Highlights
-
•
A(H7N9) patients mounted strong serum HAI antibody responses against A(H7N9) virus.
-
•
Mean A(H7N9) virus-specific HAI antibody titers remained above 80 for 11 months.
-
•
Severely ill patients mounted higher HAI antibody responses against A(H7N9) virus.
Keywords: Antibody response, Clinical severity, Follow-up, Influenza A(H7N9)
Abstract
Objectives
The long-term dynamics of antibody responses in patients with influenza A(H7N9) virus infection are not well understood.
Methods
We conducted a longitudinal serological follow-up study in patients who were hospitalized with A(H7N9) virus infection, during 2013–2018. A(H7N9) virus-specific antibody responses were assessed by hemagglutination inhibition (HAI) and neutralization (NT) assays. A random intercept model was used to fit a curve to HAI antibody responses over time. HAI antibody responses were compared by clinical severity.
Results
Of 67 patients with A(H7N9) virus infection, HAI antibody titers reached 40 on average 11 days after illness onset and peaked at a titer of 290 after three months, and average titers of ≥80 and ≥40 were present until 11 months and 22 months respectively. HAI antibody responses were significantly higher in patients who experienced severe disease, including respiratory failure and acute respiratory distress syndrome, compared with patients who experienced less severe illness.
Conclusions
Patients with A(H7N9) virus infection who survived severe disease mounted higher antibody responses that persisted for longer periods compared with those that experienced moderate disease. Studies of convalescent plasma treatment for A(H7N9) patients should consider collection of donor plasma from survivors of severe disease between 1 and 11 months after illness onset.
Introduction
Human infections with avian influenza A(H7N9) virus were first identified in the spring of 2013 in China,1 causing more human infections than any other avian influenza A virus.2 As of July 1, 2019, a total of 1568 laboratory-confirmed human cases, including 616 deaths, have been reported to the World Health Organization (WHO), primarily from China.3 , 4 During 2016–2017, A(H7N9) virus spread among poultry across China, and 30 out of 34 provinces reported human infections.5 Moreover, A(H7N9) virus strains continue to evolve and have diverged into two main lineages, the Pearl River Delta (PRD) and the Yangtze River Delta (YRD) lineages.6 YRD lineage A(H7N9) viruses evolved into antigenically distinct strains during 2016–2017, including low pathogenic avian influenza (LPAI) A(H7N9) virus strains (represented by A/Hong Kong/125/2017), and some viruses were classified as highly pathogenic avian influenza (HPAI) strains based on their genotype (represented by A/Guangdong/17SF003/2016).7 These HPAI viruses pose an increased threat to humans and agriculture.8 , 9 Although only four human cases of A(H7N9) virus infection were reported since October 2017, and virus detection has declined in poultry, the virus is far from eradication.10 Notably, A(H7N9) viruses continue to be assessed as representing a moderate to high public health risk, ranking highest among 12 influenza A viruses of animal-origin in the United States Centers for Disease Control and Prevention influenza risk assessment tool.11
Most laboratory-confirmed cases of A(H7N9) virus infection have been severely ill and have required ICU admission.1 Early neuraminidase inhibitor treatment appears to shorten the duration of viral shedding and improve outcome of A(H7N9) virus-infected patients.12 However, the emergence of neuraminidase inhibitor resistance in A(H7N9) and other avian influenza A viruses highlights the need for additional therapeutic options.13 Convalescent plasma may be a potential treatment for severe influenza,14 , 15 and was used in the treatment of one A(H7N9) patient who did not respond to oseltamivir treatment.16 Convalescent plasma or post-vaccination plasma were also used for treatment of A(H5N1) patients.17 , 18 Beigel and colleagues conducted the first randomized controlled trial of immune plasma in severe seasonal influenza and reported evidence of clinical benefit, primarily limited to participants who were treated within 4 days of illness onset.15 A larger phase III trial is now ongoing [NCT02572817].
Although promising, there are several safety concerns and practical challenges to using convalescent plasma, including defining the optimal timing for sera collection after recovery from illness, the case-to-case heterogeneity in antibody titers, and the rapid evolution of influenza viruses.19 Limited understanding of the specificity, kinetics, and duration of antibody responses in humans infected with A(H7N9) viruses therefore currently impedes the development and evaluation of convalescent plasma therapy. Ma et al. described the dynamics of the antibody response to A(H7N9) virus over a year post illness onset in 25 patients,20 they also characterized the antibody response to A(H7N9) virus up to 36 months after illness onset in a cross-sectional analysis.21 However, the longitudinal dynamics of the antibody response beyond one year have not been determined.
We conducted a longitudinal follow-up study in patients hospitalized with A(H7N9) virus infection. Our objectives were to characterize the specificity, kinetics, and duration of the antibody response in patients with A(H7N9) virus infection, to examine the association between antibody response and clinical severity, and to explore the timing for plasma collection for plasma therapy.
Materials and methods
Patient enrollment and sample collection
We recruited and followed patients with reverse transcription polymerase chain reaction (RT-PCR) confirmed influenza A(H7N9) virus infection from six provinces of China (Beijing, Shandong, Zhejiang, Hunan, Jiangxi, and Fujian) from three epidemics (2013, 2013–2014, and 2016–2017 epidemics), during hospitalization and after discharge. Patients enrolled from the 2016–2017 epidemic were from Jiangxi Province. Demographic information and clinical data during hospitalization were collected using a standardized form. Underlying medical conditions associated with high risk for complications of influenza were defined as reported previously.22 Patients were classified as having severe illness if they experienced the acute respiratory distress syndrome (ARDS) or required mechanical ventilation.
For patients who were enrolled during hospitalization, we obtained available residual sera that were collected for clinical laboratory testing; these patients were also followed up after discharge to obtain additional samples. In addition, we recruited and followed patients after discharge. Serum samples were collected at enrollment and during follow-up visits, which occurred 3, 6, 12, 18, and 65 months after illness onset. Among these serum samples, fifty-five specimens collected after hospital discharge of patients from the 2013 and 2013–2014 epidemics were tested and reported in a previous study investigating the virus-specific antibody responses and T cell memory in A(H7N9) survivors within the year following illness onset.23
The study protocol was reviewed and approved by the ethics committee of School of Public Health, Fudan University and the ethics committee of Chinese Center for Disease Control and Prevention. Written informed consent was obtained from all participants.
