Abstract
Background
Current estimates suggest that even in the most resourced settings, the aetiology of encephalitis is identified in less than half of clinical cases. It is acknowledged that filling this gap needs a combination of rigorous sampling and improved diagnostic technologies. Next generation sequencing (NGS) methods are powerful tools with the potential for comprehensive and unbiased detection of pathogens in clinical samples. We reviewed the use of this new technology for the diagnosis of suspected infectious encephalitis, and discuss the feasibility for introduction of NGS methods as a frontline diagnostic test.
Methods
A systematic literature review was performed, using MESH and text word searches for variants of “sequencing” and “encephalitis” in Medline and EMbase, and searching bibliographies and citations using the Web of Science database. Two authors independently reviewed, extracted and summarised data.
Findings
The review identified 25 articles reporting 44 case reports of patients with suspected encephalitis for whom NGS was used as a diagnostic tool. We present the data and highlight themes arising from these cases. There are no randomly controlled trials to assess the utility of NGS as a diagnostic tool.
Interpretation
There is increasing evidence of a role for NGS in the work-up of undiagnosed encephalitis. Lower costs and increasing accessibility of these technologies will facilitate larger studies of these patients. We recommend NGS should be considered as a front-line diagnostic test in chronic and recurring presentations and, given current sample-to-result turn-around times, as second-line in acute cases of encephalitis.
Keywords: Encephalitis, Deep sequencing, Metagenomics, Diagnosis, Infection
Introduction
Deficits in the current management of encephalitis
Encephalitis is defined as inflammation of brain parenchyma associated with neurological dysfunction.1, 2 It is strictly a pathological diagnosis. Recent epidemiological studies suggest that the global burden of encephalitis has been grossly underestimated, with current incidence suggested to be over 6000 annual cases in the UK, and 500,000 worldwide.3, 4
The syndrome encapsulates a myriad of diverse diseases, with distinct global distributions, presenting features and clinical courses.4, 5 Infections represent the most frequently identified aetiology, with data suggesting this accounts for 20–50% of cases.2, 6, 7 Hundreds of pathogens have been associated with encephalitis, with the most frequently identified including Herpes simplex virus (HSV), Varicella zoster virus (VZV), enteroviruses, Measles morbillivirus, Mumps virus, Japanese encephalitis virus (JEV), influenza viruses, adenoviruses and Mycoplasma pneumoniae.4 HSV, JEV and rabies are the chief causes in Europe, Asia and Africa respectively. The main alternative aetiology to infection is immune mediated, for which management includes immune suppression. It is critical to differentiate between autoimmune and infectious causes of encephalitis; immune suppression in cases where the cause is an undiscovered pathogen could be devastating.
Strikingly, more than one third of cases of encephalitis remain unidentified, even in the best-equipped medical centres.3, 6, 8 There are well-recognised challenges and inadequacies in current diagnostics and treatment, and these correspond with the poor reported outcomes.9, 10, 11, 12 Overall mortality is estimated at 30%, but is highly variable and dependent on the aetiology and access to supportive care. A high proportion is left with complex disability.11
The introduction of management guidelines for cases of acute encephalitis syndrome over the last decade has aimed to improve outcomes.8, 13, 14, 15, 16, 17, 18, 19 Notably, there are many differences between guidelines in the approach to diagnostic evaluation, such as standard and extended diagnostic testing, or whether to administer empirical Aciclovir.20 These variations may reflect geographically contrasting aetiologies, timing of publication and the rapid acceleration of technologies, as well as available resources. Further, there is a lack of a systematic approach to access to pathogen discovery methods, discussed below. It is only the newest French guidelines in which NGS is mentioned at all, and the authors reasonably state that the clinical role is still to be evaluated.19
Diagnostics for encephalitis and the role of modern technologies
Current diagnostic techniques for suspected infectious cases rely on prior knowledge of the likely causative agent. Informed by clinical presentation, epidemiological data, guidelines and local resources, a laboratory will perform targeted tests for a disease. These are largely confined to specific polymerase chain reaction (PCR) or serological assays. This approach has fundamental limitations, and contributes to the relatively high proportion of encephalitis cases that remain undiagnosed. Aside from the difficulties of testing for the myriad of rare pathogens that might be expected to cause encephalitis, this approach does not permit the identification of new or unexpected pathogens. Undeniably, methods for novel pathogen discovery such as electron microscopy and cell culture have existed for many years, however they are cumbersome, time-consuming, lack sensitivity and specificity and are often no longer routinely available. Furthermore, there are groups of patients, such as immunosuppressed patients, who frequently present with subtle or non-specific symptoms and signs, are high-risk for encephalitis, infection with unexpected or unusual pathogens, and are seen to have more severe outcomes.21
There is a need for improved diagnostic methods for encephalitis. A method which has recently been applied to pathogen detection in cases of encephalitis is metagenomic analysis using next generation sequencing (NGS). Proof of concept for its use in the diagnosis of encephalitis has been demonstrated in the literature, however its suitability for routine diagnosis has not been assessed and is the subject of this review.
An introduction to next generation sequencing
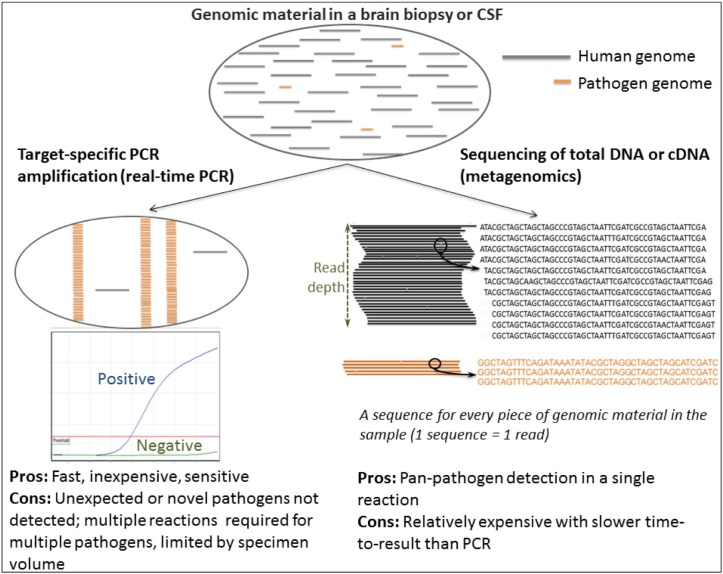
NGS, also known as deep sequencing, generates a single sequence from each fragment of DNA, or cDNA, present in a specimen. Downstream analysis allows differentiation between the origin of sequence fragments, for instance human, a specific bacterial species or a particular virus. This means mixed specimens, that contain host and microbial sequences, can be resolved (Fig. 1 ).
Fig. 1.
Schematic of pathogen-specific real-time PCR versus metagenomics for pathogen detection.
The potential application of next generation sequencing to encephalitis diagnostics
Sequencing the total DNA or RNA (known as metagenomics) from a biopsy or body fluid allows the identification of genetic material from any microorganism present in the specimen, and thus potentially causing encephalitis. This approach overcomes the limitations of targeted diagnostic methods such as PCR as it requires no prior knowledge or assumptions about the type of pathogen causing infection therefore enabling detection of novel and unexpected pathogens.
The majority of readily available sequencing methods to date are DNA based, however sequencing only total DNA would exclude detection of viruses with RNA genomes. Consequently, an alternative approach is to synthesise complementary DNA (cDNA) from total RNA, which will enable detection of viruses with RNA genomes but also the RNA transcripts of organisms with DNA genomes.
