Abstract
Leukotrienes are pro-inflammatory lipid mediators, which are biosynthesized via the lipoxygenase pathway of the arachidonic acid cascade. Lipoxygenases form a family of lipid peroxidizing enzymes and human lipoxygenase isoforms have been implicated in the pathogenesis of inflammatory, hyperproliferative (cancer) and neurodegenerative diseases. Lipoxygenases are not restricted to humans but also occur in a large number of pro- and eucaryotic organisms. Lipoxygenase-like sequences have been identified in the three domains of life (bacteria, archaea, eucarya) but because of lacking functional data the occurrence of catalytically active lipoxygenases in archaea still remains an open question. Although the physiological and/or pathophysiological functions of various lipoxygenase isoforms have been studied throughout the last three decades there is no unifying concept for the biological importance of these enzymes. In this review we are summarizing the current knowledge on the distribution of lipoxygenases in living single and multicellular organisms with particular emphasis to higher vertebrates and will also focus on the genetic diversity of enzymes and receptors involved in human leukotriene signaling.
Abbreviations: AA, arachidonic acid; EPA, 5,8,11,14,17-eicosapentaenoic acid; DHA, 4,7,10,13,16,19-docosahexaenoic acid; LOX, lipoxygenase; COX, cyclooxygenase; PG, prostaglandins; Tx, thromboxane; cPLA2, cytosolic phospholipase 2; LTA4H, leukotriene A4 hydrolase; LTC4S, leukotriene C4 synthase; GGT, gamma-glutamyltranspeptidase; DPEP, dipeptidase; Mya, million years ago; Bya, billion years ago; LX, lipoxins; RS, resolvins
Keywords: Eicosanoids, Lipoxygenase, Leukotrienes, Evolution, Gene polymorphisms, SNPs
1. Introduction
Polyunsaturated fatty acids (PUFAs) are important constituents of membrane lipids, which impact membrane functionality [1]. In addition, they also constitute substrates for the biosynthesis of signaling molecules such as prostaglandins [2], leukotrienes [3] or endocannabinoids [4]. Linoleic acid (C18:Δ2, n-6) and arachidonic acid (C20:Δ4, n-6) are abundant polyenoic fatty acids in the ester lipids of mammalian membranes but alpha- (C18:Δ3, n-6) and gamma-linolenic acid (C18:Δ3, n-3), eicosapentaenoic acid (C20:Δ5, n-3) and docosahexaenoic acid (C22:Δ6, n-3) also occur in lower abundance. However, the cellular concentrations of free polyenoic fatty acids are rather small. In order to be converted to prostaglandins, leukotrienes and related substances polyenoic fatty acids must be released from the membrane ester lipids and subsequently be oxygenated via two major alternative metabolic routes (Fig. 1 ). The cyclooxygenase (COX) pathway of the arachidonic acid cascade leads to the biosynthesis of classical prostaglandins (PGE2, PGF2, PGD2), thromboxanes (TxA2) and prostacyclin (PGI2). In contrast, leukotrienes, (LT), lipoxins (LX), resolvins (RS) and others are biosynthesized via the lipoxygenase (LOX) pathway [5]. Leukotrienes are the most extensively studied LOX products [6] and their biosynthesis involves a concerted action of several enzymes such as cytosolic phospholipase A2 (cPLA2), 5-LOX (ALOX5), Leukotriene A4 hydrolase (LTA4H), leukotriene C4 synthase (LTC4S), gamma-glutamyltranspeptidase (gGTs) and dipeptidases (DPEPs).
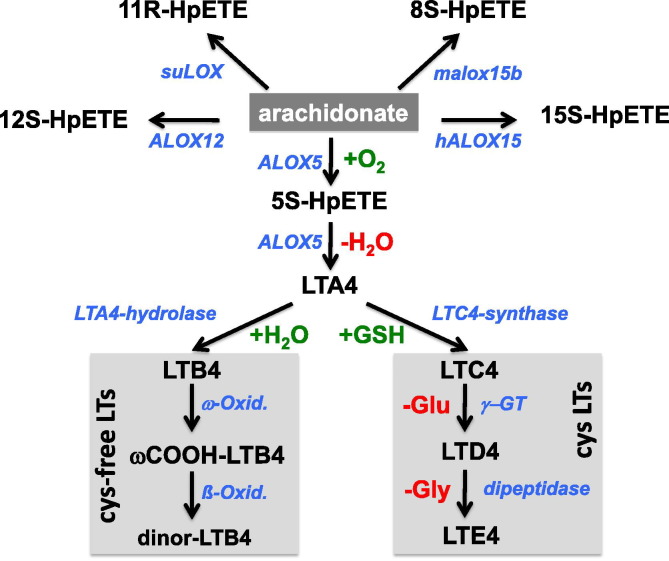
Fig. 1.
Leukotriene biosynthesis and classification. Free arachidonic acid is oxygenated by different ALOX isoforms to different hydroperoxy arachidonic acid isomers (upper part). ALOX5 converts arachidonic acid to 5S-HpETE and further to LTA4, which then gives rise to cysteinyl leukotrienes (LTC4, LTD4, LTE4) and the cysteinyl-free leukotrienes (LTB4). LTB4 is further metabolized via peroxisomal ω-oxidation to ω-carboxy-LTB4 and then by ß-oxidation to 18-carboxy,19,20-dinor-LTB4. hALOX15, human ALOX15, mAlox15b, mouse Alox15b, sea urchin 11R-LOX (suLOX).
There are several current review articles [5], [6], [7], [8], [9], [10], which together provide a detailed overview on chemistry, enzymology, biological roles and medical relevance of LOX and leukotriene signaling. However, to the best of our knowledge evolutionary aspects and genetic variability of LOX and of leukotriene signaling have never been reviewed before. This paper is aimed at filling this gap. After a short introduction into the field we will summarize our current knowledge on classification, biosynthesis and physiological importance of LOX (chapter 2) and leukotriene signaling (chapter 3). Then we will address evolutionary aspects of leukotriene signaling (chapter 4) and this part of the paper is focused on distribution of LOX isoforms and other enzymes of leukotriene biosynthesis within the three domains of life on this planet. The growing number of sequenced genomes of living and extinct organisms has led to identification of an increasing number of potential LOX and/or LOX-like sequences and it would exceed the frame of this review to refer to all of them. Instead, we selected frequently employed model organisms, which represent different stages in the evolution of life on earth and searched their genomes for putative LOX sequences. Since for most of these sequences no functional data are currently available, we used our knowledge on structural biology of these enzymes to draw functional conclusions. Finally, we will summarize the current knowledge on genetic variability of human leukotriene signaling (chapter 5). For this purpose we evaluated the public databases summarizing naturally occurring mutants of human LOX and other enzymes of leukotriene signaling. Here again, the databases are rapidly growing and it is not possible to consider all naturally occurring variants for this review. In this sense the review is somewhat selective and we are not aiming at completeness. However, we would like to apologize to those distinguished colleagues whose work we have not found space to describe and to reference.
2. Leukotrienes: classification, biosynthesis and physiological importance
Leukotrienes (LTs) are pro-inflammatory LOX products regulating the extent of the inflammatory reaction [6], [10], [11]. When formed in pathological quantities these mediators induce pain, fever, and inflammation. LTs are biosynthesized from arachidonic acid (AA) via the consecutive action of different enzymes (Fig. 1). The reaction cascade is initiated by the liberation of AA from the cellular ester lipids by cytosolic phospholipase A2 (cPLA2). There are two principle types of leukotrienes, which exhibit different bioactivities. (i) Cysteinyl-free leukotrienes (LTA4, LTB4) are biosynthesized from arachidonic acid via consecutive dioxygenation (formation of LTA4) and subsequent enzymatic epoxide hydrolysis. For these reactions the 5-lipoxygenase (ALOX5) [12] and leukotriene A4 hydrolase (LTA4H) [13] are responsible. As allylic epoxide LTA4 is very unstable and undergoes rapid hydrolysis. Thus, it may not exhibit systemic bioactivity. LTB4 functions as potent chemoattractant for inflammatory cells and induces chemokinesis and chemotaxis [14], [15]. It binds to cell surface receptors (BLT1, BLT2) and induces different intracellular signaling cascades. ii) Cysteinyl leukotrienes (LTC4, LTD4, LTE4), which are also called peptido leukotrienes, are formed when LTA4 is conjugated with reduced glutathione (GSH) to form LTC4 [3]. This reaction is catalyzed by the LTC4-synthase (LTC4S), which constitutes a special type of GSH-S-transferase [16]. When glutamate is removed from LTC4 by the catalytic activity of gamma-glutamyltranspeptidase (gamma GT) LTD4 is formed and additional removal of glycine by a dipeptidase (DPEP) yields LTE4. The cysteinyl leukotrienes are constituents of the slow reacting substance of anaphylaxis and play a major role in the pathogenesis of hyperergic diseases such as allergic rhinitis and bronchial asthma [6]. They induce airway constriction, bronchoalveolar mucus production and mucosal edema [17], [18]. Two major cysteinyl leukotriene receptors (cysLTR1, cysLTR2) have been identified and antagonists at cysLTR1 (LUKASTs) are available for prescription as anti-asthmatics [19]. In contrast, the role of cysLTR2 still remains a matter of discussion [20]. In addition to classical LT receptors (BLT1, BLT2, cysLTR1, cysLTR2), other deorphanized receptors have been shown to bind LTs but their clinical relevance remains unclear [6].
Most enzymes (cPLA2, ALOX5, LTA4H, LTA4S) involved in leukotriene biosynthesis have been well characterized with respect to their structural and functional properties and a recent review summarizes our current knowledge in this field [6]. The X-ray structures of cPLA2, ALOX5, LTA4H, and LTC4S provide a solid structural basis for a deeper understanding of the catalytic mechanisms and may serve as valuable tools for the development of specific inhibitors interfering with LT biosynthesis. ALOX5 is a key enzyme for LT biosynthesis because of several reasons: (i) It catalyzes two consecutive reactions (dioxygenation of AA to 5S-HpETE and epoxidation of 5S-HpETE to LTA4) in the biosynthetic cascade (Fig. 1). (ii) It competes with other AA metabolizing enzymes, such as other LOX- and COX-isoforms, for the fatty acid substrate and this competition is important for the equilibrium between of pro- vs. anti-inflammatory lipid mediators. (iii) It is the molecular target for zileuton (ZYFLO), a drug that is currently used for treatment of bronchial asthma. (iv) It is also a target of different regulators of LT biosynthesis such as the ALOX5 activating protein [21].
3. Lipoxygenase isoforms
In humans two COX-isoforms [22] and six LOX-isoforms [23] have been described and the corresponding genes have all been sequenced. The ALOX5 gene is localized between 45.87 and 45.94 Mb on the long arm of chromosome 10. All other human LOX genes (ALOX15, ALOX15B, ALOX12, ALOX12B, ALOXE3) were mapped at different orientations to a joint gene cluster on the short arm of chromosome 17. In mice the lipoxygenase genes are located in syntenic regions on chromosome 6 (Alox5) and 11 (Alox15, Alox15b, Alox12, Alox12b, Aloxe3, Aloxe12). The biological roles of the different LOX-isoforms have been explored in the past but for some isoforms their functionalities are not clear. Since ALOX5 constitutes the key enzyme in leukotriene biosynthesis Alox5 deficient mice show reduced responsiveness for allergen-induced asthma [23]. In humans naturally occurring mutants in the ALOX5 gene were related to an increased risk for bronchial asthma [24] but the mechanistic details for the statistical association have not been explored. In addition, ALOX5 is expressed in diseased vessel walls [25] and in Alox5 deficient mice the formation of aortic aneurysms was markedly attenuated when the mice were bred into an apolipoprotein E-deficient background [26]. These data link the Alox5 pathway to inflammatory vessel wall diseases and to the pathogenesis of aortic aneurysms. Similarly, ALOX15B is expressed in human atherosclerotic lesions [27] and thus, this enzyme has also been related to the pathogenesis of atherosclerosis [28]. For other LOX-isoforms including ALOX15 only small amounts of the corresponding mRNA was detected [25]. ALOXE3 and ALOX12B have been implicated in epidermal differentiation. In fact, mice with targeted inactivation of these two genes die shortly after birth because of excessive epidermal dehydration [29], [30]. In humans, loss-of-function mutations in the two genes have been implicated in the pathogenesis of ichthyosis, a rare skin disease [31]. ALOX12 [32] and ALOX15−/− [33] mice are viable and develop normally. However, blood platelets of ALOX12−/− mice show a defective aggregation behavior [32] and the skin of these animals has an increased water permeability [34]. ALOX15−/− mice are protected from diet induced vascular lipid deposition [35], [36] suggesting a role of this LOX-isoform in atherosclerosis. ALOX15−/− mice develop more severe symptoms in different animal arthritis models [37] and corresponding data were obtained in a mouse glomerulonephritis model, in which expression of ALOX15 was enhanced by somatic gene transfer [38]. Moreover, ALOX15−/− mice have been suggested to suffer from myeloproliferative disorders [39] but this hypothesis has recently been challenged since symptoms do not consistently develop [40]. For ALOX15B−/− mice are not available but the expression profile of this enzyme in different cells and tissues suggests a role in normal epithelial differentiation and as a suppressor of prostate cancer development [41].
In biomedical research murine disease models are frequently employed to explore the pathomechanisms of human diseases and to test potential drugs. The basis for this strategy is that in many cases the human proteins and their corresponding murine orthologs share a high degree of structural and functional similarity. Unfortunately, in the LOX field there are pronounced species specific differences between mice and men: (i) There are six functional human LOX-isoforms but mice, rats and even some higher primates (chimpanzee, bonobo) have an additional LOX gene encoding for a seventh functional LOX-isoform (Aloxe12 in mice). In humans the ALOXE12 is corrupted to a functionless pseudogene so that no catalytically active ALOXE12 is expressed. (ii) Human ALOX15 oxygenates arachidonic acid mainly to 15-HpETE and only small amounts of 12-HpETE are formed [42]. In contrast, mouse [33] and rat ALOX15 [43] orthologs are dominantly 12-lipoxygenating enzymes. The reaction specificity of LOX-isoforms is important for their bioactivity since a 15-lipoxygenating LOX-isoform is not capable of initiating biosynthesis of pro-inflammatory leukotrienes of the 5,6-series. The mechanistic basis for the different reaction specificities of ALOX15 orthologs has been explored (triad concept) and the critical amino acids have been determined [44]. It takes a single point mutation at the triad determinants (Ile418Ala or Phe353Ala) to convert the human 15-lipoxygenating ALOX15 into a 12-lipoxygenating enzyme. Applying this strategy human ALOX15 can be murinized and vice versa the murine enzyme can be humanized [44]. (iii) Human ALOX15B converts arachidonic acid almost exclusively to 15-HpETE [45]. In contrast, the mouse ortholog oxygenates the same fatty acid to 8S-HpETE. Mechanistic studies indicated that it simply requires the exchange of two critical amino acid residues to convert the 8-lipoxygenating mouse enzyme to a 15-lipoxygenating humanized isoform. The inverse mutagenesis strategy (murinizing the human enzyme) led to partial alteration of the reaction specificity [45].
4. Evolutionary aspects of lipoxygenases
4.1. Classification of living organisms
Life on earth is characterized by an enormous multiplicity and previous attempts to classify living organisms were mainly based on their phenotypic appearance. In the 18th century living organisms were classified either as vegetabilia or animalia but in the 19th century protista (microorganisms) were introduced as separate group. In 1925 an alternative concept was suggested which subclassified all living organisms into two alternative empires (procarya, eucarya). The only classification criterion for this concept was the presence or absence of a morphologically clearly defined nucleus. In the early 1990s the three-domain concept of life was developed, which classifies all living species on earth mainly on the basis of the structure of their genes encoding for ribosomal RNA (rRNA) into three domains [46]: bacteria, archaea and eucarya (Fig. 2 ). Each of the three domains consists of several kingdoms. This classification system recognizes the fundamental divide between the two procaryotic domains (archaea, bacteria) and also suggests that according to sequence comparisons archaea are more closely related to eucarya than to bacteria. Although the three-domain theory has been challenged, since sequence features and phylogenies of some highly conserved proteins are inconsistent with this concept [47], [48], it is frequently used and widely accepted [49]. The eucarya domain [50] was later on subdivided into 6 different supergroups (Excavata, Amoebozoa, Opishokonta, Rhizaria Chromalveolata, Archaeplastida). This subclassification system deliberately did not use the formal taxonomic rank “kingdom”. However, there are alternative approaches to classify living organisms. In 1998 a six-kingdom model of living organisms [51], [52] has been suggested and this concept was later on refined [50]. According to this classification living organisms should be grouped into 6 kingdoms (Bacteria, Protozoa, Chromista, Plantae, Fungi and Animalia) and this classification system does not accept the principle divide between bacteria and archaea. Here the kingdom Bacteria constitutes the sole kingdom of the procaryotic empire and it consists of two subkingdoms, which are opposed according to their membrane topologies. The bimembranous–unimembranous transition has been suggested to be far more fundamental and the long branch of genetic distance between bacteria and archaea is viewed as having no particular biological relevance [53].
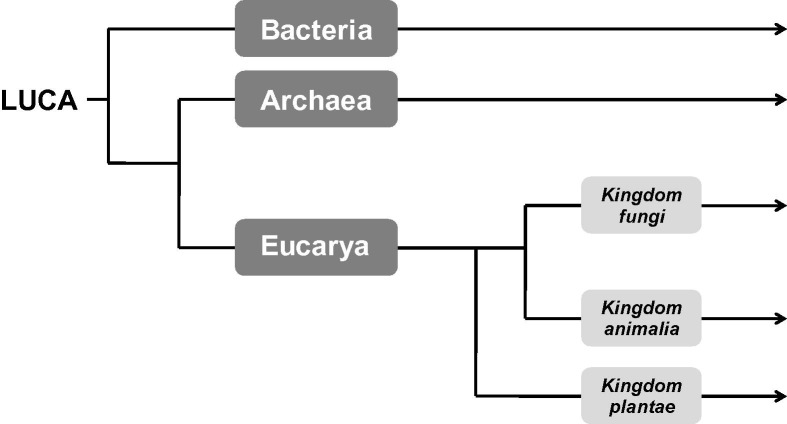
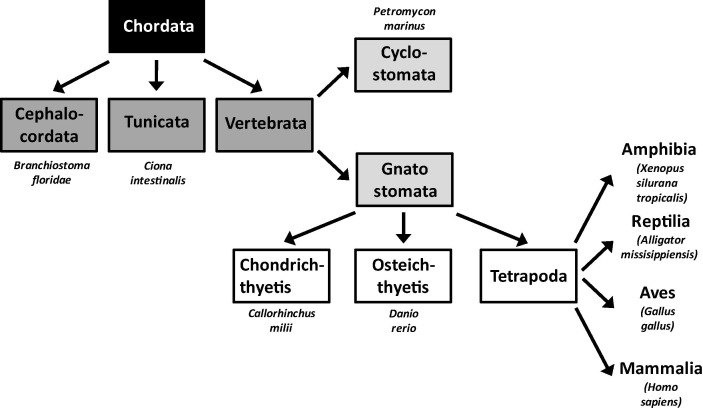
Fig. 2.
The three-domain concept of life. All living organisms on earth can be grouped according to the structure of their genes encoding for their ribosomal RNA in either of the three domains: bacteria, archaea and eucarya. Each of these domains consists of several kingdoms as shown for eucarya. However, there are alternative concepts for subclassification. LUCA, last universal common ancestor.
4.2. Lipoxygenases in viruses
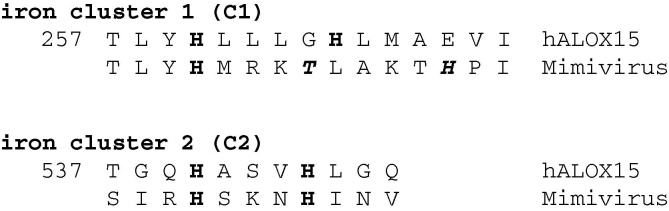
Viruses are small infectious particles that replicate only inside living cells. On their own they are not viable and thus, they have not been considered for any of the classification concepts. However, some viruses persist outside living cells for long time periods. They infect all types of cells regardless whether the host cells originate from plants, animals, bacteria or archaea [54]. Viruses are found in almost every ecosystem on earth and they are the most abundant type of biological entity [55]. For this review we carried out a PubMed search with the key words “lipoxygenase and virus” and obtained some 180 hits. However, neither paper describes a true viral LOX. We next searched the genomes of important human pathogenic viruses (SARS corona virus, human immune deficiency virus, human hepatitis B virus, human hepatitis C virus, Herpes simplex virus type 1, small pocks virus, Polio virus, Dengue virus, Ebola virus, Lyssa virus, human papilloma virus, Influenza virus B, Mumps virus, Yellow fever virus, Vaccinia virus) for LOX genes but did not find any corresponding sequences. However, when we searched all viral genomes present in the NCBI database for LOX-like sequences we obtained five hits. Three of them were eliminated from further consideration since amino acid alignments with known LOXs from organisms of different evolutionary levels (Pseudomonas aeruginosa, Plexaura homomalla, Danio rerio and Homo sapiens) did not provide meaningful results suggesting that these sequences do not encode for functional LOX-isoforms. However, in the genomes of Acanthamoeba Polyphaga Mimivirus (YP_003986553.1) and of Hirudovirus (strain Sangsue, AHA45815.1) LOX-like sequences were found. Next, we aligned on the protein level the Acanthamoeba Polyphaga Mimivirus (YP_003986553.1) sequence with different well described LOXs such as human ALOX15 (NP_001131), human ALOX5 (NP_000689), zebrafish LOX (NP_955912.1), P. homomalla 8R-LOX (AAC47283.1), P. aeruginosa LOX (AAL85880.2) and found a plausible amino acid line-up despite of low degrees of amino acid conservation (<20%). In the alignment with human ALOX15 four of the five proteinogenic iron ligands (His361, His540, H544, Ile663; numbering for human ALOX15) are strictly conserved (Fig. 3 ). His366 of human ALOX15 that constitutes the fifth proteinogenic iron ligand in the human enzyme, aligns with a Leu (no iron-coordination properties of the side chain) in the putative Mimivirus LOX. However, this Leu immediately trails a Thr and this amino acid with its free OH-group might function as iron ligand. Moreover, we found an additional His in iron cluster 1 of the Mimivirus sequence, which also might serve as iron ligand (Fig. 3). Thus, this viral sequence might indeed encode for a catalytically active LOX but to the best of our knowledge no functional data are currently available to prove this hypothesis. In a collaborative approach we recently cloned the cDNA of the putative LOX from the Anthamoeba Polyphaga Mimivirus and are now in the process of expressing and characterizing the corresponding enzyme.
Fig. 3.
Partial amino acid alignment of the iron cluster regions of human ALOX15 and with the corresponding sequences of the Acanthamoeba Polyphaga Mimivirus. The one letter amino acid code is employed and the (potential) iron ligands are indicated in bold. Amino acid numbering for hALOX15.
Virus infection impacts the LOX pathways of a number of host cells [56]. For instance, BS-C1 cells infected with cowpox or vaccinia virus exhibit enhanced 12- and 15-HETE production although the viruses do not contain any LOX genes. On the other hand, human parainfluenza type 3 or Herpes simplex virus type 1 infections did not impact the arachidonate metabolism. In other models of influenza infection lipidomic profiling demonstrated that ALOX5 metabolites correlated with the pathogenic phase of the infection, whereas ALOX15 metabolites were associated with the resolution phase [57]. Hydroxylated linoleic acid, specifically the ratio of 13- to 9-hydroxyoctadecadienoic acid, was identified as a potential biomarker for the immune status during an active infection and some of these findings were recapitulated when human nasopharyngeal lavage fluids obtained during the 2009–2011 influenza seasons were taken through the same protocol [57].
4.3. Lipoxygenases in bacteria
4.3.1. Distribution of lipoxygenase sequences in bacteria
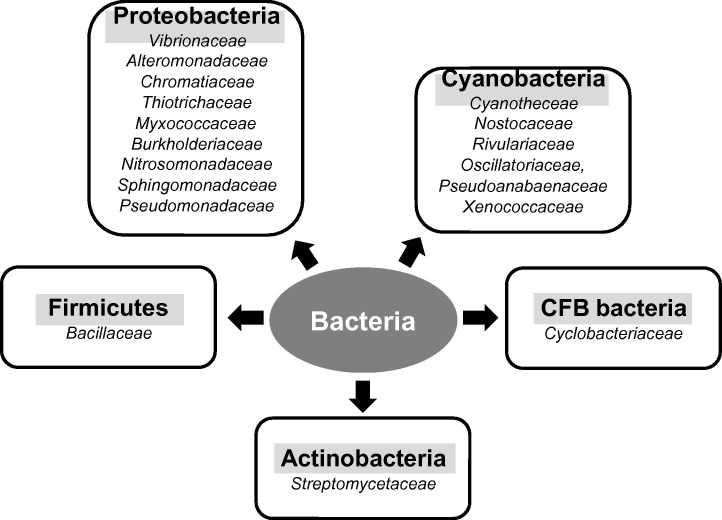
Bacteria form the most diverse domain of life on earth and based on genomic sequence comparisons a phylogeny-driven encyclopedia of bacteria has been established [58]. However, when it comes to LOXs this diversity is much less pronounced. In fact, since the discovery of the first LOXs in plants [59] and mammals [60] it has been the common understanding for many years that LOXs do not occur in bacteria. This conclusion was supported by the observation that the genome of Esche r ichia coli, which serves as major bacterial model organism, does not contain LOX-like sequences. On the other hand, there were scattered reports on the expression of LOXs in certain bacteria [61], [62] but the increasing availability of bacterial genomes put recently the search for bacterial LOXs on a more systematic basis. Today a number of catalytically active LOX-isoforms have been described in bacteria but the biological roles of them remain largely elusive [9], [63], [64], [65], [66], [67]. To obtain more detailed information on the distribution of LOX-sequences in bacteria we searched several sequence databases (Integrated Microbial Genomes, National Center for Biotechnology Information, Uniprot, Ensembl and GenBank) for potential LOX sequences in bacteria. Unfortunately, there is no strongly conserved nucleotide sequence shared by all LOXs, and thus false positive and/or false negative hits are likely. When we searched the bacterial genomes sequenced so far (∼13,000) some 60 LOX-like sequences were identified and these bacterial species include firmicutes, different types of proteobacteria, cyanobacteria, actinobacteria and representatives of the CFB group (Fig. 4 ). Although the vast majority of these potential LOX genes have not been characterized with respect to their functionality the observation that less than 0.5% of all bacterial genomes sequenced so far contain potential LOX genes suggest that these enzymes only sporadically occur in bacteria and thus, may not be essential for bacterial physiology. In fact, most human pathogenic bacteria (Table 1 ) do not carry LOX genes so that for many specific bacterial infections pathogen LOXs may not play a major role for disease development. However, host LOX may well contribute to disease progression or to the immune response. On the other hand, the opportunistic pathogen P. aeruginosa carries several well-defined LOX alleles and thus, the corresponding enzymes might be relevant for the pathogenicity of this microorganism [9], [63], [64], [65], [66], [67]. In cyanobacteria LOX sequences occur with a similarly low frequency even though these organisms are capable of producing oxygen, which is required as second substrate for the LOX reaction substrate. Bacteria living in anaerobic environments do obviously not contain LOX sequences.