Serology laboratory testing
The serological assays were performed at the Joint Institute of Virology (Shantou University-The University of Hong Kong). The antigens used were reverse genetically reconstructed viruses, including low pathogenic A(H7N9) virus strains A/Anhui/1/2013 (all genes derived from wild-type virus) and A/Hong Kong/125/2017 (YRD lineage, hemagglutinin and neuraminidase genes from wild-type virus, and six internal genes from A/Puerto Rico/8/1934) and highly pathogenic A(H7N9) virus strain A/Guangdong/17SF003/2016 (YRD lineage, with removal of the multiple basic amino acid motif at the hemagglutinin cleavage site). All three viruses are A(H7N9) candidate vaccine viruses proposed by WHO.24 Samples from patients from the 2013 and 2013–2014 epidemics were tested against A/Anhui/1/2013 virus, which represented A(H7N9) viruses isolated from humans during these epidemics.25 A(H7N9) viruses isolated from humans in Jiangxi province in the 2016–2017 epidemic belong to clade C1,26 and are antigenically similar to A/Hong Kong/125/2017 virus. To test the reactivity of human antiserum induced by A/Hong Kong/125/2017 virus to A/Anhui/1/2013 virus and A/Guangdong/17SF003/2016 virus, samples obtained from patients from the 2016–2017 epidemic were tested against the aforementioned three A(H7N9) virus strains. Hemagglutination inhibition (HAI) assay and neutralization (NT) assay were performed according to standard protocols.27 Seropositive threshold was defined as an HAI antibody titer of 40,28 or a neutralizing antibody titer of 20.29 A non-reactive titer was defined as a value of ≤10. See supplementary data for further details.
Statistical analyses
For the analyses of the kinetics of the antibody responses and association with clinical outcomes, we pooled serological data from patients with RT-PCR-confirmed A(H7N9) virus infection from three epidemics. HAI and neutralizing antibody titers were log2 transformed before analyses. Comparison of the GMTs between severely ill patients and moderately ill patients was performed with Mann–Whitney U test, clustered by sampling time from illness onset; we also used a linear regression model adjusted for sex, age, and sampling time from illness onset. Previous studies have reported that patients with A(H7N9) virus infection have lower neutralizing antibody titers than HAI antibody titers, and neutralizing antibody titers are lower in A(H7N9) patients compared to A(H5N1) patients.30, 31, 31 We also observed lower neutralizing antibody titers compared to HAI antibody titers in this study. We used a random intercept linear model with B-spline to analyze the dynamics of HAI antibody responses and neutralizing antibody responses over time in sera of A(H7N9) virus-infected patients. Degree and knots of B-spline were selected based on Akaike information criterion (AIC). A Generalized Estimating Equations (GEE) model used to fit the dynamic curve of antibody titers yielded similar results to the random intercept linear model. See supplementary data for further details.
Results
Participants and samples
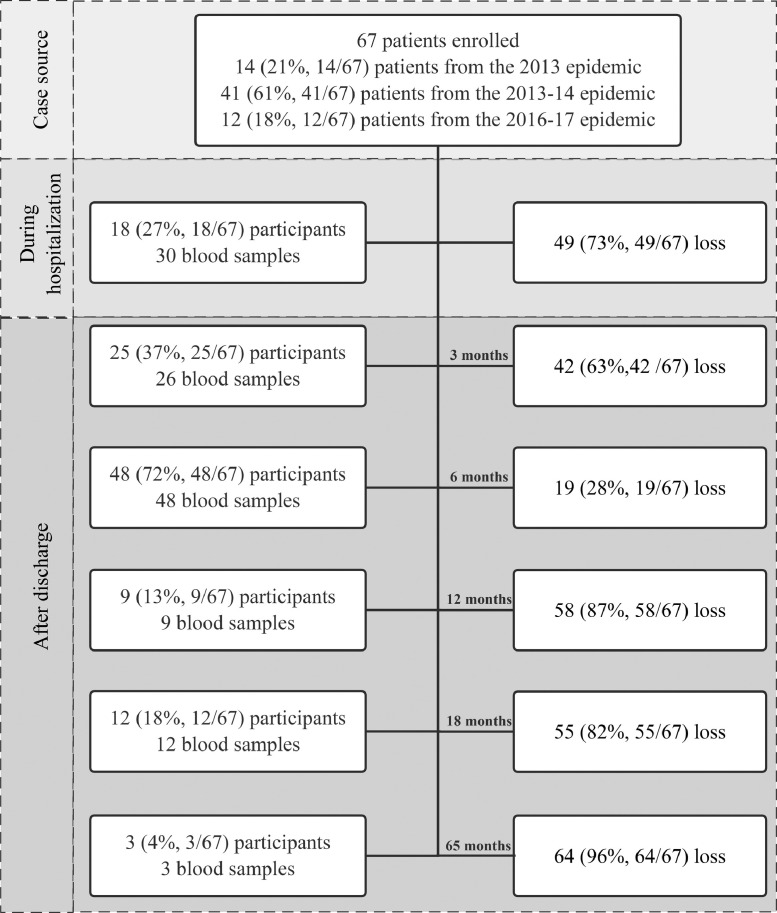
From April 2013 to September 2018, a total of 67 patients who were hospitalized with laboratory-confirmed A(H7N9) virus infection were enrolled (Supplementary Fig. 1), including fourteen participants from the 2013 epidemic, forty-one from the 2013–2014 epidemic, and twelve from the 2016–2017 epidemic (Supplementary Fig. 2A). Eighteen patients were enrolled during hospitalization, four of them died in hospital and two were lost to follow-up. Forty-nine patients were recruited and followed only after hospital discharge. Serial visits after discharge were conducted at 1–5 months, 6–8 months, 12–13 months, and 65 months after disease onset for forty-nine patients from the 2013 and 2013–14 epidemics. A single visit was conducted at 16–20 months after illness onset for patients from the 2016–2017 epidemic. Numbers of participants and blood samples at different stages are shown in Fig. 1 . A total of 128 serum samples were collected (Supplementary Fig. 2B), including one to seven specimens from each patient, and 33 patients provided at least two samples (Table 1 ).
Fig. 1.
Flow chart of enrollment of participants and collection of blood samples throughout the study.
Table 1.
Details of 128 blood samples collected from 67 A(H7N9) patients.
| No. of blood samples collected from each participant | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| No. (%) of participants | 34 (51) | 21 (31) | 3 (5) | 5 (8) | 2 (3) | 1 (1) | 1 (1) |
| No. (%) of samples | 34 (27) | 42 (33) | 9 (7) | 20 (15) | 10 (8) | 6 (5) | 7 (5) |
Characteristics of study subjects
Demographic and clinical features of enrolled patients during hospitalization are shown in Table 2 . Among these, 72% of the patients were male, and the median age was 53 years old. For those subjects with data available, 46% (27 of 59) had at least one high-risk chronic medical condition. The most common clinical complications included pneumonia (84%), respiratory failure (49%), and ARDS (46%). Forty-one percent of patients required mechanical ventilation. Forty-five percent of the patients required intensive care unit (ICU) admission. The median duration of hospitalization for all patients was 21 days (IQR, 14 to 31). No major differences in patient characteristics were observed between the three epidemics. Four patients from the 2013 epidemic died during hospitalization.
Table 2.