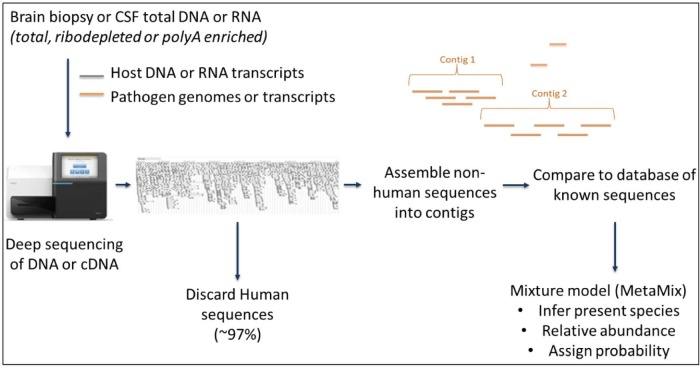
Once sequences are generated, complex downstream bioinformatic analysis is required to identify the presence of any pathogen sequences. In brief, any reads mapping to the human genome are removed, after which all remaining non-human sequences are compared to a database of known sequences to identify the provenance of the unknown sequences (Fig. 2 ).
Fig. 2.
Schematic of a typical metagenomics workflow. MetaMix22 is the analysis tool employed by our laboratory.
The possibility of incorporating unbiased pathogen discovery technology into routine diagnostics for encephalitis would represent a paradigm shift in diagnostic algorithms. We and others23 are validating the use of metagenomics for clinical use and prospective studies are already underway to examine whether application of NGS at the outset of management pathways improves patient outcome and costs, namely the Precision Diagnosis of Acute Infectious Disease (PDAID) study.24, 25 Nonetheless, there is a paucity of evidence in this field which is largely limited to case reports. We aim to perform a rigorous summary and critical review of existing evidence, to assess the utility of NGS in diagnosis of encephalitis.
Methods
Search strategy and selection criteria
Data for this review were identified by searches of Medline and EMbase using the keyword and/or MESH search terms [?sequenc* OR Metagenom OR Illumina OR RNA-seq OR RNASeq OR (Roche 454) OR (Ion torrent) OR (Proton / PGM) OR MiSeq OR HiSeq OR NextSeq OR minion OR nanopore OR pacbio] AND [Encephalitis OR Meningoencephalitis OR Brain or (Central Nervous System) OR CNS or Neur* or (Cerebrospinal) or CSF]. Only articles reporting application of NGS in CSF or brain biopsies in suspected encephalitis and published in English between January 2008 and April 2017 were included. The Web of Science database was also used to search bibliographies and citations of relevant article.
Data analysis
Two authors independently reviewed, extracted and summarised included literature. Data was extracted in the first instance by the first reviewer into a custom data extraction excel sheet with pre-defined data headings (Supplementary File 1), designed for the purpose of this review; additional miscellaneous data or observations were also noted where relevant. The extracted data and each included study were independently reviewed by the second author, with a focus on technical and scientific aspects of each study. Consensus extracted data was used in analyses; both review authors were in full agreement on the extracted consensus data. Relevant manuscripts that were not identified through the initial search, but were identified in the reference list of included literature, were also included.
Role of funding source
The funding bodies had no role in the decision to write, the analysis, manuscript preparation or the decision to submit for publication.
Results
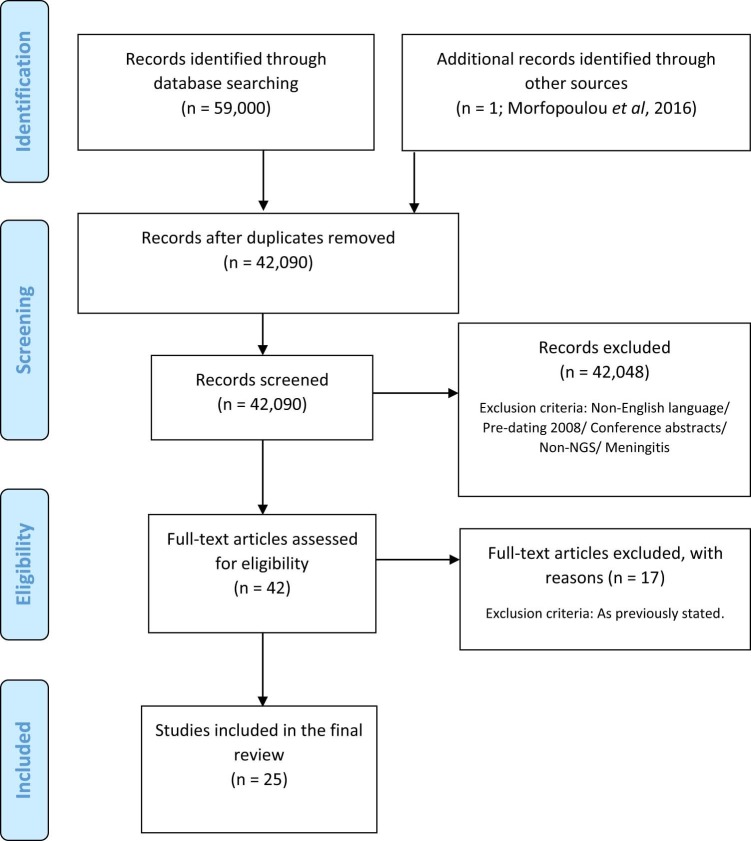
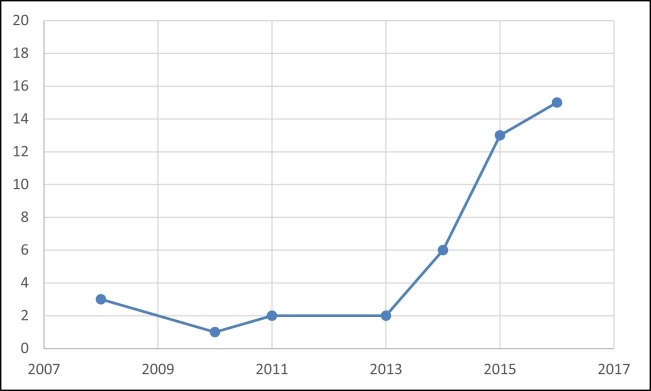
Twenty-five articles were identified from the search (Fig. 3 ). All the included articles were case reports, or case series of 1–7 patients. Altogether 44 cases were reported in which NGS provided a diagnosis in otherwise undiagnosed cases of encephalitis (Table 1 ). An exponential temporal increase in cases has been observed over the last decade (Fig. 4 ). Country of origin of cases included Australia, China, France, Germany, India, Ireland, Japan, Poland, Sri Lanka, UK, USA, and Vietnam. Samples were analysed in laboratories largely in the USA and Europe (UK, France, Germany, Poland), but also in China, Japan and Vietnam. Among the 27 cases for which age was documented, the median age was 14 years (Interquartile range 3–61). Of the 22 cases that reported immune status of the patient, 73% (16/22) were immunocompromised. There was uniformly poor reporting of encephalitis or meningoencephalitis case definitions, and limited explanation of diagnostic assays performed and algorithms used for testing. None of the studies reported adherence to published or unpublished clinical guidelines.
Fig. 3.
Study selection for inclusion in systematic review.
Table 1.
