Abstract
In the past 4 years, RNA interference (RNAi) has become widely used as an experimental tool to analyse the function of mammalian genes, both in vitro and in vivo. By harnessing an evolutionary conserved endogenous biological pathway, first identified in plants and lower organisms, double-stranded RNA (dsRNA) reagents are used to bind to and promote the degradation of target RNAs, resulting in knockdown of the expression of specific genes. RNAi can be induced in mammalian cells by the introduction of synthetic double-stranded small interfering RNAs (siRNAs) 21–23 base pairs (bp) in length or by plasmid and viral vector systems that express double-stranded short hairpin RNAs (shRNAs) that are subsequently processed to siRNAs by the cellular machinery. RNAi has been widely used in mammalian cells to define the functional roles of individual genes, particularly in disease. In addition, siRNA and shRNA libraries have been developed to allow the systematic analysis of genes required for disease processes such as cancer using high throughput RNAi screens. RNAi has been used for the knockdown of gene expression in experimental animals, with the development of shRNA systems that allow tissue-specific and inducible knockdown of genes promising to provide a quicker and cheaper way to generate transgenic animals than conventional approaches. Finally, because of the ability of RNAi to silence disease-associated genes in tissue culture and animal models, the development of RNAi-based reagents for clinical applications is gathering pace, as technological enhancements that improve siRNA stability and delivery in vivo, while minimising off-target and nonspecific effects, are developed.
Keywords: RNAi, siRNA, shRNA, Disease, Validation, Therapy
Abbreviations: AAV, adeno-associated virus; ABCA1, ATP-binding cassette, subfamily A, member 1; Bcl-2, B-cell chronic lymphocytic leukemia/lymphoma 2; BCR-ABL, breakpoint cluster region-protooncogene tyrosine-protein kinase ABL1 fusion protein; BMP4, bone morphogenetic protein 4; bp, base pairs; B-RAF, murine sarcoma viral (v-raf) oncogene homolog B1 oncogene; CCR5, chemokine (C–C motif) receptor 5; CD8, T-lymphocyte differentiation antigen T8/Leu-2; CML, chronic myelogenous leukemia; COPD, chronic obstructive pulmonary disease; Cre, causes recombination; CXCR4, chemokine (C–X–C motif), receptor 4; dsRNA, double-stranded RNA; EC, endothelial cell; EGFR, epidermal growth factor receptor; EpCAM, tumour-associated calcium signal transducer 1; ERK, extracellular signal regulated protein kinase; Fas, tumour necrosis factor receptor superfamily, member 6; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HMG-CoA, beta-hydroxy-beta-methylglutaryl coenzyme A; IFN, interferon; KC, keratinocyte-derived chemokine; K-Ras, c-Kirsten-ras protein; LacZ, β-galactosidase α-peptide; LNA, locked nucleic acid; LoxP, locus χ of crossover P1; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MDR, multidrug-resistance protein; MIP2, macrophage inflammatory protein-2; miRNA, microRNAs; NF-κB, Nuclear factor-kappaB; PEG, polyethylene glycol; PEI, polyethyleneimine; PKR, dsRNA-dependent protein kinase; RasGAP, Ras GTPase-activating protein; RdDM, RNA-directed DNA methylation; RISC, RNA-induced silencing complex; RNAi, RNA interference; Rnase III, ribonuclease III; RSV, respiratory syncytial virus; SARS, severe acute respiratory syndrome; SCID, severe combined immunodeficiency; shRNA, short hairpin RNA; siRNA, small interfering RNA; S1P, sphingosine 1-phosphate; smad 1, mothers against decapentaplegic homolog 1; smad 7, mothers against decapentaplegic homolog 7; TGFα, transforming growth factor α; TGFβ, transforming growth factor β; TLR3, toll-like receptor 3; TNFα, tumour necrosis factor α; VEGF, vascular endothelial growth factor
1. Introduction
Modern drug discovery is target-driven (Whittaker, 2003b). The identification of most of the genes in the human genome has spawned varying estimates of the number of potential new drug targets that await discovery (Drews, 2000, Hopkins & Groom, 2002). However, translating this genomic information into drug therapies is still a major challenge facing pharmaceutical companies. Providing support for the concept that drug modulation of a given target is likely to produce a therapeutic response in patients is a key step in this progression from “gene to screen” (Hardy & Peet, 2004). The main limitations are knowing which gene products are functionally involved in the pathology of a disease (target validation) and the druggability of the gene products by small molecule compounds. For over a decade now, targeting mRNA for both target validation and as a therapeutic strategy has been actively pursued using ribozymes (Breaker, 2004) or antisense (Crooke, 2004) approaches. To date, only antisense technology has produced a marketed drug: Vitravene® for cytomegalovirus-induced retinitis (Winkler, 2004). RNA interference (RNAi) is the latest entry into the field. Hailed as the “Scientific Breakthrough of the Year” for 2002 by the journal Science (Couzin, 2002), double-stranded RNA (dsRNA) reagents are used to bind to and promote the degradation of target RNAs by harnessing an endogenous biological pathway. In the context of drug discovery, the value of RNAi is based on the premise that knocking down the expression of putative drug targets can be used to rapidly and specifically simulate the biological and pharmacological effects of target inhibition by low-molecular-weight compounds in cellular assays and animal models of the disease. Although still in its infancy with respect to drug development, August 2004 brought a filing by Acuity Pharmaceuticals with the U.S. Food and Drug Administration for the first clinical trial of an RNAi therapeutic: Cand5 for treatment of a common form of blindness (http://www.acuitypharma.com/page4.html). In this article, we will review and assess the use of RNAi reagents for both target validation and their potential as therapeutics.
2. The mechanism of RNA interference
RNAi was first observed by plant biologists in the late 1980s, but its molecular mechanism remained unclear until the late 1990s, when elegant work in the nematode Caenorhabditis elegans showed that RNAi is an evolutionary conserved gene-silencing mechanism (Fire et al., 1998, Reinhart et al., 2000). The sequence-specific posttranscriptional gene silencing by double-stranded RNA is conserved in a range of organisms: plants, Neurospora, Drosophila, C. elegans, and mammals. This process is related to a normal defense against viruses and the mobilisation of transposable genetic elements (transposons; Tijsterman et al., 2002). The dsRNAs produced by integrated transposons, replicating viruses, or one of the newly identified class of regulatory noncoding microRNAs (miRNAs; Bartel, 2004) are processed into short dsRNAs. These short RNAs trigger a cascade of biochemical events (Fig. 1 ) involving a cytoplasmic ribonuclease III (RNase III)-like protein known as Dicer and the partially characterised multiprotein complex known as the RNA-induced silencing complex (RISC), leading ultimately to the degradation of mRNAs (posttranscriptional gene silencing). The naturally occurring miRNAs are synthesized in the nucleus in large precursor forms. An enzyme known as Drosha mediates the processing of the primary miRNA transcripts into pre-miRNAs (∼ 70mers), which are then exported to the cytoplasm (Lee et al., 2003). In the cytoplasm, Dicer is responsible for cleaving double-stranded molecules, whether derived from endogenous miRNAs or from replicating viruses, into small RNA duplexes of 19–25 base pairs (bp) with characteristic 3′-dinucleotide overhangs (Bernstein et al., 2001). The small interfering RNA (siRNA) duplex is incorporated into the RISC, whereupon an ATP-dependent helicase unwinds the duplex, enabling either one of the 2 strands to independently recognize mRNAs (Kisielow et al., 2002). The extent of complementarity between the guiding strand and the target mRNA determines whether mRNA silencing is achieved via site-specific cleavage of the message in the region of the siRNA–mRNA duplex (Caudy et al., 2003) or through an miRNA-like mechanism of translational repression (Doench et al., 2003). For siRNA-mediated silencing, the cleavage products are released and degraded, leaving the disengaged RISC complex to further survey the mRNA pool.
Fig. 1.

Mechanisms of siRNA-mediated gene silencing. Long (> 30 bp) dsRNA (e.g., viral), vector-based shRNAs, and endogenous miRNAs are processed by the Dicer complex to form 21–23 bp siRNA duplexes with symmetric 3′-overhangs and 5′-phosphate groups. The duplexes then associate with RISC, which contains a helicase that unwinds the duplex. Each siRNA strand incorporated RISC is then presented to the cytoplasmic mRNA pool, where it associates with its complementary mRNA strand and, depending on its complementarity, results in either targeted cleavage of the mRNA or translational arrest. In theory, the antisense strand of the siRNA duplex will target the desirable mRNA for destruction, whereas the RISC that incorporates the sense strand can potentially associate with other unintended mRNAs and cause off-target activities. More details on the mechanism can be found at the following website: http://www.nature.com/nature/focus/rnai/index.html.
3. Ribonucleic acid interference in mammalian cells
Although genomic approaches such as gene expression analysis using “gene chips” (Archacki & Wang, 2004) or disease gene mapping (Whittaker, 2003a) can associate genes with disease phenotypes, these data alone cannot define the disease-associated role for the protein encoded by the gene. Knockout mice have proved to be a powerful way of studying a gene's biological/disease relevance (Rosahl, 2003) to the extent that knockout phenotypes show a good correlation with drug efficacy (Zambrowicz et al., 2003). However, the cost and time implications of developing such models mean there has been a drive to develop methods that enable target validation in vitro (particularly when working with novel targets) before embarking on in vivo studies. Given the success of RNAi in assigning gene function in lower eukaryotes, such as the nematode worm C. elegans (Sugimoto, 2004) and the fruitfly Drosophila melanogaster (Kuttenkeuler & Boutros, 2004), it is not surprising that the approach has been adopted for analysing gene function (particularly target validation) in mammalian systems.
