Abstract
The innate immune system plays a central role in host defence against viruses. While many studies portray mechanisms in early antiviral immune responses of humans and mice, much remains to be discovered about these mechanisms in the cat. With the objective of shedding light on early host–virus interactions in felids, we have developed 12 real-time TaqMan® qPCR systems for feline genes relevant to innate responses to viral infection, including those encoding for various IFNα and IFNω subtypes, IFNβ, intracellular antiviral factor Mx, NK cell stimulator IL-15 and effectors perforin and granzyme B, as well as Toll-like receptors (TLRs) 3 and 8. Using these newly developed assays and others previously described, we measured the relative expression of selected markers at early time points after viral infection in vitro and in vivo. Feline embryonic fibroblasts (FEA) inoculated with feline leukemia virus (FeLV) indicated peak levels of IFNα, IFNβ and Mx expression already 6 h after infection. In contrast, Crandell-Rees feline kidney (CrFK) cells inoculated with feline herpes virus (FHV) responded to infection with high levels of IFNα and IFNβ only after 24 h, and no induction of Mx could be detected. In feline PBMCs challenged in vitro with feline immunodeficiency virus (FIV), maximal expression levels of IFNα, β and ω subtype genes as well as IL-15 and TLRs 3, 7 and 8 were measured between 12 and 24 h after infection, whereas expression levels of proinflammatory cytokine gene IL-6 were consistently downregulated until 48 h post inoculation. A marginal upregulation of granzyme B was also observed within 3 h after infection. In an in vivo experiment, cats challenged with FIV exhibited a 2.4-fold increase in IFNα expression in blood 1 week post infection. We furthermore demonstrate the possibility of stimulating feline immune cells in vitro with various immune response modifiers (IRMs) already known for their immunostimulatory properties in mice and humans, namely Poly IC, Resiquimod (R-848) and dSLIM™, a synthetic oligonucleotide containing several unmethylated CpG motifs. Stimulation of feline PBMCs with dSLIM™ and R-848 effectively enhanced expression of IFNα within 12 h by factors of 6 and 12, respectively, and Poly IC induced an increase in Mx mRNA expression of 28-fold. Altogether, we describe new molecular tools and their successful use for the characterization of innate immune responses against viruses in the cat and provide evidence that feline cells can be stimulated by synthetic molecules to enhance their antiviral defence mechanisms.
Keywords: Cat, Innate immune system, Virus, Immune response modifier, Real-time TaqMan® qPCR
1. Introduction
In the course of evolution, feline viruses have gained properties rendering their infections particularly challenging to prevent and treat. Presumably, the way of life of felids has driven over time the development of viruses with highly efficient viral transmission strategies and the potential to induce chronic or latent infections, thus increasing carrier populations and viral dissemination (Pontier et al., 2009). It is today widely accepted that the initial antiviral immune response of the mammalian host plays an essential role in determining the outcome of viral infection. Although extensive studies have been carried out in mouse models, the antiviral innate immune system of the cat remains poorly understood, leaving open questions regarding initial responses to viral infection as well as the possibility of manipulating innate immune mechanisms in favour of the host. In order to illustrate the current status of research concerning innate antiviral defence mechanisms in cats, we will first give a short overview of general knowledge in this field.
Studies in mice and humans have demonstrated that early pathogen recognition by the innate immune system relies on its ability to sense microbial components known as pathogen-associated molecular patterns (PAMPs) (Akira et al., 2006). With respect to viruses, the host's intrinsic defence structures recognize mostly genomic nucleic acids and replication intermediates. Three main classes of PAMPs have emerged to date: double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), and unmethylated CpG DNA. In turn, the detection of these PAMPs relies on a limited set of germline-encoded pattern recognition receptors (PRRs). Three classes of PRR molecules seem to survey various cell compartments for presence of PAMPs and promote intrinsic antiviral immunity: the Toll-like receptors (TLRs), the retinoic acid inducible gene-I (RIG-I)-like receptors (RLRs) and the cytoplasmic viral DNA sensors (Yoneyama and Fujita, 2010).
TLRs are type I integral membrane glycoproteins either expressed on the cell surface or retained in intracellular compartments. Among the 12 TLRs in mice and 10 TLRs in humans that have been identified to date (Tabeta et al., 2004), three share the ability to bind nucleic acids, namely those expressed in endosomal compartments of various subsets of dendritic cells (DCs). Thus, upon phagocytic or endocytic events, TLR3, TLR7 and TLR9 can sense viral dsRNA, ssRNA or unmethylated CpG-DNA respectively (Vaidya and Cheng, 2003). TLR8, a structurally homologous variant of TLR7, seems to be non-functional in mice, but has been described in humans as recognising similar molecular patterns to TLR7 (Jurk et al., 2002). To date, various synthetic ligands have been identified for these TLRs, among which the most popular include Poly IC, a synthetic double-stranded polyriboinosinic–polyribocytidylic acid, a well-known synthetic analogue to dsRNA and stimulator of TLR3 (Jiang et al., 2003); imidazoquinolines such as resiquimod (R-848), synthetic compounds that bind to TLR7/8 (Wagner et al., 1999); and synthetic CpG molecules, which trigger TLR9 (Vollmer et al., 2004). Such Toll-like receptor agonists have not only been used extensively for research purposes linked to the understanding of innate immune mechanisms, but have also gained much popularity in the clinical field, as treatments for various types of disease and as vaccine adjuvants (Meyer and Stockfleth, 2008, Seya and Matsumoto, 2009).
In contrast to TLRs, RLRs and DNA sensors are ubiquitously expressed in the cytoplasm and allow infected cells to detect actively replicating virus. Two main members of the RLR family, RIG-I and melanoma differentiation associated gene 5 (MDA5), have been shown to play essential roles in the recognition of cytoplasmic viral RNA (Yoneyama et al., 2004, Loo et al., 2008). Recently, additional cytoplasmic DNA sensors have been proposed to detect viral dsDNA during infection (Yoneyama and Fujita, 2010). These molecules are currently under investigation.
PAMP-triggered PRRs activate distinct signaling pathways that converge on the activation of specific transcription factors NFκB, AP-1, IFN regulatory factor (IRF) 3 and IRF7, enhancing the expression of both proinflammatory cytokines such as IL-6 and TNFα, and type I IFNs, the main players in eradication of replicating viruses in antiviral innate immunity (Schindler et al., 2007). In humans, the large family of type I IFNs primarily include IFNα, IFNβ and IFNω (Bekisz et al., 2004). IFNα consists of a group of 13 structurally related proteins, each encoded by a separate intronless gene, while only one protein each for IFNβ and IFNω have been characterized. Type I IFNs are produced by almost all cell types, with IFNβ being the main IFN secreted by fibroblasts in response to viral challenge. Essentially, the ubiquitous presence of IFN receptor complex composed of two subunits IFNAR1 and IFNAR2 (Uze et al., 2007) and the pleiotropy of these cytokines enable IFNs to initiate potent antiviral responses. First, type I IFNs activate, in uninfected target cells, the production of intracellular effectors that can interfere with several steps of virus replication cycles. Well-studied examples of these antiviral proteins include myxovirus-resistance protein (Mx) GTPase, the RNA-dependent protein kinase (PKR), the 2′,5′-oligoadenylate synthetase (OAS) and the interferon stimulated gene (ISG) 15 (mechanisms reviewed in Sadler and Williams, 2008). The Mx protein, for instance, binds to essential viral components thus blocking their intracellular transport (Haller et al., 2007). Moreover, production of this protein is widely recognized as a marker for upregulation of type I IFNs (Haller et al., 2007). Through positive feedback processes, type I IFNs were additionally shown to enhance the expression of TLRs, further sensitizing cells to microbial recognition (Siren et al., 2005). Finally, type I IFNs effectively bridge innate and adaptive immunity by promoting the differentiation and function of various immune cell populations such as DCs, NK cells, B cells, as well as CD4+ and CD8+ T cell populations (Colonna et al., 2004).
NK cells represent another important antiviral cell population (Hamerman et al., 2005). Constitutively primed to kill, these cells rapidly release IFNγ, a type II IFN affecting mainly adaptive immune cells, as well as granzyme and perforin, cytotoxic substances stored in cytosolic granules. Primary NK targets are host cells lacking MHC I molecules on their surface. As most viruses have evolved mechanisms to downregulate host cell MHC expression in order to avoid immune responses resulting from antigen presentation, virally infected cells are readily attacked by NK cells. In addition to type I IFNs, IL-15 was shown to support development and survival of NK cells as well as to stimulate their cytokine production. Liberated mainly by PAMP-activated DC, IL-15 thus also plays a crucial role in antiviral defence (Vujanovic et al., 2010).
Various essential players in the innate immune system of the cat have to date been characterized. TLRs 1–9 were shown to be differentially expressed in various feline lymphoid tissues and cell lines (Ignacio et al., 2005). Concerning TLRs 3, 7, 8 and 9, high expression levels were measured in the mesenteric lymph nodes and in the spleen. Interestingly, both B and T lymphocyte subsets purified from lymph nodes expressed TLRs and expression in feline CD4+ and CD8+ T cells was mainly restricted to these anti-viral associated TLRs. Infection with feline viruses in vitro also induced altered TLR expression levels in feline cells, with observed differences depending highly on cell type and TLR studied (Ignacio et al., 2005). The stimulation of feline TLRs with specific agonists has not yet been reported.
