Abstract
Development of effective cancer therapeutic strategies relies on our ability to interfere with cellular processes that are dysregulated in tumors. Given the essential role of the ubiquitin proteasome system (UPS) in regulating a myriad of cellular processes, it is not surprising that malfunction of UPS components is implicated in numerous human diseases, including many types of cancer. The clinical success of proteasome inhibitors in treating multiple myeloma has further stimulated enthusiasm for targeting UPS proteins for pharmacological intervention in cancer treatment, particularly in the precision medicine era. Unfortunately, despite tremendous efforts, the paucity of potent and selective UPS inhibitors has severely hampered attempts to exploit the UPS for therapeutic benefits. To tackle this problem, many groups have been working on technology advancement to rapidly and effectively screen for potent and specific UPS modulators as intracellular probes or early-phase therapeutic agents. Here, we review several emerging technologies for developing chemical- and protein-based molecules to manipulate UPS enzymatic activity, with the aim of providing an overview of strategies available to target ubiquitination for cancer therapy.
Keywords: Cancer, Drug discovery, DNA-encoded compound libraries, Target-based discovery, Phenotypic screens, Fragment-based libraries, PROTAC, Chemical genetics, Directed evolution, Ubiquitination, Phage display, Ubiquitin proteasome system, High-throughput screening
1. Introduction
Ever since the discovery of ubiquitin (Ub) four decades ago (Ciehanover, Hod, & Hershko, 1978), this small protein has been linked to multiple cellular processes, including cell proliferation, development, immune responses, and numerous human diseases including cancer (Calistri, Munegato, Carli, Parolin, & Palu, 2014; Dantuma & Bott, 2014; Senft, Qi, & Ronai, 2018; Zinngrebe, Montinaro, Peltzer, & Walczak, 2014). Indeed, protein ubiquitination, the process of attaching Ub and poly-Ub chains to cellular substrates, is an integral and essential part of the cell machinery, and serves to modify, regulate, and degrade proteins (Fig. 1 ) (Swatek & Komander, 2016). Ubiquitination initiates through the activation of Ub by a Ub activating enzyme (E1), which adenylates the Ub C-terminus using its catalytic cysteine residue, resulting in a thioester bond. Another thioester reaction is required for the transfer of Ub onto a Ub conjugating enzyme (E2). The Ub-charged E2 enzyme is then recruited by a Ub ligase (E3), which is responsible for the transfer of Ub from its E2-conjugated state to the substrate protein.
Fig. 1.
Drug targets in the ubiquitin proteasome system. (a) Schematic representation of the ubiquitination pathway. Ubiquitination involves three steps. First, E1 activates ubiquitin (Ub) with ATP, forming a thioester bond. Next, activated Ub is conjugated to the E2 enzyme via a transthiolation reaction. Finally, RING and U-box E3 ligases recruit the E2 enzyme and a substrate to facilitate transfer of Ub from the E2 to the substrate by an allosteric mechanism, whereas HECT E3 ligases directly ubiquitinate the substrate followed by the transfer of Ub from E2 to the HECT E3 ligase active site (Chen et al., 2017; Zhang & Sidhu, 2014). The diagram also illustrates drug development targets currently in the market (green dot), in clinical trials (blue dot), or still being tested in industry or academia (red dot). *Pimozide is currently used in the clinic, but not for cancer therapy. # Previously known as MLN294 (Tong et al., 2017). ** Identified within a small-molecule screen as an inhibitor of Ubc13-Uev1A E2 Ub conjugating enzyme (Pulvino et al., 2012). (b) Ub is conjugated through lysines once (monoubiquitination) or multiple times (multiubiquitination), as a single unit or as a chain (polyubiquitination). Monoubiquitination and multiubiquitination processes have been linked to protein function modification, receptor endocytosis, nuclear export, transcription and histone regulation. Lys29- and Lys48-linked polyubiquitination has been shown to lead to proteasomal degradation as part of the UPS. Lys63-linked polyubiquitination has been implicated in the lysosomal pathway, protein recruitment, membrane internalization, endosomal uptake, and autophagy. Deubiquitination may also occur (Swatek & Komander, 2016).
To date, two Ub-specific E1 activating enzymes have been identified in humans (Groettrup, Pelzer, Schmidtke, & Hofmann, 2008; McHugh et al., 2018; Schulman & Harper, 2009), and >30 E2s have been found in humans (Morreale & Walden, 2016; Mulder et al., 2016). Humans also harbour approximately 600 E3 Ub ligases, which are classified into the homologous to E6-associated protein carboxy terminus (HECT) family, the really interesting new gene (RING) family and the U-box family (Morreale & Walden, 2016; Mulder, Harari, & Simon, 2008; Zhang & Sidhu, 2014). E3 ligases bind E2 enzymes thioesterified with Ub (E2~Ub) to mediate substrate ubiquitination through different catalytic mechanisms. The HECT E3 ligases form an intermediate thioester bond with Ub to its active site cysteine residue before transferring it to substrates. In contrast, lacking a catalytic cysteine, the RING and U-box E3 ligases facilitate Ub transfer by an allosteric mechanism (Chen et al., 2017; Ozkan, Yu, & Deisenhofer, 2005; Rotin & Kumar, 2009; Zheng & Shabek, 2017). Single or multiple mono-ubiquitination and polyubiquitination at the N-terminus or lysine residues in Ub are all possible modifications of protein substrates and will determine the fate of the protein (Calistri et al., 2014). Ubiquitination at residue K11 of Ub has been linked to endoplasmic reticulum-associated protein degradation (Locke, Toth, & Petroski, 2014); K29-linked ubiquitination has been associated with protein modification (Nucifora Jr. et al., 2016); K48-linked ubiquitination is well studied for its role in proteasomal degradation of cellular substrates (Calistri et al., 2014); and, K63-linked ubiquitination results in protein recruitment, autophagy and degradation through the lysosomal pathway (Erpapazoglou, Walker, & Haguenauer-Tsapis, 2014).
On the other end of the spectrum, the process of ubiquitination can be reversed by deubiquitinating enzymes known as deubiquitinases (DUBs) (Zhang & Sidhu, 2014). Humans encode seven different structural families of DUBs, which comprise almost 100 proteins in total (Hermanns et al., 2018; Wertz & Wang, 2019). Ub-specific proteases (USPs) represent the largest and best studied family, with close to 60 USPs currently described (Yuan et al., 2018). Less research has been reported for the other families, with the next best studied being ovarian tumor Ub proteases (OTU), followed by JAB1/MPN+/Mov34 domain (JAMM) metallo-enzyme proteases, Ub C-terminal hydrolases (UCHs), and Josephin domain proteases (Yuan et al., 2018).
Targeting the ubiquitination pathway has been an attractive avenue for anti-cancer drug development, especially after the approval and success of proteasome inhibitor drugs (Bortezomib and Carfilzomib) (Fig. 1) for treatment of relapsed or refractory multiple myeloma (Richardson et al., 2003; Richardson, et al., 2005). This has propelled many studies and clinical trials to find new therapeutics targeting individual components in the ubiquitination pathway for the treatment of cancer, with the aim of providing better therapeutics with fewer side effects (Table 1 ) (Chen, Frezza, Schmitt, Kanwar, & Dou, 2011; Iida et al., 2018; Yazbeck et al., 2018). Another important aspect to consider is the selective killing of cancerous cells rather than normal cells through targeting ubiquitination, as ubiquitination is upregulated due to the error-prone nature of cancerous cells and tissues (Gallo, Ko, & Donoghue, 2017; Ge et al., 2018). Indeed, Bortezomib and Carfilzomib have higher activity in malignant cells than in normal cells and tissues (Berkers et al., 2005; Codony-Servat et al., 2006; Parlati et al., 2009). Other inhibitors of UPS components have also been shown to uniquely target cancerous cells (Fiskus et al., 2014; Li et al., 2013; Pulvino et al., 2012; Tsukamoto et al., 2008). Finally, the resistant, refractive and recurrent traits of cancer have inevitably resulted in the turnover of promising drugs like Bortezomib, due to the development of resistance in response to treatment (Hamouda et al., 2014). Consequently, finding new compounds that can target oncogenic pathways is crucial to establish effective cancer therapies. Therefore, development of innovative strategies to validate and target UPS proteins remains an important aspect of drug discovery in cancer research.
Table 1.
Advantages and disadvantages of emerging target-based drug development technologies.
| Technology | Advantages | Disadvantages | Developments | Targets | References |
|---|---|---|---|---|---|
| Target-based High Throughput Screening (HTS) |
|
|
|
|
(Chen, Dexheimer, et al., 2011; Davydov et al., 2004; Dodge, 2016; Gavory et al., 2018; Gombodorj et al., 2017; Lee et al., 2016; O'Dowd et al., 2018; Tong et al., 2017; Yang et al., 2007; Yang et al., 2005) |
| Fragment-based HTS |
|
|
|
|
|
| Proteolysis-targeting chimeric molecule (PROTAC) |
|
|
|
|
(Lebraud et al., 2016; Sakamoto et al., 2001; Sakamoto et al., 2003; Xie et al., 2014) |
| Protein design and engineering |
|
|
|
|
(Ernst et al., 2013; Gabrielsen et al., 2017; W. Zhang et al., 2016; Y. Zhang et al., 2013) |
2. Evolution of screening technologies for drug discovery
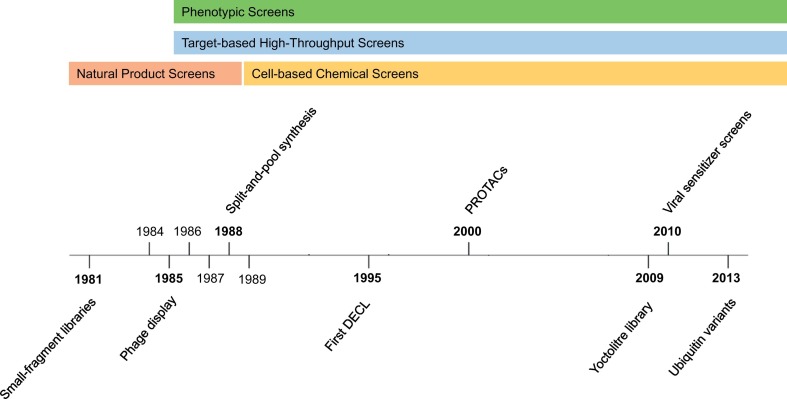
Before diving into recent advances in technology, we briefly describe the early stage of drug discovery in a historical perspective. Two crucial types of screening methods were pioneered and developed in the 1980s: target-based screens, which focus on a specific protein of interest – reviewed by Croston (2017), and phenotypic screens, which concentrate on the disease state to be treated – reviewed by Wagner (2016). Target-based high-throughput screening (HTS) strategies for drug discovery kicked off in the early 1980s (Pereira & Williams, 2007) (Fig. 2 ). Around the same time, researchers started to use phenotypic screens and genomic manipulation to paint a picture of cellular replication and the normal state of the cell, mainly in yeast as a model organism (Fraczek, Naseeb, & Delneri, 2018; Strynatka, Gurrola-Gal, Berman, & McMaster, 2018). The combination of these approaches in the late 1980s resulted in the birth of chemical genetics and led to many powerful developments such as directed evolution of enzymes, phage display of peptides and antibodies, and further refinement of target-based HTS (e.g. combinatorial chemistry, split-and-pool synthesis and fragment-based discovery) (Furka, Asgedom, & Diboa, 1988; Smith, 1985; Strynatka et al., 2018) (Fig. 2). From the late 1980s to the beginning of the 1990s, the use of animal models shifted to cell-based screens that minimized the number of reagents required and allowed the output of screens to increase (Strynatka et al., 2018) (Fig. 2). Research during these years also brought forth the generation of the first DNA-encoded compound libraries (DECLs) (Brenner & Lerner, 1992; Kinoshita & Nishigaki, 1995) (Fig. 2).
Fig. 2.
Timeline of drug development technologies in high-throughput screening. Schematic representation of advances in chemical and biochemical libraries over the past four decades.