Fig. 4.
Bacterial families carrying LOX-like sequences. Bacteria can be subdivided into four major subfamilies [cyanobacteria, proteobacteria, firmicutes, CFB group (Chlorobi, Fibrobacteres, Bacteroides)] and in each of the four subfamilies LOX positive species have been identified. Examples for LOX-positive bacteria are given in italic letters.
Table 1.
Selected human pathogenic bacteria that do not contain LOX-like genes.
| Classification | Human pathogen | Infectious diseases |
|---|---|---|
| Gram-positive | Staphylococcus aureus | Skin infections, pneumonia, sepsis |
| Streptococcus pneumoniae | Pneumonia, meningitis, sinusitis, otitis | |
| Streptococcus pyogenes | Skin infections, tonsillitis, scarlet fever | |
| Enterococcus faecium | Urinary tract infections (UTI), sepsis | |
| Bacillus anthracis | Anthrax | |
| Bacillus cereus | Food poisoning | |
| Clostridium perfringens | Gas gangrene | |
| Clostridium tetani | Tetanus | |
| Corynebacterium dyphteriae | Diphtheria | |
| Listeria monocytogenes | Listeriosis | |
| Gram-negative | Klebsiella pneumoniae | Pneumonia, UTI, sepsis |
| Vibrio cholerae | Severe diarrhea | |
| Salmonella typhi | Typhus | |
| Haemophilus influenzae | Bronchitis, sinusitis, meningitis | |
| Neisseria gonorrhoeae | Urethritis (“gonorrhea”) | |
| Brucella melitensis/abortus | Brucellosis | |
| Bordetella pertussis | Pertussis | |
| Francisella tularensis | Tularemia | |
| Helicobacter pylori | Gastric ulcer | |
| Legionella pneumophilia | Legionnaires’ disease | |
| Spirochetes | Treponema pallidum | Syphilis |
| Borrelia burgdorferi | Lyme disease | |
| Leptospira interrogans | Weil’s disease | |
| Others | Mycobacterium tuberculosis | Tuberculosis |
| Mycobacterium leprae | Lepra | |
| Mycoplasma pneumoniae | Pneumonia | |
4.3.2. Properties, structures and biological function of bacterial lipoxygenases
Most bacterial LOXs characterized so far are arachidonate 15-lipoxygenating enzymes [9]. However, the LOX from the cyanobacteria Nostoc sp. PCC 7120 [68] oxygenates linoleic acid preferentially to 9R-HpODE whereas the enzyme from Anabaena sp. oxygenates different fatty acids with variable specificity [69]. Linoleic acid is converted by the secretable P. aeruginosa LOX [64] almost exclusively to 13S-HpODE [70] and a similar product specificity was reported for two LOX-isoforms from Nostoc punctiforme [65]. LOXs in lower organisms occasionally occur as fusion proteins combining different catalytic activities within one protein. For instance, the cyanobacterium Acaryochloris marina expresses a fusion protein consisting of an N-terminal heme domain with sequence homology to catalase and a C-terminal LOX domain [71]. When the LOX-domain was separately expressed in E. coli it oxygenated omega-3 fatty acids at the n-7 position with R stereospecificity. α-Linolenic and (C18:9,12,15Δω3) and stearidonic acid (C18:6,9,12,15Δ:ω3), are converted to the corresponding 12R-hydroperoxides, eicosapentaenoic acid (C20:5,8,11,14,17Δ:ω3) to its 14R-hydroperoxide and docosahexaenoic acid (C22:4,7,10,13,13,19 Δ:ω3) to its 16R-hydroperoxide. In contrast, omega-6 polyunsaturated fatty acids were oxygenated at the n-10 position [72]. Metabolic transformation of stearidonic acid (C18:6,9,12,15Δ:ω3) by the full length fusion protein leads to the formation of an allene epoxide and the catalase-domain appears to be responsible for the secondary transformation of the primary 12R-oxygenation product [71].
LOXs of higher plants [73], [74], [75] and humans [12], [76], [77], [78], [79] consist of two domains. The C-terminal domain, which is mainly composed of α-helices, harbors the substrate-binding pocket with catalytic non-heme iron ion. The N-terminal ß-barrel domain may be of regulatory importance [44]. Truncation studies on various LOX-isoforms suggested that the N-terminal ß-barrel domains may not be essential for the catalytic activity of plant [80] and mammalian LOXs [81]. Corresponding experiments were also carried out with selected bacterial LOXs. The mini-LOX from Anabena sp. [66], the mini-9R-LOX from Nostoc sp. PCC7120 [68] and the mini-LOX from Cyanothece sp. [82] exhibit full catalytic functionality. Beside from these data the structural biology of bacterial LOXs is not well advanced. The only crystal structure solved for a bacterial LOXs is that for the secretable P. aeruginosa LOX [67]. There is no two-domain structure for this enzyme but a cluster of helices might function as lid for the entrance of the substrate into the active center. The mobility of this lid and the structural flexibility of the entire N-terminal region appear to allow substrate penetration into the active site [67].
Although our knowledge on the enzymology of bacterial LOXs is currently increasing little is known about their biological function. Polyenoic fatty acids are the natural substrates of LOXs, which can be converted to lipid mediators such as eicosanoids [83] or jasmonic acid [84] as it is the case in mammals and plants. Similar roles might be postulated for bacteria containing such lipids. However, some LOX-containing bacteria such as P. aeruginosa are devoid of polyenoic fatty acids. For these bacteria it was proposed that their LOXs might modulate host defense and inflammation during bacterial infection [64], [67]. Another interesting hypothesis for the biological role of bacterial LOXs suggests that the plasticity observed in bacteria harboring LOXs may have provided a selective advantage for adaptation to different ecological niches [9] but there is little experimental evidence for this hypothesis. Occurrence of LOX genes in photosynthesizing cyanobacteria is of particular evolutionary interest. Ancient cyanobacteria largely contributed to converting the reducing ancient atmosphere of our planet into an oxidizing environment by producing oxygen gas as by-product of photosynthesis. The enrichment of oxygen gas in the atmosphere, which started 650 million years ago (Mya), was of dual consequence: (i) It allowed oxygenation reactions, in which atmospheric oxygen is used as electron acceptor. (ii) It enabled oxidative energy metabolism, which is by at least one order of magnitude more efficient than reductive ATP biosynthesis. The oxidative energy metabolism has been suggested as major driving force of Cambrian species explosion, which dramatically increased the diversity of life on earth but also led to near-extinction of anaerobic organisms [85]. According to the endosymbiotic theory the chloroplasts found in photosynthesizing plants and algae have evolved from cyanobacterial ancestors via endosymbiosis [86]. LOXs in cyanobacteria have been implicated in the metabolism of chloroplasts, the major photosynthesizing organelles [65]. Indeed, chloroplast membranes are rich in polyenoic fatty acids making them susceptible for LOX attack. Unfortunately, there is a lack of experimental confirmation of this hypothesis.
4.3.3. Evolution of bacterial lipoxygenases
The increasing number of completely sequenced bacterial genomes allows comparative studies to explore the evolutionary relatedness of bacterial LOX-isoforms between different species [9]. Employing parsimony and distance methodology algorithms phylogenetic trees of LOX evolution can be constructed. The major disadvantage of such comparative studies is the limited number of bacterial LOX sequences, which have been confirmed to encode for functional LOXs. As indicated above only 0.5% of today’s sequenced bacterial genomes contain LOX-like sequences and for only a few of them functional data are currently available. However, comparison of true pro- and eucaryotic LOX sequences suggested that red algae LOXs were more closely related to human isoforms [9]. Moreover, these sequence comparisons suggested that introduction of LOX into P. aeruginosa, Sorangium cellulosum and N. punctiforme were not the results of a continuous evolution but rather of horizontal gene transfer events [9], [63], [65]. However, it remains unclear why these bacterial species, but not others, were susceptible for horizontal LOX gene transfer and why the transferred sequences were retained during further development. Is there any evolutionary advantage LOX expressing strains of P. aeruginosa have gained over strains, which do not carry LOX genes? On the other hand, it may well be that other bacterial species might also have picked up LOX sequences but since these enzymes did not confer an evolutionary advantage the sequences were gradually disrupted during further development. Since LOX genes from N. punctiforme share a high degree of homology with the P. aeruginosa enzymes and were closely related to LOX-like genes of nitrifying bacteria a role of these enzymes in nitrogen metabolism has been suggested [65]. However, it remains unclear whether these LOX-like sequences really encode for functional LOX-isoforms and how oxygen-metabolizing enzymes might be involved in nitrogen metabolism.
All attempts to portray a complete picture of bacterial LOX evolution suffer from two major difficulties: (i) there is only scattered sequence information on functionally well-characterized bacterial LOX-isoforms. For most genomic LOX-like sequences it remains to be shown whether or not they encode for true LOX enzymes. (ii) The methodology (neighbor joining) used for defining the evolutionary relatedness of LOX-isoforms has clear limitations. Since evolution of bacterial LOXs appears to be a complex interplay of horizontal gene transfer and lineage-specific loss amongst different bacterial families, combining the outcome of the neighbor joining methodology with other sequence- and structure-based approaches may yield a more realistic picture. Nevertheless, on the basis of the currently available sequence data bacterial LOXs are an ancient entity, which has been constantly influenced by different kinds of purifying selection pressures [9], [87]. Although, they form a joint enzyme family [9] three subfamilies (group 1: cyanobacteria LOXs, group 2A: proteobacteria LOXs, subgroup A, group 2B: proteobacteria LOXs, subgroup B) can be differentiated according to their phylogenetic relatedness. This classification underlines the fact that cyanobacterial LOXs are structurally somewhat different when compared with other bacterial LOXs.
4.4. Lipoxygenases in archaea
As bacteria archaea are procaryotes that have unique properties separating them from the representatives of other domains of life [88]. Despite the visual similarity to bacteria, archaea possess metabolic pathways that are more closely related to eucarya [89] and both domains appear to share a common ancestor (Fig. 2). Archaea are capable of employing a large diversity of energy sources including sugars, ammonia and hydrogen gas and some archaea are even capable of employing sunlight for energy metabolism. Originally, archaea were viewed as extremophiles living in harsh environments but they have also been found in more moderate biotopes including the human gastro-intestinal tract [88]. Archaea are widely distributed in the oceans and are considered to be one of the most abundant groups of organisms on the planet. Unfortunately, little is known about the occurrence of LOX in archaea.
When we searched the genomic databases of archaea we found LOX-like sequences for Methanococcus jannaschii (M. jannaschii), Methanococcus voltae (M. voltae) and Halorubrum kocurii (H. kocurii). M. voltae [90] is a methanogenic microorganism capable of reducing carbon dioxide to methane and employing this process as energy source. Methanogenes are widely distributed in nature and occur in wetlands (marsh gas production), in hot springs and hydrothermal vents. They are also found in the digestive tract of ruminants and humans where they are responsible for the methane formation. When we attempted to carry out amino acid alignments of the M. voltae LOX-like sequence with plant, mammalian and bacterial LOXs we did not obtain meaningful data. In particular, the iron ligands (His, Asn, Ile) of the well characterized LOXs were not conserved in the M. voltae sequence. Moreover, M. voltae lives under anaerobic conditions and thus, it remains unclear why it should have an oxygen consuming enzyme. M. jannaschii was discovered in the East Pacific at a white smoker. It was the first archaea species whose genome was completed and the sequence data revealed eucaryotic elements. H. kocurii are rod-shaped gram-negative, halophilic archae discovered in salt lakes. Some strains are able to grow at 25–55 °C but required at least 2.5 M NaCl for growth. Hypotonic treatment with less than 2 M NaCl caused cell lysis [91]. Here again, when we performed multiple amino acid alignments no meaningful information could be obtained and LOX iron ligands were not conserved. Taken together, these data suggest that archaea do not have true LOX genes but this conclusion might turn out wrong in the future when more archaea genomes become available.
4.5. Lipoxygenases in eucarya
4.5.1. Lipoxygenases in algae
Algae form a large and diverse group of simple organisms, which occur in unicellular or multicellular forms. Since algae do not include a common ancestor they do not constitute a homogenous phylogenetic group and there is no generally accepted definition of algae [92]. They exhibit a high degree of morphological diversity and a wide range of reproductive strategies ranging from simple asexual cell division to complex forms of sexual reproduction. Most algae are phototroph but some of them became heterotroph. Fossilized algae have been dated back to 1.6–1.7 billion years ago (Bya). Algae are considered as primary producers of polyunsaturated fatty acids [93]. Because of this metabolic property the occurrence of polyenoic fatty acid metabolizing enzymes such as LOX is likely. In fact, there is active oxylipin biosynthesis in algae [94], [95] but for a long time little was known about the presence of true LOX sequences in these organisms. This was somewhat surprising since functional LOXs have been described in cyanobacteria (see Section 4.3), which are commonly considered as procaryotic algae. In 1994 the first LOX sequences has been detected in red algae [96] and these data indicated that LOX in principle occur in these organisms. When we searched the NCBI databases for putative LOX sequences in algae we got five hits for Rhodophyta (AFQ59981.1, AAA61791.1, XP_005718273.1, AGN54275.1, AFQ59981.19), four hits for Chlorophyta (GI:159464351, XP_005643943.1, XP_005643190.1, XP_005853347.1), four hits for Phaeophyta (CBN79157.1, CBN79156.1, CBJ33123.1, CBJ33122.1) and one hit for Diatoms (DQ658366.1). However, multiple amino acid alignments suggested a low degree of homology with functional LOXs from other species and in most of these sequences the iron ligands were not conserved. Thus, although a true LOX has been detected in algae [96] these enzymes do not generally occur in these lower organisms.
4.5.2. Lipoxygenases in yeast and other fungi
Yeasts are single-cell microorganisms that are classified, along with molds and mushrooms, as members of the fungi kingdom (Fig. 2). They are evolutionary diverse and classified on the basis of their cellular characteristics as well as their cellular physiology into two separate phyla [97]: Ascomycota (sac fungi) and Basidiomycota (higher fungi). Budding yeasts, which are also referred to as “true yeasts”, are members of the phylum Ascomycota. The most famous yeast species is Saccharomyces cerevisiae. For thousands of years it has been employed for baking and alcohol production [98]. It has been selected as eucaryotic model organism in today’s cell biology research. Other yeast species as Candida albicans are opportunistic pathogens and can cause infections in immunocompromised humans [99].
On the metabolite level hydroxylated polyenoic fatty acids are present in many fungi [100] and this finding suggested a wide distribution of LOXs. However, since oxylipins can be biosynthesized via different pathways, such as LOXs, fatty acid dioxygenases, diol synthases, peroxygenases, and cytochrome P450 hydroxylases etc., the presence of oxylipins does not prove the existence of LOXs. When we searched the databases for LOX-like sequences in fungi we obtained some 40 hits, which include LOX sequences in Aspergillus fumigatus (XP_746463.1), Aspergillus oryzae (XP_001822165.2), Colletotrichum orbiculare (ENH86155.1), Magnaporthe oryzae (XP_003716006.1), Gibberella moniliformis (AY699875.1) and Neurospora crassa (EAA28941.2). Unfortunately, for most of these sequences no functional data are currently available and although some of these sequences look promising more detailed studies are required to test whether these genes encode for functional LOXs. Interestingly, the genomes of S. cerevisiae and C. albicans do not involve any LOX-like sequences.
Although LOXs also occur in several fungi their biological roles are not well understood. They have been implicated in the formation of oxylipins but the bioactivity of these lipid mediators is rather diverse and varies from species to species [101], [102]. They have been implicated in sporulation, regulate the biosynthesis of mycotoxins and aflatoxins and in the formation of sclerotia [103]. In the 1970s a heme containing fatty acid dioxygenase was characterized in Fusarium oxysporum [104], [105]. Although this enzyme catalyzes the oxygenation of polyunsaturated fatty acids to the corresponding hydroperoxides it may not qualify as true LOX because of its structural characteristics. On the other hand, LOX activities have been detected in other fungal species, such as Saprolegnia parasitica [106], Thermomyces lanuginosus [107], Pleurotus ostereatus [108], Terfezia claveryi [109] and Aspergillus flavus [110]. Recently, a non-heme iron containing LOX has been characterized in F. oxysporum [111]. The best-characterized fungal LOX is the manganese-containing LOX from Gaeumannomyces graminis. This enzyme is distinct from most other LOX isoforms since it contains catalytically active manganese instead of iron [112], [113]. The product patterns of Mn- and Fe-containing LOXs are somewhat different and so is the oxygenation mechanism [113]. For iron containing LOXs an antarafacial relation between hydrogen abstraction and oxygen insertion was observed whereas a suprafacial relation was shown for the manganese enzymes [112]. As indicated above the LOX pathway is not the only metabolic route leading to the formation of oxylipins in fungi. There are alternative enzymes such as linoleate diol synthases [114], [115], which contribute to oxylipin biosynthesis via different reaction mechanisms [101], [102].
4.5.3. Lipoxygenases in mosses and higher plants
In contrast to mammals, in which LOXs are categorized according to their reaction specificity with arachidonic acid, a linoleic acid based classification scheme has been suggested for the plant enzymes [116], [117]. The oxylipins formed via the LOX pathway function as signaling molecules and play important roles in a number of biological processes such as stress response [118], growth and development [119], germination [120], tuber formation [121], sex determination [122], senescence [123], fruit ripening [124], and aroma production [125].
Mosses (bryophyta) are small non-vascular plants. They absorb water and nutrients mainly through their leaves and produce organic material from carbon dioxide and water via photosynthesis. They differ from vascular plants in lacking water-bearing vessels. Mosses reproduce using spores (no seeds) and have no flowers. Little is known about LOX pathways in mosses. However, after the genome of Physcomitrella patens (P. patens) was published and this species became a model organism for mosses [126], [127] more detailed information on LOXs in mosses became available. The P. patens genome involves several LOX-like sequences and true LOX enzymes have been characterized in more detail [128], [129]. This data indicate that mosses have both C20 and C18 based oxylipin biosynthesizing pathways. Meanwhile the genomes of other mosses have been sequenced and the NCBI protein database involves six entries for moss LOX-like sequences. However, no functional data are currently available for these putative LOX isoforms.
In higher plants LOXs are widely distributed [116], [117], [130]. Because of the availability of the complete genome sequences of Arabidopsis thaliana, soybean, rice, wheat, maize and others the isoform pattern and the biological functions of LOX pathways in plants have extensively been studied. Soybean LOX1 was the first LOX described in the mid-thirties of the past century [59] and since then an increasing number of LOX isoforms have been identified. In general, plants show remarkable heterogeneities of their LOX families. In humans six functional LOX genes have been described which encode for six different LOX isoforms [23]. In contrast, many plant genomes contain much more LOX sequences. For instance, completion of the cucumber genome identified more than 20 candidate LOX genes [131]. Polyploidy and frequent gene duplications have contributed to the expansion of plant LOX genes. More detailed analysis of the LOX sequences found in the genomes of Glycine max and Medicago truncatula indicated 34 LOX genes, which were localized in six different gene clusters [132]. Structural comparison of these sequences suggested that the soybean LOX gene family expanded by ancient polyploidy prior to taxon divergence, followed by soybean-specific duplication. Because of the high degree of heterogeneity of plant LOX sequences classification of the corresponding enzymes is rather difficult. Nevertheless, based on selected structural parameters plant LOXs have been categorized into type-1 and type-2 enzymes [132]. Type-1 LOX share a high degree of amino acid sequence similarity (>75%) and do not have a plastidic transit peptide. Type-2 LOX have a putative chloroplast transit peptide sequence and show only moderate overall amino acid sequence similarities (∼35%).
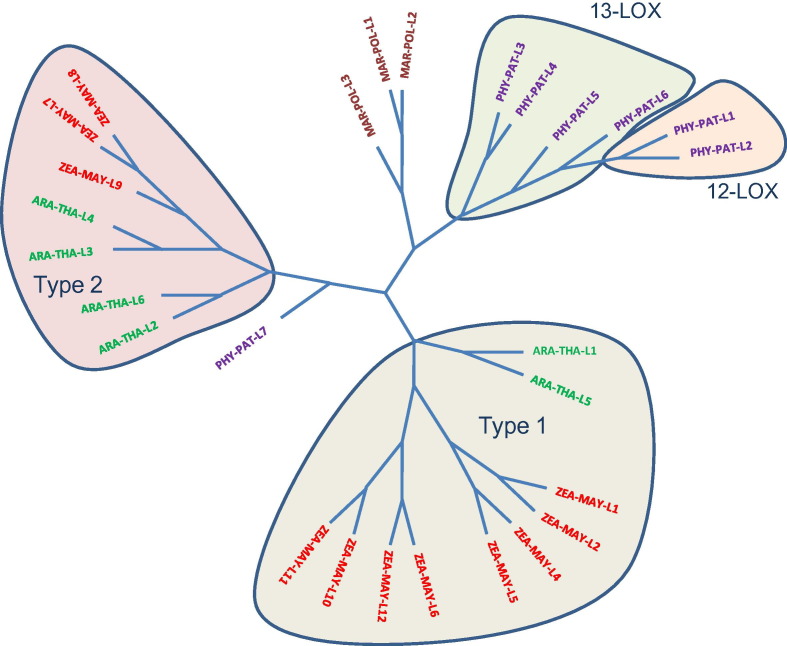
For this review we performed in silico comparison of LOX sequences from plants representing different evolutionary stages (liverworts, mosses, monocots and dicots). This phylogenetic analysis (Fig. 5 ) indicated evolutionary divergence of LOXs from higher and lower plants. In lower plants, the LOXs from liverworts and mosses formed distinct classes. In mosses, 12-LOX evolved from the 13-LOX suggesting a common ancestor. Higher plant LOXs were grouped into two distinct clusters representing the type-1 and type-2 classes. This data suggest that type 1 and type 2 LOX from monocots and dicots evolved separately confirming the previously suggested LOX classification in higher plants [133], [134].
Fig. 5.
Phylogenetic tree of plant lipoxygenases. For this analysis LOX sequences from four plant species with different evolutionary levels were selected: Eudicots – Arabidopsis thaliana (ARA-THA), Monocots – Zea mays (ZEA-MAY), mosses – Physcomitrella patens (PHY-POL), club-mosses Selaginella moellendorffii – SEL-MOE. The phylogenetic tree was constructed by the neighbor-joining method in MEGA 4.0 with bootstrap values estimated based on 5000 replications.
4.5.4. Lipoxygenases in animals
Protozoa and metazoa form the animal kingdom (Fig. 2) of life. This simple binary split of animal classification does not follow the rules of scientific taxonomy, which categorizes living species in a hierarchical order into kingdoms, phyla, classes, orders, families, species and subspecies. However, this binary split is simple and rather useful when it comes to explore the distribution of LOXs in the animal kingdom.
4.5.4.1. Lipoxygenases in protozoa
Protozoa are eucaryotic organisms containing non-filamentous structures and mainly occur in aquatic habitats. There are an estimated 30,000 protozoan species on earth and many of them live as symbionts or parasites. Most protozoa are motile since they carry moving organelles such as flagella or cilia. Alternatively, they are capable of moving by the formation of pseudopodia. According to their way of movement protozoa can be subclassified into Sporozoa (no active movement), Ciliata, (cilia as moving organelles), Flagellata (flagella as moving organelles) and Rhizipoda (formation of pseudopodia). When we searched the NCBI database for LOX-like sequences we did not get any hits for Sporozoa. For Ciliata we extracted two candidate sequences for Oxytricha (EJY76823, EJY73499). Next, we screened these sequences for additional structural parameters to increase the probability that they encode for functional LOX isoforms. For this purpose we employed a three step filtering strategy: (i) we excluded all sequences, which encode for proteins consisting of <300 amino acids. Since the two sequences encoded for 813 and 825 amino acid proteins this filtering step did not exclude either of the proteins. (ii) We searched the two sequences for the presence of the iron coordinating amino acids and excluded those sequences lacking one or both iron ligand clusters. Most functional LOX carry two iron ligand clusters [C1, His-w-x-y-z-His; C2, His-x-y-z-Asn (His, Ser, Thr)] and each cluster contains two coordinating amino acids (bold face) for the catalytic nonheme iron. In the two Oxytricha sequences both iron coordinating clusters (C1, C2) were found, which did not eliminate either sequence. (iii) For most well-characterized LOX isoforms the distance of the two iron clusters varies between 180and 200 amino acids. Longer and shorter distances decrease the probability that the corresponding sequences encode functional LOX isoforms. For the two Oxytricha sequences a distance of 200 amino acids was calculated. Summarizing these data one may conclude that these two sequences might encode for functional LOXs. Searching the Flagellata database we obtained two putative LOX sequences but both of them were excluded from further consideration because of their small size (142 and 100 amino acids). Thus, there is probably no real LOX in Flagellata. In the Rhizopoda database we found a promising sequence (ETO32332, Foraminifera) encoding for a 393 amino acid protein. In this sequence we detected the two iron coordinating clusters C1 and C2 but they were only 95 amino acids apart. This small distance and the low molecular weight suggested that there are no real LOXs in Rhizopoda.
Taken together one may conclude that for the time being there is no compelling evidence for the occurrence of true LOXs in protozoa. Even if future will reveal that the two Oxytricha sequences encode for true LOXs these enzymes are not widely distributed in Protozoa.
4.5.4.2. LOX in invertebrates
Metazoa can be subclassified as vertebrates or invertebrates according to the presence or absence of a vertebral column. This bilateral classification is rather descriptive and does not consider evolutionary relatedness. However, it is straightforward and very useful when searching for the distribution of certain proteins in different species. The overwhelming majority (95–98%) of animal species on earth are invertebrates and many individual invertebrate subcategories have a greater number of species than all vertebrates together. Morphologically, invertebrates are extremely diverse and they do not have a bony skeleton. Many of them (jelly fish, worms) have soft hydrostatic skeletons but others have hard exoskeletons. Invertebrates include multiple groups of living organisms such as insect, crap, lobster, snail, clam, octopus, starfish, sea urchin, worm and sponge. Functional LOX isoforms from various invertebrates have been characterized as fusion or nonfusion proteins in different corals [135], [136], [137], [138], [139], [140], in starfish oocytes [141], in the eggs of a sea urchin [142] and in the ovaries and oocytes of different mussels (Mylitis edulis, Spisula solidissima) [143], [144]. Recently the distribution of LOX isoforms in 14 different invertebrate species was explored [145] and the authors found putative LOX-like sequences in 10 of them (Ciona intestinalis, Saccoglossus kowalevski, Strongylocentrotus purpuratus, Crassostrea gigas, Lottia gigantea, Capitella teleta, Helobdella robusta, Acropora digitifera, Nematostella vectensis, Trichoplax adhaerens). Moreover, there is evidence for functional LOXs in Daphnia subspecies [146] but no LOX-like sequences were found in Drosophila melanogaster, Caenorhabditis elegans which constitute frequently employed model organisms for invertebrates. When we searched ENSEMBL (http://metazoa.ensembl.org) and the NCBI (www.ncbi.nim.gov) protein databases for Placozoa, Porifera, Coelenterata, Platyhelminthes, Nematoda, Mollusca, Annelida, Echinodermata, Chelicerata, Cephalochordata, Tunicata with key word “lipoxygenase” we confirmed the occurrence of LOX-like sequences in these invertebrates but got additional hits. Filtering the obtained sequences with the three-step strategy described for protozoa (search for iron coordinating clusters) we reduced the multiplicity of hits. The results of this screening strategy are shown in Table 2 and can be summarized as follows: (i) Functional LOX isoforms occur in selected invertebrates and this conclusion is consistent with previous characterization of several enzymes in corals, mussels, sea urchins and others. In many of these organisms several putative LOX sequences can be identified but for Table 2 only one example was selected. (ii) Most invertebrates do not have functional LOX genes. Of the estimated more than one million invertebrate species on earth less than 1% carries LOX-like sequences. Of course, only for a minor share of all invertebrates complete genome data are currently available and it is rather likely that more invertebrate LOX-like sequences will be identified with increasing numbers of completely sequenced invertebrate genomes.