Reports of infectious encephalitis diagnosed by metagenomic next generation sequencing (mNGS) meeting inclusion criteria.
| Study | Case No. | Age (yrs) | Immunosuppressed? | Specimen Type | Pathogen Identified | Confirmatory testing of metagenomics result | Final Diagnosis | Type of pathogen | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Palacios et al. (2008)26 | 1 | 63 | Yes (post solid organ transplantation) | Pooled RNA from brain, cerebrospinal fluid, serum, kidney, and liver | Arenavirus | Viral culture, EM, immunohistochemistry and serology. Donor IgM and IgG positive | Arenavirus Encephalitis | Novel organism | None | Died |
| 2 | 64 | Yes (post solid organ transplantation) | Pooled RNA from brain, cerebrospinal fluid, serum, kidney, and liver | Arenavirus | Viral culture, EM, immunohistochemistry and serology. Seroconversion | Arenavirus Encephalitis | Novel organism | None | Died | |
| 3 | 44 | Yes (post solid organ transplantation) | Pooled RNA from brain, cerebrospinal fluid, serum, kidney, and liver | Arenavirus | Viral culture, EM, immunohistochemistry and serology. | Arenavirus Encephalitis | Novel organism | None | Died | |
| Quan et al. (2010)27 | 4 | 15 | Yes (primary immunodeficiency disorder caused by mutations in the Btk gene, which results in absence of B lymphocytes and serum immunoglobulins) | Brain biopsy | Astrovirus VA1/HMO-C | PCR and antigen detection (IHC) | Astrovirus encephalitis | Novel organism | None | Died |
| Benjamin et al. (2011)28 | 5 | 2 | NR | CSF | Parvovirus 4 | Serum Parvovirus IgM positive, serum and CSF Parvovirus PCR positive. | Parvovirus 4 Encephalitis | Novel cause of encephalitis (known organism) | None | Discharged against medical advice on day 18 after admission |
| 6 | 3 | NR | CSF | Parvovirus 4 | CSF Parvovirus PCR positive | Parvovirus 4 Encephalitis | Novel cause of encephalitis (known organism) | None | Recovered | |
| Tan et al. (2013)29 | 7 | 31 | NR | DNAse treated CSF supernatant | Cyclovirus | PCR | None (unclear association) | Novel organism | None | Recovered |
| 8 | 1 | NR | DNAse treated CSF supernatant | Cyclovirus | PCR | None (unclear association) | Novel organism | None | Recovered | |
| Chan et al. (2014)30 | 9 | NR | NR | Brain Biopsy | Measles virus | PCR and neuropathology | Measles SSPE | Known cause of encephalitis | None | Died |
| 10 | NR | NR | Brain Biopsy | Measles virus | PCR and neuropathology | Measles SSPE | Known cause of encephalitis | None | Died | |
| 11 | NR | NR | Brain Biopsy | HSV1 | PCR and neuropathology | HSV1 Encephalitis | Known cause of encephalitis | None | Died | |
| 12 | NR | NR | Brain Biopsy | HSV1 | PCR and neuropathology | HSV1 Encephalitis | Known cause of encephalitis | None | Died | |
| 14 | NR | NR | Brain Biopsy | HSV1 | PCR and neuropathology | HSV1 Encephalitis | Known cause of encephalitis | None | Died | |
| Wilson et al. (2014)24 | 16 | 14 | Yes ((SCID); partial immune reconstitution after two BMT). | CSF and serum; DNAse treated and untreated | Leptospira santarosai | PCR | Leptospira Meningoencephalitis | Rare | High-dose intravenous penicillin G (13 million units daily). | Recovered |
| Brown et al. (2015)31 | 17 | 1 | Yes (cartilage hair hypoplasia and associated immunodeficiency due to RMRP mutations, peripheral blood stem cell transplant) | Brain Biopsy | Astrovirus VA1/HMO-C | Pan-astrovirus hemi-nested PCR, real-time PCR, IHC and EM in brain biopsy. Real-time PCR in CSF, stool, serum | Astrovirus Encephalitis | Novel organism (emerging cause of encephalitis) | None | Died |
| Frémond et al. (2015)32 | 18 | 14 | Yes (primary immunodeficiency disorder caused by mutations in the Btk gene, which results in absence of B lymphocytes and serum immunoglobulins) | Brain Biopsy | Astrovirus VA1/HMO-C | PCR positive brain (CSF negative) | Astrovirus Encephalitis | Novel organism (emerging cause of encephalitis) | IVIG, ribavirin and PEG-IFN | Some clinical improvement following tailored therapy |
| Greninger et al. (2015)33 | 19 | 15 | No (Type 1 diabetes mellitus and celiac disease) | Brain Biopsy | Balamuthia mandrillaris | PCR and histopathology | Balamuthia mandrillaris Encephalitis | Rare | None (miltefosine was requested but the patient died awaiting treatment) | Died |
| Hoffmann et al. (2015)34 | 20 | 63 | No | Brain Biopsy | Variegated Squirrel Bornavirus | Immunohistochemistry, PCR and neuropathology and serology | Bornavirus Encephalitis | Novel organism | None | Died |
| 21 | 62 | No | Brain Biopsy | Variegated Squirrel Bornavirus | Immunohistochemistry, PCR and neuropathology | Bornavirus Encephalitis | Novel organism | None | Died | |
| 22 | 72 | No | Brain Biopsy | Variegated Squirrel Bornavirus | Immunohistochemistry, PCR and neuropathology | Bornavirus Encephalitis | Novel organism | None | Died | |
| Naccache et al. (2015)35 | 23 | 42 | Yes (CLL with BMT) | Brain Biopsy and CSF (DNAse treated and untreated) | Astrovirus VA1/HMO-C | PCR and immunohistochemistry. | Astrovirus Encephalitis | Novel organism (emerging cause of encephalitis) | Ribavirin and Immunoglobulin | Died |
| Perlejewski et al. (2015)36 | 24 | 60 | No | CSF | HSV1 | PCR and seroconversion | HSV1 Encephalitis | Known cause of encephalitis | Acyclovir | Recovered |
| Phan et al. (2015)37 | 25 | NR | NR | CSF supernatant | Cyclovirus | PCR | None (unclear association) | Novel organism | Not reported | Not reported |
| 26 | NR | NR | CSF supernatant | Gemycircularvirus | PCR | None (unclear association) | Novel organism | Not reported | Not reported | |
| 27 | NR | NR | CSF supernatant | Gemycircularvirus | PCR | None (unclear association) | Novel organism | Not reported | Not reported | |
| 28 | NR | NR | CSF supernatant | Gemycircularvirus | PCR | None (unclear association) | Novel organism | Not reported | Not reported | |
| Wilson et al. (2015)38 | 29 | 74 | Yes (hydroxychloroquine for rheumatoid arthritis) | CSF | Balamuthia mandrillaris | 18S PCR positive CSF, brain biopsy and vitreous fluid; immunohistochemistry and borderline positive serology | Balamuthia mandrillaris Encephalitis | Rare | None | Died |
| Christopeit et al. (2016)39 | 30 | 65 | Yes (chemotherapy, radiotherapy, BMT) | CSF | Fungi – candida tropicalis and fusarium | Candida confirmed by PCR. Fusarium not confirmed. | Fungal encephalitis | Rare | Antifungals (caspofungin, voriconazole, amphotericin) | Died |
| Guan et al. (2016)40 | 31 | NR | NR | CSF | HSV1 | PCR | HSV 1 Encephalitis | Known cause of encephalitis | Acyclovir | Recovered |
| 32 | NR | NR | CSF | HSV1 | PCR | HSV 1 Encephalitis | Known cause of encephalitis | Acyclovir | Recovered | |
| 33 | NR | NR | CSF | HSV2 | PCR | HSV 2 Encephalitis | Known cause of encephalitis | Acyclovir | Recovered | |
| 34 | NR | NR | CSF | VZV | PCR | VZV Encephalitis | Known cause of encephalitis | Acyclovir | Recovered | |
| Kawada et al. (2016)41 | 35 | <1 | NR | CSF | Coxsachievirus A9 | PCR | Coxsachievirus A9 encephalitis | Known cause of encephalitis | Not reported | Not reported |
| 36 | 7 | NR | CSF | Coxsachievirus A9 | PCR | Coxsachievirus A9 encephalitis | Known cause of encephalitis | Not reported | Not reported | |
| 37 | 3 | NR | CSF | Mumps | PCR | Mumps encephalitis | Known cause of encephalitis | Not reported | Not reported | |
| Lum et al. (2016)42 | 38 | <1 | Yes (AML 120 days post-BMT) | FFPE Brain Biopsy | Astrovirus VA1/HMO-C | PCR | Astrovirus VA1/HMO-C encephalitis | Novel organism (emerging cause of encephalitis) | Immunosuppression reduced. | Died |
| Morfopoulou et al. (2016)43 | 39 | <1 | Yes (SCID) | Brain Biopsy | Coronavirus OC43 | PCR and immunohistochemistry. | Coronavirus OC43 Encephalitis | Novel cause of encephalitis (known organism) | Unconditioned cord-blood transplantation, which resulted in T-cell engraftment. | Died |
| Phan et al. (2016)44 | 40 | 6 | NR | Filtered CSF supernatant DNAse treated | Densovirus | PCR, including in CSF re-extracted in different lab | NMDA Encephalitis. Unclear association with densovirus. | Novel organism | High dose IV steroids and rituximab. | Recovered |