Within the span of 20 years, high-throughput technologies further advanced to include feats in protein engineering such as proteolysis-targeting chimeras (PROTACs) derived from the ubiquitination pathway (Zhou, Bogacki, McReynolds, & Howley, 2000), streamlined phage display approaches that include Ub variants (UbV) to target protein-protein interactions in the UPS (Brown et al., 2016; Ernst et al., 2013; Ernst & Sidhu, 2013; Gabrielsen et al., 2017; Ordureau et al., 2018; Zhang et al., 2016; Zhang et al., 2017; Zhang et al., 2017; Zhang & Sidhu, 2018) and cell-based pharmacological HTS assays to enhance oncolytic virus cancer-cell-killing efficiency through viral sensitizer screens (Bourgeois-Daigneault et al., 2016; Diallo et al., 2010) (Fig. 2). In less than ten years, from 1999 to 2008, HTS target-based and phenotypic approaches were pivotal for drug discovery, yielding 60% and 69% of new molecular entities and next-generation drugs tested in clinical trials, respectively (Swinney & Anthony, 2011). For instance, E2 Ub conjugating enzyme inhibitors were discovered through a phenotypic and target-based small molecule screen for tuberculosis therapeutics (Darwin, Ehrt, Gutierrez-Ramos, Weich, & Nathan, 2003) (Fig. 1a and Table 1).
Since the ubiquitination pathways and UPS targets implicated in cancer drug development have been extensively reviewed (Kar, Keskin, Fraternali, & Gursoy, 2013; Li et al., 2018; Pal, Young, & Donato, 2014; Shen, Schmitt, Buac, & Dou, 2013; Wertz & Wang, 2019; Zhang & Sidhu, 2014), here we focus on addressing emerging trends and applications that can rapidly identify modulators for proteins within the ubiquitination pathway. The topics discussed below will examine the use of high-throughput technologies, small molecule approaches, peptide mimetics, and protein engineering. In addition, we will discuss potential applications for these technologies in cancer therapeutic research.
3. Targeting UPS proteins with chemical compounds
3.1. Constructing small molecule libraries
Research on chemical space (Jencks, 1981) stimulated the development of more ambitious target-based HTS approaches (Fig. 2). Drug-protein interactions were simplified into binding events, where the probability of binding would increase with the size of the molecule (Jencks, 1981). Previous HTS relied on assaying natural or synthetic compounds, which typically originated from tedious and time-consuming reactions of individual reagents (Pereira & Williams, 2007) (Fig. 2). With the advent of combinatorial chemistry, mixtures (instead of individual reagents) began to be employed to generate many more different compounds (Furka, 1995) (Figs. 2 and 3a). Later, Furka and colleagues introduced split-and-pool synthesis (Furka et al., 1988), in which the notion of combinatorial chemistry was further exploited to include separation and ligation of compounds. These separation and ligation reactions increased the number of potential combinations (Fig. 3a), which were estimated to total approximately 60 to 120 million chemical compounds globally (Hann & Oprea, 2004). This number has been exceeded now and million- to billion-compound libraries are routinely screened in industry (Davis, Plowright, & Valeur, 2017; Goodnow Jr., Dumelin, & Keefe, 2017; Litovchick et al., 2015). However, since the potential drug-like chemical space is estimated at 1063, these numbers still barely scratch the surface (Davis et al., 2017).
Fig. 3.
Workflow of high-throughput screening for drug discovery. (a) Diagram illustrating the typical work required to identify and characterize drug candidates using a target-based high-throughout screening (HTS) or a DNA-encoded compound library (DECL) approach. Different types of chemical synthesis, such as combinatorial chemistry and split-and-pool synthesis, are depicted. (b) Schematic depicting a fragment-based approach to inhibitor development. *Not all potential combinations are represented here.
Although combinatorial chemistry and split-and-pool synthesis sampled larger chemical space and revolutionized the way HTS was conducted for target-based drug discovery (Potyrailo et al., 2011), progress remains hampered by significant bottlenecks, including the expense of setting up an HTS system, starting reagents with poor properties, and the presence of reactive and aggregator molecules (Table 1) (Erlanson, 2012). In fact, in the early 2000s, compounds with poor properties like high lipophilicity were responsible for 50% of failed HTS projects (Keseru & Makara, 2009). Starter reagents that lack qualities such as solubility result in inefficient downstream drug candidates that might be impractical in a clinical setting (Keseru & Makara, 2009). Other problems such as reactive molecules and aggregators also delayed progress in target-based HTS (Erlanson, 2012). Reactive molecules or small impurities can oxidize proteins under the conditions used during HTS, resulting in the generation of hydrogen peroxide, which can inactivate proteins and cause false positives. Reactive molecules could arise from the possible combinations in split-and-pool synthesis, or simply as a side product of combinatorial chemistry (Erlanson, 2012). Finally, aggregators are a side effect of high concentration assays. At high concentration, intermolecular interactions of a compound can result in non-specific inhibition of the target protein, but the compound itself might not be a high affinity binder (Erlanson, 2012). This effect has been documented in past studies, where as many as 95% of compounds tested were identified as aggregators (Babaoglu et al., 2008). While these pitfalls remain as intrinsic caveats for small molecule libraries, activity-based small molecule screens have identified UPS inhibitors (Chen, Dexheimer, et al., 2011; Davydov et al., 2004; Gombodorj et al., 2017; Ott et al., 2017; Pulvino et al., 2012; Yang et al., 2007; Yang et al., 2005).
3.2. Activity-based small molecule screens for UPS inhibitors
The cumbersome bottlenecks described above prompted the use of approaches to utilize chemical space in more effective ways (Erlanson, 2012; Yuen & Franzini, 2017). In the activity-based screen approach, small molecules are screened to identify high affinity binders. In these screens, small molecules are added to cells, cell lysates, or in vitro reactions with recombinant proteins to test for inhibition of activity of a target protein or a change in phenotype. This can be detected through the presence or absence of fluorescence or luminescence (Janzen, 2014). Some of the most commonly used screening technologies include imaging or detection of: binding- or cleavage-based excitation of fluorescent probe-labeled proteins, fluorescence labeled antibodies targeting a specific protein, and fluorescence resonance energy transfer (FRET) where one fluorophore emits energy and a proximal one absorbs this energy for excitation. Screens can also be conducted with the use of flow cytometry, which can measure the light scattered through a cell to determine phenotype or expression of fluorescent-labeled proteins within the cell, and with luminescence-based assays, which are similarly designed to the fluorescent imaging assays mentioned above (Janzen, 2014).
Below, we present several representative studies utilizing these screening methods to develop chemical compounds targeting UPS components of different protein families (Table 2 ). One group was able to identify two molecules, PYR-41 and HLI98 (Fig. 1), which inhibited the E1 activating enzyme Uba1 (Yang et al., 2007) and the RING-E3 ligase HDM2 (Yang et al., 2005), respectively, by first screening a commercial chemical library and then confirming the leads with purchased individual compounds (Table 2). This small-molecule library was previously developed by the Vousden group to target autoubiquitination of E3 ligases (Davydov et al., 2004). In this assay, small molecules were incubated in ubiquitination reactions with recombinant E1 and E2 (UbcH5B), E3 (HDM2), and Ub. An electrochemiluminescence (ECM) labeled antibody targeting ubiquitinated proteins was subsequently added. The authors proposed that reactions with significantly reduced ECM represented small molecule hits inhibiting HDM2 enzymatic activity (Davydov et al., 2004; Yang et al., 2005; Yang et al., 2007). During validation of these hits, PYR-41, a pyrazone derivative (Yang et al., 2007), was found to target the E1 enzyme Uba1, and inhibit its activity with an IC50 of approximately 10 μM (Yang et al., 2007). HLI98, a compound from a newly identified 7-nitro-5-deazaflavin family (Davydov et al., 2004; Yang et al., 2005), was shown to target HDM2 E3 ligase activity with an IC50 of approximately 20 μM (Yang et al., 2005). To our knowledge, off-target effects and intracellular efficacy have yet to be thoroughly assessed for HLI98. The promiscuous nature of the assay in that it detects ubiquitinated proteins and the high IC50 value suggest that other cellular targets of HLI98 may exist.
Table 2.
Ubiquitin proteasome system inhibitors identified through small molecule or fragment-based assays described in this review.
| Inhibitor | Description | Potency (IC50) | Screen | Target (s) | Testing | References |
|---|---|---|---|---|---|---|
| PYR-41 | Pyrazone derivative | 10 μM | Antibody against ubiquitinated proteins in auto-ubiquitination reaction high-throughput screen (HTS) | Uba1 (E1) | In vitro recombinant protein assay | (Davydov et al., 2004; Yang et al., 2007) |
| HLI98 | 7-nitro-5-deazaflavin compound | 20 μM | HDM2 (HECT E3) | (Davydov et al., 2004; Yang et al., 2005) | ||
| Pevonedistat (MLN-4924) | Adenosine sulfamate mimetic | > 10 nM to <28 μM | Medicinal chemistry-based fine tuning of N6-benzyl adenosine inhibitor identified via HTS | E1 pan inhibitor | Clinical trials | (Chen, Tsu, et al., 2011; Soucy et al., 2009) |
| NSC697923 | nitrofuran | ~1 μM | luciferase reporter cell line | UBE2N/Ubc13 (E2) | In vitro cellular assay | (Cheng et al., 2014; Gombodorj et al., 2017; Hodge et al., 2015; Pulvino et al., 2012) |
| Pimozide | diphenylbutylpiperidine | ~2 μM | small-molecule fluorometric assay with rhodamine-labeled Ub substrate | USP1 |
In vitro cellular assay (cancer) Clinic (Schizophrenia) |
(Chen, Dexheimer, et al., 2011; Seeman, 2002) |
| Bortezomib | Pyrazine and boronic acid derivative | ~100 nM | Target and cell-based screen | 26S proteasome inhibitor | Clinic | (Adams et al., 1999; Nocentini, Supuran, & Winum, 2018) |
| N/A (numbered, not named) | Heterocyclic Pyrimidinones |
< 10 nM to <30 nM | Small library surface plasmon resonance (SPR) screen followed by medicinal chemistry | USP7 | In vitro cellular assays | (Gavory et al., 2018; O'Dowd et al., 2018) |
Another E1-inhibitor, MLN-4924 or pevonedistat (Fig. 1 and Table 2), is an adenosine sulfamate mimetic (Chen, Tsu, et al., 2011). Penovedistat was developed from a medicinal chemistry approach aiming to improve on a previously discovered inhibitor, N6-benzyl adenosine, from a high-throughput screen (Soucy et al., 2009). Pevonedistat was originally identified as an inhibitor of NEDD8 activating E1-ubiquitin activating enzyme 3 (Uba3) complex (Soucy et al., 2009) and was later labeled as a pan-inhibitor of E1 activating enzymes (da Silva et al., 2016; Gavin et al., 2014; Wertz & Wang, 2019). Soucy et al. reported potent inhibition of Uba3 in the single-digit nanomolar range with cross-reactivity against other E1s in the low micromolar range (Soucy et al., 2009). Pevonedistat is currently being tested in clinical trials of patients with acute myeloid leukemia, where the principal side effect seems to be liver toxicity and sepsis due to disruptions in the GTPase RhoA cytoskeleton protein and tumor necrosis factor (TNF)-α (Swords et al., 2017; Swords et al., 2018).
E2 inhibitors were also identified using a luciferase reporter cell line, in which inhibitor-mediated inactivation of the target protein resulted in loss of luciferase expression (Fig. 1 and Table 2). In this study, a small molecule, the nitrofuran NSC697923, inhibited an E2 Ub conjugating enzyme, UBE2N/Ubc13 (in complex with UBE2V1/Uev1A) by preventing thioester bond formation between the E2 and Ub, and consequently, supressed NF-κB activity in active B-cell lymphoma cells (Gombodorj et al., 2017; Pulvino et al., 2012) (Fig. 1). Another study also identified NSC697923 as an inhibitor of Ubc13 in neuroblastoma cells (Cheng et al., 2014) (Fig. 1). NSC697923 has been shown to inhibit Ubc13 at concentrations of ~1 μM (Hodge et al., 2015).
Another group developed the first described fluorometric cell lysate-based assay to mirror physiological conditions for small molecule screens (Ott et al., 2017). They targeted UCHL1, a DUB that is overexpressed in many cancers (Bishop, Rocca, & Henley, 2016), as a proof of principle for developing DUB inhibitors with a cell lysate-based assay that they called AlphaLISA (Ott et al., 2017). HA-tagged UCHL1 was labeled with an alpha-streptavidin donor bead and probed with biotin-tagged Ub. Binding of Ub to the DUB would result in fluorescence, and conversely, no fluorescence would occur in the presence of an inhibitor (Ott et al., 2017). The screen yielded a series of compounds that inhibited DUB activity in the 5–20 μM range, and some had been reported previously, including isogambonic acid, celastrol, mangiferin and rifampicin (Ott et al., 2017).
