Summary
Background
Human coronavirus (HCoV) OC43 is the most prevalent HCoV in respiratory tract infections. Its molecular epidemiological characterization, particularly the genotyping, was poorly addressed.
Methods
The full-length spike (S), RNA-dependent RNA polymerase (RdRp), and nucleocapsid (N) genes were amplified from each respiratory sample collected from 65 HCoV-OC43-positive patients between 2005 and 2012. Genotypes were determined by phylogenetic analysis. Recombination was analyzed based on full-length viral genome sequences. Clinical manifestations of each HCoV genotype infection were compared by reviewing clinical records.
Results
Sixty of these 65 samples belong to genotypes B, C and D. The remaining five strains had incongruent positions in the phylogenetic trees of the S, RdRp and N genes, suggesting a novel genotype emerging, designated as genotype E. Whole genome sequencing and bootscan analysis indicated that genotype E is generated by recombination between genotypes B, C and D. Temporal analysis revealed a sequential genotype replacement of C, B, D and E over the study period with genotype D being the dominant genotype since 2007. The novel genotype E was only detected in children younger than three years suffering from lower respiratory tract infections.
Conclusions
Our results suggest that HCoV-OC43 genotypes are evolving. Such genotype shift may be an adapting mechanism for HCoV-OC43 maintaining its epidemic.
Keywords: Human coronavirus OC43, Respiratory infection, Molecular epidemiology, Genotype, Recombination
Graphical abstract
Highlights
-
•
Temporal shift of multiple human coronavirus OC43 genotypes.
-
•
Emergence of a novel genotype E by natural recombination.
-
•
Genotype D dominated HCoV-OC43 epidemic in China in recent years.
-
•
Genotype evolving plays an important role in HCoV-OC43 epidemic.
Introduction
Coronaviruses (CoVs), belonging to the family Coronavirinae, are a large group of viruses with a broad infection spectrum in human and animals. CoVs are related to respiratory tract disorders, gastroenteritis, as well as to systemic and neurological diseases.1 CoVs are the largest RNA viruses, containing a positive-sense, single-stranded RNA genome with a length of 27,000–31,500 nucletides.1, 2 Based on genome phylogeny and serological characterization, CoVs are divided into four genera, Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus(δ-CoV).1, 2, 3 Since the isolation of HCoV-229E and -OC43 in 1960s, a total of six HCoV species have been identified, including severe acute respiratory syndrome CoV (SARS-CoV) in 2003, NL63 and HKU1 in 2004, and middle east respiratory syndrome CoV (MERS-CoV) in 2012.1, 4 HCoVs belong to α- (229E and NL63) and β-genera (OC43, HKU1, SARS-CoV and MERS-CoV).
HCoVs were previously not considered to be of great importance with respect to human diseases as most HCoV-infections were thought to be associated with mild symptoms and occasional lower respiratory tract infections (LRTIs) until an outbreak of SARS in 2003. That has led to increased concerns about HCoVs, while the identification of MERS-CoV in 2012 reinforced the public health significance of HCoVs. Although SARS-CoV is no longer detected since 2004, MERS-CoV continued as an epidemic, spreading to more patients and countries. This spread indicates a high adaption capability of MERS-CoV in humans.5, 6 Insight into the epidemic characteristics of HCoVs at the molecular level will allow us to predict viral pathogenesis and transmission activities and inform HCoV prevention and control, particularly against newly emerging HCoVs.
HCoV-OC43 has been more prevalent than other common HCoVs including HCoV-229E, –NL63 and –HKU1, in pediatric and adult respiratory infections, and can also cause outbreaks in human respiratory tract infections.1, 7, 8, 9, 10 However, our understanding of the molecular epidemiology of HCoV-OC43 has been very limited. The genetic diversity of HCoV-OC43 was first reported in Belgium in 2005 and three clusters were identified based on the analysis of the spike (S) gene of the prototype strain ATCC VR-759 and seven clinical strains.11 Subsequently, Lau et al. gave the first description on the molecular epidemiology of HCoV-OC43 using sequences from 29 clinical samples in 2011.12 Four genotypes, A, B, C and D, were identified based on the viral genome and the phylogeny of the main structural genes, S, RNA-dependent RNA polymerase (RdRp), and nucleocapsid (N) genes, and genotype D was reported to have arisen due to natural recombination.12 However, these observations were based on only a limited number of HCoV-OC43 positive cases. Due to the limited availability of virus sequences, the molecular epidemiological characterization of HCoV-OC43, particularly its genotyping, was poorly deciphered.