Studies carried out in the past 20 years have mainly focused on the characterization of feline type I IFN subtypes and their potential therapeutic effects in the context of various viral diseases. Nakamura et al. cloned the first cDNA sequence for feline IFN in 1992 (Nakamura et al., 1992). The purified protein was classified as omega-type (Ueda et al., 1993b) and rapidly shown to exhibit antiviral activity both in vitro and in vivo (Table 1, Table 2 ). As a result, Nakamura's IFNω became the first feline antiviral drug available on the market, currently sold both in Japan (Intercat®, Toray Industries, Tokyo, Japan) and Europe (Virbagen®omega, Laboratoire Virbac, Carros Cedex, France) to treat feline calicivirus and canine parvovirus infections. Subsequently, therapeutic effects of this product as stand-alone or combinatorial agent were described in the context of a series of feline and canine diseases (Table 2). In recent years, nucleic acid and amino acid sequences of many feline type I IFN subtypes have been characterized. In all, 13 subtypes each of the feIFNα and feIFNω genes have been cloned and the biological antiviral properties of the purified proteins were demonstrated in vitro (Table 1). Studies concerning the structure of these IFNα and IFNω subtypes have indicated that both protein families have high homology and that feline IFNω is more similar to the IFNα than to the IFNω of other animal species. Although the individual functions of all these IFN subtypes in the cat is unknown, the impressive number of type I IFN genes identified to date underlines the necessity of broad antiviral responses in felids (Table 1).
Table 1.
Cloned feline type I IFN subtypes and the antiviral activity of their purified proteins in vitro.
| Feline type I IFN genes | Cell type/tissue for cloning | Reference | In vitro antiviral activity of purified proteins |
||
|---|---|---|---|---|---|
| Virus | Cell line | Reference | |||
| IFNωa,b | T-cell line (LSA-1) | Nakamura et al. (1992) | FCV | CrFK | Sakurai et al. (1992) |
| VSV, FCV, FHV, FCoV, FPV, rotavirus | CrFK, fcwf-4, MDCK | Mochizuki et al. (1994) | |||
| FIV | FetJ-Bang | Tanabe and Yamamoto (2001) | |||
| FHV | CrFK | Siebeck et al. (2006) | |||
| IFNα1–3, 5, 6 | Mesenteric lymph node cells | Wonderling et al. (2002) | VSV, FCV | CrFK, AH927 | Baldwin et al. (2004) |
| IFNα7–14 | Epithelial cell line (CrFK) | Nagai et al. (2004) | VSV | CrFK, fcwf-4, RK-13, MDBK, MDCK, L-929, FL | Taira et al. (2005) |
| FCoV, FCV, FHV | fcwf-4 | ||||
| IFNω1-13 | Spleen of FCoV-infected cat | Yang et al. (2007) | VSV | CrFK, MDCK, MBDK | Yang et al. (2007) |
VSV: vesicular stomatitis virus, FCV: feline calicivirus, FHV: feline herpesvirus, FCoV: feline coronavirus, FPV: feline parvovirus, FIV: feline immunodeficiency virus, AH927: feline embryo fibroblast cell line, CrFK: Crandell-Rees feline kidney cells, fcwf-4: felis catus whole fetus cells, FetJ-Bang: persistently FIV-infected feline T-cell line, FL: transformed human amnion cells, L-929: mouse fibroblast cell line, LSA-1: cells derived from thymic lymphosarkoma of feline leukemia virus positive cat, MDBK: Madin–Darby bovine kidney cells, MDCK: Madin–Darby canine cells, RK-13: rabbit kidney-derived cells.
Initially classified as IFNω, re-termed “ω-like” by Yang et al. (2007).
Purified protein commercialized and utilized in various in vivo studies (see text and Table 2).
Table 2.
In vivo studies on antiviral effects of feline IFNω (Intercat® and Virbagen®omega).
| Disease | Reference |
|---|---|
| CPV infection |
Minagawa et al. (1999) Martin et al. (2002) |
| FeLV infection FeLV/FIV co-infection | de Mari et al. (2004) |
| FeLV infection | Collado et al. (2007) |
| FPV infection | Paltrinieri et al. (2007) |
| FHV infection | Haid et al. (2007) |
| Herpes dermatitis | Gutzwiller et al. (2007) |
| Chronic gingivostomatitis | Southerden and Gorrel (2007) |
| FCV | Ohe et al. (2008) |
Regarding effector functions of feline type I IFNs, mainly the induction of intracellular antiviral proteins has been discussed. In this way, the biological activity of Nakamura's recombinant feline IFN was initially supported by its potential to modulate OAS activity in vitro (Ueda et al., 1993a). The physiological effects of oral or ocular treatment with this recombinant IFNω were later assessed by local and systemic measurement of the feline Mx protein (Bracklein et al., 2006), which had already been previously described (Horisberger et al., 1990). More recently, stimulation of feline cells with recombinant IFNω was shown to induce expression of feline ISG15. Molecular cloning of this gene enabled detection of the immunomodulatory properties of both cell-conjugated and free forms of this antiviral effector protein (Tanabe et al., 2008). Altogether, these studies illustrate the breadth of IFN-induced antiviral mechanisms in the cat. The recent cloning and molecular characterization of the feline IFNAR2 (Xue et al., 2010) may support the study of further effector functions of type I IFNs in the cat.
Finally, only few studies have targeted aspects of the feline innate immune response following viral encounter. Dean et al. reported that immunodeficiency in FIV infection also concerned early anti-pathogen defence mechanisms in vivo (Dean et al., 1998). Thus, chronically FIV-infected cats mounted weaker initial immune responses against Listeria monocytogenes, leading to a more severe form of infection. In a further study, inoculation of FIV-infected cats with modified L. monocytogenes carrying an expression vector for various feline cytokines indicated that IL-15 played a central role in restoring FIV-related innate immune disturbances and increasing the natural killer cell population (Dean et al., 2006).
Despite extensive studies on molecular structure and antiviral effects of feline IFN, much remains to be understood regarding qualitative, quantitative and timely aspects of innate immune mechanisms following viral infection in the cat. Moreover, the possibility of synthetically stimulating the intrinsic immune system has not yet been investigated in this species.
Herein, we describe the development of twelve real-time qPCR assays enabling to quantify the expression of key factors involved in feline innate immunity. Relative expression levels of various innate immune parameters were determined in viral infection both in vitro and in vivo. Furthermore, we report the quantification of innate immune responses obtained in vitro after stimulation of feline immune cells with various synthetic molecules.
2. Materials and methods
2.1. Cats, FIV infection and blood collection
Ten male specified pathogen-free (SPF) cats of 10 weeks of age and 4 male SPF cats of 4 years of age from Liberty Research Inc. (Waverly, NY, USA) were used in this study. Young and adult cats were housed separately in an animal-friendly environment and under optimal ethological conditions. All experimental procedures were reviewed and approved by the Swiss Federal Veterinary Office.
After an adaptation phase of 4 weeks, the kittens were infected intraperitoneally with 50 cat infectious doses 50 (50 CID50) of the FIV Glasgow 8 (GL8) strain, previously titrated in vivo and kindly provided by Dr. M. Hosie and Prof. O. Jarret from the University of Glasgow, Great Britain. Whole blood was collected in EDTA-supplemented evacuated tubes both on day 0 (before infection) and on day 7 post infection. 100 μl of blood was mixed with 300 μl mRNA lysis buffer (Roche Diagnostics, Rotkreuz, Switzerland) immediately after blood collection and samples were stored at −80 °C until further analysis.
EDTA-supplemented venous blood from the 4 adult SPF cats was used for the purification of PBMCs required for other experiments, namely real-time qPCR assay optimization, IRM stimulation or inoculation with FIV.
2.2. Feline PBMC isolation, cell lines and cell culture
Feline PBMCs were isolated from EDTA-supplemented whole blood by Ficoll-Hypaque density gradient centrifugation (Histopaque®-1077, Sigma–Aldrich, Buchs, Switzerland). The mononuclear cell fraction was washed once and resuspended in RPMI with Glutamax I (Gibco®, Invitrogen, San Diego, CA, USA) supplemented with 10% heat-inactivated fetal calf serum (Bioconcept, Allschwil, Switzerland), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco®, Invitrogen, San Diego, CA, USA). For the production of cDNA for real-time qPCR assay optimization, isolated PBMCs were either stimulated directly after isolation with 4 μg/ml LPS (Sigma–Aldrich, Buchs, Switzerland) or with a combination of 10 μg/ml Concavalin A (Sigma–Aldrich, Buchs, Switzerland) directly after isolation and 50 U/ml IL-2 (Sandoz Pharmaceuticals AG, Cham, Switzerland) 24 and 96 h post isolation. For experiments including IRM stimulation or FIV inoculation of feline PBMCs, the isolated cells were counted using the Sysmex XT 2000 iV (Sysmex, Norderstedt, Germany) as previously described (Weissenbacher et al., 2010) and dispersed in 96-well plates at a density of 3 × 105 cells per well in 100 μl complete RPMI.
