Highlights
-
•
PDCoV nsp15 antagonizes IFN-β production.
-
•
PDCoV nsp15 suppresses NF-κB activation other than IRF3 activation.
-
•
The endoribonuclease activity is not essential for PDCoV nsp15 to antagonize IFN-β.
Keywords: Porcine deltacoronavirus, Nonstructural protein 15 (nsp15), Endoribonuclease activity, Interferon production
Abstract
Porcine deltacoronavirus (PDCoV) is an emerging swine coronavirus causing diarrhea and intestinal damage in nursing piglets. Previous work showed that PDCoV infection inhibits type I interferon (IFN) production. To further identify and characterize the PDCoV-encoded IFN antagonists will broaden our understanding of its pathogenesis. Nonstructural protein 15 (nsp15) encodes an endoribonuclease that is highly conserved among vertebrate nidoviruses (coronaviruses and arteriviruses) and plays a critical role in viral replication and transcription. Here, we found that PDCoV nsp15 significantly inhibits Sendai virus (SEV)-induced IFN-β production. PDCoV nsp15 disrupts the phosphorylation and nuclear translocation of NF-κB p65 subunit, but not antagonizes the activation of transcription factor IRF3. Interestingly, site-directed mutagenesis found that PDCoV nsp15 mutants (H129A, H234A, K269A) lacking endoribonuclease activity also suppress SEV-induced IFN-β production and NF-κB activation, suggesting that the endoribonuclease activity is not required for its ability to antagonize IFN-β production. Taken together, our results demonstrate that PDCoV nsp15 is an IFN antagonist and it inhibits interferon-β production via an endoribonuclease activity-independent mechanism.
1. Introduction
Porcine deltacoronavirus (PDCoV), an emerging swine enteric coronavirus (CoV), belongs to the newly identified genus Deltacoronavirus, family Coronaviridae in the order Nidovirales (Ma et al., 2015; Wang et al., 2019). Clinically, PDCoV infection causes severe diarrhea and intestinal pathological damage in nursing piglets (Jung et al., 2016; Wang et al., 2016a; Zhang, 2016). PDCoV was first identified in 2012 during molecular surveillance of CoVs in mammals and birds in Hong Kong (Woo et al., 2012). In 2014, outbreaks of PDCoV were reported in some states of the United States (Chen et al., 2015; Hu et al., 2015; Marthaler et al., 2014; Wang et al., 2014). Thereafter, PDCoV was also detected in farmed pigs with diarrhea in other countries, including South Korea (Jang et al., 2017; Lee et al., 2016), China (Dong et al., 2015; Song et al., 2015; Wang et al., 2015), Thailand (Janetanakit et al., 2016), Japan (Suzuki et al., 2018), Laos, and Vietnam (Lorsirigool et al., 2016; Saeng-Chuto et al., 2017), causing great economic losses in the swine industry. Recently, Jung and co-workers reported that calves are also susceptible to infection with PDCoV (Jung et al., 2017), sparking an increased interest in studying this emerging virus.
PDCoV is single-stranded, positive-sense RNA virus with an envelope, and its full-length genome, approximately 25.4 kb, is the smallest genome among the known CoVs (Ma et al., 2015; Woo et al., 2012). The typical genome of PDCoV successively contains 5ʹUTR-ORF1a-ORF1b-S-E-M-NS6-N-NS7-NS7a-3ʹUTR, which encodes two viral replicase polyproteins that are predicted to be proteolytically processed to yield 15 replication- and transcription-associated mature nonstructural proteins (nsps), four structural proteins, and three accessory proteins (Fang et al., 2017, 2016; Woo et al., 2012). Based on studies of other known CoVs, the proteases from PDCoV nsps that are required for viral replication and transcription have been predicted, such as papain-like protease nsp3, 3C-like protease nsp5, RNA-binding protein nsp9, RNA-dependent RNA polymerase nsp12, helicase nsp13, and endoribonuclease nsp15 (Mielech et al., 2014; Ziebuhr et al., 2000; Zhu et al., 2017a, b). Of these, the endoribonuclease, termed NendoU, is highly conserved among vertebrate nidoviruses (coronaviruses and arteriviruses) but is not conserved among other RNA viruses, making it a genetic marker of this virus order (Ivanov et al., 2004; Snijder et al., 2013). NendoU, reported to have similarities to XendoU, specifically cleaves RNA at the level of 5ʹ uridines to release a 2ʹ-3ʹ-cyclic phosphate end product in the presence of Mn2+, which is considered to be an integral component of the replicase-transcriptase complex of nidoviruses and plays a critical role in viral replication and transcription (Athmer et al., 2017; Nedialkova et al., 2009).
Besides their roles in viral replication and transcription, the nsp15 of some CoVs and its orthologue nsp11 in arteriviruses have been demonstrated to antagonize innate immune responses. Overexpression of severe acute respiratory syndrome coronavirus (SARS-CoV) nsp15 significantly inhibits type I interferon (IFN) production, and the possible mechanism is via inhibiting MAVS-induced apoptosis (Frieman et al., 2009; Lei et al., 2009). However, this mechanism appears not to be shared by other CoVs (Lei et al., 2009). Porcine epidemic diarrhea virus (PEDV) nsp15 has been shown to suppress poly(I:C)-induced type I and type III IFN responses and the endoribonuclease activity appears to be not required for PEDV replication in IFN-deficient Vero cells (Zhang et al., 2016; Deng et al., 2019). A recombinant mouse hepatitis virus (MHV) strain A59 with a mutation in the active site of endoribonuclease nsp15 can stimulate an early and robust induction of type I IFN in macrophages by activating host double-stranded RNA (dsRNA) sensors, indicating that MHV A59 nsp15 may function as an IFN antagonist during MHV infection (Deng et al., 2017). Several studies have demonstrated that arterivirus nsp11 s inhibit innate immune responses and their endoribonuclease activity is required (Shi et al., 2011; Sun et al., 2016). Additionally, our previous studies showed that PDCoV infection suppresses RIG-I-mediated production of IFN-β (Luo et al., 2016). However, whether or not PDCoV nsp15 is also a type I IFN antagonist remains unclear.
