Abstract
Ti-containing α-Keggin polyoxometalates (POMs) have been proved with properties of both anti-tumor and anti-HIV (human immunodeficiency virus). The potential anti-SARS (severe acute respiratory syndrome) activity of the POMs [α-PTi2W10O40]7− isomers was investigated in this paper by molecular modeling method. The SARS 3c like protease, namely the SARS 3CLpro is the key function protease for virus replication as well as transcription and thus can be taken as one of the key targets for anti-SARS drug design. Affinity/Insight II was used to explore possible binding locations for POMs/3CLpro interaction. Charges in the POMs were obtained from density-functional theory (DFT) method. The results show that POMs bind with 3CLpro in the active site region with high affinity; POMs are more prone to bind with 3CLpro than with some organic compounds; for the POMs/3CLprocomplex, the OTi2 in POMs is the vital element for electrostatic interaction, and the electrostatic binding energy is strong enough to keep the complex stable.
Keywords: SARS, 3CLpro, Molecular dynamics, Docking, Polyoxometalates (POMs)
1. Introduction
Severe acute respiratory syndrome (SARS), a suddenly happened worldwide pandemic in 2003, was suffered by thousands of people and killed hundreds of them. Coronavirus was identified as the etiologic agent of SARS [1], [2]. SARS coronavirus (SARS-CoV) is a positive strand RNA virus. The plasmid of SARS-CoV includes six kinds of proteins: nucleocapsid (N), spike (S), membranes (M), small envelope (E), polymerase and 3c like (3CL) hydrolytic protease. There are 18 complete genome sequences from different resources that have been determined. Sequence analysis show these genomes are different but not too much, especially its main protease, the 3CL protease(3CLpro) is relatively conserved in the SARS genome and thus can be taken as one of the key targets for anti-SARS drug design [1], [2], [3], [4], [5].
The 3CLpro is the key protein during virus replication; its main function is to hydrolyze the polymerase expressed by the virus. In SARS-CoV the nonstructural gene (namely the enzyme copy gene) code for two overlapped polypeptides: pp1a (486 kDa) and ORF1b (790 kDa). They conduct almost all functions needed during the course of virus replication as well as transcription. Polypeptides with different functions are mostly produced from these two polypeptides and this kind of crack procedure is just mainly dependent on the 3CLpro [3], [4]. The catalytically active site of SARS-CoV 3CLpro has been shown to be Cys145/His41 [2], [6], [7], which is conserved in this protein. Therefore, designing drugs for this 3CLpro should inhibit SARS infection effectively.
Polyoxometalates (POMs) are oxygen rich class of inorganic cluster systems exhibiting remarkable chemical and physical properties, which have been applied to various fields such as catalysis, materials and medicines [8], [9], [10]. POMs perform significant biological activities, with high efficacy and low toxicity. In particular, the sizes and globular structural motifs of many POMs are similar, and in some cases nearly identical to some water-soluble fullerene derivatives showing fairly good anti-HIV-1P activity [11]. Liu and her co-workers have investigated POMs compounds such as PW, SiW, PMo, and SiMo. Their results show that some POMs possess lower cell virulence and higher anti-HIV activity [8], [12], [13], [14], [15], [16], [17], [18].
Among various POMs, the di-Ti-substituted α-Keggin type structure [α-PTi2W10O40]7− isomers are of particular interest because of their high catalytic activity and capability of inhibiting replication of several types of DNA or RNA viruses. Many papers report that Ti-containing α-Keggin POMs show obvious inhibition activity to HIV-1 [19], [20], [21], [22], [23], [24], [25], [26], [27]. It is very significant to explore the inhibition potency of α-Keggin POMs to SARS-CoV. Firstly, whether POMs interact with the SARS main functional protease of 3CLpro; secondly, what the interaction mechanism should be; and above all, it is of great significance to explore molecular mechanism and molecular dynamics mechanism when studying problems related to POMs. In this investigation, the α-Keggin type POM isomers including [α-1,5-PTi2W10O40]7−, [α-1,6-PTi2W10O40]7−, [α-1,11-PTi2W10O40]7−, [α-1,4-PTi2W10O40]7−, [α-1,2-PTi2W10O40]7− were selected as candidates for SARS-CoV 3CLpro inhibitors. Binding energy was compared, hydrogen bonds formation and electrostatic interaction were analyzed, active site key residues were identified and the interaction mechanism was also discussed at molecular level.