Table 2.
Lipoxygenase-like sequences obtained with our two-step searching procedure for metazoa and lower invertebrates. The size of the predicted proteins is given by the number of amino acids. Clusters 1 and 2 represent the iron ligand clusters that are explained in the text. Δ(C1-C2) quantifies the distance of the two iron ligand clusters counting the number of amino acids. In the LOX column we estimated the probability that the corresponding sequence encodes for a functional LOX (+, high probability; −, low probability). The criteria of our estimates are the following: + (size > 600 amino acids, presence of C1 and C2 in correct order, 180 < ΔC1-C2 > 200), − (absence of either C1, C2 or both or presence of C1 and C2 but ΔC1-C2 < 180). ∗Not reported in Yuan et al. (2014).
| Species | Acc. number | size | Cluster1 | Cluster2 | Δ(C1-C2) | LOX |
|---|---|---|---|---|---|---|
| Lottia gigantea | ESP02741 | 671 | + | + | 193 | + |
| Helobdella robusta | ESN98770 | 665 | + | + | 193 | + |
| Nematostella vectensis | XP_001634357 | 642 | + | + | 188 | + |
| Amphimedon queenslandica∗ | XP_003383419 | 762 | + | + | 192 | + |
| Hydra vulgaris∗ | XP_002157326 | 623 | + | + | 184 | + |
| Strongylocentrotus purpuratus | XP_782245 | 691 | + | + | 195 | + |
| Ixodes scapularis∗ | XP_002407642 | 358 | + | + | 97 | − |
| Saccoglossus kowalevskii | XP_006820345 | 664 | + | + | 193 | + |
| Crassostrea gigas | EKC38941 | 663 | + | + | 196 | + |
| Echinococcus granulosus∗ | CDJ23913 | 745 | − | + | − | |
| Schistosoma mansoni∗ | CCD75342 | 463 | + | − | − | |
| Plexaura homomalla∗ | AAC47283 | 715 | + | + | 187 | + |
| Gersemia fruticosa∗ | AAY98506 | 679 | + | + | 189 | + |
| Trichoplax adhaerens | EDV27172 | 645 | + | + | 192 | + |
| Capitella teleta | ELT89495 | 659 | + | + | 192 | + |
4.5.4.3. Lipoxygenases in Chordata
Chordates are animals possessing a notochord, which is a hollow dorsal neural organ. Taxonomically, chordates include vertebrata, tunicata and lancelets (Fig. 6 ). Primitive chordates are known from at least as early as the Cambrian explosion. Today almost 60,000 different chordate species are living on this planet and the world’s largest (blue whale) and fastest (peregrine falcon) animals belong to this phylum. Almost half of the chordate species are bony fish.
Fig. 6.
Schematic view of subclassification of Chordata. Model organisms, which are referred to in this study are indicated in italics.
4.5.4.3.1. Lipoxygenases in Cephalochordata and Tunicata
Lancelets (Branchiostoma floridae, Branchiostoma belcheri) are model organisms for Cephalochordata and they have separated from the vertebrate lineage about 520 Mya [147]. Analysis of the genome of B. belcheri indicated the presence of putative LOX sequences (BbtC2LOX1 and BbtLH2LOX2; NCBI accession numbers KJ196391 and KJ196392, respectively). However, the iron ligand clusters of mammalian LOXs are not conserved in the BbtC2LOX1 sequence suggesting that the corresponding protein may not constitute a functional LOX isoform. Searching the B. floridae genome we also found LOX-like genes (XP002601197.1, XP002606132.1), in which the iron ligands of mammalian enzymes are conserved. Phylogenetic analysis suggested a close evolutionary relation to the allene oxide synthase-lipoxygenase fusion proteins of lower marine organisms [136]. Unfortunately, to the best of our knowledge there are no functional data available demonstrating the LOX activity of these enzymes.
C. intestinalis (sea squirt) serves as a model organism for Tunicata and seven LOX-like sequences have recently been identified in this animal [145]. When we searched the NCBI and the ENSEMBL protein databases for LOX-like sequences we found three entries (XP_002123813, XP_002123202, XP_002123695). When we took these sequences through our filtering protocol we found that in the XP_002123813 sequence only the C1 iron cluster was present. Since we did not find a C2 cluster the functionality of this putative arachidonate 5-lipoxygenase-like protein is questionable. The XP_002123202 (predicted allene oxide synthase-lipoxygenase fusion protein, 766 amino acids) and XP_002123695 (arachidonate 5-lipoxygenase-like protein, 694 amino acids) sequences involve both iron clusters and the distances between them (ΔC1-C2) were 186 and 188 amino acids, respectively. Considering these structural characteristics it may well be, that these sequences encode for functional LOX isoforms.
4.5.4.3.2. Lipoxygenases in primitive Vertebrata
Vertebrata can be subclassified into Cyclostomata and Gnat h ostomata (Fig. 6) and their evolution started about 550 Mya during the Cambrian species explosion [148], [149], [150], [151], [152]. The most primitive vertebrates (hagfish, lampreys) separated from other vertebrates 550 Mya and have hardly changed since then [148], [149], [150], [151], [152]. Petromyzon marinus (sea lamprey) serves as model organism for Cyclostomata. It is considered a living fossil and has been well-characterized with respect to its morphological and metabolic properties. Its complete genome was recently sequenced [153], [154] and a search of the Uniprot database yielded one LOX-like sequence (S4RFK5). Phylogenetic analysis suggested that this sequence shared a high degree of amino acid similarity with human ALOX5 and corresponding orthologs from other mammals. This 454 amino acid fragment involved the two iron clusters C1 and C2 from the human 5-LOX (His368, His373, His551, Asn555) in a distance of 183 amino acids. As fifth iron ligand serves the C-terminal Val with its free carboxylic group. Although there are no functional data confirming the catalytic activity of the sea lamprey LOX these structural characteristics suggest a catalytically active enzyme. For the time being there are no genomic data available for hagfish and thus, it remains unclear whether these species have functional LOXs.
4.5.4.3.3. Lipoxygenases in cartilaginous and bony fish
Cartilaginous and bony fish first appeared around 420–400 Mya during the transition from the Silurian to the Devonian period [155], [156]. Sharks are representatives of cartilaginous fish and the genome of the elephant shark (Callorhinchus milii) has recently been published [157]. The genome contains several LOX-like sequences and amino acid alignments suggested that two of them (XP_007902723.1, XP_007910024.1) share a high degree of sequence similarity with ALOX5 isoforms of higher vertebrates. Other sequences (AFO96333.1, AFP03230.1) are more closely related to epidermal LOX isoforms. Three of them (XP_007902723.1, XP_007910024.1, AFO96333.1) have both iron coordination clusters C1 and C2, but in one of them (AFP03230.1) only cluster C1 is conserved. In addition, the elephant shark genome also contains a number of sequences which are important for leukotriene signaling, especially the leukotriene receptors BLT1 and CYSLTR1/2 (XP004945706.1, XP007884788.1, XP007890412). This is an interesting observation since in a previous study it was reported that the genome of the thornback ray (Raja clavata) does not contain a functional LTA4-hydrolase (LTA4H) gene and thus, it was concluded that cartilaginous fish may not have functional leukotriene signaling [158]. In the absence of more detailed experimental data such conclusion may be somewhat risky since biosynthesis of cysteinyl leukotrienes does not require a functional LTA4H and it remains unclear whether genes encoding for other epoxide hydrolases are present in the genome. Moreover, the genome involves at least one LTB4 receptor gene (XP004945706.1) suggesting functional LTB4 signaling.
Bony fish separated into ray-finned (e.g., D. rerio) and lobe-finned fish (e.g., Latimeria chalumnae) about 400 Mya and the genomes of these two bony fish species have recently been sequenced [159], [160]. When we searched the D. rerio sequence databases for potential LOX sequences we detected several LOX cDNAs (Table 3 ), which originate from different genes localized on different chromosomes [161]. Comparison of the amino acid sequences of zbfLOX1 and zbfLOX2 with the six functional human ALOX isoforms indicated that zbfLOX1 shared a similar degree (40–45%) of amino acid identity with all human ALOX isoforms. Thus, on the basis of the available sequence data it was impossible to sort out for which particular human LOX isoenzyme the zbfLOX1 was the D. rerio ortholog. Functional inactivation of zbfLOX1 causes severe impairment of embryonic development with deformation in brain, eyes and the tail region [162]. Since functional inactivation of Alox12b [29] and Aloxe3 [30] in mice also leads to structural defects in epithelial differentiation the authors concluded that zbfLOX1 might constitute the zebrafish ortholog for mouse Alox12b and/or Aloxe3 despite the low degree of amino acid identity. More detailed functional characterization of the recombinant enzyme indicated [161] that this LOX does neither follow the triad [163] nor the Ala-vs-Gly-concept [164] explaining the reaction specificity of different mammalian LOXs. For zbfLOX2 functional assignment to human ALOX isoform was less problematic since this enzyme shares a 74% of amino acid identity with the human ALOX5. Comparison with the other human ALOX isoforms revealed much lower identity degrees (37–41%). Thus, on the basis of the amino acid sequence zbfLOX2 was classified as arachidonic acid 5-LOX. Recently this LOX-isoform was expressed as recombinant His-tag fusion protein in E. coli and activity assays confirmed its reaction specificity of arachidonic acid oxygenation [165]. Unfortunately, for the other zebrafish LOX isoforms no functional data are currently available. However, the existence of a functional ALOX5 gene in the genome of D. rerio, the presence of a corresponding sequence in the L. chalumnae genome and the occurrence of sequences encoding for BLT1/2 and CYSLTR1/2 receptors suggest functional leukotriene signaling in bony fish. This conclusion is consistent with previous reports indicating the presence of LOX metabolites including LTB4 bony fish macrophages and thrombocytes [166], [167]. On the other hand, to confirm this prediction additional experimental data on the functionality of the identified sequences are required.
Table 3.
Lipoxygenase sequences in the zebrafish genome.
| Database number | Chromosome | Possible orthology based on sequence homology |
|---|---|---|
| ENSDARP00000091900 | 7 | High homology to 12/15-LOX or platelet-type 12-LOX (zbfLOX1), |
| ENSDARP00000074335 | 13 | Among zebrafish LOXs highest homology to human ALOX5 (zbfLOX2) |
| ENSDARP00000063256 | 15 | Alternative 5-LOX gene, lower homology to human ALOX5 than ENSDARP00000074335 |
| ENSDARP00000093033 | 15 | Alternative 5-LOX gene, lower homology to human ALOX5 than ENSDARP00000074335 |
| ENSDARP00000063263 | 15 | Alternative 5-LOX gene, lower homology to human ALOX5 than ENSDARP00000074335 |
| ENSDARP00000106795 | 21 | Long transcript, highest homology to human epidermal LOX isoforms (ALOX15B, ALOX12B) |
| ENSDARP00000023401 | 21 | short transcript, 100% sequence identity to ENSDARP00000106795 |
| ENSDARP00000113298 | 21 | Short transcript, possibly an epidermal LOX isoform (ALOX15B) |
| ENSDARP00000095620 | 21 | Long transcript, 99% sequence identity with ENSDARP00000113298. |
4.5.4.3.4. Lipoxygenase in amphibia
The earliest amphibian-like animals, Ichthyostega and Eusthenopteron, appeared about 370 Mya [168]. So far, a number of amphibian genomes have been published but for this review we will focus on the Western clawed frog (Xenopus Silurana tropicalis) [169]. Its genome involves at least three putative LOX genes and different sequences encoding for BLT1/2 and the CYSLTR1/2 receptors. Sequence comparison suggested that one of the putative LOX genes (XP_004815982.1) shares a high degree of amino acid similarity with human ALOX5. Both iron coordination clusters C1 and C2 are conserved suggesting a functional enzyme. This finding together with our observation that the Xenopus genome contains BLT- and CYSLT-receptor-like sequences suggests functional leukotriene signaling in this amphibian species. This conclusion is consistent with the previously reported formation of ALOX5 products (5-HETE, LTB4, LTC4) by frog leukocytes [170]. Recently, the LTA4H of the African claw toad (Xenopus laevis) was cloned and the enzyme was shown to convert LTA4 to LTB4 and Δ(6)-trans-Δ(8)-cis-LTB4 [171]. This data suggest that amphibia apparently had a functional leukotriene B4 synthesizing cascade. Moreover, leukotriene biosynthesis was observed in bullfrog erythrocytes and thrombocytes [172], [173]. In 2000, a 12-lipoxygenase was partially purified from bullfrog erythrocytes but unfortunately, no sequence data are currently available for this enzyme. Its MW of 77.7 kDa was in a normal range for LOXs [174]. Although there is no convincing evidence for the existence of an ALOX15 ortholog in frogs, 15(S)-HETE was isolated as arachidonic acid oxygenation product when frog leucocytes or oocytes were treated with exogenous arachidonic acid [170], [175]. Because of the limited experimental data on the LOX pathway of amphibians little is known about the biological function of these enzymes. Involvement of these enzymes in amphibian regeneration, wound healing [176] and oocyte maturation [177] has been suggested.
4.5.4.3.5. Lipoxygenases in reptilia and avea
Reptiles appeared on earth 320–310 Mya in the late Carboniferous period and Cassineria is one of the oldest known amniotes [178], [179]. The genome of the American alligator (Alligator missis s ippiensis) was published in 2012 [180] and we considered this vertebrate as model organism for reptiles. Searching the corresponding genome database we identified three LOX-like sequences. One of them (XP_006263756.1) shared a high degree of amino acid conservation with human ALOX5 while the others (XP_006264318.1, XP_006272458.1) were more closely related to human epidermal LOX isoforms. So far, there is no functional data for reptile ALOX isoforms but snake erythrocytes apparently contain ALOX5 metabolites [181].
Avians have emerged from feathered theropod dinosaurs during the Jurassic period [182]. We employed chicken (Gallus gallus) as model organism for birds and searching the chicken genome database [183] suggested the presence of at least two LOX-like sequences. Sequence comparison demonstrated that one of these sequences (XP_003641560.1) shares a high degree of amino acid similarity with human ALOX5 while the other (XP_42376.4) seem to be more closely related to the epidermis-type LOX isoforms. Leukotriene signaling appears to exist in birds since LTB4 receptors have been identified in avian osteoclasts [184]. Although to the best of our knowledge no bird LOX has been expressed as recombinant protein and characterized with respect to their enzymatic properties LOX metabolites have been detected in different chicken and turkey cells and tissues [185], [186], [187].
4.5.4.3.6. Lipoxygenases in mammalia
Mammals were first introduced in evolution during the mid-Permian period about 270 Mya. They evolved from the mammal-like dinosaur lineage called synapsids (later theropsids), and species like Dryolestes were closely related to nowadays placentals and marsupials whereas species like Ambondro were similar to monotremes. Later on, the eutherian and metatherian lineages separated. Eutherians are the ancestors of today’s living placentals whereas the metatherian developed to marsupials [188], [189]. The earliest known eutherian species (Juramaia sinensis) lived 160 Mya in the late Jurassic period and at that time-point the three existing mammalian subgroups, the monotremes, the marsupials and the placentals, have separated from each other [190]. For our search for mammalian LOX sequences we employed the platypus (Ornithorhynchus anatinus) and the Tasmanian devil (Sarcophilus harrisii) as model organisms for the monotremes and marsupials, respectively. The genomes of the two species have been published [191], [192] and each of them contains a number of LOX-like sequences, which share a high degree of amino acid conservation with human LOX-isoforms. Interestingly, we did not find any sequence in the Tasmanian devil exhibiting a high degree of sequence similarity with human ALOX15. This finding is rather surprising since the evolutionary more distant platypus has an ALOX15-like sequence. Many putative LOX sequences of primitive mammalian species are present as truncated versions and in the absence of direct functional data it is difficult to conclude whether the corresponding genes encode for active LOX isoforms. Placentals have developed from eutherians 160 Mya, and the genomes of many rodents and primates are currently available to search for LOX-like sequences. Rodents and primates evolved from a common ancestor called Euarchontoglires [193]. Analysis of the mouse (Mus musculus) and rat (Rattus norvegicus) genomes [194], [195] revealed seven functional LOX genes (Alox15, Alox12, Alox5, Aloxe3, Alox15b, Alox12b, and Aloxe12) in these two rodents. Since the Alox15b gene in mice encodes an 8-lipoxygenating enzyme it is sometimes referred to as Alox8 but this nomenclature is really confusing. Leukotriene signaling is functional in these mammals since ALOX5 and leukotriene receptors are present [196], [197], [198], [199], [200]. Although the mouse Alox15 and Alox15b share a high degree of amino acid sequence identity with the corresponding human orthologs they differ from the human enzymes with respect to their reaction specificity of arachidonic acid oxygenation. The structural basis for these differences will be discussed in the following chapters. The epidermal LOXs (Alox12b, Aloxe3), which have been well-characterized with respect to their enzymatic properties [201], [202], have been implicated in epidermal differentiation [29], [30] and in the formation of the epidermal water barrier [203]. Modern humans (H. sapiens) have only six functional LOX genes since the ALOXe12 gene encoding for an additional epidermis-type LOX is present in humans as corrupted pseudogene [204]. The mouse enzyme was discovered in 1995 and later on characterized in more detail [205], [206].
Lipoxygenase orthologs in mammalian evolution: As indicated above there are seven different LOX isoforms in mice and knockout studies suggested different biological roles for most of them [23]. Alox5 has been implicated in leukotriene biosynthesis [207], Alox12b, Aloxe3 and Alox12 are involved in skin development [29], [30], [34] and Alox12 might also play a role in platelet function [32]. For Alox15 and Alox15b the biological roles are still unclear. The structural multiplicity of mammalian LOXs within a single species is frequently considered a sign of biological redundancy among scientists not working in the LOX field. Consequently, inactivation of one ALOX gene might be compensated for by upregulation of the expression of other LOX isoforms. However, if one compares the enzymatic properties of the different human or mouse LOX such compensation mechanisms become questionable because of several reasons. (i) ALOX5 is the key enzyme in the biosynthesizing cascade of the 5,6-series of leukotrienes and no other LOX-isoform is capable of initiating the biosynthesis of these mediators. ALOX15 orthologs may catalyze the formation of 14,15-leukotrienes [208] but the biological role of these mediators has not been well characterized. (ii) ALOX12B has been implicated in epidermal differentiation and selective functional inactivation of the corresponding gene leads to dramatic alterations in skin development. If there is a compensatory upregulation of another LOX-isoform it appears to be insufficient to protect the animals from these defects. (iii) When we compared the expression of the mRNAs of various LOX isoforms (Alox12, Alox15b, Alox5) in different tissues of wildtype and Alox15 knockout mice we did not observe consistent indications for a major upregulation of other LOX isoforms as regulatory response towards defective Alox15 expression (Kuhn, unpublished data). These data suggest that expression regulation of these LOX isoforms may not be coupled to Alox15, which makes biological redundancy rather unlikely. Consequently, each LOX isoform is likely to have its own function (functional multiplicity of the LOX family). Unfortunately, the driving forces and the molecular mechanisms leading to the functional multiplicity remain largely elusive.
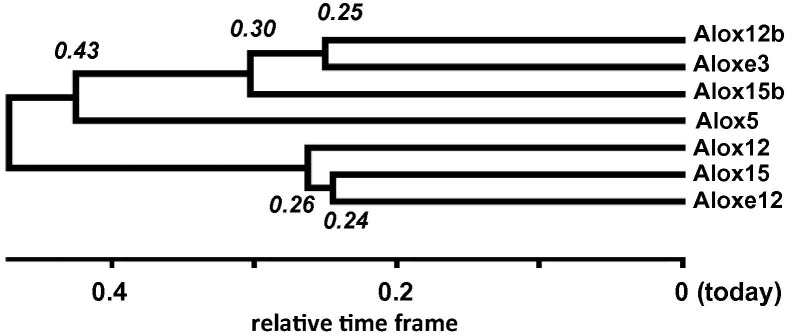
The three murine epidermal LOXs (Alox15b, Alox12b, Aloxe3) might have been evolved from a common precursor enzyme by at least two steps of gene duplications. The other murine LOX isoforms (Alox15, Alox12) should have evolved independently from another common ancestor by at least two additional steps of gene duplications to form Alox12, Aloxe12 and Alox15 (Fig. 7 ). The Alox5 evolved independently and this conclusion is consistent with a previous suggestion [209]. Although the mechanisms and the driving forces for the duplication events have not been explored in detail it is well-known that the region of chromosome 17, where all but the ALOX5 gene are localized, is a hotspot for gene duplications [210].
Fig. 7.
Gene duplication of mouse LOX isoforms. For this analysis the amino acid sequences of the seven known LOX isoforms from mouse (Mus musculus) were selected and compared with each other: Alox5 (NP_033791.1), Alox12 (NP_031466.2), Alox15 (NP_033790.3), Aloxe12 (NP_663717.1), Alox12b (NP_033789.1), Aloxe3 (NP_035916.2), and Alox15b (NP_033791.1). The amino acid sequences were aligned with MEGA 6.0 using the ClustalW package with default settings. The phylogenetic tree was constructed by the neighbor-joining method in MEGA 6.0 with bootstrap values estimated based on 5000 replications. The relative time-points for gene duplication events were calculated with MEGA 6.0 using default settings for calculation.
According to their genomic sequence data all three mammalian subgroups (monotremes, marsupials, placentals) have the genetic basis (all genes for biosynthesizing enzymes and receptors) for functional leukotriene signaling. Thus, it may be concluded that the LOX signaling cascade has been completed during transition from theropsid dinosaurs to early mammal-like animals and has been conserved since then.
Evolution of specificity determinants of mammalian ALOX15 orthologs: Human ALOX15 converts arachidonic acid to 12S- and 15S-hydroperoxy arachidonic (12S- and 15S-HpETE) acid in a ratio of about 1:10 [42]. A similar reaction specificity was observed for the ALOX15 orthologs of rabbits [211] and orangutans [163]. On the other hand, the mouse ortholog [212] forms an inverse product pattern (12-HpETE vs. 15-HpETE ratio of about 10:1) and similar results were obtained for the ortholog enzymes from rats [43], pigs [213], cattle [214] and rhesus monkeys [163]. The molecular basis for the variable positional specificity of ALOX15 orthologs has previously been studied and the triad concept has been developed [44], [215]. In 12-lipoxygenating enzymes the sequence determinants are occupied by small amino acids whereas more space filling residues are present in the 15-lipoxygenating isoforms (Table 4 ). Although the positional specificity of ALOX15 orthologs of chimpanzee, gorilla, gibbon and baboon remains to be determined experimentally 15-lipoxygenation might be predicted for the enzymes of chimpanzee and gorilla. In contrast, 12-lipoxygenation may be predicted for baboon ALOX15. For gibbon ALOX15 prediction of the reaction specificity is somewhat problematic but preliminary data from our lab suggest almost equal amounts of 12- and 15-HpETE (Adel et al., unpublished data). Taken together, these data suggest that in lower mammals (mice, rats, pigs, baboon, rhesus monkey) 12-lipoxygenating ALOX15 enzymes are expressed. In contrast, in higher mammals (orangutan, human) 15-lipoxygenating ALOX15 isoforms are present [216] and gibbons may constitute a transition species. The only exception from this rule is the rabbit but here 12- and 15-lipoxygenating ALOX15 species are expressed in a tissue specific manner [217]. Unfortunately, the biological relevance of this evolutionary switch in the reaction specificity still remains unclear.
Table 4.
Triad determinants of ALOX15 orthologs and positional specificity of arachidonic acid oxygenation. The one letter code of amino acid nomenclature was used and the triad determinants are indicated in bold face. In human ALOX15 amino acid numbering is as follows: Borngräber 1 determinant (F353), Sloane determinants (I418, M419), Borngräber 2 determinants (I593, T594). n.d., not yet determined experimentally.
| Species | Borngräber 1 determinant | Sloane determinant | Borngräber 2 determinant | Major product (predicted) | Major product (experimental) |
|---|---|---|---|---|---|
| Human (Homo sapiens) | DFQ | QIMS | SITW | 15-HpETE | 15-HpETE |
| Chimpanzee (Pan troglodytes) | DFQ | QIMS | SITW | 15-HpETE | n.d |
| Gorilla (Gorilla gorilla) | DFG | QIMS | SITW | 15-HpETE | n.d. |
| Orangutan (Pongo pygmaeus/abelii) | DFQ | QIMS | SITW | 15-HpETE | 15-HpETE |
| Gibbon (Nomascus leucogenys) | DFQ | QITS | SIVW | 12- + 15-HpETE | n.d. |
| Rhesus monkey (Macaca mulatta) | DFQ | QVVS | SITW | 12-HpETE | 12-HpETE |
| Baboon (Papio anubis) | DFQ | QVVS | SMTW | 12-HpETE | n.d. |
| Cattle (Bos taurus) | DFQ | QVVS | SITW | 12-HpETE | 12-HpETE |
| Pig (Sus scrofa) | DFQ | QVVS | SITW | 12-HpETE | 12-HpETE |
| Rat (Rattus norvegicus) | DLQ | KAMS | NVVW | 12-HpETE | 12-HpETE |
| Mouse (Mus musculus) | DLQ | KVMS | NVVW | 12-HpETE | 12-HpETE |
Evolution of specificity determinants in ALOX15B and ALOX12B orthologs: The triad concept of reaction specificity is not applicable for all LOX isoforms [163] and for ALOX15B orthologs other sequence determinants have been identified. Human ALOX15B exhibits a singular positional specificity converting arachidonic acid exclusively to 15S-HpETE [218]. In contrast, the mouse enzyme oxygenates arachidonic acid to 8S-HpETE [219] and site-directed mutagenesis indicated that substitution of Tyr603 and His604 to the corresponding residues present at these positions in human ALOX15B (Y603D+H604V) leads to complete shift in the positional specificity of arachidonic acid oxygenation from 8S- to 15S-HpETE formation. The inverse mutagenesis strategy leads to partial alterations [45]. When we compared the ALOX15B sequences of different mammals (Table 5 ) we found that chimpanzee, gorilla, orangutan, rhesus monkey, baboon, pig and even rats have an Asn-Val or Asn-Ile motif at this position and thus, they are similar to the human enzyme. In contrast, the mouse sequence has a Tyr-His motif at this position. In gibbons these positions are occupied by Leu-Gln but the functional consequence of this amino acid exchange needs to be explored experimentally.
Table 5.