In the present study, we demonstrated that PDCoV nsp15 also inhibits IFN-β production by impairing nuclear factor-κB (NF-κB) activation. Furthermore, we found that the endoribonuclease activity of PDCoV nsp15 is not essential for its ability to antagonize IFN-β production.
2. Materials and methods
2.1. Cells, viruses, and reagents
LLC-PK1 cells (porcine kidney cells) were purchased from the ATCC (ATCC CL-101) and cultured in Dulbecco's modified Eagle's medium (Invitrogen, USA) supplemented with 10% fetal bovine serum at 37 °C in a humidified 5% CO2 incubator. HEK-293T cells, obtained from the China Center for Type Culture Collection (CCTCC), were cultured in Dulbecco's modified Eagle's medium (Invitrogen, USA) supplemented with 10% fetal bovine serum under the same conditions described above. PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) was isolated from a piglet with acute diarrhea in China in 2014 (Dong et al., 2016). Sendai virus (SEV) was obtained from the Centre of Virus Resource and Information, Wuhan Institute of Virology, Chinese Academy of Sciences. Mouse monoclonal antibodies (MAbs) against hemagglutinin (HA) and β-actin were purchased from Medical and Biological Laboratories (Japan). Rabbit polyclonal antibodies directed against IRF3, phosphorylated IRF3 (p-IRF3), p65, and phosphorylated p65 (p-p65) were purchased from ABclonal (China) and Cell Signaling Technology.
2.2. Plasmids
The luciferase reporter plasmids IFN-β-Luc, IRF3-Luc (4 × PRDIII/I-Luc), and NF-κB-Luc (4 × PRDII-Luc) were described previously (Wang et al., 2016b). The nsp15 gene of PDCoV strain CHN-HN-2014 was amplified and used as the template for the mutagenesis of individual amino acid residues by overlap extension PCR using specific primers listed in Table 1 . The wild type (WT) nsp15 and its mutants (H219A, H234A, K269A) were cloned into pCAGGS-HA-C with a C-terminal HA tag to generate the eukaryotic expression constructs pCAGGS-HA-nsp15, pCAGGS-HA-H219A, pCAGGS-HA-H234A, and pCAGGS-HA-K269A. All constructs were verified by DNA sequencing.
Table 1.
Primers used for the construction of plasmids and qPCR assays.
| Primer | Nucleotide sequence (5’–3’) |
|---|---|
| nsp15-F | GCGGAATTCAACCTTGAAAACTTAGCTTACAAC |
| nsp15-R | ATACTCGAGCTGTAAGATTGGGTAGCAAGTCTT |
| H219A-F | GGTGTTGAAGCCATTATCTATGGTGATGAT |
| H219A-R | ATAGATAATGGCTTCAACACCCAGTTCCTG |
| H234A-F | GGCGGAACTGCCACACTTATCTCACTAGTT |
| H234A-R | GATAAGTGTGGCAGTTCCGCCAATGACTGG |
| K269A-F | TAACGCAAGCTCCGCGAACGTTTGCACTGT |
| K269A-R | GCAAACGTTCGCGGAGCTTGCGTTAGGTGA |
| h-IFN-β-F | TCTTTCCATGAGCTACAACTTGCT |
| h-IFN-β-R | GCAGTATTCAAGCCTCCCATTC |
| h-GAPDH-F | TCATGACCACAGTCCATGCC |
| h-GAPDH-R | GGATGACCTTGCCCACAGCC |
| p-IFN-β-F | GCTAACAAGTGCATCCTCCAAA |
| p-IFN-β-R | AGCACATCATAGCTCATGGAAAGA |
| p-GAPDH-F | ACATGGCCTCCAAGGAGTAAGA |
| p-GAPDH-R | GATCGAGTTGGGGCTGTGACT |
2.3. Luciferase reporter assay
Monolayers of LLC-PK1 or HEK-293T cells grown in 24-well plates were transiently co-transfected with 0.1 μg of reporter plasmid (IFN-β-Luc, NK-κB-Luc, or IRF3-Luc) together with 0.02 μg of the internal control plasmid pRL-TK (Promega) and the indicated expression plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 24 h after transfection, cells were mock-infected or infected with SEV (10 hemagglutinating activity units/well) for 12 h. Finally, firefly and Renilla luciferase activities were determined with the Dual-Luciferase reporter assay system according to the manufacturer’s protocol (Promega). Data are expressed as the luciferase activities of lysed cells, normalized to the activity of pRL-TK, and are representative of at least three independently conducted experiments. Data are presented as means ± standard deviations (SD).
