Abstract
Mouse models are important tools both for studying the pathogenesis of infectious diseases and for the preclinical evaluation of vaccines and therapies against a wide variety of human pathogens. The use of genetically defined inbred mouse strains, humanized mice, and gene knockout mice has allowed the research community to explore how pathogens cause disease, define the role of specific host genes in either controlling or promoting disease, and identify potential targets for the prevention or treatment of a wide range of infectious agents. This review discusses several of the most commonly used mouse model systems, as well as new resources such as the Collaborative Cross as models for studying infectious diseases.
Keywords: mouse models, infectious diseases, genetics, host/pathogen interactions
Introduction
Over the past century, advances in the development of vaccines, antibiotics/antivirals, and infection control measures have resulted in major reductions in the public health burden of infectious diseases. However, despite these advances, infectious agents continue to cause significant morbidity and mortality in human populations, as well as economic disruption in at-risk communities. The challenges associated with infectious diseases are illustrated not only by emerging pathogens, such as HIV or more recent outbreaks of Zika virus (ZIKV) or Ebola virus, but also by continuing threats from pathogens such as seasonal influenza virus, hepatitis C virus (HCV), dengue virus, or malaria. Furthermore, the emergence of antibiotic- or drug-resistant microorganisms and viruses complicates treatment strategies and further enhances the risk posed by infectious diseases. Due to these threats, there is an ongoing need for new strategies to prevent pathogen transmission, as well as the development of new vaccines and/or therapeutic agents. However, the development of new infection control strategies requires an in-depth understanding of the target pathogen's biology and disease pathogenesis, as well as appropriate systems for testing the safety and efficacy of new treatments or vaccines.
For many pathogens, much of our understanding of pathogen biology and disease pathogenesis comes from direct clinical observations in affected patient populations. This is also the case with vaccines and antimicrobial/antiviral treatments, in which clinical trials to assess safety and efficacy data can provide significant insights into correlates of protection or disease pathogenesis in human populations.1, 2, 3 Furthermore, well-controlled challenge studies have provided important insights into the host response to pathogens such as seasonal influenza viruses and rhinoviruses.4 However, several factors can confound or limit our ability to study infectious diseases in humans. Humans can vary greatly in their susceptibility to certain pathogens: some individuals may be highly susceptible, whereas others develop only mild disease or remain asymptomatic. For example, only a small fraction of West Nile virus–infected individuals become symptomatic,5 and individuals vary greatly in their susceptibility to common pathogens such as human norovirus.6 This variation confounds analysis in humans and can be driven by factors such as human or pathogen genetic variation, underlying health issues, or demographic factors such as age or sex. Furthermore, previous pathogen exposure generally results in immune-mediated protection but, in some instances, can also drive immune pathology, as illustrated by dengue hemorrhagic fever/dengue shock syndrome.7 Although it is possible to control for many of these variables, other confounding factors (eg, pathogen dose, previous exposure to related pathogens) can be more difficult to control during natural infection. In addition to the confounding effects of disease variation in human populations, it can be difficult to perform mechanistic analysis or access affected tissues in humans due to ethical limitations. Although these problems can sometimes be overcome by using primary human cells,8, 9, 10 it is difficult to accurately model complex systemic interactions between the pathogen and host by using these simplified cell culture systems.
Due to the limitations associated with studying infectious diseases in humans, the research community has relied on animal models both for studying the pathogenesis of infection and as platforms for testing the efficacy and safety of experimental vaccines and therapies before moving into human studies. These models provide several advantages, including the ability to control many of the factors that can cofound analysis in humans, such as viral dose or underlying disease states, while also allowing for much more invasive studies than would be possible in humans. A wide range of species have been used to experimentally model host/pathogen interactions, including nonvertebrate models (eg, Caenorhabditis elegans), fish (eg, zebrafish), and domesticated livestock (eg, pigs, cattle), and these systems have provided important insights into pathogen biology, zoonotic transmission, and host–pathogen interactions.11, 12, 13, 14, 15 Each of these models offers specific advantages. For example, C elegans has a short and well-defined life cycle, is easily manipulated at the genetic level, the transparent worms are easily visualized, and they can be infected by a wide variety of clinically relevant bacterial and fungal pathogens.16 Zebrafish (Danio rerio) are also experimentally tractable, visually transparent, and mount innate immune responses that are similar to those seen in humans. These traits have made zebrafish models particularly useful for visualizing host inflammatory responses and studying the role of specific pathogen determinants and host factors in regulating infection by several clinically relevant bacterial and viral pathogens, including Mycobacterium leprae, Pseudomonas aeruginosa, Shigella flexneri, and chikungunya virus.11, 17 However, the bulk of laboratory studies have relied on a more limited number of laboratory species, including mice, rats, guinea pigs, ferrets, and non-human primates (NHPs), to study the host response to specific pathogens.