Sequence determinants of ALOX15B and ALOX12B orthologs and reaction specificity of arachidonic acid oxygenation. The one letter code of amino acid nomenclature was used and the sequence determinants are indicated in bold face. In human ALOX15B amino acid numbering is D603 and V604. In human ALOX12B amino acid numbering is G441.
| Species | Jisaka determinants | Coffa/Brash determinants |
|---|---|---|
| Human (Homo sapiens) | TCDVIL | SIGRA |
| Chimpanzee (Pan troglodytes) | TCDIIL | SIGRA |
| Gorilla (Gorilla gorilla) | TCDIIL | SIGRA |
| Orangutan (Pongo abelii) | TCDVIL | SIGRA |
| Gibbon (Nomascus leucogenys) | ASLQMS | SIGRA |
| Rhesus monkey (Macaca mulatta) | TCDVIL | SIGRA |
| Baboon (Papio anubis) | TCDVIL | SIGRA |
| Cattle (Bos taurus) | TCDVVI | SIGRA |
| Pig (Sus scrofa) | TCDVVI | SIGRA |
| Rat (Rattus norvegicus) | TCDVII | SIGRA |
| Mouse (Mus musculus) | SSYHII | SIGRA |
Multiple sequence alignments of a large number of different LOX sequences [164] suggested that S-LOXs carry an Ala at a defined position (Ala402 of human ALOX15) in their primary structure. In contrast, R-LOXs carry a Gly at this position (Gly441 for human ALOX12B) and mutagenesis studies suggested that an Ala-to-Gly exchange induces major alterations in the enantioselectivity of several LOX-isoforms [164]. Although this concept was applicable for a large number of LOX isoforms [220] zebrafish LOX-1 (zbfLOX1), which contains a Gly at this position, converts arachidonic acid mainly to 12S-HpETE [161]. A sequence alignment of various ALOX12B of different mammals indicated strong conservation of the corresponding Gly residue during mammalian evolution (Table 5).
Lipoxygenases in extinct human subspecies: Homo neanderthalensis is the closest evolutionary relative of modern humans and for a long time they were widely distributed over large parts of Europe and western Asia [221]. During the later parts of their history Neanderthals settled as far east as southern Siberia and as far south as the Middle East [222]. Neanderthal individuals got in contact with modern humans and the question of whether Neanderthals have interbred with modern humans has been discussed controversially for many years [223]. Comparison of the nuclear genomes of H. neanderthalensis [224], [225] and H. sapiens suggested that Neanderthals played a role in the genetic ancestry of today’s humans outside Africa since 1–3% of the DNA of today’s Europeans originate from Neanderthals. However, it remains a matter of discussion how intense the original mixing was and where and when it took place [223]. When the first draft sequence of the H. neanderthalensis genome (1.3-fold genome coverage) was published a number of genomic regions, which are different between ancient and modern humans have been identified [224]. Among them was the cysteinyl leukotriene receptor 2 (CLTR2), the expression of which appeared to be corrupted in H. neanderthalensis [224]. In addition, when we analyzed the genomic sequence data and searched for human LOX orthologs we found [226] that the genes encoding for ALOX12 and ALOXE3 appeared to be dysfunctional because of premature stop codons interrupting the open reading frame. However, more precise genomic sequence data (42-genome coverage), which were recently obtained from a H. neanderthalensis fossil found in the Denisovan cage [224], [225], did not confirm these defects [227]. Thus, it was concluded that H. neanderthalensis had fully functional leukotriene signaling.
The Denisovan cave in the Altai Mountain in southern Siberia is a rich source of human fossils and paleontological excavations have been going on in this cave for many years. Recently, a distal manual phalanx [228], a proximal pedal phalanx [229] and a molar tooth [230] were excavated from different sublayers of layer 11 (tooth from sublayer 11/1, manual phalanx from sublayer 11/2, pedal phalanx from sublayer 11/4). This data indicate that the corresponding individuals lived in this cage about 50,000 years ago and that the finger individual was somewhat older than the two other fossils. The pedal phalanx came from the 4th or 5th toe of a human adult individual and its morphological traits linked it with both H. neanderthalensis and H. sapiens [225]. In contrast, the morphology of the two other fossils suggested an independent human subspecies, which was named H omo denisovan. Comparison of mitochondrial DNA preparations from this manual phalanx [228] and mitochondrial DNA prepared from previous H. neanderthalensis fossils [231] and from H. sapiens clearly indicated evolutionary differences between these three human subspecies but also suggested a common ancestor. Since evolutionary conclusions drawn from mitochondrial genomes are always risky, nuclear DNA was extracted from the manual phalanx and the genome was sequenced at high (>40-fold) genome coverage [232]. Taken together, high quality genomic sequences of two different archaic human subspecies (H. neanderthalensis, H. denisovan) and of modern humans (H. sapiens) are currently available for detailed structural and functional comparison. When we aligned on the amino acid level the LOX sequences of all H. sapiens LOX isoforms with those of H. denisovan we only observed subtle structural differences [227]. Neither of the detected amino acid exchanges was at positions impacting iron coordination or reaction specificity. Some of the observed mutations were at positions, for which a high degree of genetic variability (SNPs or rare mutations) has been observed among different human individuals [233]. Since only one H. denisovan individual has been sequenced so far, it remains unclear whether the observed amino acid exchanges are unique for this individual.
5. Genetic variability of human leukotriene signaling
The leukotriene signaling pathway has been introduced during evolution on the bony fish level (perhaps somewhat earlier) and has been conserved in all higher vertebrates. The modern human (H. sapiens) has six functional LOX isoforms and a number of other genes that are involved in leukotriene signaling. During the last decade various Genome-wide association studies (e.g., HapMap, 1000 Genome, COSMIC etc.) have been performed, and genetic variations in the leukotriene signaling cascade have been associated with certain diseases, such as bronchial asthma, Alzheimer’s disease, atherosclerosis and different forms of cancer. Although a large number of variations have been described only a few of them have functionally been characterized. Most alterations are either located in intronic, in the promoter, in intergenic DNA regions or they are synonymous in nature. In this part of the review we will discuss the functional consequences of gene variations involved in human leukotriene signaling. We therefore mainly focus on distribution, structural localization and functional consequences of coding non-synonymous variations in genes of the LOX pathway, and we try to summarize the current knowledge about functional relevant promoter, 5′ and 3′ UTR variations as well. In total more than 800 non-synonymous variations have been found in different genes of the LOX pathway but only 19 are defined as SNPs with an allele frequency >1% (Table 6 ).
Table 6.
Non-synonymous coding variations in enzymes and receptors of the LOX pathway.
| Enzyme/receptor (UniProt-Code) | Total | SNPs (allele frequency >1%) | Missense mutations (allele frequency <1%) | Nonsense mutations |
|---|---|---|---|---|
| Lipoxygenases | ||||
| ALOX5 (P09917) | 69 | 1 | 68 | 0 |
| ALOX15 (P16050) | 94 | 2 | 84 | 8 |
| ALOX12 (P18054) | 67 | 2 | 64 | 1 |
| ALOX15B (O15296) | 86 | 6 | 76 | 4 |
| ALOX12B (O75342) | 69 | 1 | 68 | 0 |
| ALOXE3 (Q9BYJ1) | 83 | 2 | 75 | 6 |
| Other leukotriene synthesizing enzymes | ||||
| LTA4H (P09960) | 42 | 1 | 39 | 2 |
| LTC4S (Q16873) | 10 | 0 | 10 | 0 |
| Leukotriene receptors | ||||
| CYSLTR1 (Q9Y271) | 49 | 0 | 49 | 0 |
| CYSLTR2 (Q9NS75) | 71 | 1 | 69 | 1 |
| LTB4R (Q15722) | 16 | 0 | 16 | 0 |
| LTB4R2 (Q9NPC1) | 43 | 2 | 41 | 0 |
| GPR17 (Q13304) | 67 | 0 | 66 | 1 |
| P2RY12 (Q9H244) | 35 | 1 | 34 | 0 |
| GPR99 (Q96P68) | 63 | 0 | 58 | 5 |
| Total | 864 | 19 | 817 | 28 |
5.1. Genetic variability of human lipoxygenases
5.1.1. ALOX5 (5-lipoxygenase)
The human ALOX5 gene is located on chromosome 10 and spans about 71.9 kbp consisting of 14 exonic and 13 intronic sequences [234]. It lacks TATA and CCAT boxes in the promoter but contains eight GC boxes, and five of them are arranged in tandems to be recognized by the transcription factors Sp1 and Egr1 [235], [236]. Previous clinical studies have shown that naturally occurring mutations in the ALOX5 gene can lead to the deletion or insertion of Sp1-binding sites, and the hALOX5 promoter genotype might impact anti-asthmatic therapy [237], [238]. As a possible molecular reason for this observation a reduced level of hALOX5 mRNA and LTC4 in patients with a certain genotype (non5/non5) was discussed [239]. In addition, mutations in the tandem GC boxes were associated with an increased arterial intima-media thickness and with higher plasma levels of C-reactive protein suggesting a role of this genetic variation in atherogenesis [240]. This conclusion was supported by another study, in which the Sp1 promoter variations were associated with an increased risk of coronary artery disease [241]. The molecular basis for these statistical associations has not been explored in detail. It was suggested, that the promoter variations might impact the biosynthesis of various arachidonic acid and eicosapentaenoic acid derived eicosanoids [242]. In addition, ALOX5 promoter variations have been associated with a decreased risk for colon and rectal cancer [243], [244] but there was indication for an involvement in Alzheimer’s disease as well [245].
When we quantified the genetic variability in the coding region of the human ALOX5 gene on the basis of the sequence data of the 1000 human genome project [233], we observed 69 non-synonymous coding variations but only the Glu254Lys exchange (rs2228065) had an allele frequency >1% and thus, qualifies as SNP (Table 6). This SNP has been associated with an increased risk for bronchial asthma in two independent case-control studies [246], [247]. Interestingly, this SNP is more frequent in African-Americans (allele frequency >10%) when compared with other ethnic groups and in Europeans it is much less abundant. Its low frequency in the European population appears to be the most probable reason why this SNP has never been considered for European case-control studies [248], [249], [250]. Although the Glu254Lys exchange occurs with increased frequency in patients suffering from bronchial asthma the functional basis for this relation remains unclear. It was hypothesized that the non-conservative Glu254Lys exchange destabilizes the 3D-structure [246]. When we expressed human wildtype ALOX5 and its corresponding Glu254Lys mutant as recombinant protein we found that the mutant enzyme exhibited a reduced (60% residual activity) catalytic activity [251]. On the other hand, the reaction specificity of the enzyme was not impacted by this amino acid exchange. Whether the rather subtle alterations in catalytic activity induced by the Glu254Lys exchange significantly contribute to the statistical correlation is difficult to judge, and partial functional inactivation of the enzyme should rather decrease the risk for asthma. In additional case-control studies this SNP was not significantly correlated with breast and colon cancer [243], [252].
When we localized the non-synonymous coding variations identified in the fame of the 1000 human genome project [233] in the 3D-structure of human ALOX5 [12], we found that neither the iron ligands [253] nor amino acids that are important for the reaction specificity [164], [254] were affected by genetic variations. Moreover, there are no nonsense mutations leading to interruption of the open reading frame reported in the database.
The substrate-binding pocket of hALOX5 is an elongated cavity, which appears to be sealed at both ends in the ligand free protein. Phe178 and Tyr182 (FY-cork) close the cavity at one end and Trp148 may block the opposite entrance to the cavity [12], [255]. In addition to that, the enzyme activity can be regulated by certain factors (Ca2+, phosphatidyl choline, ATP, coactosin-like protein) and for some of these ligands the binding mechanisms have been described [256], [257], [258], [259]. Neither of these functionally important amino acids were affected by any of the genetic variations [251].
Human ALOX5 can be regulated by reversible phosphorylation of Ser271, Ser523, and Ser663 [260], [261], [262], [263]. There is a rare genetic variation at Ser271 in the 1000 human genome database (TMP_ESP_10_45920561, allele frequency <0.1%), which induced on the protein level a Ser271Ile exchange. This mutation should prevent enzyme phosphorylation at this position. Since phosphorylation of Ser271 was suggested to prevent nuclear export of the protein [262], [263] this rare mutation might be of functional relevance.
5.1.2. ALOX15 (12/15-lipoxygenase)
The hALOX15 gene is located on chromosome 17 and spans about 10.7 kb consisting of 14 exonic and 13 intronic sequences. It also lacks TATA and CCAT boxes but contains several GC rich sequences [264]. A genetic promoter polymorphism (−292 C/T) leads to introduction of an additional Sp1 transcription factor-binding site. This promoter polymorphism apparently enhances transcription of the ALOX15 gene, which might be relevant for atherogenesis [265], [266]. However, in a European case-control study there was no significant association between this promoter variation and myocardial infarction [267]. Other ALOX15 promoter polymorphisms were associated with the extent of carotid plaque formation [268]. For one of these variations (rs2255888) a significant difference in the in vitro promoter activity was reported [269] but it remains unclear whether this might also be the case in vivo. Moreover, ALOX15 promoter polymorphisms appeared to be important for the pathogenesis of aspirin-exacerbated respiratory disease [270].
Human ALOX15 consists of 662 amino acids [271]. In the 1000 human genome database 94 non-synonymous coding variations were found (Table 6). Two of them, Pro617Ser (rs41432647) and Thr560Met (rs34210653), have an allele frequency of >1% (SNPs) whereas the vast majority are rare mutations. We analyzed the functional consequences of selected genetic variations. For Thr560Met (rs34210653) and Gly422Glu (rs61099320) a complete loss in catalytic activity was reported [272], [273]. More detailed analysis of the structural basis for the loss in catalytic activity suggested that the amino acid exchanges induced partial misfolding of the enzyme. For other naturally occurring rare mutations no functional consequences could be identified [251]. Heterozygous carriers of the Thr560Met mutations had an increased risk for developing coronary artery disease [267], [274] and similar effects can be expected for other hALOX15 loss-of-function variations.
The 1000 human genome database involves eight nonsense mutations in the ALOX15 gene and when these nonsense alleles occur homozygously the corresponding individuals should be devoid of ALOX15. His360, His365, His540, His544, and the C-terminal Ile662 constitute the direct iron ligands of hALOX15 [275], [276] but neither of these residues were affected by any genetic variation. Rabbit and human ALOX15 exhibit dual positional specificity of arachidonic acid oxygenation, and the amino acid residues Phe352, Ile417/Met418 and Ile592 play a major role for this enzyme property [215], [277]. Searching the 1000 human genome database we found a rare variation at Phe352, in which the bulky Phe is mutated to a less space-filling Leu (rs143365387). According to the triad concept [44] this enzyme variant should be a 12-lipoxygenating enzyme species. These data indicate that homozygous allele carriers express a 12-lipoxygenating ALOX15 (as it is the case in mice) and it would be of major interest to find out whether such individuals have any functional defects. Unfortunately, this variation has only a low allele frequency and thus, the search for homozygous allele carriers is rather laborious and the existence of such individuals is even questionable. Human ALOX15 seems to follow the Ala-to-Gly [164] concept but for the corresponding residue Ala403 there is no genetic variation described in the SNP databases.
Human and rabbit ALOX15 are capable of oxygenating membrane lipids [278]. Surface exposed hydrophobic amino acids such as Tyr15, Leu70, Leu71, Lys180 and Leu194 have been implicated in membrane binding [279] and there are genetic variations of corresponding residues in the human 1000 genome database. The Tyr15His exchange (rs13306168) has a global frequency of 0.6% whereas no frequency data are available for the Leu194Pro exchange (rs11867874). Unfortunately, there are no functional data for the corresponding mutated proteins. Mutagenesis studies and molecular dynamic simulations of enzyme-substrate complexes suggested that Arg402 and Phe414 may be involved in substrate binding [280], [281]. For Arg402 two rare mutations with a global frequency <1% have been identified in the 1000 human genome database [Arg402Gln (TMP_ESP_17_4536752) and Arg402Trp (rs144038526)]. However, the corresponding mutant protein did not exhibit major functional defects [251].
5.1.3. ALOX12 (platelet-type 12-lipoxygenase)
The human ALOX12 gene is located on chromosome 17 and spans a region of 14.6 kbp [282]. Its promoter involves five GC-boxes and carries two Sp1-binding sites [283]. Two genetic promoter variations (rs3840880, rs9897850) have been associated with an increased risk of osteoporosis [284] whereas another variation (rs11571327) was related to melanoma [285]. In addition, another ALOX12 promoter variation (rs9904779) was associated with earlier onset of menopause but there is no evidence for statistical association of this SNP with cardiovascular diseases [286], [287].
Human ALOX12 contains 663 amino acids and was the first mammalian lipoxygenase described back in 1974 [60]. In the 1000 human genome database there are 67 non-synonymous coding variations for this enzyme (Table 6). Two of them, Gln261Arg (rs28395860) and Asn322Ser (rs434473) have an allele frequency <1% [251]. The Gln261Arg SNP was associated with osteoporosis, schizophrenia and type 2 diabetes [284], [288], [289] while the Asn322Ser SNP has been related to early onset of menopause and schizophrenia [287], [289]. Unfortunately, the molecular basis for these statistical correlations remains unclear and corresponding mutagenesis studies on the recombinant enzyme did not provide convincing data for functional defects [251], [290].
When we searched the 1000 human genome database for genetic variations of functionally important amino acid residues (iron ligands, position- and stereospecificity determinants) we did not find any genetic variations at these positions. However, there is one nonsense variation (Arg348stop, rs141346813) in the 1000 human genome database and homozygous carriers of this allele should be devoid of ALOX12. As for hALOX15 [280] Arg402 of hALOX12 may play a role for substrate binding [281] and Arg402Gln exchange (rs143539715) is a naturally occurring rare enzyme variant. However, the functional consequences of this amino acid exchange have not been explored yet.
5.1.4. ALOX15B (15-lipoxygenase-2)
The gene for hALOX15B is located on chromosome 17 and spans about 10.1 kb [218]. The hALOX15B promoter has several Sp1 sites. The enzyme was suggested as negative cell-cycle regulator (tumor-suppressor activity) and Sp1-binding appeared to upregulate tumor suppressor function [291], [292].
The enzyme consists of 676 amino acids (Isoform D) and was first discovered in 1997 [218]. 86 non-synonymous coding variations have been identified in the 1000 human genome database and six of them [251] have an allele frequency >1% (Table 6): Pro77Ala (rs78230493), Ala311Val (rs148602477), Arg486His (rs9895916), Arg635Gln (rs61730298), Gln656Arg (rs4792147), and Ile676Val (rs7225107). The Arg486His and Ile676Val exchanges could not be associated to coronary artery disease [293] but heterozygous allele carriers of the Gln656Arg SNP seem to have a decreased risk for developing the disorder. However, mutagenesis studies on the recombinant protein did not provide any evidence for major functional defects of these mutations [293]. The C-terminal Ile676 is a direct iron ligand and Ile676Val is a SNP with a frequency of 9.6% but mutagenesis studies did not reveal major functional alterations [251]. There are four nonsense variations described in the 1000 genome database but neither of them had an allele frequency >1%.
The hALOX15B exhibits a singular positional specificity, as it converts arachidonic acid almost exclusively to 15-HpETE, but the mutations Asp603Tyr+Val604His convert the enzyme partially to an 8-lipoxygenating isoform [45], [218]. In the 1000 genome database there is a rare natural occurring variant of Val604 (rs192653434) with a global frequency of 0.1% but the functional consequences of this amino acid exchange have not been studied in detail. Human ALOX15B carries an Ala at the Coffa-Brash position and therefore it has been classified as S-LOX [164]. In the 1000 human genome database there is a rare genetic variation at this position (rs140152561) leading to an Ala-to-Asp exchange. We found that this amino acid exchange constitutes a loss-of-function mutation and that the catalytic inactivity of the mutant enzyme may be caused by a partial misfolding of the protein [251]. ALOX15B is highly expressed in human macrophages [294]. Lentivirus-mediated expression silencing of the enzyme in LDL-receptor deficient mice and siRNA induced down-regulation of ALOX15B expression in human primary macrophages reduced lipid deposition in the vessel wall and impaired secretion of inflammatory cytokines. Thus, the enzyme was considered a pro-atherogenic protein [295]. Since human and mouse ALOX15B orthologs exhibit different reaction specificities with arachidonic acid it remains unclear whether these data can be directly translated into the human situation.
5.1.5. ALOX12B (12R-lipoxygenase)
The gene encoding for the hALOX12B is also located on chromosome 17 and spans about 15 kb [296]. The promoters of human and mouse orthologs contain several GC boxes but lack TATA and CCAT boxes [202], [296], [297]. There is currently no information available as to the functionality of genetic promoter variations.
This LOX isoform consists of 701 amino acids and was first described in mouse and human skin [202], [296], [297]. 69 non-synonymous coding variations have been published in the 1000 human genome database (Table 6) but neither of them leads to a premature stop codon. There is a rare mutation in the ALOX12B gene (rs140063508) leading to a His398Tyr exchange. Since His398 is an immediate iron ligand this mutation might impact the catalytic activity of the enzyme. However, since Tyr carries a phenolic OH-group it might well function as iron ligand but functional studies on the corresponding mutant enzyme are still pending. There are no nonsense mutations in ALOX12B described in the 1000 human genome database and except for the iron coordinating His398 neither of the other functional important amino acid residues (other iron ligands, Coffa-Brash determinant) are affected by genetic variations.
Newborn Alox12b knockout mice die shortly after delivery because of dehydration [29] and a naturally occurring mutation in this gene induces similar skin defects [298]. In humans, genetic variations in the hALOX12B gene have been related to the development of autosomal recessive congenital ichthyosis [31], [299], [300], [301], [302]. The molecular mechanisms for the involvement of LOX isoforms in skin development are not completely understood but there is a testable mechanistic concept, which involves enzymatic oxygenation of skin specific ceramide, subsequent decomposition of the hydroperoxy derivatives and preferential hydrolysis of the oxidized lipids [303], [304].
Some of the genetic variations observed in the above referenced association studies have been picked up in human 1000 genome project but except for Pro127Ser (rs72842957), which has a global frequency of 1.1%, all others are classified as rare enzyme mutations. These data are consistent with the fact that ARCI is a rare skin disease (incident rate 1:200,000–300,000 Europeans and Northern Americans). Unfortunately, the Pro127Ser variant is not expressed in HEK 293 cells and thus, no functional data are currently available [31]. If this variation with its relatively high global allele frequency of >1% would be a strong inducer of ARCI a much higher prevalence of this skin disease should be expected. Only minor functional consequences are predicted for the Pro127Ser exchange since it is located at the enzyme surface far from the active site as suggested by structural modeling [251].
5.1.6. ALOXE3 (epidermal lipoxygenase-3)
The human ALOXE3 gene is also located on chromosome 17 and spans about 23.1 kb [305]. Similar to other LOXs the hALOXE3 promoter lacks TATA and CCAT motifs but contains several GC boxes. So far, no promoter variations have been associated with major diseases.
Like ALOX12B this LOX isoform was discovered in mouse and human skin [201], [305]. It consists of 711 amino acids and has a molecular weight of 80.6 kDa. It is an unusual LOX isoform and exhibits dominant hydroperoxide isomerase activity converting hydroperoxides (HpETEs) to specific epoxyalcohols [306], [307]. However, under certain conditions it also exhibits dioxygenase activity [308], [309].
83 non-synonymous coding variations have been published in the human 1000 genome database (Table 6) but only two of them (Ile515Val, rs3027205; Arg678Cys, rs143246503) have a global allele frequency >1% (3.0% and 1.9%). Ile515Val exchange is a conserved amino acid substitution and thus, no major functional consequences are expected. In contrast, Arg678Cys is non-conservative but its functional relevance can hardly be predicted. As for the ALOX12B gene genetic variations of ALOXE3 have been associated with autosomal recessive congenital ichthyosis [31], [299], [300], [301], [302] and some of these mutations were also found in the 1000 human genome database. The Ile515Val exchange was found with higher frequency in ichthyosis patients but it remains unclear whether this mutation impacts the enzyme functionality. Six nonsense variations have been identified in the ALOXE3 gene, which lead to C-terminally truncated enzyme variants. One of them (Arg211stop; rs141340759) has also been identified in a genome-wide melanoma study [310].
5.2. Genetic variability of other leukotriene synthesizing enzymes
5.2.1. Leukotriene A4 hydrolase (LTA4H)
The gene encoding for hLTA4H is located on chromosome 12 and spans about 42.7 kb [311], [312], [313]. Although the 5′-upstream region of the gene has not been explored for promoter activity, genetic variations in this region (rs2660845, rs2540477) have been related to an altered response to Montelukast treatment of asthma patients and to coronary artery disease [314], [315], [316]. The SNP rs2660845 was previously described in the 6-fold haplotype “HapE” and the ten-fold haplotype “HapK”. HapE is protective in female depression and in subclinical atherosclerosis [317], [318]. HapK seems to increase the risk for myocardial infarction [319].
This enzyme consists of 610 amino acids and has a MW of 69 kDa [313]. It catalyzes the final step in the biosynthesis of LTB4. In the 1000 human genome database there are 42 non-synonymous coding variations (Table 6) but only one of them (Glu481Asp, rs45630739) has a global frequency of >1% (1.6%). This is a conserved amino acid exchange and the crystal structure of hLTA4H [13] indicated that this residue is located on the enzyme surface. LTA4H is a bifunctional enzyme that has an epoxide hydrolase activity for LTA3, LTA4 and LTA5 [320], [321] but also harbors a zinc-dependent aminopeptidase activity with a zinc binding motif [322]. Three amino acids (His295, His299 and Glu318) form the Zn-binding cluster and mutagenesis studies indicated that the loss of one of these residues blocks the catalytic activity [323], [324], [325], [326]. LTA4H undergoes suicidal inactivation during LTA4 hydrolysis and as molecular reason for the loss in catalytic activity chemical modification of Tyr378 was suggested [327], [328]. Additional mechanistic studies indicated the importance of the following amino acids for the functionality of the enzyme: Gln136, Glu271, Glu296, Asp375, Arg563, and Lys565 [6], [329], [330], [331]. When we searched the 1000 human genome database we did not detect any genetic variations for these residues. However, we found two premature stop variants as rare mutations with an allele frequency of <0.1%.
5.2.2. Leukotriene C4 synthase (LTC4S)
The gene encoding for leukotriene C4 synthase is located on chromosome 5 and spans about 2.5 kb [332], [333]. The promoter involves potential cis-regulatory sequences but their in vivo functionality remains to be explored [334]. One promoter polymorphism (rs730012) was associated with a higher susceptibility of the allele carriers for ischemic stroke and with an altered response to the anti-asthma treatment with Montelukast [314], [335].
Human LTC4S is an 18 kDa enzyme with 150 amino acids which belongs to the family of membrane-associated Proteins in Eicosanoid and Glutathione (MAPEG) metabolism. It catalyzes the reaction between LTA4 and glutathione (GSH) to LTC4. 10 different non-coding variations are published in the 1000 human genome database but none of them has an allele frequency >1% (Table 6).
