Highlights
-
•
High frequency of Leptospira spp. in Brazilian bats was found with real time PCR.
-
•
The infected animals were from different municipalities and species.
-
•
None of evaluated variables was associated to Leptospira spp. positivity frequency.
-
•
The data reinforce the need for surveillance of zoonotic agents in wild animals.
Keywords: Leptospirosis, Emerging diseases, Molecular epidemiology, Zoonotic diseases, Public health
Abstract
The present study aimed to investigate the frequency of pathogenic Leptospira spp. in Brazilian bats and to determine possible risk factors associated to it. Ninety two bats of 12 species were evaluated. Whole genomic DNA from kidneys was extracted and real-time PCR specific to pathogenic Leptospira spp. was applied. Association between the frequency of specimens positive for Leptospira spp. and sex, age, bat species or family, season of collection, geographic localization and feeding habits was evaluated. The results showed that 39.13% of analyzed bats were found positive for Leptospira spp. Nine bat species had at least one positive result. There was no association among the evaluated variables and frequency of pathogenic Leptospira spp. Although the limitations due to lack of Leptospira spp. isolation, leptospiral carriage was demonstrated in bats of different species from southern Brazil, which reinforces the need for surveillance of infectious agents in wild animals.
1. Introduction
Leptospirosis is a zoonotic disease caused by pathogenic strains of Leptospira spp., which colonize host kidneys and are eliminated in urine. The transmission occurs by direct contact with contaminated urine or indirectly through contaminated water or soil [1]. Leptospirosis is worldwide distributed, and its incidence varies with climate, animal reservoirs and surveillance [2]. Rainy periods are associated to higher frequencies of disease, especially in large cities, since there is an increased chance of population contact with contaminated water in inadequate sanitation areas [3].
In Brazil, more than 60,000 human leptospirosis cases were confirmed between 2000 and 2016 and the regions with higher frequency of confirmed cases are the southern and southeastern regions [3]. The higher incidences occur in low-income populations of most populous cities, such as São Paulo or in regions with poor sanitation system [4]. Besides public health issues, leptospirosis is responsible for economic losses for the animal production sector. This is caused by costs with vaccination and decrease of production due to abortion, lower milk production and animal death [1].
An important feature of leptospirosis is the wide host range that are susceptible to disease or can serve as bacteria reservoirs [5], which hinders its control and epidemiologic understanding. Several studies have demonstrated that rodents are the main leptospirosis reservoirs [6]. However, there is a growing knowledge about the role of different wild animal species on disease cycle, which may be important due to their abundance and increasing contact with domestic animals [7], [8]. Thus, identifying wild reservoirs is a key factor in leptospirosis epidemiology knowledge [9].
Among Leptospira spp. wild reservoirs, bats are being evaluated in different parts of the world with different results according to region [10]. Bats have been implicated in epidemiological cycles of several emerging and re-emerging zoonosis, such as rabies [11], severe acute respiratory syndrome (SARS) [12], leptospirosis [8], [13] and recently Ebola in Africa [14], which points them as important key players in the epidemiology of infectious diseases. Bats are found on every continent except Antarctica [15]. In Brazil, 178 bat species have been recorded [16], of which 40 are settled in Rio Grande do Sul state, the Brazilian southern region [17]. Despite this wide range of bat species and high rates of leptospirosis, there are few Brazilian studies on this subject showing low bat leptospiral infection rates [4], [18]; however, they were performed in urban areas of Southeast region. Thus, in the present study, we sought to investigate the frequency of pathogenic Leptospira strains in bats from different areas of Rio Grande do Sul, the southern Brazilian state, which has a different ecosystem.
2. Material and methods
2.1. Ethics statement
Permission for this work on bats was granted by Ethical commission on animal experimentation of the “Instituto de Pesquisas Veterinárias Desidério Finamor” (CEUA/IPVDF) (process number 03/2012). The study did not involve any direct manipulations of live bats and relied entirely on collection of tissue samples from dead bats. All experiments were performed in compliance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series—No. 170 revised 2005) and the procedures of the Brazilian College of Animal Experimentation (COBEA).