Demographic and clinical features of A(H7N9) patients.
| Characteristic | Total (N = 67) | Epidemic Wave 1 Jan 2013–Sep 2013 (n = 14) | Epidemic Wave 2 Oct 2013–Sep 2014 (n = 41) | Epidemic Wave 5 Oct 2016–Sep 2017 (n = 12) |
|---|---|---|---|---|
| Demographic features | ||||
| Males | 48/67 (72) | 10/14 (71) | 31/41 (76) | 7/12 (58) |
| Age, median years (IQR) | 53 (36–67) | 67 (26–77) | 47 (36–64) | 55 (49–68) |
| Age group | ||||
| 0–35 years | 16 (24) | 4 (29) | 11 (27) | 1 (8) |
| 36–55 years | 20 (30) | 1 (7) | 14 (34) | 5 (42) |
| 56–65 years | 13 (19) | 2 (14) | 9 (22) | 2 (17) |
| >65 years | 18 (27) | 7 (50) | 7 (17) | 4 (33) |
| Underlying medical conditionsa | 27/59 (46) | 3/7 (43) | 18/40 (45) | 6/12 (50) |
| Complications | ||||
| Pneumonia | 48/57 (84) | 12/13 (92) | 24/32 (75) | 12/12 (100) |
| Respiratory failure | 29/59 (49) | 8/12 (67) | 16/35 (46) | 5/12 (42) |
| ARDS | 26/56 (46) | 7/12 (58) | 15/32 (47) | 4/12 (33) |
| Treatments | ||||
| Oseltamivir treatment | 53/57 (93) | 8/8 (100) | 33/37 (89) | 12/12 (100) |
| Oseltamivir initiation time (days after onset of symptoms): | ||||
| Median (IQR) | 6 (4–8) | 5 (4–7) | 6 (4–9) | 7.5 (6–9) |
| Day 1–2 | 2/39 (5) | 1/7 (14) | 1/20 (5) | 0/12 (0) |
| Day 3–5 | 13/39 (33) | 3/7 (43) | 8/20 (40) | 2/12 (17) |
| Day 6–15 | 24/39 (62) | 3/7 (43) | 11/20 (55) | 10/12 (83) |
| Corticosteroid treatment | 35/56 (63) | 8/12 (67) | 20/34 (59) | 7/10 (70) |
| Mechanical ventilation | 22/54 (41) | 5/8 (63) | 12/34 (35) | 5/12 (42) |
| ICU admission | 25/56 (45) | 3/10 (30) | 15/34 (44) | 7/12 (58) |
| Clinical course | ||||
| Hospitalization | 67/67 (100) | 14/14(100) | 41/41 (100) | 12/12 (100) |
| Median (IQR) time (days) from symptom onset to hospital admission | 4 (3–6) | 3 (1–6) | 4 (3–6) | 5 (2–6) |
| Median (IQR) time (days) of hospitalization | 21 (14–31) | 17 (13–28) | 24 (13–21) | 23 (14–33) |
| Death | 4/67 (6) | 4/14 (29) | 0/41 (0) | 0/12 (0) |
Data are no. (%) of patients, unless otherwise indicated. IQR=interquartile range. ARDS=acute respiratory distress syndrome.
Includes chronic cardiovascular disease (17), type 1 or 2 diabetes (6), chronic respiratory disease (2), neurological disease (2), chronic liver disease (1), chronic renal disease (1), and anemia (1), these underlying medical conditions are not mutually exclusive.
Specificity of antibody responses to A(H7N9) virus infection
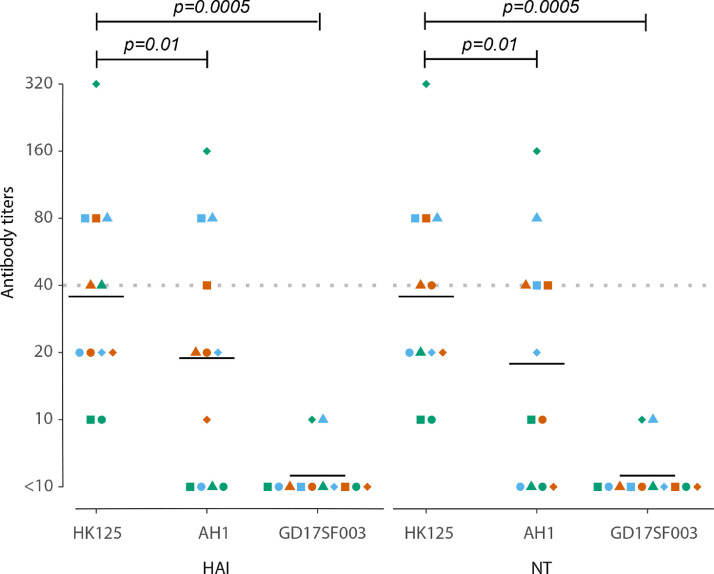
Antigenic properties of A(H7N9) viruses were evaluated by testing antibody responses against homologous A(H7N9) virus (A/Hong Kong/125/2017) and heterologous A(H7N9) viruses (A/Anhui/1/2013 and A/Guangdong/17SF003/2016) in serum samples collected 16–20 months after illness onset from patients from the 2016–2017 epidemic. These sera reacted with A/Anhui/1/2013 virus at lower titers than with A/Hong Kong/125/2017 virus, with HAI or neutralizing antibody titers that were 2- to 8-fold lower in 10 of 12 samples tested (Supplementary Table 1), and HAI and neutralizing geometric mean antibody titers (GMTs) were 47% and 50% lower, respectively (Fig. 2 ). Convalescent sera of patients from the 2016–2017 epidemic were non-reactive to the A/Guangdong/17SF003/2016 virus (Fig. 2 and Supplementary Table 1).
Fig. 2.
Antibody responses to homologous and heterologous A(H7N9) viruses. Hemagglutinin inhibition (HAI) antibody titers (left panel) and neutralizing antibody titers (right panel) against three distinct A(H7N9) viruses in 12 serum samples collected 16–20 months after illness onset from 12 patients from the 2016–2017 epidemic. A/Hong Kong/125/2017 (HK125) is a homologous virus strain, A/Anhui/1/2013 (AH1) and A/Guangdong/17SF003/2016 (GD17SF003) are heterologous virus strains. Triangles, circles and squares show results for each sample, each combination of symbol and color represents one serum sample. Black lines show geometric mean antibody titers (GMTs) of each column. The Wilcoxon matched-pairs signed-rank test was used for pairwise analyses.