Crandell-Rees feline kidney (CrFK) cells and feline embryonic fibroblasts (FEA) were maintained in RPMI supplemented as described above.
2.3. Viruses and in vitro inoculation experiments
FIV GL8 and FeLV-A/Glasgow-1 strains, generous gifts from Dr. M. Hosie and Prof. O. Jarret (University of Glasgow, Great Britain), had been previously propagated and titrated on purified feline PBMCs and FEA cells respectively. The FHV ZH5-04 strain was kindly provided by Veterinaria AG (Zurich, Switzerland), had undergone several passages in CrFK cells and was titrated in these cells before use in the present study. All viruses had been kept at −80 °C for long-term storage.
For in vitro inoculation experiments of adherent cells, 2 × 104 FEA or CrFK cells were seeded in the wells of a 96-well plate and incubated at 37 °C, 5% CO2 for 24 h. FEA cells were then inoculated with 20 tissue culture infectious dose 50 (TCID50) FeLV and CrFK cells with 50 TCID50 of FHV. For in vitro inoculation of feline PBMCs, 3 × 105 cells isolated from one individual adult cat were distributed in wells of a 96-well plate and directly infected with 50 TCID50 of FIV stock virus. The cultures were incubated at 37 °C, 5% CO2 and cells from duplicate wells were harvested at the time intervals depicted in the results of the respective experiments. For each time point, duplicate wells containing uninfected cells were included as unstimulated controls for comparison in gene expression profiles. For harvesting of adherent cells, the supernatant was removed from each well and the cells were directly lysed with 300 μl mRNA lysis buffer (mRNA isolation kit I, Roche Diagnostics). PBMCs from separate wells were first pelleted and subsequently lysed in the same manner. Lysed samples were stored at −80 °C until further analysis. In experiments with FHV and FeLV, the presence of virus-specific nucleic acids in supernatants of infected cells was confirmed by real-time qPCR 24 and 48 h post inoculation respectively using systems previously published (Vogtlin et al., 2002, Tandon et al., 2005). Measurement of FIV provirus integration in PBMCs was carried out by real time PCR as has already been described (Leutenegger et al., 1999).
2.4. IRM stimulation of feline PBMCs
Purified PBMCs from one adult cat were counted and 3 × 105 cells per well were seeded directly after isolation in a 96-well format. Cells were treated with either 144 μg/ml dSLIM™ (Mologen AG, Berlin), 20 μg/ml R-848 or 20 μg/ml Poly IC (Alexis biochemicals, Enzo Life Sciences AG, Lausen, Switzerland) and maintained at 37 °C, 5% CO2. dSLIM™ and Poly IC were solubilized in PBS, R-848 in DMSO, as recommended by the manufacturers. All solutions were diluted in PBS so that cell treatments were equi-volume. Two controls for comparison in gene expression profiles were added in which cells were treated with either an equal volume of PBS, or, for comparison with R-848 stimulation, PBS containing the corresponding concentration of DMSO. After 6, 12, 24 and 48 h, cells were harvested as described above for PBMCs.
2.5. RNA isolation and synthesis of cDNA
For real-time qPCR assay optimization experiments, total RNA was extracted manually from pellets of 2.5 × 106 PBMCs using the RNeasy®Plus Mini Kit (Qiagen AG, Hilden, Germany). Cell lysis and homogenization was thereby carried out using the QIAshredder™ and genomic DNA (gDNA) was removed with the gDNA Eliminator spin column (Qiagen AG) according to the manufacturer's recommendations. For experiments concerning the measurements of cytokine gene expression, mRNA extractions were performed with the mRNA Isolation Kit I and MagNA Pure LC Instrument (Roche Diagnostics) according to the manufacturer's instructions. In both cases, purified RNA was stored at −80 °C until further use.
First strand cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer's instructions, and samples were stored at −20 °C until use for qPCR measurements.
2.6. Real-time qPCR
2.6.1. General conditions
Real-time qPCR was carried out using a Rotor-Gene 6000 real-time rotary analyzer (Corbett, Mortlake, Australia). PCR assays comprised 5 μl of cDNA in a total volume of 25 μl per reaction using the TaqMan® Fast Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA). Thermocycling conditions consisted of an initial denaturation of 20 s at 95 °C, followed by 45 cycles of amplification by melting at 95 °C for 3 s and annealing at 60 °C for 45 s.
Systems for TLR 7 and TLR9 were used as previously described (Ignacio et al., 2005).
2.6.2. Development and validation of novel assays
Primers and probes for each gene listed in Table 3 were designed using Primer Express™ software (versions 2 and 3, Applied Biosystems). The sequences were retrieved from Ensembl (http://www.ensembl.org) and GeneBank (http://www.ncbi.nlm.nih.gov), and oligonucleotides were commercially synthesized (Microsynth, Balgach, Switzerland). All probes were labelled with the reporter dye FAM (6-carboxyfluorescein) at the 5′ end and the quencher dye TAMRA (6-carboxytetramethylrhodamine) at the 3′ end. For genes containing introns, the designed systems covered putative exon–exon junctions, in order to preclude genomic DNA amplification. Primers for each system were first tested for the generation of amplicons of expected length in PCR assays comprising cDNA synthesized from ConA-stimulated PBMCs. The PCR products were subjected to gel electrophoresis on 3% agarose gels, stained with ethidium bromide and visualized using the Chemigenius 2 BioImaging System (Syngene, Cambridge, UK). Only primers indicating appropriate specificity and amplification products of correct length were utilized for further optimization. Primer and probe concentrations for each system were optimized on five-fold serial dilutions of cDNA derived from ConA-stimulated PBMCs. cDNA was diluted in 30 μg/ml salmon sperm DNA (Invitrogen, Basel, Switzerland) and quadruplicates of each dilution were tested simultaneously. Matched forward and reverse primer concentrations of 300, 600 and 900 nM were evaluated first with a probe end concentration of 250 nM. The performance of the assays was further optimized using the best primer concentrations combined with 3 different probe end concentrations, namely 50, 150, and 250 nM. Amplification efficiencies of the newly designed assays were calculated as previously described (Klein et al., 1999) using the following equation:
Table 3.
Real-time qPCR systems for 12 feline genes related to innate immunity.
| Gene | Accession number | Oligo | Sequence | Final conc. (nM) | Amplicon size (bp) | Efficiency |
|---|---|---|---|---|---|---|
| IFNα | AY117395a | Forward | CACGTGACGAACCAGAAGATCTT | 300 | 74 | 1.04 |
| Probe | ACTTCTTCTGCACAGAGGCGTCCTCG | 300 | ||||
| Reverse | GAGGGTGGTGTTCCAAGCA | 250 | ||||
| IFNα3 | AY117393a | Forward | CGTGACGAACCAGGAGATCTTC | 900 | 72 | 1.07 |
| Probe | ACTTCTTCTGCACAGAGGCGTCCTCG | 900 | ||||
| Reverse | GAGGGTGGTGTTCCAAGCA | 250 | ||||
| IFNα7 | AB094996a | Forward | CACGTGACCAACCAGAAGATCTT | 600 | 74 | 1.13 |
| Probe | ACTTCTTCTGCACAGAGGCGTCCTCG | 600 | ||||
| Reverse | GAGGGTGGTGTTCCAAGCA | 150 | ||||
| IFNα14 | AB095003a | Forward | CGTCTGCTCTCTGGGTTGTG | 600 | 77 | 1.00 |
| Probe | CCTGCCTCAGACCCACGGCC | 600 | ||||
| Reverse | ATTTGTCCCAGGAGCGTCAA | 150 | ||||
| IFNβ | AB021707a | Forward | TGGAATGAGACCACTGTTGAGAA | 900 | 69 | 0.97 |
| Probe | CTCCTTGCGACACTCCACTGGCAG | 900 | ||||
| Reverse | GGATCGTTTCCAGGTGTTCCT | 50 | ||||
| IFNω | DQ420220a | Forward | CGCAGGTTAGCAGGGACAAC | 600 | 93 | 1.07 |
| Probe | CGGAGACTGTCCCCTTTCTTGTGCC | 600 | ||||
| Reverse | GGGAAGCGGAAGTCTTTTCTG | 50 | ||||
| Mx | NM002462a | Forward | ACCAGAGCTCGGGCAAGAG | 900 | 96 | 1.00 |
| Probe | CCTTCCCAGAGGCAGCGGTATTGTC | 900 | ||||
| Reverse | TTCAGCACCAGAGGACACCTT | 250 | ||||
| IL-15 | ENSFCAG00000011861b | Forward | AGTGATGTTCATCCCAATTGCA | 400 | 135 | 1.00 |
| Probe | TTCGCTTGAGTCCAAAAATGCGACCA | 400 | ||||
| Reverse | ACCGCTGTTTGCTAGGATAATAATG | 50 | ||||
| TLR3 | ENSFCAT00000006197b | Forward | CAACAACTTAGCACGGCTATGG | 400 | 72 | 1.07 |
| Probe | AACGTGCAAACCCTAGTGGTCCTGTTGATT | 400 | ||||
| Reverse | AATGTGGAGGTGAGAAAGACCC | 80 | ||||
| TLR8 | ENSFCAG00000007243b | Forward | GCTCCAGCTGTTTCCTCATC | 400 | 82 | 1.01 |
| Probe | CCAGTTGCTCGACTTAAGTGG | 400 | ||||
| Reverse | GAGGCTGTTGGTCAAAGAGG | 80 | ||||
| Perforin | EU032539a | Forward | TTCGCGGCCCAGAAGAC | 900 | 79 | 0.98 |
| Probe | TTCCACGACCAGTACAGCTTCAGCACTG | 900 | ||||
| Reverse | GTGAGAGCTGTAGAAGCGACATTC | 250 | ||||
| Granzyme B | EU153367a | Forward | CCACCCAGACTATAATCCAAAGAA | 600 | 77 | 0.97 |
| Probe | CCAACGACATCATGTTACTGCAGCTGG | 600 | ||||
| Reverse | CAGTCAGCTTGGCCTTTTTCA | 250 | ||||
GenBank.