2.2. Sampling
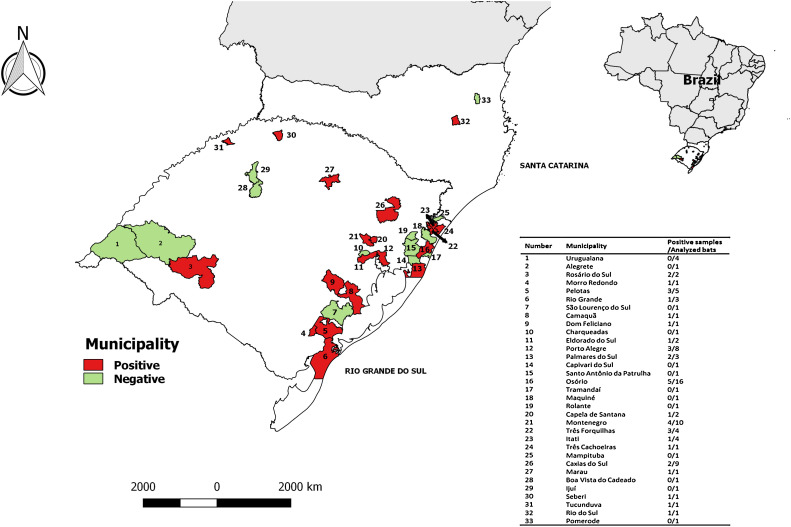
Bat samples (n = 92) from 31 and 2 municipalities of Rio Grande do Sul and Santa Catarina states, respectively (Fig. 1 ) were included in the study. The samples were sent to Instituto de Pesquisas Veterinárias Desidério Finamor for rabies diagnosis between December 2010 and December 2012. Species identification was performed based on previous studies and age determination was made through epiphysis fusion and dentition evaluation [19], [20].
Fig. 1.
Geographical distribution of sampled bats. Rio Grande do Sul, the southern Brazilian state is shown. Municipalities in red (n = 20) had at least one bat with positive result for pathogenic Leptospira spp. Municipalities in green (n = 13) had at least one evaluated bat and the results were negative. Municipalities in white were not accessed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Molecular detection of pathogenic Leptospira spp
For detection of pathogenic Leptospira spp. in bat kidneys, total DNA was extracted as previously described [21] and quantified using a spectrophotometer (L-quant, Loccus Biotechnology, Brazil). A conventional PCR for Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was performed for DNA quality confirmation [22]. About 25 ng of DNA were used as template for Taqman® real-time PCR with primers and probe targeting lipL32 as previously published [23]. All reactions were carried out in triplicate and positive results were considered if at least two reactions had detectable CT. Positive and negative controls were included in each run.
To determine the real-time PCR analytical sensitivity, a conventional PCR using the reference isolate Pomona DNA was performed and the resulting amplicon was cloned in a pCR2.1 vector (TOPO TA Cloning® kit, Invitrogen, USA). After Escherichia coli DH5-α transformation, plasmid DNA was recovered [24] and quantified with Qubit (Invitrogen, USA). The number of molecules was calculated as follows: [6.02 × 1023 molecules × cloned vector quantity (g)]/Molecular weight of cloned vector (g). Standard curve was made with amounts ranging from 109 to 10−1 leptospiral DNA molecules as templates for real-time PCR.
2.4. Statistics
In order to evaluate an association between the positivity frequency for pathogenic Leptospira spp. and the dependent variables, chi-square (sex variable) or Fisher Exact test (age, bat species or family, season of collection, geographic localization and feeding habits variables) were performed with Stata software 10.0 (Stata Corporation, College Station, Texas, USA). Statistical association was considered when p < 0.05. For geographical representation, Quantum GIS software was used.
3. Results
3.1. Sample characterization
Ninety two bat specimens were analyzed. They were classified into 12 different species, from Phyllostomidae, Molossidae and Vespertilionidae families. Most of the samples were from free tailed bats (Tadarida brasiliensis) (Table 1 ). The majority of animals were adults (82.61%). Sample distribution by sex was homogeneous and the higher frequency of collection occurred during spring and winter (Table 1). The number of analyzed bats by municipality is indicated in Fig. 1.
Table 1.