Antibody response dynamics during hospitalization and after recovery in A(H7N9) patients
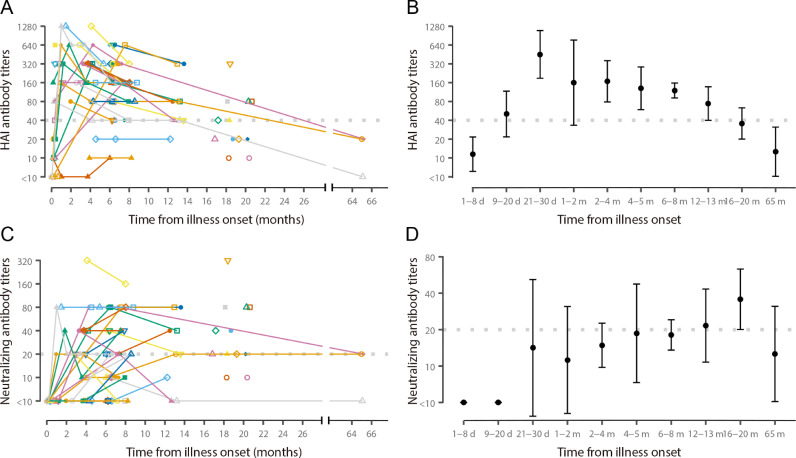
For 18 patients with serum samples collected during hospitalization, 13 (72%) developed an HAI antibody titer ≥40 during hospitalization at a median of 12 days (range 5–36 days) after illness onset; 3 (17%) patients who did not have HAI antibody titer ≥40 during hospitalization developed HAI antibody titers ≥40 after discharge, at a median of 221 days (range 126–227 days) after illness onset; and HAI antibody titers for 2 (11%) patients remained ≤10, even after discharge (on day 20 in one patient, and on days 31, 112 and 180 in another patient) (Fig. 3 (A)). A 4-fold or greater rise in HAI antibody titer was noted in seven of nine patients with paired serum samples collected during hospitalization, which occurred between a median of 5 days after illness onset for the initial specimen (range 1–13 days) and a median of 29 days (range 15 to 55 days) after illness onset for the 2nd specimen. Serum specimens collected from 4 critically ill patients with fatal outcomes had HAI antibody titers ≥40 on days 10–12 after illness onset. HAI antibody titers and neutralizing antibody titers during hospitalization and after recovery were lower in 4 moderately ill patients not treated with oseltamivir compared with 53 patients who received oseltamivir treatment, including 28 who were severely ill (Supplementary Fig. 3).
Fig. 3.
Hemagglutinin inhibition (HAI) antibody titers and neutralizing antibody titers over time in patients with A(H7N9) virus infection. (A) HAI antibody titers and (C) neutralizing antibody titers at different times after illness onset for each patient. Triangles, circles and squares show results for each sample, each combination of symbol and color represents an individual, points from the same patient are connected. (B) geometric mean HAI antibody titers and (D) geometric mean neutralizing antibody titers at indicated periods after illness onset. The dots represent geometric mean titers (GMTs) and the lines represent 95% confidence intervals (CI) for GMTs. Data from 67 participants and 128 blood samples are included in all panels, HAI and neutralizing antibody titers against A/Anhui/1/2013 virus are used for samples from patients from the 2013 and 2013–2014 epidemics, and HAI and neutralizing antibody titers against A/Hong Kong/125/2017 virus are used for samples from patients from the 2016–2017 epidemic. Gray dashed line indicates threshold for seropositive titer (HAI antibody titer ≥ 40, and neutralizing antibody titer ≥ 20).
After illness onset, HAI GMTs reached ≥40 within 9–20 days, peaked at 21–30 days with a GMT of 452.5, and persisted above 40 until 16–20 months (Fig. 3(B)). In contrast, increases of neutralizing antibody titers were delayed (Fig. 3(C) and 3(D)). Applying a cutoff value of 40, the proportion of seropositive with HAI antibodies was 58% (95% CI: 25.6–91.1) at 9–20 days after illness onset, and was greater than 89% thereafter until one year after illness onset (Supplementary Fig. 4A). Neutralizing antibodies were undetectable until a month after illness onset, and applying a cutoff value of 20, the seropositive rate increased from 40% (95% CI: 12–74) at 21–60 days to 83% (95% CI: 52–98) at 16–20 months after illness onset (Supplementary Fig. 4B). HAI antibody titers correlated with neutralizing antibody titers (rho = 0.56, p <0.01, indicating a moderately positive correlation) (Supplementary Fig. 5A). The correlation coefficient of HAI antibody titers and neutralizing antibody titers for A/Anhui/1/2013 (rho=0.64, moderately positive correlation) was lower than observed for A/Hong Kong/125/2017 (rho=0.93, strongly positive correlation) (Supplementary Figs. 5B and 5C). HAI antibody titers correlated with neutralizing antibody titers for each antigen tested for sera from patients from the 2016–2017 epidemic (rho=0.91 for A/Anhui/1/2013, rho=0.93 for A/Hong Kong/125/2017, and rho=1 for A/Guangdong/17SF003/2016, all strongly positive correlations).
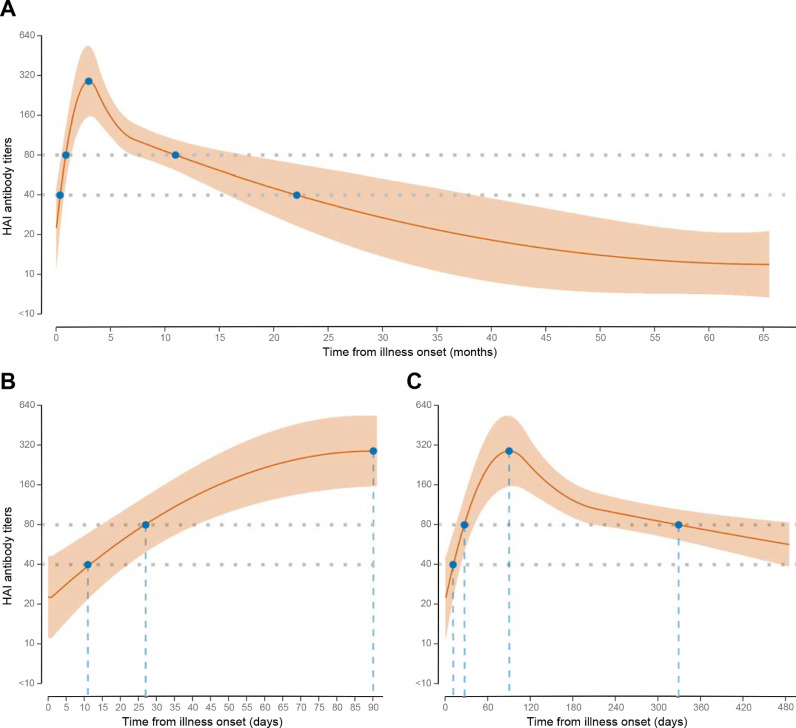
According to our model, the mean HAI antibody level reached a titer of 40 on day 11 and 80 on day 27 after illness onset (Fig. 4 (A)), peaked after three months at a GMT of 290 (Fig. 4(B)), and then declined to a titer of 80 (month 11) and 40 (month 22) (Fig. 4(A) and (C)). Neutralizing antibody titers increased slower than HAI antibody titers, reached a small peak on day 103 at a GMT of 17, decreased slightly, and then continued to increase until 35 months after illness onset, while HAI antibody titers decreased throughout this period (Supplementary Fig. 6).