Based on the crystal structures of the enzyme the catalytic importance of Arg104 was suggested [16], [336]. Interestingly, when we searched the publically available SNP databases we found a rare mutation (rs13162195), which induces an Arg104Lys exchange. Mutagenesis studies at this position have shown that exchanges of Arg104 to Ala, Ser, Thr, and Lys caused almost complete loss in catalytic activity [337]. The observation that the Arg104Lys exchange induces such dramatic functional consequences is rather surprising because of the conservative character of this amino acid substitution. Nevertheless, on the basis of this functional data one has to conclude that homozygous allele carriers of this non-synonymous SNP should completely lack functional cys-LT signaling. Thus, such individuals should be protected from bronchial asthma and other allergic diseases. However, since this SNP is a rare mutation, which only occurred once in the Human 1000 Genome Database the design of a meaningful large scale epidemiological study is somewhat problematic. In addition to Arg104, Ser28, Ser111 and Trp116 have been implicated in the catalytic cycle [16], [336], [338]. For Ser28 a rare mutation (TMP_ESP_179222609) was identified in the SNP databases and this nucleotide exchange leads to a Ser28Leu exchange. This mutation removes a hydroxyl group and thus corrupts a phosphorylation site.
5.3. Genetic variability of leukotriene receptors
5.3.1. Cysteinylleukotriene receptor 1 (CysLTR1)
The gene encoding for hCYSLTR1 is located on X chromosome and spans about 56 kbp [339], [340]. Little is known about transcriptional regulation of the gene, but a 4-fold promoter polymorphism haplotype (rs321029, rs2806489, rs7066737, and rs2637204) causes a significant decrease in transcription of the CYSLTR1 gene when compared with wildtype individuals [341]. In vitro studies indicated that the polymorphism rs2806489 reduces promoter activity by factor 2 [342] but in vivo confirmation of the effect is lacking. Other CYSLTR1 promoter polymorphisms appear to be involved in atopy and bronchial asthma [343], [344], [345].
The CYSLTR1 belongs to the family of G-protein coupled receptors (GPCRs) consisting of 337 amino acids. It contains 7-transmembrane spanning helices and binds various ligands: LTD4 ≫ LTE4 = LTC4 [346]. In the 1000 human genome database 49 non-synonymous genetic variations are described but none of them has an allele frequency >1%. Among them there is no nonsense variation (Table 6). Little is known about the molecular details of ligand binding and intracellular signaling and thus, predictions on functional consequences of structural alterations are problematic. The naturally occurring Gly300Ser exchange (CM072979) was associated with increased risks for atopy and asthma. Apparently, the Gly300Ser receptor has an increased affinity for LTD4 [347] suggesting more intense leukotriene signaling for allele carriers. Owing to its low allele frequency in the normal population this genetic variation has not been picked up in the Human 1000 Genome Project and is neither listed in the NCBI SNP database. However in the population of the Tristan da Cunha islands its allele frequency exceeds 10%. The Tristan da Cunha islands are localized in the southern Atlantic 2000 km apart from Saint Helena, 2400 km apart from the western shore of South Africa and more than 3000 km apart from South America. They are the most isolated inhabited islands on earth. The main island has a north–south length of about 11 km, and an area of 98 km2. Today 80 families are living on its main island and the total population comprises about 300 permanent residents. The genetic pool of the permanent residents has been derived from only 15 common ancestors, which makes the population an interesting group for analyzing the inheritance of certain genetic defects [348].
5.3.2. Cysteinylleukotriene receptor 2 (cysLTR2)
The gene encoding for CYSLTR2 is located on chromosome 13 and spans about 2.5 kb [200]. Although there is little knowledge about transcriptional regulation of CYSLTR2 there are genetic polymorphisms in the promoter (rs7324991), in the 3′UTR (rs912278, rs34494076) and in the downstream region of the gene (rs2407249), resulting in an increased mRNA and protein expression [349]. In vitro reporter gene experiments indicated higher activity of the mutated promoter when compared with the wildtype constructs [349]. Promoter and 3′UTR polymorphisms were associated with aspirin intolerance in asthma but the molecular basis for this interesting statistical correlation is not clear [350].
The CYSLTR2 is also a typical GPCR with 7-transmembrane spanning helices consisting of 346 amino acids. It binds Cys-LTs with different affinities in the following order: LTC4 = LTD4 ≫ LTE4 [351]. Searching the 1000 human genome database we found 71 non-synonymous genetic variations but only the Ser236Leu mutant (rs61735175) had an allele frequency >1% (Table 6). This amino acid exchange is rather conservative in nature and PolyPhen2 predictions suggested only minor effects on protein function (Horn et al., unpublished data). In addition, there is one nonsense variation (Trp104stop, COSM139999) but its low allele frequency suggests that homozygous allele carriers may hardly exist. Little is known about functionally important amino acids in CYSLTR2 but a genetic variation Met201Val (rs41347648) appears to be associated with atopy [352] and with bronchial asthma [353]. Met-to-Val exchanges are rather conservative and thus, only minor functional consequences may be predicted. However, experimental data suggested impaired receptor affinity for LTD4 [353].
5.3.3. Leukotriene B4 receptor 1 (BLT1)
The BLT1 gene is located on chromosome 14 and spans about 6.5 kb [354]. The promoter lacks typical TATA and CAAT boxes but contains one GC rich sequence, which can be activated by Sp1 [355]. No genetic promoter variations have been associated with diseases. BLT1 is a GPCR with 7-transmembrane spanning helices consisting of 352 amino acids [356]. There are 16 non-synonymous coding variations in the 1000 human genome database but none of them has an allele frequency >1%. There is no nonsense mutation leading to the formation of a truncated receptor (Table 6). Little is known about the functional relevance of important amino acids but Thr308 may be involved in BLT1 desensitization [357]. However, there is no genetic variation in the 1000 human genome database affecting this residue.
After ligand binding GPCRs activate intracellular G-proteins [358] and there are several genetic variations in the activating loop but neither of them has an allele frequency >0.1%. Helix 8 of the receptor is required for G-protein activation [359], [360] and several leucines appear to be important for receptor expression [361]. However, none of these residues is affected by any genetic variation.
5.3.4. Leukotriene B4 receptor 2 (BLT2)
The BLT2 gene is located on chromosome 14 and spans about 6.3 kb [362], [363], [364]. Little is known on transcriptional regulation of BLT2 and no promoter polymorphisms have been associated with any disease.
BLT2 is also a GPCR with 7-transmembrane spanning helices and consists of 389 amino acids [363], [364]. It binds different LTB4 derivatives [365] with high affinity (LTB4 > 12-epi-LTB4 > LTB5 > LTB3). 43 non-synonymous coding variations were found in the 1000 human genome database and two of them (Table 6), Arg80Gln (rs150416614) and Asp196Gly (rs1950504), have an allele frequency >1%. PolyPhen2 predictions suggest that Asp196Gly has only minor functional consequences while Arg80Gln might be deleterious (Horn et al., unpublished data). This residue is located in a putative cytoplasmic domain and thus, additional experiments are required to characterize the functional impact. There is no nonsense variation leading to truncated receptor.
5.3.5. Other leukotriene receptors (GPR17, P2RY12 and GPR99)
The gene encoding for GPR17 is located on chromosome 2. It spans about 6.8 kb and consists of two coding exons and an additional exon containing the 5′ UTR region [366], [367]. Little is known about the functional elements in the promoter region of this gene and currently no promoter polymorphisms have been associated with any disease. The gene encodes for a classical 7-transmembrane G-protein coupled receptor (GPCR) consisting of either 339 or 367 amino acids [367]. It selectively binds uracil nucleotides and cysLTs [368], and is a negative regulator of the cysLTR1 response to LTD4 [369]. There are 67 non-synonymous genetic variations for this receptor published in the 1000 genome database but none of them has an allele frequency >1% (Table 6). There is a single nonsense variation (Trp6stop, rs61742898) leading to a C-terminally truncated protein but it remains unclear whether this truncation is of functional importance.
The gene encoding for P2RY12 is located on chromosome 3 and spans about 47.4 kb. It consists of two exons separated by a single intron [370]. The receptor also belongs to the family of the GPCRs and consists of 342 amino acids [370]. Analyzing the 1000 genome database we found that there are 32 non-synonymous genetic variations but only one of them (Glu330Gly, rs16846673) qualifies as SNP (Table 6). However, PolyPhen2 prediction suggests that this mutation has only minor functional consequences (Horn et al., unpublished data). Mutations in the P2RY12 gene have been discovered in patients with platelet-type bleeding disorder-8 and congenital bleeding disorder [371], [372] and these mutants appear to impact receptor functionality [372]. Unfortunately, it remains unclear whether these patients have aberrant leukotriene signaling.
The GPR99 (GPR80, OXGR1) gene is located on chromosome 13 and spans about 9 kb. It consists of two exons separated by a single intron [373]. Like the P2Y receptor it belongs to the GPCRs and consists of 337 amino acids [374]. It has been implicated in citric cycle regulation [375] but it also binds LTE4 [376]. Analyzing the 1000 genome database we found that there are 58 non-synonymous genomic variations but none of them qualifies as SNP (Table 6). In addition, there are five nonsense variations [Arg148stop (COSM1677754), Tyr198stop (rs370630287), Arg240stop (ss1349524372), Gln310stop (COSM432637), and Ser315stop (COSM126856)] but neither of them has an allele frequency >1%. Unfortunately, little is known on the functionality of different amino acids for ligand binding at the GPR99 receptor and thus, it is difficult to predict the functional consequences of amino acid substitutions.
6. Conclusions
LOXs are lipid-peroxidizing enzymes and require molecular oxygen for their activity which was virtually absent at the onset of life about 3 Bya. About 650 Mya the oxygen was enriched in the atmosphere owing to the photosynthetic activity of cyanobacteria in the oceans and of primitive land plants reaching a concentration of 35% about 300 Mya. Next, the oxygen level decreased again reaching today’s steady state concentration of 21% around 250 Mya. The increase in atmospheric oxygen concentration was one of the driving forces for Cambrian species explosion, which dramatically increased the biodiversity on the planet. It might well be that LOXs as oxygen metabolizing enzymes have first been introduced during this time window of life development and the occurrence of LOXs in today’s cyanobacteria [65] might be supportive for this hypothesis. However, the currently available sequence data suggest that only a minor share of cyanobacteria contain LOX genes and thus, it must be concluded that these enzymes may not be essential for the evolution of life. Today, LOX occur in two (bacteria, eucarya) of the three domains of life but a continuous development of this class of enzymes during early evolution of life is rather unlikely. In fact, for some procaryotic LOXs horizontal gene transfer has been suggested [9], [63] and there is the possibility of lineage-specific loss of LOX genes in other bacterial species. The combination of these complex gain-of-function and loss-of-function events makes the search for the most ancient LOX isoforms problematic. In plants LOXs are widely distributed. They are already found in algae, which constitute the most primitive photosynthesizing plant-like eucarya but they also occur in fungi. Algae occurred during evolution of life already 1.6–1.5 Bya and are considered the most ancient producers of polyunsaturated fatty acids, the major substrate of LOXs. However, in this time window the oxygen concentration in our atmosphere was rather low and thus, despite the presence of PUFAs a true LOX reaction should not have taken place. Thus, oxylipin biosynthesis in algae and fungi [101], [102] should have been introduced later on. In protozoa, LOX-like sequences have been reported but functionality of the corresponding enzymes remains unclear. The low degree of LOX distribution in protozoa suggests that the enzymes may not be essential for survival of the corresponding species. In lower metazoa a number of functional LOXs have been identified as fusion and/or non-fusion proteins [135], [136], [141] and sequence searches [145] suggested the occurrence of functional LOXs in additional invertebrate species (Table 2). However, it is somewhat difficult to estimate the percentage of LOX-containing invertebrates among all invertebrates because only a limited number of completely sequenced invertebrate genomes are currently available. In Chordata LOXs are more frequently distributed. Potential LOX sequences appear to be present in primitive Chordata such as Cephalochordata and Tunicata as indicated by the genomic sequences of selected model organisms (B. floridae, C. intestinalis). However, it remains unclear whether the corresponding proteins are functional LOXs and if other representatives of these subphyla also carry LOX genes. All higher vertebrates (cartilaginous fish, bony fish, amphibia, reptilia, and birds) contain LOX-like sequences and although no functional data are currently available for most of them the existence of true LOXs in these animals are likely. In mammals LOX sequences are widely distributed and functional LOX isoforms have been identified in a large number of highly developed mammalian species including many primates.
ALOX5 constitutes the key enzyme of leukotriene biosynthesis. Since putative ALOX5-like sequences have been identified in Cephalochordata and Tunicata leukotriene signaling might already have been present in these primitive Chordata. When we searched the genome databases of B. floridae and C. intestinalis we found a CYSLTR1-like (XP002126547.2) and a LTA4H-like (XP002123481.1) sequence in the genome of C. intestinalis supporting this hypothesis. However, this is not the case for B. floridae, which is evolutionary more closely related to vertebrates. Moreover, the LOX sequences found in the genomes of these two species are more similar to coral LOXs than to ALOX5 isoforms of higher vertebrates (data not shown) and thus, leukotriene biosynthesis in C. intestinalis may be unlikely. To answer the question of whether primitive Chordata already have functional leukotriene signaling more direct experimental data and additional genomic sequences for other species representing this evolutionary stage are required. However, from the observation that both cartilaginous and bony fish (ray- and lobe-finned) appear to have all major enzymes and receptors of the leukotriene cascade it may be concluded that this signaling pathway has been introduced during evolution of life earlier than previously expected [158]. In this context it would be interesting to find out whether extinct fish species like armoured jawless fishes (Ostracodermata) or the primitive jawed fishes (Placodermata) carried functional genes for enzymes and receptors of leukotriene signaling. Genomic data on these extinct species would be helpful to close this gap of knowledge.
7. Perspectives
During the past decade the genomes of an increasing number of recent and extinct living organisms have been sequenced at high genome coverage and these high quality sequence data provide a suitable basis for detailed sequence comparisons to conclude the evolutionary relatedness of different species and their phylogenetic origin. However, a major problem for such conclusions is the lack of functional data for many of the published sequences. For instance, it remains unclear of whether or not the LOX-like sequences identified for the Acanthamoeba Polyphaga Mimivirus (YP_003986553.1) encodes a functional LOX enzyme. The sequence data are strongly supportive but sometimes it simply takes a single amino acid exchange to convert a catalytically active protein into an inactive enzyme (see Section 5.1.2). However, since the sequence is now known for this and other potential LOX isoforms the corresponding enzymes can be expressed as recombinant proteins and characterized with respect to their protein chemical and enzymatic properties. Similarly, it is of particular evolutionary interest to find out whether the LOX-like sequences identified in lower Chordata (B. floridae, B. belcheri) and in lower Vertebrata (P. marinus) encode for functional LOX isoforms. For Amphibia, Reptilia and Aves this review focused on selected model organism (Xenopus Silurana tropicalis, A. missisippiensis, G. gallus) because of space constrains. However, these restrictions might lead to misleading conclusions since other representatives of these evolutionary classes may be different. Thus, the genomes of additional class members should be screened for the presence of LOX-like sequences. The chemical stability of DNA and the improvement of the DNA sequencing techniques nowadays allows sequencing of complete genomes from excavated fossils representing extinct species. A single bone (manual phalanx) found in the Denisovan cave in south Siberia was sufficient to obtain the complete genomic sequence of a new relative of modern humans, H. denisovan [232]. This bone was conserved in the cave for more than 50,000 years but its DNA was well preserved so that a more than 40-fold genome coverage was reached during sequencing. Similar results might be obtained with even older plant and animal fossils particularly if the fossils are conserved in permafrost environments or in deserts. Sequencing the DNA remnants of Mammuthus or even of different dinosaurs will provide valuable information on the occurrence of LOX and leukotriene signaling in these animals, which dominated nature in ancient time.
In modern humans leukotrienes have been implicated in the pathogenesis of inflammatory and hyperproliferative diseases [6], [10] and thus, defects in leukotriene biosynthesis and in leukotriene signaling are expected to impact severity and duration of the disease. Consequently, naturally occurring mutations in the genes involved in leukotriene signaling are likely to impact the individual susceptibility for a specific disease and/or the responsiveness to therapeutic strategies. There are a number of correlation studies, which related specific alterations in leukotriene relevant genes to the development of a number of disorders or to the responsiveness of certain therapies. Although some studies showed significant differences, in most cases the causal chains have not been identified. The major reason for lacking causality is the fact that no reliable functional data are currently available for most genetic variations. Thus, in many cases it remains unclear of whether the naturally occurring mutants lead to defective or improved leukotriene signaling and this information is of major importance for interpretation of the clinical correlations.
Conflict of interest
The authors state that there is no conflict of interest with this work.
References
- 1.Valentine R.C., Valentine D.L. Omega-3 fatty acids in cellular membranes: a unified concept. Prog Lipid Res. 2004;43:383–402. doi: 10.1016/j.plipres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Tootle T.L. Genetic insights into the in vivo functions of prostaglandin signaling. Int J Biochem Cell Biol. 2013;45:1629–1632. doi: 10.1016/j.biocel.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Savari S., Vinnakota K., Zhang Y., Sjölander A. Cysteinyl leukotrienes and their receptors: bridging inflammation and colorectal cancer. World J Gastroenterol. 2014;20:968–977. doi: 10.3748/wjg.v20.i4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowe M.S., Nass S.R., Gabella K.M., Kinsey S.G. The endocannabinoid system modulates stress, emotionality, and inflammation. Brain Behav Immun. 2014;42C:1–5. doi: 10.1016/j.bbi.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn H., Banthiya S., van Leyen K. Mammalian lipoxygenases and their biological relevance. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbalip.2014.10.002. pii: S1388-1981(14)00211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haeggström J.Z., Funk C.D. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 7.Andreou A., Feussner I. Lipoxygenases – structure and reaction mechanism. Phytochemistry. 2009;70:1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Newcomer M.E., Gilbert N.C. Location, location, location: compartmentalization of early events in leukotriene biosynthesis. J Biol Chem. 2010;285:25109–25114. doi: 10.1074/jbc.R110.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen J., Garreta A., Benincasa M., Fusté M.C., Busquets M., Manresa A. Bacterial lipoxygenases, a new subfamily of enzymes? A phylogenetic approach. Appl Microbiol Biotechnol. 2013;97:4737–4747. doi: 10.1007/s00253-013-4887-9. [DOI] [PubMed] [Google Scholar]
- 10.Kanaoka Y., Boyce J.A. Cysteinyl leukotrienes and their receptors; emerging concepts. Allergy Asthma Immunol Res. 2014;6:288–295. doi: 10.4168/aair.2014.6.4.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh R.K., Tandon R., Dastidar S.G., Ray A. A review on leukotrienes and their receptors with reference to asthma. J Asthma. 2013;50:922–931. doi: 10.3109/02770903.2013.823447. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert N.C., Bartlett S.G., Waight M.T., Neau D.B., Boeglin W.E., Brash A.R. The structure of human 5-lipoxygenase. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thunnissen M.M., Andersson B., Samuelsson B., Wong C.H., Haeggström J. Crystal structures of leukotriene A4 hydrolase in complex with captopril and two competitive tight-binding inhibitors. FASEB J. 2002;16:1648–1650. doi: 10.1096/fj.01-1017fje. [DOI] [PubMed] [Google Scholar]
- 14.Ohnishi H., Miyahara N., Gelfand E.W. The role of leukotriene B(4) in allergic diseases. Allergol Int. 2008;57:291–298. doi: 10.2332/allergolint.08-RAI-0019. [DOI] [PubMed] [Google Scholar]
- 15.Mathis S., Jala V.R., Haribabu B. Role of leukotriene B4 receptors in rheumatoid arthritis. Autoimmun Rev. 2007;7:12–17. doi: 10.1016/j.autrev.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez Molina D., Wetterholm A., Kohl A., McCarthy A.A., Niegowski D., Ohlson E. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 17.Dahlén S.E., Björk J., Hedqvist P., Arfors K.E., Hammarström S., Lindgren J.A. Leukotrienes promote plasma leakage and leukocyte adhesion in postcapillary venules: in vivo effects with relevance to the acute inflammatory response. Proc Natl Acad Sci USA. 1981;78:3887–3891. doi: 10.1073/pnas.78.6.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peatfield A.C., Piper P.J., Richardson P.S. The effect of leukotriene C4 on mucin release into the cat trachea in vivo and in vitro. Br J Pharmacol. 1982;77:391–393. doi: 10.1111/j.1476-5381.1982.tb09310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuse H., Kohno S. Leukotriene receptor antagonists pranlukast and montelukast for treating asthma. Expert Opin Pharmacother. 2014;15:353–363. doi: 10.1517/14656566.2014.872241. [DOI] [PubMed] [Google Scholar]
- 20.Laidlaw T.M., Boyce J.A. Cysteinyl leukotriene receptors, old and new; implications for asthma. Clin Exp Allergy. 2012;42:1313–1320. doi: 10.1111/j.1365-2222.2012.03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans J.F., Ferguson A.D., Mosley R.T., Hutchinson J.H. What’s all the FLAP about?: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends Pharmacol Sci. 2008;29:72–78. doi: 10.1016/j.tips.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Rouzer C.A., Marnett L.J. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009;50(Suppl):S29–S34. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funk C.D., Chen X.S., Johnson E.N., Zhao L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 2002;68–69:303–312. doi: 10.1016/s0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 24.Bizzintino J.A., Khoo S.K., Zhang G., Martin A.C., Rueter K., Geelhoed G.C. Leukotriene pathway polymorphisms are associated with altered cysteinyl leukotriene production in children with acute asthma. Prostaglandins Leukot Essent Fatty Acids. 2009;81:9–15. doi: 10.1016/j.plefa.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Gertow K., Nobili E., Folkersen L., Newman J.W., Pedersen T.L., Ekstrand J. 12- and 15-lipoxygenases in human carotid atherosclerotic lesions: associations with cerebrovascular symptoms. Atherosclerosis. 2011;215:411–416. doi: 10.1016/j.atherosclerosis.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L., Moos M.P., Gräbner R., Pédrono F., Fan J., Kaiser B. The 5-lipoxygenase pathway promotes pathogenesis of hyperlipidemia-dependent aortic aneurysm. Nat Med. 2004;10:966–973. doi: 10.1038/nm1099. [DOI] [PubMed] [Google Scholar]
- 27.Hulten L.M., Olson F.J., Aberg H., Carlsson J., Karlstrom L., Boren J. 15-Lipoxygenase-2 is expressed in macrophages in human carotid plaques and regulated by hypoxia-inducible factor-1alpha. Eur J Clin Invest. 2010;40:11–17. doi: 10.1111/j.1365-2362.2009.02223.x. [DOI] [PubMed] [Google Scholar]
- 28.Rydberg E.K., Krettek A., Ullstrom C., Ekstrom K., Svensson P.A., Carlsson L.M. Hypoxia increases LDL oxidation and expression of 15-lipoxygenase-2 in human macrophages. Arterioscler Thromb Vasc Biol. 2004;24:2040–2045. doi: 10.1161/01.ATV.0000144951.08072.0b. [DOI] [PubMed] [Google Scholar]