In this study, we genotyped HCoV-OC43 by analyzing full-length sequences of S, RdRp, N genes and viral genomes directly from respiratory samples collected from 65 HCoV-OC43 positive patients with acute respiratory tract infections (ARTIs) recruited from 2005 to 2012. We observed a genotype shift in HCoV-OC43 over the study period and confirmed the emergence of a new genotype E arising through natural recombination.
Methods
Patients and clinical specimens
Patients suffering from ARTIs were recruited from the Beijing Children Hospital and the Peking Union Medical College Hospital in Beijing, China from March 2005 to December 2012 when they seek health care at these hospitals. Criteria for including patients in our study encompassed acute fever (body temperature ≥37.5 °C) with respiratory symptoms such as cough or wheezing, normal or low leukocyte count, and with or without radiological pulmonary abnormalities. Nasopharyngeal aspirates (NPAs) were collected from pediatric patients. Nasal and throat swabs were collected from adult patients. The respiratory samples were stored in viral transport medium (VTM) at −80 °C before use. Clinical information of each enrolled patient was recorded in standard form and reviewed retrospectively. Written informed consent was obtained from all participants or guardians on behalf of the minors/children participants. The study was approved by the Medical Ethic Review Board of the Institute of Pathogen Biology, Chinese Academy of Medical Sciences.
Molecular detection of HCoVs
Viral nucleic acids were extracted from 200 μl respiratory samples using a NucliSens easyMAG apparatus (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions and were stored at −80 °C until use. HCoV-OC43 positive respiratory samples were tested by RT-PCR with HCoV-conserved primers and were confirmed by sequencing methods as described elsewhere.13 The presence of other common respiratory viruses was also determined as described elsewhere, including influenza virus (IFV) A, B and C, human parainfluenza virus (HPIV) 1–4, adenovirus (Adv), respiratory syncytial virus (RSV) A and B, human metapneumovirus (hMPV), human bocavirus (HBoV), rhinovirus (HRV) and enterovirus (HEV).14
Sequencing of HCoV-OC43 genes and viral genome
Total RNA from respiratory specimens was converted to cDNA using combined random primers and oligo(dT) primers and the SuperScript III reverse transcription system (Invitrogen, Carlsbad, CA). The full-length S, RdRp, N genes and viral genomes were amplified from each respiratory specimen which was positive for HCoV-OC43, using specific primers (Table S1) with a genome walking method. PCR was performed using the following conditions: 94 °C for 5 min, 40 cycles of amplification at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 90 s, with a terminal elongation step at 72 °C for 10 min. PCR products were sequenced directly using an ABI 3700 DNA sequencer (Applied Biosystems, USA). Sequences were assembled manually through alignment to the reference strain HK04-02 (GenBank accession no. JN129835).
Phylogenetic analysis
All HCoV-OC43 sequences available in GenBank (www.ncbi.nlm.nih.gov) were retrieved on May 30, 2013. The background information of all the sequences used for phylogenetic analysis is summarized in Table S2. The full-length S, RdRp, N genes, and viral genomes of HCoV-OC43 were aligned using ClustalW program implemented in MEGA 5.1 with sequences deposited in GenBank.15 Pair-wise sequence identities in each region were calculated for the comparison of sequence divergence using BioEdit. Maximum likelihood (ML) trees were constructed with the best fit model of General Time Reversible with gamma-distributed rate variation across sites and 1000 bootstrap pseudo-replicates implemented in MEGA 5.1. The bovine coronavirus was used as the outgroup sequence, but is not shown in the presented figures to make the phylogenetic relationships more clear. Substitution models were selected using Modeltest (version 3.7) according to the Akaike information criterion.16 Phylogenetic trees of each gene region of HCoV-OC43 were constructed by using the neighbor-joining method with Kimura's two-parameter model and 1000 bootstrap pseudo-replicates implemented in MEGA 5.1.15 To analyze the recombination events, the genomes of HCoV-OC43 were aligned and analyzed using boot scanning method implemented in SimPlot (V3.5.1, http://sray.med.som.jhmi.edu/SCRoftware).
Statistical analysis
Distribution frequencies of HCoV-OC43 genotypes were compared by using Pearson's Chi square test or Fisher's exact test. One-way analysis of variance was used to analyze the continuous variables for population parameters. P values <0.05 were considered statistically significant.
Nucleotide sequence accession numbers
The nucleotide sequence data of S, RdRp, N genes and viral genomes of HCoV-OC43 used in this study have been lodged in GenBank and the accession numbers are shown in Table S2.
