Summary
Objective
To evaluate the prevalence and clinical manifestations of human metapneumovirus (hMPV) in immunocompetent Chinese adults with acute respiratory tract infections (ARTIs).
Methods
A reverse transcription PCR (RT-PCR) assay targeting the P gene was developed in this study and used to detect hMPV in nasal and throat swabs collected from 2936 immunocompetent adult patients with ARTIs in Beijing, China between July 2008 and June 2010.
Results
Among the 2936 patients studied, 49 (1.7%) were positive for hMPV, of whom 14 (28.6%) were positive for hMPV_A2b, 19 (38.8%) for hMPV_B1, and 16 (32.6%) for hMPV_B2. hMPV_A1 was not detected. An average detection rate of 6.6% was observed in the peak months of the two epidemic seasons studied. The hMPV prevalence was higher in the sampled elderly (>65 years, 3.2%) than in middle aged adults (25–65 years; 2.0%) and teenagers (14–25 years; 0.9%). During the study period, hMPV infections showed a biennial rhythm of seasonality, peaking from November to March in 2008/09 and from March to June in 2010.
Conclusion
hMPV infection plays an important role in immunocompetent adults in its epidemic season. The demographic and clinical data presented in this study improves our understanding of the pathogenesis and clinical burden of hMPV infection in adults.
Keywords: Human metapneumovirus, Epidemiology, Reverse transcription polymerase chain reaction, Adult, Respiratory infection
Introduction
Human metapneumovirus (hMPV) is a single-stranded, negative sense RNA virus belonging to the paramyxoviridae family and pneumovirus subfamily.41 The hMPV genome, approximately 13 kb in length, encodes six genes, which produce at least eight proteins, including nucleocapsid (N), phosphoprotein (P), matrix protein (M), fusion glycoprotein (F), transcriptional enhancer protein (M2-1), small hydrophobic protein (SH), attachment glycoprotein (G), and large polymerase protein (L). Based on the phylogeny of the G gene, hMPV is divided into two genotypes (A and B) and further into four subtypes (A1, A2, B1, and B2).3, 28 A novel sublineage A2b was identified in children in Germany in 2006.21
hMPV infection evokes incomplete immunity and re-infection can occur in people of all ages.10, 14, 15, 35, 42, 43 Prevalent worldwide, hMPV is an important respiratory pathogen that manifests in young children less than 5 years of age.4, 11, 25, 32, 34, 38, 39 Clinical symptoms caused by hMPV infection are similar to respiratory syncytial virus and include fever, cough, wheezing, dyspnea, rhinorrhea, sputum production, and asthma.6, 12, 32 Rates of hMPV infection range from 5.5% to 25% among children hospitalized with respiratory illness.18, 19, 20, 25 In adults, the clinical manifestations of hMPV infection seem to depend on age and health conditions. In young adults (aged 14–25 years), the clinical symptoms of hMPV infection are similar to those of other respiratory viral infections. In contrast, in middle aged (aged 26–65 years) and healthy elderly adults (aged > 65 years), hMPV presents similarly to influenza infections and common colds.13 Although the incidence of hMPV infection in healthy adults during winter is generally less than 6%,5, 23, 30, 42 hMPV is considered to be a major contributor to the burden of respiratory illnesses in older adults.42
Most studies of hMPV infection in adults have focused on the elderly and on immunocompromised people rather than on immunocompetent adults of all age groups. Although hMPV infection has been reported in healthy adults during winter seasons,5, 14 further study of hMPV infection in healthy adults is needed during all seasons in consecutive years to evaluate the circulation patterns, clinical features, and clinical burden of hMPV infection in a given population. In this study, we developed a reverse transcription PCR (RT-PCR) assay to evaluate the prevalence of hMPV infection in healthy adults with acute respiratory tract infections (ARTIs) in Beijing, China. Clinical presentations and phylogenetic characteristics of hMPV infection were also investigated.
Materials and methods
Clinical specimens
Nasal and throat swabs were collected from adults with suspected viral ARTIs who sought medical attention at the Fever Outpatient Clinic of the Peking Union Medical College Hospital, Beijing, China over the period between July 2008 and June 2010. Enrollment criteria of patients have been described previously.36 Briefly, patients over 14 years of age with acute respiratory symptoms, acute fever (body temperature ≧38.0 °C), and normal or low leukocyte count were enrolled. About 22 patients met these criteria each day on average (in total 16,663). Of these, 4–5 patients were enrolled daily based on random selection by clinicians in most months during the study period. A total of 2936 patients were enrolled in this study. Immediately after collection, nasal and throat swabs from the same patient were combined in one tube containing 3 ml viral transport medium (VTM) and stored at −80 °C prior to analysis. Samples were collected only once from each enrolled patient.
