Abstract
Severe acute respiratory syndrome (SARS) is a newly emergent human disease, which requires rapid diagnosis and effective therapy. Among antibody sources, immunoglobulin Y (IgY) is the major antibody found in chicken eggs and can be used as an alternative to mammalian antibodies normally used in research and immunotherapy. In this study, phage-expressing chicken monoclonal scFv antibody was chosen and characterized with phage display antibody technology. Truncated fragments of SARS-CoV spike protein were cloned in pET-21 vector and expressed in BL-21 Escherichia coli (E. coli) cells. After purification, the purity of these recombinant spike proteins was examined on SDS–PAGE and their identity verified with Western blot analysis using anti-his antibodies and sera from convalescent stage SARS-CoV-infected patients. Using these bacteria-derived proteins to immunize chickens, it was found that polyclonal IgY antibodies in the egg yolk and sera were highly reactive to the immunogens, as shown by Western blot and immunocytochemical staining analysis. A phage displaying scFv library was also established from spleen B cells of immunized chicken with 5 × 107 clones. After four panning cycles, the eluted phage titer showed a 10-fold increase. In sequence analysis with chicken germline gene, five phage clones reacted, with large dissimilarities of between 31 and 62%, in the complementarity-determining regions, one dominant phage 4S1 had strong binding to fragment Se-e, located between amino acid residues 456–650 of the spike protein and this particular phage had significantly strong binding to SARS-CoV-infected Vero E6 cells. Based on the results, we conclude that generating specific scFv-expressing phage binders with the phage display system can be successfully achieved and that this knowledge can be applied in clinical or academic research.
Keywords: SARS-CoV, Spike protein, IgY, scFv-expressing phage binder
Abbreviations: SARS-CoV, severe acute respiratory syndrome associated coronavirus; S, spike; scFv, single-chain variable fragment; RT-PCR, reverse transcription polymerase chain reaction; CDR, complementarity determining region; FR, framework region; VH, heavy chain variable region; VL, light chain variable region
1. Introduction
The severe acute respiratory syndrome (SARS) is a newly emergent disease that has threatened the human population with its epidemic potential and high mortality rate (Stadler et al., 2003). A novel SARS-associated coronavirus (SARS-CoV) was found to be the infectious agent and its genome has been completely sequenced (Ksiazek et al., 2003, Peiris et al., 2003a, Peiris et al., 2003b). SARS-CoV, an enveloped virus, replicates itself in the cytoplasm of its host cells. Its single-stranded, plus-sense RNA genome is structurally quite distinct from other coronaviruses known to infect humans or animals on the basis of their genomic sequences. Its open reading frames at the 3′-end encode four major structural proteins: spike (S) glycoprotein, membrane (M) protein, envelope (E) glycoprotein and nucleocapsid (N) protein (Marra et al., 2003, Rota et al., 2003). Many aspects of SARS are still unclear; therefore, accurate diagnosis and efficient vaccines are vital for controlling this deadly disease.
SARS-CoV S protein, a large membrane glycoprotein of 1255 amino acids (aa), has been found to be responsible for receptor binding and membrane fusion (Marra et al., 2003, Rota et al., 2003). Angiotensin-converting enzyme 2 (ACE2) has been shown to possess the functional receptor for the SARS-CoV (Li et al., 2003). The conformational changes induced by the complex of S protein and its receptor are required for membrane fusion (Dimitrov, 2003, Li et al., 2003, Wang et al., 2004). Furthermore, the high antigenicity of the S protein can efficiently elicit neutralizing antibody in mice in protecting them from infection (Bisht et al., 2004, Yang et al., 2004). Those results suggest that S protein is a good target for vaccine development, antiviral therapies and diagnostic application. Therefore, generating monoclonal antibodies specifically to recognize the S protein is needed. However, we have to overcome the difficulties associated with the traditional approach in generating monoclonal antibodies. Alternatively, the choice of the phage display system is a safe and effective in vitro procedure to enrich specific antibodies from large synthetic phage antibody libraries (Barbas et al., 1991). Domestic chicken is the simplest and easily available host choice for single-chain variable fragment (scFv) antibody library construction to screen efficient binders against various pathogen infections (Fehrsen et al., 2005, Finlay et al., 2006, Park et al., 2005).
In this study, we report the identification of 10 fragments of truncated SARS-CoV S proteins in E. coli cells to detect specific antibodies present in SARS-CoV-infected patient sera; the construction of a scFv antibody library with 5 × 107 phage clones after immunizing chickens to produce polyclonal anti-S protein IgY. We also report the characterization of one phage clone to recognize a S-protein fragment located in the amino acid residues 456–650 with the use of ELISA, Western blot analysis and immunofluorescent detection.
2. Materials and methods
2.1. Sera collection
The sera samples used in this study were provided by patients at Taipei Medical University-Wan Fang Medical Center (TMU-WFMC). Sera were prepared from convalescent SARS-CoV-infected patients and apparently healthy individuals. All collected sera were screened by a one-step anti-SARS coronavirus test kit (SD Inc., Korea) to confirm the presence or absence of anti-SARS-CoV antibodies.
