Abstract
Continuous efforts have been made to develop a prophylactic vaccine against severe acute respiratory syndrome coronavirus (SARS-CoV). In this study, two recombinant baculoviruses, vAc-N and vAc-S, were constructed, which contained the mammalian-cell activate promoter element, human elongation factor 1α-subunit (EF-1α), the human cytomegalovirus (CMV) immediate-early promoter, and the nucleocapsid (N) or spike (S) gene of bat SARS-like CoV (SL-CoV) under the control of the CMV promoter. Mice were subcutaneously and intraperitoneally injected with recombinant baculovirus, and both humoral and cellular immune responses were induced in the vaccinated groups. The secretion level of IFN-γ was much higher than that of IL-4 in vAc-N or vAc-S immunized groups, suggesting a strong Th1 bias towards cellular immune responses. Additionally, a marked increase of CD4 T cell immune responses and high levels of anti-SARS-CoV humoral responses were also detected in the vAc-N or vAc-S immunized groups. In contrast, there were significantly weaker cellular immune responses, as well as less antibody production than in the control groups. Our data demonstrates that the recombinant baculovirus can serve as an effective vaccine strategy. In addition, because effective SARS vaccines should act to not only prevent the reemergence of SARS-CoV, but also to provide cross-protection against SL-CoV, findings in this study may have implications for developing such cross-protective vaccines.
Abbreviations: N, nucleocapsid; S, spike; SARS-CoV, severe acute respiratory syndrome coronavirus; SL-CoV, SARS-like CoV
Keywords: Cellular immune response, Humoral immune response, Recombinant baculovirus, SARS-like coronavirus
1. Introduction
Baculovirus (autographa californica multiple nucleopolyhedrovirus, AcMNPV) expression systems have been widely used for producing recombinant proteins in insect cells (Hou et al., 2003, Li et al., 2003, Luckow and Summers, 1988, Miller, 1988, Ren et al., 2001), due to the high levels of expression and proper post-translational modification (Jones and Morikawa, 1996, Nakhai et al., 1991, Sridhar et al., 1994). In addition, baculoviruses can be efficiently taken up by mammalian cells without viral replication (Carbonell et al., 1985, Tjia et al., 1983, Volkman and Goldsmith, 1983). Modified AcMNPVs can express exogenous genes in mammalian cells (Boyce and Bucher, 1996, Delaney and Isom, 1998, Duisit et al., 1999, Hofmann et al., 1995, Shoji et al., 1997) when they contain promoters that are active in mammalian cells, such as Rous sarcoma virus (RSV) promoter and cytomegalovirus immediate-early (CMV-IE) promoter. For instance, AcMNPV-containing CMV-IE promoter can infect human hepatocytes and result in high-level expression of luciferase (Hofmann et al., 1995). Since it is safe, efficient, and gene delivery-specific, the use of baculoviruses may represent a novel vaccine strategy.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is the causative agent of SARS (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003, Poutanen et al., 2003). To prevent another SARS epidemic, continuous efforts have been made towards the development of a prophylactic vaccine. So far, vaccine studies have mainly focused on the two structural proteins, spike (S) and nucleocapsid (N), of SARS-CoV (Anton et al., 1996, Jackwood and Hilt, 1995). There is little knowledge regarding the immunogenicity of the N and S proteins of SARS-like coronavirus (SL-CoV) that is isolated from horseshoe bats, which are thought to be the possible animal reservoirs of SARS-CoV (Lau et al., 2005, Li et al., 2005, Ren et al., 2006). Since effective SARS vaccines should not only prevent the reemergence of SARS-CoV, but should also provide cross-protection against SL-CoV, we report the immunogenicity of N and S proteins of SL-CoV in mice using a modified baculovirus expression system.
2. Experimental
2.1. Materials and methods
2.1.1. Construction of recombinant baculoviruses
The EF-1α promoter sequence was amplified by PCR from vector pBudCE4.1 (Invitrogen, Carlsbad, CA) and the CMV promoter sequence was amplified from pcDNA3.1(+) (Invitrogen, Carlsbad, CA). The gfp gene was cloned into pFastBac DUAL vector under the control of the p10 promoter, designated pFB-GFP. The coding sequences of the N and S proteins of bat SL-CoV Rp3 strain (GenBank accession no. DQ071615) were amplified by RT-PCR and cloned into pFB-GFP under the control of the Polh promoter, respectively. The baculovirus promoters, p10 and Polh, were subsequently replaced by robust mammalian promoters, EF-1α and CMV, respectively. Recombinant viruses (vAc-N and vAc-S) were generated by transposition and transfection using the Bac-to-Bac system (Life Technologies). Virus was further amplified by propagation in Sf9 (Spodoptera frugiperda) cells, cultured in Grace's supplemented insect medium containing 10% (v/v) fetal bovine serum (HyClone). Virus titers were determined by plaque assay on Sf9 cells.