2. Models and methods
2.1. Equilibrated conformation of 3CLpro
The three dimensional crystal structures and the active site information of SARS-CoV 3CLpro were obtained by experimental methods recently [2], [6]. SARS-CoV 3CLpro consists of two promoters nearly perpendicular with each other, and each one of them has three structure domains. Domains I, II are of anti-parallel sheets, while domain O forms an anti-parallel global conformation including five α-helixes.
The crystal structure coordinate of 3CLpro was downloaded from the protein Database (PDB ID: 1UK4, Resolution = 2.5 Å). The 3CLpro is made up of two protomers: protomer A and protomer B, each one of them consists of 306 amino acids [28]. Biopolymer/Insight II module was used to repair the lost coordinates. The side residues were optimized and modified properly to decrease geometry collision with surrounding atoms. Molecular mechanics calculation was performed after molecular dynamics [29], [30], [31], [32]. After the energy converged, the conformation with the lowest energy was taken as the 3CLpro analysis structure. The final equilibrated conformation in solution of SARS 3CLpro is shown in Fig. 1 .
Fig. 1.

The equilibrated conformation in solution of SARS 3CLpro dimer. The catalytic residues of Cys145 and His41 are represented in ball-and-stick.
The SARS-CoV 3CLpro model is evaluated with the Profile-3D/Insight II module [33], and the evaluation result is shown in Fig. 2 . It is clearly shown that most residues are modeled reasonably, which indicates the structure is modeled successfully. Four residues of Arg-A60, Ser-A62, His-A64 and Lys-A236 are poorly built; the reason should be that these hydrophobic residues are not well shielded from solvent in the model. Fortunately, all the four residues locate far away from the enzyme active site, so they should have little influence on subsequent investigations.
Fig. 2.
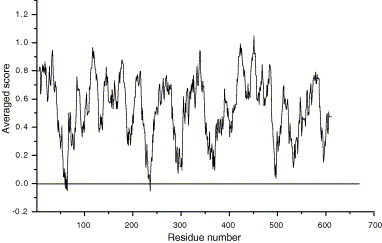
Profile-3D evaluation of the SARS-CoV 3CLpro model.
2.2. Selecting the POMs compounds
Recently, we have systematically investigated the electronic properties, redox properties, protonation, and stability of five POM [α-PTi2W10O40]7− isomers. Our results revealed that the bridging oxygens (OTi2 and OTiW) as well as the terminal oxygens (OTi), act as nucleophilic centers. In particular, the bridging oxygens bond to two titaniums (OTi2) in [α-1,4-PTi2W10O40]7− and [α-1,2-PTi2W10O40]7−exhibit strong nucleophilic activity [26]. The three dimensional structural models of the five [α-PTi2W10O40]7− isomers are shown in Fig. 3 . A short-hand notation is used for these isomers, without oxygen atoms, tungsten atoms and anion charge. (e.g., [α-1,5-PTi2W10-O40]7− = 1,5-PTi2, [α-1,6-PTi2W10O40]7− = 1,6-PTi2, [α-1,11-PTi2W10O40]7− = 1,11-PTi2, [α-1,4-PTi2W10O40]7− = 1, 4-PTi2 and [α-1,2-PTi2W10O40]7− = 1,2-PTi2).
Fig. 3.

The three-dimensional structural models of the five [α-PTi2W10O40]7− POMs.