2.2. cDNA synthesis
SARS-CoV genomic RNA was directly extracted from the sputum sample of a hospitalized patient, diagnosed as SARS-CoV infected at TMU-WFMC, as described by the manufacturer with minor modification (Trizol reagent, Invitrogen). The whole procedure was performed in a biosafety level (BSL) 2 laboratory to avoid any potential contamination. In brief, 2 ml of Trizol reagent was added to a 0.2 ml sputum sample and stored at 4 °C until used. One millilitre of virus-inactivated sample was transferred to a 1.5 ml Eppendorf tube containing 0.2 ml of chloroform. After vortexing and centrifuging at 10,000 rpm for 10 min at 4 °C, the colorless upper aqueous phase was collected and mixed with an equal volume of isopropanol to precipitate the total RNAs. We kept the mixture at room temperature (RT) for 10 min and recentrifuged it at 10,000 rpm for 10 min at 4 °C. A gel-like pellet containing RNA was formed, washed once with 1 ml of 75% ethanol and centrifuged at 10,000 rpm for 5 min. After removing the supernatant, the pellet was air-dried and dissolved in 15 μl of RNase-free water. The extracted RNA sample was used immediately for viral gene amplification.
2.3. Reverse transcription-polymerase chain reaction (RT-PCR)
To amplify truncated fragments of S gene, we designed 10 sets of primers according to the published nucleotide sequences of the SARS-CoV RNA genome (GenBank Accession NC_004718). Conditions for SARS-CoV RNA amplification were performed using a one-step RT-PCR, kit as described by the manufacturers (Qiagen, Valencia, CA, USA). In brief, 5 μl of the isolated total RNAs was used as template and added to 45 μl of a mixture of 26 μl RNase-free water, 10 μl 5× one-step RT-PCR buffer, 2 μl dNTPs mixture (10 mM/each), 2 μl one-step RT-PCR enzyme mixture, 2.5 μl/primer (10 μM/each). The amplification program consisted of a reverse-transcription reaction of 50 °C for 30 min and 95 °C for 15 min, 40 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 90 s, followed by 72 °C for 10 min. The RT-PCR products were kept at −20 °C until used.
2.4. Expression and purification of his-fused spike proteins
We amplified 10 truncated S-gene fragments of 300–750 bp in length with PCR, treated them with BamHI and XbaI and ligated them into a pET-21 expression vector (Novagen, Germany). The resultant plasmids were transformed into an E. coli BL-21 (DE3) strain. We grew clones with the respective insert in 5 ml LB medium containing ampicillin (100 μg/ml) at 37 °C overnight. The bacterial culture was 10-fold diluted in the same LB medium and grown for an additional 3–4 h until OD600 reached 0.6. To induce his-fused S protein expression, the iso-propyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM in the culture, which was further incubated for 4 h. After centrifugation, the cell pellet was resuspended in 2 ml of binding buffer (100 mM NaCl, 10 mM Tris–Cl, and 8 M urea pH 8.0) and lysed by three cycles of freezing (−70 °C) and thawing (37 °C). After centrifugation at 12,000 rpm for 10 min at 4 °C, the resulting cellular lysate was incubated with Ni2+-charged sepharose for protein purification according to the manufacturer's instructions (Amersham Biosciences, Sweden).
2.5. Western blotting
The bacteria-expressed his-fused S proteins were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). We transferred the proteins onto nitrocellulose membrane (Amersham Biosciences, UK), which was then blocked with 5% skim milk in TBST (TBS with 0.05% Tween 20) for 1 h. Polyclonal mouse anti-his antibodies (Amersham Biosciences, UK) or convalescent sera of SARS patients were added and incubated for additional 1 h. The membrane was washed with TBST three times at 5 min/each. We detected the bound antibodies by adding horseradish peroxidase (HRP)-conjugated anti-mouse antibodies or anti-human Ig antibodies (Sigma, USA). After three washings, we developed the membrane with diaminobenzidine (DAB) substrate until the desired intensity.
2.6. Chicken immunization
Female White Leghorn (Gallus domesticus) chickens were immunized with mixed recombinant S proteins (10 μg/each) in equal volume of adjuvant by intra-muscular injection. The first immunization was prepared with complete Freund's adjuvant, and three additional ones were performed with incomplete Freund's adjuvant at interval of 7 days. After each immunization, we obtained the blood and titrated it with enzyme-linked immunosorbent assay (ELISA) to determine the presence of an antigen-specific immune response. We also collected eggs for purification of immunoglobulin Y (IgY) from yolks by the standard procedure (Akita and Nakai, 1993a, Akita and Nakai, 1993b).