2.1.2. Transduction of BHK cells
Baby hamster kidney (BHK) cells were seeded in 35 mm plates for 12 h before transduction. Culture medium was replaced with virus inoculum and incubated at 37 °C for 2 h. After removal of virus, fresh medium was added and cultures were incubated at 37 °C for another 24 h before examining GFP expression under a fluorescence microscope (OLYMPUS IX51, Japan).
2.1.3. RT-PCR analysis
Total cellular RNAs from BHK cells, transduced with recombinant baculovirus, vAc-N or vAc-S, were isolated using TRIZOL® reagent (Invitrogen), and were further digested with RQ1 RNasefree DNase I (Promega) to remove residual DNA. cDNAs were synthesized with the MLV Reverse Transcriptase System, using random primers, according to the manufacture's instructions (Invitrogen). Subsequent PCR was performed, using primers specific for the S (nt 21,918–22,208) or N gene (nt 28,433–28,791) (GenBank accession no. AY278741). PCR primers were kindly provided by Dr. GuanYi, Hong Kong University. RT-PCR products were analyzed by 0.8% agarose gel. DNA maker DL2000 (D501A) was purchased form Takara.
2.1.4. Western blot analysis
After 24 h of transduction, BHK cells were harvested by centrifugation and raw proteins were extracted. These proteins were separated on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. Protein marker (SM0671) was purchased from Fermentas. The blots were blocked with 5% non-fat dry milk powder in TTBS (20 mM Tris–HCl, pH 8.0, 0.8% NaCl, 0.1% Tween-20) overnight at 4 °C, followed by 1 h incubation at room temperature with rabbit-derived antibody against SARS-CoV (kindly provided by Prof. L.F. Wang at the Australian Animal Health Laboratory). Following wash steps, the membrane was incubated with AP-conjugated anti-rabbit IgG (1:5000, Sigma) for 1 h and then developed in AP substrate for 5 min in the dark.
2.1.5. Immunization of mice
BALB/c mice (n = 5 per group) were subcutaneously and intraperitoneally immunized every 2 weeks with recombinant baculovirus, vAc-N or vAc-S, both at 7.5 × 106 pfu per dose. Mice were immunized four times in total for each condition. Ac-GFP (supernatants of Sf9 cells culture transfected with modified pFastBac DUAL, with gfp under the control of the Ef1α promoter) and PBS immunized groups were used as negative controls.
2.1.6. Enzyme-linked immunosorbent assay (ELISA)
Sera from immunized mice of different groups were collected before each immunization. The antibody titers for SARS-CoV were measured by enzyme-linked immunosorbent assay (ELISA). Briefly, 96-well microtiter plates (Costar) were coated with 200 ng per well inactivated SARS-CoV full antigens and incubated at 4 °C overnight. The plates were then blocked with PBS (pH 7.4) containing 1% BSA at room temperature for 2 h. Mouse sera, diluted 500-fold, were added to the wells and incubated at 37 °C for 45 min. Wells were rinsed five times with PBST (PBS containing 0.05% Tween-20), followed by a 30 min incubation with alkaline-phosphatase-conjugated goat-anti-mouse IgG (Sigma) (1:5000) at 37 °C. After five washings and a subsequent 15 min incubation with the substrate para-nitrophenyl phosphate (pNPP) solution at 37 °C in dark, reactions were terminated with a stop solution, and optical densities (ODs) were determined using a microplate reader set at 405 nm.
2.1.7. ELISPOT assay
Cellular immune responses to SARS-CoV were assessed by IFN-γ and IL-4 ELISPOT assays, using mouse splenocytes harvested 10 days after the final immunization. According to the instruction manual (U-CyTech, Netherlands), 96-well plates were coated overnight with 100 μl per well rat anti-mouse IFN-γ or rat anti-mouse IL-4 antibodies at 4 °C. Plates were washed three times with sterile PBS and blocked for 2 h at room temperature. Mouse splenocytes (1 × 105 per well, in triplicate) were incubated in RPMI 1640 containing purified recombinant N or S protein (2 μg/ml) as the specific stimulating antigens, respectively, for 18 h at 37 °C. Plates were then washed 10 times with PBS containing 0.05% Tween-20 and incubated with 100 μl biotinylated rat anti-mouse IFN-γ or IL-4 antibody per well for 2 h at room temperature. After washing, avidin-horseradish peroxidase was added for another 1 h incubation at room temperature. Following five washes with PBS, individual IFN-γ or IL-4 plates were developed as dark spots after a 10 min reaction with the peroxidase substrate-AEC. Reactions were stopped by rinsing plates with de-mineralized water. Plates were air-dried at room temperature and read using an ELISPOT reader (Hitech Instruments). Medium backgrounds were consistently <10 SFC per 106 splenocytes.