RT-PCR screening to detect hMPV infection
Total RNA and DNA were extracted from 200 μl VTM, using the NucliSens easyMAG™ platform (bioMérieux, Marcy 1’Etoile, France) according to the manufacturer’s instructions.8 RT-PCR was used to detect hMPV in this study. A pair of primers targeting the P gene of hMPV was designed to generate an amplicon of 240 bp. Sequences of the forward and reverse primers were: 5′-TYAACATTGCWACAGCAGGACC-3′ and 5′-CTTCWGATTCWCCRCTTGTGCT-3′, corresponding to nt 1834–1855 and nt 2052–2073 of hMPV gz01 strain (GenBank accession no GQ153651), respectively. RT-PCR was performed using the One-Step RT-PCR Kit (Invitrogen, Carlsbad, CA). Following the manufacturer’s instructions, a 5 μl aliquot of the extracted total nucleic acids was used as the template. The RT-PCR assay consisted of initial reverse transcription at 48 °C for 45 min and pre-amplification at 94 °C for 3 min, followed by 35cycles of amplification at 94 °C for 30 s, at 55 °C for 30 s, and at 72 °C for 30 s, and an additional extension at 72 °C for 10 min. All PCR products of the expected size were verified by sequence analysis. The hMPV subtypes were identified by alignment of the PCR sequence with the corresponding reference sequences from GenBank database, using BLAST program (http://blast.ncbi.nlm.nih.gov/blast.cgi). Other common respiratory viruses in clinical specimens were simultaneously detected using RT-PCR or PCR assays, as described previously,36 including influenza viruses (A, B and C), parainfluenza viruses (type 1–4), rhinovirus, enterovirus, adenovirus, coronavirus (OC43, 229E, NL63 and HKU1) and respiratory syncytial virus. To avoid potential cross-contamination, master-mix preparation, nucleic acid extraction, assembly of the PCR reaction, and detection of the PCR products were performed in separate areas. Strict controls for nucleic acid extraction and PCR amplification were also included.
Evaluation of the sensitivity and specificity of the RT-PCR assay
The full-length P gene (885 bp; GenBank accession number HQ889777) of hMPV was amplified from clinical samples by RT-PCR and cloned into the pGEM-T Easy vector (Promega, Madison, WI). Prior to in vitro transcription, the plasmid was purified and linearized using the restriction enzyme Not I (Takara, Dalian, China), and was then extracted by ethanol precipitation. The linearized plasmids were subsequently used as templates for in vitro RNA transcription using T7 RNA-polymerase (Ambion, Austin, TX). After the transcription reaction, the RNA was treated with DNase (Promega) and purified using the Transcript RNA Clean Up Kit (Takara). The concentration of the RNAs was determined using a NanoDrop ND-1000 photo spectrometer (Thermo Fisher Scientific, Wilmington, DE). The quantified RNAs were then diluted 10-fold in 2.5 μg/ml of yeast tRNA (Ambion) and used as template for one-step RT-PCR.
To evaluate the specificity of the RT-PCR assay, propagated respiratory viruses were used as negative controls. These viruses included influenza viruses A/H1N1 2009 and A/H3N2, influenza B, coronaviruses 229E and OC43, adenovirus 5, 7 and 35, and parainfluenza 3.
Phylogenetic analysis
The hMPV strains detected in this study were subtyped based on phylogenetic analysis of the amplicons of P genes using the ClustalW alignment implemented in MEGA 4.0 software.40 The neighbor-joining method with a Kimura-2 parameter model and 1000 bootstrap were applied for phylogenetic analysis.
Statistical analysis
Highest body temperature, age, clinical manifestation, laboratory parameters, and annual incidences of each hMPV subtype were compared using either the χ 2-test or Fisher’s exact test for categorical variables and using the student’s-test for continuous variables. P < 0.05 was considered significant.
Sequence accession numbers
GenBank accession numbers for the nucleotide sequences of the partial hMPV P genes identified in this study are HQ889776 through HQ889824. The GenBank accession numbers for the reference sequences of hMPV_A1 are AF371337 and AY530092. Those for hMPV_A2 are AB503857, AY297749, AY530090, AY530091, AY 530093, AY530095, DQ843659, FJ168779, GQ153651, and NC_004148. Those for hMPV_B1 are AY525843, AY530089, and AY530094. Those for hMPV_B2 are AY297748, DQ843658, EF535506, and FJ168778.