| Salzberg et al. (2016)45 | 41 | 16 | NR | Brain Biopsy | JC polyomavirus | Immunohistochemistry and neuropathology | JC encephalitis | Known cause of encephalitis | Not stated. | Not reported |
| 42 | 67 | NR | Brain Biopsy | Mycobacterium tuberculosis | Not confirmed by conventional microbiology (AFB negative) but had necrotising granulomas and responded rapidly to treatment | M. tuberculosis encephalopathy | Known cause of encephalitis | Anti-TB therapy | Recovered | |
| 43 | 44 | Yes (post solid organ transplant) | FFPE Brain Biopsy | EBV | RNA in-situ hybridisation | EBV encephalopathy | Known cause of encephalitis | Immunotherapy modulated | Not reported | |
| Sato et al. (2016)46 | 44 | 4 | Yes (congenital aplastic anaemia since birth and previous cord blood cell transplant and chronic pulmonary graft-versus host disease on immunosuppressants). | CSF | Astrovirus-MLB1 | PCR but only in stool, throat and urine. CSF and serum were PCR negative (suggest due to low viral load) | Astrovirus-MLB1 Encephalitis | Novel cause of encephalitis (known organism) | None | Recovered |
| Mongkolrattanothai et al. (2016)47 | 45 | 11 | No | CSF | Brucella melitensis | PCR and serology. 16S PCR was negative | Neurobrucellosis | Rare | Treated | Recovered |
| Morfopoulou et al. (2017)48 | 46 | 1 | Yes (SCID); BMT | Brain biopsy | Mumps vaccine strain | PCR and immunohistochemistry | Vaccine derived mumps encephalitis | Novel cause of encephalitis (known organism) | None | Died |
NR, not reported; CSF, cerebrospinal fluid; FFPE, formalin fixed paraffin embedded; SCID, severe combined immunodeficiency; BMT, bone marrow transplant; AML, acute myeloid leukaemia; CLL, chronic lymphocytic leukaemia.
Fig. 4.
Temporal trends in the Publication of Encephalitis Cases involving Next-Generation Sequencing in the last Decade.
Discussion
Improved diagnosis with NGS
In 16 of the 44 known cases, well-established causes of encephalitis were detected which could have been identified by rapid and specific primary screening methods such as PCR. These organisms included HSV, coxsackievirus A9, measles virus, VZV, mumps virus, Epstein-Barr virus, JC virus and Mycobacterium tuberculosis. However, in the remaining 28 cases novel (18/44), rare (5/44) or unexpected (5/44) organisms were detected which could not (in the case of novel organisms) or are unlikely (in the cases of rare and unexpected pathogens) to have been detected using specific PCR assays.
The five unexpected cases were known human pathogens but novel causes of encephalitis. Although diagnostic PCR assays may exist for some of these viruses, they are unlikely to have been considered in the differential diagnosis and therefore would not be routinely tested. This included two cases of human parvovirus 4 (PARV4),28 first described in 2005 when it was associated with a viraemic patient in whom an acute viral infection was suspected49; one case of human coronavirus OC-43,43 typically a human respiratory pathogen never previously described in a human case of encephalitis but known to cause encephalitis in mice50; one case of human astrovirus MLB1,46 a recently described human astrovirus strain the pathogenic role of which remains unclear; and one case of mumps vaccine virus in a child who was vaccinated prior to a primary immunodeficiency diagnosis.48
The five cases in which rare causes of encephalitis were identified were Brucella melitensis, Candida tropicalis, Leptospira santarosai and two cases of Balamuthia mandrillaris.
Eighteen cases were considered to be novel pathogens. Three (3/14) of the identified organisms were arenaviruses, three cases were a variegated squirrel bornavirus, four were a novel astrovirus (Astrovirus VA1/HMO-C), three were cycloviruses, three were gemycircuarlviruses and one was a densovirus. The three arenavirus cases occurred in three solid organ transplant recipients who all received organs from the same donor, who was later shown to be anti-arenavirus IgM and IgG seropositive. The three cases of variegated squirrel bornavirus occurred in three breeders of variegated squirrel and was retrospectively detected in one of the breeder's squirrels. The novel astrovirus VA1/HMO-C was initially detected in an adolescent with primary immunodeficiency; it has since been shown, through four further case reports identified by NGS,31, 32, 35, 42 as an emerging under recognised cause of encephalitis in immunosuppressed patients that should be included in the differential diagnosis of encephalitis in this patient group. The clinical significance of the densovirus, cyclovirus and gemycircularviruses is doubtful, and discussed in detail further on in this review.
An additional advantage of using NGS for the diagnosis of encephalitis is that, aside from pathogen identification, in instances where virus titre and read depth is high enough it is possible to generate partial or full genome sequences for the pathogen. Pathogen sequences can be used for phylogenetic analysis to elucidate the strain31, 43 or possible source of the organism, as was the case for Morfopoulou et al., who demonstrated 99.5% homology between the mumps virus found in the brain of an encephalitic child with primary immunodeficiency and the vaccine batch used to immunise the child.48
Proving causality
NGS is a powerful tool for pathogen detection, allowing us to detect organisms that may not previously have been described or associated with the disease in question. However, as with all molecular tools, detection of a microorganism does not prove causality.
To provide further evidence for an aetiological role in encephalitis of the identified pathogen, a challenge reviewed in detail elsewhere,2 some reports make use of additional clinical and laboratory indicators to exclude the possibility that the detected organism is an incidental finding. Seroconversion to the pathogen in question is highly suggestive of etiological significance of an organism51 however pre-infection or follow-up serum samples are rarely available; of the 44 cases identified in this review seroconversion was demonstrated in only two.26, 36 A further eight were able to demonstrate the presence of specific antibodies, but without knowledge of the sero-status prior to onset of symptoms. Although detection of pathogen-specific intrathecal antibodies is also highly suggestive of a causal relationship and recommended by UK guidelines,2 none of the cases identified in this review reported intrathecal antibody testing. In cases where encephalitis is caused by reactivation of a dormant pathogen, rather than primary infection, or where the pathogen does not cause a strong systemic antibody response, serology may not be useful.
In the case of a novel or emerging cause of encephalitis for which the clinical significance may be unclear, proving causality is particularly important. In this instance organism-specific immunostaining or in situ hybridisation in affected tissues will provide additional evidence of the cellular distribution of infection and exclude the possibility of reagent or tissue contamination; 18 of the reviewed cases report confirmatory immunostaining or in situ hybridisation. In this context brain biopsies are a more useful specimen than cerebrospinal fluid (CSF) since it allows immunostaining of the affected tissue. Moreover, in encephalitis caused by mutated pathogens such as in subacute sclerosing panencephalitis (SSPE) caused by chronic measles infection52 or cases of mumps vaccine encephalitis48 the pathogen may not be detected in CSF but only in brain parenchyma.
An alternative molecular method, such as PCR, can be used to confirm the presence of the detected organism and exclude the possibility that the identified organism is an artefact of the bioinformatics analysis. In this review 33/44 cases confirmed the presence of the organisms by PCR. PCR supports the identity of the organism sequenced by NGS, however does not contribute to proving causality.