Results
Genotyping of HCoV-OC43 strains
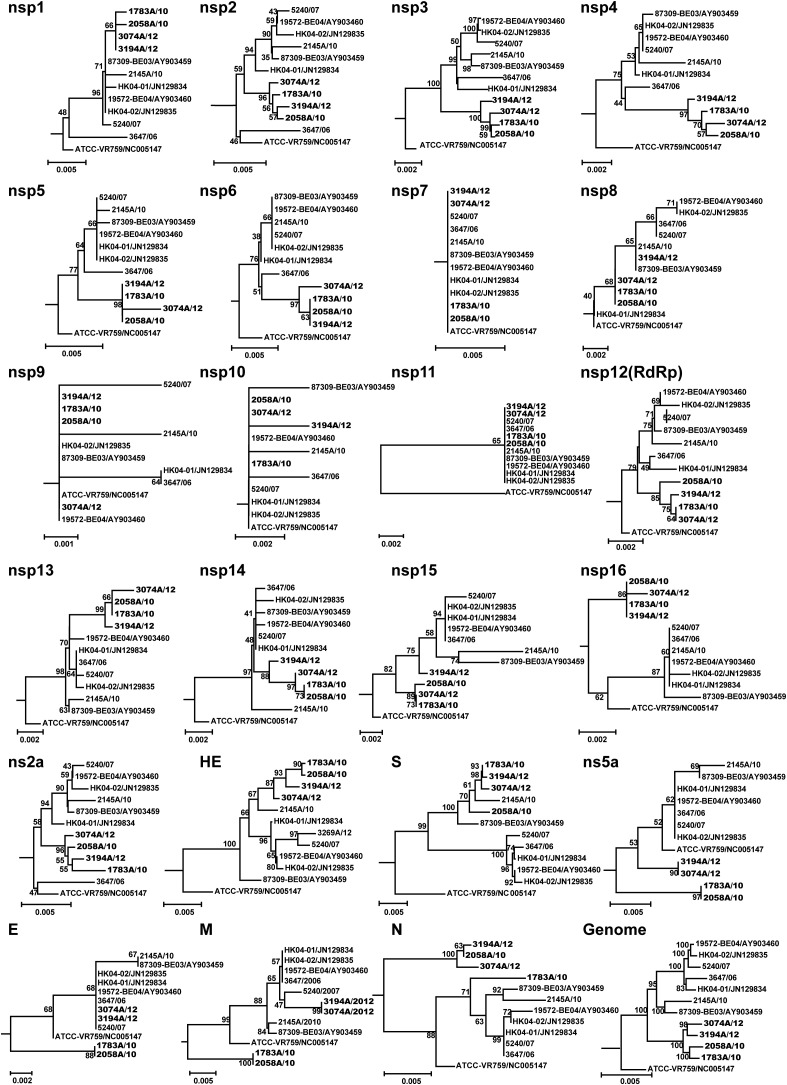
To genotype the HCoV-OC43 samples, we constructed ML trees using the full-length sequences of S, RdRp and N genes amplified from the 65 respiratory samples of HCoV-OC43 positive patients in this study and compared them to those retrieved from GenBank (Fig. 1 ). The HCoV-OC43 sequences fell into four distinct clusters on the phylogenetic tree of the S gene as reported by Lau et al.12 However, incongruities were observed in ML trees of RdRp and N genes, indicating genetic diversity. Briefly, OC43 strains identified in this study (designated CN strains) fell into three clusters in S gene, i.e., B, C and D genotypes, similar to those from Hong Kong, China (HK) and Belgium (BE). Eleven CN strains fell into genotype B together with five 2004 HK strains and the Belgium strain BE03.11, 12 The sequences of this genotype possessed nucleotide (nt) identities of 98.7%–99.6%. Three CN strains and 15 HK strains formed genotype C, possessing 99.6%–99.8% nt identities; while 51 CN strains clustered with nine HK strains and a BE04 strain to form genotype D, possessing 99.3%–100% nt identities. Genotype A contained only the cell culture strain ATCC VR-759 as previously reported.11, 12
Figure 1.
Phylogenetic analysis of different HCoV-OC43 strains based on full-length S, RdRp and N genes. Trees were constructed using the maximum-likelihood method. Strains identified in this study are presented in bold. A, B, C, D and E represent genotypes.
The strains that fell into genotype C clustered together in the ML tree of the RdRp gene, as well as in the ML tree of the S gene. Strains belonging to genotype D clustered together with strains of genotype B, and these sequences possessed 99.7%–99.8% nt identities. Notably, five CN strains (1783A/10, 2058A/10, 2941A/11, 3074A/12 and 3194A/12), which belong to genotype B in the ML tree of the S gene, formed a distinct clade in the tree of RdRp gene. Multiple alignment of RdRp results showed that these five CN strains possessed 99.5–99.6% nt identities to B_BE03, C_HK04-01 and D_HK11-01, while other B strains possessed 99.7–100% nt identities to B_BE03 (Table 1 ).
Table 1.
Nucleotide sequence identities for the S, RdRp and N genes of genotype E compared with the reference sequences of genotypes B, C and D.