Inhibitors of the DUB USP1, which has been implicated in the Fanconi anemia signalling pathway in the DNA damage response (Bergink & Jentsch, 2009; Huang et al., 2006; Nijman et al., 2005), were identified with a small-molecule fluorometric assay that employed a Ub substrate modified with rhodamine (Chen, Dexheimer, et al., 2011), which emitted fluorescence upon cleavage (Dang, Melandri, & Stein, 1998). This simple in vitro assay could be used in a high-throughput fashion to screen for both small-molecule inhibitors and previously reported larger inhibitors (Dang et al., 1998). The study identified both novel small-molecule compounds as well as approved drugs, such as pimozide (a diphenylbutylpiperidine), as inhibitors of USP1 (Chen, Dexheimer, et al., 2011) (Fig. 1 and Table 2). In non-small cell lung carcinoma cells, pimozide was shown to reverse the chemo-resistance observed with cisplatin (Chen, Dexheimer, et al., 2011), a commonly used chemotherapy drug (Bloemink & Reedijk, 1996; Dasari & Tchounwou, 2014) with known resistance in Fanconi Anemia (Wang, 2007). Interestingly, pimozide is an anti-psychotic drug commonly prescribed to patients suffering from schizophrenia (Seeman, 2002), and although it has been postulated to act as a postsynaptic dopamine receptor blocker, its mechanism remains unknown (Seeman, 2002). Another study confirmed that pimozide could inhibit USP1 and in turn decrease the clonal growth of glioblastoma cells as well as increase their sensitivity to irradiation, though they are commonly resistant to chemotherapeutic and irradiation treatments (Lee et al., 2016). Notwithstanding this success, the potency of pimozide (IC50 ~2 μM) is lower than those of clinically approved UPS inhibitor 26S proteasome inhibitor Bortezomib (IC50 ~100 nM) (Fig. 1 and Table 2), and another UPS inhibitor VLX1570, which inhibited the DUBs UCHL5 and USP14 (IC50 ~100 nM) and was previously studied in clinical trials but terminated due to limiting toxicities (study identifier NCT02372240) (ClinicalTrials.gov, 2018; Wang et al., 2016).
While in general activity- and target-based small molecule screens have provided innovative approaches that make the process of drug discovery more economical and efficient, for most UPS targets, these types of screens yielded compounds with sub-optimal pharmacological properties (e.g. biophysics, solubility, and cellular permeability) and fall short in terms of potency and specificity (Lill & Wertz, 2014). We postulate that the intrinsic structural and catalytic properties of UPS proteins pose an impediment to generation of active-site inhibitors. For example, HECT E3s contains a shallow active site and are subject to dynamic regulation including extreme inter-domain rotations accompanying catalysis (Zheng & Shabek, 2017). On the other hand, the conserved nature of the DUB catalytic pocket probably accounts for the observed low specificity of inhibitors targeting the cysteine active site (Wang et al., 2018). Indeed, almost all of the UPS small molecule inhibitors currently in the clinic or clinical trials function in an allosteric manner. For instance, Nutlins disrupt substrate (p53) binding to the RING-E3 ligase MDM2 to stabilize p53 protein level and transcriptional activity (Vassilev et al., 2004), and Smac mimetics compete with Caspase binding to the BIR domain of RING-E3 IAP (inhibitor of apoptosis protein) to stimulate apoptosis in cancer cells (Liu et al., 2000; Wu et al., 2000). Finally, other than proteasome inhibitors like Bortezomib and Carfilzomib, there is only one family of drugs (Thalidomide/Lenalidomide) targeting UPS components approved for treatment of cancer, specifically for multiple myeloma and other B-cell neoplasms. Thalidomide was once well known for its adverse effects on fetus development before re-purposing for cancer therapy through binding to an E3 ligase Cereblon (CRBN) and stimulating lymphoid transcription factor ubiquitination and degradation (Ito et al., 2010; Kronke et al., 2014; Lu et al., 2014). Therefore, further development of allosteric modulators for UPS components will likely lead to more effective and efficacious cancer therapeutics.
3.3. Fragment-based drug discovery of potent and specific USP7 inhibitors
What distinguishes fragment-based approaches from other small-molecule HTS assays is the further fine-tuning of small-molecule screens. Once fragments are identified as binders of the target protein, researchers conduct medicinal chemistry to grow, ligate and merge the fragments into potent and selective inhibitors (Erlanson, 2012; Lamoree & Hubbard, 2017) (Fig. 3b). In this approach, the focus is not on activity, but rather in discovering potential allosteric inhibitors. As such, the approach is not to detect the fragment binders in an environment where the protein might be active, but to first detect whether the fragment binds to the target protein, and customize the fragment via medicinal chemistry based on information gathered through biophysical analyses. In an elegant example, the fragment-based approach was used to develop inhibitors of USP7 (Gavory et al., 2018) (Table 2). First, surface plasmon resonance (SPR) was used to screen a small library of ~2000 molecules targeting the catalytic domain of USP7, which is responsible for regulating MDM2, the dominant Ub ligase for p53 (Gavory et al., 2018; Iwakuma & Lozano, 2003). What distinguished this work from previous studies, which pioneered the first USP7 inhibitors (Kategaya et al., 2017; Pozhidaeva et al., 2017), was the use of medicinal chemistry to tailor initial screening hits. The inhibitory fragments were used to grow six different compounds that retained the high affinity of the original fragments, setting these results apart from the poor pharmacological properties and low potencies previously obtained (Kategaya et al., 2017; Pozhidaeva et al., 2017). The new USP7 inhibitors were also free from reactive species and were not aggregators. Importantly, this work identified a new allosteric inhibitory site on USP7, implying that affinity-based screens rather than activity-based screens may reveal more druggable sites. Another group also used a similar approach to develop USP7 inhibitors with high potency (Turnbull et al., 2017). The work exemplified in these two studies is particularly relevant for producing compounds that are active against DUBs, where other methods have proven unsuccessful (Wrigley et al., 2011).
The researchers that developed the allosteric USP7 inhibitors followed up with a study in which they described in detail the fragment-based medicinal chemistry approach to identify novel USP7 inhibitors (O'Dowd et al., 2018). This study demonstrated that employing SPR, a commonly used technology to detect binding (Erlanson, 2012), can be first used to identify potential allosteric binders of the protein, before continuing with X-ray crystallography to enable structure-based design (O'Dowd et al., 2018), an approach that has been responsible for many drugs developed to date (Erlanson, 2012; Lamoree & Hubbard, 2017). With both studies, the authors showed that structure-based design can aid with the elucidation of the mechanisms of action of novel inhibitors and can provide new insights into the functions of complex proteins with additional regulatory sites beyond the active site. We believe that this will have important implications for further development of inhibitors of other DUBs.
3.4. DNA-encoded compound libraries (DECLs)
The innovation of DECLs may transform hit identification of allosteric binders of UPS proteins into a faster and more manageable process thanks to the tagging of molecules with oligonucleotides. In HTS assays where mixtures of small molecules are used, or when traditional combinatorial chemistry is employed, the disentanglement of hits is usually arduous and time consuming. However, with DECL technology, inhibitor leads can be identified by rapid and efficient methods, including the polymerase chain reaction (PCR), deep DNA sequencing, and mass spectrometry. Ensemble Therapeutics has presented data describing the use of DECLs to develop macrocycle compounds (Mullard, 2016) that selectively and potently inhibit USP9x (Dodge, 2016), a DUB that is associated with many cancers (Murtaza, Jolly, Gecz, & Wood, 2015). However, the results were presented in a conference proceeding, and without further information, the ultimate outcome of this work remains unclear. Nonetheless, we predict that there will be more screens employing DECLs for inhibition of DUBs and other UPS targets.
DECLs comprise combinatorial molecules, small molecules or fragments that are concatenated to oligonucleotides (Fig. 4 ) (Mannocci, Leimbacher, Wichert, Scheuermann, & Neri, 2011; Yuen & Franzini, 2017). The oligonucleotides can be DNA or peptide nucleic acid (PNA) (Fig. 4a). The greatest advantage of DNA is its capacity for amplification. Though not amplifiable, PNAs have the advantages over DNA of being more stable and compatible with solid-phase synthesis. PNA libraries are also more amenable to use with combinatorial chemistry and split-and-pool synthesis methods (Zambaldo, Barluenga, & Winssinger, 2015) (Fig. 4b). It has been reported that DECLs can accommodate up to 1016 compounds (Goodnow Jr. et al., 2017). The Hansen group described a yoctoliter (10−24)-scale DNA reactor to decrease the amount of material needed to deeply sample chemical space for highly efficient and unbiased molecular evolution (Hansen et al., 2009). The combinatorial chemistry field continues to produce novel strategies for generating DECLs in a cost-effective manner.
Fig. 4.
DNA-encoded compound libraries (DECLs). Approaches for using DECLs for drug discovery are similar to those developed for combinatorial chemistry and split-and-pool synthesis, as reviewed by (Favalli et al., 2018; Franzini et al., 2015; Goodnow Jr. et al., 2017; Li et al., 2015). (a) Compounds are typically developed by combinatorial or other synthesis methods, and then coupled to specific DNA sequences or PNA chains. (b) Schematic representation of DECLs. DECLs typically begin by split-and-pool synthesis and are assembled differently depending on whether they are single pharmacophores (DNA-recorded and -templated) or dual pharmacophores [Encoded Self-Assembled Chemical (ESAC)]. DNA-recording involves the coupling of small molecules to single-stranded (ss) or double-strand (ds) oligonucleotides prior to the ligation of DNA barcodes and amplification. DNA-templated approaches involve similar processes to DNA-recorded, but this time already coupled DNA-compound moieties interact with each other via complementarity; in proximity, linkers coupling the compound to the DNA are cleaved, allowing the compounds to react and generate cyclical moieties. Dual pharmacophore libraries consist of ESAC libraries. In these libraries, like in DNA-recorded, compounds are first coupled to ssDNA. Hybridization occurs thanks to the spacer region of one moiety. Amplification is then used to generate dsDNA. (c) Illustration showing a representative screen where target tagged-proteins are immobilized. Tag-based affinity methods allow for washing off impurities and non-binders to enrich for hits. Screening is conducted via amplification or mass spectrometry of barcoded compounds with DNA or PNA, respectively.
Because DECLs are subject to the same processes described above for combinatorial chemistry and split-and-pool synthesis, they are also susceptible to some of the same pitfalls, such as the presence of side reaction intermediates and by-products that can affect interactions and lead to false positives. As such, the generation and downstream purification of these libraries prior to use in HTS is of utmost importance. The chemistries applied in these processes are beyond the scope of this review, but they have been reviewed elsewhere (Favalli, Bassi, Scheuermann, & Neri, 2018; Franzini et al., 2015; Goodnow Jr. et al., 2017; Li, Zheng, Liu, & Li, 2015).
DECLs are typically made as single or dual pharmacophores (Fig. 4b). Single pharmacophores can be synthesized with DNA-recorded or DNA-templated methods. In DNA-recorded methods, compounds are coupled to single-strand (ss) or double-strand (ds) oligonucleotides, and after each coupling, additional DNA barcodes corresponding to each individual compound are ligated, followed by amplification to generate dsDNA. DNA-templating methods involve coupled DNA-compound moieties, which interact with each other via barcode complementarity; once in close proximity, coupling linkers are cleaved to release compounds that react with each other to generate cyclical compounds linked to DNA barcodes. Encoded Self-Assembled Chemical (ESAC) libraries, also known as dual or complex pharmacophore libraries, initiate with compounds coupled to ssDNA; other compounds are coupled to spacers that are used to hybridize two DNA-encoded compounds, followed by amplification that results in the formation of dual pharmacophore compounds coupled to dsDNA.
DECLs are more cost-effective than fragment-based libraries, since many of the methodologies utilized for HTS of DECLs can be performed on the bench without large, dedicated equipment (Yuen & Franzini, 2017). A tagged target protein can be immobilized to a solid support, and following incubation with the DECL, small molecules coupled to DNA or PNA that bind to the target can be enriched via tag-specific methodologies. Following elution, the small molecules can be identified via PCR or sequencing in the case of DNA-encoded libraries, or via mass spectrometry, SPR or NMR, in the case of PNA-encoded libraries (Mannocci et al., 2011) (Fig. 4c). Amplification can also be carried out in situ (Yuen & Franzini, 2017).