Ensembl (http://www.ensembl.org/index.html).
E = [10(−1/slope)] − 1.
The system for IL-6 had already been described (Taglinger et al., 2008). Optimization experiments for this assay in our laboratory indicated best results with concentrations of 800 nM for forward and reverse primers and 250 nM for the probe, rendering an efficiency of 0.99. We also designed new systems for TLR 3 and TLR8, as we were unable to obtain an adequate efficiency with those previously described (Ignacio et al., 2005).
2.6.3. Relative expression analysis of feline genes
Expression levels of selected genes were calculated using GeNorm version 3.5 (Vandesompele et al., 2002), using β-glucuronidase (GUSB) and tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) as reference genes, under conditions previously validated for the feline species (Kessler et al., 2009). For illustration of relative expressions of the various genes in the figures of the present report, a cut-off of 2-fold up- respectively downregulation measured at least at one time point in each experiment was selected.
2.7. Statistical analysis
For the measurement of the effect of FIV on cytokine expression in vivo, statistical analysis was performed with GraphPad Prism for Windows, Version 3.0 (GraphPad software, San Diego, CA, USA). Normalized cytokine expression factors were tested for statistical differences between samples of week 0 and week 1 post infection using a non-parametric Wilcoxon signed rank test for paired samples.
3. Results
3.1. Development of novel real-time qPCR systems
Primer and probe sequences for real-time qPCR systems of 12 feline genes relevant to innate immune responses against viruses were designed and validated (Table 3). The assay developed for IFNω encompasses the expression of all IFNω subtype genes simultaneously. Due to the sequence dissimilarities in IFNα subtype genes, several systems had to be created for this type I IFN family: a separate assay was designed for IFNα14 while common probes and reverse primers are shared by the IFNα3, IFNα7 and IFNα systems that recognize IFNα subtypes 3, 7 and the remaining subtypes (IFNα1, 2, 5, 6, 8–13), respectively.
Amplicon sizes of the developed real-time qPCR assays ranged from 69 to 135 bp. A single band could be visualized after initial amplification analysis of primers only, when tested with cDNA derived from purified feline PBMCs. Both primer and probe concentrations were optimized and reaction efficiencies were evaluated.
3.2. Stimulation of feline cells with various IRMs
Both immortalized and primary cells of feline origin were stimulated with 3 IRMs of different classes, namely dSLIM™, a covalently closed oligonucleotide containing several unmethylated CpG motifs (Schmidt et al., 2006), R-848 and Poly IC. The concentrations indicating optimal IFNα induction after 12 h in preliminary titration experiments were selected for use in this study (data not shown). As determined by both trypan blue exclusion and cell count measurements at regular intervals after stimulation, no evidence of cellular toxicity could be observed in CrFK cells, FEAs or purified feline PBMCs treated with dSLIM™ or R848 (data not shown). Poly IC, however, at concentrations capable of inducing innate immune responses, induced cell death in all three cell types, and housekeeping gene expression analysis in cell samples treated with this molecule repeatedly indicated an increase of 6–8 Ct values compared to untreated controls.
IRM-treated cells were systematically screened for increased expression of IFNα, Mx and IL-6 as indicators of successful stimulation. Despite measurable basal expression levels of TLRs 3, 7, 8 and 9 in CrFK and FEA cells, all 3 IRMs failed to influence the transcription of genes relevant to innate immunity (data not shown). However, differential gene profiles were observed after treatment of purified feline PBMCs with the immune-stimulating molecules (Fig. 1A–C). dSLIM™ and R-848 induced a 6- and 12-fold upregulation of IFNα expression within 12 h of stimulation. However, no increase in transcription of Mx could be measured in PBMCs treated with these two molecules. In cells pulsed with Poly IC, levels of IFNα expression could not be accurately measured due to both low basal expression levels of this cytokine and cell death induced by this IRM. Nevertheless, Poly IC induced a 28-fold increase in Mx expression that could be measured already 6 h post stimulation. Upregulation of IFNα expression was concordant with that of IL-6 after stimulation of PBMCs with Poly IC and R-848. In contrast, treatment of these cells with dSLIM™ induced significant downregulation of IL-6 expression. Finally, among all 3 IRMs, only dSLIM™ was found to modulate TLR expression, indicating a 6-fold increase of TLR9 within 12 h of stimulation.
Fig. 1.
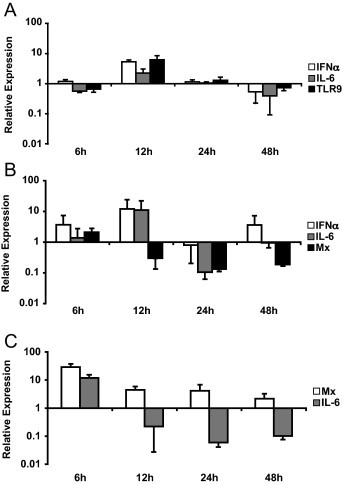
Kinetics of innate cytokine expression in feline PBMCs after IRM stimulation. Feline PBMCs of one cat were incubated for 6, 12, 24 and 48 h with (A) 144 μg/ml dSLIM™, (B) 20 μg/ml R-848 and (C) 20 μg/ml Poly IC. mRNA expression of the indicated genes was measured by real-time qPCR and normalized to expression of two feline housekeeping genes (GUSB and YWHAZ). Depicted expression factors at each time point represent the ratio of measured mRNA levels in the IRM-stimulated samples compared to samples treated with a negative control. Only genes indicating at least a 2-fold modulation in their expression at any time point are shown. Data represent means of duplicate reactions for each time point. Standard deviations are shown as an indication for reproducibility.
3.3. Early immune responses of feline cell lines after inoculation with feline viruses
The induction of host cell immune responses by FHV and FeLV were assessed in vitro on CrFK cells and FEAs respectively at regular time intervals early post infection. Productive infection of the cells was confirmed by real-time PCR of cell culture supernatants 24 and 48 h post inoculation for FHV and FeLV respectively (data not shown). Modulations in relative expressions of IFNα, IFNβ, Mx and IL-6 were utilized as parameters for immune responses by the cells. Progressively increasing levels of IFNα, IFNβ and IL-6 were measured after inoculation of CrFK cells with FHV, with highest levels corresponding to, respectively, 37-, 45- and 23-fold upregulation in gene expression measured after 24 h. No changes in Mx expression were observed (Fig. 2A). In contrast, FEA cells indicated the most potent antiviral immune response just 6 h after inoculation with FeLV, and transcription levels of all analyzed genes decreased progressively thereafter. Within 6 h post inoculation, increases of 15-fold in IFNα expression, 29-fold in IFNβ expression and 4-fold in IL-6 expression were measured. Presence of Mx mRNA was proportional to levels of IFNα and IFNβ expression, with highest levels also noted 6 h post inoculation (Fig. 2B).
Fig. 2.
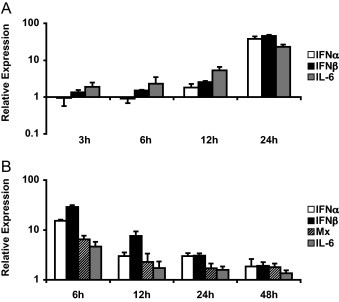
Innate immune responses to FHV and FeLV in vitro. (A) CrFK cells were inoculated with 50 TCID50 of FHV and (B) FEA cells were inoculated with 20 TCID50 of FeLV. mRNA expression of the indicated genes was measured by real-time qPCR and normalized to expression of two feline housekeeping genes (GUSB and YWHAZ) at time points indicated for each experiment. Depicted expression factors at each time point represent the ratio of measured mRNA levels in the infected samples compared to samples of non-infected cells. Only genes indicating at least a 2-fold modulation in their expression at any time point are shown. Data represent means of duplicate reactions for each time point. Standard deviations are shown as an indication for reproducibility.
