Rev. Immunol. 2014;14:495–504. doi: 10.1038/nri3689. [DOI] [PubMed] [Google Scholar]
- 41.Belyakov I.M., Ahlers J.D. What role does the route of immunization play in the generation of protective immunity against mucosal pathogens? J. Immunol. 2009;183:6883–6892. doi: 10.4049/jimmunol.0901466. [DOI] [PubMed] [Google Scholar]
- 42.Amanna I.J., Slifka M.K. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology. 2011;411:206–215. doi: 10.1016/j.virol.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 44.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesniak A., Fenaroli F., Monopoli M.P., Aberg C., Dawson K.A., Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 46.Walkey C.D., Olsen J.B., Song F., Liu R., Guo H., Olsen D.W. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 47.Walkey C.D., Chan W.C. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 48.Nel A.E., Madler L., Velegol D., Xia T., Hoek E.M.V., Somasundaran P. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2016 Mar 30 doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G., Lv L., Yin L., Li X., Luo D., Liu K. Assembly and immunogenicity of coronavirus-like particles carrying infectious bronchitis virus M and S proteins. Vaccine. 2013;31:5524–5530. doi: 10.1016/j.vaccine.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]