Regardless of the particular approach, inhibitor development with small-molecule HTS approaches depends on the solubility of chemical moieties (Erlanson, 2012). This has prompted the use of phenotypic screens to initialize screening (Wagner, 2016), and is one of the driving forces behind the field of peptidomimetics, which aims to utilize favorable qualities of peptides, including aqueous solubility, membrane permeability and internalization capacity, while maintaining the ability to interact with proteins and avoiding degradation through proteolysis (Vagner, Qu, & Hruby, 2008). Peptide libraries have also been generated via solid-phase synthesis (Bessette, Rice, & Daugherty, 2004; Kenrick & Daugherty, 2010; Ryvkin et al., 2018; Sidhu, 2000), by covalently linking peptides to a resin and conducting the synthesis in the immobilized form (Fields, 2002). Other than solid-phase peptide synthesis, peptide libraries have been typically generated with the use of bacteria or phage display. For example, antimicrobial peptides continue to be identified through screening peptides expressed in bacteria (Tucker et al., 2018). One study applied phage display to isolate potent competitive peptides for the E2 binding site of HECT E3 ligases that inhibited HECT E3 ligases by oxidizing the active site cysteine (Mund, Lewis, Maslen, & Pelham, 2014). Phage display, in particular, like DECL libraries with fragments, can be harnessed to screen for high-affinity binding polypeptides of immobilized target proteins in a high-throughput manner (Zhang, Ben-David, & Sidhu, 2017; Zhang & Sidhu, 2014).
4. Targeting UPS proteins with synthetic proteins
A major obstacle to the development of small molecule inhibitors for UPS components is the high structural similarity and functional redundancy among proteins in a particular family (Renatus et al., 2006; VanDemark & Hill, 2002). To overcome this limitation, knowledge of the Ub pathway and phage display technologies were merged to produce Ub variants (UbVs) as potent and highly selective inhibitors and activators of UPS enzymes (Ernst et al., 2013; Ernst & Sidhu, 2013; Zhang, Sartori, et al., 2017; Zhang & Sidhu, 2014).
Ub interacts weakly but specifically with thousands of structurally similar proteins, via a common binding surface (Husnjak & Dikic, 2012) and its intrinsic conformational heterogeneity (Lange et al., 2008). In addition, Ub is a highly stable protein that lacks cysteine residues and can be easily produced recombinantly, making it an ideal scaffold for generating inhibitors of proteins in the UPS (Job et al., 2015).
Via protein engineering, several groups have generated UbVs with remarkable potency, specificity and cellular activity for the modulation of the enzymatic activities of UPS proteins. Indeed, given its small size, lack of disulfide bonds, and structural stability, Ub represents an ideal scaffold for the next generation of protein therapeutics (Job et al., 2015).
4.1. UbV inhibitors of deubiquitinases
UbVs are capable of specifically inhibiting DUBs in vitro and in vivo. By mutating residues in the Ub core and on the surface, researchers at Genentech developed a UbV that bound tightly to USP7 and inhibited activity in cells, resulting in enhanced MDM2 ubiquitination and p53 stabilization (Zhang et al., 2013). Our group also developed an inhibitor of USP7 (UbV.7.2, IC50 values in the low nM range) by employing surface mutations only. Notably, UbV.7.2 proved to be more potent, and most importantly, much more selective than Genentech's UbV, showing that core mutations may reduce specificity and thus should be employed with great care. UbV.7.2 inhibited USP7 in cancer cells, and consequently, caused dramatic reductions of MDM2 levels and induction of apoptosis, in synergy with cisplatin, suggesting that UbVs could be used as enhancers of chemotherapeutic drugs (W. Zhang, Sartori, et al., 2017). We also developed a UbV targeting USP10, a second DUB that deubiquitinates p53, and showed that the UbV promoted export of p53 from the nucleus to the cytoplasm (Zhang, Sartori, et al., 2017).
Recently, we developed UbVs for all four Ub-binding domains of USP15 and engineered potent bivalent inhibitors of the enzyme by linking a UbV targeting the DUSP domain to a UbV targeting the catalytic domain (Teyra et al., 2019) (Fig. 5a). These dimeric UbVs exhibited enhanced specificity and inhibition of USP15 catalytic activity, and acted as inhibitors of the transforming growth factor β (TGF- β) signalling pathway in cells (Teyra et al., 2019). We also utilized UbVs to reduce the abundance and activity of cell surface receptors by inhibiting USP8, an enzyme that regulates levels of epidermal growth factor receptor (EGFR) and other receptors (Fig. 5b) (Ernst et al., 2013). Moreover, we recently established a Yeast-Two-Hybrid (Y2H) screening platform (Bruckner, Polge, Lentze, Auerbach, & Schlattner, 2009) for the in vivoselection of UbVs able to inhibit the cellular catalytic activity of USP2 (Pascoe et al., 2019), a DUB involved in the protection of prostate cancer from apoptosis (Priolo et al., 2006). Extending beyond USP DUBs, we targeted OTUB1, a member of the OTU DUB family, with a UbV that bound distal to the active site (Fig. 5c), but nonetheless, potently inhibited K48-linked di-Ub cleavage (Ernst et al., 2013).
Fig. 5.
Ubiquitin variants as modulators of the ubiquitin proteasome system (UPS). The cartoons depict crystal structures of UPS proteins in complex with UbVs. (a) Dimeric UbV.15.D (orange and yellow) complexed with the DUSP domain of USP15 (blue) (PDB entry 6DJ9). (b) USP8 (blue) in complex with the inhibitor UbV.8.2 (orange) (PDB entry 3N3K). (c) OTUB1 (blue) in complex with Ubv.B1.1 (orange) (PDB entry 4I6L). The structure is superposed with OTUB1 (gray) in complex with distal wild-type Ub (Ub.wt, pink) and UbcH5b E2 ligase (purple) covalently conjugated to Ub (yellow) (PDB entry 4DDG). (d) The Middle East Respiratory Syndrome (MERS) virus' USP (blue) in complex with the inhibitor UbV.ME.4 (orange) (PDB entry 5 V69). (e) HECT E3 ligase WWP1 (blue) in complex with the inhibitor UbV.P2.3 (orange) and E2 enzyme UbcH7 (gray) (PDB entry 5HPT). (f) Phosphorylated Cbl (pCbl, blue) in complex with the inhibitor UbV.pCbl (orange) (PDB entry 5O76). The phosphorylated tyrosine residue in pCbl is represented as red sticks. (g) The XIAP RING domain dimer (blue and purple) in complex with the dimeric activator UbV.XR (orange and yellow) (PDB entry 5OT). (h) Fb10-Skp1 (gray and blue, respectively) in complex with the inhibitor UbV.Fl10.1 (orange) (PDB entry 6BVA). (i) UIM-1 of yeast Vps27 (blue) in complex with UbV.v27.1 (orange) (PDB entry 5UCL).
In the past, we have also employed UbVs as inhibitors of pathogenic DUBs. In order to promote infection and replication in host cells, while evading the immune response, many human pathogenic viruses have evolved the ability to hijack and manipulate the UPS (Luo, 2016). For example, the Ub-specific protease of the herpes simplex virus 1 antagonizes NF-κB activation, whereas the E6 oncoprotein of the human papillomavirus recruits host E3 ligases to promote Ub-dependent degradation of p53 and consequent carcinogenesis (Hoppe-Seyler, Bossler, Braun, Herrmann, & Hoppe-Seyler, 2018; Ye, Su, Xu, & Zheng, 2017). We developed highly selective and potent UbVs targeting the DUBs of the Middle East respiratory syndrome coronavirus (MERS-CoV) and the Crimean-Congo hemorrhagic fever virus (CCHFV), two emergent and globally important viruses (W. Zhang, Bailey-Elkin, et al., 2017). UbV.ME.4 bound with sub-nanomolar affinity to the MERS-CoV DUB by establishing a wide network of hydrophobic packing and hydrogen bond interactions (Fig. 5d). Cell-based assays showed that UbV.ME.4 efficiently suppressed the DUB-induced activation of IFN-β promoter, thereby providing efficient blockade of viral replication and reducing MERS-CoV progeny titer by ~10,000-fold (W. Zhang, Bailey-Elkin, et al., 2017). Moreover, Han et al. recently highlighted the interplay between the HECT E3 ligase WWP1 and the VP40 matrix protein of Ebola virus in promoting viral budding from the host cell (Han et al., 2017). Therefore, Ub-based modulators of the UPS may enable the generation of novel antiviral drugs.
Saturation mutagenesis scanning experiments (Maynard, Chen, Georgiou, & Iverson, 2002) performed with distinct UbVs targeting the structurally similar catalytic domains of USP2 or USP21 enabled elucidation of the interaction landscape of Ub with the USP family (Leung, Dekel, Shifman, & Sidhu, 2016; Pascoe et al., 2019). Thus, UbVs can be used not only as inhibitors of DUBs, but also, as model molecules to identify alternative target sites for the development of small-molecule inhibitors that can target structurally conserved proteins with high specificity.
4.2. UbV inhibitors and activators of E3 ligases
HECT E3 ligases, which play crucial roles in cancer, neurological disorders and viral infections (Scheffner & Kumar, 2014), contain a HECT domain that forms a covalent bond with Ub (through a catalytic cysteine) before transferring it to a substrate protein. Understanding of the diverse HECT family has been limited by a paucity of selective and potent modulators. Despite enormous efforts, development of synthetic molecules targeting HECT E3s has been hampered by large conformational changes accompanying catalysis, a shallow active site, and dynamic regulation of activity. To overcome this limitation, we systematically developed UbV inhibitors and/or activators for 20 of the 28 human HECT E3s (Zhang et al., 2016). Structural analysis of UbVs targeting HECT E3 ligases revealed that UbV inhibitors acted by blocking the binding site for E2 ligase rather than by blocking the active site (Fig. 5e) (Zhang et al., 2016), further demonstrating that the plasticity of the Ub fold is amenable for the development of UbVs with novel biochemical properties. Surprisingly, a number of UbVs enhanced catalytic activity of HECT E3 ligases (Ernst et al., 2013; Zhang et al., 2016). For example, UbVs that bound tightly and selectively to NEDD4 increased intracellular ubiquitination of the NEDD4 substrate Ying-Yang1 (Ernst et al., 2013) and inhibited the migration of cancer cells by promoting the ubiquitination of Ras homolog gene family member B (RhoB) (Zhang et al., 2016). UbVs also modulated the activity of membrane-bound ion channels by either activating or inhibiting NEDD4L (Zhang et al., 2016). UbVs that activated or inhibited NEDD4L reduced or increased cell-surface levels of the epithelial sodium channel (ENaC), respectively (Zhang et al., 2016). Since elevated ENaC levels are associated with hypertension (Ronzaud et al., 2013), and suppression of this ion channel has been linked to Liddle's syndrome, novel therapies for hypertension could be developed by targeting NEDD4L with molecules that modulate ENaC levels at the cell surface (Aziz, Memon, Rahman, & Ali, 2016).
With ~600 members, the RING family accounts for >90% of human E3 ligases. In contrast to HECT E3s with an active catalytic cysteine, the RING domain acts as an inert scaffold that recruits E2 enzymes thioesterified with Ub (E2~Ub) and facilitates Ub transfer to substrates. Due to the lack of a catalytic cysteine, inhibition of RING function has been challenging. We identified a UbV targeting the RING domain of the E3 ligase Cbl (UbV.pCbl), which reduced the internalization of EGFR while sustaining Akt pathway activation (Fig. 5f) (Gabrielsen et al., 2017). Remarkably, UbV.pCbl bound tightly to phosphorylated Cbl (pCbl), but not to non-phosphorylated Cbl, demonstrating that UbVs can recognize post-translationally modified proteins with high specificity (Gabrielsen et al., 2017). Unexpectedly, we also isolated a dimeric UbV (UbV.XRD) that recognized the RING domain of the E3 ligase XIAP with high specificity and stimulated ligase activity in vitro and in cells (Gabrielsen et al., 2017). The structure of UbV.XRD in complex with XIAP revealed that UbV.XRD forms a domain-swapped dimer that stabilizes the E3-E2~Ub conformation, resulting in enhanced activity of the E3 ligase (Fig. 5g) (Gabrielsen et al., 2017). Additionally, we utilized the UbV technology to target a Ub-binding exosite on the RING domain of the anaphase-promoting complex subunit 11 (APC11) (Brown et al., 2016; Yamaguchi et al., 2016) to probe the biochemical mechanisms of the large, 13-subunit cell-cycle regulatory anaphase-promoting complex (APC/C). In sum, this work provided a general strategy to inhibit or activate RING E3 ligases to modulate these enzymes in order to understand their catalytic mechanisms and biological functions.