3.4. Evaluation of innate immune response parameters in feline PBMCs after in vitro infection with FIV
The influence on the expression of 14 genes including receptors and cytokines relevant to innate immunity was tested in feline PBMCs at regular time points after inoculation with FIV. Effective integration of FIV provirus was confirmed by real-time PCR 48 h after infection of the cells (data not shown). Modulations in the expression of the tested factors are depicted in Fig. 3 . All genes tested were influenced within the first 48 h of stimulation, with the exception of TLR9 (not shown). The strongest response was measured 12 h after infection, with substantial modulation at this time point in mRNA levels of 9 out of 10 cytokines tested. Type I IFN genes were upregulated as of 6 h post inoculation. Peak levels of 374-fold and 121-fold inductions in gene expression of IFNα and IFNω were achieved at 12 h post infection, while strongest induction of IFNβ, namely by a factor of 114, was measured 24 h after FIV inoculation. Although maximal gene expression levels were slightly lower for the individual IFNα subtypes tested (65-fold, 67-fold and 24-fold for IFNs 3, 7 and 14 respectively), a similar pattern of stimulation was observed, with highest expression 12 h after infection. Mx and IL-15 genes were induced in a manner proportional to the type I IFN genes at all time points measured, with highest increases at 12 h post inoculation of 38-fold and 7-fold, respectively. During the first 48 h, FIV infection downregulated IL-6 expression attaining lowest levels within 6 h after inoculation, when a 60-fold decrease in expression was observed. Genes encoding for contents of NK cell cytotoxic granules appeared only marginally affected when their expression was measured in PBMCs. Both perforin and granzyme B mRNA levels were maximally increased 3 h post infection by 1.8-fold and 2-fold, respectively. Similarly, the expression of TLR genes in PBMCs was only slightly influenced by FIV infection. Transcription of TLRs 3, 7 and 8 was however induced in a delayed manner when compared to the cytokines, attaining maximal levels 24 h after infection with induction factors ranging from 2 to 2.6.
Fig. 3.
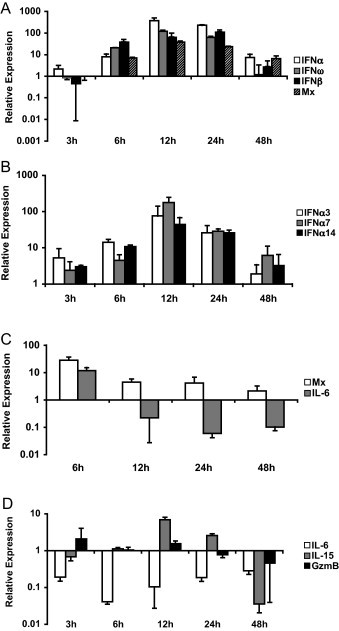
Innate immune responses to FIV infection of feline PBMCs in vitro. PBMCs of one cat were inoculated with 50 TCID50 of FIV. mRNA expression of the indicated genes was measured 3, 6, 12, 24 and 48 h after inoculation and normalized to expression of two feline housekeeping genes (GUSB and YWHAZ) at each time point. Depicted expression factors at each time point represent the ratio of measured mRNA levels in the IRM-stimulated samples compared to samples treated with a negative control. Only genes indicating at least a 2-fold modulation in their expression at any time point are shown. Data represent means of duplicate reactions for each time point. Standard deviations are shown as an indication for reproducibility. GzmB = Granzyme B.
3.5. Measurement of type I IFN in cats after FIV challenge in vivo
10 SPF cats were subjected to FIV infection with the highly virulent GL8 strain. Expression levels of feline IFNα were compared in whole blood samples collected from the cats immediately before and 1 week after FIV challenge infection. 9 out of 10 cats displayed higher IFNα expression after infection. An average of 2.4-fold induction in expression of this cytokine could be measured in the cats after infection (p = 0.0059) (Fig. 4 ).
Fig. 4.
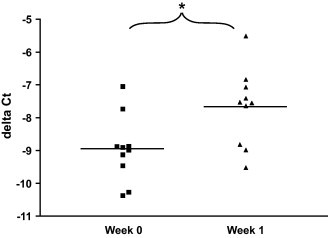
Measurement of IFNα expression levels in blood 1 week after FIV challenge infection in vivo. mRNA expression of IFNα and two housekeeping genes (GUSB and YWHAZ) from whole blood samples of 10 cats collected before and 1 week after challenge infection with FIV was measured by real-time qPCR. DeltaCt values depicted were calculated by substracting the average of 45-Ct values for two housekeeping genes (GUSB and YWHAZ) from 45-Ct values for IFNα of the corresponding sample. *p = 0.0059.
4. Discussion
Innate immune responses to invading pathogens play a key role in the outcome of infection. Over the course of time, mammalian hosts have developed the capacity to counterbalance virus attacks through sensing of viral signatures by specialized immune cells and immediate activation of the type I IFN system. Within minutes to hours, this family of antiviral cytokines initiates an explosion of potent defence mechanisms, leading to the suppression of viral replication and protection of the host. These intrinsic immune responses remain poorly understood in felids, a species that seems particularly sensitive to viral infections, with sporadic viral outbreaks compromising endangered populations (Evermann et al., 1988, Roelke-Parker et al., 1996, Cunningham et al., 2008, Meli et al., 2010). Until now, the appropriate tools for characterizing innate immune mechanisms in this species were unavailable. In order to better understand the early host-pathogen interactions occurring in feline viral infections, we describe herein the development of 12 real-time qPCR systems to measure the expression of feline genes related to innate antiviral defence mechanisms. Through a series of experiments, we demonstrate the possibility of monitoring key events in innate immune responses both in vitro and in vivo. We further demonstrate the initiation of antiviral responses in feline immune cells upon stimulation with various IRMs.
The strength, extent and kinetics of immune responses elaborated by feline cells upon viral inoculation in vitro were evaluated. In these experiments, expression levels of IFNα and IL-6 were systematically analyzed as indicators for the initiation of innate immune mechanisms. As Mx expression is strictly regulated by type I IFN (Holzinger et al., 2007), biological functionality of the induced immune responses was assessed by simultaneous analysis of this factor. In all models of infection used in this study, we observed the characteristic transient expression of type I IFN genes, with a duration ranging from 6 to 12 h in CrFK cells and FEAs inoculated with FHV and FeLV respectively to 36 h in PBMCs infected with FIV. The presence of specialized cells for the production of type I IFNs in the blood is most likely linked to the longer expression of these innate cytokines in PBMCs (Colonna et al., 2004). Moreover, IFNs α, β and ω were expressed at differential levels in feline cells upon infection; while PBMCs heavily upregulated expression of IFNα and IFNω genes, cell lines retaining epithelial and fibroblast properties preferentially increased production of IFNβ mRNA levels. Time points at which highest expression of antiviral genes was measured also greatly varied with cell type and virus. Although FEA cells responded to presence of FeLV within 6 h, induction of innate immune gene expression could only be observed after 24 h following FHV infection in CrFK cells. Peak antiviral response in PBMCs was measured 12 h after infection, with evidence for progressive development and rapid decrease of the response. Further experiments are necessary to determine the significance of individual properties of cells and tissues as well as virus-related factors such as source and infectious dose in these observations.
Interestingly, although high levels of type I IFN expression were observed 24 h after FHV infection of CrFK cells, no induction of Mx was measured, indicating that this virus may have evolved mechanisms to interrupt IFN-induced host defence mechanisms in infected cells. Indeed, many viruses have evolutionarily acquired sophisticated strategies to counteract the IFN system (Katze et al., 2002). Various members of the herpesvirus family have been reported to directly inhibit the proteins that mediate the antiviral state (Elia et al., 1996), interfere with the expression of signaling molecules downstream of the IFNAR (Miller et al., 1998), or hypothetically support the disassembly of nuclear structures to get rid of antiviral components (Van Sant et al., 2001). These mechanisms have been linked to persistence of infections induced by some viruses of this family. Although there is no evidence for a specific viral inhibitor of Mx proteins so far, viruses can subvert the Mx system through exceptionally fast growth in host cells (Haller et al., 2007) or by affecting signal transduction upstream of IFN-regulated gene transcription. The presence of other antiviral factors in FHV-infected feline cells could shed light on possible mechanisms linked to these findings.