3.1. Small interfering RNA
The specific and effective silencing of genes by RNAi in C. elegans and D. melanogaster can be achieved using long (> 500 bp) dsRNAs (Mello & Conte, 2004). Furthermore, dsRNAs can be introduced into worms by direct injection, feeding them bacteria expressing dsRNA, or by soaking the worms in a solution of dsRNA (Wang & Barr, 2005). However, this is not the case in mammalian systems. Firstly, dsRNAs > 30 bp trigger the γ-interferon (IFN) pathway (the interferon pathway is one arm of the innate immune system triggered partly by dsRNA, a common replicative intermediate in viral infections), thus, 21–23 bp double-stranded small interfering RNAs (siRNAs) generated by chemical synthesis (Elbashir et al., 2001), enzymatic cleavage (Kittler et al., 2004), or expression systems (Zheng et al., 2004) have to be used. These molecules elude the stress response by mimicking Dicer products and entering the RNAi pathway further downstream. Secondly, the siRNA duplexes have to be transfected into mammalian cells using either lipid-based formulations (Brazas & Hagstrom, 2005), electroporation (Gresch et al., 2004), or by linking to peptides (Muratovska & Eccles, 2004). As siRNAs have become more widely used, the basic structure of effective siRNAs has been defined (Fig. 2 ), including the need for a 19-bp RNA duplex with a 2-nucleotide overhang on the 3′-ends. It has also become clear that the effectiveness of siRNA silencing is sequence specific, hence, rules for siRNA design have been developed (Mittal, 2004, Reynolds et al., 2004). Despite this, siRNA duplexes still have to be experimentally assessed and optimised for the knockdown of individual mRNA molecules, as not all duplexes matching the design criteria are potent (Kumar et al., 2003, Hsieh et al., 2004). It should also be emphasised that gene expression is knocked down rather than knocked out. siRNA has been successfully applied to mammalian cell lines (Caplen et al., 2001, Elbashir et al., 2001), primary cells (Kao et al., 2004, Krick et al., 2005), and embryonic stem cells (Ikeda et al., 2004, Hoelters et al., in press).
Fig. 2.
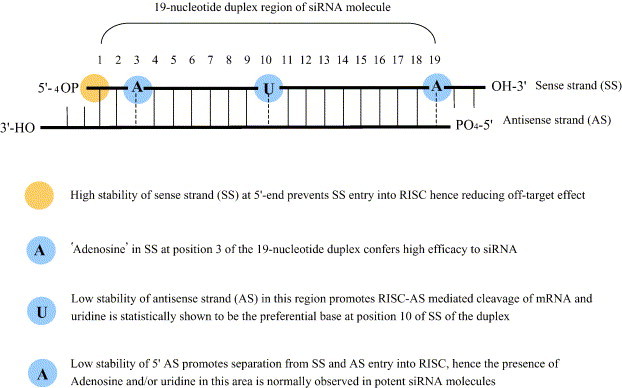
The design of an effective siRNA molecule. Modified from Mittal (2004).
3.2. Stable induction of small interfering RNA by short hairpin RNA (shRNA)
In contrast to C. elegans, where RNAi effects are stable, long lasting, and are passed onto the offspring (Grishok & Mello, 2002), gene silencing by transfected siRNA duplexes in mammalian cells is transient. This is because mammalian cells lack the RNA-dependent RNA polymerases that amplify siRNAs in C. elegans. As a result, gene silencing is dependent on the number of siRNA molecules transfected into the cells and the duplexes become progressively diluted as cells divide. The persistence of siRNA activity in mammalian cells varies with the proliferative status of the cells, such that siRNA activity lasts for 3–7 days in proliferating cells, but can persist for 3 weeks or more in terminally differentiated cells, such as neurons (Omi et al., 2004). To get around this problem, vector-based systems for the introduction and stable expression of siRNA in target cells have been developed (Tuschl, 2002). These vectors contain RNA polymerase III promoters that either express sense and antisense strands from separate promoters (tandem type) or express short hairpin RNAs (shRNAs) that are cleaved by the Dicer to produce siRNA (shRNA type; Fig. 3 ). Stably transfected cell lines can be generated by selecting for a drug resistance marker (expressed either on the vector or on a co-transfected plasmid). Such vector systems have been successfully used to obtain efficient and stable knockdown of target genes in mammalian cells (Mittal, 2004). Although it is difficult to say which of these 2 vector systems are more efficient at silencing gene expression, work does suggest that the shRNA system is more effective than the tandem system is (Miyagishi & Taira, 2003). Recent work also indicates that shRNAs are more potent inducers of RNAi than is siRNA (Siolas et al., 2005).
Fig. 3.
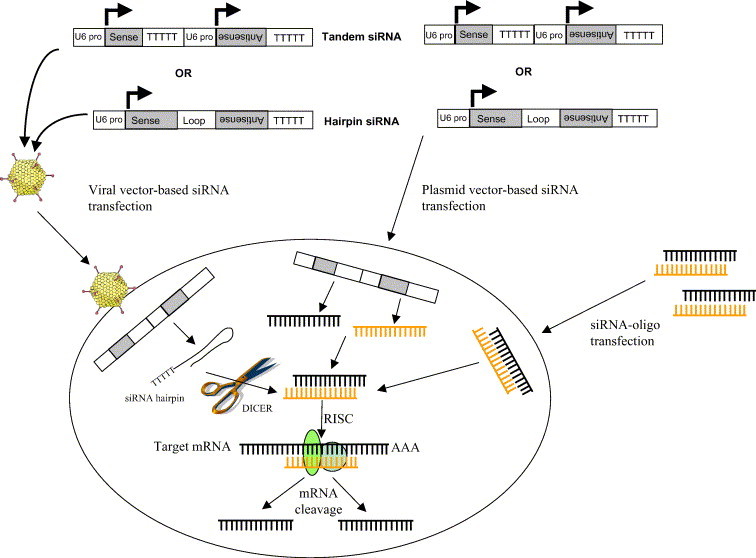
Tandem and hairpin-type expression vectors for RNAi. In tandem-type vectors, the sense and antisense strands are expressed by separate polymerase III promoters and the strands anneal inside the cells to form duplex siRNA molecules. In the hairpin-type vectors, sense and antisense strands are connected by a loop and are expressed as a single molecule, which rapidly forms a hairpin structure with a stem and a loop that are processed to siRNA by Dicer.
The limitations of using plasmid vectors in terms of efficiency of transfection and difficulty in transfecting primary cells (Dykxhoorn et al., 2003) have resulted in workers developing retroviral (Brummelkamp et al., 2002), lentiviral (Rubinson et al., 2003), and adenoviral (Arts et al., 2003) vector systems for shRNA delivery. Viral vectors permit the efficient delivery and stable expression of shRNA constructs in a range of mammalian cells (including primary cells) and a variety of animal species. Retroviral vectors are based on murine stem cell virus or Moloney murine leukemia virus and permit the stable introduction of shRNA into dividing cells (transformed and primary). They have been used to suppress gene expression in stem cells and reconstituted organs derived from those cells (Hemann et al., 2003). Lentiviral vectors are derived from human immunodeficiency virus (HIV)-1 and can infect both dividing and nondividing postmitotic cells (e.g., neurones) and have been used to generate transgenic animals that display loss-of-function phenotypes and vector transmission to offspring (Rubinson et al., 2003, Tiscornia et al., 2003). Adenoviral vectors based on adeno-associated viruses (AAV) can infect both dividing and nondividing cells, and because they integrate site specifically into the AAVS1 region of chromosome 19, they are safer than retroviral or lentiviral vectors, which are associated with insertional mutagenesis (Mittal, 2004). AAV vectors have been successfully used to silence genes, both in vitro and in vivo (Xia et al., 2002, Xia et al., 2004, Boden et al., 2004).
3.3. RNA interference in vivo
RNAi is being used for the knockdown of gene expression in animals and promises to provide a quicker and cheaper way to generate knockout animals. The delivery of siRNA into model mammalian organisms has been achieved by intravenous injection or by electroporation of synthetic siRNAs directly into target tissues and organs (Dillon et al., 2004). However, efficient and stable silencing has been most effectively obtained using retroviral and lentiviral vectors to transduce stem cells and embryos (Hemann et al., 2003, Rubinson et al., 2003). The results of experiments in a number of laboratories show that siRNA-mediated gene silencing functions in a range of cell and tissue types from blastocysts to adult animals and is both heritable and stable. Kunath et al. (2003) transfected mouse ES cells with constructs expressing shRNAs for Ras GTPase-activating protein (RasGAP) and were able to produce mice that died during embryogenesis, with defects similar to those of RasGAP knockout mice produced by homologous recombination. In another study, CD8 mRNA levels in T-cells were reduced in transgenic mice made using a lentiviral vector expressing a hairpin (Rubinson et al., 2003). Generating these mice is technically easier than generating knockout mice by the traditional route. Furthermore, the finding that the approach can be applied to species not amenable to embryonic stem cell gene targeting, such as the rat (Hasuwa et al., 2002), is particularly useful. Indications that the knockdown phenotype produced by RNAi may be more “variegated” than the traditional knockout phenotype (Kunath et al., 2003, Rubinson et al., 2003) may be useful in modelling complex human diseases, where susceptibility or resistance is encoded by gene variants that display relative rather than absolute differences in expression.
Cell-specific targeting of si/shRNA expression is an important consideration, particularly when considering RNAi for therapeutic applications (van Haaften et al., 2004) or as an alternative to low-molecular-weight molecules for target validation in vivo. To get around the lethal effects of knocking out genes that are crucial for development, inducible knockout systems are used (Kwak et al., 2004).Vectors that carry inducible promoters to allow the controlled expression of shRNA in response to an inducer based on the pol III promoters controlled by tetracycline (Matsukura et al., 2003), ecdysone (Gupta et al., 2004), or causes recombination (Cre) recombinase (Wiznerowicz & Trono, 2003, Fritsch et al., 2004, Kasim et al., 2004, Tiscornia et al., 2004) have been developed. An siRNA system that combines Cre–loxP for tissue-specific expression and tetracycline-on for inducible expression has been described recently (Chang et al., 2004). By mating one mouse (carrying the siRNA/loxP plasmid) with another mouse carrying the Cre recombinase in specific tissues, siRNA in the offspring is induced by tetracycline. Using this method, an ABCA1-deficient mouse line that mimicked Tangier disease was produced, without the need to use embryonic stem cells or gene targeting.
