Abstract
Severe acute respiratory syndrome (SARS) is a newly emerging infectious disease caused by a novel coronavirus (SARS-CoV), which has overwhelmed more than 30 countries claiming nearly 8400 cases with over 800 fatalities. Thanks to the unprecedented international collaboration, the whole-genomes of SARS-CoVs were successfully deciphered shortly after the identification of the causative pathogen for outbreak of SARS in southern China, in 2003. Hitherto, the SARS-CoV, as a viral paradigm of emerging infectious entities, has been extensively studied that has ranged from epidemiology, molecular virology/immunology to structural genomics. Also, several lines of breakthroughs have been record-brokenly obtained, that included the finding of ACE2, a functional receptor for the SARS-CoV, solution of the 3CLpro structure, a first crystal structure of SARS-related macromolecules, revealing of bats as natural reservoirs for SARS-like viruses and the possible involvement of civet cats in the SARS emergence. This review intends to outline the major progress in the journey of SARS-related exploration, by emphasizing those inaugurated studies with milestone-like significance contributed by Chinese research groups.
Keywords: Severe acute respiratory syndrome (SARS), Coronavirus (CoV), SARS-CoV, Molecular epidemiology, Natural reservoir, Receptor, T cell immunity, CTL epitopes, Structural genomics
Résumé
Le syndrome respiratoire aigu sévère (SRAS ou SARS), une maladie infectieuse émergeante causée par un nouveau coronavirus (SARS-CoV), toucha plus de 8400 personnes causant 800 morts dans 30 pays différents. Grâce à la mise en place d’une collaboration internationale sans précédent le décryptage du génome complet du SARS-CoV fut réalisé très peu de temps après l’identification de l’agent pathogène suite au déclenchement de l’épidémie dans le sud de la Chine en 2003. Le SARS-CoV en temps que paradigme de maladies infectieuses émergeantes a fait l’objet d’une étude complète en passant de l’épidémiologie, la virologie/immunologie moléculaire à la génomique structurale. Ainsi, c’est en un temps record qu’ont été obtenues des données majeures incluant la découverte de ACE2, un récepteur fonctionnel pour le SARS-CoV, la structure du 3CLpro première structure cristallographique d’une macromolécule du SARS, révélant que la chauve-souris était le réservoir naturel de virus du type SARS, et que la civette était certainement à l’origine de l’émergence du SRAS. Cette revue vise à souligner les progrès majeurs réalisés dans l’exploration du SRAS, en soulignant l’importance des travaux primordiaux et précoces réalisés par les chercheurs chinois.
Mots-cléfs: Syndrome Respiratoire Aigu Sévère (SRAS), Coronavirus (CoV), SARS-CoV, Épidémiologie moléculaire, Réservoir naturel, Récepteur, Immunité des cellules T, Épitopes CTL, Génomiques structurales
1. Introduction
Severe acute respiratory syndrome (SARS), also initially called “atypical pneumonia” in China, is recognized as an emerging infectious disease that ever posed a great threat to public health and even resulted in a global panic to some extent [1], [2], [3], [4], [5], [6], [7], [8], [9]. This unusual disease was first recognized in Guangdong Province, China, in November 2002 [3], [4], [7], [9], [10], [11], and characterized by a series of clinical manifestations including high fever, shortness of breath, cough, and pneumonia [1], [7]. Subsequently, it spread to other Asian countries, North America, and Europe [2], [3], [7], [9], [10], [11], [12]. As of July 3, 2003, this epidemic had affected at least 30 countries across 5 continents, infecting over 8400 persons with 812 announced fatalities [2], [3], [7], [9], [10], [11], [12], [13], [14]. Of being mentioned, after the first epidemic of SARS ended in July 2003, as announced by World Health Organization (WHO), sporadic new cases were reported, but was really rare [7], [15]. Unlike the cases of laboratory infections reported from Singapore, Taiwan, and Beijing, the four confirmed SARS patients of the 2003–2004 episode in Guangzhou, China, were all community-infected cases without obvious human-to-human contact history related to SARS [16]. Collectively, SARS did devastate profoundly the social and economic activities regionally as well as globally, however, it was also quickly defeated by the great efforts of the international community [1], [5], [11], [12], [16].
Thanks to the unprecedented global collaboration coordinated by WHO, SARS-CoV, a novel member of Coronaviridae family, was rapidly confirmed to be the aetiological agent for the SARS epidemic [1], [11], [17]. More excitedly, its complete genome was sequenced by two different groups within weeks of isolation of the virus [17], [18]. Subsequently, molecular epidemiology of the SARS-CoVs based on genomic approaches and bioinformatics indicated that the co-evolution episodes are ongoing during the period of their circulation, with several typical molecular characteristics [5], [8], [10], [11], [12], [16], [19]. Meanwhile, classic epidemiological surveys implied that the SARS-CoV may be of animal origin, and its precursor is still present in animal population [3], [7], [10]. Furthermore, the live-animal markets in southern China were suggested to provide the animal–human interface allowing this precursor virus to adapt to human–human interspecies transmission [3], [7], [10].