The choice of a specific model system for studying a given pathogen can be driven by a number of factors, including permissiveness to infection, physiologic conservation with humans, reproducibility, ease of manipulation, safety, and/or cost. However, it is also generally true that no model organism perfectly reproduces the response to infection seen in humans, and in many cases, different animal models can be used in complementary ways to address specific questions related to pathogen-induced disease. For example, NHP species, due to their physiologic similarity and evolutionary conservation with humans, are commonly used for studying viral pathogens such as Ebola virus, Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV), and influenza viruses.18, 19, 20 However, due to factors such as the expense associated with NHP studies, as well as their limited availability, long reproduction times, and difficulties in manipulating NHPs at the genetic level, other model systems are used to study these same pathogens. This is nicely illustrated by influenza virus, in which NHPs can be used to study viral pathogenesis.20 However, ferrets are highly permissive to a wide variety of human respiratory viruses, including influenza A virus, and the ferret respiratory tract is anatomically very similar to humans.21 Therefore, viral transmission and pathogenesis studies are often performed in ferret models, which in addition to reproducing many aspects of human influenza pathogenesis, are lower cost and more readily accessible than NHPs.20 Furthermore, although mice are generally not useful for studying influenza virus transmission, they have historically been the preferred model system for studying influenza-specific immunity due in part to the availability of well-defined immune reagents and gene-specific knockout mice.20, 22
Although different animal models are appropriate for the study of specific pathogens, or for studying specific aspects of pathogen biology or disease pathogenesis, the laboratory mouse is one of the most commonly used animal models for studying infectious diseases. The dominance of the mouse as a model system is driven by several factors, including their relatively low cost, ease of housing, and their rapid reproduction times and large litter sizes.23 Importantly, the availability of inbred mouse strains, in which every mouse in the strain is essentially genetically identical, as well as genetically modified mice lacking specific host genes,24, 25 has allowed the scientific research community to study how specific host genes affect the response to pathogen challenge while also promoting understanding of the host immune system and its role in protecting from pathogen challenge or mediating vaccine-induced immunity.26
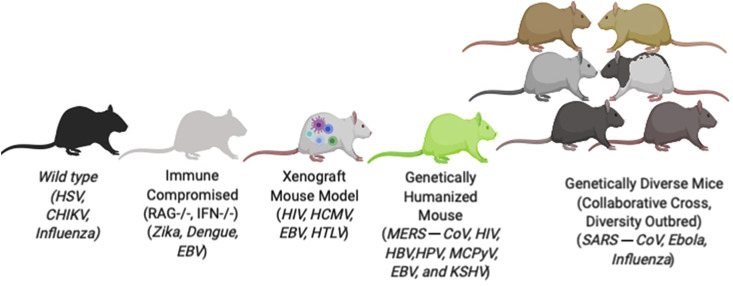
Mouse models do have limitations, including physiologic differences that limit how well mice reproduce key aspects of host–pathogen interactions, such as efficient influenza virus transmission.22 Genetic differences between mice and humans can also interfere with a pathogen's ability to replicate or cause human-like disease outcomes in mice.27, 28, 29 However, despite these limitations, mouse models provide a number of advantages for studying host–pathogen interactions. Furthermore, as discussed later, in some instances in which standard mouse models are not useful for working with a specific pathogen, the availability of genetically modified mice, humanized mouse models, or genetically diverse mouse populations can sometimes be used to overcome these difficulties and allow mice to be useful tools for studying host–pathogen interactions. Therefore, given the importance of the laboratory mouse as a model system for studying infectious diseases, the remainder of this review focuses on some of the major types of mouse models and their utility for studying infectious diseases (Figure 1 ).
Figure 1.
Examples of mouse model platforms used in infectious disease research.
Standard Inbred Mouse Strains
As noted earlier, one of the major advantages provided by inbred mouse strains is their reproducibility. Therefore, inbred strains such as C57BL/6 or BALB/c mice can be used to study pathogen tropism and replication kinetics, or compare the virulence of different pathogen strains in genetically identical sets of mice. This enables investigators to assess experimental reproducibility both within and between laboratories, while controlling for the effect of host genetics on the response to infection. For infectious disease studies, the availability of inbred strains allows investigators to perform replicate studies, compare results between laboratories, or investigate how different pathogens, alterations in pathogen dose, or different treatments affect genetically identical individuals.23 Another advantage provided by inbred mouse strains is the fact that a wide variety of immune reagents exist for quantifying host immune responses. These include pathogen-specific, major histocompatibility complex–restricted T-cell epitopes and tetramer reagents that enable quantification of pathogen-specific T cells after infection or vaccination in specific mouse genetic backgrounds. Therefore, inbred mouse strains have also been an invaluable resource for studying T-cell responses against viruses such as lymphocytic choriomeningitis virus or influenza A virus,30, 31 and these systems have been instrumental in advancing our understanding of the immune system.30 In addition to providing reproducible, well-characterized models for studying the biology of infection and pathogen-specific immunity, the availability of gene-specific knockouts on defined mouse genetic backgrounds (eg, C57BL/6) has been used by the research community for >25 years to study the role of specific host genes in the response to infection.32, 33, 34, 35 By comparing wild-type mice versus mice knocked out for a specific gene on the same genetic background, investigators can test the specific effect of that gene on pathogen-induced disease or immunity while controlling for effects of the remainder of the genome.