Six cases identified novel small circular ssDNA viruses in the CSF of patients with encephalitis of unknown aetiology; three cycloviruses29, 37 and three gemycircularviruses.37 However, the clinical significance of these is doubtful. Phan et al.37 confirmed, via repeat DNA extraction using an alternative method, that the source of the cyclovirus is not reagent contamination. Nevertheless, whilst screening CSF samples for cyclovirus by PCR, Tan et al.29 detected cyclovirus in the CSF of patients in whom the aetiology of their central nervous system (CNS) disease had already been confirmed as Japanese encephalitis, dengue virus or bacterial meningitis. The only identified cellular host for gemycircularviruses is fungi. In the absence of other evidence of pathogenicity, such as seroconversion or demonstration of the pathogen within cells in the brain, the probability remains that detection of cyclovirus or gemycircularvirus in CSF is an incidental finding. Similarly the detection of densovirus, a small linear ssDNA virus, in CSF may be incidental since it was detected in a case with confirmed N-methyl D-aspartate (NMDA)-receptor encephalitis.44 Autoimmune antibodies have previously been co-detected with herpesvirus DNA in CSF from cases of encephalitis,53 however the host range of densoviruses is to date exclusively invertebrates; the authors suggest a possible explanation for detection in CSF is the passive transfer of virus from an insect bite or CSF contamination from skin flora or an environmental source. In these cases, further evidence is required before assigning a pathogenic role.
As with other molecular tests, including PCR which has become the gold standard of virological diagnostics, results from metagenomics applied to cases of encephalitis should be interpreted in the context of other clinical and laboratory findings, particularly when a novel or unexpected organism is detected.
Diagnostic yield
The majority of reports concerning the use of metagenomics for diagnosis of encephalitis are comprised of single case reports, therefore it is difficult to assess the diagnostic yield (number of positive results/number of cases tested), and thus utility, of metagenomics for encephalitis.
Five reports included testing multiple cases of encephalitis; in these the diagnostic yield was 0% (0/36),7 1.6% (2/125),29 6% (4/62),37 19% (3/16)41 and 30% (3/10).45 The first three studies with low diagnostic yield of 0–6% tested only CSF supernatant which is cell-free, therefore only cell free viruses or cell-free microbial nucleic acid can be detected; moreover CSF often contains a lower pathogen load then brain biopsies and so pathogen detection is more challenging. The aforementioned studies also included only samples for which primary routine diagnostic testing using standard methods was negative therefore the utility of NGS as a first-line test cannot be assessed. A higher diagnostic yield was reported where specimens were tested using metagenomics as a first-line screening tool; 19% using whole CSF41 and 30% using brain biopsies.45 The use of brain tissue rather than CSF may increase diagnostic yield. In three cases a pathogen was detected in brain biopsy but not in CSF32, 35, 48 while the opposite was not observed, although in one instance the proportion of pathogen reads was greater in CSF than brain biopsy.33
In our hands, the diagnostic yield for metagenomics in encephalitis is, to date, 50% (8/16) all of which were brain biopsies. The eight identified pathogens were coronavirus OC-43,43 two cases of vaccine derived mumps virus,48, 54 Toxoplasma gondii (unpublished) and four cases of astrovirus VA1/HMO-C31, 42 (two cases unpublished). All of our positive results were in immunocompromised patients and in the majority of cases there was a high index of suspicion of infection, suggesting metagenomics may be best applied to a targeted population in whom it will be most rewarding.
Feasibility of NGS for routine diagnostic use
Quality assurance
Prior to providing a new diagnostic test, extensive validation must be undertaken to ensure the service is fit for purpose, such as determining the specificity and sensitivity of an assay, the purpose of which is to ensure a robust, accurate and reproducible result all of which is part of quality assurance. The regulatory requirements that should be fulfilled for the validation of metagenomics for pathogen detection is discussed in detail elsewhere,23 however one aspect that must continue beyond the validation stage is the use of positive and negative controls.
In the context of metagenomics for pathogen detection a positive control is a specimen (real or constructed) that is known to be positive for one or multiple organisms; a negative control is one that is known to be negative for any pathogens. These should be included in every sequencing run; if the positive control fails (i.e. the known pathogen/s is not detected) this invalidates the results of all clinical specimens processed in parallel for which no pathogen was identified. Conversely if the negative control fails (i.e. an unexpected organism is identified in the sequence data) this could indicate reagent contamination or a problem with the analysis pipeline and thus positive results from samples processed in parallel are invalidated and should be repeated. This having been said, of 44 cases of encephalitis identified in this review (Table 1), only 5 included positive controls and 15 included negative controls. The accuracy of a positive result is critical for patient management; however a negative result can also be useful to exclude infection, particularly where anti-inflammatory and immunosuppressive treatments are being considered. Consequently prior to provision of a clinical service appropriate controls must be in place to ensure results are reliable and therefore clinically actionable.
The use of controls is aptly demonstrated by Mongkolrattanothai et al.,47 who not only included positive and negative controls but also implemented defined criteria in their analysis pipeline that dictates any viruses detected in a clinical specimen should not be detected in the negative controls and, moreover, bacteria detected in a clinical specimen should only be reported as a significant finding if detected with a reads per million (RPM) ratio ≥10 (RPM ratio = RPM sample / RPM negative control). This approach overcomes the common problem of reagent contamination with microbial nucleic acids; of 33 cases in which the presence or absence of contaminating reads was reported, 22 cases reported the presence of environmental bacteria, plant viruses, bacteriophages and/or avian retroviruses.24, 29, 31, 33, 35, 36, 38, 39, 40, 41, 43, 45, 47, 48
Turn-around times
Only 5 cases reported the time from specimen collection to pathogen identification; for these the turn-around time was 2–6 days, with a median time of 6 days. Due to limitations of currently available sequence library preparation methods and sequencing chemistries a sample-to-answer turn-around time as short as 48 hours24 is only achievable with a fast downstream analysis pipeline; in the reported case analysis took only 97 minutes compared to up to two days in other, computationally intensive, pipelines. Nevertheless it serves as proof of principle that relatively short turn-around times are achievable, even though up to 6 days is more common.
The sample-to-answer turn-around time of specific real-time PCR, which is the current gold standard for diagnosis of viral infections, is often less than 12 hours in a clinical laboratory and potentially less than 4 hours without batch processing. Consequently NGS cannot yet offer the same speed of result as PCR, which could delay the diagnosis in instances of encephalitis caused by well-known pathogens that are detectable by PCR. Nonetheless with rapidly increasing library preparation and sequencing speeds, turn-around times are likely to significantly improve in coming years.
Limitations of metagenomics for diagnosis of encephalitis
Metagenomics for pan-pathogen detection has the potential to revolutionise the diagnosis of encephalitis and other difficult-to-diagnose infections; however, there are some limitations of the technique that should be considered.
The sensitivity and limit of detection (LOD) of metagenomics can be determined for model organisms that represent major pathogen groups, such as DNA and RNA viruses, gram positive and negative bacteria, fungi and parasites; however, the broad-range nature of the technique makes it impossible to determine the sensitivity or LOD for every possible organism. This is also a recognised problem with pan-bacterial PCR detection which is used clinically,55 however can be confounded in metagenomics by differences between specimens and specimen types (for instance tissue biopsies versus CSF) in the quantity of genomic material, the ratio of host:pathogen sequences and, depending on the sequencing chemistry and degree of specimen multiplexing, the sequencing yield. Some of these limitations may be overcome by the careful use of processing and sequencing controls, as discussed elsewhere.23
Whilst metagenomics will produce a sequence for every fragment of DNA or RNA in a specimen, only pathogens with homology to known organisms in the sequence database of choice will be identified. If an organism is missing from the database, or if the pathogen causing infection is novel with no homology to known organisms, it will not be identified. Consequently, a negative result obtained by metagenomics, whilst reducing the likelihood of an infectious cause, cannot unequivocally exclude infection.