Shortly after the definition of the SARS-CoV entity, multiple lines of investigations at the molecular level were carried out with the aim of developing effective approaches for diagnostics, control and prevention of SARS as well as presenting a comprehensive knowledge on the interplay between SARS-CoV and its host [5], [10], [12], [13], [14], [15], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]. Of being noteworthy, several milestone findings have been harvested, which included angiotensin-converting enzyme 2 (ACE2), a functional receptor utilized for SARS-CoV entry into host cells [37], [38], [39], [40], the crystal structure of 3CLpro, a key enzyme involved in SARS-CoV life cycle, as an attractive drug target for SARS therapeutics [41], molecular co-evolution of SARS-CoV [10], [12] and the bat, a natural host reservoir for SARS-CoV [42], [43]. Up to now (at the time of writing this review, October 2006), the number of literatures concerning SARS has expanded to 4651 items in PubMed (www.ncbi.nlm.nih.gov), indicating strongly that serious attentions have ever been paid to this emerging infectious disease. Luckily, the disease “mysteriously disappeared” with our great efforts.
So far, there have been several excellent reviews documenting the issue of SARS [7], [15], [22], [34], [38], therefore readers are kindly reminded for these. Here, we attempt to provide an outline of SARS research from epidemiology to molecular virology, and to vaccine development. Major progress/breakthroughs contributed by Chinese scientists are especially being emphasized in this review.
2. General aspects of SARS
2.1. Clinical manifestations of SARS
The sharply accumulated knowledge on SARS has profoundly facilitated to perform clinical diagnosis of the suspected cases during the period of atypical pneumonia [7], [11]. To better define SARS cases, the following criteria were proposed by WHO: (1) having close contact with a patient or having infected other people, (2) fever (>38 °C) and symptoms of respiratory illness, (3) leukocyte count <10.0×109/L, (4) radiographic evidence of infiltrates consistent with pneumonia or respiratory distress syndrome on chest X-ray, and (5) no response to antimicrobial drug treatments (within 72 h) [1], [2], [7], [11], [14], [24]. In general, those people were diagnosed to be suspected SARS patients if they met criteria 1–4 or 2–5, but excluded if an alternative diagnosis could fully explain their illness.
The typical incubation period of SARS ranges from 2 to 10 days but may occasionally be as long as 16 days [1], [7], [14]. The prodrome includes influenza-like symptoms, such as fever, myalgia, headache, and diarrhea [1], [7]. After onset of the disease, most cases progress to a moderate-severe variant characterized by a more serious later respiratory phase with dyspnea on exertion or at rest and hypoxia [1], [7], [22]. The case-fatality rate during these outbreaks was nearly 10% (range, 0–40%) [1], [22].
In general, pulmonary lesions in SARS patients can be frequently diagnosed as diffuse alveolar damage. The following histological changes are included, which are composed of desquamation of pneumocytes, formation of hyaline membranes, flooding of alveolar lumina with oedema fluid mixed with inflammatory cells, and the presence of enlarged pneumocytes and syncytia. Also, alveolar walls are thickened by mild mononuclear infiltrate, and in later stages air spaces contain fibromyxoid-organizing exudates [1], [7], [22], [24]. At the aid of transmission electron microscopy, virus-like particles can be observed in the cells from a lung biopsy specimen and bronchoalveolar lavage sample, and in pneumocytes from a postmortem lung samples [11], [44].
2.2. Identification of a new coronavirus (SARS-CoV) as the causative agent of SARS
Judging based on Koch's postulates, the causal pathogen responsible for the atypical pneumonia was quickly determined as a novel member of group II coronaviruses, called SARS-CoV (severe acute respiratory syndrome-related coronavirus), in the family Coronaviridae (Fig. 1C ) [2], [4], [7], [17]. It is an enveloped, positive-sense (+), single-stranded (ss) RNA virus featuring a large genome of approximate 29.7 kb (Fig. 1A and B) [7], [45]. Subsequently, its genomic annotation revealed that it may consist of 14 functional open reading frames (ORFs) encoding three classes of proteins: two large polyproteins (pp1a and pp1ab), which are cleaved into 16 non-structural proteins (nsps) essential for viral RNA synthesis; four structural proteins (surface (S), envelope (E), matrix (M), and nucleocapsid (N) proteins), required for viral assembly; and eight accessory proteins, which may confer a selective advantage in the infected host (Table 1 and Fig. 1) [38], [46].
Fig. 1.
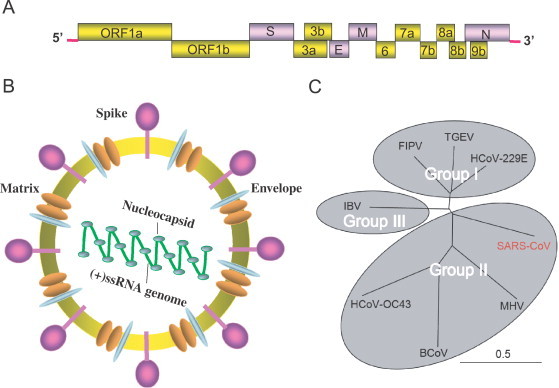
Characterization of the SARS-CoV. (A) Genomic structure of the SARS-CoV. (B) Schematic representation of the SARS-CoV virion. (C) Phylogenetic tree showing the relationships among the members of Family Coronaviridae based on the amino acid sequences of the spike proteins. MHV, HCoV-OC43, and BCoV belong to Group I of coronavirus, FIPV, TGEV, HCoV-229E, and SARS-CoV are classified into Group II of coronavirus. The SARS-CoV was highlighted in red. IBV forms an independent unique group, Group III. MHV, mouse hepatitis virus; HCoV-OC43, human coronavirus isolate OC43; BCoV, bovine coronavirus; FIPV, feline infectious peritonitis virus; TGEV, transmissible gastroenteritis virus; HCoV-229E, human coronavirus isolate 229E; IBV, infectious bronchitis virus.
Table 1.