Results
Sensitivity and specificity of the RT-PCR assay
To evaluate the prevalence of hMPV infection in adults, we developed an RT-PCR assay targeting the hMPV P gene in order to detect the hMPV genotypes A and B. The limit of detection (LOD) of the RT-PCR assay was 52 molecules per reaction when the RNA templates of the full-length P gene were obtained from in vitro transcription and used as standards for quantification. No cross reactivity of the developed primers was observed when stocks of other respiratory viruses, including influenza viruses A/H1N1 2009 and A/H3N2, influenza B, coronaviruses 229E and OC43, adenovirus 5, 7 and 35, and parainfluenza 3, were tested (data not shown). In addition, our sequence analysis showed that all amplicons generated from the clinical specimens collected for this study were specific for the hMPV P gene, further indicating the specificity of this RT-PCR assay.
Prevalence of hMPV infection
A total of 2936 patients ranging from 14 to 90 years old (median 29 years; mean 33.9 years) were enrolled in this study. Of those patients, 1347 were males (46%) and 1589 were females (54%). hMPV RNA was detected in 49 (1.7%) patients, of whom 14 (28.6%) were positive for hMPV_A2, 19 (38.8%) for hMPV_B1, and 16 (32.6%) for hMPV_B2. hMPV_A1 was not detected in any of the patients. The detection rate of hMPV was less than that of influenza virus (27.6%), rhinovirus (6.6%), enterovirus (2.6%), and parainfluenza virus (2.1%), but higher than that of respiratory syncytial virus (1.5%), adenovirus (1.3%), and coronavirus (1.2%).
The hMPV-positive patients ranged from 16 to 90 years of age (median 42 years; mean 44 years). The ratio of hMPV-positive male–female patients was 1.18:2.07 (χ 2 = 3.47, P = 0.06). Thus, hMPV infection does not show a gender preference in the healthy adults screened. The overall detection rate of hMPV did not vary significantly between the two study years of July 2008–June 2009 and July 2009–June 2010 (χ 2 = 0.01, P = 0.93; Table 1 ).
Table 1.
Detection of hMPV in adult patients.
| Subtype of hMPV | Year (July to June) |
|
|---|---|---|
| 2008–2009 (n = 1395) | 2009–2010 (n = 1541) | |
| A2 | 13 (0.9)a | 1 (0.1) |
| B1 | 6 (0.4) | 13 (0.8) |
| B2 | 5 (0.4) | 11 (0.7) |
| Total cases | 24 (1.7) | 25 (1.6) |
Numbers in parentheses are the percentage of positive infection in total samples.
hMPV infection was detected in all age groups sampled (14–25 years, 26–65 years, and >65 years). However, the detection rates of hMPV in different age groups varied significantly (χ 2 = 7.19, P = 0.03), with the elderly (>65 years) showing the highest detection rate of 3.2% (Table 2 ); the detection rate in middle aged adults (26–65 years, 2.0%) was significantly higher than that in teenagers (14–25 years, 0.9%; χ 2 = 4.98, P = 0.03).13
Table 2.
Age distribution of adults infected with hMPV.
| 14–25years | 26–65years | >65years | Total | |
|---|---|---|---|---|
| No. of detected | 1016 | 1766 | 154 | 2936 |
| A2 | 3 (0.3)a | 7 (0.4) | 4 (2.6) | 14 (0.5) |
| B1 | 2 (0.2) | 16 (0.9) | 1 (0.6) | 19 (0.6) |
| B2 | 4 (0.4) | 12 (0.7) | 0 | 16 (0.5) |
| Total | 9 (0.9) | 35 (2.0) | 5 (3.2) | 49 (1.7) |
Numbers in parentheses are the percentage of positive infection in total samples.
Other common respiratory viruses were simultaneously screened. Co-detection of hMPV with other viruses was found in six (12.2%) patients, five with upper and one with lower respiratory tract infection. Influenza virus type A and enterovirus were co-detected with hMPV_B1. Influenza virus types A and B were co-detected with hMPV_B2. No other respiratory viruses were co-detected with hMPV_A2 (Table 3 ).
Table 3.
Clinical manifestations of patients with hMPV infection.