2.4. RNA extraction and quantitative real-time PCR
LLC-PK1 or HEK-293T cells in 24-well plates were transfected with 1 μg of empty vector or the indicated expression plasmid. After 24 h, the cells were left untreated or infected with SEV for 12 h. Total cellular RNA was extracted from the cells using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA by avian myeloblastosis virus reverse transcriptase (TaKaRa, Japan). The above cDNA (1 μl of the 20 μl RT reaction mixture) were used as templates and subjected to SYBR green PCR assays (Applied Biosystems) at least three times. The IFN-β mRNA expression levels were normalized to the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers were designed with Primer Premier 5 software, and the sequences are listed in Table 1.
2.5. Western blot analysis
LLC-PK1 or HEK-293T cells were cultured in 60-mm dishes, transfected with the indicated plasmids for 24 h, and then infected or mock-infected with SEV (50 hemagglutinating activity units/dish) for 8 h. The cells were harvested by adding lysis buffer (4% SDS, 3% dithiothreitol [DTT], 0.065 mM Tris-HCl [pH 6.8], 30% glycerin) supplemented with a protease inhibitor cocktail, phenylmethylsulfonyl fluoride (PMSF), and a phosphatase inhibitor cocktail (Sigma). The samples were subjected to separation by 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA) to determine protein expression. The endogenous expression of p-IRF3, p65, p-p65, and IRF3 proteins were analyzed with the indicated antibodies, anti-p-IRF3, anti-p65 (ABclonal), anti-p-p65, and anti-IRF3 (Cell Signaling Technology), respectively. The overexpression of a plasmid with a HA tag was evaluated with an anti-HA antibody (MBL), and the expression levels of β-actin were detected with an anti-β-actin monoclonal antibody (MBL). The detections of similar amounts among groups were taken to indicate equal protein sample loading.
2.6. Indirect immunofluorescence assay (IFA)
IFAs were performed to examine the subcellular localization of endogenous IRF3 and NF-κB subunit p65 in LLC-PK1 or HEK-293T cells. Cells seeded on microscope coverslips in 24-well plates were transfected with the indicated expression plasmid when the cells reached approximately 80% confluence. After 24 h, the cells were mock-infected or infected with SEV for 6 h. The cells were fixed with 4% paraformaldehyde for 15 min and then permeated with methyl alcohol for 10 min at room temperature. After three washes with PBST, the cells were sealed with PBST containing 5% bovine serum albumin (BSA) for 1 h, followed by incubation separately with a rabbit polyclonal antibody against IRF3 (1:200) or against p65 (1:200) or a mouse anti-HA antibody (1:200) for 1 h at room temperature. The cells were then treated with secondary antibodies Alexa Fluor 594-conjugated donkey anti-mouse IgG and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Santa Cruz Biotechnology) for 1 h at 37 °C, followed by treatment with 4ʹ, 6-diamidino-2-phenylindole (DAPI) (Beyotime, China) for 15 min at room temperature. After the samples were washed with PBST, fluorescent images were visualized and examined with a confocal laser scanning microscope (Fluoviewver. 3.1; Olympus, Japan).
2.7. IFN bioassay
To evaluate the effect of PDCoV nsp15 on the amount of IFN production by LLC-PK1 or HEK-293T cells following stimulation by SEV, IFN bioassays were performed as described previously (Fang et al., 2018).
2.8. Statistical analysis
All experiments were performed in triplicate. Significant differences were determined using Student’s t-tests, and p values of <0.05 were considered statistically significant.
3. Results
3.1. PDCoV nsp15 inhibits SEV-induced IFN-β production
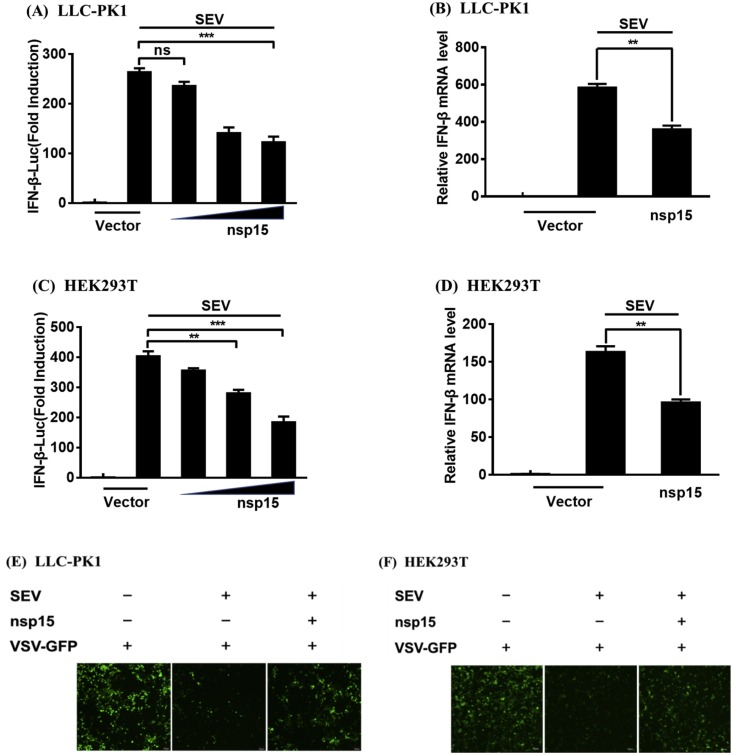
To explore whether or not PDCoV nsp15 antagonizes IFN-β production, LLC-PK1 or HEK-293T cells were co-transfected with increasing amounts of a PDCoV nsp15 expression plasmid (pCAGGS-HA-nsp15) or an empty vector and the IFN-β-Luc reporter plasmid, along with pRL-TK (an internal control plasmid), and then infected with SEV for 12 h. The cell lysates were harvested, and the IFN-β promoter-driven luciferase activity and IFN-β mRNA expression levels were measured by dual-luciferase reporter assays and real-time RT-PCR analyses, respectively. As shown in Fig. 1 , ectopic expression of PDCoV nsp15 significantly inhibited SEV-induced IFN-β promoter activity in both LLC-PK1 (Fig. 1 A) and HEK-293T (Fig. 1 C) cells. The mRNA expression levels of IFN-β were also decreased compared with the control group in both cell lines (Fig. 1 B and D). To further verify the inhibition of IFN-β production on protein level by PDCoV nsp15, the IFN bioassay was performed by using an IFN-sensitive vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP). As shown in Fig. 1 E and F, the cellular supernatants from SEV-infected LLC-PK1 and HEK-293T cells notably suppressed the replication of VSV-GFP in both cell lines. However, the replication of VSV-GFP was partially restored by the presence of supernatants from cells expressing PDCoV nsp15 compared to that of supernatants from empty vector-transfected cells. These results suggest that PDCoV nsp15 is an IFN-β antagonist.