Summary of SARS-CoV-related proteins with known structures
| Proteins | Protein size (aa) | Location in the genome (bp) | Description of the related structure | References |
|---|---|---|---|---|
| Structural proteins (2/4) | ||||
| S | 1255 | ORF2 (21492–25259) | Fusion core, RBD-ACE2 complex, RBD-Ab complex | [38], [39], [51], [76] |
| N | 422 | ORF9a (28120–29388) | N-terminal domain | [75] |
| Nsps (7/16) | ||||
| Nsp3 | 1922 | ORF1a (2719–8484) | PL2pro | [82] |
| Nsp5 | 308 | ORF1a (9985–10902) | +, 3CLpro | [41] |
| Nsp7 | 83 | ORF1a (11773–12021) | + | [46] |
| Nsp8 | 198 | ORF1a (12022–12615) | + | [46] |
| Nsp9 | 113 | ORF1a (12616–12954) | +, ssRNA binding subunit | [61], [83] |
| Nsp10 | 139 | ORF1a (12955–13371) | + | [79], [81] |
| Nsp15 | 346 | ORF1b (19551–20588) | +, endoribonuclease | [80] |
| Accessory proteins (2/8) | ||||
| ORF7a | 122 | ORF7a (27273–27261) | +, Ig-like | [85] |
| ORF9b | 98 | ORF9b (28130–28426) | + | [86] |
Similar to that of most other coronaviruses, S protein of the SARS-CoV can be divided into amino (S1) and carboxyl (S2) regions [38], [47], [48], [49]. The S1 subunit mediates the binding of ACE2 receptor, and the S2 subunit is responsible for the membrane fusion between the virus and its host cell [38], [40], [49], [50], [51], [52], [53], [54], [55]. As such, S protein of SARS-CoV is also suggested to account for inducing host immune responses, production of neutralizing antibodies, high virulence, etc. [38], [47], [49], [56], [57].
It is no doubt that the SARS-CoV has emerged as an infectious entity and evolved to possess the capability to overcome the interspecies barriers, causing serious panic worldwide in the field of public health [3], [19], [42]. However, it remains unclear what disrupted the natural ecological balance system, making the virus jump off their natural reservoir and adapt to human hosts [3], [42]. The molecular mechanism underlying this mystery is in great need from the scientific community.
3. Epidemiology of SARS-CoV
It seems there is a common notion that SARS originated from Guangdong Province of southern China [9], [17], and the worldwide spread was triggered by a single individual (so-called “super-spreader” at the time) infected from Guangdong who spent some time in Hong Kong before succumbing to SARS [3], [11], [45]. During that period, he unwittingly infected others that in turn caused a series of SARS outbreaks in Toronto, Canada and other areas around the world. Eventually, in nearly 9 months, it traveled in no less than 30 countries and areas of the world, and was passed over several generations as reflected by the genetic variation of SARS-CoVs. Here, we attempted to discuss the tracing to its origin of SARS-CoVs as well as the general rules of their molecular evolution during the epidemics.
3.1. Animal origin of SARS-CoV
Retrospectively, several index SARS cases, the earliest of which was recorded from Foshan City in Guangdong Province, China, on November 16, 2002, were found to share a common feature with a typical history of contacting animals in the food industry. This critical clue prompted the initiation of field epidemiological investigation mainly on live-animal markets in Guangdong Province. Unexpectedly, from those live-animal markets examined, 13–40% of wild animal traders and slaughterers were revealed to be seropositive for the SARS-CoV, which led to a speculation that SARS may be zoonotic from an unknown animal reservoir [3], [7]. Subsequent suspicion was focused on palm civets, just due to the fact that above 70% of SARS seropositives were among the traders who were primarily in trading of masked palm civets [7]. Fortunately, SARS-like viruses were successfully isolated from Himalayan palm civets (Paguma larvata) and a raccoon dog (Nyctereutes procyonoides) at an animal market in Shenzhen City, China [3]. Moreover, further analysis of viral genomes showed they are of above 99% homology to the SARS-CoV [3]. These findings established strongly the basis for animal origin of SARS-CoV, and also resulted in a large-scale culling of civets to prevent re-emergence of SARS in the area [6], [10].
However, subsequent survey failed to observe the evidence that supports the widespread infection of SARS-CoV in wild and/or farmed civets [3], [9]. In addition, experimental infection of civets with human isolates of the SARS-CoV reproduced overt clinical symptoms similar to that of SARS patients, ruling out the possibility of civets playing a role of natural reservoir host for SARS-CoVs, rather a susceptible host [10]. Nevertheless, civets may have ever been the carrier source of SARS-like virus precipitating the SARS outbreak, and the existence of the viruses in this and other common animals within wet-market systems in southern China, seemed to be more likely a reflection of an “artificial” market cycle in native species than an indication of the natural reservoir of this virus. Thereby, endless search for the origin of SARS-CoVs was ongoing again, which paid much attention to other candidate animals [6]. Excitedly, the mysterious mask was recently unveiled by Li et al. that bats function as the natural reservoirs for SARS-like coronaviruses (Fig. 2 ) [42]. In fact, bats have ever been recognized as natural hosts for several zoonotic viruses such as Hendra and Nipah viruses, which recently emerged in Australia and East Asia, respectively [42], [43]. Also, bats rarely display clinical symptoms even though being in persistent infection with multiple viruses. Moreover, increasing utilization of bats or its products in food and traditional medicine markets especially in southern China and elsewhere in Asia makes it possible that SARS-like viruses jump into humans from bats, subsequently adapt to humans, and finally trigger human-to-human transmission resulting in global outbreak of SARS [42], [43].