UbV inhibitors have also been developed for members of the multi-subunit Skp1-Cul1-F-box (SCF) E3 ligase family, and these have been shown to affect the mitotic cell cycle progression of cancer cells (Fig. 5h) (Gorelik et al., 2016; Gorelik et al., 2018). Structural analysis of a UbV in complex with its target Skp1-Fbw7 revealed extensive contacts with both Skp1 and the F-box and showed that the UbV inhibited activity by blocking the binding site for Cul1 (Gorelik et al., 2016). Structure-based engineering of a loop on the UbV enabled the development of specific inhibitors for 17 different members of the SCF E3 ligase family (Gorelik et al., 2018). Finally, the UbV technology has been extended to target non-catalytic Ub-binding domains, exemplified by the small, helical Ub-interacting motif of the yeast protein Vps27 (Fig. 5i) (Manczyk et al., 2017).
4.3. UbV inhibitors of non-UPS proteins
Engineered UbVs have also been developed to target proteins that are not known to associate with Ub. Hoffmann et al. used ribosome display (Zahnd, Amstutz, & Pluckthun, 2007) to develop a UbV that bound to and inhibited the activity of tumor necrosis factor alpha (Hoffmann et al., 2012). Similarly, we recently isolated UbVs that bound specifically to the human epidermal growth factor receptor 3 (HER3) and the growth factor receptor-bound protein 2 (Grb2) and showed that UbVs acted as potent antagonists of Grb2-mediated cell signalling (Leung, Jarvik, & Sidhu, 2017).
We also developed a potent and highly selective UbV inhibitor of the tumor suppressor p53-binding protein 1 (53BP1), which recognizes ubiquitinated histone H2A and acts as a central regulator of the double-strand break repair pathway (Fradet-Turcotte et al., 2013). Expression of the UbV in either human or mouse cells prevented the accumulation of 53BP1 at double-strand break sites, stimulating gene conversion and enhancing the efficiency of CRISPR-Cas9 dependent gene editing (Canny et al., 2018). This work highlighted the robustness of protein design and engineering with the Ub scaffold and the potential for targeting DNA damage signalling proteins for applications beyond the UPS.
In summary, UbV technology represents a robust platform for the rapid development of intracellular probes for target discovery and validation. Delivery of UbVs to cells represents a major obstacle for their direct use as therapeutics, but the constant advancements in delivery technologies for folded proteins has the capacity to unlock the potential of UbVs as drug candidates.
5. Harnessing the power of ubiquitination for selective target degradation
5.1. Proteolysis-targeting chimera (PROTAC)
Drug discovery campaigns traditionally focus on the development of tight and specific small molecules that act as inhibitors by blocking binding pockets or active sites of disease-associated proteins. Although this approach has proven successful (Salami & Crews, 2017), it has been limited by the number of druggable protein targets, accounting for only ~3% of the human proteome (Dixon & Stockwell, 2009; Landry & Gies, 2008; Rask-Andersen, Almen, & Schioth, 2011; Santos et al., 2017; Uhlen et al., 2015). In fact, while nuclear receptors, enzymes, ion channels and G-protein-coupled receptors represent the majority of current drug targets, the human proteome is mainly composed of proteins involved in the organization, maintenance and assembly/disassembly of protein complexes that regulate signal transduction events and other critical functions. Therefore, to expand the druggable protein space, researchers have employed alternative approaches that allow targeting of transient protein-protein interactions rather than deep grooves to which small molecules can bind.
This paradigm shift focuses on the use of small molecules to control the cellular levels of drug targets through their precise degradation rather than the inhibition of their activity. This shift from target inhibition to induced target destruction is exemplified by proteolysis-targeting chimeras (PROTACs), small molecules that recruit a ubiquitin E3 ligase for protein degradation (Lucas & Ciulli, 2017; Neklesa, Winkler, & Crews, 2017; Toure & Crews, 2016). In fact, PROTACs are hetero-bifunctional molecules composed of a ligand for a target protein (warhead) and a ligand for an E3 Ub ligase (Toure & Crews, 2016) that mediate the formation of a complex between the drug target and an E3 ligase promoting ubiquitination and subsequent degradation of the target (Fig. 6a).
Fig. 6.
Methods for targeted protein degradation. (a) Representation of the proteolysis-targeting chimera (PROTAC) technology. PROTACs are heterodimeric molecules composed of a protein-targeting warhead connected to an E3 Ub ligase binder via a chemical linker. PROTACs mediate the formation of a stable complex between the target protein and an E3 ligase, triggering ubiquitination and proteosomal degradation of the target protein. (b) Click-formed proteolysis targeting chimeras (CLIPTACs) exploit the bio-orthogonal click reaction between a trans-cyclooctene-tagged binder for a target protein and a tetrazine-containing E3 Ub ligase binder. The two molecules are administered individually and, upon cell penetration, form a complete PROTAC that is able to induce protein degradation. (c) Ubiquitibodies are constructed by fusing a scFv to the CHIP E3 Ub ligase. The scFv enables specific recognition of a target protein and the E3 ligase facilitates ubiquitination to trigger degradation. (d) A target protein was fused to a HaloTag to achieve hydrophobic protein tagging via site-specific and covalent conjugation with a HaloTagged-adamantyl moiety (England, Luo, & Cai, 2015). The adamantyl moiety displayed on the target protein mimics the partially unfolded state of proteins, triggering the activity of cellular quality control machinery, which recruits the UPS for protein degradation. (e) A hydrophobic tag was developed by modifying an inhibitor of a target protein with an hydrophobic (Boc3Arg) moiety (Long et al., 2012). Upon binding to the target protein, the modified inhibitor enabled recruitment to the proteasome and subsequent degradation.
The first PROTAC was built by linking target protein inhibitors to peptides that recruited the SCFβ-TRCP E3 ligase (Sakamoto et al., 2001). Although these protein degraders induced rapid depletion of targets such as the methionine aminopeptidase-2, the estrogen receptor and androgen receptors (Sakamoto et al., 2001; Sakamoto et al., 2003), the peptidic nature of the E3 ligase engager and its intrinsic instability posed limits for the application of the technology. A breakthrough in the PROTAC technology occurred with the use of the non-peptidic inhibitor Nutlin as the engager of the E3 ligase MDM2, resulting in partial degradation of androgen receptors in cells (Schneekloth, Pucheault, Tae, & Crews, 2008). However, due to the limited activity and complex chemical composition of this second generation of PROTACs, researchers were motivated to further improve stability and pharmacokinetic properties by decreasing size and identifying novel E3 ligase recruiters (Buckley et al., 2012).These efforts led to the development of current PROTAC technologies.
PROTACs rely on degradation rather than inhibition of target proteins, showing a remarkable efficacy at doses 1000 times lower than conventional drugs (Bondeson et al., 2015), a feature that has the potential to reduce systemic toxicity in vivo. Recently, a systematic analysis of the mode of action of PROTACs revealed further benefits of this technology (Bondeson et al., 2018). This study showed that the stability of the target:PROTAC:E3 ternary complex rather than the affinity of the target:PROTAC interaction is the main factor responsible for activity (Bondeson et al., 2018). This finding opens the possibility of turning low affinity small molecules into potent protein degraders, potentially allowing the rapid development of new drugs.
Comparison between the degrading activities of PROTACs with different E3 ligase ligands demonstrated that the Ub ligase engager not only dictates the efficacy of these molecules, but also controls the target specificity (Bondeson et al., 2018). In fact, PROTACs based on promiscuous kinase inhibitors (Bondeson et al., 2018; H. T. Huang et al., 2018) were rendered more selective simply by engaging different E3 ligases, suggesting that the specificity of degradation induced by a PROTAC is mainly driven by the endogenous ligand specificity of the E3 ligase. Therefore, PROTACs hold promise for the development of highly specific protein degraders even when using non-specific warheads to target proteins of interest. Thus far, only a limited number of E3 ligases have been exploited for the generation of PROTACs. The technology can be further advanced by the development of novel ligands to recruit other Ub ligases among the ~600 human E3 enzymes (Nakayama & Nakayama, 2006).
The importance of each PROTAC component - warhead, linker and E3 engager - for achieving optimal target degradation has been described (Bondeson et al., 2015; Lai et al., 2016; Maniaci et al., 2017; Zengerle, Chan, & Ciulli, 2015), but the prominent role played by the linker geometry has emerged only recently (Burslem et al., 2018; Gadd et al., 2017). For example, the short linker connecting the promiscuous bromo- and extra-terminal (BET) JQ1 ligand and the potent VH032 von Hippel–Lindau target's E3 ligase (VHL) engager, allowed for the formation of a stable target:VHL interface, ensuring the efficient and specific degradation of Brd4 over the highly homologous Brd2 and Brd3 proteins (Gadd et al., 2017).
PROTACs offer numerous advantages over conventional small molecules and provide an opportunity to expand the druggable protein target space. However, a number of questions remain to be addressed. Despite the modular nature of PROTACs, the efficacy and specificity of new PROTACs needs to be assessed empirically, as changes in the PROTAC chemical composition and geometry might affect activity and specificity. Therefore, the development of high throughput methods for synthesis and characterization of PROTAC molecules will accelerate the development of new inhibitors. Limited in vivo data are available (Qin et al., 2018; Saenz et al., 2017); toxicity, pharmacokinetics, bioavailability, tissue distribution and metabolism of PROTAC molecules will require extensive investigation. The large size of PROTACs (~1000 Da) might affect cell permeability, potentially resulting in sub-optimal presentation of a target to the E3 ligase; smaller PROTACs could improve cellular uptake and potency. To overcome this obstacle, Lebraud et al. developed in-cell click-formed proteolysis targeting chimeras (CLIPTACs): PROTACs that are formed intracellularly upon click chemistry–mediated self assembly of the target warhead and an E3 ligase engager (Lebraud, Wright, Johnson, & Heightman, 2016) (Fig. 6b).
5.2. Other strategies for targeted degradation
Similar to the PROTAC technology, chimeric proteins formed by a single-chain variable fragment (scFv) of an antibody or a fibronectin type III domain monobody fused to the E3 Ub ligase carboxyl terminus of Hsc70-interacting protein (CHIP) have also been used to degrade target proteins (Portnoff, Stephens, Varner, & DeLisa, 2014). Although this strategy enabled the efficient depletion of target proteins in cells, its application has been hindered by the correct folding of disulfide bond-containing scFvs in the reducing environment of the cell cytoplasm (Fig. 6c).
Another method to trigger selective protein degradation via the UPS is represented by the modification of proteins with hydrophobic tags (Neklesa & Crews, 2012). Covalent modification of proteins with hydrophobic tags induces the recruitment of molecular chaperones, which promote the ubiquitination and proteolytic removal of the target by the proteasome 26S (Neklesa et al., 2011) (Fig. 6d). This approach achieved selective depletion of HER3 and reduced cancer cell proliferation (Xie et al., 2014). Similarly, linkage of small molecule inhibitors to a hydrophobic tert-butyl carbamate-protected arginine (Boc3Arg) moiety induced selective degradation of target proteins (Long, Gollapalli, & Hedstrom, 2012) (Fig. 6e). In contrast to hydrophobic tags, the (Boc3Arg) moiety enabled the efficient depletion of both glutathione S-transferase (GST) and bacterial dihydrofolate reductase (DHFR) in a manner independent of Ub and ATP, suggesting that degradation was mediated by a direct interaction between the (Boc3Arg) moiety and the proteasome subunit 20S (Long et al., 2012), without requiring target ubiquitination. These recent developments demonstrate that induced protein degradation has been significantly improved over recent years and is now well positioned to expand the druggable protein space.
6. Summary
The UPS provides a wide canvas for developing next-generation cancer therapeutic agents. While advances in HTS technologies have decreased the time and cost needed to carry out drug discovery studies, activity-based small molecule screens have not yielded a satisfying number of viable drug leads. Recent developments in fragment-based medicinal chemistry methodologies offer a promising approach to tackle the small molecule-based screen problems. The advent of UbVs also represents an exciting new technology that can rapidly provide highly potent and selective inhibitors and activators of enzymes that have resisted conventional approaches. Combined with fragment-based drug discovery methods and recent development in DECLs, small molecule screens for chemical compounds that can displace UbVs from target proteins could enable deep and effective sampling of chemical space to modulate the UPS. Finally, PROTAC technology represents another promising means for cancer therapy. Indeed, it seems likely that the most effective cancer therapies in the future could well rely on the combination of these diverse innovations to develop drug-like entities with activities well beyond the scope of current drugs.