Description of bat samples collected in Southern Brazil.
| Variable | Number of bats (% of total) | |
|---|---|---|
| Sex | Female | 45 (48.9) |
| Male | 47 (51.1) | |
| Age | Newborn | 2 (2.2) |
| Juvenile | 8 (8.7) | |
| Adult | 76 (82.6) | |
| Old | 3 (3.3) | |
| Unclassified | 3 (3.3) | |
| Species | Eptesicus diminutus | 3 (3.3) |
| Eptesicus furinalis | 1 (1.1) | |
| Eumops auripendulus | 5 (5.4) | |
| Eumops patagonicus | 3 (3.3) | |
| Glossophaga soricina | 2 (2.2) | |
| Histiotus velatus | 2 (2.2) | |
| Lasiurus ega | 1 (1.1) | |
| Molossus molossus | 19 (20.7) | |
| Molossus rufus | 8 (8.7) | |
| Myotis nigricans | 1 (1.1) | |
| Sturnira lilium | 1 (1.1) | |
| Tadarida brasiliensis | 46 (50.0) | |
| Season of collection | Spring | 39 (42.4) |
| Summer | 8 (8.7) | |
| Autumn | 4 (4.4) | |
| Winter | 41 (44.6) | |
| Feeding habits | Insectivorous | 89 (96.7) |
| Nectarivorous | 2 (2.2) | |
| Frugivorous | 1 (1.1) | |
| Family | Molossidae | 81 (88.0) |
| Vespertilionidae | 8 (8.7) | |
| Phyllostomidae | 3 (3.3) | |
3.2. Molecular analysis
The real-time PCR was able to detect 10 genome copies of Leptospira spp. with 103.42% efficiency (R2 = 0.99) (Fig. 2 .), which was the same detection capacity than the previous study [23]. All DNA amplified the GAPDH gene, showing that the samples were in appropriate conditions for molecular analysis. Of these, 36 (39.13%) had positive results for pathogenic Leptospira (Table 2 ) and 9 species had at least one positive bat (Table 2).
Fig. 2.
Standard curve of real-time PCR to detect pathogenic Leptospira spp. Determination of the detection threshold by titration of plasmids contaning lipL32 sequence. The detection limit was 10 DNA molecules with 103.42% efficiency.
Table 2.
Bat species and frequencies of real-time PCR results for pathogenic Leptospira spp.
| Family | Bat species | Feeding habits | No. collected | No. positives | Overall positivity frequency (%) | Positivity frequency within species (%) | Positivity frequency within family (%) | |
|---|---|---|---|---|---|---|---|---|
| Vespertilionidae | Eptesicus diminutus | Insectivorous | 3 | 2 | 2.2 | 66.7 | 75.0 | |
| Eptesicus furinalis | 1 | 0 | 0.0 | 0.0 | ||||
| Histiotus velatus | 2 | 2 | 2.2 | 100.0 | ||||
| Lasiurus ega | 1 | 1 | 1.1 | 100.0 | ||||
| Myotis nigricans | 1 | 1 | 1.1 | 100.0 | ||||
| Molossidae | Eumops auripendulus | Insectivorous | 5 | 2 | 2.2 | 40.0 | 35.8 | |
| Eumops patagonicus | 3 | 0 | 0.0 | 0.0 | ||||
| Molossus molossus | 19 | 4 | 4.4 | 21.1 | ||||
| Molossus rufus | 8 | 2 | 2.2 | 25.0 | ||||
| Tadarida brasiliensis | 46 | 21 | 22.8 | 45.7 | ||||
| Phyllostomidae | Glossophaga soricina | Nectarivorous | 2 | 1 | 1.1 | 50.0 | 33.3 | |
| Sturnira lilium | Frugivorous | 1 | 0 | 0.0 | 0.0 | |||
| Total | 92 | 36 | 39.1 | |||||
Among the species with higher number of analyzed specimens, Tadarida brasiliensis had almost half of the animals positive for leptospiral genome carriage (21/46; 45.65%; Table 2) and Molossus molossus had a lower positivity rate (4/19; 21.05%; Table 2). A low number of other bat species has also been surveyed and their infection rates varied from 0% (Eptesicus furinalis, Eumops patagonicus and Sturnira lilium) to 100% (Histiotus velatus (2/2), Lasiurus ega (1/1) and Myotis nigricans (1/1)) (Table 2).
In 33 municipalities where the samples were collected, 20 had at least, one positive bat (Fig. 1.). Factors such bat species, Leptospira spp. serovar, age and breeding season may influence bat leptospiral infection rates. In the present study, there was no association among age (p = 0.502), sex (p = 0.867), species (p = 0.139), season of collection (p = 0.838), location (p = 0.477), feeding habits (p = 1.000) or family (p = 0.09) and the frequency of positive results for pathogenic Leptospira.