Fig. 2.
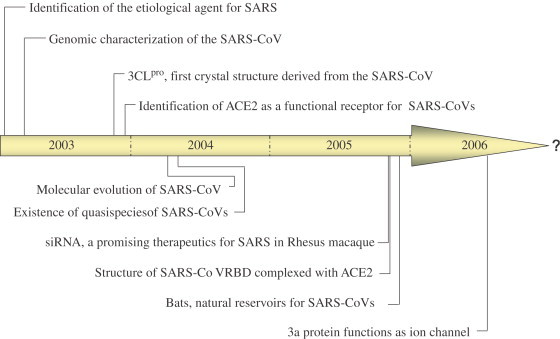
Landmark events in the SARS-related research field.
3.2. Molecular evolution of SARS-CoV isolates
Multiple lines of investigations focusing on molecular epidemiology of SARS-CoVs have presented a general rule of the SARS-CoV during the different courses of the SARS epidemics.
Based on a wide range of sequence analysis of different SARS-CoV isolates, Guan's group postulated that SARS-CoVs may be totally divided into two distinct clusters [11]. One genotype was linked with infections originating from Hotel M in Hong Kong, the other one comprises isolates from Hong Kong, Guangdong and Beijing that had no association with Hotel M [3], [11]. To date, there is no further information as to whether different SARS-CoV strains may have different degrees of virulence.
In addition, Zhao's group carried out intensive comparative genomic studies by sequencing the whole genomes of 61 isolates of SARS-CoVs derived from the early, middle, and late phases of SRAS epidemics as well as two viral genomes from palm civets [12]. The exciting finding showed that (1) the earliest genotype is similar to the animal SARS-like coronaviruses; (2) major deletions in orf8 can be observed at both the start and the end of the epidemics; (3) the neutral mutation rate of the genome is constant but the amino acid substitution rate of the CDS slows during the epidemic period; (4) S protein presents the robust initial response to positive selection pressure, followed by selection purification and eventual stabilization [12]. In general, this comprehensive analysis did provide crucial clues to the mysterious evolutionary tale of SARS-CoVs, and greatly strengthened the arguments for animal origin of the human SARS epidemics [10], [12].
Another interesting result of SARS-CoV variation came from Wang's group [58]. A concept of quasispecies was introduced into SARS-CoVs, which is originally well-known in hepatitis C virus (HCV), and human immunodeficiency virus (HIV) [58], [59]. The experimental evidence suggested that SARS-associated virus may consist of complex and dynamic distributions of mutants in vivo, rather than a single, defined genome sequence [19], [58], [59]. Actually, it is also reasonable that genetic variants of SARS-CoV can form a pool of heterogeneous virus in individual patients owing to poor fidelity of its RNA polymerase [58], [59].
Recently, Zhao et al. further delineated molecular insights into cross-host evolution of SARS-CoV in palm civet and human [16]. First, genomic sequences of SARS-CoVs from human and palm civet of the 2003/2004 outbreak in Guangzhou City, China, were nearly identical, suggesting an independent viral invasion from animal to human in this new episode. Second, viruses in palm civets were suggested to undergo a rapid evolving process, much like their adaptation in the human host in the early 2002–2003 epidemic. Third, major genetic variations particularly in Spike gene, might contribute greatly to the transition from animal-to-human transfer to human-to-human transmission, responsible for the first SARS outbreak of 2002/2003 [16].
4. Molecular and immunopathological aspects of SARS
In the context of molecular interplay between SARS-CoVs and their hosts, multiple lines of issues would be paid much attention to, including immunological response, pathological response, etc. Here, we would focus on partners of N protein, epitope identification, and apoptosis.
4.1. Host partners interacting with N protein
N protein is a major structural component of virions, which appears to be involved in multiple functions such as viral RNA replication and translation. So far, several host factors have been demonstrated to bind to the N protein [20], [21], [60], [61].
First, human cyclophilin A (hCypA) has been demonstrated by surface plasmon resonance (SPR) technology to be an interacting partner. Furthermore, its probable binding model was proposed and validated by mutagenesis experiments [21]. It may provide a new hint for facilitating the understanding of another possible SARS-CoV infection pathway against human cell [21].
Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) has also been discovered to exhibit high binding affinity to the N protein, which is originally related to the pre-mRNA splicing in the nucleus and translation regulation in the cytoplasm. Yeast two-hybrid assays combined with GST pull-down experiment further finely mapped each binding domains in N protein and hnRNP A1 [20].
Very recently, our group reported that hUbc9, a ubiquitin conjugating enzyme of sumoylation system, can interact specifically with the N protein, implying the post-translational sumoylation of the N protein [36]. The availability of host factors interacting with the N protein will facilitate to better understand the molecular pathogenesis of SARS-CoV, indicating several new approaches for preventions and disruption of SARS-CoV infections.
4.2. Identification of CTL epitopes
T cells are essential for adaptive immunity against viral infections in vivo, and CD8+ cytotoxic T lymphocytes (CTLs) can kill virus-infected host cells through recognizing viral epitopes presented by MHC Class I (HLA) molecules. Thus, identification of natural CTL epitopes from SARS-CoVs would facilitate to better understand the immunological response to the SARS-CoV infection in humans, especially towards virus clearance [62], [63].
To date, those CTL epitopes identified, mainly originated from the S or N protein of SARS-CoVs. An HLA A2-restricted CTL epitope (amino acid residues: 1167–1175) has been identified in the S protein of SARS-CoV [63]. The synthetic epitope polypeptide (RLNEVAKL) also can stimulate the production of peptide-specific CTLs in vivo (transgenic mice) and in vitro (human PBLs).