Immunodeficient Mouse Models
Although many medically relevant pathogens are capable of replicating and causing disease in standard laboratory mouse strains, many pathogens, including viruses such as ZIKV, human norovirus, and Crimean-Congo hemorrhagic fever virus, replicate poorly and/or fail to cause disease in standard mouse strains.36, 37, 38 For a subset of these pathogens, it is possible to use mice that lack key aspects of the host immune system, which can range from broadly immunodeficient mouse strains such as SCID or RAG-1 knockout mice that lack functional T and B cells,36 to mice that lack specific immune pathways such as the type I interferon system.38, 39 The utility of broadly immunodeficient models for studying infectious diseases is illustrated by human norovirus, a pathogen for which animal models were lacking until the demonstration that the virus can replicate in mice lacking a functional adaptive immune system.36 Furthermore, mice that specifically lack a functional type I interferon system have been used to develop models for a number of important human viruses,39 including ZIKV, Crimean-Congo hemorrhagic fever virus, dengue virus, and yellow fever virus.40, 41, 42, 43 For example, ZIKV, which caused a major epidemic in 2015–2016 associated with severe birth defects (reviewed elsewhere44), fails to replicate efficiently in wild-type mice due to the virus's inability to efficiently antagonize the STAT2 signaling protein in the mouse type I interferon system.27 To overcome this limitation, investigators have taken advantage of mice that lack a functional type I interferon system due to the ablation of the type I interferon receptor or essential interferon signaling molecules such as STAT1.40 Unlike wild-type mice, these interferon-deficient mouse strains are susceptible to ZIKV infection and have been useful as platforms for studying the pathogenesis of ZIKV-induced birth defects or for testing potential ZIKV therapies.45, 46
Genetically Humanized or Human Tissue Xenografted Mice
Although immunodeficient mice are useful systems for studying viruses such as ZIKV, in other instances it is necessary to introduce specific human genes or engraft human cells or organ systems into the mouse to generate infectious disease models. In cases in which mice are nonpermissive for infection by a pathogen, the introduction of one or a small number of pro-viral genes, such as mouse entry receptors, may be sufficient to allow pathogen replication and disease. These types of “genetically humanized” mice have been used for a number of viruses, including measles virus and the MERS-CoV.47, 48, 49 In the case of MERS-CoV, mouse cells are nonpermissive for viral infection,50 and this host range restriction can be overcome by expressing the human version of the viral receptor protein, dipeptidyl peptidase IV (DPP4).51, 52, 53, 54, 55 Two amino acid differences between mouse and human DPP4 are responsible for this difference in receptor function,29 and the expression of a full-length human DPP4 molecule in the mouse or introducing the 2 human amino acid changes into the mouse DPP4 molecule makes mice permissive for MERS-CoV replication.51, 52, 53, 54, 55
Although simply expressing a single human gene is sufficient to make mice permissive for MERS-CoV, for some of these models, it is still necessary to allow the virus to adapt to the mouse to achieve full virulence.52 In other cases, such as measles virus, it is necessary to introduce a human receptor molecule (CD150), while also ablating components of the host antiviral response to make mice fully permissive for infection.49 Alternatively, for viruses such as ZIKV, in which the block on viral replication in the mouse is due to an inability of ZIKV to efficiently antagonize the STAT2 component of the type I interferon signaling pathway,27 replacing the mouse STAT2 gene with the human STAT2 gene was used to generate a fully immunocompetent mouse model of ZIKV infection and disease.56
Although genetically humanized mouse strains can be powerful tools for modeling infection with some viral pathogens, for other viruses, such as HCV, several genes regulate host range, making it difficult to fully recapitulate the human factors that promote pathogen replication or disease in either standard or genetically humanized mice.57, 58 Furthermore, differences between the mouse and human immune system may limit the utility of standard or modified mouse strains as models for studying host immune responses or disease pathogenesis.59 To overcome these problems, the research community has used mice engrafted with human tissues to model human responses to infection. These human tissue xenografted mice take advantage of the fact that mice lacking a functional adaptive immune system (eg, SCID or RAG-1 knockout mice) are permissive for engraftment by human immune cells, while removal of other mouse genes, such as the common gamma chain of the interleukin-2 receptor, results in improved engraftment. These models can be used to study multiple aspects of the host innate and adaptive immune response, and can also be engrafted with solid organ tissues from humans.59, 60 Furthermore, because immune cells and other tissues from the same individual can be used to generate cohorts of humanized mice, it is possible to test responses in sets of individuals carrying tissues from the same genetic background.61 Although a detailed description of the different types of humanized mouse models, and the methods used in their development is beyond the scope of the present review, we refer the reader to several recent reviews that nicely summarize the current state of xenograft mouse development.59, 62