Analysis of the N genes showed that the strains that belong to genotype B (other than the five distinct CN stains) in the ML tree of the S gene clustered together, while the strains belonged to genotype C and D in the ML tree of the S gene clustered together. The aforementioned five distinct CN strains were separated from all the known genotypes and formed two clades. Multiple alignment results were consistent with our phylogenetic analysis as the five distinct CN strains had lower nt identities with representatives of B, C and D genotypes than other genotype B strains had with the reference strain, including B_BE03 (97.6–98.7%), C_HK04-01 (97.6–99.1%) and D_HK11-01 (97.5–99.0%) (Table 1). Taken together, the incongruities in the phylogenetic trees together with the analysis of nt identities showed that a novel genotype, may have arisen, which we designated as genotype E.
Recombination analysis of genotype E strains
The incongruent phylogenetic pattern of the S, RdRp and N genes in the five genotype E strains, particularly the drop-out of 1783A/10 from the linage formed by other genotype E strains in the phylogenetic tree of N genes, indicate the occurrence of potential recombination events. To further demonstrate the emergence of genotype E strains, we amplified the whole viral genome sequences directly from respiratory samples. We obtained the whole genome sequences of four of the five distinct strains (1783A/10, 2058A/10, 3074A/12 and 3194A/12; 2194A/11 was not available due to the very low viral load in the specimen). We then analyzed the potential recombination by constructing the phylogenetic trees of all known 23 gene regions of these four strains. Ten other whole genome sequences of OC43 were used as reference strains, including BE03 and 2145A/10 (genotype B), HK04-01 and 3647/06 (genotype C), BE04, HK04-02 and 5240/07 (genotype D), and the ATCC strain (genotype A) (Fig. 2 ). Bovine CoV (accession no. U00735) was used as outgroup sequence, which was not displayed in the figure to save spaces. We found that these four strains form a separate linage (genotype E) in the phylogenetic trees of complete genome, S, RdRp and most of the nonstructual protein (nsp) genes. These findings further confirmed that these distinct CN strains belong to a novel genotype E, despite the incongruent phylogenetic pattern was observed in ns5a, E, M and N genes.
Figure 2.
Phylogenetic analysis of multiple gene regions of HCov-OC43 strains. A total of 23 gene regions are analyzed, including nsp1 to nsp16, ns2α, HE, S, NS5α, E, M and N of ten genomes of HCoV-OC43. The neighbor-joining method (Kimura's two-parameter) was used to construct the trees with 1000 bootstrap values.