FIV can readily infect feline immune cells in vitro, offering a system that enables the study of innate antiviral immune reactions in a heterogeneous population of cells that actively take part in complex networks of immune interactions both in vitro and in vivo. Consequently, we studied the effect of viral inoculation in PBMCs on the expression of all cytokines and receptors for which we had developed real-time qPCR systems. High levels of type I IFN expression were measured in response to infection, again most probably linked to the activation of professional dendritic cells producing extraordinary amounts of these cytokines (Colonna et al., 2004). Similar expression patterns were noted for all IFNα gene subtypes analyzed. Lower induction rates of IFNα3, 7 and 14 subtypes are most likely due to the measurement of individual genes versus the simultaneous quantification of many subtypes with the real-time assay developed for the remaining IFNα subtypes. Although the induction of type I IFN genes largely overlap, the array of IFNα and IFNω subtypes produced upon stimulation seems to be determined by cell-specific levels of transcription factors IRF3 and IRF7 (Sato et al., 2000). Furthermore, the extent of biological activity conferred by each individual protein depends on a variety of factors. Thus, the differential affinity of each subtype to the subunits of the IFNAR, the surface expression level of IFNAR1 and IFNAR2 in target cells, as well as the lifetime and stability of the ligand–receptor complex all play a role in the extent of the host response to infection (Genin et al., 2009). Different pathogens may influence, in this way, the phenotype of the developing immune response by shaping the profile of IFN subtypes induced (Foster et al., 2004). As previously shown for the human IFN system (Loseke et al., 2003), the expression analysis of individual subtypes by real-time qPCR can provide valuable information on host innate immune responses to pathogens. We have initiated the development of appropriate tools to study these mechanisms in more detail in feline species.
Infection of feline PBMCs with FIV led to a substantial downregulation of the expression of the proinflammatory cytokine gene IL-6. These findings are reminiscent of previous experiments in which a 100 to 100,000-fold reduction of IL-6 transcription was observed in monocytes from FIV-infected cats (Kipar et al., 2004). Downregulation of this cytokine was reported to be more intense when infection of the monocyte fraction itself could not be demonstrated (Kipar et al., 2004). Hypothetically, factors liberated by primarily infected lymphocytes could affect function and immune responses of monocytic cells, the main producers of IL-6 in blood. Thus, further investigations on PBMC subpopulations are required to determine the role of preferential cell tropism of certain viral strains on measured cytokine responses.
Our results further indicate a type I IFN-regulated pattern of NK cell stimulator IL-15 gene expression, supporting the previously described role of these IFNs in stimulation of NK cell activity (Vujanovic et al., 2010). In contrast, effectors of NK cytotoxicity were only marginally induced as measured in PBMCs acutely infected with FIV in this study. Although low levels of increase in perforin and granzyme B mRNA production have been described upon stark stimulation, preformed transcripts for these proteins have been detected in resting NK cells (Fehniger et al., 2007). Gene expression analysis in separate immune cell populations as well as specific cytotoxicity assays would confer additional information on the biological value of our observations for these NK cell related cytokines.
Analysis of the expression of TLR genes demonstrated slight enhancement in mRNA levels of TLRs 3, 7 and 8 during the peak time point of innate immune response against FIV. The ssRNA phenotype of this virus coupled with the presence of dsRNA after reverse transcription during intracellular replication support the requirement for selectively higher levels of these TLRs upon infection. The relatively low levels of induction observed could be explained by the presence of a pool of receptors in the endoplasmatic reticulum that is actively transported to the endosomes following stimulation of the cell (Latz et al., 2004). Although upregulation of TLR9 after FIV inoculation of CrFK cells has been reported (Ignacio et al., 2005), our observations regarding mRNA expression of this TLR gene in acutely FIV-infected PBMCs do not corroborate with the results previously published.
To our knowledge, this is the first report showing successful stimulation of innate mechanisms in feline immune cells by various IRMs. Both kinetics and potency of responses could be measured. Since mRNA expression of proinflammatory cytokine gene IL-6 had been previously used as readout for stimulation through TLRs in pancreatic islet cells (Franchini et al., 2010), we included this cytokine as comparative indicator for biological activity of TLR agonist. Our results indicate that IFNα is a more reliable marker for TLR stimulation in immune cells due to higher stability in expression over time and apparent higher expression of these cytokines.
Both R-848 and dSLIM™ could efficiently enhance IFNα expression within 12 h. This level of stimulation was likely insufficient to induce measurable increase in Mx expression, as induction of the gene encoding for this antiviral factor was repeatedly 5–10 times lower than that of IFNα in our experiments.
dSLIM™ is a non-coding DNA molecule that contains several unmethylated CpG motifs and possesses the broad spectrum immunomodulatory properties of Class C CpG ODNs (Schmidt et al., 2006). In vitro and in vivo studies have demonstrated immunomodulatory effects and safety of dSLIM™ both in humans and mice (Wittig et al., 2001, Kochling et al., 2003). Moreover, this molecule has exhibited protective immune stimulation in human colon cancer and is currently undergoing second phase clinical trials (Weihrauch et al., 2005). In addition to positively influencing IFNα expression in feline immune cells, dSLIM™ was the only tested IRM that affected TLR mRNA levels, leading to a 6-fold upregulation of TLR9. Further experiments will determine whether such properties could support combinatorial use of this molecule to enhance effects of other IRMs.
Poly IC was toxic to feline cells already at the lowest concentrations capable of inducing innate immune responses. Toxicity of this product has long been acknowledged and studied both in vitro and in vivo (Homan et al., 1972, Lv and Bao, 2009). Mechanisms by which cell death is induced remain unclear, however it has been suggested that pathways leading to the induction of type I IFN genes are uncoupled from apoptotic pathways (Han et al., 2004). We measured high induction of Mx in Poly IC-treated PBMCs after just 6 h post inoculation. While the possibility that Mx proteins fulfil a cellular function involved in cellular trafficking and/or in stress responses has been postulated (Horisberger, 1992) it remains more likely that presence of this antiviral factor is indicative of IFNα function.
Finally, the experiments here described were carried out with PBMCs from only one cat in order to ensure comparability of both the effects of infection and IRMs at different time points and the relation between expression levels of the cytokines tested. Admittedly, innate immune responses may vary considerably between individuals of an outbred species; however, when purified PBMCs from 8 adult SPF cats were previously utilized in similar smaller experiments, only slight variation was observed in their response to IRM stimulation and FIV infection (data not shown). Studies with cells from cats in the field would most likely give relevant information about individuality in responses after IRM stimulation or infection.
Altogether, we describe the development of tools to measure antiviral innate immune responses in the cat and show their successful utilization in the context of various virus infections in vitro and in vivo. Moreover, we provide initial evidence of the possibility of inducing innate immune responses in feline immune cells by stimulation with various IRMs. Future plans include screening of a vast array of IRMs for optimal manipulation of the feline innate immune system and enhancing the resistance of felids to viral infection.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
This work was supported by the Research Commission and the Young Academic Support Committee of the University of Zürich.
Laboratory work was performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich. R.H. L. is the recipient of a professorship by the Swiss National Science Foundation (PP00P3-119136).