2.1.8. Flow cytometry analysis
Splenocytes were harvested 10 days after the final immunization. Prior to intracellular cytokine staining, 2 × 105 pooled splenocytes/well from different groups of vaccinated mice were seeded in triplicate in 96-well plates and incubated for 5 h. A mixture of 5 μg per well of corresponding purified protein and anti-CD28 mAb (final concentration of 1 μg/ml) was added to each well. In each group, as a negative control, 1 μg/ml of anti-CD28 mAb was added without stimulation of the purified specific protein. After incubation, mononsesin (1.5 μl/ml) was added to each well and blocked at 37 °C for 3 h. The splenocytes in each well were then washed and stained with phycoerythrin (PE)-conjugated anti-CD4 and PECy5-conjugated anti-CD8 antibodies for 20 min at 4 °C. The cells were then washed, fixed and permeabilized. Intracellular IFN-γ and IL-2 (both fluorescein isothiocyanate (FITC)-conjugated) staining and flow cytometry analysis (Beckman, EPICS ALTRA Idaho, USA) were performed in succession. Data were analyzed using EXOPO analysis software. A minimum of 10,000 events were collected and analyzed for each sample. All of the antibodies were from BD PharMingen. Isotype-matched controls were included in each staining.
3. Results
3.1. Construction of recombinant baculoviruses, vAc-N and vAc-S
Full-length n and s genes of SL-CoV were amplified and confirmed through sequence analysis and further cloned into pFastBac DUAL vector, respectively. Promoters EF-1α and CMV promoted high levels of protein expression in a wide range of mammalian cells, and were therefore used to replace the promoters p10 and Polh in the pFastBac DUAL, respectively. Following transfection, two recombinant baculoviruses, vAc-N and vAc-S, were successfully constructed to express the N or S protein of SL-CoV. The plaque assay resulted in a titer of 8.2 × 106 and 7.5 × 106 pfu/ml, respectively.
3.2. Expression of N and S proteins in recombinant virus-transduced cells
Baculoviruses are capable of transducing a variety of dividing and non-dividing mammalian cells with significant efficiency in vitro, which, depending on the promoter used, can result in foreign gene expression (Boyce and Bucher, 1996, Ojala et al., 2001, Pieroni et al., 2001, Shoji et al., 1997, Song et al., 2003). In our study, GFP expression was observed in BHK cells 24 h post-transduction with vAc-N or vAc-S at a multiplicity of infection (MOI) rate of 100 pfu per cell. The GFP signal lasted until 72 h post-transduction (data not shown). The control group, BHK cells transduced with vAc-GFP, also expressed GFP.
To confirm expression of the N and S proteins at the transcriptional level, BHK cells were transduced with vAc-N or vAc-S for 24 h and subsequently harvested for RT-PCR analysis. The primers used for RT-PCR detection were designed from Urbani strain AY27874 and the primer sequences were highly conserved between SARS-CoV and bat SL-CoV. These primers were successfully used to detect SARS-like coronavirus in bats (Li et al., 2005) and other wild animals. As shown in Fig. 1A, either N or S mRNA-specific bands were detected in cells transduced with vAc-N or vAc-S; however, no mRNA was detected in non-transduced cells or cells transduced with Ac-GFP. In agreement with the sizes of the S and N proteins of SL-CoV, an ∼180 kDa band was detected by western blot in vAc-S-transduced cells, while an ∼50 kDa band was visualized in those cells transduced with vAc-N (Fig. 1B). No band was observed in control groups (non-transduced cells or cells transduced with Ac-GFP).
Fig. 1.
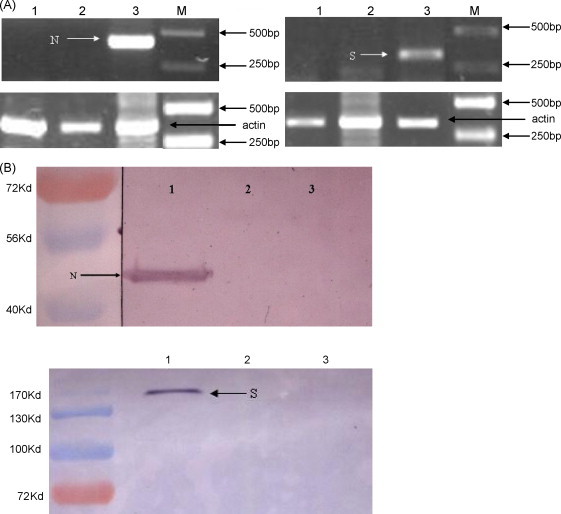
Expression of the N or S protein in recombinant baculovirus-transduced BHK cells (A) BHK cells were transduced with recombinant or control viruses for 24 h and total RNAs were extracted for RT-PCR analysis. Lane 1: non-transduced BHK cells; Lane 2: BHK cells transduced with Ac-GFP; Lane 3: BHK cells transduced with vAc-N or vAc-S. Arrows indicate the specific PCR products. (B) BHK cells were transduced with recombinant viruses for 24 h and raw proteins were extracted for western blot analysis. Lane 1: BHK cells transduced with vAc-N or vAc-S; Lane 2: non-transduced BHK cells; Lane 3: BHK cells transduced with Ac-GFP. Arrows indicate the specific proteins. One out of three experiments is shown.