2.3. Docking study of POMs with SARS-CoV 3CLpro
Charge properties of POMs were obtained from quantum chemical calculation employing the density-functional theory (DFT) method. The parameters for atomic point charges were not available for heavy metal atoms of the tungsten atom and titanium atom in POMs, this may affect the precise computation value of charge distribution and absolute values of binding energy. However, the general interaction position of POMs with 3CLpro should not change because it was based on energy difference instead of the absolute values. An assumption made in this study was that neither the enzyme nor the inhibitors undergo major structural changes on binding. The five POMs of 1,5-PTi2, 1,6-PTi2, 1,11-PTi2, 1,4-PTi2, and 1,2-PTi2 were docked respectively in the active site of SARS 3CLpro. More than ten slightly different docking positions were examined across the Affinity/Insight II [34]. The enzyme–inhibitor complex with the lowest energy was chosen for further examination.
3. Results and discussion
Docking simulations of the five POM isomers of 1,6-PTi2, 1,5-PTi2, 1,11-PTi2, 1,4-PTi2, 1,2-PTi2 with SARS 3CLpro were carried out using Affinity/Insight II, the results are shown in Fig. 4 and Table 1 .
Fig. 4.

The representative structure of POM/SARS-CoV 3CLpro complex. The 3CLpro is represented in cartoon, while binding site and POM are represented in ball-and-stick.
Table 1.
Binding energy between ligand and receptor (KJ/mol)
| EPot | Δ | ENonb | Δ1 | Δ2(Δ + Δ1) | |
|---|---|---|---|---|---|
| 3CLpro | −9728.458 | −13646.040 | |||
| 1,2-PTi2/3CLpro | −9690.838 | 37.620 | −13755.647 | −109.607 | −71.987 |
| 1,4-PTi2/3CLpro | −9689.251 | 39.207 | −13753.616 | −107.576 | −68.369 |
| 1,11-PTi2/3CLpro | −9688.869 | 39.589 | −13748.752 | −102.712 | −63.123 |
| 1,5-PTi2/3CLpro | −9691.295 | 37.163 | −13749.754 | −103.714 | −66.551 |
| 1,6-PTi2/3CLpro | −9691.009 | 37.449 | −13749.227 | −103.187 | −65.738 |
Note: Δ is the energy change originating from the steric hindrance.
Δ1 is the energy change of electrostatic energy.
Δ2 is the total energy change of electrostatic energy plus steric hindrance.
The representative structure of POM/3CLpro complexes is shown in Fig. 4. The results show that all the five POM ligands interact with the 3CLpro receptor just in the active site region including the catalytic residues. Hydrogen bonds are formed between POMs and several key residues including the catalyst residues of 3CLpro His41/Cys145. The ligand–receptor interaction of POMs/3CLpro generally causes energy decreasing and conformation changing; accordingly these changes should influence the enzyme catalytic activity of the SARS-CoV 3CLpro.
Binding energy of the five Ti-containing α-Keggin POMs of 1,5-PTi2, 1,6-PTi2, 1,11-PTi2, 1,4-PTi2 and 1,2-PTi2 with the SARS-CoV 3CLpro receptor are summarized in Table 1. Where “Δ” represents energy difference originated from steric hindrance between POMs and 3CLpro, “Δ 1” represents electrostatic energy changing of the complex, and “Δ 2” is the total energy changing. As is shown in the results that, for all these five ligand–receptor complexes, the electrostatic energy (Δ 1) both decrease remarkably, which indicates that the highly negative charged POM inhibitors interact with the 3CLpro and therefore cause electrostatic energy to decrease. Furthermore, the electrostatic interaction is strong enough to overcome the unfavorable steric effects, therefore results in total negative energy (Δ 2) in all the five complexes. Binding energy is an important criterion for reliable virtual screening. Several researches have performed virtual screening study of SARS-CoV 3CLpro using small molecular like AG7088 as reference molecule [35], [36]. As is shown in our investigation that, binding energy of POM/3CLpro complex is much higher than that of some small organic molecules. This indicates that POMs may even more prone to bind with 3CLpro than small organic molecules. What is more, an important conclusion can be drawn from our study is that, the total binding energies of the five complex systems follow the order of 1,2-PTi2/3CLpro > 1,4-PTi2/3CLpro > 1,5-PTi2/3CLpro > 1,6-PTi2/3CLpro > 1,11-PTi2/3CLpro, which can provide perfectly well interpretation on the relative stability of the five POM/3CLpro complexes.