Disclosure statement
The authors declare no conflicts of interests.
Acknowledgments
We sincerely apologize to the researchers whose work was not included in this review due to limited space. SSS is supported by a Genome Canada Disruptive Innovation in Genomics grant (OGI-119) and a CIHR operating grant (MOP#136956). WZ is the recipient of the Cancer Research Society/ BMO Bank of Montreal Scholarship for the Next Generation of Scientists.
References
- Adams J., Palombella V.J., Sausville E.A., Johnson J., Destree A., Lazarus D.D.…Elliott P.J. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Research. 1999;59:2615–2622. [PubMed] [Google Scholar]
- Aziz D.A., Memon F., Rahman A., Ali M. Liddle's Syndrome. Journal of Ayub Medical College, Abbottabad. 2016;28:809–811. [PubMed] [Google Scholar]
- Babaoglu K., Simeonov A., Irwin J.J., Nelson M.E., Feng B., Thomas C.J.…Shoichet B.K. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. Journal of Medicinal Chemistry. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S., Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Berkers C.R., Verdoes M., Lichtman E., Fiebiger E., Kessler B.M., Anderson K.C.…Galardy P.J. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nature Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- Bessette P.H., Rice J.J., Daugherty P.S. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Engineering, Design & Selection. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- Bishop P., Rocca D., Henley J.M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. The Biochemical Journal. 2016;473:2453–2462. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemink M.J., Reedijk J. Cisplatin and derived anticancer drugs: mechanism and current status of DNA binding. Metal Ions in Biological Systems. 1996;32:641–685. [PubMed] [Google Scholar]
- Bondeson D.P., Mares A., Smith I.E., Ko E., Campos S., Miah A.H.…Crews C.M. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature Chemical Biology. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson D.P., Smith B.E., Burslem G.M., Buhimschi A.D., Hines J., Jaime-Figueroa S.…Crews C.M. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chemical Biology. 2018;25(78–87) doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois-Daigneault M.C., St-Germain L.E., Roy D.G., Pelin A., Aitken A.S., Arulanandam R.…Bell J.C. Combination of Paclitaxel and MG1 oncolytic virus as a successful strategy for breast cancer treatment. Breast Cancer Research. 2016;18:83. doi: 10.1186/s13058-016-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Lerner R.A. Encoded combinatorial chemistry. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.G., VanderLinden R., Watson E.R., Weissmann F., Ordureau A., Wu K.P.…Schulman B.A. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell. 2016;165:1440–1453. doi: 10.1016/j.cell.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner A., Polge C., Lentze N., Auerbach D., Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. International Journal of Molecular Sciences. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley D.L., Gustafson J.L., Van Molle I., Roth A.G., Tae H.S., Gareiss P.C.…Crews C.M. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angewandte Chemie (International Ed. in English) 2012;51:11463–11467. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., McQuaid D.C., Bondeson D.P.…Crews C.M. The advantages of targeted protein degradation over inhibition: An RTK case study. Cell Chemical Biology. 2018;25(67–77) doi: 10.1016/j.chembiol.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri A., Munegato D., Carli I., Parolin C., Palu G. The ubiquitin-conjugating system: multiple roles in viral replication and infection. Cells. 2014;3:386–417. doi: 10.3390/cells3020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny M.D., Moatti N., Wan L.C.K., Fradet-Turcotte A., Krasner D., Mateos-Gomez P.A.…Durocher D. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nature Biotechnology. 2018;36:95–102. doi: 10.1038/nbt.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Frezza M., Schmitt S., Kanwar J., Dou Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Current Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Dexheimer T.S., Ai Y., Liang Q., Villamil M.A., Inglese J.…Zhuang Z. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chemistry & Biology. 2011;18:1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J., Tsu C.A., Gavin J.M., Milhollen M.A., Bruzzese F.J., Mallender W.D.…Dick L.R. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. The Journal of Biological Chemistry. 2011;286:40867–40877. doi: 10.1074/jbc.M111.279984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Jiang H., Xu W., Li X., Dempsey D.R., Zhang X.…Cole P.A. A Tunable Brake for HECT Ubiquitin Ligases. Molecular Cell. 2017;66 doi: 10.1016/j.molcel.2017.03.020. 345–357.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Fan Y.H., Xu X., Zhang H., Dou J., Tang Y.…Yang J. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciehanover A., Hod Y., Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochemical and Biophysical Research Communications. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov . In: Identifier NCT02372240, A Study of VLX1570 and Dexamethasone in Myeloma Patients - Full Text View - ClinicalTrials.gov. U. S. N. L. o. Medicine, editor. Vol. 2018. National Institutes of Health; Bethesda (MD): 2018. [Google Scholar]
- Codony-Servat J., Tapia M.A., Bosch M., Oliva C., Domingo-Domenech J., Mellado B.…Albanell J. Differential cellular and molecular effects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Molecular Cancer Therapeutics. 2006;5:665–675. doi: 10.1158/1535-7163.MCT-05-0147. [DOI] [PubMed] [Google Scholar]
- Croston G.E. The utility of target-based discovery. Expert Opinion on Drug Discovery. 2017;12:427–429. doi: 10.1080/17460441.2017.1308351. [DOI] [PubMed] [Google Scholar]
- Dang L.C., Melandri F.D., Stein R.L. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- Dantuma N.P., Bott L.C. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Frontiers in Molecular Neuroscience. 2014;7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K.H., Ehrt S., Gutierrez-Ramos J.C., Weich N., Nathan C.F. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Dasari S., Tchounwou P.B. Cisplatin in cancer therapy: molecular mechanisms of action. European Journal of Pharmacology. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.M., Plowright A.T., Valeur E. Directing evolution: the next revolution in drug discovery? Nature Reviews. Drug Discovery. 2017;16:681–698. doi: 10.1038/nrd.2017.146. [DOI] [PubMed] [Google Scholar]
- Davydov I.V., Woods D., Safiran Y.J., Oberoi P., Fearnhead H.O., Fang S.…Vousden K.H. Assay for ubiquitin ligase activity: high-throughput screen for inhibitors of HDM2. Journal of Biomolecular Screening. 2004;9:695–703. doi: 10.1177/1087057104267956. [DOI] [PubMed] [Google Scholar]
- Diallo J.S., Le Boeuf F., Lai F., Cox J., Vaha-Koskela M., Abdelbary H.…Bell J.C. A high-throughput pharmacoviral approach identifies novel oncolytic virus sensitizers. Molecular Therapy. 2010;18:1123–1129. doi: 10.1038/mt.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Stockwell B.R. Identifying druggable disease-modifying gene products. Current Opinion in Chemical Biology. 2009;13:549–555. doi: 10.1016/j.cbpa.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge D. In Targeting the Ubiquitin Proteasome System. Discovery on Target; Boston, Massachusetts, United States of America: 2016. Discovery of Highly Selective Macrocyclic Inhibitors of DUBs: USP9x as a Case Study. [Google Scholar]
- England C.G., Luo H., Cai W. HaloTag technology: a versatile platform for biomedical applications. Bioconjugate Chemistry. 2015;26:975–986. doi: 10.1021/acs.bioconjchem.5b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson D.A. Introduction to fragment-based drug discovery. Topics in Current Chemistry. 2012;317:1–32. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- Ernst A., Avvakumov G., Tong J., Fan Y., Zhao Y., Alberts P.…Sidhu S.S. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A., Sidhu S.S. Engineering ubiquitin to modulate the ubiquitin proteosome system. Cell Cycle. 2013;12:1651–1652. doi: 10.4161/cc.24985. [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z., Walker O., Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014;3:1027–1088. doi: 10.3390/cells3041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favalli N., Bassi G., Scheuermann J., Neri D. DNA-encoded chemical libraries - achievements and remaining challenges. FEBS Letters. 2018;592:2168–2180. doi: 10.1002/1873-3468.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields G.B. Introduction to peptide synthesis. Current Protocols in Protein Science. 2002 doi: 10.1002/0471140864.ps1801s26. (Chapter 18, Unit 18 11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W., Saba N., Shen M., Ghias M., Liu J., Gupta S.D.…Bhalla K.N. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Research. 2014;74:2520–2532. doi: 10.1158/0008-5472.CAN-13-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczek M.G., Naseeb S., Delneri D. History of genome editing in yeast. Yeast. 2018;35:361–368. doi: 10.1002/yea.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Diaz C., Orthwein A., Leung C.C., Huang H.…Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini R.M., Biendl S., Mikutis G., Samain F., Scheuermann J., Neri D. “Cap-and-Catch” purification for enhancing the quality of libraries of DNA conjugates. ACS Combinatorial Science. 2015;17:393–398. doi: 10.1021/acscombsci.5b00072. [DOI] [PubMed] [Google Scholar]
- Furka A. History of Combinatorial Chemistry. Drug Development Research. 1995;36:1–12. [Google Scholar]
- Furka A.S.F., Asgedom M., Diboa G. 1st ed. Vol. 5. VSP; Utrecht, The Netherlands: 1988. Highlights of Modern Biochemistry, Proceedings of the 14th International Congress of Biochemistry. [Google Scholar]
- Gabrielsen M., Buetow L., Nakasone M.A., Ahmed S.F., Sibbet G.J., Smith B.O.…Huang D.T. A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants. Molecular Cell. 2017;68:456–470. doi: 10.1016/j.molcel.2017.09.027. e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd M.S., Testa A., Lucas X., Chan K.H., Chen W., Lamont D.J.…Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nature Chemical Biology. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo L.H., Ko J., Donoghue D.J. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin J.M., Hoar K., Xu Q., Ma J., Lin Y., Chen J.…Chen J.J. Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway. The Journal of Biological Chemistry. 2014;289:22648–22658. doi: 10.1074/jbc.M114.573972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavory G., O'Dowd C.R., Helm M.D., Flasz J., Arkoudis E., Dossang A.…Harrison T. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nature Chemical Biology. 2018;14:118–125. doi: 10.1038/nchembio.2528. [DOI] [PubMed] [Google Scholar]
- Ge Z., Leighton J.S., Wang Y., Peng X., Chen Z., Chen H.…Liang H. Integrated genomic analysis of the ubiquitin pathway across cancer types. Cell Reports. 2018;23:213–226. doi: 10.1016/j.celrep.2018.03.047. e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombodorj N., Yokobori T., Yoshiyama S., Kawabata-Iwakawa R., Rokudai S., Horikoshi I.…Nakano T. Inhibition of Ubiquitin-conjugating enzyme E2 may activate the degradation of hypoxia-inducible factors and, thus, overcome cellular resistance to radiation in colorectal cancer. Anticancer Research. 2017;37:2425–2436. doi: 10.21873/anticanres.11582. [DOI] [PubMed] [Google Scholar]
- Goodnow R.A., Jr., Dumelin C.E., Keefe A.D. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nature Reviews. Drug Discovery. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Gorelik M., Manczyk N., Pavlenco A., Kurinov I., Sidhu S.S., Sicheri F. A structure-based strategy for engineering selective ubiquitin variant inhibitors of Skp1-Cul1-F-Box ubiquitin ligases. Structure. 2018;26:1226–1236. doi: 10.1016/j.str.2018.06.004. e1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik M., Orlicky S., Sartori M.A., Tang X., Marcon E., Kurinov I.…Sidhu S.S. Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:3527–3532. doi: 10.1073/pnas.1519389113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M., Pelzer C., Schmidtke G., Hofmann K. Activating the ubiquitin family: UBA6 challenges the field. Trends in Biochemical Sciences. 2008;33:230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Hamouda M.A., Belhacene N., Puissant A., Colosetti P., Robert G., Jacquel A.…Luciano F. The small heat shock protein B8 (HSPB8) confers resistance to bortezomib by promoting autophagic removal of misfolded proteins in multiple myeloma cells. Oncotarget. 2014;5:6252–6266. doi: 10.18632/oncotarget.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Sagum C.A., Takizawa F., Ruthel G., Berry C.T., Kong J.…Harty R.N. Ubiquitin Ligase WWP1 Interacts with Ebola Virus VP40 To Regulate Egress. Journal of Virology. 2017;91 doi: 10.1128/JVI.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann M.M., Oprea T.I. Pursuing the leadlikeness concept in pharmaceutical research. Current Opinion in Chemical Biology. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Hansen M.H., Blakskjaer P., Petersen L.K., Hansen T.H., Hojfeldt J.W., Gothelf K.V., Hansen N.J. A yoctoliter-scale DNA reactor for small-molecule evolution. Journal of the American Chemical Society. 2009;131:1322–1327. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- Hermanns T., Pichlo C., Woiwode I., Klopffleisch K., Witting K.F., Ovaa H.…Hofmann K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nature Communications. 2018;9:799. doi: 10.1038/s41467-018-03148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.D., Edwards R.A., Markin C.J., McDonald D., Pulvino M., Huen M.S.…Glover J.N. Covalent inhibition of Ubc13 affects ubiquitin signaling and reveals active site elements important for targeting. ACS Chemical Biology. 2015;10:1718–1728. doi: 10.1021/acschembio.5b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Kovermann M., Lilie H., Fiedler M., Balbach J., Rudolph R., Pfeifer S. New binding mode to TNF-alpha revealed by ubiquitin-based artificial binding protein. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031298. e31298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe-Seyler K., Bossler F., Braun J.A., Herrmann A.L., Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends in Microbiology. 2018;26:158–168. doi: 10.1016/j.tim.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Huang H.T., Dobrovolsky D., Paulk J., Yang G., Weisberg E.L., Doctor Z.M.…Gray N.S. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chemical Biology. 2018;25:88–99. doi: 10.1016/j.chembiol.2017.10.005. e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W.…D'Andrea A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biology. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Husnjak K., Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual Review of Biochemistry. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Iida S., Ichinohe T., Shinagawa A., Suzuki K., Takezako N., Aoki M. Safety and efficacy of daratumumab in combination with bortezomib and dexamethasone in Japanese patients with relapsed or refractory multiple myeloma. International Journal of Hematology. 2018;107:460–467. doi: 10.1007/s12185-017-2390-2. [DOI] [PubMed] [Google Scholar]
- Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y.…Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Iwakuma T., Lozano G. MDM2, an introduction. Molecular Cancer Research. 2003;1:993–1000. [PubMed] [Google Scholar]
- Janzen W.P. Screening technologies for small molecule discovery: the state of the art. Chemistry & Biology. 2014;21:1162–1170. doi: 10.1016/j.chembiol.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Jencks W.P. On the attribution and additivity of binding energies. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job F., Settele F., Lorey S., Rundfeldt C., Baumann L., Beck-Sickinger A.G.…Bosse-Doenecke E. Ubiquitin is a versatile scaffold protein for the generation of molecules with de novo binding and advantageous drug-like properties. FEBS Open Bio. 2015;5:579–593. doi: 10.1016/j.fob.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar G., Keskin O., Fraternali F., Gursoy A. Emerging role of the ubiquitin-proteasome system as drug targets. Current Pharmaceutical Design. 2013;19:3175–3189. doi: 10.2174/1381612811319180002. [DOI] [PubMed] [Google Scholar]
- Kategaya L., Di Lello P., Rouge L., Pastor R., Clark K.R., Drummond J.…Wertz I.E. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–538. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- Kenrick S.A., Daugherty P.S. Bacterial display enables efficient and quantitative peptide affinity maturation. Protein Engineering, Design & Selection. 2010;23:9–17. doi: 10.1093/protein/gzp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseru G.M., Makara G.M. The influence of lead discovery strategies on the properties of drug candidates. Nature Reviews. Drug Discovery. 2009;8:203–212. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Nishigaki K. Enzymatic synthesis of code regions for encoded combinatorial chemistry (ECC) Nucleic Acids Symposium Series. 1995:201–202. [PubMed] [Google Scholar]
- Kronke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConkey M.…Ebert B.L. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E.…Crews C.M. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angewandte Chemie (International Ed. in English) 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoree B., Hubbard R.E. Current perspectives in fragment-based lead discovery (FBLD) Essays in Biochemistry. 2017;61:453–464. doi: 10.1042/EBC20170028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry Y., Gies J.P. Drugs and their molecular targets: an updated overview. Fundamental & Clinical Pharmacology. 2008;22:1–18. doi: 10.1111/j.1472-8206.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- Lange O.F., Lakomek N.A., Fares C., Schroder G.F., Walter K.F., Becker S.…de Groot B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- Lebraud H., Wright D.J., Johnson C.N., Heightman T.D. Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. ACS Central Science. 2016;2:927–934. doi: 10.1021/acscentsci.6b00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K., Chang N., Yoon Y., Yang H., Cho H., Kim E.…Lee J. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro-Oncology. 2016;18:37–47. doi: 10.1093/neuonc/nov091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung I., Dekel A., Shifman J.M., Sidhu S.S. Saturation scanning of ubiquitin variants reveals a common hot spot for binding to USP2 and USP21. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:8705–8710. doi: 10.1073/pnas.1524648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung I., Jarvik N., Sidhu S.S. A highly diverse and functional naive ubiquitin variant library for generation of intracellular affinity reagents. Journal of Molecular Biology. 2017;429:115–127. doi: 10.1016/j.jmb.2016.11.016. [DOI] [PubMed] [Google Scholar]
- Li G., Zheng W., Liu Y., Li X. Novel encoding methods for DNA-templated chemical libraries. Current Opinion in Chemical Biology. 2015;26:25–33. doi: 10.1016/j.cbpa.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Li X., Elmira E., Rohondia S., Wang J., Liu J., Dou Q.P. A patent review of the ubiquitin ligase system: 2015–2018. Expert Opinion on Therapeutic Patents. 2018;28:919–937. doi: 10.1080/13543776.2018.1549229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu S., Huang H., Liu N., Zhao C., Liao S.…Liu J. Gambogic acid is a tissue-specific proteasome inhibitor in vitro and in vivo. Cell Reports. 2013;3:211–222. doi: 10.1016/j.celrep.2012.11.023. [DOI] [PubMed] [Google Scholar]
- Lill J.R., Wertz I.E. Toward understanding ubiquitin-modifying enzymes: from pharmacological targeting to proteomics. Trends in Pharmacological Sciences. 2014;35:187–207. doi: 10.1016/j.tips.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Litovchick A., Dumelin C.E., Habeshian S., Gikunju D., Guie M.A., Centrella P.…Clark M.A. Encoded library synthesis using chemical ligation and the discovery of seh inhibitors from a 334-million member library. Scientific Reports. 2015;5:10916. doi: 10.1038/srep10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sun C., Olejniczak E.T., Meadows R.P., Betz S.F., Oost T.…Fesik S.W. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Locke M., Toth J.I., Petroski M.D. Lys11- and Lys48-linked ubiquitin chains interact with p97 during endoplasmic-reticulum-associated degradation. The Biochemical Journal. 2014;459:205–216. doi: 10.1042/BJ20120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M.J., Gollapalli D.R., Hedstrom L. Inhibitor mediated protein degradation. Chemistry & Biology. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Middleton R.E., Sun H., Naniong M., Ott C.J., Mitsiades C.S.…Kaelin W.G., Jr. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas X., Ciulli A. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Current Opinion in Structural Biology. 2017;44:101–110. doi: 10.1016/j.sbi.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Luo H. Interplay between the virus and the ubiquitin-proteasome system: Molecular mechanism of viral pathogenesis. Current Opinion in Virology. 2016;17:1–10. doi: 10.1016/j.coviro.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczyk N., Yates B.P., Veggiani G., Ernst A., Sicheri F., Sidhu S.S. Structural and functional characterization of a ubiquitin variant engineered for tight and specific binding to an alpha-helical ubiquitin interacting motif. Protein Science. 2017;26:1060–1069. doi: 10.1002/pro.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniaci C., Hughes S.J., Testa A., Chen W., Lamont D.J., Rocha S.…Ciulli A. Homo-PROTACs: Bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nature Communications. 2017;8:830. doi: 10.1038/s41467-017-00954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannocci L., Leimbacher M., Wichert M., Scheuermann J., Neri D. 20 years of DNA-encoded chemical libraries. Chemical Communication (Cambridge) 2011;47:12747–12753. doi: 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- Maynard J.A., Chen G., Georgiou G., Iverson B.L. Vol. 182. Humana Press; Totowa, NJ: 2002. In Vitro Mutagenesis Protocols. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- McHugh A., Fernandes K., South A.P., Mellerio J.E., Salas-Alanis J.C., Proby C.M.…Saville M.K. Preclinical comparison of proteasome and ubiquitin E1 enzyme inhibitors in cutaneous squamous cell carcinoma: The identification of mechanisms of differential sensitivity. Oncotarget. 2018;9:20265–20281. doi: 10.18632/oncotarget.24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreale F.E., Walden H. Types of ubiquitin ligases. Cell. 2016;165:248. doi: 10.1016/j.cell.2016.03.003. e241. [DOI] [PubMed] [Google Scholar]
- Mulder L.C., Harari A., Simon V. Cytidine deamination induced HIV-1 drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M.P., Witting K., Berlin I., Pruneda J.N., Wu K.P., Chang J.G.…Ovaa H. A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes. Nature Chemical Biology. 2016;12:523–530. doi: 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. DNA tags help the hunt for drugs. Nature. 2016;530:367–369. doi: 10.1038/530367a. [DOI] [PubMed] [Google Scholar]
- Mund T., Lewis M.J., Maslen S., Pelham H.R. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16736–16741. doi: 10.1073/pnas.1412152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M., Jolly L.A., Gecz J., Wood S.A. La FAM fatale: USP9X in development and disease. Cellular and Molecular Life Sciences. 2015;72:2075–2089. doi: 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K.I., Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nature Reviews. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Neklesa T.K., Crews C.M. Chemical biology: Greasy tags for protein removal. Nature. 2012;487:308–309. doi: 10.1038/487308a. [DOI] [PubMed] [Google Scholar]
- Neklesa T.K., Tae H.S., Schneekloth A.R., Stulberg M.J., Corson T.W., Sundberg T.B.…Crews C.M. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nature Chemical Biology. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacology & Therapeutics. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D'Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Molecular Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Nocentini A., Supuran C.T., Winum J.Y. Benzoxaborole compounds for therapeutic uses: a patent review (2010–2018) Expert Opinion on Therapeutic Patents. 2018;28:493–504. doi: 10.1080/13543776.2018.1473379. [DOI] [PubMed] [Google Scholar]
- Nucifora F.C., Jr., Nucifora L.G., Ng C.H., Arbez N., Guo Y., Roby E.…Ross C.A. Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nature Communications. 2016;7:11792. doi: 10.1038/ncomms11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dowd C.R., Helm M.D., Rountree J.S.S., Flasz J.T., Arkoudis E., Miel H.…Harrison T. Identification and structure-guided development of Pyrimidinone based USP7 inhibitors. ACS Medicinal Chemistry Letters. 2018;9:238–243. doi: 10.1021/acsmedchemlett.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., Paulo J.A., Zhang W., Ahfeldt T., Zhang J., Cohn E.F.…Harper J.W. Dynamics of PARKIN-dependent mitochondrial Ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Molecular Cell. 2018;70 doi: 10.1016/j.molcel.2018.03.012. 211-227.e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C.A., Baljinnyam B., Zakharov A.V., Jadhav A., Simeonov A., Zhuang Z. Cell lysate-based AlphaLISA Deubiquitinase assay platform for identification of small molecule inhibitors. ACS Chemical Biology. 2017;12:2399–2407. doi: 10.1021/acschembio.7b00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan E., Yu H., Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A., Young M.A., Donato N.J. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Research. 2014;74:4955–4966. doi: 10.1158/0008-5472.CAN-14-1211. [DOI] [PubMed] [Google Scholar]
- Parlati F., Lee S.J., Aujay M., Suzuki E., Levitsky K., Lorens J.B.…Bennett M.K. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439–3447. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]
- Pascoe N., Seetharaman A., Teyra J., Manczyk N., Satori M.A., Tjandra D.…Sidhu S. Yeast two-hybrid analysis for ubiquitin-variant inhibitors of human deubiquitinases. Journal of Molecular Biology. 2019 doi: 10.1016/j.jmb.2019.02.007. (PMID: 30763569) [DOI] [PubMed] [Google Scholar]
- Pereira D.A., Williams J.A. Origin and evolution of high throughput screening. British Journal of Pharmacology. 2007;152:53–61. doi: 10.1038/sj.bjp.0707373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoff A.D., Stephens E.A., Varner J.D., DeLisa M.P. Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. The Journal of Biological Chemistry. 2014;289:7844–7855. doi: 10.1074/jbc.M113.544825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potyrailo R., Rajan K., Stoewe K., Takeuchi I., Chisholm B., Lam H. Combinatorial and high-throughput screening of materials libraries: Review of state of the art. ACS Combinatorial Science. 2011;13:579–633. doi: 10.1021/co200007w. [DOI] [PubMed] [Google Scholar]