3.3. Antibody responses in vAc-N or vAc-S recombinant baculovirus-vaccinated mice
We then proceeded to measure the antibody activity of sera from vAc-S or vAc-N recombinant baculovirus-immunized mice. Because an in vitro culture model for SL-CoV was not available, we were unable to directly detect the antibody activity of mouse sera against SL-CoV. Instead, we examined whether mouse sera were capable of recognizing SARS-CoV. The titers from mouse sera collected on days 0, 14, 28, and 42 were determined using inactivated SARS-CoV as the captured antigen. As shown in Fig. 2 , mice immunized with either vAc-N or vAc-S showed detectable antibody responses to SARS-CoV on day 14, and the titers significantly increased following two additional injections. By day 42, both vAc-N and vAc-S immunized groups had an antibody response to SARS-CoV that was ∼9-fold greater than the background (PBS control group). There was no significant difference in antibody titer detected from the vAc-N and vAc-S immunized groups. It should be noted that sera from the Ac-GFP-immunized group showed a detectable response to SARS-CoV antigens, although the response was significantly lower than from the vAc-N or vAc-S immunized groups. This cross-activity was very likely due to “non-specific” antibodies induced by baculoviral antigens, which can also interact with SARS-CoV proteins, because pre-incubation with Ac-GFP completely abrogated such cross-activity (data not shown).
Fig. 2.
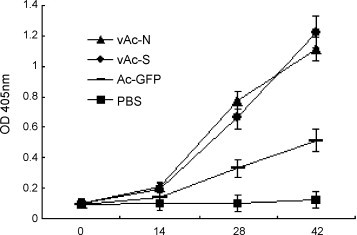
Examination of antibody responses in immunized mice. Mice were immunized with vAc-N, vAc-S, Ac-GFP or PBS. Sera of immunized mice from different groups were collected before each immunization. The titers of antibodies interacting with SARS-CoV were measured by ELISA. Data shown are the mean ± S.D. of three experiments from different animals.
3.4. Cellular immune responses in vAc-N or vAc-S recombinant baculovirus-vaccinated mice
The type of immune response elicited by either vAc-N or vAc-S immunized mice was evaluated by measuring IFN-γ and IL-4 secretion in mouse splenocytes. As indicated in Fig. 3A, the vAc-N vaccination of mice resulted in the highest number of IFN-γ-secreting splenocytes. The number of IFN-γ-secreting splenocytes in both the vAc-N and vAc-S groups was at least threefold higher than that of the Ac-GFP group, and 20-fold higher than that of the PBS control group. As for IL-4 secreting splenocytes, at least a fivefold increase was achieved in vAc-N and vAc-S groups when compared to the Ac-GFP and PBS groups (Fig. 3B). It is worthy to note that the IFN-γ secretion level was much higher than that of IL-4 in both the vAc-N and vAc-S immunized groups, suggesting a strong Th1 bias towards cellular immune responses. Th1 cells can elicit phagocyte-mediated defense against infections; therefore, Th1-dominated immune responses, elicited by vAc-N and vAc-S, may be important for virus control.
Fig. 3.
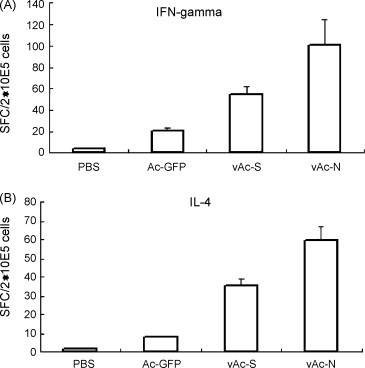
Determination of IFN-γ or IL-4 producing splenocytes in immunized mice. Mouse splenocytes were harvested 10 days after the final immunization. ELISPOT assay was used to determine IFN-γ (A) or IL-4 (B) producing cells. Data shown are the mean ± S.D. of three experiments from different animals.
Th1 cytokine IFN-γ and IL-2 positive cells in CD4+ or CD8+ cell populations were analyzed by flow cytometry. As shown in Fig. 4A and B, the number of IFN-γ-positive or IL-2 positive cells in the CD4+ T cell population was significantly higher than that of the CD8+ T cells in each vaccination group. vAc-N-immunized mice produced higher IFN-γ-positive or IL-2 positive cell numbers, in both CD4+ and CD8+ T cell populations, than the vAc-S-immunized group. In contrast, the Ac-GFP and PBS groups produced no significant amount of IFN-γ or IL-2 producing CD4+ and CD8+ T cells.
Fig. 4.