Fig. 1.
PDCoV nsp15 antagonizes SEV-induced IFN-β production. (A, C) LLC-PK1 cells (A) or HEK-293T cells (C) cultured in 24-well plates were co-transfected with IFN-β-Luc and pRL-TK together with increasing amounts (0.1, 0.5, and 1.0 μg) of pCAGGS-HA-nsp15 or empty vector. After 24 h, the cells were mock-infected or infected with SEV (10 hemagglutinating activity units/well) for 12 h and subjected to a dual-luciferase reporter assay. The relative firefly luciferase activity was normalized to the Renilla luciferase activity with the untreated empty vector control value set to 1. (B, D) LLC-PK1 cells (B) or HEK-293T cells (D) cultured in 24-well plates were co-transfected with 1.0 μg of pCAGGS-HA-nsp15 or empty vector for 24 h, and then left untreated or infected with SEV (10 hemagglutinating activity units/well). At 8 h after infection, the cells were collected, and total RNA was extracted to detect the expression levels of IFN-β and GAPDH by SYBR Green PCR assay. (E, F) LLC-PK1 (E) or HEK-293T (F) cells were transfected with 1.0 μg of pCAGGS-HA-nsp15 or empty vector. At 24 h after transfection, both cell lines were infected with SEV for 12 h and the cell supernatants were collected. The UV-irradiated cell supernatants were overlaid onto fresh LLC-PK1 or HEK-293T cells in 24-well plates. After 24 h of incubation, cells were infected with VSV-GFP for 12 h. The replication of VSV-GFP was detected via fluorescence microscopy. Data are representative of three independent experiments. ***, p < 0.001; **, p < 0.01; ns, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. PDCoV nsp15 inhibits SEV-induced activation of NF-κB
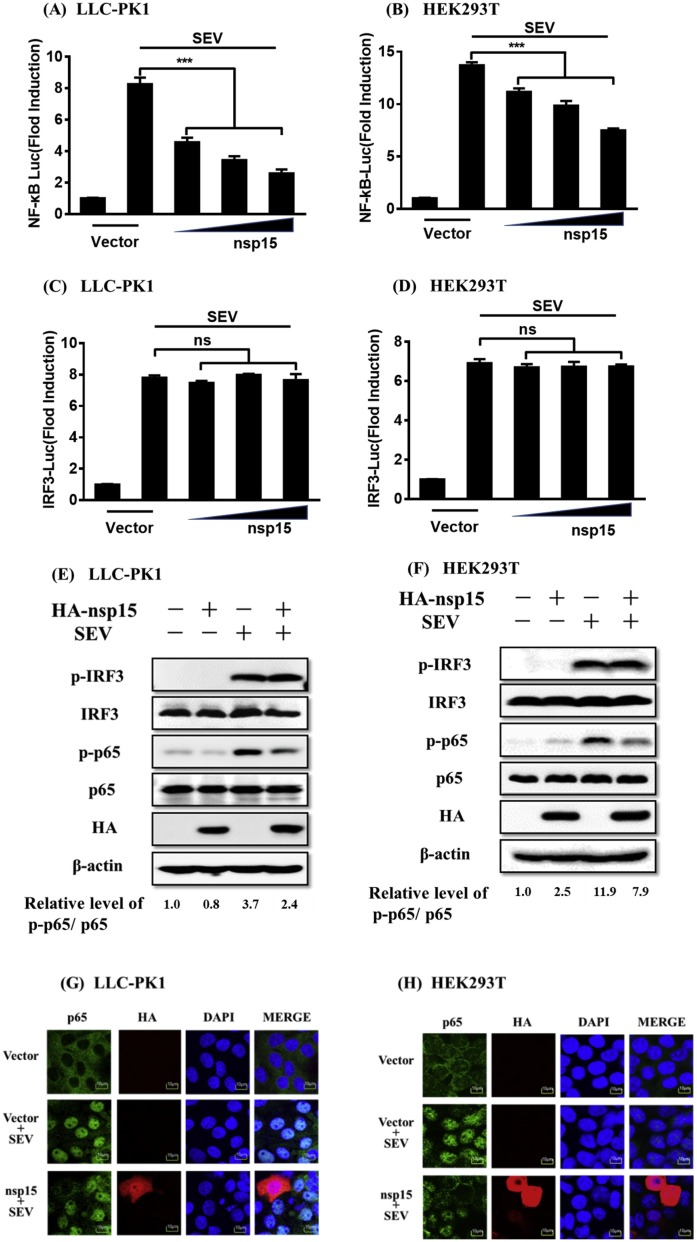
It is well known that the interferon regulatory factor 3 (IRF3) and NF-κB are critical transcription factors for induction of type I IFN (Wathelet et al., 1998). Since the above results indicate that PDCoV nsp15 is an IFN-β antagonist, we thus next investigated if PDCoV nsp15 affects the SEV-induced activation of IRF3 and NF-κB. LLC-PK1 or HEK-293T cells were co-transfected with pCAGGS-HA-nsp15 and the IRF3-Luc or NF-κB-Luc luciferase reporter plasmid together with pRL-TK, after which they were infected with SEV for 12 h. The results of luciferase reporter assays showed that PDCoV nsp15 blocked the SEV-induced promoter activity of NF-κB in a dose-dependent manner in both LLC-PK1 (Fig. 2 A) and HEK-293T cells (Fig. 2 B), but it had no effect on the SEV-induced promoter activity of IRF3 in both LLC-PK1 (Fig. 2 C) and HEK-293T cells (Fig. 2 D).