Sequencing total DNA or RNA will inevitably include sequencing host DNA or RNA transcripts which can result in >99% of the sequence data generated mapping to the human genome. The consequence of this is wasted cost, as the sequencing reaction is dominated by host rather than pathogen sequences, and also has implications for the turn-around times and sensitivity of pathogen detection. In order to detect pathogen sequences, which can be as few as nine in 68 million reads,30 vast sequencing read depths are required; very high throughput sequencing platforms with only a very limited number of samples sequenced in parallel are required to achieve this. To overcome this, depletion of host DNA or RNA prior to sequencing is required; however the options for this are currently limited and are not all suitable for detection of viral pathogens. Improved methods for host DNA and RNA depletion would considerably reduce the cost and time to result and improve the sensitivity of metagenomics for diagnosis of encephalitis.
Finally it is important to remember that in some cases of encephalitis, the pathology is due to the immune response and the pathogen may be rarely detected if at all. For example in Japanese encephalitis virus (JEV) infection, the commonest cause of encephalitis in Asia, the most sensitive RT-qPCR detects RNA in less than 10% of cases, and the mainstay of diagnosis is serology.56 In these cases, NGS is unlikely to significantly improve the diagnostic yield.
Recommendations for the use of NGS in diagnosis of encephalitis
This systematic literature review and case series suggests there is preliminary evidence to support a role for NGS in the management of undiagnosed encephalitis. Undeniably, the research is limited to case reports, with poor reporting of clinical case definitions of encephalitis, baseline tests performed, or adherence to clinical guidelines. Nonetheless, current epidemiological data suggests that the cause of encephalitis remains unknown in 30–60% of cases8, 21; and NGS has striking potential to identify undiagnosed pathogens and thus reduce the number of cases with unknown aetiology. NGS also has utility for pathogen detection in other clinical syndromes, such as respiratory infections,57 therefore the implementation of this technique in clinical laboratories would have wider implications for diagnosis of infection beyond encephalitis.
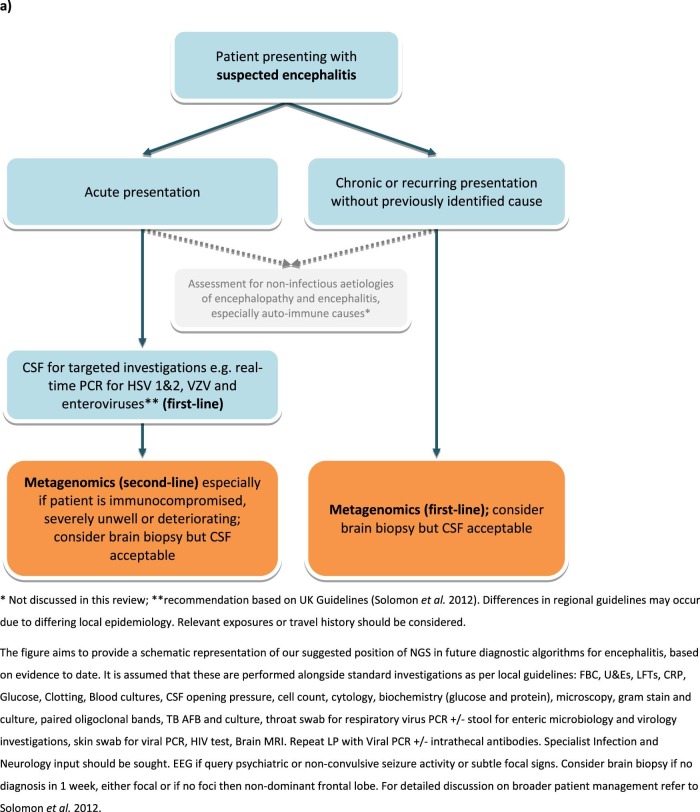
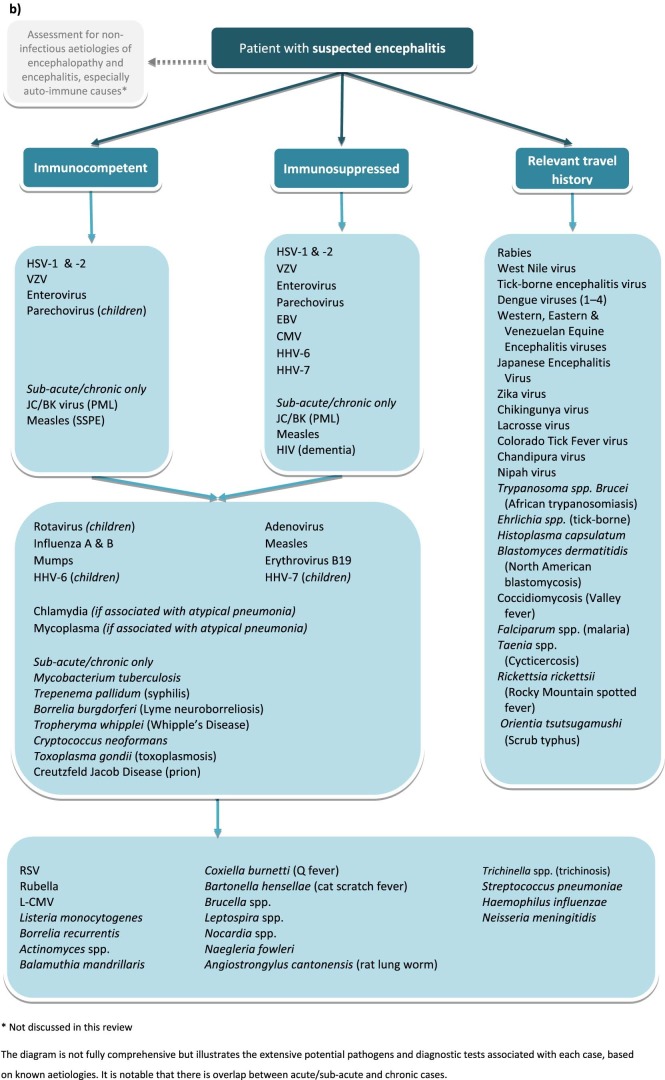
Notably, current turn-around-times prevent the replacement of routine methods, such as PCR, for the diagnosis of acute encephalitis. For these reasons, the role of NGS in clinical algorithms is still to be delineated. At this point in time, we suggest NGS is routinely applied for the diagnosis of acute cases of encephalitis for which no cause is found after targeted investigations using PCR. Recommendations for first-line targeted testing are discussed in detail elsewhere14 but in the UK should include PCR for HSV, VZV and enteroviruses. However due to differing local epidemiology clinicians should consult the relevant national guidelines.8, 13, 14, 16, 17 In immunocompromised patients however, metagenomics ought to be considered earlier; 73% of case reports in this review involved immunocompromised patients. This is the population at most risk of infection with novel and unexpected organisms and, moreover, may present with a more chronic or insidious clinical history58 in which a one-week turn-around-time is more acceptable. Given that the causative pathogen is not always detected in CSF, in all cases of encephalitis in which diagnosis by NGS is being sought the preferred specimen type is brain biopsy. Nevertheless CSF samples are acceptable if it is the only specimen available. Our recommendations for the use of NGS in diagnosis of microbial causes of encephalitis are summarised in Fig. 5a , with the contrasting algorithm for targeted testing summarised in Fig. 5b. This review was limited to pathogen detection by NGS in brain biopsies or CSF. There may also be a role for testing other specimens, such as throat samples and urine. This was recently shown in an encephalitis case diagnosed by NGS of urine, identifying a case of Japanese encephalitis virus.59
Fig. 5.
(A) Recommendations for microbial investigations in the diagnosis of encephalitis incorporating NGS metagenomics, allowing detection of unexpected or novel organisms; (B) Recommendations for targeted microbial investigations (modified from Solomon et al. 2012) without NGS metagenomics, which will not allow detection of unexpected or novel organisms and for which there is unlikely to be sufficient specimen for exhaustive testing.