Very recently, our group reported a novel HLA-A*0201-restricted decameric epitope, P15 (S411–S420, KLPDDFMGCV) that localizes within the ACE2 receptor-binding domain (RBD) of the S protein [62]. P15 can significantly enhance the expression of HLA-A*0201 molecules on the T2 cell surface, stimulate IFN-gamma-producing CTLs from the PBMCs of the recovered SARS patients, and induce specific CTLs from P15-immunized HLA-A2.1 transgenic mice in vivo. Moreover, significant P15-specific CTLs were induced from HLA-A2.1-transgenic mice immunized by a DNA vaccine encoding the S protein; suggesting that P15 is a naturally processed epitope.
4.3. SARS-CoV induced apoptosis
Apoptosis is one of the key issues to better understand its molecular pathogenesis of SARS. SARS-CoV can induce apoptosis in the cultured cells and in the infected patient [44], [57], [64], [65], [66], [67], [68]. A series of experimental evidence have indicated that several viral components from SARS-CoV are involved in apoptosis, one of consequences of the interplay between SARS-CoVs and its host cells [57], [64], [65], [66], [67], [68], [69], [70], [71]. First, three structural proteins (S, N and M) have been suggested to contribute to inducing of apoptosis in different cell lines [57], [65], [68], [70]. Second, of 16 nsps of SARS-CoV, only 3CLpro (nsp3) is pointed out to function as an inducer of apoptosis in Vero-E6 cells [64]. Third, three accessory proteins have been identified to have roles in apoptosis [66], [67], [69], [71]. Very recently, the 3a protein, a transmembrane protein, was unveiled to function as an ion channel, which might be similar to M2 protein of influenza virus, indicating its possible role in the release and package of mature virus particles [72], [73], [74]. Also, it can trigger apoptosis in Vero E6 cells, which is supported by in vitro experiments of chromatin condensation and DNA fragmentation [66]. The 3b protein (ORF4), with poor documentations till now, was recently suggested to have a link to G0/G1 arrest, apoptosis, and even necrosis in the following transfected cells such as Vero E6, 293T and COS-7 cells [69], [71]. Overexpression of 7a induces apoptosis via a caspase-dependent pathway in cell lines derived from different organs, including lung, kidney, and liver [67].
5. Structural insights into SARS-CoV
The increasingly accumulative structural information concerning SARS-CoV has enabled greatly the elucidation of their crucial roles in its life cycle, e.g. spike protein-mediated membrane fusion, proteolytic processing of two replicase polyproteins (pp1a and pp1ab), and replication/transcription. So far, the crystal structures of nearly 11 SARS-derived proteins have been obtained (Table 1) [38].
5.1. Structural proteins
SARS-CoV harbors four structural proteins that include S protein, M protein, N protein and E protein. However, only partial structures of the S protein and N protein have been solved so far [39], [51], [75], [76].
S protein, a member of class I virus fusion proteins, can be characterized by the presence of two heptad repeat (HR) regions, HR1 and HR2. It attaches the virus to its cellular receptor, ACE2, subsequently triggers the membrane fusion between virus and its host cell [50], [51], [52], [53], [55]. Presently, the crystal structure of fusion core has been determined, representing a typical 6-helix bundle with three HR2 helices packed against the hydrophobic grooves on the surface of central coiled-coil formed by three parallel HR1 helices in an oblique anti-parallel manner [51]. The structural availability of fusion core makes it possible to design small molecule or polypeptide drugs for SARS therapeutics that aims to inhibiting viral entry by blocking membrane fusion, which has ever been in successful application for HIV therapeutics such as T20 of Enfuvertide. More recently, two major progresses in S protein have been achieved. (1) Li et al. [39] solved the complex structure of RBD of S protein bound with the peptidase domain of human ACE2, indicating potential development direction of SARS vaccines or inhibiting small molecules. (2) Crystal structure of S protein RBD complexed with a neutralizing monoclonal antibody, m396, was well defined, providing a structural rationale for the major determinant of immunogenicity and neutralization of SARS-CoV [76].
N protein plays an essential role in SARS-CoV genome packaging and virion assembly [60], [75], [77]. As previous data indicated that it forms a dimer in solution through its C-terminal domain, the crystal structure of N protein dimerization domain, consisting of residues 270–370, shows a dimer with extensive interactions between the two subunits, suggesting that the dimeric form of the N protein is the functional unit in vivo [75], [77]. Interestingly, although lacking significant sequence similarity, it presents a folding mode similar to that of N protein of the porcine reproductive and respiratory syndrome virus (PRRSV), i.e. it may be pivotal structural evidence of the evolutionary link between Coronaviridae and Arteriviridae, implying that the N proteins of both viruses have a common origin [75].
5.2. Non-structural proteins
In total, the replicase gene of SARS-CoV encodes 16 nsps with multiple enzymatic activities. They are composed of an RNA-dependent RNA polymerase (RdRp, nsp12), a chymotrypsin-like (3C-like) serine protease (3CLpro or Mpro, nsp5), a papain-like protease 2 (PL2pro, nsp3), etc. [41], [78]. Here, we discussed 7 nsps with available structures, of which nsp5 is the most extensively characterized one [41], [46], [61], [79], [80], [81], [82], [83].