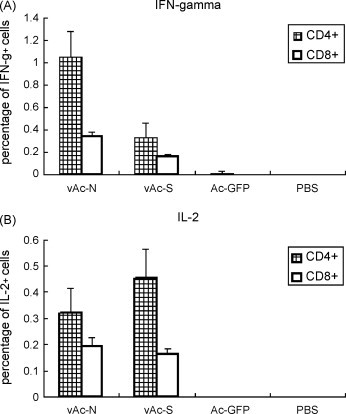
Analysis of IFN-γ and IL-2 positive cells in CD4+ or CD8+ splenocytes from immunized mice. Mouse splenocytes were harvested 10 days after the final immunization. IFN-γ and IL-2 positive cells in CD4+ or CD8+ cell populations were analyzed by flow cytometry. Data shown are the mean ± S.D. of three experiments from different animals.
4. Discussion
Since the outbreak of SARS and the identification of SARS-CoV as the causative agent, much effort has been devoted to understanding viral pathogenesis and to developing treatment strategies. The isolation of homologous viruses from other animal species and the completion of viral genome sequences make it possible to develop effective vaccines, in particular subunit and DNA-based vaccines. These advances will hopefully help to prevent the reemergence of this life-threatening disease.
To date, several vaccines have been tested in pre-clinical or clinical studies, including vector-based DNA vaccines (Bisht et al., 2004, He et al., 2005, Okada et al., 2005, Wang et al., 2005, Yang et al., 2004), combination of whole killed virus and DNA vaccines (Zakhartchouk et al., 2005), inactivated whole virus vaccines (He et al., 2004, Spruth et al., 2006), and other recombinant proteins and their fragments. The majority of these vaccines target SARS-CoV structural proteins, particularly the S and N proteins. The S protein is essential for viral infection, while the N protein is important for the transcription of viral RNA. Full-length S or S1 subunit was shown to induce effective immune response against SARS-CoV and to elicit neutralizing antibodies that protect against virus challenge (Bukreyev et al., 2004, Gao et al., 2003, Yang et al., 2004). Anti-N antibodies were present at high levels in SARS patients and persisted for a long time. In agreement, vectors that expressed full-length or truncated N proteins were highly immunogenic (Chen et al., 2004, Guan et al., 2004, Huang et al., 2004, Leung et al., 2004, Tan et al., 2004, Woo et al., 2004). Hence, the S and N proteins may be the most promising vaccine candidates against SARS-CoV. SL-CoV isolated from bats is ∼92% homologous to human SARS-CoV (Li et al., 2005, Ren et al., 2006), suggesting that bats may serve as a natural reservoir of SARS-CoV. Therefore, bat SL-CoV should be taken into consideration for developing a cross-protective vaccine.
We chose baculoviruses for gene transfer and expression systems for several reasons. Baculoviruses are not toxic, even at high multiplicity of infection, and are considered to be safe, since they do not replicate in mammalian cells (Volkman and Goldsmith, 1983). In addition, baculoviruses are capable of carrying large DNA insertions and their recombinant forms are straightforward to construct (Cheshenko et al., 2001). Moreover, engineered baculoviruses that contain mammalian-cell activate promoter elements have been demonstrated to efficiently transfer genes and stably express proteins in mammalian cells (Cheng et al., 2004, Condreay et al., 1999, Hofmann et al., 1995, Kost and Condreay, 2002). In order to express the N and S proteins of SL-CoV in mammalian cells, we replaced the primary promoters, Polh and p10, in the pFastBac DUAL vector with other promoters, CMV and EF-1α, respectively. The GFP gene was constructed as a reporter under the control of EF-1α. According to Cheng et al. (2004), incubation of target mammalian cells with the culture supernatant of infected Sf9 cells (1 × 107 pfu/ml) for 12 h (MOI = 50) resulted in the highest gene transfer efficiency, the highest protein expression, and the least impairment to cell viability (Cheng et al., 2004). In our study, the titers of the two recombinant baculoviruses, vAc-S and vAc-N, did not reach 1 × 107 pfu/ml. We, therefore, prolonged the incubation time to 24 h by transducing BHK cells with 100 MOI of recombinant virus, vAc-N or vAc-S. Under these conditions, fluorescence was observed at 24 h post-transduction and lasted until 72 h post-transduction, indicating that the modified baculoviruses successfully expressed exogenous genes in mammalian cells.
Previous investigations have demonstrated that both cellular and humoral immunity contributed to the long-term protection against coronaviruses (Sestak et al., 1999). Studies on infectious bronchitis virus (Cavanagh et al., 1986, Kapczynski et al., 2003, Song et al., 1998), transmissible gastroenteritis virus (Anton et al., 1996), and SARS-CoV (Bukreyev et al., 2004, Gao et al., 2003, Yang et al., 2004) showed that immunization with recombinant S protein or plasmid encoding S protein elicited high titers of neutralizing antibodies and provided protection. Our data indicated that mice immunized with either vAc-N or vAc-S produced sera that strongly interacted with SARS-CoV. Since an in vitro cell culture model of bat SL-CoV has not yet been established, we were unable to determine the serum neutralizing activity that was specific to SL-CoV. Given that the S protein is the key protein of viral entry and contains neutralizing epitopes, as demonstrated in other coronavirus studies, antibodies raised against the S protein of SL-CoV will also likely have the capability of neutralizing this virus.