| Parameters | hMPV subtypes |
||
|---|---|---|---|
| A2 | B1 | B2 | |
| No. of positive patients | 14 | 19 | 16 |
| Age range (year) | 16–90 | 24–63 | 17–65 |
| Mean/median age (year) | 51.3/53 | 45.6/52.5 | 35.5/32 |
| Gender (M/F) | 5/9 | 5/14 | 6/10 |
| URTI | 14 (93.3)a | 18 (94.7) | 16 (100) |
| LRTI | 1 (6.7) | 1 (5.3) | 0 |
| Clinical symptoms | |||
| Cough | 9 (64.3) | 12 (63.2) | 9 (56.3) |
| Sputum production | 7 (50) | 9 (47.4) | 9 (56.3) |
| Headache | 8 (57.1) | 14 (73.7) | 14 (87.5) |
| Muscle pain | 6 (42.9) | 13 (68.4)c | 5 (31.3) |
| Sore throat | 11 (78.6) | 13 (68.4) | 9 (56.3) |
| Chills | 10 (71.4) | 16 (84.2) | 11 (68.8) |
| Vomiting | 0 | 0 | 1 (6.25) |
| Rhinorrhea | 10 (71.4) | 14 (73.7) | 6 (37.5) |
| Sneezing | 8 (57.1) | 12 (63.2) | 5 (31.3) |
| Peripheral blood tests | |||
| Mean leukocyte count ( × 109/L) | 6.8 ± 1.7 | 7.0 ± 2.0 | 6.9 ± 1.4 |
| Co-detection | 3 | 3 | |
| IFVA (n = 2) | IFVA (n = 2) | ||
| EV (n = 1) | IFVB (n = 1) | ||
LRTI, lower respiratory tract infection; URTI, upper respiratory tract infection; IFV, influenza virus; EV, enterovirus.
The number in parentheses indicates the percentages of positive infection in total patients.
Seasonality of hMPV infection in adults
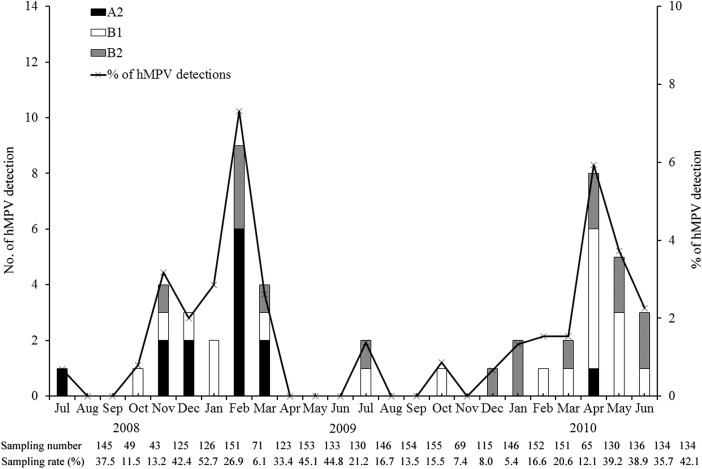
The seasonal distribution of hMPV infection varied during the two study years of July 2008June 2009 and July 2009–June 2010 (Fig. 1 ). During the study year of July 2008–June 2009, the peak circulation of hMPV occurred from November through March of the winter-spring season, with the highest detection rate of 7.3% in March 2009. During the study year of June 2009–July 2010, the peak circulation occurred from March through June of the spring-summer season, with the highest detection rate of 5.9% in April 2010. The seasonality analysis of hMPV subtypes shows that hMPV_A2 was most prevalent between July 2008 and March 2009, with the highest detection rate in February 2009. We did not detect any hMPV_A2 between April 2009 and March 2010, but detected hMPV_B1 and hMPV_B2 in both study years with an approximately 2-fold higher detection rate for 2009–2010 than for 2008–2009 (Fig. 1).
Figure 1.
Number of samples positive for human metapneumovirus and detection rate in adult respiratory samples. The bar graph indicates the number of hMPV A1, A2, B1, and B2 positive cases, and the line graph indicates the total detection rates of human metapneumovirus in each month. The sampling number and the portion of sampling are indicated for each month below the x-axis.
Clinical characteristics of hMPV infection in adults
Among the 49 hMPV-positive patients, 47 (95.9%) were diagnosed with an upper respiratory tract infection and 2 (4.1%) were diagnosed with a lower respiratory tract infection. No hMPV-positive patients enrolled in this study were admitted to hospital. Clinical symptoms included myalgia (49.0%), rhinorrhea (61.2%), sneezing (51.0%), cough (61.2%), sputum production (51.0%), sore throat (67.3%), chills (83.7%), headache (71.4%), muscle pain (44.9%), and sneezing (51.0%). Vomiting was observed in only one patient infected with hMPV_B2 (Table 3). Body temperatures were similar among patients infected with different hMPV subtypes (data not shown). Laboratory examination showed no significant difference in the granulocytes, mean lymphocyte count, percentage of neutrophilic granulocytes, or percentage of lymphocytes between patients positive for different hMPV subtypes.