2.7. Construction of phage displaying chicken scFv library
We constructed the library according to the published protocols with minor modifications (Andris-Widhopf et al., 2000). Briefly, 7 days following the final immunization, chickens were euthanized and spleens were harvested. Spleens were placed immediately in Trizol reagent for homogenization. The extraction of total RNAs was carried out and 10 μg of RNAs was reversely transcribed to synthesize the first-strand cDNA using a SuperScript RT kit (Invitrogen). After PCR amplification with chicken specific primers, we obtained gene products of heavy and light chain variable regions (VH and VL). For full-length single chain (sc) Fv construction, we connected VH and VL fragments with a peptide linker (GGSSRSS) by an overlap extension PCR, which was further cloned into a pComb3H vector. The recombinant DNAs were transformed into E. coli XL-1 strain by electroporation. Recombinant phage production was initiated by the addition of helper phage VCS-M13 (Barbas et al., 1991), precipitated with 4% polyethylglycol 8000 and 3% NaCl (w/v), resuspended in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and stored at 4 °C.
2.8. Biopanning and phage clone preparation
We coated microtitre plates with equal amounts of the mixed S-protein fragments (0.5 μg/well) at 4 °C overnight. After being blocked with 3% BSA at 37 °C for 1 h and washed with TBST, we added more than 1011 pfu of recombinant phages from above to each well and the plates were incubated at 37 °C for 2 h; unbound phages were removed and the wells were washed vigorously with TBST buffer. Bound phages were eluted with 0.1 M HCl/glycine (pH 2.2)/0.1% BSA, and neutralized with Tris base buffer. Eluted phages were used to infect E. coli XL-1 strain at log phase for amplification and rescued with 4% polyethylglycol 8000 and 3% NaCl for the next round of selection. Phage preparation and further panning on wells were repeated four times. For preparing single phage clone, individual colonies from the final panning cycle were grown in 10 ml SB medium containing ampicillin (100 μg/ml) at 37 °C with agitation for 4–6 h. VCS-M13 helper phage was added to the cultures and shaken for another 2 h. Kanamycin was added to a final concentration of 70 μg/ml. Individual phage clones were rescued, as above, and tested for their binding ability.
2.9. ELISA and epitopic mapping
We used microtitre plates precoated with SARS-CoV-infected Vero E6 cell lysates to detect the binding ability of scFv-expressing phage to S protein, as shown in the anti-SARS-CoV ELISA kit (Euroimmun, Lubeck, Germany). After rinsing with TBST in the plates, we distributed 1010 phage particles dissolved in PBS to wells in duplicate and incubated them at 37 °C for 1 h. After washing with PBST (PBS with 0.05% Tween 20), we detected bound phage particles with HRP-labeled mouse anti-M13 antibody (Amersham Biosciences). In the epitopic mapping assay, we coated 10 purified his-fused S-protein fragments in microtitre plates at 0.5 μg/well individually. After being blocked with 3% BSA, we distributed 1010 phage particles to each well in duplicate and incubated them at 37 °C for 1 h. After washing, the detection was processed as described above.
2.10. Immunocytochemical staining analysis
Biochip slides (Euroimmun, Lubeck, Germany) were used to test the binding reactivity of antibodies in sera or scFv-expressing phage to S protein in SARS-CoV-infected Vero E6 cells. After rinsed with TBST, slides were incubated with either 1011 phage particles, immunized chicken sera or human sera for 1 h at RT. After washed with TBST, the bound antibodies or phage were detected with mouse anti-M13 antibodies or corresponding secondary antibodies (Sigma, USA) conjugated with fluorescein isothiocyanate (FITC). Slides were coated with mounting oil and inspected with an immunofluorescent microscope.
2.11. Sequence analysis
The nucleotide sequences of VH and VL genes from selected clones were determined by an auto-sequencer (ABI PRISM 377; Perkin-Elmer) using ompseq (5′-AAGACAGCTATCGCGATTGCAGTG-3′) and HRML-F (5′-GGTGGT-TCCTCTAGATCTTCC-3′) primers.
3. Results
3.1. Production, purification and characterization of truncated S proteins
To produce antigens for animal immunization and epitopic mapping, 10 fragments of S protein gene were PCR-amplified, cloned into pET-21 vector and expressed as his-fused recombinant proteins. The locations and predicted gene lengths of these fragments are shown in Fig. 1A and B. After induction, these fragments were successfully expressed in E. coli BL-21 cells with molecular weights of 12–30 kDa. Individual fusion protein was purified by Ni2+-charged sepharose and analyzed on SDS–PAGE (Fig. 2A). In Fig. 2B, anti-his antibodies were used to further confirm their identity on Western blot.
Fig. 1.
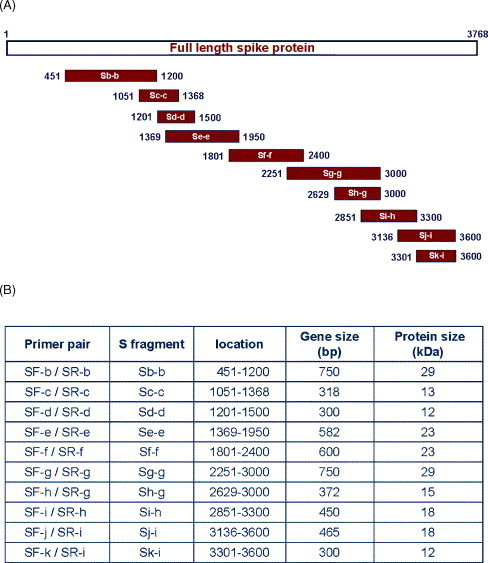
(A) Schematic drawing of cloning strategy for 10 truncated S genes. (B) Amino acid residual locations and predicted molecular weights of these truncated proteins expressed in E. coli cells.