Mice with xenografted human immune systems have been used to study the pathogenesis of a wide range of infectious agents, including Plasmodium falciparum (malaria), Mycobacterium tuberculosis, dengue virus, and influenza virus.59, 61 These models have been particularly useful for studying HIV, including analysis of viral and host factors that promote viral replication, HIV interactions with the host immune response, and as platforms for testing therapeutic approaches for controlling or curing HIV infection.62 Although the majority of studies using xenografted mice have focused on human immune cells, these models can also be used to evaluate pathogen interactions with solid organs. Human liver xenograft mice have been shown to support replication of hepatitis viruses B, C, D, and E, and these models are useful for studying the pathogenesis of virus-induced liver injury, and as platforms for evaluating potential therapies to treat acute and chronic HCV and hepatitis B virus infection.59 Likewise, human intestinal xenograft models have been used to study Entamoeba histolytica infection, while lung xenografts have been shown to support Nipah virus infection.60, 63 Despite their utility, human xenograft models do have limitations, including issues with engraftment efficiency/tissue rejection and an inability to fully recapitulate human immune responses, such as antigen-specific antibody responses.59 However, despite these limitations, these model systems represent an important set of tools for modeling pathogen interactions with human cells and tissues in vivo, and as these models continue to improve, they are likely to gain even greater use within the infectious disease research community.
The Collaborative Cross Genetic Reference Population
As discussed earlier, one of the major reasons that inbred mouse strains and their genetically modified derivatives are such powerful systems for studying infectious diseases is their reproducibility. The ability to use sets of genetically identical animals from an individual inbred mouse strain (eg, C57BL/6J) allows for replicate experiments and comparison of responses across different treatments (eg, pathogen dose, strain, therapies). However, the reliance on one or even a few mouse strains means that investigators are evaluating pathogen infection in a limited number of genetic backgrounds, which is unlikely to recapitulate the phenotypic diversity seen within genetically diverse human populations upon pathogen exposure. Therefore, to model how pathogens interact with genetically diverse populations, a number of investigators have used genetically diverse mouse populations such as the Collaborative Cross (CC).
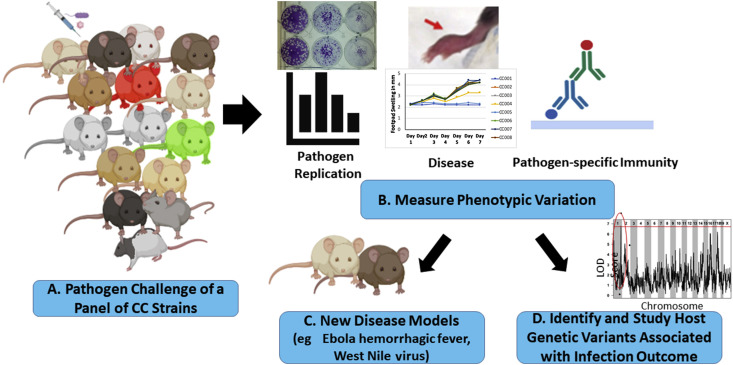
The CC is a panel of recombinant inbred mouse strains designed to model the genetic diversity present in human populations and promote the identification of polymorphic host genes that contribute to phenotypic variation across the population.64 The CC is derived from 8 parental mouse strains, including 5 classical laboratory strains (C57BL/6J, 129S1/SvImJ, A/J, NOD/ShiLtJ, and NZO/HiLtJ), as well as 3 wild derived strains from 3 Mus musculus subspecies (domesticus [WSB/EiJ], casteneus [CAST/EiJ], and musculus [PWK/PhJ]), which provide the majority of the genetic diversity found in the CC.65, 66 The 8 founder strains were intercrossed by using a funnel breeding system to produce progeny mice that were genetic mosaics of the original 8 founder strains. These mice were then bred to homozygosity to create recombinant inbred CC mouse strains.64 There are currently >60 inbred CC strains that are available to use by the scientific community (https://csbio.unc.edu/CCstatus/index.py). Because the CC strains are inbred, they allow investigators to perform the same types of comparative studies across reproducible sets of animals that are conducted with standard inbred strains (ie, C57BL/6). However, because each CC strain possesses a unique combination of genetic material from the original 8 founder strains, CC strains often exhibit unique phenotypes not observed in the original 8 founder strains,64 resulting in the development of new mouse models that better reproduce specific aspects of human disease (Figure 2 ). Furthermore, by comparing phenotypes between CC strains, investigators can test how host genetic variation affects specific phenotypes, including the response to pathogen challenge,64 and then map the specific genetic loci responsible for this variation.