Nsp5, also known as 3CLpro, mediates extensive proteolysis of replicase polyproteins to produce multiple functional subunits, which are involved in the generation of replication complex [60], [84]. Its functional importance in the viral life cycle and the lack of closely related cellular homologues have made it an attractive target for the development of drugs against SARS infections [60]. In 2003, shortly after the peak of the SARS outbreak, Rao's group reported, for the first time, the crystal structures of SARS-CoV 3CLpro and its complex with an inhibitor, which provided important structural basis for rational drug design, and also posed extensive concern worldwide [41].
Similar to 3CLpro, nsp3 (PL2pro), the other viral cysteine proteases of SARS-CoV, serves as a potential anti-SARS drug target. Most recently, the crystal structure of its catalytic core was reported [78]. Although its overall architecture adopts a fold similar to that of known deubiquitinating enzymes, it can also be distinguished from them by its unique features, including an intact zinc-binding motif, an unobstructed catalytically competent active site, and the presence of an intriguing, ubiquitin-like N-terminal domain [82].
Nsp9, a unique single-stranded RNA-binding protein, has been implicated into viral RNA synthesis of SARS-CoV [61], [83]. The solved structure pinpointed that it is a dimer, and comprises a single beta-barrel with a previously unknown fold. Further analysis suggested that the fold superficially resembles an OB-fold with a C-terminal extension and is related to both of the two sub-domains of the SARS-CoV 3CLpro, i.e. it is of possibility that nsp9 has, presumably, evolved from a protease [61].
Surprisingly, the crystal structure of the hexadecameric nsp7/nsp8 supercomplex from SARS-CoV has been successfully solved [46]. First, nsp8 possesses a novel “golf-club” fold with two conformations. Second, the complex is a unique hollow, cylinder-like structure assembled from 8 copies of nsp8 and held tightly together by 8 copies of nsp7. Third, the central channel shares the dimensions and positive electrostatic properties which in turn favor the nucleic-acid binding, implying its role in the replication and transcription machinery of SARS-CoV.
Nsp10 is newly considered as a transcription factor in SARS-CoV. Its crystal structures from two independent research groups revealed a novel fold and is the first structural representative of zinc finger proteins found so far exclusively in coronaviruses [79], [81]. This novel fold means that 12 identical subunits assemble into a unique spherical dodecameric architecture. Moreover, Gel shift assays indicated that nsp10 binds single- and double-stranded RNA and DNA with high-micromolar affinity and without obvious sequence specificity. Thus, the structural and nucleic-acid binding information together with the high conservation of the zinc finger motifs among coronaviruses, make it reasonable that the functional role of a transcription factor was assigned to nsp10 [79].
NendoU (nsp15) is a Mn (2+)-dependent, uridylate-specific enzyme, which leaves 2′-3′-cyclic phosphates 5′ to the cleaved bond [80]. Very recently, nsp15 of SARS-CoV was unveiled to exhibit a unique fold and assemble into a toric hexamer with six potentially active, peripheric catalytic sites [80]. The structure combined with the spatial arrangement of the catalytic residues into an RNase A-like active site define a separate endonuclease family, endoU, representing an example of convergent evolution toward an enzymatic function that is critically involved in the coronavirus replication cycle [80].
5.3. Accessary proteins
Eight novel ORFs (ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b) at its 3′ end are only present in SARS-CoVs, not in other members of coronavirus, suggesting their unique functions that might be advantageous to the SARS-CoV replication, assembly or virulence [17], [18], [38]. To date, only two proteins among these so-called accessory proteins with poor function assignation have been elucidated with their structure information that included ORF-7a and ORF-9b [85], [86].
ORF-7a encodes a unique type I transmembrane protein of unknown function [67]. The crystal structure of the N-terminal ectodomain of orf7a has been determined, revealing a compact seven-stranded beta sandwich, unexpectedly similar in fold and topology to members of the Ig superfamily [85].
ORF-9b, an alternative ORF within N gene from SARS-CoV, has been suggested to encode a lipid binding protein [86]. The protein has been demonstrated to share a novel fold, a dimeric tent-like beta structure with an amphipathic surface, and a central hydrophobic cavity that binds lipid molecules. Also, ORF-9b associates with intracellular vesicles, consistent with a role in the virus assembly [86].
6. Vaccines and therapeutics for SARS
6.1. Vaccines
The severe morbidity and mortality associated with SARS makes it imperative to develop an effective and safe vaccine to prevent its re-emergence and epidemics [15], [34], [87]. The inactivated vaccine may be the first one available for clinical application because it is easy to generate, however, its safety is of the major concern [13]. In fact, the inactivated SARS-CoV vaccine has been demonstrated to elicit potent spike protein-specific neutralizing antibodies that block receptor binding and virus entry [13]. At present, the RBD of S protein is identified to contain multiple conformational neutralizing epitopes, implying its potential of serving as an ideal vaccine candidate [54], [88], [89]. A DNA vaccine encoding S glycoprotein of the SARS-CoV can induce T cell and neutralizing antibody responses, as well as protective immunity, in a mouse model [23], [24], [54], [62], [87], [90], [91]. More excitedly, by using recombinant adeno-associated virus (rAAV) delivery system, RBD-rAAV has been shown to be a promising SARS vaccine [88]. Similarly, transgenic plant (e.g. tomato) producing S1 antigen was successfully used in a mouse model [87].
6.2. Therapeutics
To date, no effective and specific therapeutic measures can be used to treat SARS patients [22], [31], [92]. RNA interference (RNAi), a process by which the introduced small interfering RNA (siRNA) could cause the degradation of the specific mRNA, has been suggested to work well for inhibition of SARS-CoV in cultured cells and in animals [25], [26], [27], [28], [29], [30], [31], [32], [33], [93].