4. Discussion
In the present study, bats from Southern Brazil were shown to be Leptospira spp. carriers. The data showed higher frequency than previous Brazilian studies – 39.1% vs 7.8% and 1.75% in Botucatu and São Paulo, respectively [25], [26]. This may be related to the environment in which the bats live, as in the present study bats from South Brazilian areas, most of them distant from urban centers, were evaluated.
A variable number of bats from the surveyed species were analyzed; however, it was possible to observe an equal distribution of positive and negative results for Tadarida brasiliensis, the species presenting a higher number of analyzed specimens; and a lower rate of positive results for Molossus molossus, the second species with more samples. Considering the family, there seems to be higher frequency of positive results within Vespertilionidae family, although there was no statistical significance; thus, further studies are needed to confirm that different bat families have different degrees of susceptibility to Leptospira spp. infection. The leptospiral host specificity could not be accessed, since there was failure to amplify the secY gene fragment. This may have been occurred due to low bacterial loads or poor DNA quality, as the bats arrived in the laboratory days after death. Previous studies also report difficulty to amplify larger amplicons from kidney samples positive for leptospiral real-time PCR, even though the bat specimens were captured avoiding DNA quality issues [27], [28].
Regarding the feed behavior, the presence of leptospiral DNA was detected in insectivorous species, which refutes the hypothesis that sharing food with rodents increases the likelihood of bats to carry Leptospira spp. [29]. As the bats’ habitats were not accessed, it was not possible to determine the routes by which the animals were probably infected. Taking into account the Leptospira spp. transmission routes, the contact with contaminated soil or water could be considered.
No statistical difference was observed regarding Leptospira spp. positivity frequency when analyzing sex, age, local or season of collection. Some of these variables have been described to influence leptospiral carriage in rodents [30]; however, the observed results may be related to low sample number and the way of selection, which was by convenience. The majority of bats evaluated in the present study were found distant from large urban centers and many of them in unusual situations, such as in daylight, sick, or dead. This can indicate increased risk of co-infection with other infectious agents; however, to test this hypothesis, new studies with animals randomly selected should be performed.
Currently, urban sprawl has been responsible for the closer contact of humans and wild animals. Bats often live at roofs, gaps between buildings and urban vegetation, which may have impact on disease transmission to humans and domesticated animals. As the number of analyzed specimens by municipality varied and in general was low, it was not possible to associate bat leptospiral carriage and human cases of leptospirosis; moreover, it was not possible to access the leptospiral serovars infecting the bats. These limitations precluded an analysis about the role of these animals on human leptospirosis transmission, which needs to be further investigated.
There are differences associated with social, health and environmental conditions regarding infectious diseases transmissibility, which are still of major concerns worldwide. Since in the last decades some important infectious diseases have emerged or re-emerged, challenging the global public health security, a better knowledge on the ecology of different infectious agents becomes important. The present study, even though presenting limitations due to lack of Leptospira spp. isolation and genotyping, brings new knowledge on leptospiral carriage in Brazilian bats. The data reinforces the need for surveillance of infectious agents, especially the zoonotic ones which are hosted by wild animals.
Acknowledgments
Emily Marques dos Reis was recipient of FINEP/CNPq scholarship and André Vinícius Andrade Bezerra was recipient of FAPERGS scholarship. This project was financially supported by FINEP (Grant number SANIMARS 01100783-00). The sponsors had no role in the study design, in the collection, analysis and interpretation of data, in the writing of the manuscript and in the decision to submit the manuscript for publication.