Figure 2.
Strategies for using the Collaborative Cross (CC) in infectious disease studies. (A) An overview of potential strategies for using the CC mouse population for developing new animal models or studying how host genetic factors affect disease pathogenesis. (B) Sets of mice from a panel of CC strains (eg, 8–16 strains) can be infected with a pathogen of interest and the strains evaluated for variation in their susceptibility to pathogen-induced disease, their ability to support pathogen replication, or variation in the host response to infection. (C) If strains exhibit variation in the phenotype(s) of interest (eg, pathogen-induced disease), investigators can select a CC strain or strains with the desired phenotype to develop a new model system.75, 79 (D) Alternatively, investigators can take advantage of the phenotypic variation in the CC to perform genetic mapping studies to identify and study polymorphic host genes associated with variation in infection outcomes.71, 73, 74.
The CC was designed to map quantitative trait loci (QTL) associated with phenotypic variation across the CC strains, and several groups have taken advantage of this capability to map QTL associated with variation in the host response to infection. During the development of the CC, several groups used incompletely inbred populations of CC mice known as the pre-CC to assess variation in susceptibility to viral, bacterial, and fungal pathogens.64 The first study examining infectious disease susceptibility in the pre-CC was by Durrant et al,67 who analyzed a panel of pre-CC lines for variation in susceptibility to Aspergillus fumigatus. These studies found high levels of variation in survival time across pre-CC lines, and mapped several QTL associated with A fumigatus susceptibility. The pre-CC, as well as fully inbred CC strains, have also been used to study the impact of host genetic variation on bacterial infections. Vered et al68 evaluated pre-CC lines for variation in susceptibility to Klebsiella pneumoniae, and identified 3 QTL associated with variation in K pneumoniae survival. A second study, by Zhang et al,69 evaluated variation in susceptibility to Salmonella enterica challenge in a panel of fully inbred CC strains and identified several genetic loci associated with variation in bacterial load. Several studies have also used either the pre-CC or F1 crosses between fully inbred CC strains to map QTL associated with variation in virus-induced disease. Studies detailed by Maurizio et al,70 Ferris et al,71 and Bottomly et al72 reported high levels of variation in influenza virus susceptibility that was highly heritable. Ferris et al71 went on to identify several QTL associated with variation in influenza-induced disease susceptibility, including the identification of previously unidentified variants in the influenza resistance gene, Mx1. The pre-CC was also used by Gralinski et al73 to identify 4 unique genetic loci associated with variation in susceptibility to the SARS-coronavirus (SARS-CoV). Detailed analysis of one of these loci, a QTL on chromosome 3 associated with variation in vascular cuffing in the lungs of SARS-CoV–infected mice, identified the gene Trim55 as a high-priority candidate gene under the locus. Validation studies using Trim55 knockout mice showed that TRIM55 plays an important role in regulating virus-induced inflammation within the lungs of SARS-CoV–infected mice.73 Follow-up studies by Gralinski et al74 used F2 mapping approaches to identify additional genetic loci regulating SARS-CoV susceptibility in susceptible or resistant CC strains. This analysis identified 5 additional genetic loci associated with variation in SARS-CoV–induced disease, and identified and validated the gene for the TLR4 adaptor Ticam2, as a contributor to variation in SARS-CoV–induced disease susceptibility.
In addition to utilizing the CC for identifying polymorphic host genes that regulate pathogen susceptibility, investigators have taken advantage of the high levels of phenotypic variation in CC strains to develop improved models of pathogen-induced disease. This is perhaps best illustrated by the studies in Rasmussen et al,75 who used the CC to develop new mouse models of Ebola virus–induced hemorrhagic disease. Ebola virus causes a wide range of symptoms in humans, including hemorrhagic disease; however, although the use of mouse-adapted Ebola virus in standard mouse strains has been useful for studying disease pathogenesis and testing vaccines/therapies, these models do not fully reproduce the disease signs seen in infected humans.76 Rasmussen et al evaluated a panel of F1 crosses between fully inbred CC strains for susceptibility to challenge with mouse-adapted Ebola virus. These studies found high levels of variation in disease susceptibility, including the identification of CC strains that developed severe disease with hemorrhagic signs and liver damage.75 Studies by several other groups have identified CC strains that exhibit novel susceptibility profiles to a wide range of pathogens, including M tuberculosis, West Nile virus, SARS-CoV, and influenza virus.71, 73, 77, 78, 79 These results indicate that by taking advantage of the genetic diversity present in the CC, investigators can develop novel models of pathogen-induced disease that more closely represent disease phenotypes observed in human populations. Furthermore, once mouse strains with novel disease phenotypes have been identified, it is possible to identify the genetic loci responsible for these novel disease outcomes71, 73 and gain additional insights into the mechanisms underlying disease pathogenesis.