Fig. 2.
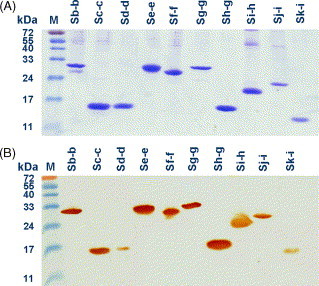
Coomassie blue staining and Western blotting of various his-fused S fragments. Ten recombinant his-fused S fragments (Sb-b to Sk-i) were purified from E. coli cellular lysates using Ni2+-charged sepharose and visualized on SDS–PAGE (A). The molecular weights of the recombinant S fragments were 12–29 kDa, as expected. The proteins were transferred onto nitrocellular paper and analyzed by anti-his antibodies conjugated with horseradish peroxidase (B).
3.2. Determination of binding activity
Western blot analysis was used to determine antigenic reactivity of 10 SARS-CoV-infected patients’ sera and three normal control sera. As shown in Table 1 , fragment Sg-g had apparent binding reactivity with all patients’ sera, while three other fragments, Sb-b, Se-e and Sf-f, were recognized by 70% of patients’ sera. Only 20% of patients’ sera recognized fragments Sc-c and Sh-g. Four remaining fragments showed no binding reactivity to all patients’ sera as well as to three normal sera. The representative Western blots of these results are shown in Fig. 3A (reacted with one patient's serum) and Fig. 3B (with one normal serum). On contrast to normal sera, most patients’ sera could clearly recognize the fragments of Sb-b, Se-e, Sf-f and Sg-g. The results suggested that fragment Sg-g alone or together with the three other fragments may serve as a good target to detect anti-SARS-CoV S protein antibodies. To generate S protein-specific phage clone, 10 S fragments (15 μg/fragment) were mixed to immunize chicken from which spleens had been taken for scFv antibody library construction.
Table 1.
Reactivity of his-fused S proteins with patient and normal sera
| Truncated spike protein fragments |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sb-b | Sc-c | Sd-d | Se-e | Sf-f | Sg-g | Sh-g | Si-h | Sj-i | Sk-i | |
| Patient sera | ||||||||||
| 1 | + | − | − | + | + | + | − | − | − | − |
| 2 | − | − | − | − | − | + | − | − | − | − |
| 3 | + | − | − | + | + | + | + | − | − | − |
| 4 | + | − | − | + | + | + | + | − | − | − |
| 5 | + | + | − | + | + | + | − | − | − | − |
| 6 | − | − | − | − | − | + | − | − | − | − |
| 7 | + | + | − | + | + | + | − | − | − | |
| 8 | + | − | − | + | + | + | − | − | − | − |
| 9 | − | − | − | + | + | + | − | − | − | − |
| 10 | + | − | − | − | − | + | − | − | − | − |
| Positive rates (%) | 70 | 20 | 0 | 70 | 70 | 100 | 20 | 0 | 0 | 0 |
| Normal sera | ||||||||||
| 1 | − | − | − | − | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | − | − | − | − |
| 3 | − | − | − | − | − | − | − | − | − | − |
Fig. 3.
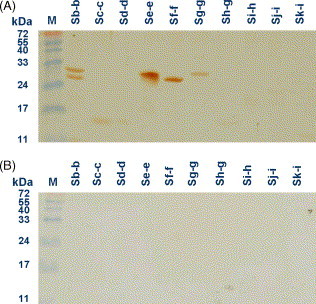
Western blotting of truncated S fragments with patients’ sera. Sera of one patient who recovered from SARS-CoV infection (A) and an un-infected individual (B) were tested for the presence of specific anti-SARS-CoV antibodies. Secondary antibodies were anti-human Ig antibodies conjugated with horseradish peroxidase.
3.3. Library construction and clone characterization
The amplification of scFv immunoglobulin genes in chicken was performed by two consecutive PCR steps to produce final products with bands of the expected 750 base pair (bp). In the primary PCR, VH and VL gene were amplified as 400 and 350 bp in size, respectively. Difference in intensity was observed, which may reflect variation in the gene expression and biased amplification efficiency intrinsic to each chicken (data not shown). Thereafter, a library (4S) containing about 5 × 107 clones was constructed. Restriction analysis of 10 randomly chosen clones from the library indicated that 8/10 (80%) of clones contained scFv gene inserts (data not shown). The SARS-CoV S protein-specific binders were enriched by four rounds of panning. Then, individual phage clones were amplified and analyzed for their binding activity by phage ELISA. The results in Fig. 4 showed that the randomly chosen phage clones exhibited significant binding activity (OD between 0.7 and 1.2) and six clones showed intermediate level of binding activity (OD between 0.4 and 0.6) against SARS-CoV-infected Vero E6 cell lysates. In control samples, both wild-type M13 phage and irrelevant scFv-expressing phage showed weak reactivity (OD below 0.2). Accordingly, three highly and two moderately positive clones, including 4S1, 4S3, 4S4, 4S12 and 4S15, were further characterized. To understand the scFv gene diversity of these clones, VL and VH genes were sequenced by two designed sense primers (Barbas et al., 2001). After aligning with the chicken germline gene, the five chosen clones revealed 6–12% and 31–62% variation in framework (FR) and CDR regions, respectively, leading to a 13–25% mutation rate in the entire V region (Fig. 5 and Table 2 ).