References
- 1.Faine S., Faine S., Adler B., Adler B., Bolin C., Bolin C., Perolat P., Perolat P. 1999. Leptospira and Leptospirosis. [Google Scholar]
- 2.Slack A., Symonds M., Dohnt M., Harris C., Brookes D., Smythe L. Evaluation of a modified Taqman assay detecting pathogenic Leptospira spp. against culture and Leptospira-specific IgM enzyme-linked immunosorbent assay in a clinical environment. Diagn. Microbiol. Infect. Dis. 2007;57:361–366. doi: 10.1016/j.diagmicrobio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.2014. Brasil, Portal Da Saúde. [Google Scholar]
- 4.Bessa T.Á.F., Spichler A., Berardis Chapola É.G., Husch A.C., De Almeida M.F., Sodré M.M., Mouriz Savani E.S.M., Veiga Sacramento D.R., Vinetz J.M. The contribution of bats to leptospirosis transmission in São Paulo City, Brazil. Am. J. Trop. Med. Hyg. 2010;82:315–317. doi: 10.4269/ajtmh.2010.09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler B. Pathogenesis of leptospirosis: cellular and molecular aspects. Vet. Microbiol. 2014;172:353–358. doi: 10.1016/j.vetmic.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Bharti A.R., Nally J.E., Ricaldi J.N., Matthias M.A., Diaz M.M., Lovett M.A., Levett P.N., Gilman R.H., Willig M.R., Gotuzzo E., Vinetz J.M. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 2003;3:757–771. doi: 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 7.Petrakovsky J., Bianchi A., Fisun H., Nájera-Aguilar P., Pereira M. Animal leptospirosis in latin america and the caribbean countries: reported outbreaks and literature review (2002–2014) Int. J. Environ. Res. Public Health. 2014;11:10770–10789. doi: 10.3390/ijerph111010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthias M.A., Díaz M.M., Campos K.J. Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by Bayesian phylogenetic analysis of 16S ribosomal DNA sequences. Am. J. 2005;73:964–974. [PMC free article] [PubMed] [Google Scholar]
- 9.Tulsiani S.M., Graham G.C., Dohnt M.F., Burns M., Craig S.B. Maximizing the chances of detecting pathogenic leptospires in mammals: the evaluation of field samples and a multi-sample-per-mammal, multi-test approach. Ann. Trop. Med. Parasitol. 2011;105:145–162. doi: 10.1179/136485911X12899838683205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich M., Wilkinson D.A., Benlali A., Lagadec E., Ramasindrazana B., Dellagi K., Tortosa P. Leptospira and Paramyxovirus infection dynamics in a bat maternity enlightens pathogen maintenance in wildlife. Environ. Microbiol. 2015;17(11):4280–4289. doi: 10.1111/1462-2920.12766. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi Y., Ogawa A., Sato G., Sato T., Itou T., Samara S.I., Carvalho A.A., Nociti D.P., Ito F.H., Sakai T. Geographical distribution of vampire bat-related cattle rabies in Brazil. J. Vet. Med. Sci. 2006;68:1097–1100. doi: 10.1292/jvms.68.1097. [DOI] [PubMed] [Google Scholar]
- 12.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 13.Smythe L.D., Field H.E., Barnett L.J., Smith C.S., Dohnt M.F., Symonds M.L., Moore M.R., Rolfe P.F. Leptospiral antibodies in flying foxes in Australia. J. Wildl. Dis. 2002;38:182–186. doi: 10.7589/0090-3558-38.1.182. [DOI] [PubMed] [Google Scholar]
- 14.Saéz A.M., Weiss S., Nowak K., Lapeyre V., Kaba M., Regnaut S., Zimmermann F., Düx A., Ku H.S., Merkel K., Sachse A., Thiesen U., Villányi L., Boesch C., Dabrowski P.W., Nitsche A., Leendertz S.A.J., Petterson S., Becker S., Krähling V., Couacy-hymann E., Akoua-Koffi C., Weber N., Schaade L., Fahr J., Borchert M., Gogarten J.F., Calvignac-spencer S., Leendertz F.H., Saez A.M., Weiss S., Nowak K., Lapeyre V., Zimmermann F., Dux A., Kuhl H.S., Kaba M., Regnaut S., Merkel K., Sachse A., Thiesen U., Villanyi L., Boesch C., Dabrowski P.W., Radonic A., Nitsche A., Leendertz S.A.J., Petterson S., Becker S., Krahling V., Couacy-hymann E., Akoua-Koffi C., Weber N., Schaade L., Fahr J., Borchert M., Gogarten J.F., Calvignac-spencer S., Leendertz F.H. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol. Med. 2014;7:17–23. doi: 10.15252/emmm.201404792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogueira M.R., de Lima I.P., Moratelli R., da C. Tavares V., Gregorin R., Peracchi A.L. Checklist of Brazilian bats, with comments on original records. Check List. 2014;10:808–821. doi: 10.15560/10.4.808. [DOI] [Google Scholar]