Wang et al. [32] reported that plasmid-based siRNAs, which specifically target the viral RNA polymerase, can block effectively the cytopathic effects of SARS-CoV on Vero cells. Effective and specific expression inhibition of S and N protein were observed in SARS-CoV-infected cells [27], [30], [33].
Zhong's group revealed that siRNA duplexes targeting SARS-CoV genomic RNA are potent agents for inhibition of the viral infection and replication in the non-human primate cells [25], [31]. More recently, they further evaluated siRNA inhibitors of SARS for efficacy and safety in a rhesus macaque (Macaca mulatta) SARS model, marking siRNA-based SARS-CoV inhibitors as one of useful therapeutic agents [25].
Utilizing human monoclonal antibodies (mAbs) in anti-SARS therapeutics is also a promising direction [23]. Experimental animal data show that protection against SARS-CoV infection with human mAbs is feasible, e.g. the combination of two non-competing human mAbs CR3014 and CR3022 potentially controls immune escape and extends the breadth of protection in infected ferrets [23].
Meanwhile, several substantial progresses have also been achieved in designing polypeptides or screening small molecules drugs against SARS [35], [53], [94]. Deng's and Gao's group have identified two polypeptide derived from HR regions of SARS-CoVs, HR1-1 and HR2-18, presenting robust inhibitory activity for SARS-CoV entry [52]. Recently, Gao and Tien's group also developed two recombinant protein inhibitors (HR121 and HR212), exhibiting high stability and potent inhibitory activity on entry of the HIV/SARS pseudoviruses [53]. Interestingly, two kinds of Chinese herbal medicine-derived small molecules (tetra-O-galloyl-beta-d-glucose (TGG) and luteolin) were identified, whose anti-SARS-CoV activities were confirmed using a wild-type SARS-CoV infection system [35]. It is noteworthy to mention that, Rao's group have made a major progress in designing the wide-spectrum inhibitors targeting Coronavirus with SARS-CoV included [94].
7. Concluding remarks
SARS is a quickly conquered new infectious disease, attracting much attention worldwide. Thus it has been extensively documented within 4 years since its first discovery in China [1]. Moreover, the knowledge on SARS up to now has been enriched unprecedentedly, among which several events are in the significance of landmarks (Fig. 2). However, there are still lots of unclear SARS-related issues that desiderated to be solved as always a case for any virus we encounter.
As previously indicated, reverse genetics have contributed to functional characterization of those accessory proteins such as 3a [57], [66], [67]. Further investigations may focus on the interaction between accessory proteins and other viral proteins as well as host factors. It will lead to a better understanding of intricate interplay between the SARS-CoV and its human host. Also, it may contribute to the development of antiviral therapeutics and design of efficient strategies for disease control [25], [29].
Structural biology combined with X-ray crystallography has been demonstrated to be fruitful in addressing the bio-macromolecules of SARS-CoV [38]. Typically, the first structure of SARS-CoV worldwide, 3CLpro can be available shortly after SARS outbreak in 2003 [41], as well as fusion core [51]. Future prospects for SARS structural biology include the structures of other nsps and protein–protein complexes, favoring to understand the sophisticated function and assembly of the replication/transcription machinery [46].
Thus far, no effective vaccines or antiviral drugs is available for SARS therapeutics [31], [92]. Fortunately, there are several candidate remedies exemplified with siRNA that has shown promising effect in an animal model (rhesus macaque) [25]. However, the safety and effect in clinical treatments of SARS patients still need further evaluation.
The molecular and social trigger of the SARS-CoV appearance and disappearance in the community remains a dogma which needs attract our attention for the prevention of both the SARS and any other emerging viruses worldwide.
Acknowledgments
We would like to thank Dr. Di Liu for his kind help in preparing the phylogenetic tree. This work was supported by National Basic Research Program (Project 973) of China (2005CB523001) and Beijing Natural Science Foundation (5062028). GFG is a distinguished young investigator of National Natural Science Foundation of China (NSFC) (Grant No. 30525010).