Fig. 4.
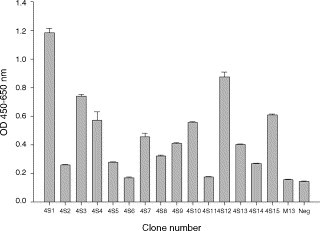
Binding activity of randomly chosen phages from library 4S after the fourth panning cycle. Fifteen phage clones (4S1 to 4S15) were amplified and prepared to test their binding to SARS-CoV-infected Vero E6 cell lysates in an ELISA test. Wild-type M13 phage and one irrelevant phage expressing anti-Eno1 scFv were used as negative controls. Binding phages were detected by anti-M13 phage antibodies conjugated with horseradish peroxidase. The color was developed and absorbance was measured at 450–650 nm.
Fig. 5.

Sequence analysis of scFv genes of five potentially positive clones. The amino acid sequences of light and heavy chain variable regions (VL and VH) of scFv fragments were compared to those of the chicken germline gene. Sequence gaps were introduced to maximize alignment and indicated by blank spaces. FR and CDR boundaries were indicated above germline sequence.
Table 2.
The mutation rate of five scFv amino acid sequences
| scFv IgY | CDRs |
FRs |
Entire V region |
|||
|---|---|---|---|---|---|---|
| Exp/gl V | Dif/total | % | Dif/total | % | Dif/total | % |
| 4S1 | ||||||
| λ/gl VL | 10/24 | 42% | 7/78 | 9% | 17/102 | 17% |
| h/gl VH | 22/38 | 58% | 6/87 | 7% | 28/125 | 22% |
| 4S3 | ||||||
| λ/gl VL | 14/28 | 50% | 9/78 | 12% | 23/106 | 22% |
| h/gl VH | 19/36 | 53% | 7/87 | 8% | 26/123 | 21% |
| 4S4 | ||||||
| λ/gl VL | 12/27 | 44% | 7/78 | 9% | 19/105 | 18% |
| h/gl VH | 19/37 | 51% | 7/87 | 8% | 26/124 | 21% |
| 4S12 | ||||||
| λ/gl VL | 16/28 | 57% | 8/78 | 10% | 24/106 | 23% |
| h/gl VH | 24/39 | 62% | 7/87 | 8% | 31/126 | 25% |
| 4S15 | ||||||
| λ/gl VL | 8/26 | 31% | 6/78 | 8% | 14/104 | 13% |
| h/gl VH | 19/40 | 48% | 5/87 | 6% | 24/127 | 19% |
Abbreviations: Dif/total denote [no. of different bp]/[no. of total bp]. Exp/gl V denote the expressed V gene/the germline V gene.
3.4. Immunocytochemical determination with scFv-expressing phages
We used immunocytochemical staining to determine binding ability of scFv-expressing phage 4S1 to the SARS-CoV-infected Vero E6 cells (Fig. 6 ). Four other phage clones, positive in the ELISA method, were also examined; however, they only had weak reactions (data not shown). Bound phages were detected with FITC-labeled anti-M13 antibodies. Patient and normal human sera were used as positive and negative controls. Sera from immunized and pre-immunized chickens were also included. Staining data clearly showed that both polyclonal IgY antibodies (Fig. 6C) and phage 4S1 (Fig. 6E) bind to SARS-CoV-infected Vero E6 cells, whose binding activity is comparable to that of convalescent patient serum (Fig. 6A). Nevertheless, all negative controls, including normal human serum (Fig. 6B), pre-immunized serum (Fig. 6D) and wild M13 phage (Fig. 6F), exhibited no or little binding signal. These results suggested that the bacteria-derived S protein fragments were able to mimic the conformation of native S protein and induced the humoral immune response in chicken. Moreover, specific scFv antibody on phages could be generated to recognize the native S protein expressed on the SARS-CoV-infected Vero E6 cells.
Fig. 6.