- Pozhidaeva A., Valles G., Wang F., Wu J., Sterner D.E., Nguyen P.…Bezsonova I. USP7-specific inhibitors target and modify the Enzyme's active site via distinct chemical mechanisms. Cell Chemical Biology. 2017;24:1501–1512. doi: 10.1016/j.chembiol.2017.09.004. e1505. [DOI] [PubMed] [Google Scholar]
- Priolo C., Tang D., Brahamandan M., Benassi B., Sicinska E., Ogino S.…Loda M. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Research. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- Pulvino M., Liang Y., Oleksyn D., DeRan M., Van Pelt E., Shapiro J.…Zhao J. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood. 2012;120:1668–1677. doi: 10.1182/blood-2012-02-406074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Hu Y., Zhou B., Fernandez-Salas E., Yang C.Y., Liu L.…Wang S. Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the Bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. Journal of Medicinal Chemistry. 2018;61:6685–6704. doi: 10.1021/acs.jmedchem.8b00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M., Almen M.S., Schioth H.B. Trends in the exploitation of novel drug targets. Nature Reviews. Drug Discovery. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Renatus M., Parrado S.G., D'Arcy A., Eidhoff U., Gerhartz B., Hassiepen U.…Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Barlogie B., Berenson J., Singhal S., Jagannath S., Irwin D.…Anderson K.C. A phase 2 study of bortezomib in relapsed, refractory myeloma. The New England Journal of Medicine. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Sonneveld P., Schuster M.W., Irwin D., Stadtmauer E.A., Facon T.…Assessment of Proteasome Inhibition for Extending Remissions, I Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. The New England Journal of Medicine. 2005;352:2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- Ronzaud C., Loffing-Cueni D., Hausel P., Debonneville A., Malsure S.R., Fowler-Jaeger N.…Staub O. Renal tubular NEDD4-2 deficiency causes NCC-mediated salt-dependent hypertension. The Journal of Clinical Investigation. 2013;123:657–665. doi: 10.1172/JCI61110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nature Reviews. Molecular Cell Biology. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Ryvkin A., Ashkenazy H., Weiss-Ottolenghi Y., Piller C., Pupko T., Gershoni J.M. Phage display peptide libraries: Deviations from randomness and correctives. Nucleic Acids Research. 2018;46 doi: 10.1093/nar/gky077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz D.T., Fiskus W., Qian Y., Manshouri T., Rajapakshe K., Raina K.…Bhalla K.N. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017;31:1951–1961. doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Verma R., Ransick A., Stein B., Crews C.M., Deshaies R.J. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Molecular & Cellular Proteomics. 2003;2:1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- Salami J., Crews C.M. Waste disposal-an attractive strategy for cancer therapy. Science. 2017;355:1163–1167. doi: 10.1126/science.aam7340. [DOI] [PubMed] [Google Scholar]
- Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G.…Overington J.P. A comprehensive map of molecular drug targets. Nature Reviews. Drug Discovery. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Kumar S. Mammalian HECT ubiquitin-protein ligases: Biological and pathophysiological aspects. Biochimica et Biophysica Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Schneekloth A.R., Pucheault M., Tae H.S., Crews C.M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorganic & Medicinal Chemistry Letters. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman B.A., Harper J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nature Reviews. Molecular Cell Biology. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. Atypical antipsychotics: Mechanism of action. Canadian Journal of Psychiatry. 2002;47:27–38. [PubMed] [Google Scholar]
- Senft D., Qi J., Ronai Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nature Reviews. Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Schmitt S., Buac D., Dou Q.P. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opinion on Therapeutic Targets. 2013;17:1091–1108. doi: 10.1517/14728222.2013.815728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu S.S. Phage display in pharmaceutical biotechnology. Current Opinion in Biotechnology. 2000;11:610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- da Silva S.R., Paiva S.L., Bancerz M., Geletu M., Lewis A.M., Chen J.…Gunning P.T. A selective inhibitor of the UFM1-activating enzyme, UBA5. Bioorganic & Medicinal Chemistry Letters. 2016;26:4542–4547. doi: 10.1016/j.bmcl.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Smith G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S.…Langston S.P. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Strynatka K.A., Gurrola-Gal M.C., Berman J.N., McMaster C.R. How surrogate and chemical genetics in model organisms can suggest therapies for human genetic diseases. Genetics. 2018;208:833–851. doi: 10.1534/genetics.117.300124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek K.N., Komander D. Ubiquitin modifications. Cell Research. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D.C., Anthony J. How were new medicines discovered? Nature Reviews. Drug Discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Swords R.T., Coutre S., Maris M.B., Zeidner J.F., Foran J.M., Cruz J.…Savona M.R. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood. 2018;131:1415–1424. doi: 10.1182/blood-2017-09-805895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords R.T., Watts J., Erba H.P., Altman J.K., Maris M., Anwer F.…DeAngelo D.J. Expanded safety analysis of pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukemia and myelodysplastic syndromes. Blood Cancer Journal. 2017;7 doi: 10.1038/bcj.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyra J., Singer A.U., Schmitges F.W., Jaynes P., Kit Leng Lui S., Polyak M.J.…Sidhu S.S. Structural and functional characterization of ubiquitin variant inhibitors of USP15. Structure. 2019 doi: 10.1016/j.str.2019.01.002. (PMID: 30713027) [DOI] [PubMed] [Google Scholar]
- Tong S., Si Y., Yu H., Zhang L., Xie P., Jiang W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Scientific Reports. 2017;7:5599. doi: 10.1038/s41598-017-06098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure M., Crews C.M. Small-molecule PROTACS: New approaches to protein degradation. Angewandte Chemie (International Ed. in English) 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S., Takeuchi T., Rotinsulu H., Mangindaan R.E., van Soest R.W., Ukai K.…Yokosawa H. Leucettamol a: a new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. Microrhaphis. Bioorganic & Medicinal Chemistry Letters. 2008;18:6319–6320. doi: 10.1016/j.bmcl.2008.10.110. [DOI] [PubMed] [Google Scholar]
- Tucker A.T., Leonard S.P., DuBois C.D., Knauf G.A., Cunningham A.L., Wilke C.O.…Davies B.W. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 2018;172:618–628. doi: 10.1016/j.cell.2017.12.009. e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A.P., Ioannidis S., Krajewski W.W., Pinto-Fernandez A., Heride C., Martin A.C.L.…Komander D. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–486. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A.…Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vagner J., Qu H., Hruby V.J. Peptidomimetics, a synthetic tool of drug discovery. Current Opinion in Chemical Biology. 2008;12:292–296. doi: 10.1016/j.cbpa.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark A.P., Hill C.P. Structural basis of ubiquitylation. Current Opinion in Structural Biology. 2002;12:822–830. doi: 10.1016/s0959-440x(02)00389-5. [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z.…Liu E.A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Wagner B.K. The resurgence of phenotypic screening in drug discovery and development. Expert Opinion on Drug Discovery. 2016;11:121–125. doi: 10.1517/17460441.2016.1122589. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature Reviews. Genetics. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Wang X., Mazurkiewicz M., Hillert E.K., Olofsson M.H., Pierrou S., Hillertz P.…D'Arcy P. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Scientific Reports. 2016;6:26979. doi: 10.1038/srep26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang Y., Ding S., Li J., Song N., Ren Y.…Mei Z. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Research. 2018;28:1186–1194. doi: 10.1038/s41422-018-0091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., Wang X. From discovery to the bedside: Targeting the ubiquitin system. Cell Chemical Biology. 2019;26 doi: 10.1016/j.chembiol.2018.10.022. [DOI] [PubMed] [Google Scholar]
- Wrigley J.D., Eckersley K., Hardern I.M., Millard L., Walters M., Peters S.W.…Hudson K. Enzymatic characterisation of USP7 deubiquitinating activity and inhibition. Cell Biochemistry and Biophysics. 2011;60:99–111. doi: 10.1007/s12013-011-9186-4. [DOI] [PubMed] [Google Scholar]
- Wu G., Chai J., Suber T.L., Wu J.W., Du C., Wang X., Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Xie T., Lim S.M., Westover K.D., Dodge M.E., Ercan D., Ficarro S.B.…Gray N.S. Pharmacological targeting of the pseudokinase Her3. Nature Chemical Biology. 2014;10:1006–1012. doi: 10.1038/nchembio.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., VanderLinden R., Weissmann F., Qiao R., Dube P., Brown N.G.…Schulman B.A. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Molecular Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Kitagaki J., Dai R.M., Tsai Y.C., Lorick K.L., Ludwig R.L.…Weissman A.M. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Research. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ludwig R.L., Jensen J.P., Pierre S.A., Medaglia M.V., Davydov I.V.…Vousden K.H. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Yazbeck V., Shafer D., Perkins E.B., Coppola D., Sokol L., Richards K.L.…Holkova B. A phase II trial of Bortezomib and Vorinostat in mantle cell lymphoma and diffuse large B-cell lymphoma. Clinical Lymphoma, Myeloma & Leukemia. 2018;18:569–575. doi: 10.1016/j.clml.2018.05.023. e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Su C., Xu H., Zheng C. Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-kappaB activation in DNA sensing signal pathway. Journal of Virology. 2017;91 doi: 10.1128/JVI.02417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T., Yan F., Ying M., Cao J., He Q., Zhu H., Yang B. Inhibition of ubiquitin-specific proteases as a novel anticancer therapeutic strategy. Frontiers in Pharmacology. 2018;9:1080. doi: 10.3389/fphar.2018.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen L.H., Franzini R.M. Achievements, challenges, and opportunities in DNA-encoded library research: An academic point of view. Chembiochem. 2017;18:829–836. doi: 10.1002/cbic.201600567. [DOI] [PubMed] [Google Scholar]
- Zahnd C., Amstutz P., Pluckthun A. Ribosome display: Selecting and evolving proteins in vitro that specifically bind to a target. Nature Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- Zambaldo C., Barluenga S., Winssinger N. PNA-encoded chemical libraries. Current Opinion in Chemical Biology. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Zengerle M., Chan K.H., Ciulli A. Selective small molecule induced degradation of the BET Bromodomain protein BRD4. ACS Chemical Biology. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Bailey-Elkin B.A., Knaap R.C.M., Khare B., Dalebout T.J., Johnson G.G.…Sidhu S.S. Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLoS Pathogens. 2017;13 doi: 10.1371/journal.ppat.1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ben-David M., Sidhu S.S. Engineering cell signaling modulators from native protein-protein interactions. Current Opinion in Structural Biology. 2017;45:25–35. doi: 10.1016/j.sbi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sartori M.A., Makhnevych T., Federowicz K.E., Dong X., Liu L.…Sidhu S.S. Generation and validation of intracellular ubiquitin variant inhibitors for USP7 and USP10. Journal of Molecular Biology. 2017;429:3546–3560. doi: 10.1016/j.jmb.2017.05.025. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sidhu S.S. Development of inhibitors in the ubiquitination cascade. FEBS Letters. 2014;588:356–367. doi: 10.1016/j.febslet.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sidhu S.S. Drug development: Allosteric inhibitors hit USP7 hard. Nature Chemical Biology. 2018;14:110–111. doi: 10.1038/nchembio.2557. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wu K.P., Sartori M.A., Kamadurai H.B., Ordureau A., Jiang C.…Sidhu S.S. System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Molecular Cell. 2016;62:121–136. doi: 10.1016/j.molcel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhou L., Rouge L., Phillips A.H., Lam C., Liu P.…Corn J.E. Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7. Nature Chemical Biology. 2013;9:51–58. doi: 10.1038/nchembio.1134. [DOI] [PubMed] [Google Scholar]
- Zheng N., Shabek N. Ubiquitin ligases: Structure, function, and regulation. Annual Review of Biochemistry. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- Zhou P., Bogacki R., McReynolds L., Howley P.M. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Molecular Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Zinngrebe J., Montinaro A., Peltzer N., Walczak H. Ubiquitin in the immune system. EMBO Reports. 2014;15:28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]