Although the production of transgenic mice by RNAi potentially provides a speedy way of producing knockout animals, it is not as well characterised as the knockout approach. It is unclear if the technology is robust enough to be a general approach for in vivo studies. Detailed analyses have not been performed to determine how reliably gene function can be knocked down in all tissues at all times throughout the life of the animal and what level of knockdown is required to result in a phenotype. The lentiviral approach often results in multiple integration events (Rubinson et al., 2003, Tiscornia et al., 2003), and mosaic expression of the shRNA might be expected.
3.4. High-throughput analysis of gene function
One of the most exciting opportunities offered by RNAi is the facility to identify all the genes required for certain physiological processes using genome-wide RNAi screens (Carpenter & Sabatini, 2004, Silva et al., 2004). High-throughput screens to identify genes involved in development and carcinogenesis have been successfully carried out in C. elegans (Lettre et al., 2004, Poulin et al., 2004) and D. melanogaster (Lum et al., 2003, Dasgupta & Perrimon, 2004), and web databases that archive and distribute RNAi data for these organisms have been developed (www.RNAi.org and www.flyRNAi.org).The ability to extend such screens to mammalian systems is potentially very powerful, as most genes have now been identified to some level of accuracy, hence, it should be possible to define the role of genes in cell-based phenotypic assays by systematic inhibition of gene expression. Although synthetic siRNAs are compatible with high-throughput formats (Vanhecke & Janitz, 2004a), genome-scale libraries of chemically synthesized siRNA have not been reported, however, smaller collections for the screening of between 30 and 500 genes have been (Aza-Blanc et al., 2003, Hsieh et al., 2004). An siRNA library that targeted > 8000 human genes based on an expression system in which siRNA duplexes are generated upon transfection into mammalian cells was used to identify regulators of nuclear factor-kappaB (NF-κB) signalling (Zheng et al., 2004).
Most of the efforts in this area have concentrated on the use of vector-based shRNA libraries (Vanhecke & Janitz, 2005). Large-scale gene-knockdown studies using retroviral vectors to express shRNAs have been reported in mammalian cells (Berns et al., 2004, Paddison et al., 2004). Both studies included “molecular bar codes” in the vectors so that individual shRNAs in the cells could be easily detected. The screens identified new members of the pathways analysed, however, the failure to detect certain known members of the pathways highlights the current limitations of these approaches. Adenoviral vectors have been used to generate an shRNA library aimed at knocking down nearly 5000 transcripts encoding “druggable” proteins (http://www.galadeno.com). Potentially, genome-wide screens could be carried out for any process for which a tissue culture model exists. Furthermore, the development of transfected cell arrays in which si/shRNAs are printed onto a modified glass surface (Vanhecke & Janitz, 2004b) provides a way of reducing reagent usage and costs in such large screens (Erfle et al., 2004). The large amounts of phenotypic data arising from such large-scale knockdown studies will require the development of bioinformatics tools to transform these complex datasets into “digitised” formats that can be readily mined (Gunsalus & Piano, 2005).
4. The use of RNA interference for the study of disease-associated genes
siRNA has been widely used in mammalian cells to define the functional roles of individual genes, particularly in disease (Dykxhoorn et al., 2003, Dillon et al., 2004). siRNAs have been extensively used in cell-based studies for pathway dissection (Semizarov et al., 2004, Sun et al., 2004, Siripurapu et al., 2005) and to knockdown the expression of genes involved in a range of cellular processes, including endocytosis (Huang et al., 2004a), signal transduction (Debes et al., 2002, Shu et al., 2002), apoptosis (Kartasheva et al., 2002, Lassus et al., 2002), and the cell cycle (Chen et al., 2002), as well as genes relevant to neurodegenerative disease (Alberi et al., 2004, Buckingham et al., 2004, Li et al., 2004, Thakker et al., 2004, Yokota et al., 2004) and xenotransplantation (Karlas et al., 2004). In this section, we will concentrate on the use of RNAi to study disease-relevant genes in cancer, infection, and respiratory disease.
4.1. Cancer
The success of the drug Gleevec (imatinib) has shown the effectiveness of directly targeting cancer-promoting proteins for cancer therapy by revolutionizing drug therapy of chronic myelogenous leukemia (CML; Deininger et al., 2004). However, drug resistance is common in advanced phases of the disease because of mutations in the kinase domain of the BCR–ABL fusion protein that impair imatinib binding. These fusion proteins are attractive candidates for RNAi. It has been shown that siRNA molecules targeting the junction fusion sequence attenuated the proliferation of transformed haematopoietic cells, sensitised cells to imatinib, and also inhibited an imatinib mutant (Chen et al., 2004). The ability of siRNAs to silence specific oncogenic variants while sparing the wild-type products of genes has been demonstrated for K-RasV12 in human pancreatic carcinoma (Brummelkamp et al., 2002). Similarly, a single point mutation in the tumour suppressor p53 was effectively discriminated using siRNAs (Martinez et al., 2002), resulting in the targeted destruction of the mutant, but not the wild-type product. In uterine cancers, specific knockdown of Akt isoforms 2 and 3, but not 1 using siRNA technology, was shown to significantly reduce the resistance of the endometrial KLE cells to cisplatin-induced apoptosis (Gagnon et al., 2004).
Receptors associated with certain mitogenic pathways are known to promote malignancy. For example, stimulation of the gonadotropin receptor activates the mitogen-activated protein kinase (MAPK) cascade, and this has been shown to enhance the development of a wide range of gynaecological malignancies, including ovarian and breast endometrial carcinomas (Freimann et al., 2004). When the chemokine receptor chemokine (C–X–C motif), receptor 4 (CXCR4) on the breast cancer cell line MDA-MB-231 was specifically targeted by siRNA, the rate of cell expansion slowed dramatically (Lapteva et al., 2004). Furthermore, α6β4 integrin (Lipscomb et al., 2003) and EpCAM (Osta et al., 2004c) receptors on the same cells were identified by RNA interference as being involved in cellular invasion and metastasis. In prostate cancers, the androgen receptor was shown to be essential in the androgen-induced down-regulation of oncogene B-cell chronic lymphocytic leukemia/lymphoma 2 (Bcl-2) expression using siRNAs directed against several genes encoding proteins involved in the androgen signalling pathway (Huang et al., 2004b).
The selective regulation of the expression of cancer-associated genes is evolving as another potential therapeutic approach. Clusterin is an antiapoptotic protein commonly expressed in cancer cells that are refractory to conventional chemotherapy. Using A549 cells, July et al. (2004) showed a strong correlation between siRNA-mediated reduction of clusterin protein and the chemosensitivity of the cells in vitro. Perhaps, the most well-studied member of this class of antiapoptotic proteins is survivin, whose expression levels have been shown to be directly proportional to tumour grade, recurrence risk, and survival of patients with bladder cancer (Ambrosini et al., 1997). Ning et al. (2004) demonstrated that survivin siRNAs promoted apoptosis and inhibited the survival of bladder cancer cells. Genes that encode the multidrug-resistance protein (MDR; pumps chemotherapeutic drugs out of tumours; Stege et al., 2004), telomerase (involved in overcoming the chromosomal shortening that occurs with each cell division; Ma et al., 2004), and Bcl-2 (makes cells resistant to caspase-mediated apoptosis; Yin et al., 2004) are further examples of genes that are not mutated in cancer but are overexpressed in a variety of cancers and represent attractive targets for silencing. DNA repair mechanisms are crucial for the maintenance of genomic stability and are emerging as potential therapeutic targets for cancer. Chow et al. (2004) used siRNA to identify endo-exonuclease (a protein involved in the recombination repair process of the DNA double-stranded break pathway, which is overexpressed in a variety of cancer cells), as a potential anticancer target.
Signalling molecules have been intensively researched as potential therapeutic targets in some cancers. For instance, B-RAF is a serine/threonine-specific protein kinase that is mutated in ∼ 70% of human melanomas, and B-RAF linked extracellular signal regulated protein kinase (ERK) signalling promotes proliferation and is antiapoptotic (Wellbrock et al., 2004). Karasarides et al. (2004) showed that B-RAF depletion by siRNA blocked ERK activity, inhibited DNA synthesis, and induced apoptosis in three melanoma cell lines. Many studies indicate that Rho GTPases are important in malignant transformation and angiogenesis and represent good anticancer targets (Aznar et al., 2004). Pille et al. (2005) used anti-RhoA and anti-RhoC siRNAs to block the Rho-signalling pathway and demonstrated that the siRNAs inhibited cell proliferation and invasion more effectively than do conventional blockers of Rho signalling (e.g., HMG-CoA reductase inhibitors), both in vitro and in vivo.
Reports on in vivo experiments of siRNA in cancer are growing. When nude mice were implanted with colon adenocarcinoma cells, the survival of these mice was greatly prolonged by pretreating the cells with siRNA against β-catenin (Verma et al., 2003). Similarly, silencing of the oncogene H-Ras led to the inhibition of in vivo tumour growth of human ovarian cancer in mice (Yang et al., 2003).
4.2. Infectious diseases
4.2.1. Viruses and bacteria
The availability of the genome sequences of a range of pathogenic viruses (http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/viruses.html) provides the basis for the development of new antiviral therapies (DeFilippis et al., 2003, Wang et al., 2003). Attractive targets are viral genes that are essential for virus replication and host genes that are essential for virus entry or that play an essential role in the virus life cycle. For RNA viruses such as HIV, potentially any region of the viral genome can be targeted by siRNA/shRNA. Silencing of genes relevant to viral diseases, such as HIV (Dave & Pomerantz, 2004), hepatitis (Wilson et al., 2003), and severe acute respiratory syndrome (SARS)-associated coronavirus (Zhang et al., 2004a), has been achieved using RNAi. In the case of HIV, silencing of the primary HIV receptor chemokine (C–C motif) receptor 5 (CCR5) by siRNA resulted in the prevention of viral entry into human peripheral blood lymphocytes (Qin et al., 2003) and primary haematopoietic stem cells (Li et al., 2003). In hepatitis C, siRNA silencing led to a 98% reduction in detectable virus in infected cells (Randall et al., 2003, Randall & Rice, 2004). Using siRNA targeted to the surface antigen region of the hepatitis B virus (HBV), Giladi et al. (2003) showed that the viral mRNA, viral antigens, and viral genomic DNA were significantly reduced in vitro and in vivo. Similarly, shRNAs have been used to inhibit the expression of hepatitis B virus surface antigen in HepG2 cells and decrease the levels of secreted antigen (Yang et al., 2005). In the case of SARS, it has recently been shown that SARS-associated coronavirus replication can be efficiently inhibited using siRNAs against two SARS viral polymerases (Wang et al., 2004). Similarly, Zhang et al. (2003) used the cytotoxicity of Vero cells as a surrogate marker of SARS-virus infection to show that siRNA transfection could effectively inhibit coronavirus replication.