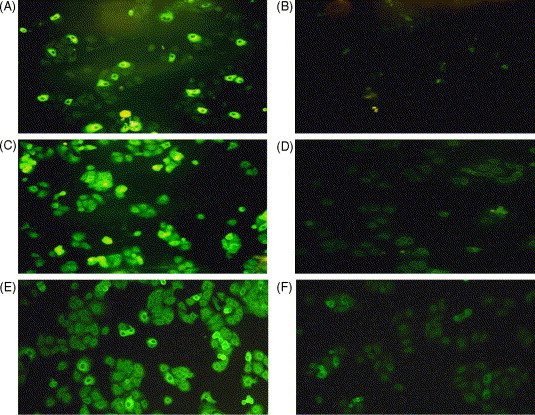
Binding activity of 4S1 phage binder to native S protein. Commercially available Biochips, precoated with SARS-CoV-infected Vero E6 cells were immunostained, respectively, with sera from a convalescent-stage patient (A), un-infected person (B), post- (C) and pre-immunized (D) chickens, 4S1 phage (E) and wild-type M13 phage (F).
3.5. Epitope mapping of antigenic site on S protein
The 4S1 phage binder was used to define the possible antigenic site on the S protein. After being reacted with individual truncated S protein fragments, 4S1 phage showed distinct binding activity against Se-e fragment, suggesting a dominant epitope existed in this location (Fig. 7A). On the contrary, an irrelevant phage clone expressing anti-ENO1 scFv showed no apparent binding activity (OD below 0.2) in the negative control test (Fig. 7B).
Fig. 7.
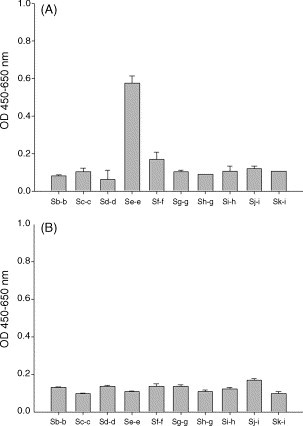
Epitope mapped by 4S1 phage binder. 4S1 phage (A) and an anti-ENO1 phage (B) were examined for their binding activity to 10 purified and truncated S protein fragments coated on ELISA plates.
4. Discussions and conclusion
In this study, we successfully constructed a scFv-displaying phage library (5 × 107 total transformants) from a chicken immunized with 10 truncated S proteins of SARS-CoV from E. coli cells. We did four rounds of panning and enriched specific phage binders 11.6-fold. In general, an enrichment value larger than 50-fold implies the success of the panning process (Barbas et al., 1991). At lower enrichment levels, nonspecific and weak affinity binders could cause outgrowth in the output phages after amplification. The observation by Barbas et al. (1991) is consistent with our phage ELISA result, i.e. that the affinity in more than half of the chosen clones was weak. In this study, we recognized the potential problems in selecting processes, especially to get a specific phage clone with efficient binding ability against the S protein. The result in our panning process showed an increase after the third cycle, although the phage output slightly dropped in the second round (data not shown). Our findings agrees with other studies (Hoe et al., 2005, Richard et al., 2006).
Epitope mapping is an efficient tool to find antigenic sites for the development of preventive agents against viral infection. Because the SARS-CoV S protein can induce neutralizing antibodies, it is a suitable candidate for developing vaccine and diagnostic applications. Our mapping results showed that fragments Se-e (aa 456–650), Sf-f (aa 600–800) and Sg-g (aa 750–1000), reacting with 70, 70 and 100% of patients’ sera, respectively (Table 1), covered most potential epitopic regions reported elsewhere (Chou et al., 2005, Greenough et al., 2005, He et al., 2004, Hua et al., 2005, Ren et al., 2003, Wang et al., 2005, Zhi et al., 2005). We also identified, for the first time, an additional, highly antigenic fragment Sb-b (aa 150–400), which was recognized by 70% of patients’ sera. Notably, the carboxyl terminus of fragment Sb-b in a receptor-binding region of S protein is responsible for SARS-CoV infection to its host cells (Li et al., 2003, van den Brink et al., 2005). Moreover, fragment Sg-g, which is recognized by all the examined sera, overlaps with a neutralizing domain located at residues 797–1192. This domain, which has been previously reported, can induce a protective antibody response against SARS-CoV infection (Chou et al., 2005, Wang et al., 2005). Taken together, we suggest that these E. coli-derived truncated S fragments are excellent targets for diagnostic and therapeutic applications.
The phage display system is a fast and valuable technique for producing monoclonal antibodies against broad viral infection (Cortay et al., 2006, Karle et al., 2004, Tarr et al., 2006). Of the various recombinant antibody formats, scFv is the most commonly used because not only can be rapidly constructed and well-expressed in E. coli, but it also has high affinity and relative stability (Bird et al., 1988, Huston et al., 1988). Among all diagnostic tests, the phage-based method is relatively easy to perform and can be adopted as a quick test for screening many laboratory samples (Lee et al., 2006). Using specific biotin-tagged phage conjugated with streptavidin-coated quantum-dot, it has the power to detect as few as 10 cells/ml (Edgar et al., 2006). In this study, we found that 4S1 posessed the greatest binding ability detected on both phage ELISA and immunocytochemical staining when 1010–1011 phage particles were used. Lu et al. (2004) have previously developed an ELISA assay using phage particles, which is about 100 times more sensitive than the conventional immune-absorbent assay. Our results are comparable to those obtained by Lu et al. (2004), indicating that 4S1 phage is a suitable candidate for developing a phage-based diagnostic assay.