Conclusions
Animal models have been, and continue to be, an important resource for investigating the mechanisms underlying infectious disease pathogenesis and as platforms for testing potential vaccines and therapies. Mouse models have been a particularly important resource for studying infection by a wide range of human pathogens and are widely used for preclinical screening of vaccines/therapies due to their reproducibility, low cost, and ease of experimental manipulation. Although mouse models are not appropriate for studying some aspects of pathogen-induced disease, the use of genetically modified animals and humanized mice allows investigators to study pathogens that will not replicate efficiently in species other than humans or chimpanzees (eg, HCV) and perform mechanistic studies that cannot be conducted in humans. Furthermore, the availability of genetically diverse mouse resources such as the CC allows investigators to test how host genetic diversity affects the response to infection, while also taking advantage of the phenotypic diversity in these resources to develop model systems that better reproduce aspects of pathogen-induced diseases seen in humans.
Acknowledgements
This work was supported by the National Institute for Allergy and Infectious Diseases (NIAID) /National Institutes of Health (NIH), U.S.A. grants U19 AI100625 and U01 AI149644. The figures were developed in part using a license from BioRender.
Dr. Sankar wrote and edited the manuscript and prepared the figure; and Dr. Heise wrote and edited the manuscript.
Disclosures
The authors have indicated that they have no conflicts of interest regarding the content of this article. The funding agency sponsoring this work, the NIAID/NIH, had no involvement in the preparation of this manuscript or the decision to submit the manuscript for publication.
References
- 1.Edelman R., Tacket C.O., Wasserman S.S., Bodison S.A., Perry J.G., Mangiafico J.A. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am J Trop Med Hyg. 2000;62:681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- 2.Gao Y., McKay P.F., Mann J.F.S. 2018. Advances in HIV-1 Vaccine Development. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day T.A., Kublin J.G. Lessons learned from HIV vaccine clinical efficacy trials. Curr HIV Res. 2013;11:441–449. doi: 10.2174/1570162x113116660051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambkin-Williams R., Noulin N., Mann A., Catchpole A., Gilbert A.S. The human viral challenge model: accelerating the evaluation of respiratory antivirals, vaccines and novel diagnostics. Respir Res. 2018;19:123. doi: 10.1186/s12931-018-0784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mostashari F., Bunning M.L., Kitsutani P.T. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet. 2001;358:261–264. doi: 10.1016/S0140-6736(01)05480-0. [DOI] [PubMed] [Google Scholar]
- 6.Le Pendu J., Ruvoen-Clouet N., Kindberg E., Svensson L. Mendelian resistance to human norovirus infections. Semin Immunol. 2006;18:375–386. doi: 10.1016/j.smim.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slon Campos J.L., Mongkolsapaya J., Screaton G.R. The immune response against flaviviruses. Nat Immunol. 2018;19:1189–1198. doi: 10.1038/s41590-018-0210-3. [DOI] [PubMed] [Google Scholar]
- 8.Pyrc K., Sims A.C., Dijkman R. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol. 2010;84:11255–11263. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramani S., Crawford S.E., Blutt S.E., Estes M.K. Human organoid cultures: transformative new tools for human virus studies. Curr Opin Virol. 2018;29:79–86. doi: 10.1016/j.coviro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torraca V., Mostowy S. Zebrafish infection: from pathogenesis to cell biology. Trends Cel Biol. 2018;28:143–156. doi: 10.1016/j.tcb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avadhanula V., Weasner B.P., Hardy G.G., Kumar J.P., Hardy R.W. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. Plos Pathog. 2009;5 doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez Hernandez Y., Yero D., Pinos-Rodriguez J.M., Gibert I. Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front Microbiol. 2015;6:38. doi: 10.3389/fmicb.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buddle B.M., Vordermeier H.M., Hewinson R.G. Experimental infection models of tuberculosis in domestic livestock. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.TBTB2-0017-2016. [DOI] [PubMed] [Google Scholar]
- 15.Caine E.A., Jagger B.W., Diamond M.S. 2018. Animal Models of Zika Virus Infection during Pregnancy. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balla K.M., Troemel E.R. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol. 2013;15:1313–1322. doi: 10.1111/cmi.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palha N., Guivel-Benhassine F., Briolat V. Real-time whole-body visualization of chikungunya virus infection and host interferon response in zebrafish. Plos Pathog. 2013;9 doi: 10.1371/journal.ppat.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett R.S., Huzella L.M., Jahrling P.B., Bollinger L., Olinger G.G., Jr., Hensley L.E. Nonhuman primate models of ebola virus disease. Curr Top Microbiol Immunol. 2017;411:171–193. doi: 10.1007/82_2017_20. [DOI] [PubMed] [Google Scholar]