Phylogenetic analysis of hMPV strains
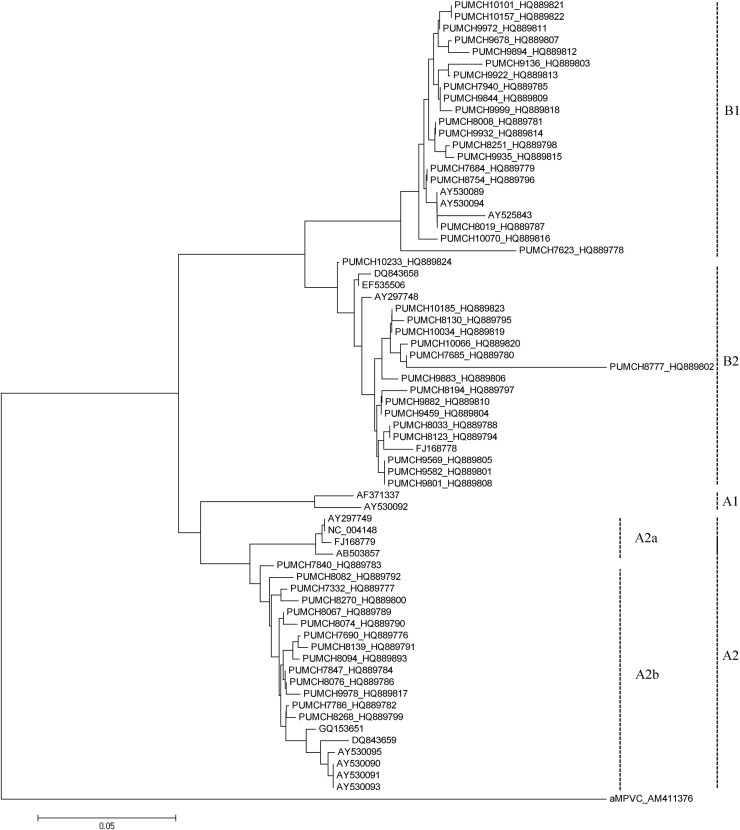
A phylogenetic tree was constructed for all strains of hMPV_A2, hMPV_B1, and hMPV_B2 identified in this study, based on clustalW alignment (implemented in MEGA) of the 240 bp amplicons of the hMPV P gene. Strains of each subtype clustered together (Fig. 2 ), indicating that the target region of the established RT-PCR assay contains enough variability for hMPV genotyping. Moreover, all hMPV_A2 strains, which can be divided into hMPV_A2a and A2b clusters,22 were only found in the hMPV_A2b cluster in this study. Pairwise distance compute of the targeting P gene between our sample sequences and reference sequences from GenBank (see Fig. 2 legend) shows that hMPV_A2 strains identified in this study had 97.4%–100% identity of both nucleotide sequences and amino acid sequences. hMPV_B1 had 93.1%–100% and 93.3%–100% identity of nucleotide sequences and amino acid sequences whereas hMPV_B2 had 90.6%–100% and 88.2%–100% nucleotide and amino acid identity, respectively.
Figure 2.
Phylogenetic analysis of hMPV based on partial sequences of the P gene. Fragments of the P genes of hMPV_A2, hMPV_B1, and hMPV_B2 were amplified by RT-PCR and analyzed with MEGA 4.0 software using the distance method and the neighbor-joining algorithm with Kimura-2 parameters. Each strain from this study is indicated by a specific identification code (PUMCH) followed by the patient number and GenBank accession number. The GenBank accession numbers of the reference sequences of hMPV_A1 are AF371337 and AY530092. Those for hMPV_A2 are AB503857, AY297749, AY530090, AY530091, AY 530093, AY530095, DQ843659, FJ168779, GQ153651, and NC_004148. Those for hMPV_B1 are AY525843, AY530089, and AY530094. Those for hMPV_B2 are AY297748, DQ843658, EF535506, and FJ168778. Scale bar indicates nucleotide substitutions per site.
Discussion
In this study, we used an RT-PCR assay to screen clinical specimens collected from adults with ARTIs. The hMPV P gene is an ideal molecular target for detection and genotyping of hMPV, because its conserved sequences allow specific detection using RT-PCR and enough variability to permit distinction between genotypes.3, 28 Four subtypes of hMPV and two clusters of hMPV_A2 were clearly distinguished through our phylogenetic analysis of hMPV P gene fragments. All strains of hMPV_A2 were found to be hMPV_A2b.
The sensitivity of the RT-PCR assay was determined to be 52 copies per reaction, which is at least as sensitive as reported for real-time RT-PCR assays previously.24, 29 The primer pair used in this study can also be applied to an SYBR Green RT-PCR system for quick screening of clinical specimens for hMPV, based on data obtained from about 400 clinical specimens comparing SYBR Green RT-PCR and standard RT-PCR analysis (data not shown). No cross reactivity was observed for other propagated respiratory viruses, indicating high specificity of the primer pair under the RT-PCR conditions used.