We also found that four other phage clones (4S3, 4S4, 4S12 and 4S15) had superior reactivity on phage ELISA but weak binding signals on immunocytochemical staining. We offer two possible explanations for these conflicting results: (1) truncated S fragments do not completely mimic some conformational epitopes of native S protein secreted by SARS-CoV, or (2) the antigenic sites of S protein recognized by these clones are highly glycosylated on native S protein.
Research to produce polyclonal antibodies in birds and extract specific IgY from egg yolk has increased (Abouzid et al., 2005, LeClaire et al., 2002). The purification process to mass produce IgY with stable, specific poly-IgY activity in eggs is simple (Mine and Kovacs-Nolan, 2002), but monoclonal antibodies are needed to obtain higher specificity. In the chicken antibody gene, both the light and heavy chains have single V and J regions (heavy chain has an additional D region). Based on single VL and VH genes in chicken immunoglobulins, researchers have found that one set of antibody gene primers is convenient for library construction (Andris-Widhopf et al., 2000, Davies et al., 1995). Using this distinctive feature, we have rearranged and generated a diverse antibody repertoire from chicken. To our knowledge, we are the first to use chicken in generating a scFv library directed against the SARS-CoV S protein. After comparing the findings in sequence analysis of chosen phage clones with chicken germline, we found large dissimilarities, typically in the CDR regions of both VL and VH (Fig. 5), revealing a 31–57% and 48–62% mutation rate, respectively (Table 2). Gearhart and Bogenhagen (1983) reported somatic hypermutations to increase antibody affinity occurred more often in the CDR than FR regions of the rearranged V gene. Thus, the Ig genes of our chosen clones from hyperimmunized chickens had accumulated more mutations in CDR regions as a result of affinity selection of B cells.
In conclusion, our data indicate that immunizing chicken and using phage display technology offers an alternative method of producing poly- and mono-specific antibodies. This knowledge may be helpful in the development of a rapid diagnostic and therapeutic agent for clinical and scientific application.
Acknowledgments
This study was supported by Grants SKH-TMU-93-45 and NSC 92-2751-B-038-001-Y from National Science Council (NSC) in Taiwan. Professor Winston W. Shen made some editorial comments in an earlier version of this manuscript.
References
- Abouzid K., Ndeboko B., Durantel S., Jamard C., Zoulim F., Buronfosse T., Cova L. Genetic vaccination for production of DNA-designed antibodies specific to Hepadnavirus envelope proteins. Vaccine. 2005;24:4615–4617. doi: 10.1016/j.vaccine.2005.08.085. [DOI] [PubMed] [Google Scholar]
- Akita E.M., Nakai S. Comparison of four purification methods for the production of immunoglobulins from eggs laid by hens immunized with an enterotoxigenic E. coli strain. J. Immunol. Methods. 1993;160:207–214. doi: 10.1016/0022-1759(93)90179-b. [DOI] [PubMed] [Google Scholar]
- Akita E.M., Nakai S. Production and purification of Fab’ fragments from chicken egg yolk immunoglobulin Y (IgY) J. Immunol. Methods. 1993;162:155–164. doi: 10.1016/0022-1759(93)90380-p. [DOI] [PubMed] [Google Scholar]
- Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F., III Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods. 2000;242:159–181. doi: 10.1016/s0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Barbas C.F., III, Kang A.S., Lerner R.A., Benkovic S.J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C.F., III, Burton D.R., Scott J.K., Silverman G.J. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2001. Phage Display: A Laboratory Manual. [Google Scholar]
- Bird R.E., Hardman K.D., Jacobson J.W., Johnson S., Kaufman B.M., Lee S.M., Lee T., Pope S.H., Riordan G.S., Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.H., Wang S., Sakhatskyy P.V., Mboudjeka I., Lawrence J.M., Huang S., Coley S., Yang B., Li J., Zhu Q., Lu S. Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2005;334:134–143. doi: 10.1016/j.virol.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortay J.C., Gerlier D., Iseni F. Selection of single-chain antibodies that specifically interact with vesicular stomatitis virus (VSV) nucleocapsid and inhibit viral RNA synthesis. J. Virol. Methods. 2006;131:16–20. doi: 10.1016/j.jviromet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Davies E.L., Smith J.S., Birkett C.R., Manser J.M., Anderson-Dear D.V., Young J.R. Selection of specific phage-display antibodies using libraries derived from chicken immunoglobulin genes. J. Immunol. Methods. 1995;186:125–135. doi: 10.1016/0022-1759(95)00143-x. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., McKinstry M., Hwang J., Oppenheim A.B., Fekete R.A., Giulian G., Merril C., Nagashima K., Adhya S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrsen J., van Wyngaardt W., Mashau C., Potgieter A.C., Chaudhary V.K., Gupta A., Jordaan F.A., du Plessis D.H. Serogroup-reactive and type-specific detection of bluetongue virus antibodies using chicken scFvs in inhibition ELISAs. J. Virol. Methods. 