Notably, in the phylogenetic trees of the nsp2-nsp6 genes, these four genotype E strains were closely related to genotype C; while clustered more closely with the strains of genotype B in the trees of nsp1, nsp8, hemagglutinin-esterase (HE) and the S genes. Strains 3074A/12 and 3194A/12 were also clustered together with genotype D in envelope (E) and membrane (M) genes. These results support our hypothesis that recombination events occur among OC43 genotypes.
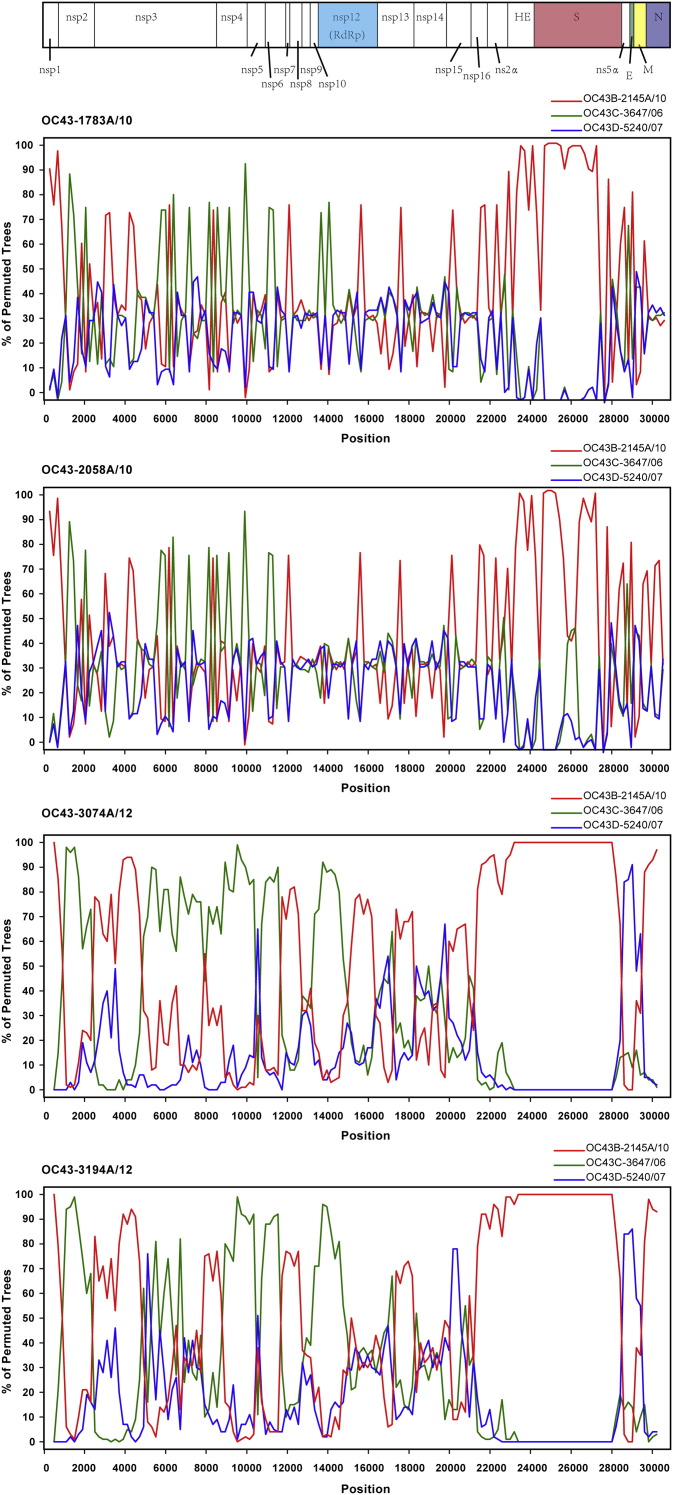
To verify these findings, we then carried out boot scanning analysis and the genome sequences of B_2145A/10, C_3647/06 and D_5240/07 were used as references. When the genomes of 1783A/10, 2058A/10, 3074A/12 and 3194A/12 were used as query sequences, we identified several potential recombination sites in the viral genomes of genotype E (Fig. 3 ). Here 3074A/12 was used as an example to show the recombination analysis results. From positions nt 1000 to 14,500, most of the region of 3074A/12 were closely related to C_3647/06, except positions upstream of nt 1,000, nt 2500 to 4,500, and nt 11,500 to 12,500, where 3074A/12 was closely related to B_2145A/10. From positions of nt 14,500 to nt 28,000, most of the region was closely related to B_2145A/10. From positions nt 28,000 to the 3′ end of the viral genome, most of the region was closely related to D_5240/07. Potential recombination sites were at the junctions of nsp2/nsp3, nsp6/nsp7, nsp9/nsp10, nsp12/nsp13, ns5α/E and M/N corresponding to the schematic diagram of the whole viral genome (Fig. 3). These findings were consistent with the observations in phylogenetic analysis of S, RdRp and N genes described above. Similar boot-scanning results were obtained when 3194/12 was used as query strain. Most of the recombination sites were also found when 1783A/10 and 2058A/10 were used as query strains. However, lower similarities were found in ns5α, M and N gene regions between 1783A/10, 2058A/10 sequences and references, which indicates the diversity of parent strains of recombination.
Figure 3.
Recombination analyses of HCoV-OC43 genomes. Bootscan plot analysis of the whole genome sequences identified in this study in comparison to reference strains in genotypes B, C and D. Graphs were generated using 1783A/10, 2058A/10, 3074A/12 and 3194A/12 as query sequences. The bootstrap value is 1000 for a window of 200 bp.
Taken together, these findings indicate that natural recombination events led to the emergence of novel genotype E and suggest complicated recombination events in the circulation of HCoV-OC43 strains in nature.