The targeting of other infectious viruses using RNAi is also being pursued. For instance, it is possible to induce apoptosis in primary patient tumour samples by targeting the E6 gene of human papilloma virus (Butz et al., 2003). Plasmid-driven siRNA specific to West Nile Virus was effective in suppressing viral transcripts and infectious virion production in cell lines (McCown et al., 2003). In addition, siRNAs specific for the conserved regions of the influenza A virus genome can protect cell lines and embryonated chicken eggs against infection (Ge et al., 2004a). Respiratory syncytial virus (RSV) is ubiquitous in the environment and is the common cause of bronchiolitis-associated hospitalization of children and immunocompromised adults (McBride, 1999). Monick et al. (2004) showed that RSV infection activates ERK via epidermal growth factor receptor (EGFR), leading to pronounced inflammation and prolonged survival of infected cells. The inflammation was resolved, and apoptosis was induced in both infected immortalized (A549) and primary lung epithelial cells by targeting EGFR using siRNA. In another study, Rudd et al. (2005) showed that toll-like receptor 3 (TLR3) mediates inflammatory cytokine and chemokine production in RSV-infected epithelial cells using siRNAs targeting toll-like receptor 3 (TLR3). As a complement to the experiments in vitro, Zhang et al. (2005) showed that mice treated intranasally with siRNA nanoparticles targeting the viral NS1 gene before or after infection with RSV showed substantially decreased virus titers in the lung and decreased inflammation and airway reactivity compared with the controls.
In contrast to viruses, bacteria are not generally amenable to silencing by siRNA because they mainly replicate outside of host cells and lack the necessary machinery (Lieberman et al., 2003). However, it might still be possible to reduce morbidity and mortality from life-threatening bacterial infections by silencing host genes involved in aspects of the immune response that lead to adverse consequences, or host genes involved in mediating bacterial invasion. For instance, reducing the expression of proinflammatory cytokines such as tumour necrosis factor α (TNFα) lessened septic shock in mice treated with lipopolysaccharide (LPS) without jeopardizing the development of protective immunity (Sorensen et al., 2003). Knockdown of caveolin-2 in murine lung epithelial cells inhibited Pseudomonas aeruginosa (the major pulmonary pathogen in cystic fibrosis patients) invasion by a lipid raft-dependent mechanism (Zaas et al., 2005).
4.2.2. Parasites
Despite the rapid expansion of medical technologies in recent years, Malaria still claims an estimated 2.7 million deaths per year, with 90% of the casualties occurring in sub-Saharan Africa (Vernick & Waters, 2004). During the latter part of the 20th century, there was an alarming increase in the number of cases of Malaria reported in the Indian subcontinent, Southeast Asia, and South America (Malaney et al., 2004). Chloroquine remains the gold standard treatment for Malaria today. However, chloroquine resistance is a growing concern (Verdrager, 1995). The causative agent of the disease, Plasmodium falciparum, is a member of the intracellular protozoan family Apicomplexa (Kumar et al., 2002). Kumar et al. (2002) silenced a PP1 serine/threonine protein phosphatase in the parasite and showed that the enzyme plays an essential role in its life cycle, therefore offering a potential target for drug development. The downside to this approach is that PP1 is highly conserved throughout evolution, and toxic side effects can be expected unless specific variants of the protein are identified. The finding that the machinery for RNAi exists in Anopheles gambiae (the principal malaria vector in Africa; Hoa et al., 2003) has allowed insights into the effects of mosquito genes on Plasmodium development (Osta et al., 2004a). In addition, it has opened up the possibility of developing transgenic insect vectors that have innate resistance to the development and growth of P. falciparum (Osta et al., 2004b).
Another protozoan parasite, Entamoeba histolytica, causes human amoebiasis (Vayssie et al., 2004)—the second leading cause of death worldwide due to protozoan infection. Once inside its host, the parasite invades the intestinal mucosa, causing dysentery, and travels through the circulatory system to the liver, where it causes the development of abscesses (Petri, 2002). Using RNAi knockdown, Vayssie et al. (2004) demonstrated that γ-tubulin is essential for microtubule nucleation and cycling of the parasite. Importantly, the primary amino acid sequence of the protein is homologous (46%) but not identical to its human homolog. Therefore, specific siRNA may be developed to destroy the parasite's γ-tubulin while leaving the host's counterpart untouched.
4.3. Respiratory diseases
The discovery and development of inhaled antisense oligonucleotides (Ball et al., 2003) suggest that the delivery of siRNA molecules to the lung might be a viable approach to the treatment of respiratory diseases such as asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis.
COPD is a global health problem and is the fourth leading cause of death in the United States alone (Murphy, 2000). Goblet cell hyperplasia and mucus hypersecretion are prominent features of COPD, especially during exacerbations (Pistelli et al., 2003). Shao et al. (2004) showed that by specifically knocking down transforming growth factor (TGF)-α production via siRNA in NCI-H292 human airway epithelial cells, the expression of epidermal growth factor receptor (EGFR) was significantly reduced, leading to diminished mucus production. Another clinical manifestation of asthma and COPD is chronic inflammation and injury of both the airways and the parenchymal structures of the lung, in which TGF-β is thought to play a key role (Rennard, 1999, Sacco et al., 2004). TGF-β mediates a diverse range of cellular processes, including cell proliferation and differentiation, wound healing, and angiogenesis (Blobe et al., 2000). In a murine lung fibrosis model (Gurujeyalakshmi & Giri, 1995), interferon (IFN)-γ down-regulated TGF-β expression and improved fibrosis. Wen et al. (2004) further identified that TGF-β suppression was mediated via Smad 7 proteins, as targeted knockdown of Smad 7 by siRNA reversed the suppression. Jeffery et al. (2005) used Smad 1 siRNA to show that the antiproliferative and prodifferentiation effects of bone morphogenetic protein 4 (BMP4) on lung fibroblasts were Smad1 dependent. Fibroblast proliferation, differentiation, and migration contribute to the characteristic pulmonary vascular remodeling seen in primary pulmonary hypertension (PPH).
The recruitment of neutrophils to the lung is central to host defense against invading pathogens. However, in trauma patients exposed to a secondary infectious/septic challenge, neutrophil targeting and sequestration can result in acute lung injury (ALI) and contribute to the high morbidity/mortality seen in this population. ALI is a progressive syndrome in which an exaggerated inflammatory response is central to causing lung injury. Lomas-Neira et al. (in press) used a mouse model of haemorrhagic shock to analyse the effects of postshock/sepsis intratracheal instillation of siRNA targeting keratinocyte-derived chemokine (KC) and macrophage-inflammatory protein-2 (MIP-2) on the development of ALI. The results showed that the local production of MIP-2 was a regulator of neutrophil influx. Increased endothelial cell (EC) permeability is central to the pathophysiology of ALI. By silencing sphingosine 1-phosphate (S1P) expression using siRNA, Finigan et al. (2005) showed that activated protein C mediates EC permeability via the transactivation of S1P1, the potent barrier-enhancing receptor for S1P.
The airway epithelium provides a barrier against the external and internal milieu in the lung and is responsive to various environmental factors, as well as being an active participant in inflammatory and immune responses. Epithelial cells respond to cytokines, chemokines, hormones, growth factors, and inflammatory mediators, and it has been postulated that they play a crucial role in airway diseases such as asthma (Cookson, 2004). However, the intracellular signalling pathways involved in the regulation of epithelial cells are poorly understood. Using siRNA targeting Syk tyrosine kinase, Ulanova et al. (2005) showed that Syk (which is regarded as an attractive antiinflammatory target) participates in β1-integrin signalling and inflammatory responses in airway epithelial cells.
Cell-specific targeting of siRNA is an important issue to consider when considering therapy. With regards to the lung, Gou et al. (2004) were able to get RNAi silencing specifically in rat lung alveolar epithelial type II cells using adenoviral vectors containing various shRNAs under the control of the surfactant protein C promoter, both in vitro and in vivo.
5. siRNA molecules as therapeutics: from cells to man
Given the ability of RNAi to silence disease-associated genes, including allelic variants (Dykxhoorn et al., 2003), in tissue culture and animal models, it is not surprising that there has been a lot of interest in developing RNAi-based reagents for clinical applications (Wall & Shi, 2003). Currently, there are a dozen or so biotechnology companies developing clinical applications of siRNA in various human diseases (Howard, 2003; Table 1 ). In addition to the ease of synthesis and low production costs (compared with protein or antibody therapies), data indicate that siRNA has favourable pharmacokinetic properties and can be delivered to a wide range of organs (Braasch et al., 2004). However, blood stability, delivery, poor intracellular uptake, and nonspecific immune stimulation still present significant challenges for the development of RNAi reagents for clinical use.
Table 1.