We observed an annual incidence of 1.7% for hMPV infection in immunocompetent adults with ARTIs, sampled over a period of two consecutive years in China. Similar annual incidences have been reported in the United States of America,19 the United Kingdom,16 and in Cambodia.4 Although the winter-spring season is traditionally regarded as the typical epidemic season of hMPV in temperate regions,5, 7, 14, 18, 23, 30, 42 the results of our study show that the hMPV epidemic peaked during the winter–spring season (November–February) of 2008–2009 and the spring-summer season (March–June) of 2010. The hMPV seasonality observed in this study is comparable to that reported in Japan,31 which showed a biennial rhythm of hMPV seasonality, with the epidemic peaking between November and March in odd years and between March and June in the successive even years.1, 2, 31 Our results indicate that hMPV seasonality in adults is similar to that in children.
The reason for this differing seasonality is unclear. Previous studies have shown that climate can affect seasonality.4, 7, 17, 33, 37 However, the climate records for Beijing do not indicate significant variability between the same months of each study year (data not shown). It is possible that the pandemic of A/H1N1/2009 affected the circulation of hMPV during the 2010 season. Based on our findings, the surveillance of hMPV infection in immunocompetent adults should be extended from the winter season to year round to improve our understanding of the burden of hMPV infection.
hMPV_A1 was not detected in the patients sampled for this study. The absence of hMPV_A1 in this study is consistent with other reports.4, 9, 26, 27 Indeed, circulation of hMPV genotypes has been reported to vary annually, with replacement of predominant genotypes occurring every 1–3 years in a given population. Such genotype replacement is believed to result from adaptive immunity of a population to the predominant circulating genotype.1, 17, 22 In this study, analysis of the incidences of hMPV subtypes shows that hMPV_A (represented by hMPV_A2) was declining whereas hMPV_B (hMPV_B1 and B2) was rising from July 2008 to June 2010. Thus, it is possible that hMPV_A1 was in its troughs during the study period. A longer study period would be required to investigate the prevalence patterns of hMPV subtypes.
Children and elderly adults (>65 years) are reportedly at higher risk of hMPV infection due to incomplete immunity.7, 13, 14 In this study, the detection rate of hMPV (3.2%) was highest in the elderly (>65 years) compared to that in other adult age groups, consistent with previous reports.12, 14, 23 In contrast to previous reports,13, 14, 35, 42 the detection rate of hMPV in teenagers (14–25 year, 0.9%) was significantly lower than that in middle aged adults (26–65 years, 2.0%) in our study. The reason for this disparity is unclear. Further studies are needed to clarify the age distribution of hMPV infection in adults.
We observed a moderate detection rate of hMPV in Chinese adults, lower than that of influenza virus, enterovirus, rhinovirus, and parainfluenza virus, but higher than that of respiratory syncytial virus, adenovirus, and coronavirus. If the observed data is extrapolated from the sampled subjects to the whole population, 1.7% could be translated to a significant number of hMPV ARTI cases in China every year.
In summary, this study reports the detection of hMPV and a large-scale, detailed analysis of the prevalence and clinical symptoms of hMPV infection in healthy adults with ARTIs in Beijing, China. A biennial rhythm of hMPV seasonality in immunocompetent adults was observed, as was a higher detection rate of hMPV in adults aged >65 compared to those in other adult age groups screened in this study. Our results improve current understanding of the pathogenesis of hMPV in healthy adults. However, as this study covered a limited population and a relatively short time period, further investigations are required to characterize the infection of different hMPV subtypes in adults more thoroughly.
Acknowledgments
We thank the clinicians of Peking Union Medical College Hospital for their assistance in sample collection. This study was supported in part by the National S & T Major Project for the Control and Prevention of Major Infectious Diseases in China (2008ZX10004-002 and 2009ZX10004-206).
Contributor Information
Qi Jin, Email: zdsys@vip.sina.com.
Jianwei Wang, Email: wangjw28@163.com.