- 19.van Doremalen N., Munster V.J. Animal models of Middle East respiratory syndrome coronavirus infection. Antivir Res. 2015;122:28–38. doi: 10.1016/j.antiviral.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margine I., Krammer F. Animal models for influenza viruses: implications for universal vaccine development. Pathogens. 2014;3:845–874. doi: 10.3390/pathogens3040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enkirch T., von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479–480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouvier N.M., Lowen A.C. Animal models for influenza virus pathogenesis and transmission. Viruses. 2010;2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masopust D., Sivula C.P., Jameson S.C. Of mice, dirty mice, and men: using mice to understand human immunology. J Immunol. 2017;199:383–388. doi: 10.4049/jimmunol.1700453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doetschman T., Gregg R.G., Maeda N. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 25.Thomas K.R., Capecchi M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 26.Bouabe H., Okkenhaug K. Gene targeting in mice: a review. Methods Mol Biol. 2013;1064:315–336. doi: 10.1007/978-1-62703-601-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant A., Ponia S.S., Tripathi S. Zika virus targets human STAT2 to inhibit Type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck K.M., Cockrell A.S., Yount B.L., Scobey T., Baric R.S., Heise M.T. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J Virol. 2015;89:4696–4699. doi: 10.1128/JVI.03445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cockrell A.S., Peck K.M., Yount B.L. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88:5195–5199. doi: 10.1128/JVI.03764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Ramachandran S., Mann M., Popkin D.L. Role of lymphocytic choriomeningitis virus (LCMV) in understanding viral immunology: past, present and future. Viruses. 2012;4:2650–2669. doi: 10.3390/v4112650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hufford M.M., Kim T.S., Sun J., Braciale T.J. The effector T cell response to influenza infection. Curr Top Microbiol Immunol. 2015;386:423–455. doi: 10.1007/82_2014_397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann S.H., Ladel C.H. Application of knockout mice to the experimental analysis of infections with bacteria and protozoa. Trends Microbiol. 1994;2:235–242. doi: 10.1016/0966-842x(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 33.Bender B.S., Croghan T., Zhang L., Small P.A., Jr. Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J Exp Med. 1992;175:1143–1145. doi: 10.1084/jem.175.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinhoff U., Muller U., Schertler A., Hengartner H., Aguet M., Zinkernagel R.M. Antiviral protection by vesicular stomatitis virus-specific antibodies in alpha/beta interferon receptor-deficient mice. J Virol. 1995;69:2153–2158. doi: 10.1128/jvi.69.4.2153-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang S., Hendriks W., Althage A. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 36.Taube S., Kolawole A.O., Hohne M. A mouse model for human norovirus. MBio. 2013;4 doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrison A.R., Smith D.R., Golden J.W. Vol. 11. 2019. Animal Models for Crimean-Congo Hemorrhagic Fever Human disease. Viruses. pii: E590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison T.E., Diamond M.S. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol. 2017;91 doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong G., Qiu X.G. Type I interferon receptor knockout mice as models for infection of highly pathogenic viruses with outbreak potential. Zool Res. 2018;39:3–14. doi: 10.24272/j.issn.2095-8137.2017.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazear H.M., Govero J., Smith A.M. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier K.C., Gardner C.L., Khoretonenko M.V., Klimstra W.B., Ryman K.D. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. Plos Pathog. 2009;5 doi: 10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shresta S., Kyle J.L., Snider H.M., Basavapatna M., Beatty P.R., Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bereczky S., Lindegren G., Karlberg H., Akerstrom S., Klingstrom J., Mirazimi A. Crimean-Congo hemorrhagic fever virus infection is lethal for adult type I interferon receptor-knockout mice. J Gen Virol. 2010;91:1473–1477. doi: 10.1099/vir.0.019034-0. [DOI] [PubMed] [Google Scholar]
- 44.Lazear H.M., Diamond M.S. Zika virus: new clinical syndromes and its emergence in the Western Hemisphere. J Virol. 2016;90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long F., Doyle M., Fernandez E. Structural basis of a potent human monoclonal antibody against Zika virus targeting a quaternary epitope. Proc Natl Acad Sci U S A. 2019;116:1591–1596. doi: 10.1073/pnas.1815432116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caine E.A., Scheaffer S.M., Arora N. Interferon lambda protects the female reproductive tract against Zika virus infection. Nat Commun. 2019;10:280. doi: 10.1038/s41467-018-07993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baseler L., de Wit E., Feldmann H. A comparative review of animal models of Middle East Respiratory Syndrome coronavirus infection. Vet Pathol. 2016;53:521–531. doi: 10.1177/0300985815620845. [DOI] [PubMed] [Google Scholar]