Clinical pipelines in RNAi therapeutics
| Company | Disease | Status | Technology |
|---|---|---|---|
| Acuity Pharmaceuticals | Age-related Macular degeneration (AMD) | Phase I Clinical | modified siRNA |
| Alnylam Pharmaceuticals | AMD | Phase I Clinical | direct-siRNA |
| Alnylam Pharmaceuticals | Respiratory Syncytial Virus (RSV) | Phase I Clinical in 2006 | direct-siRNA |
| Alnylam Pharmaceuticals | Spinal Cord Injury | Preclinical | direct-siRNA |
| Alnylam Pharmaceuticals | Parkinson's disease | Preclinical | direct-siRNA |
| Benitec | Hepatitis C | Not available | DNA-direct siRNA |
| Benitec | Human immunodeficiency virus (HIV) | Phase I Clinical in 2005 | Multiple siRNA |
| Intradigm Corporation | Solid Tumour | Preclinical | nanoparticle-siRNA |
| Sirna Therapeutics | Age-related Macular degeneration (AMD) | Phase I Clinical | modified siRNA |
| Sirna Therapeutics | Hepatitis C | Preclinical | modified siRNA |
| Sirna Therapeutics | Asthma | Phase I Clinical in 2006 | systemic/aerosol siRNA |
| Sirna Therapeutics | Huntington's disease | Preclinical | AAV-delivered siRNA |
AAV, Adeno-associated Virus. The corresponding company websites are:www.acuitypharmaceuticals.com; www.alynylam.com; www.benitec.com.au; www.intradigm.com and www.sirna.com.
5.1. Stability
Although siRNA molecules appear to be more resistant to nuclease degradation than antisense molecules (Bertrand et al., 2002), some serum nucleases can degrade siRNAs (Paroo & Corey, 2004, Dykxhoorn & Lieberman, 2005). As a result, a number of groups have investigated the use of chemical modifications that improve stability and protect against nuclease degradation (Fougerolles et al., 2005). Although phosphothioate modification has been important for the success of antisense applications in vivo (Crooke, 2004), studies have shown that toxicity and loss of silencing activity can be a problem with phosphothioate siRNAs (Braasch et al., 2003, Harborth et al., 2003). As a result, several groups have investigated the use of alternative backbone and nucleotide modifications to address these problems. Hall et al. (2004) showed that boranophosphate siRNAs were more effective at silencing than phosphothioate siRNAs and were 10 times more nuclease resistant than are unmodified siRNAs. In addition, boranophosphate siRNAs were more potent than unmodified siRNAs and appeared to act through the standard RNAi pathway. Elmen et al. (2005) modified siRNAs with the synthetic RNA-like high-affinity nucleotide analogue, locked nucleic acid (LNA), and showed that the incorporation of a modest number of LNA modifications significantly enhanced the serum half-life of siRNAs and stabilized the duplex structure while not affecting gene silencing. Similarly, Allerson et al. (2005) showed that siRNAs containing 2′-O-methyl and 2′-fluoro nucleotides not only displayed enhanced stability but also showed increased potency. Experiments indicate that gene silencing by 2′-modified siRNAs in vivo is not impaired (Layzer et al., 2004).
As an alternative to backbone modifications, other groups have improved the stability of siRNAs and improved their delivery in vivo by complexing them with polyethyleneimine (PEI; Urban-Klein et al., 2005), atelocollagen (Minakuchi et al., 2004), or cholesterol (Soutschek et al., 2004). In another approach, biodegradable poly (d,l-lactide-co-glycolide) copolymer microspheres were shown to give sustained release of siRNA molecules at the site of administration in mice even after 7 days postadministration (Khan et al., 2004).
5.2. Delivery
A number of different approaches have been developed for the in vivo delivery of siRNA (Fig. 4 ). Among the approaches tried in rodents, rapid infusion by hydrodynamic injection of siRNA achieves the best delivery, albeit to a limited set of vascularized tissues (Lewis & Wolff, 2005). The primary recipient tissue is the liver, where successful transgene delivery and expression in up to 40% of hepatocytes can be obtained (Liu et al., 1999). Several groups have used this technique to successfully introduce siRNA- or shRNA-expressing constructs into mice, achieving efficient delivery and subsequent silencing of target genes (McCaffrey et al., 2002, Giladi et al., 2003, Zender et al., 2003, Hamar et al., 2004, Xu et al., 2005). A key constraint to this method is that delivery tends to be restricted to highly vascularized tissues, such as the liver, spleen, or kidneys. In addition, this technique is currently not a viable method for delivery in human clinical studies.
Fig. 4.
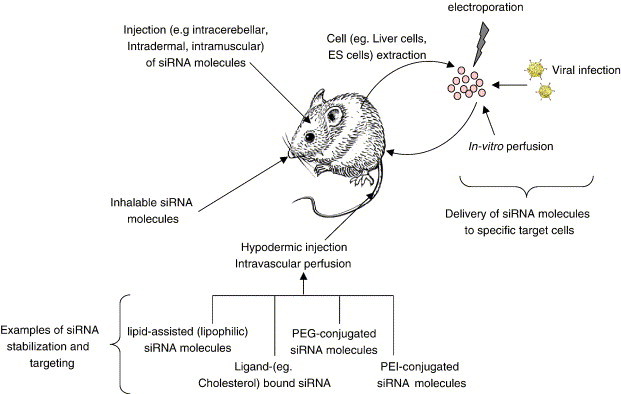
Strategies for the delivery of RNAi molecules in vivo.
In an effort to develop methods for targeted, tissue-specific delivery, lipid-based strategies for in vivo applications have been devised (Sioud & Sorensen, 2004). While lipid-based formulations have been used extensively for cell culture experiments, the attributes for optimal uptake in cell culture are different from those in vivo. The principal issue is that the cationic nature of the lipids used in cell culture leads to aggregation when used in animals and results in rapid serum clearance and lung accumulation. Despite this, there are an increasing number of reports citing success with lipid-mediated delivery of siRNAs in vivo (Ge et al., 2004b, Hassani et al., 2004, Zhang et al., 2004b). In a subcutaneous mouse tumour model, intraperitoneal administration of siRNAs complexed with PEI led to the delivery of intact siRNAs into tumours (Urban-Klein et al., 2005). Schiffelers et al. (2004) constructed self-assembling nanoparticles with siRNA and PEI PEGylated with an Arg-Gly-Asp peptide ligand attached to the distal end of the polyethylene glycol (PEG) as a means to target tumour neovasculature-expressing integrins. Intravenous administration into tumour-bearing mice resulted in selective tumour uptake, siRNA sequence-specific inhibition of protein expression within the tumour, and inhibition of both tumour angiogenesis and growth rate. The results show that siRNA can be targeted at two levels: tumour tissue-selective delivery via the nanoparticle ligand and gene pathway selectivity via the siRNA oligonucleotide. To improve the delivery of siRNA into human liver cells without transfection agents, Lorenz et al. (2004) synthesized lipophilic siRNAs conjugated with derivatives of cholesterol, lithocholic acid, or lauric acid. The lipid moieties were covalently linked to the 5′-ends of the RNAs using phosphoramidite chemistry and were shown to down-regulate the expression of a LacZ expression construct. By conjugating cholesterol to the 3′-end of the sense strand of siRNA by means of a pyrrolidine linker, Soutschek et al. (2004) markedly improved the pharmacological properties of siRNA molecules. Besides being more resistant to nuclease degradation, the cholesterol attachment stabilized the siRNA molecules in the blood by increasing binding to human serum albumin and increased uptake of siRNA molecules by the liver. Massaro et al. (2004) showed that siRNA could be effectively delivered to pulmonary alveoli in mice using pulmonary surfactant (a lipoprotein that lines the alveoli) as the delivery vehicle.
Intravascular delivery of siRNA molecules is attractive because of its inherent simplicity and has been used to deliver siRNA in mice. In an attempt to protect mice from fulminant hepatitis using siRNAs against Fas receptors, Song et al. (2003) administered Fas siRNA by intravenous injection into mice over a 24-hr period. The effects persisted for 10 days and protected mice against experimentally induced liver fibrosis. Hagstrom et al. (2004) injected naked siRNA into a distal vein of a limb transiently isolated by tourniquet or blood pressure cuff and were able to show an efficient and repeatable delivery of nucleic acids to muscle cells (myofibers) throughout the limb muscles of mammals. The rapid injection of a sufficient volume enabled the extravasation of siRNA into myofibers. High levels of transgene expression in skeletal muscle were achieved in both small and large animals with apparently minimal toxicity.
Other approaches for the delivery of siRNAs in vivo have been described. Local delivery of siRNAs into the eye has been used to show that siRNAs targeting the vascular endothelial growth factor (VEGF) pathway could be therapeutically beneficial in neovascularization-related eye diseases (Kim et al., 2004). Electroporation has been used to deliver siRNAs into the kidney (Takabatake et al., in press), brain (Akaneya et al., 2005), eyes (Matsuda & Cepko, 2004), muscles (Golzio et al., 2005), and skin (Zhang et al., 2002) of rodents. Topical gels have also been used to deliver siRNAs to cells and could open the way for dermatological applications, as well as the treatment of cervical cancer (Jiang et al., 2004). Intradermal administration of nucleic acids via gene gun has been used to deliver siRNA in vivo to enhance cancer vaccine potency (Kim et al., 2005). Ultrasound (Taniyama et al., 2002) could be also be potentially used to deliver siRNA, although there are no reports of this currently. For clinical applications, gentle methods for the delivery of siRNA molecules similar to those that have been developed for antisense molecules (inhaled, cream formulation, and oral; Crooke, 2004) would be preferable.
Several groups have investigated the use of direct injection of viral vectors for the in vivo delivery of siRNA. An AAV shRNA vector injected directly into the midbrain neurons of adult mice resulted in the silencing of the tyrosine hydroxylase gene near the site of injection for several weeks (Hommel et al., 2003). This resulted in behavioural changes, including a motor performance deficit and reduced response to a psychostimulant. Intracerebellar injection of an AAV vector expressing shRNAs directed against a mutant variant of ataxin-1 profoundly improved motor coordination, restored cerebellar morphology, and resolved characteristic ataxin-1 inclusions in Purkinje cells in a mouse model of the neurodegenerative disorder spinocerebellar ataxia (Xia et al., 2004). As an alternative to injection, Banerjea et al. (2003) used an ex vivo approach to generate HIV-1-resistant lymphocytes and macrophages. Using a lentiviral vector, an anti-rev siRNA construct was introduced into CD34(+) hematopoietic progenitor cells. The siRNA-transduced progenitor cells were allowed to mature into macrophages in vitro and T-cells in vivo in severe combined immunodeficiency (SCID) mice transplanted with human thymus and fetal liver grafts. Phenotypically normal T-cells and macrophages displaying characteristic surface markers were obtained. In vitro HIV-1 challenge of the siRNA-expressing macrophages and T-cells with macrophage-tropic and T-cell-tropic HIV-1, respectively, showed marked viral resistance. Despite these successes, reports of viral-mediated insertional mutagenesis in patients receiving retrovirus-mediated gene therapy for treatment of X-linked severe combined immunodeficiency (X-SCID; Hacein-Bey-Abina et al., 2003, Marshall, 2003) have raised concerns over the safety of using viral vectors for therapy.