- 29.Epp N., Fürstenberger G., Müller K., de Juanes S., Leitges M., Hausser I. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol. 2007;177:173–182. doi: 10.1083/jcb.200612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieg P., Rosenberger S., de Juanes S., Latzko S., Hou J., Dick A. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol. 2013;133:172–180. doi: 10.1038/jid.2012.250. [DOI] [PubMed] [Google Scholar]
- 31.Eckl K.M., Krieg P., Küster W., Traupe H., André F., Wittstruck N. Mutation spectrum and functional analysis of epidermis-type lipoxygenases in patients with autosomal recessive congenital ichthyosis. Hum Mutat. 2005;26:351–361. doi: 10.1002/humu.20236. [DOI] [PubMed] [Google Scholar]
- 32.Johnson E.N., Brass L.F., Funk C.D. Increased platelet sensitivity to ADP in mice lacking platelet-type 12-lipoxygenase. Proc Natl Acad Sci USA. 1998;95:3100–3105. doi: 10.1073/pnas.95.6.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D., Funk C.D. Disruption of 12/15-lipoxygenase expression in peritoneal macrophages. Enhanced utilization of the 5-lipoxygenase pathway and diminished oxidation of low density lipoprotein. J Biol Chem. 1996;271:24055–24062. [PubMed] [Google Scholar]
- 34.Johnson E.N., Nanney L.B., Virmani J., Lawson J.A., Funk C.D. Basal transepidermal water loss is increased in platelet-type 12-lipoxygenase deficient mice. J Invest Dermatol. 1999;112:861–865. doi: 10.1046/j.1523-1747.1999.00595.x. [DOI] [PubMed] [Google Scholar]
- 35.Cyrus T., Witztum J.L., Rader D.J., Tangirala R., Fazio S., Linton M.F. Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E-deficient mice. J Clin Invest. 1999;103:1597–1604. doi: 10.1172/JCI5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George J., Afek A., Shaish A., Levkovitz H., Bloom N., Cyrus T. 12/15-Lipoxygenase gene disruption attenuates atherogenesis in LDL receptor-deficient mice. Circulation. 2001;104:1646–1650. doi: 10.1161/hc3901.095772. [DOI] [PubMed] [Google Scholar]
- 37.Krönke G., Katzenbeisser J., Uderhardt S., Zaiss M.M., Scholtysek C., Schabbauer G. 12/15-Lipoxygenase counteracts inflammation and tissue damage in arthritis. J Immunol. 2009;183:3383–3389. doi: 10.4049/jimmunol.0900327. [DOI] [PubMed] [Google Scholar]
- 38.Munger K.A., Montero A., Fukunaga M., Uda S., Yura T., Imai E. Transfection of rat kidney with human 15-lipoxygenase suppresses inflammation and preserves function in experimental glomerulonephritis. Proc Natl Acad Sci USA. 1999;96:13375–13380. doi: 10.1073/pnas.96.23.13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middleton M.K., Zukas A.M., Rubinstein T., Jacob M., Zhu P., Zhao L. Identification of 12/15-lipoxygenase as a suppressor of myeloproliferative disease. J Exp Med. 2006;203:2529–2540. doi: 10.1084/jem.20061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor P.R., Heydeck D., Jones G.W., Krönke G., Funk C.D., Knapper S. Development of myeloproliferative disease in 12/15-lipoxygenase deficiency. Blood. 2012;119:6173–6174. doi: 10.1182/blood-2012-02-410928. [author reply 4–5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shappell S.B., Boeglin W.E., Olson S.J., Kasper S., Brash A.R. 15-Lipoxygenase-2 (15-LOX-2) is expressed in benign prostatic epithelium and reduced in prostate adenocarcinoma. Am J Pathol. 1999;155:235–245. doi: 10.1016/S0002-9440(10)65117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kühn H., Barnett J., Grunberger D., Baecker P., Chow J., Nguyen B. Overexpression, purification and characterization of human recombinant 15-lipoxygenase. Biochim Biophys Acta. 1993;1169:80–89. doi: 10.1016/0005-2760(93)90085-n. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe T., Haeggström J.Z. Rat 12-lipoxygenase: mutations of amino acids implicated in the positional specificity of 15- and 12-lipoxygenases. Biochem Biophys Res Commun. 1993;192:1023–1029. doi: 10.1006/bbrc.1993.1519. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov I., Heydeck D., Hofheinz K., Roffeis J., O’Donnell V.B., Kuhn H. Molecular enzymology of lipoxygenases. Arch Biochem Biophys. 2010;503:161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Jisaka M., Kim R.B., Boeglin W.E., Brash A.R. Identification of amino acid determinants of the positional specificity of mouse 8S-lipoxygenase and human 15S-lipoxygenase-2. J Biol Chem. 2000;275:1287–1293. doi: 10.1074/jbc.275.2.1287. [DOI] [PubMed] [Google Scholar]
- 46.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta R.S. Life’s third domain (Archaea): an established fact or an endangered paradigm? Theor Popul Biol. 1998;54:91–104. doi: 10.1006/tpbi.1998.1376. [DOI] [PubMed] [Google Scholar]
- 48.Mayr E. Two empires or three? Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagan T., Roettger M., Bryant D., Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol. 2010;2:379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adl S.M., Simpson A.G., Farmer M.A., Andersen R.A., Anderson O.R., Barta J.R. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 51.Cavalier-Smith T. A revised six-kingdom system of life. Biol Rev Camb Philos Soc. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- 52.Cavalier-Smith T. Only six kingdoms of life. Proc Biol Sci. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin E.V., Senkevich T.G., Dolja V.V. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edwards R.A., Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 56.Palumbo G.J., Glasgow W.C., Buller R.M. Poxvirus-induced alteration of arachidonate metabolism. Proc Natl Acad Sci USA. 1993;90:2020–2024. doi: 10.1073/pnas.90.5.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tam V.C., Quehenberger O., Oshansky C.M., Suen R., Armando A.M., Treuting P.M. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154:213–227. doi: 10.1016/j.cell.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu D., Hugenholtz P., Mavromatis K., Pukall R., Dalin E., Ivanova N.N. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andre D., Hou K. The presence of a lipoid oxidase in soybean. Glycine soya. C R Acad Sci. 1932;194:645–647. [Google Scholar]
- 60.Hamberg M., Samuelsson B. Prostaglandin endoperoxides. Novel transformations of arachidonic acid in human platelets. Proc Natl Acad Sci USA. 1974;71:3400–3404. doi: 10.1073/pnas.71.9.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimahara K. Peroxidation of soybean oil by lipoxygenase-forming bacteria (Gram-negative, rod-shaped) isolated from garbage. J Soc Chem Ind Jpn. 1964;67:1164–1168. [Google Scholar]
- 62.Iny D., Grossman S., Pinsky A. Lipoxygenase of the thermophilic bacteria Thermoactinomyces vulgaris—properties and study on the active site. Int J Biochem. 1993;25:1325–1330. [Google Scholar]
- 63.Porta H., Rocha-Sosa M. Lipoxygenase in bacteria: a horizontal transfer event? Microbiology. 2001;147:3199–3200. doi: 10.1099/00221287-147-12-3199. [DOI] [PubMed] [Google Scholar]
- 64.Vance R.E., Hong S., Gronert K., Serhan C.N., Mekalanos J.J. The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proc Natl Acad Sci USA. 2004;101:2135–2139. doi: 10.1073/pnas.0307308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koeduka T., Kajiwara T., Matsui K. Cloning of lipoxygenase genes from a cyanobacterium, Nostoc punctiforme, and its expression in Escherichia coli. Curr Microbiol. 2007;54:315–319. doi: 10.1007/s00284-006-0512-9. [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y., Boeglin W.E., Schneider C., Brash A.R. A 49-kDa mini-lipoxygenase from Anabaena sp. PCC 7120 retains catalytically complete functionality. J Biol Chem. 2008;283:5138–5147. doi: 10.1074/jbc.M705780200. [DOI] [PubMed] [Google Scholar]
- 67.Garreta A., Val-Moraes S.P., García-Fernández Q., Busquets M., Juan C., Oliver A. Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa. FASEB J. 2013;27:4811–4821. doi: 10.1096/fj.13-235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andreou A.Z., Vanko M., Bezakova L., Feussner I. Properties of a mini 9R-lipoxygenase from Nostoc sp. PCC 7120 and its mutant forms. Phytochemistry. 2008;69:1832–1837. doi: 10.1016/j.phytochem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Wang X., Lu F., Zhang C., Lu Y., Bie X., Ren D. Peroxidation radical formation and regiospecificity of recombinated Anabaena sp. lipoxygenase and its effect on modifying wheat proteins. J Agric Food Chem. 2014;62:1713–1719. doi: 10.1021/jf405425c. [DOI] [PubMed] [Google Scholar]
- 70.Lu X., Zhang J., Liu S., Zhang D., Xu Z., Wu J. Overproduction, purification, and characterization of extracellular lipoxygenase of Pseudomonas aeruginosa in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:5793–5800. doi: 10.1007/s00253-012-4457-6. [DOI] [PubMed] [Google Scholar]
- 71.Gao B., Boeglin W.E., Zheng Y., Schneider C., Brash A.R. Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the cyanobacterium Acaryochloris marina. J Biol Chem. 2009;284:22087–22098. doi: 10.1074/jbc.M109.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao B., Boeglin W.E., Brash A.R. Omega-3 fatty acids are oxygenated at the n-7 carbon by the lipoxygenase domain of a fusion protein in the cyanobacterium Acaryochloris marina. Biochim Biophys Acta. 2010;1801:58–63. doi: 10.1016/j.bbalip.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyington J.C., Gaffney B.J., Amzel L.M. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 74.Minor W., Steczko J., Stec B., Otwinowski Z., Bolin J.T., Walter R. Crystal structure of soybean lipoxygenase L-1 at 1.4 Å resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 75.Youn B., Sellhorn G.E., Mirchel R.J., Gaffney B.J., Grimes H.D., Kang C. Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins. 2006;65:1008–1020. doi: 10.1002/prot.21182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gillmor S.A., Villaseñor A., Fletterick R., Sigal E., Browner M.F. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 77.Choi J., Chon J.K., Kim S., Shin W. Conformational flexibility in mammalian 15S-lipoxygenase: reinterpretation of the crystallographic data. Proteins. 2008;70:1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- 78.Xu S., Mueser T.C., Marnett L.J., Funk M.O. Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure. 2012;20:1490–1497. doi: 10.1016/j.str.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kobe M.J., Neau D.B., Mitchell C.E., Bartlett S.G., Newcomer M.E. The structure of human 15-lipoxygenase-2 with a substrate mimic. J Biol Chem. 2014;289:8562–8569. doi: 10.1074/jbc.M113.543777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maccarrone M., Salucci M.L., van Zadelhoff G., Malatesta F., Veldink G., Vliegenthart J.F. Tryptic digestion of soybean lipoxygenase-1 generates a 60 kDa fragment with improved activity and membrane binding ability. Biochemistry. 2001;40:6819–6827. doi: 10.1021/bi010187m. [DOI] [PubMed] [Google Scholar]
- 81.Walther M., Hofheinz K., Vogel R., Roffeis J., Kühn H. The N-terminal β-barrel domain of mammalian lipoxygenases including mouse 5-lipoxygenase is not essential for catalytic activity and membrane binding but exhibits regulatory functions. Arch Biochem Biophys. 2011;516:1–9. doi: 10.1016/j.abb.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Andreou A., Göbel C., Hamberg M., Feussner I. A bisallylic mini-lipoxygenase from cyanobacterium Cyanothece sp. that has an iron as cofactor. J Biol Chem. 2010;285:14178–14186. doi: 10.1074/jbc.M109.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korotkova M., Jakobsson P.J. Persisting eicosanoid pathways in rheumatic diseases. Nat Rev Rheumatol. 2014;10:229–241. doi: 10.1038/nrrheum.2014.1. [DOI] [PubMed] [Google Scholar]
- 84.Ghasemi Pirbalouti A., Sajjadi S.E., Parang K. A review (research and patents) on jasmonic acid and its derivatives. Arch Pharm (Weinheim) 2014;347:229–239. doi: 10.1002/ardp.201300287. [DOI] [PubMed] [Google Scholar]
- 85.Seilacher A. Great revolutions in the history of life. J Nat Hist Mus Inst Chiba. 1997;4:87–91. [PubMed] [Google Scholar]
- 86.Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 87.Busquets M., Carpena X., Fita I., Fusté C., Garreta A., Manresa Á. Transworld Research Network; Trivandrum, India: 2011. Crystallization of the lipoxygenase of Pseudomonas aeruginosa 42A2, evolution and phylogenetic study of the subfamilies of the lipoxygenases. [Google Scholar]
- 88.DeLong E.F. Everything in moderation: archaea as ‘non-extremophiles’. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 89.Kelman L.M., Kelman Z. Archaea: an archetype for replication initiation studies? Mol Microbiol. 2003;48:605–615. doi: 10.1046/j.1365-2958.2003.03369.x. [DOI] [PubMed] [Google Scholar]
- 90.Jarrell K.F., Koval S.F. Ultrastructure and biochemistry of Methanococcus voltae. Crit Rev Microbiol. 1989;17:53–87. doi: 10.3109/10408418909105722. [DOI] [PubMed] [Google Scholar]
- 91.Gutiérrez M.C., Castillo A.M., Pagaling E., Heaphy S., Kamekura M., Xue Y. Halorubrum kocurii sp. nov., an archaeon isolated from a saline lake. Int J Syst Evol Microbiol. 2008;58:2031–2035. doi: 10.1099/ijs.0.65840-0. [DOI] [PubMed] [Google Scholar]
- 92.Palmer J.D., Soltis D.E., Chase M.W. The plant tree of life: an overview and some points of view. Am J Bot. 2004;91:1437–1445. doi: 10.3732/ajb.91.10.1437. [DOI] [PubMed] [Google Scholar]
- 93.Harwood J.L., Guschina I.A. The versatility of algae and their lipid metabolism. Biochimie. 2009;91:679–684. doi: 10.1016/j.biochi.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 94.Gerwick W.H. Structure and biosynthesis of marine algal oxylipins. Biochim Biophys Acta. 1994;1211:243–255. doi: 10.1016/0005-2760(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 95.Gerwick W.H., Roberts M.A., Vulpanovici A., Ballantine D.L. Biogenesis and biological function of marine algal oxylipins. Adv Exp Med Biol. 1999;447:211–218. doi: 10.1007/978-1-4615-4861-4_20. [DOI] [PubMed] [Google Scholar]
- 96.Liu Q.Y., Reith M.E. Isolation of a gametophyte-specific cDNA encoding a lipoxygenase from the red alga Porphyra purpurea. Mol Mar Biol Biotechnol. 1994;3:206–209. [PubMed] [Google Scholar]
- 97.Kurtzman C.P. Molecular taxonomy of the yeasts. Yeast. 1994;10:1727–1740. doi: 10.1002/yea.320101306. [DOI] [PubMed] [Google Scholar]
- 98.Legras J.L., Merdinoglu D., Cornuet J.M., Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 99.Alam M.Z., Alam Q., Jiman-Fatani A., Kamal M.A., Abuzenadah A.M., Chaudhary A.G. Candida identification: a journey from conventional to molecular methods in medical mycology. World J Microbiol Biotechnol. 2014;30:1437–1451. doi: 10.1007/s11274-013-1574-z. [DOI] [PubMed] [Google Scholar]
- 100.Van Dyk M.S., Kock J.L., Botha A. Hydroxy long-chain fatty acids in fungi. World J Microbiol Biotechnol. 1994;10:495–504. doi: 10.1007/BF00367653. [DOI] [PubMed] [Google Scholar]
- 101.Andreou A., Brodhun F., Feussner I. Biosynthesis of oxylipins in non-mammals. Prog Lipid Res. 2009;48:148–170. doi: 10.1016/j.plipres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 102.Brodhun F., Feussner I. Oxylipins in fungi. FEBS J. 2011;278:1047–1063. doi: 10.1111/j.1742-4658.2011.08027.x. [DOI] [PubMed] [Google Scholar]
- 103.Scarpari M., Punelli M., Scala V., Zaccaria M., Nobili C., Ludovici M. Lipids in Aspergillus flavus–maize interaction. Front Microbiol. 2014;5:74. doi: 10.3389/fmicb.2014.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matsuda Y., Satoh T., Beppu T., Arima K. Purification and properties of Co2+ requiring heme protein having lipoxygenase activity from Fusarium oxysporum. Agric Biol Chem. 1976;40:963–976. [Google Scholar]
- 105.Matsuda Y., Beppu T., Arima K. Crystallization and positional specificity of hydroperoxidation of Fusarium lipoxygenase. Biochim Biophys Acta. 1978;530:439–450. [PubMed] [Google Scholar]
- 106.Hamberg M. Isolation and structures of lipoxygenase products from Saprolegnia parasitica. Biochem Biophys Acta. 1986;876:688–692. [Google Scholar]
- 107.Li D.-C., Lui Z.-W., Lu J. Purification and characterization of lipoxygenase from the thermophilic fungus Thermomyces lanuginosus. Mycol Res. 2001;105:190–194. [Google Scholar]
- 108.Kuribayashi T., Kaise H., Uno C., Hara T., Hayakawa T., Joh T. Purification and characterization of lipoxygenase from Pleurotus ostreatus. J Agric Food Chem. 2002;50:1247–1253. doi: 10.1021/jf0112217. [DOI] [PubMed] [Google Scholar]
- 109.Pérez-Gilabert M., Sánchez-Felipe I., García-Carmona F. Purification and partial characterization of lipoxygenase from desert truffle (Terfezia claveryi Chatin) ascocarps. J Agric Food Chem. 2005;53:3666–3671. doi: 10.1021/jf048087l. [DOI] [PubMed] [Google Scholar]
- 110.Horowitz Brown S., Zarnowski R., Sharpee W.C., Keller N.P. Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl Environ Microbiol. 2008;74:5674–5685. doi: 10.1128/AEM.00565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brodhun F., Cristobal-Sarramian A., Zabel S., Newie J., Hamberg M., Feussner I. An iron 13S-lipoxygenase with an α-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS ONE. 2013;8:e64919. doi: 10.1371/journal.pone.0064919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su C., Oliw E.H. Manganese lipoxygenase. Purification and characterization. J Biol Chem. 1998;273:13072–13079. doi: 10.1074/jbc.273.21.13072. [DOI] [PubMed] [Google Scholar]
- 113.Hamberg M., Su C., Oliw E. Manganese lipoxygenase. Discovery of a bis-allylic hydroperoxide as product and intermediate in a lipoxygenase reaction. J Biol Chem. 1998;273:13080–13088. doi: 10.1074/jbc.273.21.13080. [DOI] [PubMed] [Google Scholar]
- 114.Garscha U., Jernerén F., Chung D., Keller N.P., Hamberg M., Oliw E.H. Identification of dioxygenases required for Aspergillus development. Studies of products, stereochemistry, and the reaction mechanism. J Biol Chem. 2007;282:34707–34718. doi: 10.1074/jbc.M705366200. [DOI] [PubMed] [Google Scholar]
- 115.Hörnsten L., Su C., Osbourn A.E., Garosi P., Hellman U., Wernstedt C. Cloning of linoleate diol synthase reveals homology with prostaglandin H synthases. J Biol Chem. 1999;274:28219–28224. doi: 10.1074/jbc.274.40.28219. [DOI] [PubMed] [Google Scholar]
- 116.Siedow J.N. Plant lipoxygenase: structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:145–188. [Google Scholar]
- 117.Veldink G.A., Hilbers M.P., Nieuwenhuizen W.F., Viegenthart J.F.G. Plant lipoxygenase: structure and mechanism. In: Rowley A.F., Kühn H., Schewe T., editors. Eicosanoids and related compounds in plants and animals. Portland Press; London: 1998. [Google Scholar]
- 118.Grebner W., Stingl N.E., Oenel A., Mueller M.J., Berger S. Lipoxygenase6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013;161:2159–2170. doi: 10.1104/pp.113.214544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vellosillo T., Martínez M., López M.A., Vicente J., Cascón T., Dolan L. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007;19:831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Feussner I., Kühn H., Wasternack C. Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci. 2001;6:268–273. doi: 10.1016/s1360-1385(01)01950-1. [DOI] [PubMed] [Google Scholar]
- 121.Kolomiets M.V., Hannapel D.J., Chen H., Tymeson M., Gladon R.J. Lipoxygenase is involved in the control of potato tuber development. Plant Cell. 2001;13:613–626. doi: 10.1105/tpc.13.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 123.Prakash T.R., Swamy P.M., Suguna P., Reddanna P. Characterization and behaviour of 15-lipoxygenase during peanut cotyledonary senescence. Biochem Biophys Res Commun. 1990;172:462–470. doi: 10.1016/0006-291x(90)90695-j. [DOI] [PubMed] [Google Scholar]
- 124.Zhang B., Chen K., Bowen J., Allan A., Espley R., Karunairetnam S. Differential expression within the LOX gene family in ripening kiwifruit. J Exp Bot. 2006;57:3825–3836. doi: 10.1093/jxb/erl151. [DOI] [PubMed] [Google Scholar]
- 125.Shen J., Tieman D., Jones J.B., Taylor M.G., Schmelz E., Huffaker A. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J Exp Bot. 2014;65:419–428. doi: 10.1093/jxb/ert382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lang D., Zimmer A.D., Rensing S.A., Reski R. Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 2008;13:542–549. doi: 10.1016/j.tplants.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 128.Senger T., Wichard T., Kunze S., Göbel C., Lerchl J., Pohnert G. A multifunctional lipoxygenase with fatty acid hydroperoxide cleaving activity from the moss Physcomitrella patens. J Biol Chem. 2005;280:7588–7596. doi: 10.1074/jbc.M411738200. [DOI] [PubMed] [Google Scholar]
- 129.Anterola A., Göbel C., Hornung E., Sellhorn G., Feussner I., Grimes H. Physcomitrella patens has lipoxygenases for both eicosanoid and octadecanoid pathways. Phytochemistry. 2009;70:40–52. doi: 10.1016/j.phytochem.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 130.Feussner I., Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 131.Liu S.Q., Liu X.H., Jiang L.W. Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genet Mol Res. 2011;10:2613–2636. doi: 10.4238/2011.October.25.9. [DOI] [PubMed] [Google Scholar]
- 132.Shin J.H., Van K., Kim D.H., Kim K.D., Jang Y.E., Choi B.S. The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 2008;8:133. doi: 10.1186/1471-2229-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bannenberg G., Martínez M., Hamberg M., Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44:85–95. doi: 10.1007/s11745-008-3245-7. [DOI] [PubMed] [Google Scholar]
- 134.Feng B., Dong Z., Xu Z., An X., Qin H., Wu N. Molecular analysis of lipoxygenase (LOX) genes in common wheat and phylogenetic investigation of LOX proteins from model and crop plants. J Cereal Sci. 2010;52:12–15. [Google Scholar]
- 135.Bundy G.L., Nidy E.G., Epps D.E., Mizsak S.A., Wnuk R.J. Discovery of an arachidonic acid C-8 lipoxygenase in the gorgonian coral Pseudoplexaura porosa. J Biol Chem. 1986;261:747–751. [PubMed] [Google Scholar]
- 136.Koljak R., Boutaud O., Shieh B.H., Samel N., Brash A.R. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 137.Boutaud O., Brash A.R. Purification and catalytic activities of the two domains of the allene oxide synthase-lipoxygenase fusion protein of the coral Plexaura homomalla. J Biol Chem. 1999;274:33764–33770. doi: 10.1074/jbc.274.47.33764. [DOI] [PubMed] [Google Scholar]
- 138.Mortimer M., Järving R., Brash A.R., Samel N., Järving I. Identification and characterization of an arachidonate 11R-lipoxygenase. Arch Biochem Biophys. 2006;445:147–155. doi: 10.1016/j.abb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 139.Neau D.B., Gilbert N.C., Bartlett S.G., Boeglin W., Brash A.R., Newcomer M.E. The 1.85 Å structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity. Biochemistry. 2009;48:7906–7915. doi: 10.1021/bi900084m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eek P., Järving R., Järving I., Gilbert N.C., Newcomer M.E., Samel N. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J Biol Chem. 2012;287:22377–22386. doi: 10.1074/jbc.M112.343285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brash A.R., Hughes M.A., Hawkins D.J., Boeglin W.E., Song W.C., Meijer L. Allene oxide and aldehyde biosynthesis in starfish oocytes. J Biol Chem. 1991;266:22926–22931. [PubMed] [Google Scholar]
- 142.Hawkins D.J., Brash A.R. Eggs of the sea urchin, Strongylocentrotus purpuratus, contain a prominent (11R) and (12R) lipoxygenase activity. J Biol Chem. 1987;262:7629–7634. [PubMed] [Google Scholar]
- 143.Coffa G., Hill E.M. Discovery of an 11(R)- and 12(S)-lipoxygenase activity in ovaries of the mussel Mytilus edulis. Lipids. 2000;35:1195–1204. doi: 10.1007/s11745-000-0636-5. [DOI] [PubMed] [Google Scholar]
- 144.Hada T., Swift L.L., Brash A.R. Discovery of 5R-lipoxygenase activity in oocytes of the surf clam Spisula solidissima. Biochim Biophys Acta. 1997;1346:109–119. doi: 10.1016/s0005-2760(96)00179-8. [DOI] [PubMed] [Google Scholar]
- 145.Yuan D., Zou Q., Yu T., Song C., Huang S., Chen S. Ancestral genetic complexity of arachidonic acid metabolism in Metazoa. Biochim Biophys Acta. 2014;1841:1272–1284. doi: 10.1016/j.bbalip.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 146.Heckmann L.H., Sibly R.M., Timmermans M.J., Callaghan A. Outlining eicosanoid biosynthesis in the crustacean Daphnia. Front Zool. 2008;5:11. doi: 10.1186/1742-9994-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Putnam N.H., Butts T., Ferrier D.E., Furlong R.F., Hellsten U., Kawashima T. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 148.Bockelie T., Fortey R.A. An early Ordovician vertebrate. Nature. 1976;94:36–38. [Google Scholar]
- 149.Volff J.N. Genome evolution and biodiversity in teleost fish. Heredity (Edinb) 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- 150.Smith M.P., Sansom I.J., Repetski J.E. Histology of the first fish. Nature. 1996:702–704. [Google Scholar]
- 151.Shu D.G., Luo H.L., Conway Morris S., Zhang X.L., Hu S.X., Chen L. Lower Cambrian vertebrates from south China. Nature. 1999;402:42–46. [Google Scholar]
- 152.Shu D.G., Morris S.C., Han J., Zhang Z.F., Yasui K., Janvier P. Head and backbone of the Early Cambrian vertebrate Haikouichthys. Nature. 2003;421:526–529. doi: 10.1038/nature01264. [DOI] [PubMed] [Google Scholar]
- 153.Gess R.W., Coates M.I., Rubidge B.S. A lamprey from the Devonian period of South Africa. Nature. 2006;443:981–984. doi: 10.1038/nature05150. [DOI] [PubMed] [Google Scholar]
- 154.Smith J.J., Kuraku S., Holt C., Sauka-Spengler T., Jiang N., Campbell M.S. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhu M., Zhao W., Jia L., Lu J., Qiao T., Qu Q. The oldest articulated osteichthyan reveals mosaic gnathostome characters. Nature. 2009;458:469–474. doi: 10.1038/nature07855. [DOI] [PubMed] [Google Scholar]
- 156.Zhu M., Yu X., Ahlberg P.E., Choo B., Lu J., Qiao T. A Silurian placoderm with osteichthyan-like marginal jaw bones. Nature. 2013;502:188–193. doi: 10.1038/nature12617. [DOI] [PubMed] [Google Scholar]