Fig. 4.
Dynamic of hemagglutinin inhibition (HAI) antibody titers in patients with A(H7N9) virus infection. A, average curve covers the whole study period (0–65 months after illness onset); B, curve between 0 and 90 days after illness onset; and C, curve for 0–480 days after illness onset are shown. Dark orange lines: average HAI antibody curve; light orange zones: 95% confidence interval around average antibody curve. Gray dash lines: threshold titers at 40 and 80.
Association between clinical outcomes and antibody responses
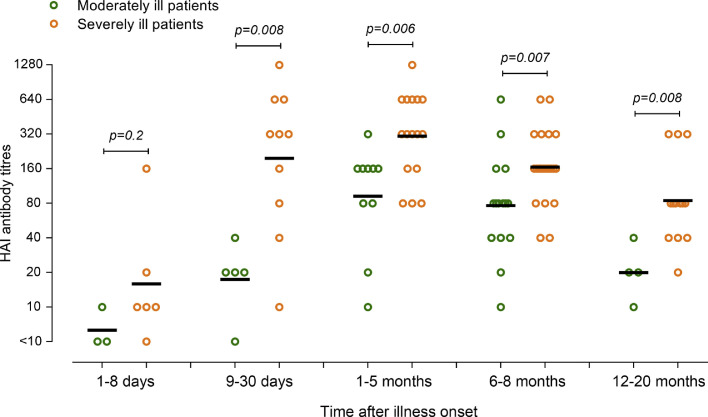
From 9 days to 20 months after illness onset, HAI antibody titers in severely ill patients who progressed to ARDS or required mechanical ventilation were significantly higher in unadjusted analyses (Fig. 5 ), and in the model adjusted for sex, age, and time of sampling from illness onset (p = 0.007). Association between other factors and clinical outcome were also analyzed. Age older than 65 years and corticosteroid treatment were significantly associated with severe illness in the univariate analyses, while only corticosteroid treatment was associated with severe illness in multivariable analyses (Supplementary Table 2).
Fig. 5.
Relationship between hemagglutinin inhibition (HAI) antibody response and disease severity. Serum samples were grouped by time of sampling (1–8 days, 9–30 days, 1–5 months, 6–8 months, and 12–20 months after illness onset), and clinical outcomes [moderately ill: patients without ARDS and did not receive mechanical ventilation (n = 25), severely ill: patients progressed to ARDS or required invasive mechanical ventilation (n = 32)]. HAI antibody responses between moderately ill and severely ill patients were compared by using Mann–Whitney U test.
Discussion
In this study, patients hospitalized with virologically-confirmed A(H7N9) virus infections during the 2013, 2013–2014, and 2016–2017 epidemics mounted strong serum HAI antibody responses against A(H7N9) virus during hospitalization and after recovery. We estimated that mean A(H7N9) virus-specific HAI antibody titers peaked three months after illness onset and remained above 80 for 11 months. Severely ill patients mounted higher antibody responses during hospitalization that persisted for a long period after recovery. Therefore, studies of convalescent plasma treatment for A(H7N9) patients should consider collection of donor plasma from survivors of severe disease between 1 and 11 months after illness onset.
The kinetics of A(H7N9) virus-specific HAI antibody responses during hospitalization in these patients were similar to those described in other studies of A(H7N9) patients.33 , 34 In this cohort, most patients who provided blood samples in hospital had HAI antibody titers ≥40 during hospitalization. In the study by Ma et al., 2 of 25 patients did not have an HAI antibody titer ≥40 during hospitalization or after recovery.20 Therefore, most, but not all patients with A(H7N9) virus infection who survive severe disease are able to mount detectable HAI antibody levels. The differences in peak time of HAI antibody titers between analyses of GMTs and model analysis might be due to inadequate sampling. The contacts of these virologically-confirmed cases were not investigated by serology in this study. No patient had serologic evidence of A(H7N9) virus re-infection after recovery. Epidemiological investigations of patients who survived A(H7N9) virus infections across five epidemics by our group also found no reoccurrence of A(H7N9) related-illness1; re-infection of A(H7N9) virus has not been reported in other longitudinal sero-epidemiology studies.35 , 36
Our analysis estimated that mean HAI antibody titers remained ≥40 until 22 months after illness onset. Ma et al. reported that HAI GMTs declined to lower than 40 at 10 months after illness onset.20 However, they used a 2013 human A(H7N9) virus isolate to test serum samples from patients from the 2016–2017 epidemic, which might result in an underestimate of antibody titers. In another cross-sectional study, Ma et al. reported that HAI GMT was near 40 at about 26 months after illness onset, which was similar to our findings.21
For the pooled analyses of antibody kinetics, data for antigens that were antigenically-matched to circulating strains are preferred. However, there might be antigen-specific differences in HAI titers, which could affect the results. We adopted two statistical approaches to investigate this question: (1) we evaluated the main effect and interactive effect of each antigen in the model and found that the effects were not statistically significant; (2) we constructed the model with the HAI results for A/Anhui/1/2013 virus only, with the same formula and knots as the model for pooled analysis (Supplementary Fig. 7). Before 7 months after illness onset, the trend of the average curve of the A/Anhui/1/2013 virus-only model was similar to that of the model with pooled results, and subsequently the former curve decreased slower than the later one. However, due to the lack of sera available between 14 months and 65 months after illness onset for testing in the A/Anhui/1/2013 virus-only model, we viewed the model with pooled data to be more reliable. We did not find that antigen-specific differences in HAI responses affected the model significantly, but this could be due to small sample size and additional investigations are needed when more data are available for the different antigens.
In our study, the small number of cases with fatal outcomes precluded statistical comparisons of antibody titers of fatal cases vs. nonfatal cases. For A(H7N9) virus-specific antibody responses in patients during hospitalization, Zhang et al. reported that neutralizing antibody titers increased faster in survivors than in fatal cases34; however, no statistically significant difference between antibody titers for fatal vs. nonfatal cases was found in another patient cohort.37 For A(H7N9) virus-specific antibody responses after discharge in survivors, Ma et al. found no association between disease severity and antibody response in one study,20 while reported that severe disease was associated with higher HAI titer after discharge in another study.21 Antibody responses to A(H5N1) virus infection after recovery have been reported to be lower in patients with asymptomatic infection than in those with severe illness.30 Previous studies in A(H7N9) virus infected patients indicate that effective cross-reactive T cell memory is important for protection against severe influenza disease caused by newly emerging influenza A viruses.38, 39, 40 Therefore, lack of sufficient T cell response might result in prolonged viral replication and cytokine dysregulation, more severe disease and higher antibody responses in most patients.