5.3. Off-target and nonspecific effects
A careful selection of sequences is needed to maximise gene silencing and minimise off-target and nonspecific effects. Although initially thought to have laser-like specificity, the off-target effects of siRNA have become evident (Jackson et al., 2003, Couzin, 2004, Jackson & Linsley, 2004, Scacheri et al., 2004). An siRNA duplex may target more than 1 mRNA molecule because of sequence homologies. It is now widely observed that most siRNAs can tolerate 1 mismatch to the mRNA target and at the same time retain good silencing capacity (Yu et al., 2002, Pusch et al., 2003, Vickers et al., 2003). In some cases, siRNAs can tolerate several mismatches (Saxena et al., 2003) or even tolerate mismatches while acting as a single-stranded antisense siRNA (Holen et al., 2003). In addition, some domains of the siRNAs tolerate more mismatches than others do (Schwarz et al., 2003). Saxena et al. (2003) also demonstrated tolerance for G/U wobble pairing between the RNA oligo and the target RNA. To further investigate the effect of mismatches of siRNAs on off-target activity, Snove and Holen (2004) analysed 359 published siRNA sequences and found that around 75% could potentially elicit nonspecific effects. Microarray technology has been used to examine off-target effects, but results have been contradictory, with some studies finding a high specificity of siRNA effects (Chi et al., 2003, Semizarov et al., 2004), but others not (Jackson et al., 2003, Persengiev et al., 2004). Another consideration is that some off-target activities might actually represent genuine physiological knock-on effects of specific target knock-down in certain biochemical pathways/cascades. Therefore, such activity should be interpreted carefully in the context of the biological system in question, and the results of RNAi studies should be confirmed using complementary approaches or the use of multiple siRNA duplexes.
Recent data suggest that siRNAs and shRNAs can induce subsets of genes involved in the interferon response in mammalian cells. Sledz et al. (2003) showed that 21-bp siRNAs activate the Jak–Stat pathway and induce a global up-regulation of IFN-stimulated genes, mediated by the dsRNA-dependent protein kinase PKR. However, their findings also confirmed that siRNA-induced knockdown is independent of the IFN system. Another report (Bridge et al., 2003) linked the activation of interferon to the expression vectors carrying shRNA hairpins, while siRNA alone did not elicit such response. In a complementary study, Heidel et al. (2004) showed that it was possible to administer naked synthetic DNA to mice and down-regulate an endogenous or exogenous target without inducing the IFN response. In addition to the IFN response, it has been reported that si/shRNAs initiated immune activation in macrophages and dendritic cells through toll-like receptor 3 (toll receptors recognize exogenous nucleic acids; Kariko et al., 2004). Currently, it is unclear how often si/shRNAs trigger such effects in cells and what conditions favour these responses. Immunosuppression, selection of different administration routes, and dose of the vectors, using tissue-specific promoters and vector modification, are being investigated (Zhou et al., 2004).
Although RNAi in plants is associated with de novo RNA-directed DNA methylation (RdDM; Hamilton et al., 2002), experiments suggest that this may not be the case in mammalian cells (Park et al., 2004), although effects on other epigenetic modifications such as histone modification have yet to be assessed.
6. Conclusions and future prospects
In a relatively short time, RNAi has rapidly progressed to become a key experimental tool for the analysis of gene function and target validation in mammalian systems, both in vitro and in vivo. As a result, we have only been able to cite a small portion of the thousands of papers that have already been published in this area. However, we hope that we have been sufficiently thorough in our coverage to give readers an insight into the many uses to which this important technology is being put, as well as highlight current limitations and the potential of RNAi as a therapeutic strategy. The effectiveness of RNAi reagents will improve, as more is gleaned about the biology of RNAi in mammalian systems, and improvements in the stability, delivery, and reduction of off-target and nonspecific effects are made. Although still a fledgling area, clinical applications for RNAi will build on the lessons learned from clinical use of antisense molecules. The delivery (viral or otherwise) and target specificity of siRNA will be the keys to unlock this transition. RNAi-based gene therapy has great potential in cancer and infectious disease, as well as in genetic diseases that are due to a dominant genetic effect (e.g., Huntington's chorea and amyotrophic lateral sclerosis). The use of inducible RNAi systems will be particularly attractive in the clinical setting, where the ability to reverse siRNA treatment by simply withdrawing the inducing agent would allow treatment to be terminated as appropriate.
References
- Akaneya Y., Jiang B., Tsumoto T. RNAi-induced gene silencing by local electroporation in targeting brain region. J Neurophysiol. 2005;93(1):594–602. doi: 10.1152/jn.00161.2004. [DOI] [PubMed] [Google Scholar]
- Alberi L., Sgado P., Simon H.H. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131(13):3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J Med Chem. 2005;48(4):901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Ambrosini G., Adida C., Altieri D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Archacki S., Wang Q. Expression profiling of cardiovascular disease. Hum Genomics. 2004;1(5):355–370. doi: 10.1186/1479-7364-1-5-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts G.J., Langemeijer E., Tissingh R., Ma L., Pavliska H., Dokic K. Adenoviral vectors expressing SiRNAs for discovery and validation of gene function. Genome Res. 2003;13(10):2325–2332. doi: 10.1101/gr.1332603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Cooper C.L., Wagner K., Batalov S., Deveraux Q.L., Cooke M.P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12(3):627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- Aznar S., Fernandez-Valeron P., Espina C., Lacal J.C. Rho GTPases: potential candidates or anticancer therapy. Cancer Lett. 2004;206(2):181–191. doi: 10.1016/j.canlet.2003.08.035. [DOI] [PubMed] [Google Scholar]
- Ball H.A., Sandrasagra A., Tang L., Van Scott M., Wild J., Nyce J.W. Clinical potential of respirable antisense oligonucleotides (RASONs) in asthma. Am J Pharmacogenomics. 2003;3(2):97–106. doi: 10.2165/00129785-200303020-00003. [DOI] [PubMed] [Google Scholar]
- Banerjea A., Li M.J., Bauer G., Remling L., Lee N.S., Rossi J. Inhibition of HIV-1 by lentiviral vector-transduced SiRNAs in T lymphocytes differentiated in SCID-Hu mice and CD34+ progenitor cell-derived macrophages. Mol Ther. 2003;8(1):62–71. doi: 10.1016/s1525-0016(03)00140-0. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M. A large-scale RNAi screen in human cells identifies new components of he p53 pathway. Nature. 2004;428(6981):431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bertrand J.R., Pottier M., Vekris A., Opolon P., Maksimenko A., Malvy C. Comparison of antisense oligonucleotides and SiRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296(4):1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- Blobe G.C., Schiemann W.P., Lodish H.F. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342(18):1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Efficient gene transfer of HIV-1-specific short hairpin RNA into human lymphocytic cells using recombinant adeno-associated virus vectors. Mol Ther. 2004;9(3):396–402. doi: 10.1016/j.ymthe.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42(26):7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Braasch D.A., Paroo Z., Constantinescu A., Ren G., Oz O.K., Mason R.P. Biodistribution of phosphodiester and phosphorothioate SiRNA. Bioorg Med Chem Lett. 2004;14(5):1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- Brazas R.M., Hagstrom J.E. Delivery of small interfering RNA to mammalian cells in culture by using cationic lipid? Polymer-based transfection reagents. Methods Enzymol. 2005;392:112–124. doi: 10.1016/S0076-6879(04)92007-1. [DOI] [PubMed] [Google Scholar]
- Breaker R.R. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432(7019):838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2(3):243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Buckingham S.D., Esmaeili B., Wood M., Sattelle D.B. RNA interference: from model organisms towards therapy for neural and neuromuscular disorders. Hum Mol Genet. 2004;13(Spec No 2):R275–R288. doi: 10.1093/hmg/ddh224. [DOI] [PubMed] [Google Scholar]
- Butz K., Ristriani T., Hengstermann A., Denk C., Scheffner M., Hoppe-Seyler F. SiRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22(38):5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Parrish S., Imani F., Fire A., Morgan R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci U S A. 2001;98(17):9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Sabatini D.M. Systematic genome-wide screens of gene function. Nat Rev Genet. 2004;5(1):11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425(6956):411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Chang H.S., Lin C.H., Chen Y.C., Yu W.C. Using SiRNA technique to generate transgenic animals with spatiotemporal and conditional gene knockdown. Am J Pathol. 2004;165(5):1535–1541. doi: 10.1016/S0002-9440(10)63411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., Dynlacht B.D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3(3):339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Chen J., Wall N.R., Kocher K., Duclos N., Fabbro D., Neuberg D. Stable expression of small interfering RNA sensitizes TEL-PDGFbetaR to inhibition with imatinib or rapamycin. J Clin Invest. 2004;113(12):1784–1791. doi: 10.1172/JCI20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J.T., Chang H.Y., Wang N.N., Chang D.S., Dunphy N., Brown P.O. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100(11):6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow T.Y., Alaoui-Jamali M.A., Yeh C., Yuen L., Griller D. The DNA double-stranded break repair protein endo-exonuclease as a therapeutic target for cancer. Mol Cancer Ther. 2004;3(8):911–919. [PubMed] [Google Scholar]