In addition to the antibody response, mice that were immunized with either vAc-N or vAc-S also demonstrated cellular immune responses. Th1 (IFN-γ) and Th2 (IL-4) cytokines were examined by ELISPOT and a strong bias towards Th1 production was detected. Our results are consistent with those observed in other SARS DNA vaccine studies (Huang et al., 2006, Kong et al., 2005, Woo et al., 2005, Yang et al., 2004, Zakhartchouk et al., 2005). The balance between Th1 and Th2 responses to an infectious agent can influence both pathogen growth and immunopathology. Th1 cells are responsible for cell-mediated immunity, while Th2 cells are responsible for humoral immunity (Bottomly, 1988, Mosmann and Cofman, 1989). The principal function of Th1 cells is to elicit phagocyte-mediated defense against infections, and Th1-dominant immune responses are often associated with inflammation and tissue injury. Humoral immune responses could be induced against virus; however, this type of response alone may not be sufficient for protection and clearing of the virus. T cell responses, especially Th1-mediated cellular immunity, may be a crucial factor for long-term protection. The Th1 cytokine, IFN-γ, can activate macrophages and stimulate the production of IgG antibodies (Wu et al., 2002), whereas IL-2 is required for the growth of T cells and may even be required for naive T cells to produce IL-4 (Powers et al., 1988). In the present study, although cellular immune responses were observed in both CD4+ and CD8+ T cells, there was a greater number of IFN-γ and IL-2-secreting CD4 T cells detected. Given that CD4 T cells can increase the number of immune memory cells that respond rapidly when re-exposed to pathogens, and thus play a vital role in protection against virus challenge (Bourgeois et al., 2002a, Bourgeois et al., 2002b), the marked increase of CD4 T cell in mice immunized with the recombinant baculoviruses, vAc-N or vAc-S, may prove to be significant in the protection against SARS-CoV and SL-CoV infection.
5. Conclusions
In summary, we have successfully expressed the S and N proteins of bat SL-CoV using a baculovirus expression system. Mice injected with these two recombinant baculoviruses elicited an antibody response that interacted with SARS-CoV and demonstrated a Th1-dominant immune response. Our results indicate that recombinant baculovirus can be transduced into mammalian cells by direct injection, and such transductions can elicit immune responses. This strategy is considered to be safe, since baculoviruses do not replicate in mammalian cells and do not induce a cytopathic effect (CPE). To our knowledge, this is the first report characterizing the N and S proteins of bat SL-CoV in a mouse model. Because it is adjuvant-free and there is no need to purify proteins, the use of recombinant baculoviruses may represent a convenient strategy for studying the immunogenicity of genes in animal models.
Conflict of interest
The authors do not have a commercial association, or other, that might pose a conflict of interest.
Acknowledgements
This work was supported in part by a National “973 Project” (2005CB523000) from the Ministry of Science and Technology, People's Republic of China, and by a grant (KSCXL-YW-R-07) from the Chinese Academy of Sciences.
We thank Prof. Wang L.F. at the Australian Animal Health Laboratory for kindly providing the rabbit-derived anti-SARS-CoV antibody, and Prof. Shi Z.L. at the Wuhan Institute of Virology for providing the N and S gene of bat SL-CoV Rp3 strain.
References
- Anton I.M., Gonzalez S., Bullido M.J., Corsin M., Risco C., Langeveld J.P., Enjuanes L. Cooperation between transmissible gastroenteritis coronavirus (TGEV) structural proteins in the in vitro induction of virus-specific antibodies. Virus Res. 1996;46:111–124. doi: 10.1016/S0168-1702(96)01390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly K. A functional dichotomy in CD4+ T lymphocytes. Immunol. Today. 1988;9:268–274. doi: 10.1016/0167-5699(88)91308-4. [DOI] [PubMed] [Google Scholar]
- Bourgeois C., Rocha B., Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- Bourgeois C., Veiga-Fernandes H., Joret A.M., Rocha B., Tanchot C. CD8 lethargy in the absence of CD4 help. Eur. J. Immunol. 2002;32:2199–2207. doi: 10.1002/1521-4141(200208)32:8<2199::AID-IMMU2199>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Boyce F.M., Bucher N.L. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell L.F., Klowden M.J., Miller L.K. Baculovirus-mediated expression of bacterial genes in dipteran and mammalian cells. J. Virol. 1985;56:153–160. doi: 10.1128/jvi.56.1.153-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67:1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H., Zhou D., Zhai J., Du Z., Li B., Qiu M., Han Y., Guo Z., Yang R. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 2004;50:988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Xu C.Y., Wang Y.B., Chen M., Wu T., Zhang J., Xia N.S. A rapid and efficient method to express target genes in mammalian cells by baculovirus. World J. Gastroenterol. 2004;10:1612–1618. doi: 10.3748/wjg.v10.i11.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshenko N., Krougliak N., Eisensmith R.C., Krougliak V.A. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001;8:846–854. doi: 10.1038/sj.gt.3301459. [DOI] [PubMed] [Google Scholar]