- 48.Rall G.F., Manchester M., Daniels L.R., Callahan E.M., Belman A.R., Oldstone M.B. A transgenic mouse model for measles virus infection of the brain. Proc Natl Acad Sci U S A. 1997;94:4659–4663. doi: 10.1073/pnas.94.9.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno S., Ono N., Seki F. Measles virus infection of SLAM (CD150) knockin mice reproduces tropism and immunosuppression in human infection. J Virol. 2007;81:1650–1659. doi: 10.1128/JVI.02134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coleman C.M., Matthews K.L., Goicochea L., Frieman M.B. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol. 2014;95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li K., Wohlford-Lenane C.L., Channappanavar R. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cockrell A.S., Yount B.L., Scobey T. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao X., Garron T., Agrawal A.S. Characterization and demonstration of the value of a lethal mouse model of Middle East Respiratory Syndrome coronavirus infection and disease. J Virol. 2016;90:57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Li K., Wohlford-Lenane C. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascal K.E., Coleman C.M., Mujica A.O. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorman M.J., Caine E.A., Zaitsev K. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe. 2018;23:672–685.e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaska J.M., Ding Q., Ploss A. Mouse models for studying HCV vaccines and therapeutic antibodies. Methods Mol Biol. 2019;1911:481–503. doi: 10.1007/978-1-4939-8976-8_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaska J.M., Balev M., Ding Q., Heller B., Ploss A. Vol. 8. Elife; 2019. Differences across cyclophilin A orthologs contribute to the host range restriction of hepatitis C virus. pii: e44436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh N.C., Kenney L.L., Jangalwe S. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017;12:187–215. doi: 10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valbuena G., Halliday H., Borisevich V., Goez Y., Rockx B. A human lung xenograft mouse model of Nipah virus infection. Plos Pathog. 2014;10 doi: 10.1371/journal.ppat.1004063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnakumar V., Durairajan S.S.K., Alagarasu K., Li M., Dash A.P. Vol. 11. 2019. Recent Updates on Mouse Models for Human Immunodeficiency, Influenza, and Dengue Viral Infections. Viruses. pii: E252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsden M.D., Zack J.A. Humanized mouse models for human immunodeficiency virus infection. Annu Rev Virol. 2017;4:393–412. doi: 10.1146/annurev-virology-101416-041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seydel K.B., Li E., Swanson P.E., Stanley S.L., Jr. Human intestinal epithelial cells produce proinflammatory cytokines in response to infection in a SCID mouse-human intestinal xenograft model of amebiasis. Infect Immun. 1997;65:1631–1639. doi: 10.1128/iai.65.5.1631-1639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noll K.E., Ferris M.T., Heise M.T. The Collaborative Cross: a systems genetics resource for studying host-pathogen interactions. Cell Host Microbe. 2019;25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D.W. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collaborative Cross C. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durrant C., Tayem H., Yalcin B. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21:1239–1248. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vered K., Durrant C., Mott R., Iraqi F.A. Susceptibility to Klebsiella pneumonaie infection in collaborative cross mice is a complex trait controlled by at least three loci acting at different time points. BMC Genomics. 2014;15:865. doi: 10.1186/1471-2164-15-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J., Malo D., Mott R., Panthier J.J., Montagutelli X., Jaubert J. Identification of new loci involved in the host susceptibility to Salmonella Typhimurium in collaborative cross mice. BMC Genomics. 2018;19:303. doi: 10.1186/s12864-018-4667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maurizio P.L., Ferris M.T., Keele G.R. Bayesian Diallel analysis reveals Mx1-dependent and Mx1-independent effects on response to influenza A virus in mice. G3 (Bethesda) 2018;8:427–445. doi: 10.1534/g3.117.300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferris M.T., Aylor D.L., Bottomly D. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. Plos Pathog. 2013;9 doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bottomly D., Ferris M.T., Aicher L.D. Expression quantitative trait Loci for extreme host response to influenza a in pre-collaborative cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gralinski L.E., Ferris M.T., Aylor D.L. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. Plos Genet. 2015;11 doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gralinski L.E., Menachery V.D., Morgan A.P. Allelic variation in the Toll-Like receptor adaptor protein Ticam2 contributes to SARS-Coronavirus pathogenesis in mice. G3 (Bethesda) 2017;7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rasmussen A.L., Okumura A., Ferris M.T. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siragam V., Wong G., Qiu X.G. Animal models for filovirus infections. Zool Res. 2018;39:15–24. doi: 10.24272/j.issn.2095-8137.2017.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith C.M., Proulx M.K., Olive A.J. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. MBio. 2016;7 doi: 10.1128/mBio.01516-16. pii: e01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graham J.B., Swarts J.L., Wilkins C. A mouse model of chronic West Nile virus disease. Plos Pathog. 2016;12 doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graham J.B., Thomas S., Swarts J. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6 doi: 10.1128/mBio.00493-15. e00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]