- Cookson W. The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol. 2004;4(12):978–988. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- Couzin J. Breakthrough of the year. Small RNAs make big splash. Science. 2002;298(5602):2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- Couzin J. Molecular biology. RNAi shows cracks in its armor. Science. 2004;306(5699):1124–1125. doi: 10.1126/science.306.5699.1124. [DOI] [PubMed] [Google Scholar]
- Crooke S.T. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- Dasgupta R., Perrimon N. Using RNAi to catch Drosophila genes in a web of interactions: insights into cancer research. Oncogene. 2004;23(51):8359–8365. doi: 10.1038/sj.onc.1208028. [DOI] [PubMed] [Google Scholar]
- Dave R.S., Pomerantz R.J. Antiviral effects of human immunodeficiency virus type 1-specific small interfering RNAs against targets conserved in select neurotropic viral strains. J Virol. 2004;78(24):13687–13696. doi: 10.1128/JVI.78.24.13687-13696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes J.D., Schmidt L.J., Huang H., Tindall D.J. P300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Res. 2002;62(20):5632–5636. [PubMed] [Google Scholar]
- DeFilippis V., Raggo C., Moses A., Fruh K. Functional genomics in virology and antiviral drug discovery. Trends Biotechnol. 2003;21(10):452–457. doi: 10.1016/S0167-7799(03)00207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2004;105(7):2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- Dillon C.P., Sandy P., Nencioni A., Kissler S., Rubinson D.A., Van Parijs L. RNAi as an experimental and therapeutic tool to study and regulate physiological and disease processes. Annu Rev Physiol. 2004;67:147–173. doi: 10.1146/annurev.physiol.67.040403.130716. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. SiRNAs can function as MIRNAS. Genes Dev. 2003;17(4):438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews J. Drug discovery: a historical perspective. Science. 2000;287(5460):1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4(6):457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elmen J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y. Locked nucleic acid (LNA) mediated improvements in SiRNA stability and functionality. Nucleic Acids Res. 2005;33(1):439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfle H., Simpson J.C., Bastiaens P.I, Pepperkok R. SiRNA cell arrays for high-content screening microscopy. Biotechniques. 2004;37(3):454–458. doi: 10.2144/04373RT01. [DOI] [PubMed] [Google Scholar]
- Finigan J.H., Dudek S.M., Singleton P.A., Chiang E.T., Jacobson J.R., Camp S.M. Activated protein C mediates novel lung endothelial barrier enhancement: role of sphingosine 1-phosphate receptor transactivation. J Biol Chem. 2005;280(17):17286–17293. doi: 10.1074/jbc.M412427200. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fougerolles A., Manoharan M., Meyers R., Vornlocher H.P. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- Freimann S., Ben Ami I., Hirsh L., Dantes A., Halperin R., Amsterdam A. Drug development for ovarian hyper-stimulation and anti-cancer treatment: blocking of gonadotropin signaling for epiregulin and amphiregulin biosynthesis. Biochem Pharmacol. 2004;68(6):989–996. doi: 10.1016/j.bcp.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Fritsch L., Martinez L.A., Sekhri R., Naguibneva I., Gerard M., Vandromme M. Conditional gene knock-down by cre-dependent short interfering RNAs. EMBO Rep. 2004;5(2):178–182. doi: 10.1038/sj.embor.7400064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon V., Mathieu I., Sexton E., Leblanc K., Asselin E. AKT involvement in cisplatin chemoresistance of human uterine cancer cells. Gynecol Oncol. 2004;94(3):785–795. doi: 10.1016/j.ygyno.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Ge Q., Eisen H.N., Chen J. Use of SiRNAs to prevent and treat influenza virus infection. Virus Res. 2004;102(1):37–42. doi: 10.1016/j.virusres.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Ge Q., Filip L., Bai A., Nguyen T., Eisen H.N., Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004;101(23):8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H., Ketzinel-Gilad M., Rivkin L., Felig Y., Nussbaum O., Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8(5):769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Golzio M., Mazzolini L., Moller P., Rols M.P., Teissie J. Inhibition of gene expression in mice muscle by in vivo electrically mediated SiRNA delivery. Gene Ther. 2005;12(3):246–251. doi: 10.1038/sj.gt.3302405. [DOI] [PubMed] [Google Scholar]
- Gou D., Narasaraju T., Chintagari N.R., Jin N., Wang P., Liu L. Gene silencing in alveolar type II cells using cell-specific promoter in vitro and in vivo. Nucleic Acids Res. 2004;32(17):e134. doi: 10.1093/nar/gnh129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresch O., Engel F.B., Nesic D., Tran T.T., England H.M., Hickman E.S. New non-viral method for gene transfer into primary cells. Methods. 2004;33(2):151–163. doi: 10.1016/j.ymeth.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Grishok A., Mello C.C. RNAi (Nematodes: Caenorhabditis elegans) Adv Genet. 2002;46:339–360. doi: 10.1016/s0065-2660(02)46012-9. [DOI] [PubMed] [Google Scholar]
- Gunsalus K.C., Piano F. RNAi as a tool to study cell biology: building the genome–phenome bridge. Curr Opin Cell Biol. 2005;17(1):3–8. doi: 10.1016/j.ceb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Gupta S., Schoer R.A., Egan J.E., Hannon G.J., Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(7):1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurujeyalakshmi G., Giri S.N. Molecular mechanisms of antifibrotic effect of interferon gamma in bleomycin-mouse model of lung fibrosis: downregulation of TGF-beta and procollagen I and III gene expression. Exp Lung Res. 1995;21(5):791–808. doi: 10.3109/01902149509050842. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L. A facile nonviral method for delivering genes and SiRNAs to skeletal muscle of mammalian limbs. Mol Ther. 2004;10(2):386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Hall A.H., Wan J., Shaughnessy E.E., Ramsay S.B., Alexander K.A. RNA interference using boranophosphate SiRNAs: structure–activity relationships. Nucleic Acids Res. 2004;32(20):5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamar P., Song E., Kokeny G., Chen A., Ouyang N., Lieberman J. Small interfering RNA targeting fas protects mice against renal ischemia–reperfusion injury. Proc Natl Acad Sci U S A. 2004;101(41):14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21(17):4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13(2):83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- Hardy L.W., Peet N.P. The multiple orthogonal tools approach to define molecular causation in the validation of druggable targets. Drug Discov Today. 2004;9(3):117–126. doi: 10.1016/S1359-6446(03)02969-6. [DOI] [PubMed] [Google Scholar]
- Hassani Z., Lemkine G.F., Erbacher P., Palmier K., Alfama G., Giovannangeli C. Lipid-mediated SiRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. J Gene Med. 2004;7(2):198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- Hasuwa H., Kaseda K., Einarsdottir T., Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532(1–2):227–230. doi: 10.1016/s0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- Heidel J.D., Hu S., Liu X.F., Triche T.J., Davis M.E. Lack of interferon response in animals to naked SiRNAs. Nat Biotechnol. 2004;22(12):1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- Hemann M.T., Fridman J.S., Zilfou J.T., Hernando E., Paddison P.J., Cordon-Cardo C. An Epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33(3):396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Hoa N.T., Keene K.M., Olson K.E., Zheng L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem Mol Biol. 2003;33(9):949–957. doi: 10.1016/s0965-1748(03)00101-2. [DOI] [PubMed] [Google Scholar]
- Hoelters, J., Ciccarella, M., Drechsel, M., Geissler, C., Gulkan, H., & Bocker, W., et al. (in press). Nonviral genetic modification mediates effective transgene expression and functional RNA interference in human mesenchymal stem cells.J Gene Med. [DOI] [PubMed]
- Holen T., Amarzguioui M., Babaie E., Prydz H. Similar behaviour of single-strand and double-strand SiRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31(9):2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel J.D., Sears R.M., Georgescu D., Simmons D.L., DiLeone R.J. Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med. 2003;9(12):1539–1544. doi: 10.1038/nm964. [DOI] [PubMed] [Google Scholar]
- Hopkins A.L., Groom C.R. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Howard K. Unlocking the money-making potential of RNAi. Nat Biotechnol. 2003;21(12):1441–1446. doi: 10.1038/nbt1203-1441. [DOI] [PubMed] [Google Scholar]
- Hsieh A.C., Bo R., Manola J., Vazquez F., Bare O., Khvorova A. A library of SiRNA duplexes targeting the phosphoinositide 3-kinase pathway: determinants of gene silencing for use in cell-based screens. Nucleic Acids Res. 2004;32(3):893–901. doi: 10.1093/nar/gkh238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Khvorova A., Marshall W., Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279(16):16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- Huang H., Zegarra-Moro O.L., Benson D., Tindall D.J. Androgens repress bcl-2 expression via activation of the retinoblastoma (rb) protein in prostate cancer cells. Oncogene. 2004;23(12):2161–2176. doi: 10.1038/sj.onc.1207326. [DOI] [PubMed] [Google Scholar]
- Ikeda R., Yoshida K., Tsukahara S., Sakamoto Y., Tanaka H., Furukawa K.I. PLZF promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. J Biol Chem. 2004;280(9):8523–8530. doi: 10.1074/jbc.M409442200. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Linsley P.S. Noise amidst the silence: off-target effects of SiRNAs? Trends Genet. 2004;20(11):521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jeffery T.K., Upton P.D., Trembath R.C., Morrell N.W. BMP4 inhibits proliferation and promotes myocyte differentiation of lung fibroblasts via Smad1 and JNK pathways. Am J Physiol Lung Cell Mol Physiol. 2005;288(2):L370–L378. doi: 10.1152/ajplung.00242.2004. [DOI] [PubMed] [Google Scholar]