- Condreay J.P., Witherspoon S.M., Clay W.C., Kost T.A. Transient and stable gene expression in mammalian-cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. U.S.A. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney W.E., Isom H.C. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28:1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duisit G., Saleun S., Douthe S., Barsoum J., Chadeuf G., Moullier P. Baculovirus vector requires electrostatic interactions including heparan sulfate for efficient gene transfer in mammalian cells. J. Gene Med. 1999;1:93–102. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<93::AID-JGM19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stöhr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Foo S.Y., Tan Y.J., Goh P.Y., Wee S.H. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 2004;11:287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 2004;325:445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Tang Y., Qin X., Xu W., Wang Y., Liu X., Liu X., Xiong S., Li J., Zhang M., Duan M. Construction of a eukaryotic expression plasmid encoding partial S gene fragments of the SARS-CoV and its potential utility as a DNA vaccine. DNA Cell Biol. 2005;24:516–520. doi: 10.1089/dna.2005.24.516. [DOI] [PubMed] [Google Scholar]
- Hofmann C., Sandig V., Jennings G., Rudolph M., Schlag P., Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L.H., Du G.X., Guan R.B., Tong Y.G., Wang H.T. In vitro assay for HCV serine proteinase expressed in insect cells. World J. Gastroenterol. 2003;9:1629–1632. doi: 10.3748/wjg.v9.i7.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.R., Chiu C.M., Yeh S.H., Huang W.H., Hsueh P.R., Yang W.Z., Yang J.Y., Su I.J., Chang S.C., Chen P.J. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J. Med. Virol. 2004;73:338–346. doi: 10.1002/jmv.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ma R., Wu C.Y. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24:4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A. Production and immunogenicity of multiple antigenic peptide (MAP) constructs derived from the S1 glycoprotein of infectious bronchitis virus (IBV) Adv. Exp. Med. Biol. 1995;380:213–219. doi: 10.1007/978-1-4615-1899-0_35. [DOI] [PubMed] [Google Scholar]
- Jones I., Morikawa Y. Baculovirus vectors for expression in insect cells. Curr. Opin. Biotechnol. 1996;7:512–516. doi: 10.1016/s0958-1669(96)80054-1. [DOI] [PubMed] [Google Scholar]
- Kapczynski D.R., Hilt D.A., Shapiro D., Sellers H.S., Jackwood M.W. Protection of chickens from infectious bronchitis by in Ovo and intramuscular vaccination with a DNA vaccine expressing the S1 glycoprotein. Avian Dis. 2003;47:272–285. doi: 10.1637/0005-2086(2003)047[0272:POCFIB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kong W.P., Xu L., Stadler K., Ulmer J.B., Abrignani S., Rappuoli R., Nabel G.J. Modulation of the immune response to the severe acute respiratory syndrome spike glycoprotein by gene-based and inactivated virus immunization. J. Virol. 2005;11:13915–13923. doi: 10.1128/JVI.79.22.13915-13923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost T.A., Condreay J.P. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol. 2002;20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H., Tam J.S., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Wu H.Y., Qian X.P., Li Y., Chen W.F. Expression, purification and serological analysis of hepatocellular carcinoma associated antigen HCA587 in insect cells. World J. Gastroenterol. 2003;9:678–682. doi: 10.3748/wjg.v9.i4.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Luckow V.A., Summers M.D. Signals important for high-level expression of foreign genes in Autographa californica nuclear polyhedrosis virus expression vectors. Virology. 1988;167:56–71. doi: 10.1016/0042-6822(88)90054-2. [DOI] [PubMed] [Google Scholar]
- Miller L.K. Baculoviruses for foreign gene expression in insect cells. Biotechnology. 1988;10:457–465. doi: 10.1016/b978-0-409-90042-2.50029-5. [DOI] [PubMed] [Google Scholar]
- Mosmann T.R., Cofman R.L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rew. Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Nakhai B., Pal R., Sridhar P., Talwar G.P., Hasnain S.E. The alpha subunit of human chorionic gonadotropin hormone synthesized in insect cells using a baculovirus vector is biologically active. FEBS Lett. 1991;283:104–108. doi: 10.1016/0014-5793(91)80564-j. [DOI] [PubMed] [Google Scholar]