References
- 1.Aberle J.H., Aberle S.W., Redlberger-Fritz M., Sandhofer M.J., Popow-Kraupp T. Human metapneumovirus subgroup changes and seasonality during epidemics. Pediatr Infect Dis J. 2010;29:1016–1018. doi: 10.1097/INF.0b013e3181e3331a. [DOI] [PubMed] [Google Scholar]
- 2.Aberle S.W., Aberle J.H., Sandhofer M.J., Pracher E., Popow-Kraupp T. Biennial spring activity of human metapneumovirus in Austria. Pediatr Infect Dis J. 2008;27:1065–1068. doi: 10.1097/INF.0b013e31817ef4fd. [DOI] [PubMed] [Google Scholar]
- 3.Agapov E., Sumino K.C., Gaudreault-Keener M., Storch G.A., Holtzman M.J. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis. 2006;193:396–403. doi: 10.1086/499310. [DOI] [PubMed] [Google Scholar]
- 4.Arnott A., Vong S., Sek M., Naughtin M., Beaute J., Rith S. Genetic variability of human metapneumovirus amongst an all ages population in Cambodia between 2007 and 2009. Infect Genet Evol. 2011 doi: 10.1016/j.meegid.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenfanger J., Mueller T., O’Brien J., Drake C., Lawhorn J. Prevalence of human metapneumovirus in central illinois in patients thought to have respiratory viral infection. J Clin Microbiol. 2008;46:1489–1490. doi: 10.1128/JCM.02283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien N., Normand S., Taylor T., Ward D., Peret T.C., Boivin G. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 2003;93:51–62. doi: 10.1016/S0168-1702(03)00065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastien N., Ward D., Van Caeseele P., Brandt K., Lee S.H., McNabb G. Human metapneumovirus infection in the Canadian population. J Clin Microbiol. 2003;41:4642–4646. doi: 10.1128/JCM.41.10.4642-4646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., Zhang Z.Y., Zhao Y., Liu E.M., Zhao X.D. Acute lower respiratory tract infections by human metapneumovirus in children in Southwest China: a 2-year study. Pediatr Pulmonol. 2010;45:824–831. doi: 10.1002/ppul.21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Ding R., Yu J.X., Yang J.C., Sun C.W. Epidemiological feature and clinical symptoms association with human metapeumovirus in all age groups of Xuzhou city. Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27:827. [PubMed] [Google Scholar]
- 11.Debur M.C., Vidal L.R., Stroparo E., Nogueira M.B., Almeida S.M., Takahashi G.A. Impact of human metapneumovirus infection on in and outpatients for the years 2006–2008 in Southern Brazil. Mem Inst Oswaldo Cruz. 2010;105:1010–1018. doi: 10.1590/s0074-02762010000800010. [DOI] [PubMed] [Google Scholar]
- 12.El Sayed Zaki M., Raafat D., El-Metaal A.A., Ismail M. Study of human metapneumovirus-associated lower respiratory tract infections in Egyptian adults. Microbiol Immunol. 2009;53:603–608. doi: 10.1111/j.1348-0421.2009.00162.x. [DOI] [PubMed] [Google Scholar]
- 13.Falsey A.R. Human metapneumovirus infection in adults. Pediatr Infect Dis J. 2008;27:S80–S83. doi: 10.1097/INF.0b013e3181684dac. [DOI] [PubMed] [Google Scholar]
- 14.Falsey A.R., Erdman D., Anderson L.J., Walsh E.E. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 15.Foulongne V., Lechiche C., Reynes J., Segondy M. Human metapneumovirus pneumonia in an adult patient hospitalized for suspected severe acute respiratory syndrome (SARS) Presse Med. 2004;33:1006–1007. doi: 10.1016/S0755-4982(04)98824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaunt E., McWilliam-Leitch E.C., Templeton K., Simmonds P. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; assessment of its importance as a diagnostic screening target. J Clin Virol. 2009;46:318–324. doi: 10.1016/j.jcv.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Gerna G., Campanini G., Rovida F., Sarasini A., Lilleri D., Paolucci S. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter-spring seasons. Brief report. Arch Virol. 2005;150:2365–2375. doi: 10.1007/s00705-005-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gioula G., Chatzidimitriou D., Melidou A., Exindari M., Kyriazopoulou-Dalaina V. Contribution of human metapneumovirus to influenza-like infections in North Greece, 2005–2008. Euro Surveill. 2010;15(9) doi: 10.2807/ese.15.09.19499-en. pii = 19499. [DOI] [PubMed] [Google Scholar]
- 19.Gray G.C., Capuano A.W., Setterquist S.F., Erdman D.D., Nobbs N.D., Abed Y. Multi-year study of human metapneumovirus infection at a large US Midwestern Medical Referral Center. J Clin Virol. 2006;37:269–276. doi: 10.1016/j.jcv.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamelin M.E., Abed Y., Boivin G. Human metapneumovirus: a new player among respiratory viruses. Clin Infect Dis. 2004;38:983–990. doi: 10.1086/382536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huck B., Scharf G., Neumann-Haefelin D., Puppe W., Weigl J., Falcone V. Novel human metapneumovirus sublineage. Emerg Infect Dis. 2006;12:147–150. doi: 10.3201/eid1201.050772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hustedt J.W., Vazquez M. The changing face of pediatric respiratory tract infections: how human metapneumovirus and human bocavirus fit into the overall etiology of respiratory tract infections in young children. Yale J Biol Med. 2010;83:193–200. [PMC free article] [PubMed] [Google Scholar]