Relative information about hydrogen bonds interaction is listed in Table 2 . There are eight hydrogen bonds formed in the 1,6-PTi2/3CLpro complex, and six hydrogen bonds in the 1,2-PTi2/3CLpro complex. Moreover, it is clearly shown that the OTi2of POMs contributes much to hydrogen bond formation within all the five ligand-receptor complexes. Our former DFT calculation have shown that in these POM isomers, the charge loading of OTi2 is much stronger than that of other bridging oxygen and terminal oxygen. That should be the reason why the two systems of the 1,2-PTi2/3CLpro complexes and the 1,4-PTi2/3CLpro complex are much more stable than the others. It should be noticed that there are only six hydrogen bonds in 1,11-PTi2/3CLpro and the terminal oxygen also attends hydrogen bonds formation. Our quantum chemistry calculation also proved that the charge loading of the terminal oxygen is smaller than that of OW2. Therefore, it is easy to interpret why the complex of 1,11-PTi2/3CLpro is the least stable one among all the five ligand–receptor complex systems.
Table 2.
Binding site and H-bond formation (H-bond length: Å)
| Complex | 3CLpro residue | Ligand atom | Distance |
|---|---|---|---|
| 1,2-PTi2/3CLpro | A46:O | O11 | 2.29 |
| A145:O | O16 | 2.75 | |
| A164:O | O22 | 1.75 | |
| A41:ND1 | O22 | 2.87 | |
| A41:ND1 | O23 | 1.40 | |
| A41:ND1 | O25 | 2.56 | |
| A189:OE1 | O28 | 0.76 | |
| A26:O | O41 | 1.96 | |
| 1,4-PTi2/3CLpro | A166:O | O6 | 2.04 |
| A41:HE2 | O20 | 2.11 | |
| A166:OE2 | O44 | 2.98 | |
| A41:ND1 | O47 | 2.35 | |
| A143:O | O50 | 1.84 | |
| A145:HN | O50 | 2.47 | |
| 1,11-PTi2/3CLpro | A25:OG1 | O12 | 1.47 |
| A41:O | O12 | 2.96 | |
| A26:O | O19 | 2.51 | |
| A187:O | O22 | 2.92 | |
| A166:O | O27 | 2.86 | |
| A142:OD1 | O42 | 2.51 | |
| 1,5-PTi2/3CLpro | A119:OD1 | O9 | 2.37 |
| A145:HN | O9 | 2.13 | |
| A166:OE1 | O19 | 1.39 | |
| A26:O | O22 | 1.44 | |
| A46:O | O29 | 1.69 | |
| A41:O | O36 | 1.48 | |
| A164:O | O39 | 1.92 | |
| A166:O | O42 | 2.76 | |
| A41:HE2 | O43 | 2.07 | |
| 1,6-PTi2/3CLpro | A25:O | O45 | 2.28 |
| A26:O | O52 | 2.13 | |
| A26:HN | O39 | 0.67 | |
| A41:O | O50 | 2.53 | |
| A41:ND1 | O42 | 2.65 | |
| A41:HE2 | O33 | 1.96 | |
| A44:O | O47 | 2.93 | |
| A142:O | O48 | 2.84 |
In addition, it should be noticed from the hydrogen bonding interaction shown in Fig. 5 that, POMs interact with 3CLpro in the active site region with several positive charged amino residues including the active site catalysis residues. Those key residues for ligand–receptor interaction including Thr26, His41, Cys44, Asn142, Cys145, Clu166, Asp187 and Gln189, are all polarity residues, this make them easy to form hydrogen bonds meshwork and increase the complex stability. What is more, during the course of POMs/SARS-CoV 3CLpro complex formation, the atom charge loading properties and the electrostatic characteristic are very vital elements that keep the enzyme–inhibitor interaction. Therefore, the atom charge loading property and the electrostatic characteristic are two vital elements keeping POM/3CLpro complex stable and deserve paying much more attention when consider POMs/enzyme interactions. POMs interact with 3CLpro in the active site region with obvious electrostatic property compensation adjustment, accordingly it may preclude the hydrolysis of the two polypeptides expressed by SARS virus, and therefore may cut off the approach of virus replication. The kinetics and binding studies, conducted after molecular modeling, are both in remarkable agreement with relative results.