- 157.Venkatesh B., Lee A.P., Ravi V., Maurya A.K., Lian M.M., Swann J.B. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Morgan E.L., Maskrey B.H., Rowley A.F. At what stage in metazoan evolution did leukotriene generation first appear? – key insights from cartilaginous fish. Dev Comp Immunol. 2005;29:53–59. doi: 10.1016/j.dci.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 159.Amemiya C.T., Alföldi J., Lee A.P., Fan S., Philippe H., Maccallum I. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jansen C., Hofheinz K., Vogel R., Roffeis J., Anton M., Reddanna P. Stereocontrol of arachidonic acid oxygenation by vertebrate lipoxygenases: newly cloned zebrafish lipoxygenase 1 does not follow the Ala-versus-Gly concept. J Biol Chem. 2011;286:37804–37812. doi: 10.1074/jbc.M111.259242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Haas U., Raschperger E., Hamberg M., Samuelsson B., Tryggvason K., Haeggström J.Z. Targeted knock-down of a structurally atypical zebrafish 12S-lipoxygenase leads to severe impairment of embryonic development. Proc Natl Acad Sci USA. 2011;108:20479–20484. doi: 10.1073/pnas.1117094108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vogel R., Jansen C., Roffeis J., Reddanna P., Forsell P., Claesson H.E. Applicability of the triad concept for the positional specificity of mammalian lipoxygenases. J Biol Chem. 2010;285:5369–5376. doi: 10.1074/jbc.M109.057802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Coffa G., Brash A.R. A single active site residue directs oxygenation stereospecificity in lipoxygenases: stereocontrol is linked to the position of oxygenation. Proc Natl Acad Sci USA. 2004;101:15579–15584. doi: 10.1073/pnas.0406727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Adel S., Hofheinz K., Heydeck D., Kuhn H., Häfner A.K. Phosphorylation mimicking mutations of ALOX5 orthologs of different vertebrates do not alter reaction specificities of the enzymes. Biochim Biophys Acta. 2014;1841:1460–1466. doi: 10.1016/j.bbalip.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 166.Pettitt T.R., Rowley A.F., Barrow S.E., Mallet A.I., Secombes C.J. Synthesis of lipoxins and other lipoxygenase products by macrophages from the rainbow trout, Oncorhynchus mykiss. J Biol Chem. 1991;266:8720–8726. [PubMed] [Google Scholar]
- 167.Hill D.J., Griffiths D.H., Rowley A.F. Trout thrombocytes contain 12- but not 5-lipoxygenase activity. Biochim Biophys Acta. 1999;1437:63–70. doi: 10.1016/s1388-1981(98)00007-9. [DOI] [PubMed] [Google Scholar]
- 168.Jarvik E. The Devonian tetrapod Ichthyostega. Fossils Strata. 1996;40:1–213. [Google Scholar]
- 169.Hellsten U., Harland R.M., Gilchrist M.J., Hendrix D., Jurka J., Kapitonov V. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Green F.A., Herman C.A., Herman R.P., Claesson H.E., Hamberg M. Leukotrienes and related eicosanoids are produced by frog leukocytes. Biochem Biophys Res Commun. 1987;142:309–314. doi: 10.1016/0006-291x(87)90274-9. [DOI] [PubMed] [Google Scholar]
- 171.Stsiapanava A., Tholander F., Kumar R.B., Qureshi A.A., Niegowski D., Hasan M. Product formation controlled by substrate dynamics in leukotriene A4 hydrolase. Biochim Biophys Acta. 2014;1844:439–446. doi: 10.1016/j.bbapap.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 172.Gronert K., Virk S.M., Herman C.A. Endogenous sulfidopeptide leukotriene synthesis and 12-lipoxygenase activity in bullfrog (Rana catesbeiana) erythrocytes. Biochim Biophys Acta. 1995;1255:311–319. doi: 10.1016/0005-2760(94)00248-w. [DOI] [PubMed] [Google Scholar]
- 173.Gronert K., Virk S.M., Herman C.A. Thrombocytes are the predominant source of endogenous sulfidopeptide leukotrienes in the American bullfrog (Rana catesbeiana) Biochim Biophys Acta. 1995;1259:203–210. doi: 10.1016/0005-2760(95)00160-3. [DOI] [PubMed] [Google Scholar]
- 174.Shen K., Herman C.A. Partial purification and characterization of 12-lipoxygenase in bullfrog erythrocytes. Comp Biochem Physiol B: Biochem Mol Biol. 2000;127:563–573. doi: 10.1016/s0305-0491(00)00288-1. [DOI] [PubMed] [Google Scholar]
- 175.Hawkins D.J., Brash A.R. Lipoxygenase metabolism of polyunsaturated fatty acids in oocytes of the frog Xenopus laevis. Arch Biochem Biophys. 1989;268:447–455. doi: 10.1016/0003-9861(89)90312-3. [DOI] [PubMed] [Google Scholar]
- 176.Menger B., Vogt P.M., Allmeling C., Radtke C., Kuhbier J.W., Reimers K. AmbLOXe – an epidermal lipoxygenase of the Mexican axolotl in the context of amphibian regeneration and its impact on human wound closure in vitro. Ann Surg. 2011;253:410–418. doi: 10.1097/SLA.0b013e318207f39c. [DOI] [PubMed] [Google Scholar]
- 177.Ortiz M.E., Bühler M.I., Zelarayán L.I. Involvement of PLA2, COX and LOX in Rhinella arenarum oocyte maturation. Zygote. 2013:1–6. doi: 10.1017/S096719941200069X. [DOI] [PubMed] [Google Scholar]
- 178.Paton R.L., Smithson T.R., Clack J.A. An amniote-like skeleton from the Early Carboniferous of Scotland. Nature. 1999;398:508–513. [Google Scholar]
- 179.Laurin M., Reisz R.R. A reevaluation of early amniote phylogeny. Zool J Linn Soc. 1995;113:165–223. [Google Scholar]
- 180.St John J.A., Braun E.L., Isberg S.R., Miles L.G., Chong A.Y., Gongora J. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Herman C.A., Wang X., Gronert K. Biosynthesis of thromboxane by snake (Elaphe obsoleta) erythrocytes and the requirement of eicosanoid production for blood clotting. Biochim Biophys Acta. 1997;1334:23–27. doi: 10.1016/s0304-4165(96)00122-5. [DOI] [PubMed] [Google Scholar]
- 182.Hou L., Martin L.D., Zhou Z., Feduccia A. Early ADAPTIVE RADIATION OF BIRDS: EVIDENCE FROM FOSSILS FROM Northeastern China. Science. 1996;274:1164–1167. doi: 10.1126/science.274.5290.1164. [DOI] [PubMed] [Google Scholar]
- 183.Consortium ICGS Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 184.Flynn M.A., Qiao M., Garcia C., Dallas M., Bonewald L.F. Avian osteoclast cells are stimulated to resorb calcified matrices by and possess receptors for leukotriene B4. Calcif Tissue Int. 1999;64:154–159. doi: 10.1007/s002239900595. [DOI] [PubMed] [Google Scholar]
- 185.Einzig S., Noren G.R., Staley N.A., White J.G., Rao G.H. Arachidonic acid metabolism in thrombocytes and vascular tissues of turkeys. Prostaglandins. 1985;30:999–1017. doi: 10.1016/0090-6980(85)90172-8. [DOI] [PubMed] [Google Scholar]
- 186.Greenberg-Levy S.H., Budowski P., Grossman S. Lipoxygenase activity in the brain regions of young chicks: isolation and some properties. Int J Biochem. 1992;24:1607–1614. doi: 10.1016/0020-711x(92)90177-3. [DOI] [PubMed] [Google Scholar]
- 187.Grossman S., Bergman M., Sklan D. Lipoxygenase in chicken muscle. J Agric Food Chem. 1988;36:1268–1270. [Google Scholar]
- 188.Rougier G.W., Martinelli A.G., Forasiepi A.M., Novacek M.J. American Museum Novitates; Argentina: 2007. New Jurassic mammals from Patagonia. A reappraisal of Australosphenidan morphology and interrelationships. p. 1–54. [Google Scholar]
- 189.Waggoner B. University of California Museum of Paleontology; 2012. Introduction to the Synapsida. [Google Scholar]
- 190.Luo Z.X., Yuan C.X., Meng Q.J., Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 191.Warren W.C., Hillier L.W., Marshall Graves J.A., Birney E., Ponting C.P., Grützner F. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Murchison E.P., Schulz-Trieglaff O.B., Ning Z., Alexandrov L.B., Bauer M.J., Fu B. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.O’Leary M.A., Bloch J.I., Flynn J.J., Gaudin T.J., Giallombardo A., Giannini N.P. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science. 2013;339:662–667. doi: 10.1126/science.1229237. [DOI] [PubMed] [Google Scholar]
- 194.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 195.Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 196.Balcarek J.M., Theisen T.W., Cook M.N., Varrichio A., Hwang S.M., Strohsacker M.W. Isolation and characterization of a cDNA clone encoding rat 5-lipoxygenase. J Biol Chem. 1988;263:13937–13941. [PubMed] [Google Scholar]
- 197.Chen X.S., Naumann T.A., Kurre U., Jenkins N.A., Copeland N.G., Funk C.D. CDNA cloning, expression, mutagenesis, intracellular localization, and gene chromosomal assignment of mouse 5-lipoxygenase. J Biol Chem. 1995;270:17993–17999. doi: 10.1074/jbc.270.30.17993. [DOI] [PubMed] [Google Scholar]
- 198.Hui Y., Yang G., Galczenski H., Figueroa D.J., Austin C.P., Copeland N.G. The murine cysteinyl leukotriene 2 (CysLT2) receptor. cDNA and genomic cloning, alternative splicing, and in vitro characterization. J Biol Chem. 2001;276:47489–47495. doi: 10.1074/jbc.M107556200. [DOI] [PubMed] [Google Scholar]
- 199.Martin V., Sawyer N., Stocco R., Unett D., Lerner M.R., Abramovitz M. Molecular cloning and functional characterization of murine cysteinyl-leukotriene 1 (CysLT(1)) receptors. Biochem Pharmacol. 2001;62:1193–1200. doi: 10.1016/s0006-2952(01)00774-2. [DOI] [PubMed] [Google Scholar]
- 200.Takasaki J., Kamohara M., Matsumoto M., Saito T., Sugimoto T., Ohishi T. The molecular characterization and tissue distribution of the human cysteinyl leukotriene CysLT(2) receptor. Biochem Biophys Res Commun. 2000;274:316–322. doi: 10.1006/bbrc.2000.3140. [DOI] [PubMed] [Google Scholar]
- 201.Kinzig A., Fürstenberger G., Bürger F., Vogel S., Müller-Decker K., Mincheva A. Murine epidermal lipoxygenase (Aloxe) encodes a 12-lipoxygenase isoform. FEBS Lett. 1997;402:162–166. doi: 10.1016/s0014-5793(96)01517-7. [DOI] [PubMed] [Google Scholar]
- 202.Sun D., McDonnell M., Chen X.S., Lakkis M.M., Li H., Isaacs S.N. Human 12(R)-lipoxygenase and the mouse ortholog. Molecular cloning, expression, and gene chromosomal assignment. J Biol Chem. 1998;273:33540–33547. doi: 10.1074/jbc.273.50.33540. [DOI] [PubMed] [Google Scholar]
- 203.Muñoz-Garcia A., Thomas C.P., Keeney D.S., Zheng Y., Brash A.R. The importance of the lipoxygenase-hepoxilin pathway in the mammalian epidermal barrier. Biochim Biophys Acta. 2014;1841:401–408. doi: 10.1016/j.bbalip.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sun D., Elsea S.H., Patel P.I., Funk C.D. Cloning of a human “epidermal-type” 12-lipoxygenase-related gene and chromosomal localization to 17p13. Cytogenet Cell Genet. 1998;81:79–82. doi: 10.1159/000014993. [DOI] [PubMed] [Google Scholar]
- 205.van Dijk K.W., Steketee K., Havekes L., Frants R., Hofker M. Genomic and cDNA cloning of a novel mouse lipoxygenase gene. Biochim Biophys Acta. 1995;1259:4–8. doi: 10.1016/0005-2760(95)00158-9. [DOI] [PubMed] [Google Scholar]
- 206.Bürger F., Krieg P., Marks F., Fürstenberger G. Positional- and stereo-selectivity of fatty acid oxygenation catalysed by mouse (12S)-lipoxygenase isoenzymes. Biochem J. 2000;348(Pt 2):329–335. [PMC free article] [PubMed] [Google Scholar]
- 207.Chen X.S., Sheller J.R., Johnson E.N., Funk C.D. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372:179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- 208.Sailesh S., Kumar Y.V., Prasad M., Reddanna P. Sheep uterus dual lipoxygenase in the synthesis of 14,15-leukotrienes. Arch Biochem Biophys. 1994;315:362–368. doi: 10.1006/abbi.1994.1512. [DOI] [PubMed] [Google Scholar]
- 209.Toh H., Yokoyama C., Tanabe T., Yoshimoto T., Yamamoto S. Molecular evolution of cyclooxygenase and lipoxygenase. Prostaglandins. 1992;44:291–315. doi: 10.1016/0090-6980(92)90004-d. [DOI] [PubMed] [Google Scholar]
- 210.Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bryant R.W., Bailey J.M., Schewe T., Rapoport S.M. Positional specificity of a reticulocyte lipoxygenase. Conversion of arachidonic acid to 15-S-hydroperoxy-eicosatetraenoic acid. J Biol Chem. 1982;257:6050–6055. [PubMed] [Google Scholar]
- 212.Freire-Moar J., Alavi-Nassab A., Ng M., Mulkins M., Sigal E. Cloning and characterization of a murine macrophage lipoxygenase. Biochim Biophys Acta. 1995;1254:112–116. doi: 10.1016/0005-2760(94)00199-9. [DOI] [PubMed] [Google Scholar]
- 213.Yoshimoto T., Suzuki H., Yamamoto S., Takai T., Yokoyama C., Tanabe T. Cloning and sequence analysis of the cDNA for arachidonate 12-lipoxygenase of porcine leukocytes. Proc Natl Acad Sci USA. 1990;87:2142–2146. doi: 10.1073/pnas.87.6.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.De Marzo N., Sloane D.L., Dicharry S., Highland E., Sigal E. Cloning and expression of an airway epithelial 12-lipoxygenase. Am J Physiol. 1992;263:1. doi: 10.1152/ajplung.1992.262.2.L198. [preceding L] [DOI] [PubMed] [Google Scholar]
- 215.Borngräber S., Browner M., Gillmor S., Gerth C., Anton M., Fletterick R. Shape and specificity in mammalian 15-lipoxygenase active site. The functional interplay of sequence determinants for the reaction specificity. J Biol Chem. 1999;274:37345–37350. doi: 10.1074/jbc.274.52.37345. [DOI] [PubMed] [Google Scholar]
- 216.Johannesson M., Backman L., Claesson H.E., Forsell P.K. Cloning, purification and characterization of non-human primate 12/15-lipoxygenases. Prostaglandins Leukot Essent Fatty Acids. 2010;82:121–129. doi: 10.1016/j.plefa.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 217.Berger M., Schwarz K., Thiele H., Reimann I., Huth A., Borngräber S. Simultaneous expression of leukocyte-type 12-lipoxygenase and reticulocyte-type 15-lipoxygenase in rabbits. J Mol Biol. 1998;278:935–948. doi: 10.1006/jmbi.1998.1737. [DOI] [PubMed] [Google Scholar]
- 218.Brash A.R., Boeglin W.E., Chang M.S. Discovery of a second 15S-lipoxygenase in humans. Proc Natl Acad Sci USA. 1997;94:6148–6152. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jisaka M., Kim R.B., Boeglin W.E., Nanney L.B., Brash A.R. Molecular cloning and functional expression of a phorbol ester-inducible 8S-lipoxygenase from mouse skin. J Biol Chem. 1997;272:24410–24416. doi: 10.1074/jbc.272.39.24410. [DOI] [PubMed] [Google Scholar]
- 220.Schneider C., Pratt D.A., Porter N.A., Brash A.R. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hublin J.J. Out of Africa: modern human origins special feature: the origin of Neanderthals. Proc Natl Acad Sci USA. 2009;106:16022–16027. doi: 10.1073/pnas.0904119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Krause J., Orlando L., Serre D., Viola B., Prüfer K., Richards M.P. Neanderthals in central Asia and Siberia. Nature. 2007;449:902–904. doi: 10.1038/nature06193. [DOI] [PubMed] [Google Scholar]
- 223.Gibbons A. Human evolution. Oldest Homo sapiens genome pinpoints Neandertal input. Science. 2014;343:1417. doi: 10.1126/science.343.6178.1417. [DOI] [PubMed] [Google Scholar]
- 224.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chaitidis P., Adel S., Anton M., Heydeck D., Kuhn H., Horn T. Lipoxygenase pathways in Homo neanderthalensis: functional comparison with Homo sapiens isoforms. J Lipid Res. 2013;54:1397–1409. doi: 10.1194/jlr.M035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Adel S., Kakularam K.R., Horn T., Reddanna P., Kuhn H., Heydeck D. Leukotriene signaling in the extinct human subspecies Homo denisovan and Homo neandertal. Arch Biochem Biophys. 2014;565C:17–24. doi: 10.1016/j.abb.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 228.Krause J., Fu Q., Good J.M., Viola B., Shunkov M.V., Derevianko A.P. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mednikova M.B. A proximal pedal phalanx of a Paleolithic hominin from Denisova Cave, Altai. Archaeol Ethnol Anthropol Eurasia. 2011;39:129–138. [Google Scholar]
- 230.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Green R.E., Malaspinas A.S., Krause J., Briggs A.W., Johnson P.L., Uhler C. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Funk C.D., Hoshiko S., Matsumoto T., Rdmark O., Samuelsson B. Characterization of the human 5-lipoxygenase gene. Proc Natl Acad Sci USA. 1989;86:2587–2591. doi: 10.1073/pnas.86.8.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hoshiko S., Rådmark O., Samuelsson B. Characterization of the human 5-lipoxygenase gene promoter. Proc Natl Acad Sci USA. 1990;87:9073–9077. doi: 10.1073/pnas.87.23.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Silverman E.S., Du J., De Sanctis G.T., Rådmark O., Samuelsson B., Drazen J.M. Egr-1 and Sp1 interact functionally with the 5-lipoxygenase promoter and its naturally occurring mutants. Am J Respir Cell Mol Biol. 1998;19:316–323. doi: 10.1165/ajrcmb.19.2.3154. [DOI] [PubMed] [Google Scholar]
- 237.In K.H., Asano K., Beier D., Grobholz J., Finn P.W., Silverman E.K. Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J Clin Invest. 1997;99:1130–1137. doi: 10.1172/JCI119241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Drazen J.M., Yandava C.N., Dubé L., Szczerback N., Hippensteel R., Pillari A. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22:168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 239.Kalayci O., Birben E., Sackesen C., Keskin O., Tahan F., Wechsler M.E. ALOX5 promoter genotype, asthma severity and LTC production by eosinophils. Allergy. 2006;61:97–103. doi: 10.1111/j.1398-9995.2006.00979.x. [DOI] [PubMed] [Google Scholar]
- 240.Dwyer J.H., Allayee H., Dwyer K.M., Fan J., Wu H., Mar R. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N Engl J Med. 2004;350:29–37. doi: 10.1056/NEJMoa025079. [DOI] [PubMed] [Google Scholar]
- 241.Todur S.P., Ashavaid T.F. Association of Sp1 tandem repeat polymorphism of ALOX5 with coronary artery disease in Indian subjects. Clin Transl Sci. 2012;5:408–411. doi: 10.1111/j.1752-8062.2011.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Stephensen C.B., Armstrong P., Newman J.W., Pedersen T.L., Legault J., Schuster G.U. ALOX5 gene variants affect eicosanoid production and response to fish oil supplementation. J Lipid Res. 2011;52:991–1003. doi: 10.1194/jlr.P012864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Goodman J.E., Bowman E.D., Chanock S.J., Alberg A.J., Harris C.C. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25:2467–2472. doi: 10.1093/carcin/bgh260. [DOI] [PubMed] [Google Scholar]
- 244.Kleinstein S.E., Heath L., Makar K.W., Poole E.M., Seufert B.L., Slattery M.L. Genetic variation in the lipoxygenase pathway and risk of colorectal neoplasia. Genes Chromosom Cancer. 2013;52:437–449. doi: 10.1002/gcc.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Alvarez V., González P., Corao A.I., Menéndez M., Lahoz C.H., Martínez C. The Sp1/Egr1-tandem repeat polymorphism in the 5-lipoxygenase gene promoter is not associated with late onset Alzheimer disease. Alzheimer Dis Assoc Disord. 2008;22:177–180. doi: 10.1097/WAD.0b013e3181572046. [DOI] [PubMed] [Google Scholar]
- 246.Bai C., Matsui E., Ohnishi H., Kimata K., Kasahara K., Kaneko H. A novel polymorphism, E254K, in the 5-lipoxygenase gene associated with bronchial asthma. Int J Mol Med. 2008;21:139–144. [PubMed] [Google Scholar]
- 247.Bai C., Yu X., Yun R., Shi T., Zhang C., Zhou J. Association of 5-lipoxygenase gene polymorphisms with bronchial asthma. Exp Ther Med. 2012;4:967–971. doi: 10.3892/etm.2012.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Foroud T., Ichikawa S., Koller D., Lai D., Curry L., Xuei X. Association studies of ALOX5 and bone mineral density in healthy adults. Osteoporos Int. 2008;19:637–643. doi: 10.1007/s00198-007-0484-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Küry S., Buecher B., Robiou-du-Pont S., Scoul C., Colman H., Le Neel T. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008;8:326. doi: 10.1186/1471-2407-8-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Crosslin D.R., Shah S.H., Nelson S.C., Haynes C.S., Connelly J.J., Gadson S. Genetic effects in the leukotriene biosynthesis pathway and association with atherosclerosis. Hum Genet. 2009;125:217–229. doi: 10.1007/s00439-008-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Horn T., Reddy Kakularam K., Anton M., Richter C., Reddanna P., Kuhn H. Functional characterization of genetic enzyme variations in human lipoxygenases. Redox Biol. 2013;1:566–577. doi: 10.1016/j.redox.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang J., John E.M., Ingles S.A. 5-Lipoxygenase and 5-lipoxygenase-activating protein gene polymorphisms, dietary linoleic acid, and risk for breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2748–2754. doi: 10.1158/1055-9965.EPI-08-0439. [DOI] [PubMed] [Google Scholar]
- 253.Ishii S., Noguchi M., Miyano M., Matsumoto T., Noma M. Mutagenesis studies on the amino acid residues involved in the iron-binding and the activity of human 5-lipoxygenase. Biochem Biophys Res Commun. 1992;184:1133–1134. doi: 10.1016/0006-291x(92)90710-3. [DOI] [PubMed] [Google Scholar]
- 254.Schwarz K., Walther M., Anton M., Gerth C., Feussner I., Kuhn H. Structural basis for lipoxygenase specificity. Conversion of the human leukocyte 5-lipoxygenase to a 15-lipoxygenating enzyme species by site-directed mutagenesis. J Biol Chem. 2001;276:773–779. doi: 10.1074/jbc.M005114200. [DOI] [PubMed] [Google Scholar]
- 255.Gilbert N.C., Rui Z., Neau D.B., Waight M.T., Bartlett S.G., Boeglin W.E. Conversion of human 5-lipoxygenase to a 15-lipoxygenase by a point mutation to mimic phosphorylation at Serine-663. FASEB J. 2012;26:3222–3229. doi: 10.1096/fj.12-205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rådmark O., Samuelsson B. Regulation of the activity of 5-lipoxygenase, a key enzyme in leukotriene biosynthesis. Biochem Biophys Res Commun. 2010;396:105–110. doi: 10.1016/j.bbrc.2010.02.173. [DOI] [PubMed] [Google Scholar]
- 257.Hammarberg T., Provost P., Persson B., Rådmark O. The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J Biol Chem. 2000;275:38787–38793. doi: 10.1074/jbc.M006136200. [DOI] [PubMed] [Google Scholar]
- 258.Kulkarni S., Das S., Funk C.D., Murray D., Cho W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem. 2002;277:13167–13174. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- 259.Esser J., Rakonjac M., Hofmann B., Fischer L., Provost P., Schneider G. Coactosin-like protein functions as a stabilizing chaperone for 5-lipoxygenase: role of tryptophan 102. Biochem J. 2010;425:265–274. doi: 10.1042/BJ20090856. [DOI] [PubMed] [Google Scholar]
- 260.Werz O., Bürkert E., Fischer L., Szellas D., Dishart D., Samuelsson B. Extracellular signal-regulated kinases phosphorylate 5-lipoxygenase and stimulate 5-lipoxygenase product formation in leukocytes. FASEB J. 2002;16:1441–1443. doi: 10.1096/fj.01-0909fje. [DOI] [PubMed] [Google Scholar]
- 261.Werz O., Bürkert E., Fischer L., Szellas D., Dishart D., Samuelsson B. 5-Lipoxygenase activation by MAPKAPK-2 and ERKs. Adv Exp Med Biol. 2003;525:129–132. doi: 10.1007/978-1-4419-9194-2_26. [DOI] [PubMed] [Google Scholar]
- 262.Luo M., Jones S.M., Flamand N., Aronoff D.M., Peters-Golden M., Brock T.G. Phosphorylation by protein kinase a inhibits nuclear import of 5-lipoxygenase. J Biol Chem. 2005;280:40609–40616. doi: 10.1074/jbc.M507045200. [DOI] [PubMed] [Google Scholar]
- 263.Flamand N., Luo M., Peters-Golden M., Brock T.G. Phosphorylation of serine 271 on 5-lipoxygenase and its role in nuclear export. J Biol Chem. 2009;284:306–313. doi: 10.1074/jbc.M805593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kelavkar U., Wang S., Montero A., Murtagh J., Shah K., Badr K. Human 15-lipoxygenase gene promoter: analysis and identification of DNA binding sites for IL-13-induced regulatory factors in monocytes. Mol Biol Rep. 1998;25:173–182. doi: 10.1023/a:1006813009006. [DOI] [PubMed] [Google Scholar]
- 265.Wittwer J., Marti-Jaun J., Hersberger M. Functional polymorphism in ALOX15 results in increased allele-specific transcription in macrophages through binding of the transcription factor SPI1. Hum Mutat. 2006;27:78–87. doi: 10.1002/humu.20273. [DOI] [PubMed] [Google Scholar]
- 266.Wittwer J., Bayer M., Mosandl A., Muntwyler J., Hersberger M. The c.-292C > T promoter polymorphism increases reticulocyte-type 15-lipoxygenase-1 activity and could be atheroprotective. Clin Chem Lab Med. 2007;45:487–492. doi: 10.1515/CCLM.2007.103. [DOI] [PubMed] [Google Scholar]
- 267.Hersberger M., Müller M., Marti-Jaun J., Heid I.M., Coassin S., Young T.F. No association of two functional polymorphisms in human ALOX15 with myocardial infarction. Atherosclerosis. 2009;205:192–196. doi: 10.1016/j.atherosclerosis.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 268.McCaskie P., Beilby J., Hung J., Chapman C.L., McQuillan B., Powell B. 15-Lipoxygenase gene variants are associated with carotid plaque but not carotid intima-media thickness. Hum Genet. 2008;123:445–453. doi: 10.1007/s00439-008-0496-6. [DOI] [PubMed] [Google Scholar]
- 269.Samanta S., Anderson K., Moran S., Hawke D., Gorenstein D., Fornage M. Characterization of a human 12/15-lipoxygenase promoter variant associated with atherosclerosis identifies vimentin as a promoter binding protein. PLoS One. 2012;7:e42417. doi: 10.1371/journal.pone.0042417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Song Y.S., Yang E.M., Kim S.H., Jin H.J., Park H.S. Effect of genetic polymorphism of ALOX15 on aspirin-exacerbated respiratory disease. Int Arch Allergy Immunol. 2012;159:157–161. doi: 10.1159/000335681. [DOI] [PubMed] [Google Scholar]
- 271.Sigal E., Craik C.S., Highland E., Grunberger D., Costello L.L., Dixon R.A. Molecular cloning and primary structure of human 15-lipoxygenase. Biochem Biophys Res Commun. 1988;157:457–464. doi: 10.1016/s0006-291x(88)80271-7. [DOI] [PubMed] [Google Scholar]
- 272.Schurmann K., Anton M., Ivanov I., Richter C., Kuhn H., Walther M. Molecular basis for the reduced catalytic activity of the naturally occurring T560M mutant of human 12/15-lipoxygenase that has been implicated in coronary artery disease. J Biol Chem. 2011;286:23920–23927. doi: 10.1074/jbc.M110.211821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Horn T., Ivanov I., Di Venere A., Kakularam K.R., Reddanna P., Conrad M.L. Molecular basis for the catalytic inactivity of a naturally occurring near-null variant of human ALOX15. Biochim Biophys Acta. 2013;1831:1702–1713. doi: 10.1016/j.bbalip.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 274.Assimes T.L., Knowles J.W., Priest J.R., Basu A., Borchert A., Volcik K.A. A near null variant of 12/15-LOX encoded by a novel SNP in ALOX15 and the risk of coronary artery disease. Atherosclerosis. 2008;198:136–144. doi: 10.1016/j.atherosclerosis.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kuban R.J., Wiesner R., Rathman J., Veldink G., Nolting H., Solé V.A. The iron ligand sphere geometry of mammalian 15-lipoxygenases. Biochem J. 1998;332(Pt 1):237–242. doi: 10.1042/bj3320237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kühn H., Kuban R., Walther M., Veldink G.A. X-ray absorption studies into the iron ligand sphere of plant and animal lipoxygenases. Adv Exp Med Biol. 1999;469:99–104. doi: 10.1007/978-1-4615-4793-8_15. [DOI] [PubMed] [Google Scholar]