Fig. 2.
PDCoV nsp15 impairs SEV-induced activation of NF-κB. (A–D) LLC-PK1 cells (A, C) or HEK-293T cells (B, D) grown in 24-well plates were co-transfected with NF-κB-Luc (A, B) or IRF3-Luc (C, D) and pRL-TK together with increasing quantities (0.1, 0.5, and 1.0 μg) of PDCoV nsp15 expression plasmid for 24 h, followed by infection with SEV or mock-infection for 12 h before luciferase reporter assays were performed. The averages of data from three independent experiments are shown. ***, p < 0.001. (E, F) LLC-PK1 cells (E) or HEK-293T cells (F) cultured in 60-mm dishes were mock-transfected or transfected with increasing amounts (3, 5, and 7 μg) of pCAGGS-HA-nsp15 for 24 h before infection or mock-infection with SEV for 8 h. Western blot analyses with antibodies against p-IRF3 (Ser386), IRF3, p-p65 (Ser536), p65, HA, or β-actin were performed on lysates from these cells. The relative levels of rate of p-p65 /p65 in comparison to the control group are shown as fold values below the images via Image J software analysis. (G, H) LLC-PK1 cells (G) or HEK-293T cells (H) cultured in 24-well plates were transfected with 1.0 μg of pCAGGS-HA-nsp15, followed by SEV infection for 6 h, and subjected to indirect immunofluorescence assays to detect endogenous p65 (green) and PDCoV nsp15 (red) with rabbit anti-p65, and mouse anti-HA antibodies, respectively. The cell nuclei (blue) were stained with DAPI. Fluorescent images were acquired with a confocal laser scanning microscope (Fluoviewver. 3.1; Olympus, Japan). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The phosphorylation and nuclear translocation of IRF3 and the NF-κB p65 subunit are the hallmarks for activation of IRF3 and NF-κB, respectively, so we next investigated the effects of PDCoV nsp15 on the phosphorylation and nuclear translocation of IRF3 and p65. To this end, LLC-PK1 or HEK-293T cells were transfected with pCAGGS-HA-nsp15 or empty vector, followed by SEV infection. Western blot assays were performed on lysates of these cells to analyze the phosphorylation levels of IRF3 and p65. As shown in Fig. 2 E and F, the phosphorylation levels of endogenous IRF3 and p65 were both markedly higher following SEV infection, compared with the levels in mock-infected cells. In line with our previous results from luciferase reporter assays, the SEV-induced p65 phosphorylation in nsp15-expressing cells was notably inhibited, but nsp15 failed to impede the IRF3 phosphorylation induced by SEV in both LLC-PK1 (Fig. 2 E) and HEK-293T cells (Fig. 2 F). The results of nuclear translocation assays of IRF3 and p65 also showed that nsp15 expression inhibited the SEV-induced nuclear translocation of p65 in both LLC-PK1 (Fig. 2 G) and HEK-293T cells (Fig. 2 H), but not that of IRF3 in both cell lines (data not shown). Overall, these results demonstrate that PDCoV nsp15 antagonizes SEV-induced IFN-β production mainly by inhibiting the activation of NF-κB.
3.3. PDCoV nsp15 inhibits IFN-β production independently of its endoribonuclease activity
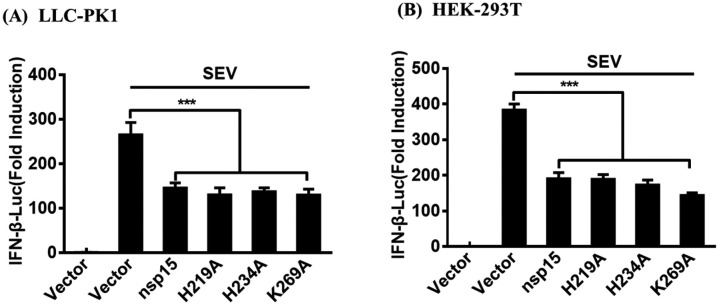
Previous study has demonstrated that PDCoV nsp15 His219 (H219), H234, and Lys269 (K269) are the critical sites for its endoribonuclease activity (Zheng et al., 2018). To investigate whether or not PDCoV nsp15 mutants lacking endoribonuclease activity possess the ability to antagonize IFN-β production. LLC-PK1 cells were co-transfected with expression plasmids encoding PDCoV WT nsp15 or one of its mutants (H219A, H234A, or K269A), together with IFN-β-Luc and pRL-TK, followed by stimulation with SEV. The results of dual-luciferase reporter assays showed that all three mutants significantly inhibited the SEV-induced IFN-β promoter activity, similar to the WT nsp15. (Fig. 3 A). We also used HEK-293T cells to repeat the experiments and similar results were observed (Fig. 3 B). Based on these results, we can conclude that the endoribonuclease activity of PDCoV nsp15 is not essential for inhibiting IFN-β production.