Temporal evolution of HCoV-OC43 genotypes
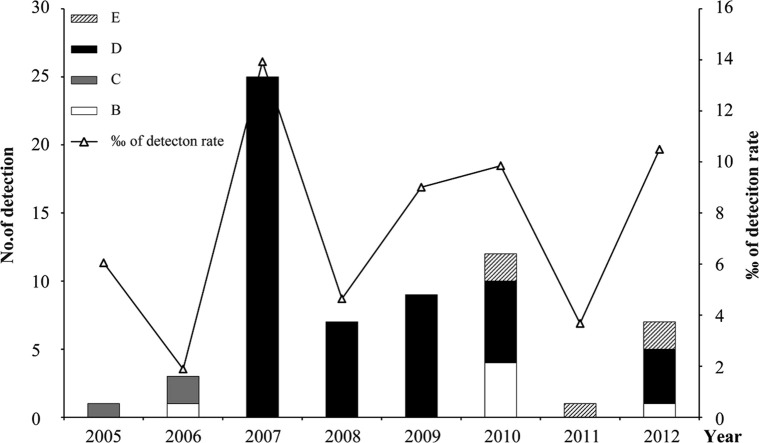
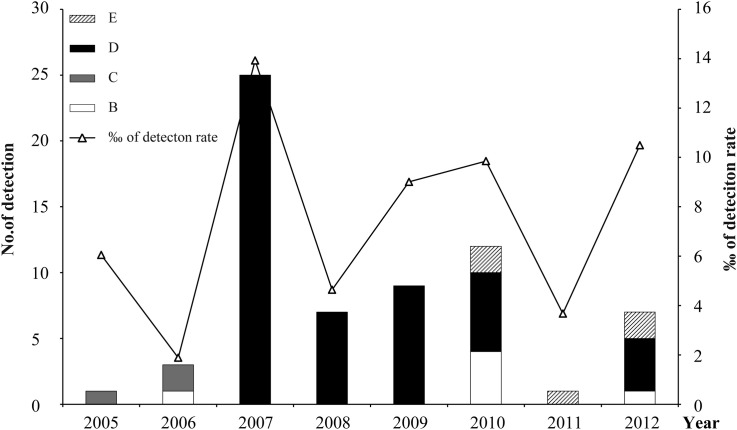
Genotype shift plays an important role in virus adaption to hosts.17, 18, 19 To determine whether genotype shift occurred in HCoV-OC43, the yearly distribution of genotypes during the study period (2005–2012) were determined. HCoV-OC43 positive cases were identified for each year analyzed, and their detection rate ranged from 1.9‰ to 13.9‰ with the highest detection rates in 2007 (Fig. 4 ). We found that the detection rate of HCoV-OC43 spiked every other year except in 2010. Shifts of HCoV-OC43 genotypes over time were observed. After a low level epidemic of genotypes C and B, genotype D became the major epidemic since 2007, with the highest detection rate of HCoV-OC43 during the study period, and dominated between 2007 and 2009. Genotype B re-occurred in 2010 together with the novel genotype E. The co-epidemics of genotype B, D and E were observed in 2010 and 2012. Genotype C has not been detected since 2006.
Figure 4.
Yearly distribution of HCoV-OC43 genotypes during 2005–2012. The left and right vertical axis show the number of HCoV-OC43 cases and the detection rate among the recruited cases.
Clinical characteristics of HCoV-OC43 genotype infections
To characterize the clinical manifestations of different HCoV-OC43 genotypes, the clinical data of the 65 HCoV-OC43 positive cases were analyzed (Detailed information of each patient is summarized in Table S3). Of the 65 cases, 28 were children less than 14 years old, one was a 16-year-old teenager, and 36 were adults more than 16 years old. Patient age ranged from 0.2 to 90 years old (mean 29.6 years; median 20 years), with 33 males and 32 females (Table 2 ). In 17 (26.2%) of all patients an additional virus was co-detected. Each of the genotypes showed co-detection except genotype C. The most frequent co-detected viruses were RSV and HRV. The age distributions in different genotypes differed significantly (One-way analysis of variance, P = 0.0094). Genotype D was detected in patients with a broad age range (0.2–90 year old), although the majority (35 out of 51 cases) occurred in children and adults less than 50 years old. Genotype B was detected in one young adult (21 years old) with URTI in 2006, and in five children with LRTIs after 2010. Genotype C was detected in three older adults aged 58, 72 and 88 years with URTIs, whereas genotype E was only detected in five children less than 3 years of age (0.8–2.7 years old) with LRTIs.
Table 2.
Characteristics of HCoV-OC43 positive cases.
| Parameters | Genotype |
Total | |||
|---|---|---|---|---|---|
| B | C | D | E | ||
| Positive cases | 6 (9.2)a | 3 (4.6) | 51 (78.5) | 5 (7.7) | 65 |
| Age, years | |||||
| Rangeb | 0.6–21.0 | 58.0–88.0 | 0.2–90.0 | 0.8–2.7 | 0.2–90.0 |
| Mean/Median | 5.3/1.8 | 72.7/72.0 | 32.5/23.0 | 2.5/1.1 | 29.6/20.0 |
| Gender (M/F) | 4/2 | 1/2 | 26/25 | 2/3 | 33/32 |
| Diagnosis | |||||
| URTIs | 1 (16.7) | 3 (100) | 33 (64.7) | 0 | 37 (56.9) |
| LRTIs | 5 (83.3) | 0 | 18 (35.3) | 5 (100) | 28 (43.1) |
| Co-detected cases | 3 (50) | 0 | 12 (23.5) | 2 (40.0) | 17 (26.2) |
| 1 virus | 3 (50) | 0 | 8 (15.7) | 1 (20.0) | 12 (18.5) |
| 2 virus | 0 | 0 | 2 (3.9) | 1 (20.0) | 3 (4.6) |
| 3 virus | 0 | 0 | 2 (3.9) | 0 | 2 (3.1) |
| Frequency of co-detected respiratory viruses | |||||
| RSVA/B | 1 (16.7) | 0 | 5 (9.8) | 2 (40.0) | 8 (12.3) |
| HRV | 2 (33.3) | 0 | 5 (9.8) | 0 | 7 (10.8) |
| IFVA | 0 | 0 | 2 (3.9) | 1 (20.0) | 3 (4.6) |
| HPIVs | 0 | 0 | 3 (5.9) | 0 | 3 (4.6) |
| HBoV | 0 | 0 | 2 (3.9) | 0 | 2 (3.1) |
| hMPV | 0 | 0 | 1 (2.0) | 0 | 1 (1.5) |
M, male. F, female. URTIs, upper respiratory tract infections. LRTIs, lower respiratory tract infections. RSV, respiratory syncytial virus. HRV, rhinovirus. IFVA, influenza viurs type A. HPIVs, human parainfluenza virus. hMPV, human metapneumovirus. HBoV, human bocaviurs.