- 23.Johnstone J., Majumdar S.R., Fox J.D., Marrie T.J. Human metapneumovirus pneumonia in adults: results of a prospective study. Clin Infect Dis. 2008;46:571–574. doi: 10.1086/526776. [DOI] [PubMed] [Google Scholar]
- 24.Kuypers J., Wright N., Corey L., Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33:299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legrand L., Vabret A., Dina J., Petitjean-Lecherbonnier J., Stephanie G., Cuvillon D. Epidemiological and phylogenic study of human metapneumovirus infections during three consecutive outbreaks in Normandy, France. J Med Virol. 2011;83:517–524. doi: 10.1002/jmv.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo L.H., Tan B.H., Ng L.M., Tee N.W., Lin R.T., Sugrue R.J. Human metapneumovirus in children, Singapore. Emerg Infect Dis. 2007;13:1396–1398. doi: 10.3201/eid1309.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luciani F., Alizon S. The evolutionary dynamics of a rapidly mutating virus within and between hosts: the case of hepatitis C virus. PLoS Comput Biol. 2009;5:e1000565. doi: 10.1371/journal.pcbi.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay I.M., Bialasiewicz S., Waliuzzaman Z., Chidlow G.R., Fegredo D.C., Laingam S. Use of the P gene to genotype human metapneumovirus identifies 4 viral subtypes. J Infect Dis. 2004;190:1913–1918. doi: 10.1086/425013. [DOI] [PubMed] [Google Scholar]
- 29.Maertzdorf J., Wang C.K., Brown J.B., Quinto J.D., Chu M., de Graaf M. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–986. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manuel A. Human metapneumovirus: a new threat? Thorax. 2009;64:475. [PubMed] [Google Scholar]
- 31.Mizuta K., Abiko C., Aoki Y., Ikeda T., Itagaki T., Katsushima N. Endemicity of human metapneumovirus subgenogroups A2 and B2 in Yamagata, Japan, between 2004 and 2009. Microbiol Immunol. 2010;54:634–638. doi: 10.1111/j.1348-0421.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 32.Moattari A., Aleyasin S., Arabpour M., Sadeghi S. Prevalence of human metapneumovirus (hMPV) in children with wheezing in Shiraz-Iran. Iran J Allergy Asthma Immunol. 2010;9:250–254. [PubMed] [Google Scholar]
- 33.Noyola D.E., Alpuche-Solis A.G., Herrera-Diaz A., Soria-Guerra R.E., Sanchez-Alvarado J., Lopez-Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. J Med Microbiol. 2005;54:969–974. doi: 10.1099/jmm.0.46052-0. [DOI] [PubMed] [Google Scholar]
- 34.Pitoiset C., Darniot M., Huet F., Aho S.L., Pothier P., Manoha C. Human metapneumovirus genotypes and severity of disease in young children (n = 100) during a 7-year study in Dijon hospital, France. J Med Virol. 2010;82:1782–1789. doi: 10.1002/jmv.21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prins J.M., Wolthers K.C. Human metapneumovirus: a new pathogen in children and adults. Neth J Med. 2004;62:177–179. [PubMed] [Google Scholar]
- 36.Ren L., Gonzalez R., Wang Z., Xiang Z., Wang Y., Zhou H. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect. 2009;15:1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarasini A., Percivalle E., Rovida F., Campanini G., Genini E., Torsellini M. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter–spring season. J Clin Virol. 2006;35:59–68. doi: 10.1016/j.jcv.2005.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J., Zhu Q.R., Zeng M., Yu H., Wang X.H. Molecular epidemiology of human metapneumovirus in children with acute lower respiratory tract infections in Shanghai. Zhonghua Liu Xing Bing Xue Za Zhi. 2010;31:447–450. [PubMed] [Google Scholar]
- 39.Sloots T.P., Mackay I.M., Bialasiewicz S., Jacob K.C., McQueen E., Harnett G.B. Human metapneumovirus, Australia, 2001–2004. Emerg Infect Dis. 2006;12:1263–1266. doi: 10.3201/eid1208.051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh E.E., Peterson D.R., Falsey A.R. Human metapneumovirus infections in adults: another piece of the puzzle. Arch Intern Med. 2008;168:2489–2496. doi: 10.1001/archinte.168.22.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams J.V. Human metapneumovirus: an important cause of respiratory disease in children and adults. Curr Infect Dis Rep. 2005;7:204–210. doi: 10.1007/s11908-005-0036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]