HPAI A(H7N9) viruses are antigenically distinct from other LPAI A(H7N9) viruses, including A/Hunan/2650/2016 and A/Anhui/1/2013, as determined by using post-infection ferret antisera.7 Lu et al. reported that sera collected 2–3 weeks after illness onset from patients with LPAI A(H7N9) clade C1 virus infection reacted with HPAI A(H7N9) human isolates, although at lower titers than those against homologous virus.26 Since we lacked virological data to identify the specific virus strains infecting the patients, we tested serum samples from patients from the 2016–2017 epidemic against three distinct A(H7N9) virus strains. Those patients with positive serological responses were likely infected by LPAI A(H7N9) viruses. Convalescent sera from these patients collected at 16–20 months after illness did not react with HPAI A(H7N9) virus. These data from human samples verified the antigenic difference among distinct A(H7N9) viruses.
Development of neutralizing antibody responses was delayed compared to HAI antibody responses, and neutralizing antibody titers continue to increase for longer periods after recovery. Zhang et al. reported increase of neutralizing antibody response within 2 weeks from illness onset by using a pseudovirus-based neutralization assay.34 However, results of pseudovirus-based neutralization assay might not correlate well with results of an authentic virus-based neutralization test.41 In the study by Guo et al., no neutralizing antibodies were detected in samples collected before day 28 after illness onset, which is consistent with our findings.31 Ma et al. also observed delayed neutralizing antibody response after infection, with GMTs increasing overtime, peaking on day 200.20 We observed discordance in H7N9 virus HAI and neutralizing antibody responses with the HAI antibody response decreasing after a peak at 3 months while the neutralizing antibody response increased long after clinical recovery. The correlation of HAI and neutralizing antibody responses varied by A(H7N9) virus antigen, with greater correlation for the more recent A/Hong Kong/125/2017 virus compared with the older antigenically distinct A/Anhui/1/2013 virus. A(H7N9) vaccines induce comparable levels of HA-reactive IgG antibody as seasonal influenza vaccines in mice, but neutralizing titers are low, suggesting a large component of the induced antibody response is non-neutralizing42; this phenomenon might be attributed to the internal genes of A(H7N9) virus.32 Nevertheless, both neutralizing and non-neutralizing antibodies induced by A(H7N9) vaccination of humans protected mice in vivo against a stringent lethal challenge.43 Therefore, neutralizing antibody titers may underestimate the protective effect of antibody responses following A(H7N9) virus infection.
Convalescent plasma therapy has shown promise for the treatment of a variety of emerging infectious diseases,44 including 1918 H1N1 pandemic influenza,14 influenza A(H5N1),17 , 18 influenza A(H1N1)pdm09,45 and severe acute respiratory syndrome (SARS).46 Timely treatment of patients with severe A(H1N1)pdm09 illness with hyperimmune IV immunoglobulin (H-IVIG) reduced mortality.47 The time-to-treatment effect requires timely availability of virus specific convalescent plasma with high antibody titers. It has been shown that implementation of a passive-immunotherapy program is logistically feasible during an influenza pandemic.48 Kreil et al. demonstrated the feasibility of producing H-IVIG preparation at large scale relatively rapidly, which could be further improved by reducing low-titer donations.49 , 50 To be used for convalescent plasma transfusion or H-IVIG production, a HAI antibody titer of ≥80 in donor plasma is needed,15 and a dose of 0.5 g/kg would be needed for H-IVIG treatment.49 Up to three plasmapheresis donations with donation of 500 ml plasma each time could produce enough plasma for the treatment of two to three patients.15 , 51
The median duration of A(H7N9) viral shedding was 15.5 days from illness onset,52 and nonfatal A(H7N9) patients recovered at a median of 26 days,53 which indicates that convalescent plasma could be collected a month after illness onset. Collecting blood from survivors of severe illness from A(H7N9) virus infection at 1 to 11 months after illness onset should yield convalescent plasma with HAI antibody titer above 80, suitable for convalescent plasma therapy and H-IVIG production. Among the patients enrolled in this study, 64% (18/28) of the severely ill survivors were aged 18–65 years old, making them suitable potential blood donors for plasma therapy.54 Another potential source of convalescent plasma is from participants in clinical trials of A(H7N9) vaccines.55 However, since A(H7N9) viruses continue to evolve into antigenically distinct strains, regardless of the source of the plasma, it will be important to determine the levels of cross-reactive antibodies in the convalescent plasma against a patient's virus infection to understand whether treatment is likely to be beneficial. Furthermore, administration of convalescent plasma treatment for patients with A(H7N9) virus infection should ideally be done through a controlled clinical trial.
Our study had several limitations. First, all participants had symptomatic disease requiring hospitalization. The magnitude and kinetics of the antibody responses in patients with milder illness or asymptomatic infections might be different. Second, clinical data were not available for all patients during their hospitalization, which might impact some analyses. Third, we were not able to collect serial serum specimens from patients during hospitalization and were limited to testing remaining available sera. Survivors were not all followed up at the same designated time points. Fourth, we did not have respiratory specimens to characterize the A(H7N9) viruses that infected the patients, or to determine viral load, duration of shedding and associations with disease severity and antibody responses. Finally, the A(H7N9) patients followed up in this study may not be representative of all survivors of A(H7N9) virus infection in China; however, we included patients from 6 provinces and from 3 epidemics since 2013.
In conclusion, we have established the largest cohort of A(H7N9) survivors to date, followed them for up to 5 years and determined the specificity, kinetics and longevity of A(H7N9) virus-specific antibodies. If convalescent plasma treatment is pursued in clinical trials, our findings indicate that plasma could be prepared from blood from survivors of severe disease with homologous A(H7N9) virus strain-specific infection collected approximately 1–11 months after illness onset.
CRediT authorship contribution statement
Junbo Chen: Data curation, Formal analysis, Writing - original draft. Huachen Zhu: Data curation, Writing - review & editing. Peter W. Horby: Conceptualization, Writing - original draft. Qianli Wang: Data curation. Jiaxin Zhou: Formal analysis. Hui Jiang: . Liwei Liu: . Tianchen Zhang: . Yongli Zhang: . Xinhua Chen: Data curation. Xiaowei Deng: Formal analysis. Birgit Nikolay: . Wei Wang: Formal analysis. Simon Cauchemez: Writing - review & editing. Yi Guan: Funding acquisition, Writing - review & editing. Timothy M. Uyeki: Writing - original draft. Hongjie Yu: Conceptualization, Funding acquisition, Supervision, Writing - original draft.