Fig. 3.
PDCoV nsp15 inhibits IFN-β production independently of its endoribonuclease activity. LLC-PK1 cells (A) or HEK-293T cells (B) cultured in 24-well plates were transfected with 1.0 μg of expression plasmid (PDCoV nsp15, H219A, H234A, or K269A) or empty vector, along with IFN-β-Luc and pRL-TK for 24 h. The cells were then mock-infected or infected with SEV for 12 h and subjected to dual-luciferase reporter assays. Data are means ± SD from three independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
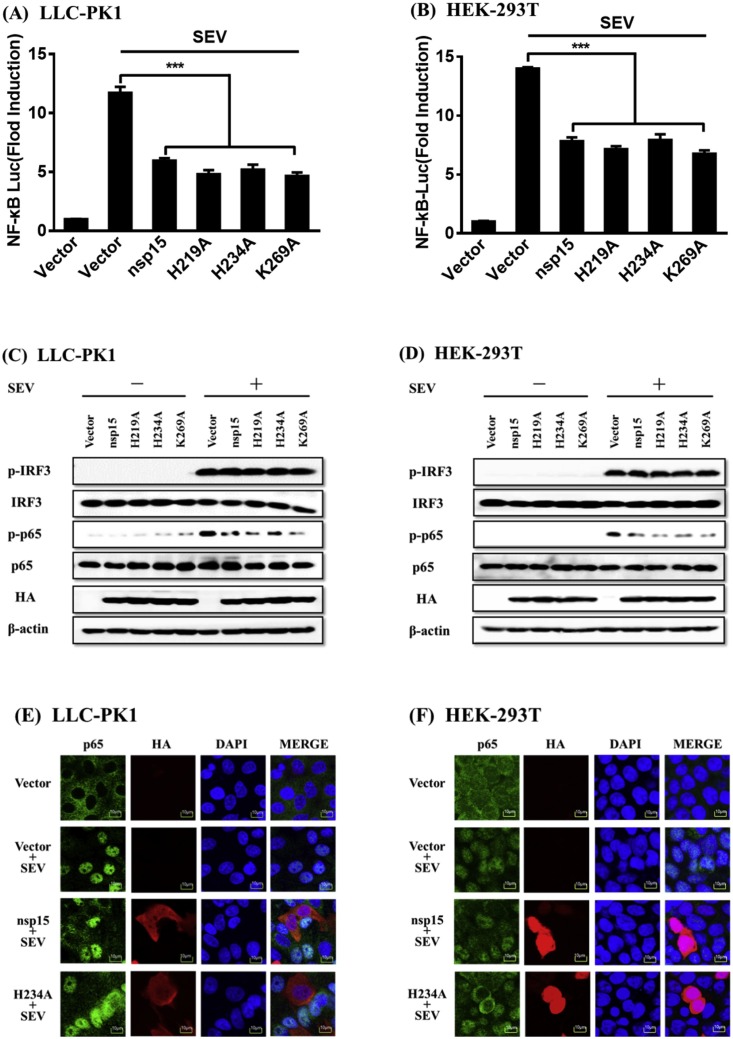
3.4. The mutants of PDCoV nsp15 lacking endoribonuclease activity significantly inhibit SEV-induced NF-κB activation
Because PDCoV nsp15 mutants lacking endoribonuclease activity can still antagonize IFN-β production, we further investigated if, similar to the WT nsp15, the nsp15 mutants could also impair NF-κB activation. To this end, LLC-PK1 or HEK-293T cells were co-transfected with expression plasmids encoding one of the nsp15 mutants (H219A, H234A, or K269A) and the NF-κB-Luc luciferase reporter plasmid, together with the internal control plasmid pRL-TK, and then infected with SEV. A plasmid encoding the WT nsp15 was used as a control. The results of dual-luciferase reporter assays revealed that all mutants of PDCoV nsp15 suppressed the SEV-induced promoter activity of NF-κB to a similar degree as the WT nsp15 in both LLC-PK1 (Fig. 4 A) and HEK-293T cells (Fig. 4 B). We also analyzed the phosphorylation levels of endogenous IRF3 and p65 after the overexpression of nsp15 mutants in both LLC-PK1 (Fig. 4 C) and HEK-293T (Fig. 4 D) cells, and the results demonstrated that cells transfected with any of the three mutants, similar to the WT nsp15, markedly decreased p65 phosphorylation compared to the cells transfected with empty vector, while the expression levels of total IRF3 and p65 and the phosphorylation level of IRF3 were similar among all groups (Fig. 4 C and D). Using the mutant H234A as a representative, we analyzed the nuclear translocation of endogenous IRF3 and p65 after the overexpression of a nsp15 mutant. Similar to the WT nsp15, when the mutant H234A was ectopically expressed, it also inhibited the SEV-induced nuclear translocation of p65 in both LLC-PK1 (Fig. 4 E) and HEK-293T cells (Fig. 4 F), but did not affect IRF3 translocation in both cell lines (data not shown). Together, our results suggest that the nsp15 mutants lacking endoribonuclease activity utilized a similar mechanism as the WT nsp15 to antagonize IFN-β production by impairing NF-κB activation.