Fig. 5.

Key residues and hydrogen bond interactions of POM/3CLpro complex.
4. Conclusions
Docking investigations on five POM ([α-PTi2W10O40]7−) isomers with SARS-CoV 3CLpro were performed using computer assisted molecular simulation technology. The investigation results show that: (1) POMs interact with the 3CLpro receptor in enzyme active site region, with several polarized residues including the enzyme catalyst residues of His41/Cys145; (2) The total binding energies of the five POM/SARS-CoV 3CLpro complexes lead to the following order of complex stability: 1,2-PTi2/3CLpro > 1,4-PTi2/3CLpro > 1,5-PTi2/3CLpro > 1,6-PTi2/3CLpro > 1,11-PTi2/3CLpro. (3) Electrostatic energy and hydrogen bond interaction contribute much to the enzyme–inhibitor complex formation between POMs and SARS-CoV 3CLpro. The atom charge loading property and electrostatic characteristic are very vital elements that keep the enzyme–inhibitor interaction. In POMs, the electronegative OTi2 oxygen contributes much for electrostatic energy interactions. As a result, those POMs with OTi2 oxygen should form relative stable complex with protein receptors. The simulation results provide important information for POMs/SARS-CoV 3CLpro interaction, and may provide insights for anti-SARS drug design.
5. Abbreviations
- POMs
Polyoxometalates
- HIV
human immunodeficiency virus
- SARS-CoV
SARS coronavirus
- SARS 3CLpro
SARS 3c like protease
- DFT
density-functional theory
- 1,5-PTi2
[α-1,5-PTi2W10O40]7−
- 1,6-PTi2
[α-1,6-PTi2W10O40]7−
- 1,11-PTi2
[α-1,11-PTi2W10O40]7−
- 1,4PTi2
[α-1,4-PTi2W10O40]7−
- 1,2-PTi2
[α-1,2-PTi2W10O40]7−
Acknowledgements
We gratefully acknowledge the financial support from the National Natural Science Foundation of China (Projects 20373009 and 20573016) and Youth Fund of Northeast Normal University (No. 111494017, 20060305). All the work including molecular modeling, molecular dynamics and molecular docking are carried out on SGI GRAPHICS work station in State Key Laboratory of Theoretical and Computational Chemistry of Jilin University, using Insight II molecular simulating software package of MSI, with modules including Biopolymer, Homology, Discover3, Docking/Affinity, and Delphi [37].
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jinorgbio.2006.08.013.
Appendix A. Supplementary data
Supplementary table.