- 277.Sloane D.L., Leung R., Barnett J., Craik C.S., Sigal E. Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: the side-chain geometry of amino acids 417 and 418 determine positional specificity. Protein Eng. 1995;8:275–282. doi: 10.1093/protein/8.3.275. [DOI] [PubMed] [Google Scholar]
- 278.Kuhn H., Belkner J., Wiesner R., Brash A.R. Oxygenation of biological membranes by the pure reticulocyte lipoxygenase. J Biol Chem. 1990;265:18351–18361. [PubMed] [Google Scholar]
- 279.Walther M., Wiesner R., Kuhn H. Investigations into calcium-dependent membrane association of 15-lipoxygenase-1. Mechanistic roles of surface-exposed hydrophobic amino acids and calcium. J Biol Chem. 2004;279:3717–3725. doi: 10.1074/jbc.M309564200. [DOI] [PubMed] [Google Scholar]
- 280.Gan Q.F., Browner M.F., Sloane D.L., Sigal E. Defining the arachidonic acid binding site of human 15-lipoxygenase. Molecular modeling and mutagenesis. J Biol Chem. 1996;271:25412–25418. doi: 10.1074/jbc.271.41.25412. [DOI] [PubMed] [Google Scholar]
- 281.Toledo L., Masgrau L., Maréchal J.D., Lluch J.M., González-Lafont A. Insights into the mechanism of binding of arachidonic acid to mammalian 15-lipoxygenases. J Phys Chem B. 2010;114:7037–7046. doi: 10.1021/jp912120n. [DOI] [PubMed] [Google Scholar]
- 282.Funk C.D., Funk L.B., FitzGerald G.A., Samuelsson B. Characterization of human 12-lipoxygenase genes. Proc Natl Acad Sci USA. 1992;89:3962–3966. doi: 10.1073/pnas.89.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Liu Y.W., Arakawa T., Yamamoto S., Chang W.C. Transcriptional activation of human 12-lipoxygenase gene promoter is mediated through Sp1 consensus sites in A431 cells. Biochem J. 1997;324(Pt 1):133–140. doi: 10.1042/bj3240133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Harsløf T., Husted L.B., Nyegaard M., Carstens M., Stenkjær L., Brixen K. Polymorphisms in the ALOX12 gene and osteoporosis. Osteoporos Int. 2011;22:2249–2259. doi: 10.1007/s00198-010-1472-2. [DOI] [PubMed] [Google Scholar]
- 285.Breunis W.B., Tarazona-Santos E., Chen R., Kiley M., Rosenberg S.A., Chanock S.J. Influence of cytotoxic T lymphocyte-associated antigen 4 (CTLA4) common polymorphisms on outcome in treatment of melanoma patients with CTLA-4 blockade. J Immunother. 2008;31:586–590. doi: 10.1097/CJI.0b013e31817fd8f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Burdon K.P., Rudock M.E., Lehtinen A.B., Langefeld C.D., Bowden D.W., Register T.C. Human lipoxygenase pathway gene variation and association with markers of subclinical atherosclerosis in the diabetes heart study. Mediators Inflamm. 2010;2010:170153. doi: 10.1155/2010/170153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Liu P., Lu Y., Recker R.R., Deng H.W., Dvornyk V. ALOX12 gene is associated with the onset of natural menopause in white women. Menopause. 2010;17:152–156. doi: 10.1097/gme.0b013e3181b63c68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Liu Y., Freedman B.I., Burdon K.P., Langefeld C.D., Howard T., Herrington D. Association of arachidonate 12-lipoxygenase genotype variation and glycemic control with albuminuria in type 2 diabetes. Am J Kidney Dis. 2008;52:242–250. doi: 10.1053/j.ajkd.2007.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kim T., Kim H.-J., Park J., Kim J., Chung J.-H. Association between polymorphisms of arachidonate 12-lipoxygenase (ALOX12) and schizophrenia in a Korean population. Behav Brain Funct. 2010;6:44. doi: 10.1186/1744-9081-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aleem A.M., Wells L., Jankun J., Walther M., Kühn H., Reinartz J. Human platelet 12-lipoxygenase: naturally occurring Q261/R261 variants and N544L mutant show altered activity but unaffected substrate binding and membrane association behavior. Int J Mol Med. 2009;24:759–764. doi: 10.3892/ijmm_00000289. [DOI] [PubMed] [Google Scholar]
- 291.Tang S., Bhatia B., Maldonado C.J., Yang P., Newman R.A., Liu J. Evidence that arachidonate 15-lipoxygenase 2 is a negative cell cycle regulator in normal prostate epithelial cells. J Biol Chem. 2002;277:16189–16201. doi: 10.1074/jbc.M111936200. [DOI] [PubMed] [Google Scholar]
- 292.Tang S., Bhatia B., Zhou J., Maldonado C.J., Chandra D., Kim E. Evidence that Sp1 positively and Sp3 negatively regulate and androgen does not directly regulate functional tumor suppressor 15-lipoxygenase 2 (15-LOX2) gene expression in normal human prostate epithelial cells. Oncogene. 2004;23:6942–6953. doi: 10.1038/sj.onc.1207913. [DOI] [PubMed] [Google Scholar]
- 293.Wuest S.J., Horn T., Marti-Jaun J., Kühn H., Hersberger M. Association of polymorphisms in the ALOX15B gene with coronary artery disease. Clin Biochem. 2014;47:349–355. doi: 10.1016/j.clinbiochem.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 294.Wuest S.J., Crucet M., Gemperle C., Loretz C., Hersberger M. Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis. 2012;225:121–127. doi: 10.1016/j.atherosclerosis.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 295.Magnusson L.U., Lundqvist A., Karlsson M.N., Skålén K., Levin M., Wiklund O. Arachidonate 15-lipoxygenase type B knockdown leads to reduced lipid accumulation and inflammation in atherosclerosis. PLoS One. 2012;7:e43142. doi: 10.1371/journal.pone.0043142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Boeglin W.E., Kim R.B., Brash A.R. A 12R-lipoxygenase in human skin: mechanistic evidence, molecular cloning, and expression. Proc Natl Acad Sci USA. 1998;95:6744–6749. doi: 10.1073/pnas.95.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Krieg P., Siebert M., Kinzig A., Bettenhausen R., Marks F., Fürstenberger G. Murine 12(R)-lipoxygenase: functional expression, genomic structure and chromosomal localization. FEBS Lett. 1999;446:142–148. doi: 10.1016/s0014-5793(99)00196-9. [DOI] [PubMed] [Google Scholar]
- 298.Moran J.L., Qiu H., Turbe-Doan A., Yun Y., Boeglin W.E., Brash A.R. A mouse mutation in the 12R-lipoxygenase, Alox12b, disrupts formation of the epidermal permeability barrier. J Invest Dermatol. 2007;127:1893–1897. doi: 10.1038/sj.jid.5700825. [DOI] [PubMed] [Google Scholar]
- 299.Jobard F., Lefèvre C., Karaduman A., Blanchet-Bardon C., Emre S., Weissenbach J. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet. 2002;11:107–113. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 300.Yu Z., Schneider C., Boeglin W.E., Brash A.R. Mutations associated with a congenital form of ichthyosis (NCIE) inactivate the epidermal lipoxygenases 12R-LOX and eLOX3. Biochim Biophys Acta. 2005;1686:238–247. doi: 10.1016/j.bbalip.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 301.Eckl K.M., de Juanes S., Kurtenbach J., Nätebus M., Lugassy J., Oji V. Molecular analysis of 250 patients with autosomal recessive congenital ichthyosis: evidence for mutation hotspots in ALOXE3 and allelic heterogeneity in ALOX12B. J Invest Dermatol. 2009;129:1421–1428. doi: 10.1038/jid.2008.409. [DOI] [PubMed] [Google Scholar]
- 302.Lesueur F., Bouadjar B., Lefèvre C., Jobard F., Audebert S., Lakhdar H. Novel mutations in ALOX12B in patients with autosomal recessive congenital ichthyosis and evidence for genetic heterogeneity on chromosome 17p13. J Invest Dermatol. 2007;127:829–834. doi: 10.1038/sj.jid.5700640. [DOI] [PubMed] [Google Scholar]
- 303.Zheng Y., Yin H., Boeglin W.E., Elias P.M., Crumrine D., Beier D.R. Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: a proposed role in releasing omega-hydroxyceramide for construction of the corneocyte lipid envelope. J Biol Chem. 2011;286:24046–24056. doi: 10.1074/jbc.M111.251496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Krieg P., Fürstenberger G. The role of lipoxygenases in epidermis. Biochim Biophys Acta. 2014;1841:390–400. doi: 10.1016/j.bbalip.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 305.Krieg P., Marks F., Fürstenberger G. A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: cloning, physical mapping, and expression. Genomics. 2001;73:323–330. doi: 10.1006/geno.2001.6519. [DOI] [PubMed] [Google Scholar]
- 306.Yu Z., Schneider C., Boeglin W.E., Marnett L.J., Brash A.R. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci USA. 2003;100:9162–9167. doi: 10.1073/pnas.1633612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yu Z., Schneider C., Boeglin W.E., Brash A.R. Human and mouse eLOX3 have distinct substrate specificities: implications for their linkage with lipoxygenases in skin. Arch Biochem Biophys. 2006;455:188–196. doi: 10.1016/j.abb.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zheng Y., Brash A.R. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J Biol Chem. 2010;285:39866–39875. doi: 10.1074/jbc.M110.155374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Zheng Y., Brash A.R. On the role of molecular oxygen in lipoxygenase activation: comparison and contrast of epidermal lipoxygenase-3 with soybean lipoxygenase-1. J Biol Chem. 2010;285:39876–39887. doi: 10.1074/jbc.M110.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wei X., Walia V., Lin J.C., Teer J.K., Prickett T.D., Gartner J. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Minami M., Ohno S., Kawasaki H., Rådmark O., Samuelsson B., Jörnvall H. Molecular cloning of a cDNA coding for human leukotriene A4 hydrolase. Complete primary structure of an enzyme involved in eicosanoid synthesis. J Biol Chem. 1987;262:13873–13876. [PubMed] [Google Scholar]
- 312.Funk C.D., Rådmark O., Fu J.Y., Matsumoto T., Jörnvall H., Shimizu T. Molecular cloning and amino acid sequence of leukotriene A4 hydrolase. Proc Natl Acad Sci USA. 1987;84:6677–6681. doi: 10.1073/pnas.84.19.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mancini J.A., Evans J.F. Cloning and characterization of the human leukotriene A4 hydrolase gene. Eur J Biochem. 1995;231:65–71. doi: 10.1111/j.1432-1033.1995.tb20671.x. [DOI] [PubMed] [Google Scholar]
- 314.Lima J.J., Zhang S., Grant A., Shao L., Tantisira K.G., Allayee H. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hartiala J., Li D., Conti D.V., Vikman S., Patel Y., Tang W.H. Genetic contribution of the leukotriene pathway to coronary artery disease. Hum Genet. 2011;129:617–627. doi: 10.1007/s00439-011-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Kotani H., Kishi R., Mouri A., Sashio T., Shindo J., Shiraki A. Influence of leukotriene pathway polymorphisms on clinical responses to montelukast in Japanese patients with asthma. J Clin Pharm Ther. 2012;37:112–116. doi: 10.1111/j.1365-2710.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- 317.Zhao J., Quyyumi A.A., Patel R., Zafari A.M., Veledar E., Onufrak S. Sex-specific association of depression and a haplotype in leukotriene A4 hydrolase gene. Psychosom Med. 2009;71:691–696. doi: 10.1097/PSY.0b013e3181b05c57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhao J., Goldberg J., Vaccarino V. Leukotriene A4 hydrolase haplotype, diet and atherosclerosis: a twin study. Atherosclerosis. 2013;226:238–244. doi: 10.1016/j.atherosclerosis.2012.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Helgadottir A., Manolescu A., Helgason A., Thorleifsson G., Thorsteinsdottir U., Gudbjartsson D.F. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38:68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- 320.Evans J.F., Nathaniel D.J., Zamboni R.J., Ford-Hutchinson A.W. Leukotriene A3. A poor substrate but a potent inhibitor of rat and human neutrophil leukotriene A4 hydrolase. J Biol Chem. 1985;260:10966–10970. [PubMed] [Google Scholar]
- 321.Ohishi N., Izumi T., Minami M., Kitamura S., Seyama Y., Ohkawa S. Leukotriene A4 hydrolase in the human lung. Inactivation of the enzyme with leukotriene A4 isomers. J Biol Chem. 1987;262:10200–10205. [PubMed] [Google Scholar]
- 322.Vallee B.L., Auld D.S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 323.Haeggström J.Z., Wetterholm A., Vallee B.L., Samuelsson B. Leukotriene A4 hydrolase: an epoxide hydrolase with peptidase activity. Biochem Biophys Res Commun. 1990;173:431–437. doi: 10.1016/s0006-291x(05)81076-9. [DOI] [PubMed] [Google Scholar]
- 324.Haeggström J.Z., Wetterholm A., Shapiro R., Vallee B.L., Samuelsson B. Leukotriene A4 hydrolase: a zinc metalloenzyme. Biochem Biophys Res Commun. 1990;172:965–970. doi: 10.1016/0006-291x(90)91540-9. [DOI] [PubMed] [Google Scholar]
- 325.Minami M., Ohishi N., Mutoh H., Izumi T., Bito H., Wada H. Leukotriene A4 hydrolase is a zinc-containing aminopeptidase. Biochem Biophys Res Commun. 1990;173:620–626. doi: 10.1016/s0006-291x(05)80080-4. [DOI] [PubMed] [Google Scholar]
- 326.Medina J.F., Wetterholm A., Rådmark O., Shapiro R., Haeggström J.Z., Vallee B.L. Leukotriene A4 hydrolase: determination of the three zinc-binding ligands by site-directed mutagenesis and zinc analysis. Proc Natl Acad Sci USA. 1991;88:7620–7624. doi: 10.1073/pnas.88.17.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Mueller M.J., Wetterholm A., Blomster M., Jörnvall H., Samuelsson B., Haeggström J.Z. Leukotriene A4 hydrolase: mapping of a henicosapeptide involved in mechanism-based inactivation. Proc Natl Acad Sci USA. 1995;92:8383–8387. doi: 10.1073/pnas.92.18.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Blomster M., Wetterholm A., Mueller M.J., Haeggström J.Z. Evidence for a catalytic role of tyrosine 383 in the peptidase reaction of leukotriene A4 hydrolase. Eur J Biochem. 1995;231:528–534. doi: 10.1111/j.1432-1033.1995.0528d.x. [DOI] [PubMed] [Google Scholar]
- 329.Maycock A.L., Anderson M.S., DeSousa D.M., Kuehl F.A. Leukotriene A4: preparation and enzymatic conversion in a cell-free system to leukotriene B4. J Biol Chem. 1982;257:13911–13914. [PubMed] [Google Scholar]
- 330.Rudberg P.C., Tholander F., Andberg M., Thunnissen M.M., Haeggström J.Z. Leukotriene A4 hydrolase: identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates. J Biol Chem. 2004;279:27376–27382. doi: 10.1074/jbc.M401031200. [DOI] [PubMed] [Google Scholar]
- 331.Tholander F., Muroya A., Roques B.P., Fournié-Zaluski M.C., Thunnissen M.M., Haeggström J.Z. Structure-based dissection of the active site chemistry of leukotriene A4 hydrolase: implications for M1 aminopeptidases and inhibitor design. Chem Biol. 2008;15:920–929. doi: 10.1016/j.chembiol.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 332.Lam B.K., Penrose J.F., Freeman G.J., Austen K.F. Expression cloning of a cDNA for human leukotriene C4 synthase, an integral membrane protein conjugating reduced glutathione to leukotriene A4. Proc Natl Acad Sci USA. 1994;91:7663–7667. doi: 10.1073/pnas.91.16.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Penrose J.F., Spector J., Baldasaro M., Xu K., Boyce J., Arm J.P. Molecular cloning of the gene for human leukotriene C4 synthase. Organization, nucleotide sequence, and chromosomal localization to 5q35. J Biol Chem. 1996;271:11356–11361. doi: 10.1074/jbc.271.19.11356. [DOI] [PubMed] [Google Scholar]
- 334.Zhao J.L., Austen K.F., Lam B.K. Cell-specific transcription of leukotriene C(4) synthase involves a Kruppel-like transcription factor and Sp1. J Biol Chem. 2000;275:8903–8910. doi: 10.1074/jbc.275.12.8903. [DOI] [PubMed] [Google Scholar]
- 335.Wang G., Zhang J., Sun H., Cao W., Wang Y., Xiao H. Genetic variation in members of the leukotrienes biosynthesis pathway confers risk of ischemic stroke in Eastern Han Chinese. Prostaglandins Leukot Essent Fatty Acids. 2012;87:169–175. doi: 10.1016/j.plefa.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 336.Ago H., Kanaoka Y., Irikura D., Lam B.K., Shimamura T., Austen K.F. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature. 2007;448:609–612. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 337.Rinaldo-Matthis A., Wetterholm A., Martinez Molina D., Holm J., Niegowski D., Ohlson E. Arginine 104 is a key catalytic residue in leukotriene C4 synthase. J Biol Chem. 2010;285:40771–40776. doi: 10.1074/jbc.M110.105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Schröder O., Sjöström M., Qiu H., Stein J., Jakobsson P.J., Haeggström J.Z. Molecular and catalytic properties of three rat leukotriene C(4) synthase homologs. Biochem Biophys Res Commun. 2003;312:271–276. doi: 10.1016/j.bbrc.2003.10.115. [DOI] [PubMed] [Google Scholar]
- 339.Lynch K.R., O’Neill G.P., Liu Q., Im D.S., Sawyer N., Metters K.M. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature. 1999;399:789–793. doi: 10.1038/21658. [DOI] [PubMed] [Google Scholar]
- 340.Woszczek G., Pawliczak R., Qi H.Y., Nagineni S., Alsaaty S., Logun C. Functional characterization of human cysteinyl leukotriene 1 receptor gene structure. J Immunol. 2005;175:5152–5159. doi: 10.4049/jimmunol.175.8.5152. [DOI] [PubMed] [Google Scholar]
- 341.Sokolowska M., Wodz-Naskiewicz K., Cieslak M., Seta K., Bednarek A.K., Pawliczak R. Variable expression of cysteinyl leukotriene type I receptor splice variants in asthmatic females with different promoter haplotypes. BMC Immunol. 2009;10:63. doi: 10.1186/1471-2172-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Duroudier N.P., Sayers I., Castagna C.C., Fenech A.G., Halapi E., Swan C. Functional polymorphism and differential regulation of CYSLTR1 transcription in human airway smooth muscle and monocytes. Cell Biochem Biophys. 2007;47:119–130. doi: 10.1385/cbb:47:1:119. [DOI] [PubMed] [Google Scholar]
- 343.Kim S.H., Oh J.M., Kim Y.S., Palmer L.J., Suh C.H., Nahm D.H. Cysteinyl leukotriene receptor 1 promoter polymorphism is associated with aspirin-intolerant asthma in males. Clin Exp Allergy. 2006;36:433–439. doi: 10.1111/j.1365-2222.2006.02457.x. [DOI] [PubMed] [Google Scholar]
- 344.Hao L., Sayers I., Cakebread J.A., Barton S.J., Beghé B., Holgate S.T. The cysteinyl-leukotriene type 1 receptor polymorphism 927T/C is associated with atopy severity but not with asthma. Clin Exp Allergy. 2006;36:735–741. doi: 10.1111/j.1365-2222.2006.02511.x. [DOI] [PubMed] [Google Scholar]
- 345.Duroudier N.P., Strachan D.P., Blakey J.D., Hall I.P. Association of the cysteinyl leukotriene receptor 1 gene with atopy in the British 1958 birth cohort. J Allergy Clin Immunol. 2009;124:566–572. doi: 10.1016/j.jaci.2009.06.004. [72.e1-3] [DOI] [PubMed] [Google Scholar]
- 346.Sarau H.M., Ames R.S., Chambers J., Ellis C., Elshourbagy N., Foley J.J. Identification, molecular cloning, expression, and characterization of a cysteinyl leukotriene receptor. Mol Pharmacol. 1999;56:657–663. doi: 10.1124/mol.56.3.657. [DOI] [PubMed] [Google Scholar]
- 347.Thompson M.D., Capra V., Takasaki J., Maresca G., Rovati G.E., Slutsky A.S. A functional G300S variant of the cysteinyl leukotriene 1 receptor is associated with atopy in a Tristan da Cunha isolate. Pharmacogenet Genomics. 2007;17 doi: 10.1097/FPC.0b013e328012d0bf. [DOI] [PubMed] [Google Scholar]
- 348.Soodyall H., Nebel A., Morar B., Jenkins T. Genealogy and genes: tracing the founding fathers of Tristan da Cunha. Eur J Hum Genet. 2003;11:705–709. doi: 10.1038/sj.ejhg.5201022. [DOI] [PubMed] [Google Scholar]
- 349.Shin J.A., Chang H.S., Park S.M., Jang A.S., Park S.W., Park J.S. Genetic effect of CysLTR2 polymorphisms on its mRNA synthesis and stabilization. BMC Med Genet. 2009;10:106. doi: 10.1186/1471-2350-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Park J.S., Chang H.S., Park C.S., Lee J.H., Lee Y.M., Choi J.H. Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet Genomics. 2005;15:483–492. doi: 10.1097/01.fpc.0000166456.84905.a0. [DOI] [PubMed] [Google Scholar]
- 351.Heise C.E., O’Dowd B.F., Figueroa D.J., Sawyer N., Nguyen T., Im D.S. Characterization of the human cysteinyl leukotriene 2 receptor. J Biol Chem. 2000;275:30531–30536. doi: 10.1074/jbc.M003490200. [DOI] [PubMed] [Google Scholar]
- 352.Thompson M.D., Gravesande K.S., Galczenski H., Siminovitch K.A., Zamel N., Slutsky A. A cysteinyl leukotriene 2 receptor variant is associated with atopy in the population of Tristan da Cunha. Pharmacogenetics. 2003;13:641–649. doi: 10.1097/00008571-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 353.Pillai S.G., Cousens D.J., Barnes A.A., Buckley P.T., Chiano M.N., Hosking L.K. A coding polymorphism in the CYSLT2 receptor with reduced affinity to LTD4 is associated with asthma. Pharmacogenetics. 2004;14:627–633. doi: 10.1097/00008571-200409000-00007. [DOI] [PubMed] [Google Scholar]
- 354.Owman C., Nilsson C., Lolait S.J. Cloning of cDNA encoding a putative chemoattractant receptor. Genomics. 1996;37:187–194. doi: 10.1006/geno.1996.0541. [DOI] [PubMed] [Google Scholar]
- 355.Kato K., Yokomizo T., Izumi T., Shimizu T. Cell-specific transcriptional regulation of human leukotriene B(4) receptor gene. J Exp Med. 2000;192:413–420. doi: 10.1084/jem.192.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Yokomizo T., Izumi T., Chang K., Takuwa Y., Shimizu T. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature. 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- 357.Gaudreau R., Le Gouill C., Venne M.H., Stankova J., Rola-Pleszczynski M. Threonine 308 within a putative casein kinase 2 site of the cytoplasmic tail of leukotriene B(4) receptor (BLT1) is crucial for ligand-induced, G-protein-coupled receptor-specific kinase 6-mediated desensitization. J Biol Chem. 2002;277:31567–31576. doi: 10.1074/jbc.M202723200. [DOI] [PubMed] [Google Scholar]
- 358.Kuniyeda K., Okuno T., Terawaki K., Miyano M., Yokomizo T., Shimizu T. Identification of the intracellular region of the leukotriene B4 receptor type 1 that is specifically involved in Gi activation. J Biol Chem. 2007;282:3998–4006. doi: 10.1074/jbc.M610540200. [DOI] [PubMed] [Google Scholar]
- 359.Okuno T., Ago H., Terawaki K., Miyano M., Shimizu T., Yokomizo T. Helix 8 of the leukotriene B4 receptor is required for the conformational change to the low affinity state after G-protein activation. J Biol Chem. 2003;278:41500–41509. doi: 10.1074/jbc.M307335200. [DOI] [PubMed] [Google Scholar]
- 360.Okuno T., Yokomizo T., Hori T., Miyano M., Shimizu T. Leukotriene B4 receptor and the function of its helix 8. J Biol Chem. 2005;280:32049–32052. doi: 10.1074/jbc.R500007200. [DOI] [PubMed] [Google Scholar]
- 361.Gaudreau R., Beaulieu M.E., Chen Z., Le Gouill C., Lavigne P., Stanková J. Structural determinants regulating expression of the high affinity leukotriene B4 receptor: involvement of dileucine motifs and alpha-helix VIII. J Biol Chem. 2004;279:10338–10345. doi: 10.1074/jbc.M309207200. [DOI] [PubMed] [Google Scholar]
- 362.Nilsson N.E., Tryselius Y., Owman C. Genomic organization of the leukotriene B(4) receptor locus of human chromosome 14. Biochem Biophys Res Commun. 2000;274:383–388. doi: 10.1006/bbrc.2000.3153. [DOI] [PubMed] [Google Scholar]
- 363.Tryselius Y., Nilsson N.E., Kotarsky K., Olde B., Owman C. Cloning and characterization of cDNA encoding a novel human leukotriene B(4) receptor. Biochem Biophys Res Commun. 2000;274:377–382. doi: 10.1006/bbrc.2000.3152. [DOI] [PubMed] [Google Scholar]
- 364.Yokomizo T., Kato K., Terawaki K., Izumi T., Shimizu T. A second leukotriene B(4) receptor, BLT2. A new therapeutic target in inflammation and immunological disorders. J Exp Med. 2000;192:421–432. doi: 10.1084/jem.192.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kamohara M., Takasaki J., Matsumoto M., Saito T., Ohishi T., Ishii H. Molecular cloning and characterization of another leukotriene B4 receptor. J Biol Chem. 2000;275:27000–27004. doi: 10.1074/jbc.C000382200. [DOI] [PubMed] [Google Scholar]
- 366.Raport C.J., Schweickart V.L., Chantry D., Eddy R.L., Jr., Shows T.B., Godiska R. New members of the chemokine receptor gene family. J Leukoc Biol. 1996;59:18–23. doi: 10.1002/jlb.59.1.18. [DOI] [PubMed] [Google Scholar]
- 367.Blasius R., Weber R.G., Lichter P., Ogilvie A. A novel orphan G protein-coupled receptor primarily expressed in the brain is localized on human chromosomal band 2q21. J Neurochem. 1998;70:1357–1365. doi: 10.1046/j.1471-4159.1998.70041357.x. [DOI] [PubMed] [Google Scholar]
- 368.Ciana P., Fumagalli M., Trincavelli M.L., Verderio C., Rosa P., Lecca D. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006;25:4615–4627. doi: 10.1038/sj.emboj.7601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Maekawa A., Balestrieri B., Austen K.F., Kanaoka Y. GPR17 is a negative regulator of the cysteinyl leukotriene 1 receptor response to leukotriene D4. Proc Natl Acad Sci USA. 2009;106:11685–11690. doi: 10.1073/pnas.0905364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Fontana P., Gaussem P., Aiach M., Fiessinger J.N., Emmerich J., Reny J.L. P2Y12 H2 haplotype is associated with peripheral arterial disease: a case-control study. Circulation. 2003;108:2971–2973. doi: 10.1161/01.CIR.0000106904.80795.35. [DOI] [PubMed] [Google Scholar]
- 371.Hollopeter G., Jantzen H.M., Vincent D., Li G., England L., Ramakrishnan V. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. 2001;409:202–207. doi: 10.1038/35051599. [DOI] [PubMed] [Google Scholar]
- 372.Cattaneo M., Zighetti M.L., Lombardi R., Martinez C., Lecchi A., Conley P.B. Molecular bases of defective signal transduction in the platelet P2Y12 receptor of a patient with congenital bleeding. Proc Natl Acad Sci USA. 2003;100:1978–1983. doi: 10.1073/pnas.0437879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Lee D.K., Nguyen T., Lynch K.R., Cheng R., Vanti W.B., Arkhitko O. Discovery and mapping of ten novel G protein-coupled receptor genes. Gene. 2001;275:83–91. doi: 10.1016/s0378-1119(01)00651-5. [DOI] [PubMed] [Google Scholar]
- 374.Wittenberger T., Hellebrand S., Munck A., Kreienkamp H.J., Schaller H.C., Hampe W. GPR99, a new G protein-coupled receptor with homology to a new subgroup of nucleotide receptors. BMC Genomics. 2002;3:17. doi: 10.1186/1471-2164-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.He W., Miao F.J., Lin D.C., Schwandner R.T., Wang Z., Gao J. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–193. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 376.Kanaoka Y., Maekawa A., Austen K.F. Identification of GPR99 protein as a potential third cysteinyl leukotriene receptor with a preference for leukotriene E4 ligand. J Biol Chem. 2013;288:10967–10972. doi: 10.1074/jbc.C113.453704. [DOI] [PMC free article] [PubMed] [Google Scholar]