- 17.Pacheco S.M., Sekiama M.L., a De Oliveira K.P., Quintela F., Weber M.M., Marques R.V., Geiger D., Silveira D. Biogeografia de quirópteros da região sul. Ciência E Ambiente. 2007;35:181–202. [Google Scholar]
- 18.Zetun C.B., Hoffmann J.L., Silva R.C., Souza L.C., Langoni H. Leptospira spp. and Toxoplasma gondii antibodies in vampire bats (Desmodus rotundus) in Botucatu region, SP Brazil. J. Venomous Anim. Toxins Incl. Trop. Dis. 2009;15:546–552. [Google Scholar]
- 19.Gregorin R., Taddei V.A. Chave artificial para determinação de molossídeos brasileiros (Mammalia: Chiroptera) J. Neotrop. Mammal. 2002;9:13–32. [Google Scholar]
- 20.Barquez R.M., Díaz M.M. Los Murciélagos de Argentina: Clave de Identificación, in: Publicación Especial N° 1 PCMA. Tucumán. 2016;2009:p84. [Google Scholar]
- 21.Singh K.K., Muralidhar M., Kumar A., Chattopadhyaya T.K., Kapila K., Singh M.K., Sharma S.K., Jain N.K., Tyagi J.S. Comparison of in house polymerase chain reaction with conventional techniques for the detection of Mycobacterium tuberculosis DNA in granulomatous lymphadenopathy. J. Clin. Pathol. 2000;53:355–361. doi: 10.1136/jcp.53.5.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerva C., Bremm C., dos Reis E.M., Bezerra A.V.A., Loiko M.R., da Cruz C.E.F., Cenci A., Mayer F.Q. Food safety in raw milk production: risk factors associated to bacterial DNA contamination. Trop. Anim. Health Prod. 2014;46:877–882. doi: 10.1007/s11250-014-0580-y. [DOI] [PubMed] [Google Scholar]
- 23.Stoddard R.A., Gee J.E., Wilkins P.P., McCaustland K., Hoffmaster A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009;64:247–255. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 24.J. Sambrook, D.W. Russel, Preparation and transformation of competent E. coli using calcium chloride, in: J. Sambrook, D.W. Russel (Eds.), Molecular Cloning: A Laboratory Manual, 3rd ed., New York, 2001: pp. 116–118.
- 25.Álvares Franco Bessa T., Spichler A., Berardis Chapola E.G., Husch A.C., Fernandes de Almeida M., Martos Sodré M., Mouriz Savani E.S.M., Veiga Sacramento D.R., Vinetz J.M. The contribution of bats to leptospirosis transmission in Sao Paulo City, Brazil. Am. J. Trop. Med. Hyg. 2010;82:315–317. doi: 10.4269/ajtmh.2010.09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zetun C., Hoffmann J., Silva R., Souza L., Langoni H. Leptospira spp. and Toxoplasma gondii antibodies in vampire bats (Desmodus rotundus) in Botucatu region, SP, Brazil. J. Venomous Anim. Toxins Incl. Trop. Dis. 2009;15:546–552. doi: 10.1590/S1678-91992009000300014. [DOI] [Google Scholar]
- 27.Gomard Y., Dietrich M., Wieseke N., Ramasindrazana B., Lagadec E., Goodman S.M., Dellagi K., Tortosa P. Malagasy bats shelter a considerable genetic diversity of pathogenic Leptospira suggesting notable host-specificity patterns. FEMS Microbiol. Ecol. 2016;92:1–12. doi: 10.1093/femsec/fiw037. [DOI] [PubMed] [Google Scholar]
- 28.Lagadec E., Gomard Y., Guernier V., Dietrich M., Pascalis H., Temmam S., Ramasindrazana B., Goodman S.M., Tortosa P., Dellagi K. Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg. Infect. Dis. 2012;18:1696–1698. doi: 10.3201/eid1810.111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harkin K.R., Hays M., Davis R., Moore M. Use of PCR to identify leptospira in kidneys of big brown bats (Eptesicus fuscus) in Kansas and Nebraska, USA. J. Wildl. Dis. 2014;50:651–654. doi: 10.7589/2013-08-201. [DOI] [PubMed] [Google Scholar]
- 30.Mason M., Encina C., Gonzalez M., Berg S. Household characteristics associated with rodent presence and leptospira infection in rural and urban communities from southern Chile. Am. J. Trop. Med. Hyg. 2014;90:497–506. doi: 10.4269/ajtmh.13-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]