References
- 1.Berger A., Drosten C., Doerr H.W., Sturmer M., Preiser W. Severe acute respiratory syndrome (SARS)—paradigm of an emerging viral infection. J Clin Virol. 2004;29:13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Liu W., Tang F., Fontanet A., Zhan L., Wang T.B., Zhang P.H. Molecular epidemiology of SARS-associated coronavirus, Beijing. Emerg Infect Dis. 2005;11:1420–1424. doi: 10.3201/eid1109.040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu H., Zhao Y., Zhang J., Wang Y., Li W., Zhu X. Date of origin of the SARS coronavirus strains. BMC Infect Dis. 2004;4:3. doi: 10.1186/1471-2334-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poon L.L., Guan Y., Nicholls J.M., Yuen K.Y., Peiris J.S. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsui S.K., Chim S.S., Lo Y.M. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N Engl J Med. 2003;349:187–188. doi: 10.1056/NEJM200307103490216. [DOI] [PubMed] [Google Scholar]
- 9.Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y., Peiris J.S., Zheng B., Poon L.L., Chan K.H., Zeng F.Y. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 13.He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun. 2004;325:445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie Y., Wang G., Shi X., Zhang H., Qiu Y., He Z. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190:1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang S., He Y., Liu S. SARS vaccine development. Emerg Infect Dis. 2005;11:1016–1020. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 18.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Lim S.L., Ruan Y., Ling A.E., Ng L.F., Drosten C. SARS transmission pattern in Singapore reassessed by viral sequence variation analysis. PLoS Med. 2005;2:e43. doi: 10.1371/journal.pmed.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H., Chen Q., Chen J., Chen K., Shen X., Jiang H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem Biophys Res Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J Gen Virol. 2004;85:3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- 25.Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni B., Shi X., Li Y., Gao W., Wang X., Wu Y. Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA. Antivir Ther. 2005;10:527–533. [PubMed] [Google Scholar]
- 27.Zhao P., Qin Z.L., Ke J.S., Lu Y., Liu M., Pan W. Small interfering RNA inhibits SARS-CoV nucleocapsid gene expression in cultured cells and mouse muscles. FEBS Lett. 2005;579:2404–2410. doi: 10.1016/j.febslet.2005.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y., Yang D.H., Xiong J., Jia J., Huang B., Jin Y.X. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 2005;15:193–200. doi: 10.1038/sj.cr.7290286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C.J., Huang H.W., Liu C.Y., Hong C.F., Chan Y.L. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin Z.L., Zhao P., Zhang X.L., Yu J.G., Cao M.M., Zhao L.J. Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293 T cells. Biochem Biophys Res Commun. 2004;324:1186–1193. doi: 10.1016/j.bbrc.2004.09.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng B.J., Guan Y., Tang Q., Du C., Xie F.Y., He M.L. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir Ther. 2004;9:365–374. [PubMed] [Google Scholar]
- 32.Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X. Silencing SARS-CoV spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhi Y., Wilson J.M., Shen H. SARS vaccine: progress and challenge. Cell Mol Immunol. 2005;2:101–105. [PubMed] [Google Scholar]
- 35.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Z., Zhuo Y., Tan X., Zhou Z., Yuan J., Qiang B. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J Med Virol. 2006;78:1365–1373. doi: 10.1002/jmv.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W. Expression cloning of functional receptor used by SARS coronavirus. Biochem Biophys Res Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr Opin Struct Biol. 2005;15:664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 40.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 43.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K. SARS coronavirus induces apoptosis in Vero E6 cells. J Med Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chim S.S., Tsui S.K., Chan K.C., Au T.C., Hung E.C., Tong Y.K. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362:1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J Biol Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y., Zhou Y., Wu H., Luo B., Chen J., Li W. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem Biophys Res Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni L., Zhu J., Zhang J., Yan M., Gao G.F., Tien P. Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. Biochem Biophys Res Commun. 2005;330:39–45. doi: 10.1016/j.bbrc.2005.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nie Y., Wang P., Shi X., Wang G., Chen J., Zheng A. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem Biophys Res Commun. 2004;321:994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai C.W., Chan Z.R., Yang D.G., Lo W.H., Lai Y.K., Chang M.D. Accelerated induction of apoptosis in insect cells by baculovirus-expressed SARS-CoV membrane protein. FEBS Lett. 2006;580:3829–3834. doi: 10.1016/j.febslet.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu D., Zhang Z., Wang F.S. SARS-associated coronavirus quasispecies in individual patients. N Engl J Med. 2004;350:1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- 59.Tang J.W., Cheung J.L., Chu I.M., Sung J.J., Peiris M., Chan P.K. The large 386-nt deletion in SARS-associated coronavirus: evidence for quasispecies? J Infect Dis. 2006;194:808–813. doi: 10.1086/507044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo H., Ye F., Chen K., Shen X., Jiang H. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 61.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 63.Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chow K.Y., Yeung Y.S., Hon C.C., Zeng F., Law K.M., Leung F.C. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 2005;579:6699–6704. doi: 10.1016/j.febslet.2005.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 67.Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan S., Fielding B.C., Tan T.H., Chou C.F., Shen S., Lim S.G. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006 doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao G., Shi S.Q., Yang Y., Peng J.P. M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol Toxicol. 2006;22:313–322. doi: 10.1007/s10565-006-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 74.Wang C., Takeuchi K., Pinto L.H., Lamb R.A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J Biol Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo H., Ye F., Sun T., Yue L., Peng S., Chen J. In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys Chem. 2004;112:15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Brooun A., Griffith M. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure (Camb) 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan J., Verschueren K.H., Anand K., Shen J., Yang M., Xu Y. pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J Mol Biol. 2005;354:25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pogrebnyak N., Golovkin M., Andrianov V., Spitsin S., Smirnov Y., Egolf R. Severe acute respiratory syndrome (SARS) S protein production in plants: development of recombinant vaccine. Proc Natl Acad Sci USA. 2005;102:9062–9067. doi: 10.1073/pnas.0503760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Du L., He Y., Wang Y., Zhang H., Ma S., Wong C.K. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: implication for developing SARS vaccines. Virology. 2006;353:6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Y., Li J., Du L., Yan X., Hu G., Zhou Y. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X., Ni B., Du X., Zhao G., Gao W., Shi X. Protection of mammalian cells from severe acute respiratory syndrome coronavirus infection by equine neutralizing antibody. Antivir Ther. 2005;10:681–690. [PubMed] [Google Scholar]
- 91.Tarnovitski N., Matthews L.J., Sui J., Gershoni J.M., Marasco W.A. Mapping a neutralizing epitope on the SARS coronavirus spike protein: computational prediction based on affinity-selected peptides. J Mol Biol. 2006;359:190–201. doi: 10.1016/j.jmb.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qin C., Wang J., Wei Q., She M., Marasco W.A., Jiang H. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J Pathol. 2005;206:251–259. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li T., Zhang Y., Fu L., Yu C., Li X., Li Y. siRNA targeting the leader sequence of SARS-CoV inhibits virus replication. Gene Ther. 2005;12:751–761. doi: 10.1038/sj.gt.3302479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]