Fig. 4.
PDCoV nsp15 mutants lacking endoribonuclease activity also significantly inhibit SEV-induced activation of NF-κB. (A, B) LLC-PK1 cells (A) or HEK-293T cells (B) grown in 24-well plates were co-transfected with NF-κB–Luc together with pRL-TK and 1.0 μg of expression plasmid (PDCoV nsp15, H219A, H234A, K269A) or empty vector for 24 h, followed by stimulation with SEV for 12 h before luciferase reporter assays were performed. ***, p < 0.001; *, p < 0.05; ns, not significant. (C, D) LLC-PK1 cells (C) or HEK-293T cells (D) were transfected with expression plasmid (PDCoV nsp15, H219A, H234A, K269A) or empty vector and infected with SEV, and cells were then collected for western blot analyses using antibodies against p-IRF3, IRF3, p-p65, p65, HA, and β-actin. (E, F) Nsp15 H234A acts as a representative mutant protein to analyze its effect on nuclear translocation of p65. LLC-PK1 cells (E) or HEK-293T cells (F) were transfected with PDCoV nsp15 or H234A expression plasmid and then infected with SEV. Immunofluorescence analyses to detect endogenous p65 (green) and PDCoV nsp15/H234A (red) were performed with rabbit anti-p65, and mouse anti-HA antibodies, respectively. The nuclei (blue) were stained with DAPI. Cells transfected with an empty vector or mock-infected with SEV were used as negative controls. Fluorescent images were acquired with a confocal laser scanning microscope (Fluoviewver. 3.1; Olympus, Japan). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
To combat the antiviral effects of IFN, many viruses have evolved elaborate strategies to inhibit IFN production. For PDCoV, previous studies have revealed that PDCoV infection suppresses the RIG-I-mediated production of type I IFN (Luo et al., 2016). To date, nsp5 and accessory protein NS6 encoded by PDCoV have been confirmed to be antagonists IFN production (Zhu et al., 2017a; Fang et al., 2018). Whether other proteins from PDCoV have also been responsible for the inhibition of RIG-I-mediated production of IFN-β remains to be unclear. To further identify and characterize the PDCoV-encoded IFN antagonists will broaden our understanding of its infection and pathogenesis.
As a unique endoribonuclease encoded by nidoviruses, the functions of nsp15 of CoVs (nsp11 in arteriviruses) have been received more attentions. Previous studies showed that the nsp15 encoded by SARS-CoV (Frieman et al., 2009), MHV (Deng et al., 2017), PEDV (Zhang et al., 2016; Deng et al., 2019) and the nsp11 encoded by PRRSV (Shi et al., 2011; Sun et al., 2016) can antagonize antiviral innate immune responses. Our present study also found that the nsp15 of PDCoV antagonizes type I IFN production, suggesting that it may be a common property for the endoribonuclease of nidoviruses to antagonizes IFN production. However, it is appears that different mechanisms are utilized by the endoribonucleases from different nidoviruses. For example, the nsp15 s of MHV and HCoV-229E prevent the activation of IFN response by mediating the evasion of host recognition of viral dsRNA (Deng et al., 2017; Kindler et al., 2017); PRRSV nsp11, the orthologue of CoV nsp15, antagonizes type I IFN production by suppressing both MAVS and RIG-I expression (Sun et al., 2016). SARS-CoV nsp15 antagonizes type I IFN production by inhibiting MAVS-induced apoptosis (Lei et al., 2009), however, MERS-CoV nsp15 does not utilize this strategy although it also inhibits type I IFN production with a yet-to-be identified mechanism. More recently, PEDV nsp15 has been demonstrated to be involved in IFN antagonize through a possible mechanism similar to the nsp15 of MHV and HCoV-229E (Deng et al., 2019). Although different mechanisms are utilized by the endoribonucleases from different nidoviruses, it appears that all tested endoribonucleases inhibit the activations of both IRF3 and NF-κB, the two critical transcription factors for induction of type I IFN production. In this study, we found that PDCoV nsp15 antagonizes IFN-β production by impairing the activation of transcription factor NF-κB, but not IRF3, which is a different strategy from other reported endoribonucleases of nidoviruses.