References
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Baldwin S.L., Powell T.D., Sellins K.S., Radecki S.V., Cohen J.J., Milhausen M.J. The biological effects of five feline IFN-alpha subtypes. Vet Immunol Immunopathol. 2004;99:153–167. doi: 10.1016/j.vetimm.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Bekisz J., Schmeisser H., Hernandez J., Goldman N.D., Zoon K.C. Human interferons alpha, beta and omega. Growth Factors. 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- Bracklein T., Theise S., Metzler A., Spiess B.M., Richter M. Activity of feline interferon-omega after ocular or oral administration in cats as indicated by Mx protein expression in conjunctival and white blood cells. Am. J. Vet. Res. 2006;67:1025–1032. doi: 10.2460/ajvr.67.6.1025. [DOI] [PubMed] [Google Scholar]
- Collado V.M., Gomez-Lucia E., Tejerizo G., Miro G., Escolar E., Martin S., Domenech A. Effect of type I interferons on the expression of feline leukaemia virus. Vet Microbiol. 2007;123:180–186. doi: 10.1016/j.vetmic.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Colonna M., Trinchieri G., Liu Y.J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- de Mari K., Maynard L., Sanquer A., Lebreux B., Eun H.M. Therapeutic effects of recombinant feline interferon-omega on feline leukemia virus (FeLV)-infected and FeLV/feline immunodeficiency virus (FIV)-coinfected symptomatic cats. J Vet Intern Med. 2004;18:477–482. doi: 10.1892/0891-6640(2004)18<477:teorfi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Cunningham M.W., Brown M.A., Shindle D.B., Terrell S.P., Hayes K.A., Ferree B.C., McBride R.T., Blankenship E.L., Jansen D., Citino S.B., Roelke M.E., Kiltie R.A., Troyer J.L., O’Brien S.J. Epizootiology and management of feline leukemia virus in the Florida puma. J. Wildl. Dis. 2008;44:537–552. doi: 10.7589/0090-3558-44.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G.A., Bernales J.A., Pedersen N.C. Effect of feline immunodeficiency virus on cytokine response to Listeria monocytogenes in vivo. Vet. Immunol. Immunopathol. 1998;65:125–138. doi: 10.1016/s0165-2427(98)00148-2. [DOI] [PubMed] [Google Scholar]
- Dean G.A., LaVoy A., Yearley J., Stanton C. Cytokine modulation of the innate immune response in feline immunodeficiency virus-infected cats. J. Infect. Dis. 2006;193:1520–1527. doi: 10.1086/503873. [DOI] [PubMed] [Google Scholar]
- Elia A., Laing K.G., Schofield A., Tilleray V.J., Clemens M.J. Regulation of the double-stranded RNA-dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein-Barr virus genome. Nucleic Acids Res. 1996;24:4471–4478. doi: 10.1093/nar/24.22.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evermann J.F., Heeney J.L., Roelke M.E., McKeirnan A.J., O’Brien S.J. Biological and pathological consequences of feline infectious peritonitis virus infection in the cheetah. Arch. Virol. 1988;102:155–171. doi: 10.1007/BF01310822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger T.A., Cai S.F., Cao X., Bredemeyer A.J., Presti R.M., French A.R., Ley T.J. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Foster G.R., Masri S.H., David R., Jones M., Datta A., Lombardi G., Runkell L., de Dios C., Sizing I., James M.J., Marelli-Berg F.M. IFN-alpha subtypes differentially affect human T cell motility. J. Immunol. 2004;173:1663–1670. doi: 10.4049/jimmunol.173.3.1663. [DOI] [PubMed] [Google Scholar]
- Franchini M., Zini E., Osto M., Jablonski K., Kaufmann K., Lutz T.A., Reusch C.E., Ackermann M. Feline pancreatic islet-like clusters and insulin producing cells express functional Toll-like receptors (TLRs) Vet. Immunol. Immunopathol. 2010 doi: 10.1016/j.vetimm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Genin P., Vaccaro A., Civas A. The role of differential expression of human interferon—a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20:283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Gutzwiller M.E., Brachelente C., Taglinger K., Suter M.M., Weissenbock H., Roosje P.J. Feline herpes dermatitis treated with interferon omega. Vet Dermatol. 2007;18:50–54. doi: 10.1111/j.1365-3164.2007.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid C., Kaps S., Gonczi E., Hassig M., Metzler A., Spiess B.M., Richter M. Pretreatment with feline interferon omega and the course of subsequent infection with feline herpesvirus in cats. Vet Ophthalmol. 2007;10:278–284. doi: 10.1111/j.1463-5224.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- Haller O., Kochs G., Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman J.A., Ogasawara K., Lanier L.L. NK cells in innate immunity. Curr. Opin. Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Han K.J., Su X., Xu L.G., Bin L.H., Zhang J., Shu H.B. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J. Biol. Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- Holzinger D., Jorns C., Stertz S., Boisson-Dupuis S., Thimme R., Weidmann M., Casanova J.L., Haller O., Kochs G. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J. Virol. 2007;81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan E.R., Zendzian R.P., Schott L.D., Levy H.B., Adamson R.H. Studies on poly I:C toxicity in experimental animals. Toxicol. Appl. Pharmacol. 1972;23:579–588. doi: 10.1016/0041-008x(72)90098-1. [DOI] [PubMed] [Google Scholar]
- Horisberger M.A. Interferon-induced human protein MxA is a GTPase which binds transiently to cellular proteins. J. Virol. 1992;66:4705–4709. doi: 10.1128/jvi.66.8.4705-4709.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger M.A., Schrenk R., Staiger S., Leyvraz A.R., Martinod S. Induction of Mx-related protein in cat peripheral blood mononuclear cells after administration of recombinant human interferon hybrid. Antiviral Res. 1990;13:53–59. doi: 10.1016/0166-3542(90)90021-x. [DOI] [PubMed] [Google Scholar]
- Ignacio G., Nordone S., Howard K.E., Dean G.A. Toll-like receptor expression in feline lymphoid tissues. Vet. Immunol. Immunopathol. 2005;106:229–237. doi: 10.1016/j.vetimm.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Zamanian-Daryoush M., Nie H., Silva A.M., Williams B.R., Li X. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J. Biol. Chem. 2003;278:16713–16719. doi: 10.1074/jbc.M300562200. [DOI] [PubMed] [Google Scholar]
- Jurk M., Heil F., Vollmer J., Schetter C., Krieg A.M., Wagner H., Lipford G., Bauer S. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- Katze M.G., He Y., Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kessler Y., Helfer-Hungerbuehler A.K., Cattori V., Meli M.L., Zellweger B., Ossent P., Riond B., Reusch C.E., Lutz H., Hofmann-Lehmann R. Quantitative TaqMan real-time PCR assays for gene expression normalisation in feline tissues. BMC Mol. Biol. 2009;10:106. doi: 10.1186/1471-2199-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Boretti F.S., Meli M.M., Failing K., Reinacher M., Lutz H. Reduced constitutive cytokine transcription in isolated monocytes of clinically healthy cats, infected with an FIV strain of low pathogenicity. Vet. Immunol. Immunopathol. 2004;98:215–221. doi: 10.1016/j.vetimm.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Klein D., Janda P., Steinborn R., Muller M., Salmons B., Gunzburg W.H. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis. 1999;20:291–299. doi: 10.1002/(SICI)1522-2683(19990201)20:2<291::AID-ELPS291>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kochling J., Konig-Merediz S.A., Stripecke R., Buchwald D., Korte A., Von Einsiedel H.G., Sack F., Henze G., Seeger K., Wittig B., Schmidt M. Protection of mice against Philadelphia chromosome-positive acute lymphoblastic leukemia by cell-based vaccination using nonviral, minimalistic expression vectors and immunomodulatory oligonucleotides. Clin. Cancer Res. 2003;9:3142–3149. [PubMed] [Google Scholar]
- Latz E., Schoenemeyer A., Visintin A., Fitzgerald K.A., Monks B.G., Knetter C.F., Lien E., Nilsen N.J., Espevik T., Golenbock D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Leutenegger C.M., Klein D., Hofmann-Lehmann R., Mislin C., Hummel U., Boni J., Boretti F., Guenzburg W.H., Lutz H. Rapid feline immunodeficiency virus provirus quantitation by polymerase chain reaction using the TaqMan fluorogenic real-time detection system. J. Virol. Methods. 1999;78:105–116. doi: 10.1016/s0166-0934(98)00166-9. [DOI] [PubMed] [Google Scholar]
- Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., Garcia-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loseke S., Grage-Griebenow E., Wagner A., Gehlhar K., Bufe A. Differential expression of IFN-alpha subtypes in human PBMC: evaluation of novel real-time PCR assays. J. Immunol. Methods. 2003;276:207–222. doi: 10.1016/s0022-1759(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Lv Y., Bao E. Apoptosis induced in chicken embryo fibroblasts in vitro by a polyinosinic:polycytidylic acid copolymer. Toxicol. In Vitro. 2009;23:1360–1364. doi: 10.1016/j.tiv.2009.06.026. [DOI] [PubMed] [Google Scholar]
- Martin V., Najbar W., Gueguen S., Grousson D., Eun H.M., Lebreux B., Aubert A. Treatment of canine parvoviral enteritis with interferon-omega in a placebo-controlled challenge trial. Vet Microbiol. 2002;89:115–127. doi: 10.1016/s0378-1135(02)00173-6. [DOI] [PubMed] [Google Scholar]
- Meli M.L., Cattori V., Martinez F., Lopez G., Vargas A., Palomares F., Lopez-Bao J.V., Hofmann-Lehmann R., Lutz H. Feline leukemia virus infection: a threat for the survival of the critically endangered Iberian lynx (Lynx pardinus) Vet. Immunol. Immunopathol. 2010;134:61–67. doi: 10.1016/j.vetimm.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T., Stockfleth E. Clinical investigations of Toll-like receptor agonists. Expert Opin. Investig. Drugs. 2008;17:1051–1065. doi: 10.1517/13543784.17.7.1051. [DOI] [PubMed] [Google Scholar]