Declaration of Competing Interest
H. Y. has received investigator-initiated research funding from Sanofi Pasteur, GlaxoSmithKline, and Yichang HEC Changjiang Pharmaceutical Company. None of those research funds are related to research on human infections with avian influenza A viruses. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
We thank staff of the Centers for Disease Control and Prevention in Beijing, Shandong, Zhejiang, Hunan, Jiangxi, and Fujian provinces for providing assistance in patient recruitment, administration and data collection. The findings and conclusions reported here are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
Funding
This work was supported by grants from the National Science Fund for Distinguished Young Scholars (81525023), Program of Shanghai Academic/Technology Research Leader (18XD1400300), National Science and Technology Major Project of China (2017ZX10103009-005, 2018ZX10201001-010), National Key Plan (2016YFD0500302, 2017YFE0190800) and the Shenzhen Peacock Program (KQTD201203). BN and SC acknowledge financial support from the AXA Research Fund, the Investissement d'Avenir program, the Laboratoire d'Excellence Integrative Biology of Emerging Infectious Diseases program (Grant ANR-10-LABX-62-IBEID) and the Models of Infectious Disease Agent Study of the National Institute of General Medical Sciences. The funding bodies had no role in study design, data collection and analysis, preparation of the manuscript, or the decision to publish.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2019.11.024.
Appendix. Supplementary materials
References
- 1.Wang X., Jiang H., Wu P., Uyeki T.M., Feng L., Lai S. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013–17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17:822–832. doi: 10.1016/s1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao G.F. From "A"IV to "Z"IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu D., Xiang G., Zhu W., Lei X., Li B., Meng Y. The re-emergence of highly pathogenic avian influenza H7N9 viruses in humans in Mainland China, 2019. Euro Surveill Bull Eur Maladies Transm = Eur Commun Dis Bull. 2019:24. doi: 10.2807/1560-7917.es.2019.24.21.1900273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Record. 2019;94:151–160. [Google Scholar]
- 5.Kile J.C., Ren R., Liu L., Greene C.M., Roguski K., Iuliano A.D. Update: increase in human infections with novel Asian lineage avian influenza A(H7N9) viruses during the fifth epidemic – China, October 1, 2016-August 7, 2017. MMWR Morb Mortal Wkly Rep. 2017;66:928–932. doi: 10.15585/mmwr.mm6635a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Yang L., Zhu W., Zhang Y., Zou S., Bo H. Two outbreak sources of influenza a (H7N9) viruses have been established in china. J Virol. 2016;90:5561–5573. doi: 10.1128/jvi.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Zoonotic influenza viruses: antigenic and genetic characteristics and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec. 2017;92:129–144. [PubMed] [Google Scholar]
- 8.Shi J., Deng G., Kong H., Gu C., Ma S., Yin X. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017;27:1409–1421. doi: 10.1038/cr.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Wong G., Yang L., Tan S., Li J., Bai B. Comparison between human infections caused by highly and low pathogenic H7N9 avian influenza viruses in wave five: clinical and virological findings. J Infect. 2019;78:241–248. doi: 10.1016/j.jinf.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Shi J., Deng G., Ma S., Zeng X., Yin X., Li M. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe. 2018;24:558–568. doi: 10.1016/j.chom.2018.08.006. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Summary of influenza risk assessment tool (IRAT) results. Available at:https://www.cdc.gov/flu/pandemic-resources/monitoring/irat-virus-summaries.htm. Accessed 1 July 2019.
- 12.Zheng S., Tang L., Gao H., Wang Y., Yu F., Cui D. Benefit of early initiation of neuraminidase inhibitor treatment to hospitalized patients with avian influenza A(H7N9) virus. Clin Infect Dis Off Publ Infect Dis Soc Am. 2018;66:1054–1060. doi: 10.1093/cid/cix930. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y., Lu S., Song Z., Wang W., Hao P., Li J. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381:2273–2279. doi: 10.1016/s0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 14.Luke T.C., Kilbane E.M., Jackson J.L., Hoffman S.L. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann Intern Med. 2006;145:599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 15.Beigel J.H., Tebas P., Elie-Turenne M.C., Bajwa E., Bell T.E., Cairns C.B. Immune plasma for the treatment of severe influenza: an open-label, multicentre, phase 2 randomised study. Lancet Respir Med. 2017;5:500–511. doi: 10.1016/s2213-2600(17)30174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X.X., Gao H.N., Wu H.B., Peng X.M., Ou H.L., Li L.J. Successful treatment of avian-origin influenza A (H7N9) infection using convalescent plasma. Int J Infect Dis IJID: Off Publ Int Soc Infect Dis. 2015;41:3–5. doi: 10.1016/j.ijid.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Yu H., Gao Z., Feng Z., Shu Y., Xiang N., Zhou L. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One. 2008;3:e2985. doi: 10.1371/journal.pone.0002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Feng Z., Shu Y., Yu H., Zhou L., Zu R. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet. 2008;371:1427–1434. doi: 10.1016/s0140-6736(08)60493-6. [DOI] [PubMed] [Google Scholar]
- 19.Lee N., Hui D.S.C. Potential and challenges of serotherapy for severe influenza. Lancet Respir Med. 2017;5:e27. doi: 10.1016/s2213-2600(17)30266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma M.J., Liu C., Wu M.N., Zhao T., Wang G.L., Yang Y. Influenza A(H7N9) virus antibody responses in survivors 1 year after infection, China, 2017. Emerging Infect Dis. 2018;24 doi: 10.3201/eid2404.171995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma M.J., Wang X.X., Wu M.N., Wang X.J., Bao C.J., Zhang H.J. Characterization of antibody and memory T-cell response in H7N9 survivors: a cross-sectional analysis. Clin Microbiol Infect. 2019 doi: 10.1016/j.cmi.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiore A.E., Shay D.K., Broder K., Iskander J.K., Uyeki T.M., Mootrey G., et al. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. Morb Mortal Wkly Rep Recommendations and Reports 2009; 58:1–52. [PubMed]
- 23.Zhao M., Chen J., Tan S., Dong T., Jiang H., Zheng J. Prolonged evolution of virus-specific memory T cell immunity post severe avian influenza A (H7N9) virus infection. J Virol. 2018 doi: 10.1128/jvi.01024-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO Zoonotic influenza viruses: antigenic and genetic characteristics and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec. 2017;92:633–648. [PubMed] [Google Scholar]
- 25.WHO Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec. 2014;89:457–464. [PubMed] [Google Scholar]
- 26.Lu J., Raghwani J., Pryce R., Bowden T.A., Theze J., Huang S. Molecular evolution, diversity, and adaptation of influenza A(H7N9) viruses in China. Emerging Infect Dis. 2018;24:1795–1805. doi: 10.3201/eid2410.171063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. WHO manual on animal influenza diagnosis and surveillance. 2002; Available at:http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf. Accessed 1 July 2019.