It is expected that over the next few years, the cost and time-to-result of metagenomics will reduce, and with this, it is foreseen that it will be possible to offer this as the first-line diagnostic test. This depends on the ability to deplete host DNA and RNA prior to sequencing, reduced read depth requirement and faster sequencing and bioinformatics technologies.
The role of autoimmune encephalitis, in some instances triggered by an infection,60 is beyond the scope of this review. However, since up to 20% of cases of encephalitis are caused by autoimmune disorders, in which the immune system attacks specific host proteins,21, 61 a comprehensive diagnostic service should include antibody mediated, as well as infectious, causes of encephalitis. The clinical presentation of autoimmune (non-infectious) and infectious encephalitis are similar,62 however the treatment is often opposing. Non-infectious causes may require immunosuppressive therapy63; however administering immunosuppressive therapy where the cause is infectious exacerbates the infection, with potentially fatal results. A conclusive diagnosis of the causative agent of encephalitis would improve differentiation between infectious and auto-immune causes thus appropriate management of immunosuppression.
In addition to the direct impact on individual patients, improving the diagnosis of encephalitis will increase our understanding of the causes of encephalitis generally. This knowledge is critical for future development of fast point-of-care tests and to develop clinical algorithms that minimise the time to diagnosis and treatment, thus maximising the chances of recovery.
Contributors
TB and JRBrown conducted the systematic review and prepared the manuscript, figures and table. JBreuer contributed to manuscript preparation.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
JR Brown is supported by a paediatric research grant from the Great Ormond Street Hospital Children's Charity (“Diagnosis of encephalitis by deep sequencing”, V4317). JBreuer receives funding from the UCL/UCLH NIHR Biomedical Research Centre.
All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
The corresponding author (JBrown) had access to all the data and had final responsibility for the decision to submit for publication.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jinf.2017.12.014.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Supplementary File 1.
References
- 1.Venkatesan A., Tunkel A., Bloch K.C., Lauring A.S., Sejvar J., Bitnun A. Case definitions, diagnostic algorithms, and priorities in encephalitis: consensus statement of the international encephalitis consortium. Clin Infect Dis. 2013;57(8):1114–1128. doi: 10.1093/cid/cit458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Granerod J., Cunningham R., Zuckerman M., Mutton K., Davies N.W., Walsh A.L. Causality in acute encephalitis: defining aetiologies. Epidemiol Infect. 2010;138:783–800. doi: 10.1017/S0950268810000725. [DOI] [PubMed] [Google Scholar]
- 3.Granerod J., Crowcroft N.J. The epidemiology of acute encephalitis. Neuropsychol Rehabil. 2007;17:406–428. doi: 10.1080/09602010600989620. [DOI] [PubMed] [Google Scholar]
- 4.Granerod J., Tam C.C., Crowcroft N.S., Davies N.W.S., Borchert M., Thomas S.L. Challenge of the unknown: A systematic review of acute encephalitis in non-outbreak situations. Neurology. 2010;75:924–932. doi: 10.1212/WNL.0b013e3181f11d65. [DOI] [PubMed] [Google Scholar]
- 5.Marra C.W.R., Scheld M. 4th ed. Lippincott Williams and Wilkins; Philadelphia: 2014. Approach to the patient with central nervous system infection; pp. 1–3. [Google Scholar]
- 6.Glaser C.A., Honarmand S., Anderson L.J., Schnurr D.P., Forghani B., Cossen C.K. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43:1565–1577. doi: 10.1086/509330. [DOI] [PubMed] [Google Scholar]
- 7.Ambrose H.E., Granerod J., Clewley J.P., Davies N.W., Keir G., Cunningham R. Diagnostic strategy used to establish etiologies of encephalitis in a prospective cohort of patients in England. J Clin Microbiol. 2011;49:3576–3583. doi: 10.1128/JCM.00862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesan A., Tunkel A.R., Bloch K.C., Lauring A.S., Sejvar J., Bitnun A. Case definitions, diagnostic algorithms, and priorities in encephalitis: consensus statement of the international encephalitis consortium. Clin Infect Dis. 2013;57:1114–1128. doi: 10.1093/cid/cit458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nath A. Neuroinfectious diseases: a crisis in neurology and a call for action. JAMA Neurol. 2015;72:143–144. doi: 10.1001/jamaneurol.2014.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly C., Sohal A., Michael B.D., Riordan A., Solomon T., Kneen R. Suboptimal management of central nervous system infections in children: a multi-centre retrospective study. BMC Pediatr. 2012;12:145. doi: 10.1186/1471-2431-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mailles A., De Broucker T., Costanzo P., Martinez-Almoyna L., Vaillant V., Stahl J.P. Long-term outcome of patients presenting with acute infectious encephalitis of various causes in France. Clin Infect Dis. 2012;54:1455–1464. doi: 10.1093/cid/cis226. [DOI] [PubMed] [Google Scholar]
- 12.Venkatesan A. Epidemiology and outcomes of acute encephalitis. Curr Opin Neurol. 2015;28:277–282. doi: 10.1097/WCO.0000000000000199. [DOI] [PubMed] [Google Scholar]
- 13.Tunkel A.R., Glaser C.A., Bloch K.C., Sejvar J.J., Marra C.M., Roos K.L. The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis. 2008;47:303–327. doi: 10.1086/589747. [DOI] [PubMed] [Google Scholar]
- 14.Solomon T., Michael B.D., Smith P.E., Sanderson F., Davies N.W., Hart I.J. Management of suspected viral encephalitis in adults—Association of British Neurologists and British Infection Association National Guidelines. J Infect. 2012;64:347–373. doi: 10.1016/j.jinf.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Misra U.K., Mani V.E., Kalita J. A cost-effective approach to the diagnosis and management of acute infectious encephalitis. Eur Neurol. 2017;77:66–74. doi: 10.1159/000453662. [DOI] [PubMed] [Google Scholar]
- 16.Britton P.N., Eastwood K., Brew B.J., Nagree Y., Jones C.A. Consensus guidelines for the investigation and management of encephalitis. Med J Aust. 2015;202:576–577. doi: 10.5694/mja14.01042. [DOI] [PubMed] [Google Scholar]
- 17.Stahl J., Mailles A., Vaillant V., Floret D. Les encéphalites infectieuses aiguës : recommandations pour un diagnostic étiologique. Réanimation. 2007;16:485–489. [Google Scholar]
- 18.Fillatre P., Crabol Y., Morand P., Piroth L., Honnorat J., Stahl J.P. Infectious encephalitis: Management without etiological diagnosis 48hours after onset. Med Mal Infect. 2017 doi: 10.1016/j.medmal.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl J.P., Azouvi P., Bruneel F., De Broucker T., Duval X., Fantin B. Guidelines on the management of infectious encephalitis in adults. Med Mal Infect. 2017 doi: 10.1016/j.medmal.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Kulik D.M., Mekky M., Yang M., Bitnun A., Parkin P.C. Should a hospitalized child receive empiric treatment with acyclovir? Ital J Pediatr. 2012;38:72. doi: 10.1186/1824-7288-38-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granerod J., Ambrose H.E., Davies N.W., Clewley J.P., Walsh A.L., Morgan D. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10:835–844. doi: 10.1016/S1473-3099(10)70222-X. [DOI] [PubMed] [Google Scholar]