- Miller D.M., Rahill B.M., Boss J.M., Lairmore M.D., Durbin J.E., Waldman J.W., Sedmak D.D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa T., Ishiwata K., Kajimoto T. Feline interferon-omega treatment on canine parvovirus infection. Vet Microbiol. 1999;69:51–53. doi: 10.1016/s0378-1135(99)00087-5. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Nakatani H., Yoshida M. Inhibitory effects of recombinant feline interferon on the replication of feline enteropathogenic viruses in vitro. Vet Microbiol. 1994;39:145–152. doi: 10.1016/0378-1135(94)90095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A., Taira O., Ishikawa M., Hiramatsu K., Hohdatsu T., Koyama H., Arai S., Sato H., Nakano K., Maehara N. Cloning of cDNAs encoding multiple subtypes of feline interferon-alpha from the feline epitherial cell line. J Vet Med Sci. 2004;66:725–728. doi: 10.1292/jvms.66.725. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Sudo T., Matsuda S., Yanai A. Molecular cloning of feline interferon cDNA by direct expression. Biosci. Biotechnol. Biochem. 1992;56:211–214. doi: 10.1271/bbb.56.211. [DOI] [PubMed] [Google Scholar]
- Ohe K., Takahashi T., Hara D., Hara M. Sensitivity of FCV to recombinant feline interferon (rFeIFN) Vet Res Commun. 2008;32:167–174. doi: 10.1007/s11259-007-9019-5. [DOI] [PubMed] [Google Scholar]
- Paltrinieri S., Crippa A., Comerio T., Angioletti A., Roccabianca P. Evaluation of inflammation and immunity in cats with spontaneous parvovirus infection: consequences of recombinant feline interferon-omega administration. Vet Immunol Immunopathol. 2007;118:68–74. doi: 10.1016/j.vetimm.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D., Fouchet D., Bahi-Jaber N., Poulet H., Guiserix M., Natoli E., Sauvage F. When domestic cat (Felis silvestris catus) population structures interact with their viruses. C. R. Biol. 2009;332:321–328. doi: 10.1016/j.crvi.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke-Parker M.E., Munson L., Packer C., Kock R., Cleaveland S., Carpenter M., O’Brien S.J., Pospischil A., Hofmann-Lehmann R., Lutz H., Mwamengele G.L., Mgasa M.N., Machange G.A., Summers B.A., Appel M.J. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;379:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T., Ueda Y., Sato M., Yanai A. Feline interferon production in silkworm by recombinant baculovirus. J Vet Med Sci. 1992;54:563–565. doi: 10.1292/jvms.54.563. [DOI] [PubMed] [Google Scholar]
- Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Schindler C., Levy D.E., Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Anton K., Nordhaus C., Junghans C., Wittig B., Worm M. Cytokine and Ig-production by CG-containing sequences with phosphorodiester backbone and dumbbell-shape. Allergy. 2006;61:56–63. doi: 10.1111/j.1398-9995.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- Seya T., Matsumoto M. The extrinsic RNA-sensing pathway for adjuvant immunotherapy of cancer. Cancer Immunol. Immunother. 2009;58:1175–1184. doi: 10.1007/s00262-008-0652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeck N., Hurley D.J., Garcia M., Greene C.E., Kostlin R.G., Moore P.A., Dietrich U.M. Effects of human recombinant alpha-2b interferon and feline recombinant omega interferon on in vitro replication of feline herpesvirus-1. Am J Vet Res. 2006;67:1406–1411. doi: 10.2460/ajvr.67.8.1406. [DOI] [PubMed] [Google Scholar]
- Siren J., Pirhonen J., Julkunen I., Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J. Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- Southerden P., Gorrel C. Treatment of a case of refractory feline chronic gingivostomatitis with feline recombinant interferon omega. J Small Anim Pract. 2007;48:104–106. doi: 10.1111/j.1748-5827.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- Tabeta K., Georgel P., Janssen E., Du X., Hoebe K., Crozat K., Mudd S., Shamel L., Sovath S., Goode J., Alexopoulou L., Flavell R.A., Beutler B. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U S A. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglinger K., Van Nguyen N., Helps C.R., Day M.J., Foster A.P. Quantitative real-time RT-PCR measurement of cytokine mRNA expression in the skin of normal cats and cats with allergic skin disease. Vet. Immunol. Immunopathol. 2008;122:216–230. doi: 10.1016/j.vetimm.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Taira O., Suzuki M., Takeuchi Y., Aramaki Y., Sakurai I., Watanabe T., Motokawa K., Arai S., Sato H., Maehara N. Expression of feline interferon-alpha subtypes in Esherichia coli, and their antiviral activity and animal species specificity. J Vet Med Sci. 2005;67:543–545. doi: 10.1292/jvms.67.543. [DOI] [PubMed] [Google Scholar]
- Tanabe T., Shimoda M., Soeno T., Suzuki M., Tajima M., Sato H. Molecular cloning and sequence analysis of feline interferon-stimulated gene 15. Vet. Immunol. Immunopathol. 2008;126:20–26. doi: 10.1016/j.vetimm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Tanabe T., Yamamoto J.K. Feline immunodeficiency virus lacks sensitivity to the antiviral activity of feline IFN-gamma. J Interferon Cytokine Res. 2001;21:1039–1046. doi: 10.1089/107999001317205169. [DOI] [PubMed] [Google Scholar]
- Tandon R., Cattori V., Gomes-Keller M.A., Meli M.L., Golder M.C., Lutz H., Hofmann-Lehmann R. Quantitation of feline leukaemia virus viral and proviral loads by TaqMan real-time polymerase chain reaction. J. Virol. Methods. 2005;130:124–132. doi: 10.1016/j.jviromet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Ueda Y., Sakurai T., Kasama K., Satoh Y., Atsumi K., Hanawa S., Uchino T., Yanai A. Pharmacokinetic properties of recombinant feline interferon and its stimulatory effect on 2′,5′-oligoadenylate synthetase activity in the cat. J. Vet. Med. Sci. 1993;55:1–6. doi: 10.1292/jvms.55.1. [DOI] [PubMed] [Google Scholar]
- Ueda Y., Sakurai T., Yanai A. Homogeneous production of feline interferon in silkworm by replacing single amino acid code in signal peptide region in recombinant baculovirus and characterization of the product. J. Vet. Med. Sci. 1993;55:251–258. doi: 10.1292/jvms.55.251. [DOI] [PubMed] [Google Scholar]
- Uze G., Schreiber G., Piehler J., Pellegrini S. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- Vaidya S.A., Cheng G. Toll-like receptors and innate antiviral responses. Curr. Opin. Immunol. 2003;15:402–407. doi: 10.1016/s0952-7915(03)00070-0. [DOI] [PubMed] [Google Scholar]
- Van Sant C., Hagglund R., Lopez P., Roizman B. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U S A. 2001;98:8815–8820. doi: 10.1073/pnas.161283098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtlin A., Fraefel C., Albini S., Leutenegger C.M., Schraner E., Spiess B., Lutz H., Ackermann M. Quantification of feline herpesvirus 1 DNA in ocular fluid samples of clinically diseased cats by real-time TaqMan PCR. J. Clin. Microbiol. 2002;40:519–523. doi: 10.1128/JCM.40.2.519-523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer J., Weeratna R., Payette P., Jurk M., Schetter C., Laucht M., Wader T., Tluk S., Liu M., Davis H.L., Krieg A.M. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur. J. Immunol. 2004;34:251–262. doi: 10.1002/eji.200324032. [DOI] [PubMed] [Google Scholar]
- Vujanovic L., Szymkowski D.E., Alber S., Watkins S.C., Vujanovic N.L., Butterfield L.H. Virally infected and matured human dendritic cells activate natural killer cells via cooperative activity of plasma membrane-bound TNF and IL-15. Blood. 2010;116:575–583. doi: 10.1182/blood-2009-08-240325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.L., Ahonen C.L., Couture A.M., Gibson S.J., Miller R.L., Smith R.M., Reiter M.J., Vasilakos J.P., Tomai M.A. Modulation of TH1 and TH2 cytokine production with the immune response modifiers, R-848 and imiquimod. Cell. Immunol. 1999;191:10–19. doi: 10.1006/cimm.1998.1406. [DOI] [PubMed] [Google Scholar]
- Weihrauch M.R., Ansen S., Jurkiewicz E., Geisen C., Xia Z., Anderson K.S., Gracien E., Schmidt M., Wittig B., Diehl V., Wolf J., Bohlen H., Nadler L.M. Phase I/II combined chemoimmunotherapy with carcinoembryonic antigen-derived HLA-A2-restricted CAP-1 peptide and irinotecan, 5-fluorouracil, and leucovorin in patients with primary metastatic colorectal cancer. Clin. Cancer Res. 2005;11:5993–6001. doi: 10.1158/1078-0432.CCR-05-0018. [DOI] [PubMed] [Google Scholar]
- Weissenbacher S., Riond B., Hofmann-Lehmann R., Lutz H. Evaluation of a novel haematology analyser for use with feline blood. Vet J. 2010;187:381–387. doi: 10.1016/j.tvjl.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Wittig B., Marten A., Dorbic T., Weineck S., Min H., Niemitz S., Trojaneck B., Flieger D., Kruopis S., Albers A., Loffel J., Neubauer A., Albers P., Muller S., Sauerbruch T., Bieber T., Huhn D., Schmidt-Wolf I.G. Therapeutic vaccination against metastatic carcinoma by expression-modulated and immunomodified autologous tumor cells: a first clinical phase I/II trial. Hum. Gene Ther. 2001;12:267–278. doi: 10.1089/10430340150218404. [DOI] [PubMed] [Google Scholar]
- Wonderling R., Powell T., Baldwin S., Morales T., Snyder S., Keiser K., Hunter S., Best E., McDermott M.J., Milhausen M. Cloning, expression, purification, and biological activity of five feline type I interferons. Vet Immunol Immunopathol. 2002;89:13–27. doi: 10.1016/s0165-2427(02)00188-5. [DOI] [PubMed] [Google Scholar]
- Xue Q., Yang L., Liu X., Liu W. Molecular characterization of feline type I interferon receptor 2. J. Interferon Cytokine Res. 2010;30:81–88. doi: 10.1089/jir.2009.0031. [DOI] [PubMed] [Google Scholar]
- Yang L.M., Xue Q.H., Sun L., Zhu Y.P., Liu W.J. Cloning and characterization of a novel feline IFN-omega. J Interferon Cytokine Res. 2007;27:119–127. doi: 10.1089/jir.2006.0094. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]