- 28.Coudeville L., Bailleux F., Riche B., Megas F., Andre P., Ecochard R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med Res Methodol. 2010;10:18. doi: 10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong L., Bo H., Bai T., Gao R., Dong J., Zhang Y. A combination of serological assays to detect human antibodies to the avian influenza A H7N9 virus. PLoS One. 2014;9:e95612. doi: 10.1371/journal.pone.0095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchy P., Vong S., Chu S., Garcia J.M., Hien T.T., Hien V.M. Kinetics of neutralizing antibodies in patients naturally infected by H5N1 virus. PLoS One. 2010;5:e10864. doi: 10.1371/journal.pone.0010864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L., Zhang X., Ren L., Yu X., Chen L., Zhou H. Human antibody responses to avian influenza A(H7N9) virus, 2013. Emerging Infect Dis. 2014;20:192–200. doi: 10.3201/eid2002.131094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee A.C., Zhu H., Zhang A.J., Li C., Wang P., Li C. Suboptimal humoral immune response against influenza A(H7N9) virus is related to its internal genes. Clin Vacc Immunol: CVI. 2015;22:1235–1243. doi: 10.1128/cvi.00443-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S., Chen Y., Cui D., Yao H., Lou J., Huo Z. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: a serological study. J Infect Dis. 2014;209:265–269. doi: 10.1093/infdis/jit430. [DOI] [PubMed] [Google Scholar]
- 34.Zhang A., Huang Y., Tian D., Lau E.H., Wan Y., Liu X. Kinetics of serological responses in influenza A(H7N9)-infected patients correlate with clinical outcome in China, 2013. Euro Surveill Bull Eur Mal Transm = Eur Commun Dis Bull. 2013;18:20657. doi: 10.2807/1560-7917.es2013.18.50.20657. [DOI] [PubMed] [Google Scholar]
- 35.Yang P., Ma C., Cui S., Zhang D., Shi W., Pan Y. Avian influenza A(H7N9) and (H5N1) infections among poultry and swine workers and the general population in Beijing, China, 2013–2015. Sci Rep. 2016;6:33877. doi: 10.1038/srep33877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma M.J., Zhao T., Chen S.H., Xia X., Yang X.X., Wang G.L. Avian influenza A virus infection among workers at live poultry markets, China, 2013–2016. Emerging Infect Dis. 2018;24:1246–1256. doi: 10.3201/eid2407.172059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman G., Cowling B.J. Serological responses following influenza A(H7N9) virus infection. J Infect Dis. 2014;209:2018–2019. doi: 10.1093/infdis/jiu021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Wan Y., Qiu C., Quinones-Parra S., Zhu Z., Loh L. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun. 2015;6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., Zhu L., Nguyen T.H.O., Wan Y., Sant S., Quinones-Parra S.M. Clonally diverse CD38(+)HLA-DR(+)CD8(+) T cells persist during fatal H7N9 disease. Nat Commun. 2018;9:824. doi: 10.1038/s41467-018-03243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Li X., Tian L., Zheng S., Yang S., Dong Y. Dynamic behavior of lymphocyte subgroups correlates with clinical outcomes in human H7N9 infection. J Infect. 2014;69:358–365. doi: 10.1016/j.jinf.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Wang W., Peng H., Tao Q., Zhao X., Tang H., Tang Z. Serologic assay for avian-origin influenza A (H7N9) virus in adults of Shanghai, Guangzhou and Yunnan, China. J Clin Virol: Off Publ Pan Am Soc Clin Virol. 2014;60:305–308. doi: 10.1016/j.jcv.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Kamal R.P., Blanchfield K., Belser J.A., Music N., Tzeng W.P., Holiday C. Inactivated H7 influenza virus vaccines protect mice despite inducing only low levels of neutralizing antibodies. J Virol. 2017;91 doi: 10.1128/jvi.01202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henry Dunand C.J., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casadevall A., Dadachova E., Pirofski L.A. Passive antibody therapy for infectious diseases. Nat Rev Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 45.Hung I.F., To K.K., Lee C.K., Lee K.L., Chan K., Yan W.W. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2011;52:447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y., Wong R., Soo Y.O., Wong W.S., Lee C.K., Ng M.H. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis: Off Publ Eur Soc Clin Microbiol. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung I.F.N., To K.K.W., Lee C.K., Lee K.L., Yan W.W., Chan K. Hyperimmune IV immunoglobulin treatment: a multicenter double-blind randomized controlled trial for patients with severe 2009 influenza A(H1N1) infection. Chest. 2013;144:464–473. doi: 10.1378/chest.12-2907. [DOI] [PubMed] [Google Scholar]
- 48.Wu J.T., Lee C.K., Cowling B.J., Yuen K.Y. Logistical feasibility and potential benefits of a population-wide passive-immunotherapy program during an influenza pandemic. Proc Natl Acad Sci USA. 2010;107:3269–3274. doi: 10.1073/pnas.0911596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreil T.R., Mc Vey J.K., Lei L.S., Camacho L., Wodal W., Kerschbaum A. Preparation of commercial quantities of a hyperimmune human intravenous immunoglobulin preparation against an emerging infectious disease: the example of pandemic H1N1 influenza. Transfusion. 2012;52:803–809. doi: 10.1111/j.1537-2995.2011.03347.x. [DOI] [PubMed] [Google Scholar]
- 50.Khalenkov A., He Y., Reed J.L., Kreil T.R., McVey J., Norton M. Characterization of source plasma from self-identified vaccinated or convalescent donors during the 2009 H1N1 pandemic. Transfusion. 2018;58:1108–1116. doi: 10.1111/trf.14530. [DOI] [PubMed] [Google Scholar]
- 51.Wong H.K., Lee C.K., Hung I.F., Leung J.N., Hong J., Yuen K.Y. Practical limitations of convalescent plasma collection: a case scenario in pandemic preparation for influenza A (H1N1) infection. Transfusion. 2010;50:1967–1971. doi: 10.1111/j.1537-2995.2010.02651.x. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y., Guo Q., Yan Z., Zhou D., Zhang W., Zhou S. Factors associated with prolonged viral shedding in patients with avian influenza A(H7N9) virus infection. J Infect Dis. 2018;217:1708–1717. doi: 10.1093/infdis/jiy115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y., Zhong H., Song T., He J., Guo L., Tan X. Epidemiological and clinical characteristics of humans with avian influenza A (H7N9) infection in Guangdong, China, 2013-2017. Int J Infect Dis IJID: Off Publ Int Soc Infect Dis. 2017;65:148–155. doi: 10.1016/j.ijid.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 54.World Health Organization. Blood donor selection: guidelines on assessing donor suitability for blood donation. 2012; Available at:http://www.who.int/iris/handle/10665/76724. Accessed 1 July 2019. [PubMed]
- 55.Jackson L.A., Campbell J.D., Frey S.E., Edwards K.M., Keitel W.A., Kotloff K.L. Effect of varying doses of a monovalent H7N9 influenza vaccine with and without AS03 and MF59 adjuvants on immune response: a randomized clinical trial. JAMA. 2015;314:237–246. doi: 10.1001/jama.2015.7916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.