- Ojala K., Mottershead D.G., Suokko A., Oker-Blom C. Specific binding of baculoviruses displaying gp64 fusion proteins to mammalian cells. Biochem. Biophys. Res. Commun. 2001;284:777–784. doi: 10.1006/bbrc.2001.5048. [DOI] [PubMed] [Google Scholar]
- Okada M., Takemoto Y., Okuno Y., Hashimoto S., Yoshida S., Fukunaga Y., Tanaka T., Kita Y., Kuwayama S., Muraki Y., Kanamaru N., Takai H., Okada C., Sakaguchi Y., Furukawa I., Yamada K., Matsumoto M., Kase T., Demello D.E., Peiris J.S., Chen P.J., Yamamoto N., Yoshinaka Y., Nomura T., Ishida I., Morikawa S., Tashiro M., Sakatani M. The development of vaccines against SARS corona virus in mice and SCID-PBL/hu mice. Vaccine. 2005;23:2269–2272. doi: 10.1016/j.vaccine.2005.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieroni L., Maione D., La Monica N. In vivo gene transfer in mouse skeletal muscle mediated by baculovirus vectors. Hum. Gene Ther. 2001;12:871–881. doi: 10.1089/104303401750195845. [DOI] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Powers G.D., Abbas A.K., Miller R.A. Frequencies of IL-2- and IL-4-secreting T cells in naive and antigen stimulated lymphocyte populations. J. Immunol. 1988;140:3352–3357. [PubMed] [Google Scholar]
- Ren H., Zhu F.L., Zhu S.Y., Song Y., Qi Z.T. Immunogenicity of HGV NS5 protein expressed from Sf9 insect cells. World J. Gastroenterol. 2001;7:98–101. doi: 10.3748/wjg.v7.i1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Li W., Yu M., Hao P., Zhang Y., Zhou P., Zhang S., Zhao G., Zhong Y., Wang S., Wang L.F., Shi Z. Full-length genome sequences of two SARS-like corona viruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- Sestak K., Meister R.K., Hayes J.R., Kim L., Lewis P.A., Myers G., Saif L.J. Active immunity and T-cell populations in pigs intraperitoneally inoculated with baculovirus-expressed transmissible gastroenteritis virus structural proteins. Vet. Immunol. Immunopathol. 1999;70:203–221. doi: 10.1016/S0165-2427(99)00074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji I., Aizaki H., Tani H., Ishii K., Chiba T., Saito I., Miyamura T., Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 1997;78:2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- Song C.S., Lee Y.J., Lee C.W., Sung H.W., Kim J.H., Mo I.P., Izumiya Y., Jang H.K., Mikami T. Induction of protective immunity in chickens vaccinated with infectious bronchitis virus S1 glycoprotein expressed by a recombinant baculovirus. J. Gen. Virol. 1998;79:719–723. doi: 10.1099/0022-1317-79-4-719. [DOI] [PubMed] [Google Scholar]
- Song S.U., Shin S.H., Kim S.K., Choi G.S., Kim W.C., Lee M.H., Kim S.J., Kim I.H., Choi M.S., Hong Y.J., Lee K.H. Effective transduction of osteogenic sarcoma cells by a baculovirus vector. J. Gen. Virol. 2003;84:697–703. doi: 10.1099/vir.0.18772-0. [DOI] [PubMed] [Google Scholar]
- Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M., Bruhl P., Grillberger L., Reiter M., Tauer C., Mundt W., Barrett P.N. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24:652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar P., Awasthi A.K., Azim C.A., Burma S., Habib S., Jain A., Mukherjee B., Ranjan A., Hashain S.E. Baculovirus vector-mediated expression of heterologous genes in insect cells. J. Biosci. 1994;19:603–614. [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjia S.T., zu Altenschildesche G.M., Doerfler W. Autographa californica nuclear polyhedrosis virus (AcNPV) DNA does not persist in mass cultures of mammalian cells. Virology. 1983;125:107–117. doi: 10.1016/0042-6822(83)90067-3. [DOI] [PubMed] [Google Scholar]
- Volkman L.E., Goldsmith P.A. In vitro survey of Autographa californica nuclear polyhedrosis virus interaction with nontarget vertebrate host cells. Appl. Environ. Microbiol. 1983;45:1085–1093. doi: 10.1128/aem.45.3.1085-1093.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chou T.H., Sakhatskyy P.V., Huang S., Lawrence J.M., Cao H., Huang X., Lu S. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 2005;79:1906–1910. doi: 10.1128/JVI.79.3.1906-1910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Tsoi H.W., Fung A.M., Chan K.H., Tam V.K., Peiris J.S., Yuen K.Y. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2004;42:2306–2309. doi: 10.1128/JCM.42.5.2306-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chen Z.W., Wong B.H., Zhang L., Chan J.K., Wong L.P., He W., Ma C., Chan K.H., Ho D.D., Yuen K.Y. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine. 2005;23:4959–4968. doi: 10.1016/j.vaccine.2005.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Kirman J.R., Rotte M.J., Davey D.F., Perfetto S.P., Rhee E.G., Freidag B.L., Hill B.J., Douek D.C., Seder R.A. Distinct lineages of TH1 cells have differential capacities for memory cell generation in vivo. Nat. Immunol. 2002;3:852–858. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Liu Q., Petric M., Babiuk L.A. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine. 2005;23:4385–4391. doi: 10.1016/j.vaccine.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