Although all tested endoribonuclease of nidoviruses have been demonstrated to antagonize IFN production, it is still inconclusive whether this function fully depends on its endoribonuclease activity. Several studies showed that overexpression of WT nsp15 s of CoV or nsp11 s of arterivirus significantly suppressed IFN production, while the mutants lacking endoribonuclease activity lost this capacity (Kindler et al., 2017; Shi et al., 2011; Sun et al., 2016). These evidences appear to indicate that the endoribonuclease activity is essential for nsp15/nsp11-mediated inhibition of IFN production. However, the WT nsp15 s or nsp11 s have been demonstrated to be extremely toxic to prokaryotic and eukaryotic cells, on the contrary, no cytotoxicity could be observed in cells overexpressing nsp15/nsp11 mutants lacking endoribonuclease activity (Deng and Baker, 2018; Sun et al., 2016). Thus, many researchers suspected that the suppression of IFN production by WT nsp15/nsp11 is due to its cytotoxicity (Deng and Baker, 2018; Shi et al., 2016). In addition, although MHV A59 mutant expressing a catalysis-deficient nsp15 induces robust type I IFN production compared to wild-type MHV in the infected macrophages, however, another mutant expressing an unstable nsp15 also exhibits similar property (Deng et al., 2017). Thus, no direct evidence for the linkage between endoribonuclease activity and type I IFN antagonism has been obtained. In this study, we found that PDCoV nsp15 antagonizes IFN production independent on its endoribonuclease activity. Both WT nsp15 and its mutants lacking endoribonuclease activity inhibited type I IFN production to a similar degree. These results indicated that PDCoV nsp15 has evolved a novel mechanism for the antagonism of IFN production in an endoribonuclease activity-independent manner, distinct from that reported previously for the inhibition of IFN-β production by other CoV nsp15 s or arterivirus nsp11 s. The reason that results in these differences remains unclear. In terms of structure, the active form of endoribonuclease from PDCoV nsp15 is different from that of other CoV nsp15 s. PDCoV nsp15 is a mixture of dimers and monomers, while the nsp15 s of SARS-CoV, MHV, HCoV-229E, as well as MERS-CoV, function as a hexamer (Ricagno et al., 2006; Xu et al., 2006; Zhang et al., 2018; Zheng et al., 2018). Whether these structure differences result in different mechanisms used to antagonize IFN production remains further study. In addition, it should be noted that this study involved the individual expression of PDCoV nsp15, outside the context of infection. However, we also compared the inhibitory effects of PEDV WT nsp15 and its mutant (nsp15 H226A) lacking endoribonuclease activity in overexpression system, and found that the PEDV WT nsp15 significantly inhibited IFN-β production, while such an inhibitory effect was not observed in cells overexpressing mutant nsp15 H226A (data not shown). Because the reverse genetics system of PDCoV is not available nowadays, it is difficult to investigate the IFN antagonist activity of PDCoV nsp15 in the context of virus infection. We are now working to establish a reverse genetics system of PDCoV, which will allow the generation of mutant or deletion viruses to investigate the precise mechanism of PDCoV nsp15-mediated inhibition of type I IFN.
In summary, our present study reveals that PDCoV nsp15 acts an IFN antagonist. Distinct from the endoribonucleases of other nidoviruses, PDCoV nsp15 impairs NF-κB activation rather than IRF3 activation. Furthermore, PDCoV nsp15 inhibits type I IFN production independent its endoribonuclease activity. These results indicated that PDCoV nsp15 may utilizes a distinct mechanism to antagonize type I IFN production.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31730095, U1704231), the National Key R&D Program of China (2016YFD0500103), and the Special Project for Technology Innovation of Hubei Province (2017ABA138).
References
- Athmer J., Fehr A.R., Grunewald M., Smith E.C., Denison M.R., Perlman S. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. MBio. 2017;8 doi: 10.1128/mBio.02320-16. e02320-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Baker S.C. An "old" protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163. doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2017;114:e4251–e4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., van Geelen A., Buckley A.C., O’Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93 doi: 10.1128/JVI.02000-18. e02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Ren J., Hong Y., Liu X., Zhao Y., Wang D., Peng G., Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018;92 doi: 10.1128/JVI.00712-18. e00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U S A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Lee K.K., Kim S.H., Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound. Emerg. Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M., V’kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., van Kuppeveld F.J., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Chung H.C., Nguyen V.G., Moon H.J., Kim H.K., Park S.J., Lee C.H., Lee G.E., Park B.K. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound. Emerg. Dis. 2016;63:248–252. doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.P. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A., Saeng-Chuto K., Temeeyasen G., Madapong A., Tripipat T., Wegner M., Tuntituvanont A., Intrakamhaeng M., Nilubol D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 2016;161:2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Fang L., Dong N., Fang P., Ding Z., Wang D., Chen H., Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D.T., Stott C.J., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W., Tantituvanont A., Kaewprommal P., Piriyapongsa J., Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis. 2017;64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Wang L., Li X., Zhang G., Guo J., Zhao D., Chai S., Deng R. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-beta induction. Mol. Immunol. 2011;48:1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Li Y., Lei Y., Ye G., Shen Z., Sun L., Luo R., Wang D., Fu Z.F., Xiao S., Peng G. A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J. Virol. 2016;90:4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ke H., Han M., Chen N., Fang W., Yoo D. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus suppresses both MAVS and RIG-I expression as one of the mechanisms to antagonize type I interferon production. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Imai N., Yamamoto T., Ohashi S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 2018;61:176–182. doi: 10.1016/j.meegid.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3 doi: 10.1128/genomeA.00945-15. e00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Sarver C., Byrum B., Zhang Y. Porcine deltacoronavirus: histological lesions and genetic characterization. Arch. Virol. 2016;161:171–175. doi: 10.1007/s00705-015-2627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Lin C.H., Parekh B.S., Ronco L.V., Howley P.M., Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li L., Yan L., Ming Z., Jia Z., Lou Z., Rao Z. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by middle east respiratory syndrome coronavirus. J. Virol. 2018;92 doi: 10.1128/JVI.00893-18. e00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A., Shi Y., Shen Z., Wang G., Shi J., Xiong Q., Fang L., Xiao S., Fu Z.F., Peng G. Insight into the evolution of nidovirus endoribonuclease based on the finding that nsp15 from porcine Deltacoronavirus functions as a dimer. J. Biol. Chem. 2018;293:12054–12067. doi: 10.1074/jbc.RA118.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Fang L., Wang D., Yang Y., Chen J., Ye X., Foda M.F., Xiao S. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology. 2017;502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., Lv M., Ye X., Peng G., Fang L., Xiao S. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017;91 doi: 10.1128/JVI.00003-17. e00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]