Numbers in parentheses indicate the percentages of positive detections reported to the total number of positive samples.
P = 0.0094.
Discussion
As an important human respiratory virus, the epidemic features of HCoV-OC43 at molecular level have not been well addressed. In this study, we describe the molecular epidemiological features of HCoV-OC43 in detail based on 65 cases. Our results showed marked variations of HCoV-OC43 genotype prevalence from year to year, similar to that observed in other HCoVs.7, 20, 21, 22 In line with previous reports,11, 12 genotypes B and C were detected before 2006, but disappeared after 2006. However, genotype B re-emerged in 2010 in our study, which has not been reported before. Genotype D, generated from recombination and first identified in 2004, was the dominant genotype for epidemics starting in 2007. This findings overlap with those of Lau et al., who reported that eight HCoV-OC43 strains detected between 2008 and 2011 all belonged to genotype D.12 In our study, genotype D was not detected in 2011 but in 2012, albeit at lower numbers (four out of seven HCoV-OC43 positive samples). It seems that immunity developed in the human population after the wide-spread of genotype D had blocked its epidemic as the overall prevalence of genotype D showed decreased over time. Additional analysis of the evolution of antigenic genes, particularly the S gene will help to further our understanding of the adaption of viral genotypes.
Recombination is a common phenomenon among coronaviruses. A special random template switching mechanisms can be used during RNA replication.23, 24 The high frequency of homologous recombination together with the high mutation rates of the genome may lead to the adaptation of CoVs and allow the generation of new strains and genotypes.25, 26, 27, 28, 29 For example, recombination has been reported to generate new genotypes and to contribute to the genetic diversity in HCoV-HKU1 and -NL63, with recombination sites on nsp6/nsp7, nsp16/HE, and nsp3, and the S genes, respectively.17, 26 Our work, which included a larger number of samples over a longer surveillance period than previous studies, shows that a novel genotype E emerged in 2010. This highlights again the role of recombination in the evolution of HCoV-OC43. Based on nucleotide identity comparison, phylogenetic analysis of different genes, and boot scanning analysis, genotype E might be generated from a recombination between genotypes B, C and D. Potential recombination sites may be at the junctions of nsp2/nsp3, nsp6/nsp7, nsp9/nsp10, nsp12/nsp13, ns5α/E and M/N gene. However, these observations need to be clarified based on more whole genome sequences of OC43. In addition, our results together with those of previous reports on the recombination analysis of HCoVs, indicate that the amplification of genes including at least nsp2/nsp3, nsp12/nsp13 (corresponding to pol gene) and the S and N genes is needed for genotyping and recombination analysis.12, 20, 26
The association of HCoV-OC43 genotypes with disease severity has not been well defined. A previous study found that among eight genotype D positive patients, seven were diagnosed with pneumonia.12 However, in our study, genotype D showed no associations with severe symptoms, as most of the patients suffered from URTIs. This difference in results may be attributed to the studied cohort and number of positive cases. However, host immune pressure in response to genotype D during a long epidemic period may also affect virulence.
All cases of the novel genotype E and those of genotype B identified after 2010 were found in children younger than three years with LRTIs, but not detected in adults with LRTIs or URTIs. However, as the number of positive cases was limited, it is unclear whether the association of genotypes with LRTIs and special age groups is significant. This association may require further investigations for a larger number of samples. In addition, it should be further investigated whether the genetic configuration of genotype E allow it to spread rapidly, leading to the replacement of other genotypes such as genotype D.