References
- 1.Rota P.A., Oberste M.S., Monroe S.S. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 2.Anand K., Ziebuhr J., Wadhwani P. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 3.Liu H.L., Lin J.C., Yih H. Chem. Phys. Lett. 2005;401:24–29. doi: 10.1016/j.cplett.2004.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris J.S.M., Lai S.T., Poon L.L.M. Lancet. 2004;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan Y.C., Liu H.F., Shih Y.P. Infect. Gene. Evol. 2005;5:261–269. doi: 10.1016/j.meegid.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand K., Palm G.J., Mesters J.R. EMBO. J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poon L.L.M., Leung C.S.W., Tashiro M. Clin. Chem. 2004;50:1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang M.L., Xiao S.X., Yuan Z.R. Res. Appl. 2000;2:621–625. [Google Scholar]
- 9.Erik D.C. Rev. Med. Virol. 2000;10:255–257. [Google Scholar]
- 10.Erik D.C. Med. Res. Rev. 2002;22:531–565. doi: 10.1002/med.10021. [DOI] [PubMed] [Google Scholar]
- 11.Judd D.A., Nettles J.H., Nevins N. J. Am. Chem. Soc. 2001;123:886–897. doi: 10.1021/ja001809e. [DOI] [PubMed] [Google Scholar]
- 12.Liu S.X., Wang L., Wang E.B. J. Chin. Rare. Earth. Soc. 1997;15:59–63. [Google Scholar]
- 13.Chen C.N., Lin C.P.C., Huang K.K. Altern. Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang M.L., Zhao Z.G., Yuan Z.R. J. Biomed. Eng. 2002;2:291–297. [Google Scholar]
- 15.He Y., Lu H., Siddiqui P. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 16.Liu S.X., Li Y.G., Han Z.B. Chem. J. Chin. Univ. 2002;23:778–782. [Google Scholar]
- 17.Zhai H.J., Liu S.X., Li Y.X. Chin. J. Inorg. Chem. 2005;21:1798–1802. [Google Scholar]
- 18.Liu S.X., Wang C.L., Yu M. Acta. Chin. Sinica. 2005;63:1069–1074. [Google Scholar]
- 19.Take Y., Tokutake Y., Inouye Y. Antiv. Res. 1991;15:113–124. doi: 10.1016/0166-3542(91)90029-q. [DOI] [PubMed] [Google Scholar]
- 20.Inouye Y., Fujimoto Y., Sugiyama M. Bio. Pharm. Bull. 1995;18:996–1000. doi: 10.1248/bpb.18.996. [DOI] [PubMed] [Google Scholar]
- 21.Inouye Y., Tokutake Y., Kunihara J. Chem. Pharm. Bull. 1992;40:805–807. doi: 10.1248/cpb.40.805. [DOI] [PubMed] [Google Scholar]
- 22.Rhule J.T., Hill C.L., Judd D.A. Chem. Rev. 1998;98:327–357. doi: 10.1021/cr960396q. [DOI] [PubMed] [Google Scholar]
- 23.Witvrouw M., Weigold H., Pannecouque C. J. Med. Chem. 2000;43:778–783. doi: 10.1021/jm980263s. [DOI] [PubMed] [Google Scholar]
- 24.Shigeta S., Mori S., Kodama E. Antiv. Res. 2003;58:265–271. doi: 10.1016/s0166-3542(03)00009-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Wang E.B., Ji L.N. Prog. Chem. 2006;18:114–119. [Google Scholar]
- 26.Guan W., Yan L.K. Z. M. Su. Inorg. Chem. 2005;44:100–109. doi: 10.1021/ic049830u. (the nomenclature is incorrect) [DOI] [PubMed] [Google Scholar]
- 27.Shao C., Hu D.H., Su Z.M. Chem. J. Chin. Univ. 2005;26:1512–1516. [Google Scholar]
- 28.Yang H.T., Yang M.J., Ding Y. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C.Z., Xuan Z.Y., Chen R.S. Chin. J. Biochem. Mol. Biol. 2002;18:499–505. [Google Scholar]
- 30.Barlow S., Rohl A.L., Shi S. J. Am. Chem. Soc. 1996;188:7578–7592. [Google Scholar]
- 31.Luthy R., Bowie J.U., Eisenberg D. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 32.Peiris J., Lai S., Poon L. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Discover 3 User Guide, San Diego, MSI, USA, 1999.
- 34.Greenidge P.A., Carlsson B., Bland L.G. J. Med. Chem. 1998;41:2503–2512. doi: 10.1021/jm9708691. [DOI] [PubMed] [Google Scholar]
- 35.Chen L., Gui C., Luo X. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C.W., Tsai C.H., Tsai F.J. FEBS Lett. 2004;574:131–137. doi: 10.1016/j.febslet.2004.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Insight II, Version 98.0, San Diego, MSI, 1998.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table.