2005;129:31–39. doi: 10.1016/j.jviromet.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Finlay W.J., Shaw I., Reilly J.P., Kane M. Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl. Environ. Microbiol. 2006;72:3343–3349. doi: 10.1128/AEM.72.5.3343-3349.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart P.J., Bogenhagen D.F. Clusters of point mutations are found exclusively around rearranged antibody variable genes. Proc. Natl. Acad. Sci. U.S.A. 1983;80:3439–3443. doi: 10.1073/pnas.80.11.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough T.C., Babcock G.J., Roberts A., Hernandez H.J., Thomas W.D., Jr., Coccia J.A., Graziano R.F., Srinivasan M., Lowy I., Finberg R.W., Subbarao K., Vogel L., Somasundaran M., Luzuriaga K., Sullivan J.L., Ambrosino D.M. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Hoe L.N., Wan K.L., Nathan S. Construction and characterization of recombinant single-chain variable fragment antibodies against Toxoplasma gondii MIC2 protein. Parasitology. 2005;131:759–768. doi: 10.1017/S0031182005008450. [DOI] [PubMed] [Google Scholar]
- Hua R.H., Wang Y.F., Bu Z.G., Zhou Y.J., Ge J.Y., Wang X.J., Tong G.Z. Identification and antigenic epitope mapping of immunodominant region amino residues 510 to 672 on the spike protein of the severe acute respiratory syndrome coronavirus. DNA Cell Biol. 2005;24:503–509. doi: 10.1089/dna.2005.24.503. [DOI] [PubMed] [Google Scholar]
- Huston J.S., Levinson D., Mudgett-Hunter M., Tai M.S., Novotny J., Margolies M.N., Ridge R.J., Bruccoleri R.E., Haber E., Crea R., Oppermann H. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karle S., Planque S., Nishiyama Y., Taguchi H., Zhou Y.X., Salas M., Lake D., Thiagarajan P., Arnett F., Hanson C.V., Paul S. Cross-clade HIV-1 neutralization by an antibody fragment from a lupus phage display library. Aids. 2004;18:329–331. doi: 10.1097/00002030-200401230-00026. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- LeClaire R.D., Hunt R.E., Bavari S. Protection against bacterial superantigen staphylococcal enterotoxin B by passive vaccination. Infect. Immun. 2002;70:2278–2281. doi: 10.1128/IAI.70.5.2278-2281.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.C., Yusoff K., Nathan S., Tan W.S. Detection of virulent Newcastle disease virus using a phage-capturing dot blot assay. J. Virol. Methods. 2006 doi: 10.1016/j.jviromet.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Weiss P., Block T. A phage with high affinity for hepatitis B surface antigen for the detection of HBsAg. J. Virol. Methods. 2004;119:51–54. doi: 10.1016/j.jviromet.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mine Y., Kovacs-Nolan J. Chicken egg yolk antibodies as therapeutics in enteric infectious disease: a review. J. Med. Food. 2002;5:159–169. doi: 10.1089/10966200260398198. [DOI] [PubMed] [Google Scholar]
- Park K.J., Park D.W., Kim C.H., Han B.K., Park T.S., Han J.Y., Lillehoj H.S., Kim J.K. Development and characterization of a recombinant chicken single-chain Fv antibody detecting Eimeria acervulina sporozoite antigen. Biotechnol. Lett. 2005;27:289–295. doi: 10.1007/s10529-005-0682-8. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Yuen K.Y., Osterhaus A.D., Stohr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Ren Y., Zhou Z., Liu J., Lin L., Li S., Wang H., Xia J., Zhao Z., Wen J., Zhou C., Wang J., Yin J., Xu N., Liu S. A strategy for searching antigenic regions in the SARS-CoV spike protein. Genom. Proteom. Bioinform. 2003;1:207–215. doi: 10.1016/S1672-0229(03)01026-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Perreault J., Gane P., el Nemer W., Cartron J.P., St-Louis M. Phage-derived monoclonal anti-Lu. Transfusion. 2006;46:1011–1017. doi: 10.1111/j.1537-2995.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS–beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr A.W., Owsianka A.M., Timms J.M., McClure C.P., Brown R.J., Hickling T.P., Pietschmann T., Bartenschlager R., Patel A.H., Ball J.K. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 2006;43:592–601. doi: 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- van den Brink E.N., Ter Meulen J., Cox F., Jongeneelen M.A., Thijsse A., Throsby M., Marissen W.E., Rood P.M., Bakker A.B., Gelderblom H.R., Martina B.E., Osterhaus A.D., Preiser W., Doerr H.W., de Kruif J., Goudsmit J. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chou T.H., Sakhatskyy P.V., Huang S., Lawrence J.M., Cao H., Huang X., Lu S. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 2005;79:1906–1910. doi: 10.1128/JVI.79.3.1906-1910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H., Schumer G., Gao G., Boyer J.L., Crystal R.G., Wilson J.M. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335:34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]