In summary, our results on the evolving genotypes of HCoV-OC43 and the emergence of a novel genotype E indicate that genotype shift may be one of the major ways for HCoV-OC43 to maintain its epidemic. Our findings provide insight into the evolution of HCoVs and its epidemicity, and can help inform CoV surveillance and control in humans and animals.
Conflicts of interest
The authors have declared that no competing interests exist.
Acknowledgments
We thank Dr. Patrick CY Woo (The University of Hong Kong, Hong Kong, China) for providing sequence and primer information and for helpful discussions. This study was supported in part by the National Major Science & Technology Project for Control and Prevention of Major Infectious Diseases in China (2012ZX10004-206, 2014ZX10004-001), the Program for New Century Excellent Talents in University (3332013127), the China National Funds for Distinguished Young Scientists (81225014), Program for Changjiang Scholars and Innovative Research Team in University (IRT13007), and Fondation Mérieux. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding was received for this study.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jinf.2014.12.005
Contributor Information
Lili Ren, Email: renliliipb@163.com.
Jianwei Wang, Email: wangjw28@163.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields virology. 6th ed. Lippincott Williams &Wilkins; Philadelphia: 2013. pp. 825–854. [Google Scholar]
- 2.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Committee on Taxonomy of Viruses. Virus taxonomy: 2013 Release. http://ictvonline.org/virusTaxonomy.asp [accessed 20.05.14].
- 4.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Bialek S.R., Allen D., Alvarado-Ramy F., Arthur R., Balajee A., Bell D. First confirmed cases of middle east respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities – May 2014. MMWR Morb Mortal Wkly Rep. 2014;63(19):431–436. [PMC free article] [PubMed] [Google Scholar]
- 6.Premila Devi J., Noraini W., Norhayati R., Chee Kheong C., Badrul A., Zainah S. Laboratory-confirmed case of middle east respiratory syndrome coronavirus (MERS-CoV) infection in Malaysia: preparedness and response, April 2014. Euro Surveill. 2014;19(18) doi: 10.2807/1560-7917.es2014.19.18.20797. pii: 20797. [DOI] [PubMed] [Google Scholar]
- 7.Ren L., Gonzalez R., Xu J., Xiao Y., Li Y., Zhou H. Prevalence of human coronaviruses in adults with acute respiratory tract infections in Beijing, China. J Med Virol. 2011;83(2):291–297. doi: 10.1002/jmv.21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol. 2012;53(2):135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venter M., Lassaunière R., Kresfelder T.L., Westerberg Y., Visser A. Contribution of common and recently described respiratory viruses to annual hospitalizations in children in South Africa. J Med Virol. 2011;83(8):1458–1468. doi: 10.1002/jmv.22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 2003;36(8):985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijgen L., Keyaerts E., Lemey P., Moës E., Li S., Vandamme A.M. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology. 2005;337(1):85–92. doi: 10.1016/j.virol.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S.K., Lee P., Tsang A.K., Yip C.C., Tse H., Lee R.A. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 2011;85(21):11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren L., Gonzalez R., Wang Z., Xiang Z., Wang Y., Zhou H. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect. 2009;15(12):1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum Parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 17.van Niekerk S., Venter M. Replacement of previously circulating respiratory syncytial virus subtype B strains with the BA genotype in South Africa. J Virol. 2011;85(17):8789–8797. doi: 10.1128/JVI.02623-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westgeest K.B., Russell C.A., Lin X., Spronken M.I., Bestebroer T.M., Bahl J. Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J Virol. 2014;88(5):2844–2857. doi: 10.1128/JVI.02163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull R.A., Eden J.S., Rawlinson W.D., White P.A. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010;6(3):e1000831. doi: 10.1371/journal.ppat.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez S.R., Sims G.E., Wentworth D.E., Halpin R.A., Robinson C.C., Town C.D. Genomic analysis of 16 Colorado human NL63 coronaviruses identifies a new genotype, high sequence diversity in the N-terminal domain of the spike gene and evidence of recombination. J Gen Virol. 2012;93(Pt 11):2387–2398. doi: 10.1099/vir.0.044628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44(6):2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: how to make sense...? J Gen Virol. 2006;87(Pt 6):1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 24.Masters P.S. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo P.C., Lau S.K., Yuen K.Y. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr Opin Infect Dis. 2006;19(5):401–407. doi: 10.1097/01.qco.0000244043.08264.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80(14):7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72(5):4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motokawa K., Hohdatsu T., Aizawa C., Koyama H., Hashimoto H. Molecular cloning and sequence determination of the peplomer protein gene of feline infectious peritonitis virus type I. Arch Virol. 1995;140(3):469–480. doi: 10.1007/BF01718424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J., Hon C.C., Li Y., Wang D., Xu G., Zhang H. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol. 2010;91(Pt 4):1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.