- 22.Morfopoulou S., Plagnol V. Bayesian mixture analysis for metagenomic community profiling. Bioinformatics. 2015;31:2930–2938. doi: 10.1093/bioinformatics/btv317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 24.Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu C.Y., Coffey L.L., Murkey J., Symmes K., Sample H.A., Wilson M.R. Diagnosis of fatal human case of St. Louis encephalitis virus infection by metagenomic sequencing, California, 2016. Emerg Infect Dis. 2017;23:1964–1968. doi: 10.3201/eid2310.161986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 27.Quan P.L., Wagner T.A., Briese T., Torgerson T.R., Hornig M., Tashmukhamedova A. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamin L.A., Lewthwaite P., Vasanthapuram R., Zhao G., Sharp C., Simmonds P. Human parvovirus 4 as potential cause of encephalitis in children, India. Emerg Infect Dis. 2011;17:1484–1487. doi: 10.3201/eid1708.110165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan le V., van Doorn H.R., Nghia H.D., Chau T.T., Tu le T.P., de Vries M. Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. MBio. 2013;4:e00231. doi: 10.1128/mBio.00231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan B.K., Wilson T., Fischer K.F., Kriesel J.D. Deep sequencing to identify the causes of viral encephalitis. PLoS ONE. 2014;9:e93993. doi: 10.1371/journal.pone.0093993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.R., Morfopoulou S., Hubb J., Emmett W.A., Ip W., Shah D. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis. 2015;60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fremond M.L., Perot P., Muth E., Cros G., Dumarest M., Mahlaoui N. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc. 2015;4:e53–e57. doi: 10.1093/jpids/piv040. [DOI] [PubMed] [Google Scholar]
- 33.Greninger A.L., Messacar K., Dunnebacke T., Naccache S.N., Federman S., Bouquet J. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med. 2015;7:113. doi: 10.1186/s13073-015-0235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann B., Tappe D., Hoper D., Herden C., Boldt A., Mawrin C. A Variegated squirrel bornavirus associated with fatal human encephalitis. N Engl J Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 35.Naccache S.N., Peggs K.S., Mattes F.M., Phadke R., Garson J.A., Grant P. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis. 2015;60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlejewski K., Popiel M., Laskus T., Nakamura S., Motooka D., Stokowy T. Next-generation sequencing (NGS) in the identification of encephalitis-causing viruses: Unexpected detection of human herpesvirus 1 while searching for RNA pathogens. J Virol Methods. 2015;226:1–6. doi: 10.1016/j.jviromet.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Phan T.G., Mori D., Deng X., Rajindrajith S., Ranawaka U., Fan Ng T.F. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology. 2015;482:98–104. doi: 10.1016/j.virol.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson M.R., Shanbhag N.M., Reid M.J., Singhal N.S., Gelfand J.M., Sample H.A. Diagnosing balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann Neurol. 2015;78:722–730. doi: 10.1002/ana.24499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christopeit M., Grundhoff A., Rohde H., Belmar-Campos C., Grzyska U., Fiehler J. Suspected encephalitis with Candida tropicalis and Fusarium detected by unbiased RNA sequencing. Ann Hematol. 2016;95:1919–1921. doi: 10.1007/s00277-016-2770-3. [DOI] [PubMed] [Google Scholar]
- 40.Guan H., Shen A., Lv X., Yang X., Ren H., Zhao Y. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22:240–245. doi: 10.1007/s13365-015-0390-7. [DOI] [PubMed] [Google Scholar]
- 41.Kawada J., Okuno Y., Torii Y., Okada R., Hayano S., Ando S. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep. 2016;6:33452. doi: 10.1038/srep33452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lum S.H., Turner A., Guiver M., Bonney D., Martland T., Davies E. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis. 2016;18:960–964. doi: 10.1111/tid.12607. [DOI] [PubMed] [Google Scholar]
- 43.Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 44.Phan T.G., Messacar K., Dominguez S.R., da Costa A.C., Deng X., Delwart E. A new densovirus in cerebrospinal fluid from a case of anti-NMDA-receptor encephalitis. Arch Virol. 2016;161:3231–3235. doi: 10.1007/s00705-016-3002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salzberg S.L., Breitwieser F.P., Kumar A., Hao H., Burger P., Rodriguez F.J. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol Neuroimmunol Neuroinflamm. 2016;3:e251. doi: 10.1212/NXI.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato M., Kuroda M., Kasai M., Matsui H., Fukuyama T., Katano H. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol. 2016;78:66–70. doi: 10.1016/j.jcv.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Mongkolrattanothai K., Naccache S.N., Bender J.M., Samayoa E., Pham E., Yu G. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatric Infect Dis Soc. 2017 doi: 10.1093/jpids/piw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morfopoulou S., Mee E.T., Connaughton S.M., Brown J.R., Gilmour K., Chong W.K. Deep sequencing reveals persistence of cell-associated mumps vaccine virus in chronic encephalitis. Acta Neuropathol. 2017;133:139–147. doi: 10.1007/s00401-016-1629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones M.S., Kapoor A., Lukashov V.V., Simmonds P., Hecht F., Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talbot P.J., Desforges M., Brison E., Jacomy H. Coronaviruses as encephalitis-inducing infectious agents. In: Tkachev S., editor. Non-flavivirus encephalitis. InTech; 2011. [DOI] [Google Scholar]
- 51.Rivers T.M. Viruses and Koch's postulates. J Bacteriol. 1937;33:1–12. doi: 10.1128/jb.33.1.1-12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anlar B. Subacute sclerosing panencephalitis and chronic viral encephalitis. Handb Clin Neurol. 2013;112:1183–1189. doi: 10.1016/B978-0-444-52910-7.00039-8. [DOI] [PubMed] [Google Scholar]
- 53.Linnoila J.J., Binnicker M.J., Majed M., Klein C.J., McKeon A. CSF herpes virus and autoantibody profiles in the evaluation of encephalitis. Neurol Neuroimmunol Neuroinflamm. 2016;3 doi: 10.1212/NXI.0000000000000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan C.J., Mohamad S.M., Young D.F., Skelton A.J., Leahy T.R., Munday D.C. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci Transl Med. 2015;7:307ra154. doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris K.A., Hartley J.C. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J Med Microbiol. 2003;52:685–691. doi: 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]
- 56.World Health Organization S-EAatWP, World Health Organisation. Japanese encephalitis surveillance standards. In: WHO-recommended standards for surveillance of selected vaccine-preventable diseases WHO/V&B/03.01.
- 57.Lewandowska D.W., Schreiber P.W., Schuurmans M.M., Ruehe B., Zagordi O., Bayard C. Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS ONE. 2017;12:e0177340. doi: 10.1371/journal.pone.0177340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saylor D., Thakur K., Venkatesan A. Acute encephalitis in the immunocompromised individual. Curr Opin Infect Dis. 2015;28:330–336. doi: 10.1097/QCO.0000000000000175. [DOI] [PubMed] [Google Scholar]
- 59.Mai N.T.H., Phu N.H., Nhu L.N.T., Hong N.T.T., Hanh N.H.H., Nguyet L.A. Central nervous system infection diagnosis by next-generation sequencing: a glimpse into the future? Open Forum Infect Dis. 2017;4:ofx046. doi: 10.1093/ofid/ofx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbagallo M., Vitaliti G., Pavone P., Romano C., Lubrano R., Falsaperla R. Pediatric autoimmune encephalitis. J Pediatr Neurosci. 2017;12:130–134. doi: 10.4103/jpn.JPN_185_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh T.D., Fugate J.E., Rabinstein A.A. The spectrum of acute encephalitis: causes, management, and predictors of outcome. Neurology. 2015;84:359–366. doi: 10.1212/WNL.0000000000001190. [DOI] [PubMed] [Google Scholar]
- 62.Sejvar J.J., Kohl K.S., Bilynsky R., Blumberg D., Cvetkovich T., Galama J. Encephalitis, myelitis, and acute disseminated encephalomyelitis (ADEM): case definitions and guidelines for collection, analysis, and presentation of immunization safety data. Vaccine. 2007;25:5771–5792. doi: 10.1016/j.vaccine.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 63.Graus F., Titulaer M.J., Balu R., Benseler S., Bien C.G., Cellucci T. A clinical approach to diagnosis of autoimmune encephalitis. Lancet Neurol. 2016;15:391–404. doi: 10.1016/S1474-4422(15)00401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1.