- Jiang M., Rubbi C.P., Milner J. Gel-based application of SiRNA to human epithelial cancer cells induces RNAi-dependent apoptosis. Oligonucleotides. 2004;14(4):239–248. doi: 10.1089/oli.2004.14.239. [DOI] [PubMed] [Google Scholar]
- July L.V., Beraldi E., So A., Fazli L., Evans K., English J.C. Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol Cancer Ther. 2004;3(3):223–232. [PubMed] [Google Scholar]
- Kao S.C., Krichevsky A.M., Kosik K.S., Tsai L.H. BACE1 suppression by RNA interference in primary cortical neurons. J Biol Chem. 2004;279(3):1942–1949. doi: 10.1074/jbc.M309219200. [DOI] [PubMed] [Google Scholar]
- Karasarides M., Chiloeches A., Hayward R., Niculescu-Duvaz D., Scanlon I., Friedlos F. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23(37):6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- Kariko K., Bhuyan P., Capodici J., Ni H., Lubinski J., Friedman H. Exogenous SiRNA mediates sequence-independent gene suppression by signaling through toll-like receptor 3. Cells Tissues Organs. 2004;177(3):132–138. doi: 10.1159/000079987. [DOI] [PubMed] [Google Scholar]
- Karlas A., Kurth R., Denner J. Inhibition of porcine endogenous retroviruses by RNA interference: increasing the safety of xenotransplantation. Virology. 2004;325(1):18–23. doi: 10.1016/j.virol.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Kartasheva N.N., Contente A., Lenz-Stoppler C., Roth J., Dobbelstein M. P53 induces the expression of its antagonist p73 delta N, establishing an autoregulatory feedback loop. Oncogene. 2002;21(31):4715–4727. doi: 10.1038/sj.onc.1205584. [DOI] [PubMed] [Google Scholar]
- Kasim V., Miyagishi M., Taira K. Control of SiRNA expression using the cre-loxp recombination system. Nucleic Acids Res. 2004;32(7):e66. doi: 10.1093/nar/gnh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Benboubetra M., Sayyed P.Z., Ng K.W., Fox S., Beck G. Sustained polymeric delivery of gene silencing antisense ODNs, SiRNA, DNAzymes and ribozymes: in vitro and in vivo studies. J Drug Target. 2004;12(6):393–404. doi: 10.1080/10611860400003858. [DOI] [PubMed] [Google Scholar]
- Kim B., Tang Q., Biswas P.S., Xu J., Schiffelers R.M., Xie F.Y. Inhibition of ocular angiogenesis by SiRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165(6):2177–2185. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., He L., Boyd D.A., Hardwick J.M., Hung C.F. Modification of professional antigen-presenting cells with small interfering rna in vivo to enhance cancer vaccine potency. Cancer Res. 2005;65(1):309–316. [PubMed] [Google Scholar]
- Kisielow M., Kleiner S., Nagasawa M., Faisal A., Nagamine Y. Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem J. 2002;363(Pt 1):1–5. doi: 10.1042/0264-6021:3630001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D. An endoribonuclease-prepared SiRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432(7020):1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Krick S., Eul B.G., Hanze J., Savai R., Grimminger F., Seeger W. Role of HIF-1{Alpha} in hypoxia-induced apoptosis of primary alveolar epithelial type II cells. Am J Respir Cell Mol Biol. 2005;32(5):395–403. doi: 10.1165/rcmb.2004-0314OC. [DOI] [PubMed] [Google Scholar]
- Kumar R., Adams B., Oldenburg A., Musiyenko A., Barik S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: demonstration of its essential role using RNA interference. Malar J. 2002;1(1):5. doi: 10.1186/1475-2875-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Conklin D.S., Mittal V. High-throughput selection of effective RNAi probes for gene silencing. Genome Res. 2003;13(10):2333–2340. doi: 10.1101/gr.1575003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Gish G., Lickert H., Jones N., Pawson T., Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol. 2003;21(5):559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- Kuttenkeuler D., Boutros M. Genome-wide RNAi as a route to gene function in Drosophila. Brief Funct Genomic Proteomic. 2004;3(2):168–176. doi: 10.1093/bfgp/3.2.168. [DOI] [PubMed] [Google Scholar]
- Kwak I., Tsai S.Y., DeMayo F.J. Genetically engineered mouse models for lung cancer. Annu Rev Physiol. 2004;66:647–663. doi: 10.1146/annurev.physiol.66.032102.134301. [DOI] [PubMed] [Google Scholar]
- Lapteva N., Yang A.G., Sanders D.E., Strube R.W., Chen S.Y. CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo. Cancer Gene Ther. 2004;12(1):84–89. doi: 10.1038/sj.cgt.7700770. [DOI] [PubMed] [Google Scholar]
- Lassus P., Opitz-Araya X., Lazebnik Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 2002;297(5585):1352–1354. doi: 10.1126/science.1074721. [DOI] [PubMed] [Google Scholar]
- Layzer J.M., McCaffrey A.P., Tanner A.K., Huang Z., Kay M.A., Sullenger B.A. In vivo activity of nuclease-resistant SiRNAs. RNA. 2004;10(5):766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J. The nuclear RNase III drosha initiates MicroRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lettre G., Kritikou E.A., Jaeggi M., Calixto A., Fraser A.G., Kamath R.S. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C Elegans. Cell Death Differ. 2004;11(11):1198–1203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- Lewis D.L., Wolff J.A. Delivery of SiRNA and SiRNA expression constructs to adult mammals by hydrodynamic intravascular injection. Methods Enzymol. 2005;392:336–350. doi: 10.1016/S0076-6879(04)92020-4. [DOI] [PubMed] [Google Scholar]
- Li M.J., Bauer G., Michienzi A., Yee J.K., Lee N.S., Kim J. Inhibition of HIV-1 infection by lentiviral vectors expressing pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8(2):196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- Li Y., Yokota T., Matsumura R., Taira K., Mizusawa H. Sequence-dependent and independent inhibition specific for mutant ataxin-3 by small interfering RNA. Ann Neurol. 2004;56(1):124–129. doi: 10.1002/ana.20141. [DOI] [PubMed] [Google Scholar]
- Lieberman J., Song E., Lee S.K., Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol Med. 2003;9(9):397–403. doi: 10.1016/S1471-4914(03)00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb E.A., Dugan A.S., Rabinovitz I., Mercurio A.M. Use of RNA interference to inhibit integrin (Alpha6beta4)-mediated invasion and migration of breast carcinoma cells. Clin Exp Metastasis. 2003;20(6):569–576. doi: 10.1023/a:1025819521707. [DOI] [PubMed] [Google Scholar]
- Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Lomas-Neira, J. L., Chung, C. S., Wesche, D. E., Perl, M., & Ayala, A., (in press). In vivo gene silencing (With SiRNA) of pulmonary expression of MIP-2 versus KC results in divergent effects on hemorrhage-induced, neutrophil-mediated septic acute lung injury.J Leukoc Biol. [DOI] [PubMed]
- Lorenz C., Hadwiger P., John M., Vornlocher H.P., Unverzagt C. Steroid and lipid conjugates of SiRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14(19):4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Lum L., Yao S., Mozer B., Rovescalli A., Von Kessler D., Nirenberg M. Identification of hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299(5615):2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Ma J.P., Zhan W.H., Wang J.P., Peng J.S., Gao J.S., Yin Q.W. Specific inhibition of HTERT gene expression by short interfering RNAs in gastric cancer SGC7901 cell. Zhonghua Wai Ke Za Zhi. 2004;42(22):1372–1376. [PubMed] [Google Scholar]
- Malaney P., Spielman A., Sachs J. The Malaria Gap. Am J Trop Med Hyg. 2004;71(2 Suppl):141–146. [PubMed] [Google Scholar]
- Marshall E. Gene therapy. Second child in french trial is found to have leukemia. Science. 2003;299(5605):320. doi: 10.1126/science.299.5605.320. [DOI] [PubMed] [Google Scholar]
- Martinez L.A., Naguibneva I., Lehrmann H., Vervisch A., Tchenio T., Lozano G. Synthetic small Inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore P53 pathways. Proc Natl Acad Sci U S A. 2002;99(23):14849–14854. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro D., Massaro G.D., Clerch L.B. Noninvasive delivery of small inhibitory RNA and other reagents to pulmonary alveoli in mice. Am J Physiol Lung Cell Mol Physiol. 2004;287(5):L1066–L1070. doi: 10.1152/ajplung.00067.2004. [DOI] [PubMed] [Google Scholar]
- Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101(1):16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S., Jones P.A., Takai D. Establishment of conditional Vectors for hairpin SiRNA knockdowns. Nucleic Acids Res. 2003;31(15):e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride J.T. Pulmonary function changes in children after respiratory syncytial virus infection in infancy. J Pediatr. 1999;135(2 Pt 2):28–32. [PubMed] [Google Scholar]
- McCaffrey A.P., Ohashi K., Meuse L., Shen S., Lancaster A.M., Lukavsky P.J. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5(6):676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- McCown M., Diamond M.S., Pekosz A. The utility of SiRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology. 2003;313(2):514–524. doi: 10.1016/s0042-6822(03)00341-6. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431(7006):338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Minakuchi Y., Takeshita F., Kosaka N., Sasaki H., Yamamoto Y., Kouno M. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32(13):e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V. Improving the efficiency of RNA interference in mammals. Nat Rev Genet. 2004;5(5):355–365. doi: 10.1038/nrg1323. [DOI] [PubMed] [Google Scholar]
- Miyagishi M., Taira K. Strategies for generation of an SiRNA expression library directed against the human genome. Oligonucleotides. 2003;13(5):325–333. doi: 10.1089/154545703322617005. [DOI] [PubMed] [Google Scholar]
- Monick M.M., Cameron K., Staber J., Powers L.S., Yarovinsky T.O., Koland J.G. Activation of the epidermal growth factor receptor by respiratory syncytial virus results in increased inflammation and delayed apoptosis. J Biol Chem. 2004;280(3):2147–2158. doi: 10.1074/jbc.M408745200. [DOI] [PubMed] [Google Scholar]
- Muratovska A., Eccles M.R. Conjugate for efficient delivery of short interfering RNA (SiRNA) into mammalian cells. FEBS Lett. 2004;558(1–3):63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- Murphy S.L. Deaths: final data for 1998. Natl Vital Stat Rep. 2000;48(11):1–105. [PubMed] [Google Scholar]
- Ning S., Fuessel S., Kotzsch M., Kraemer K., Kappler M., Schmidt U. SiRNA-mediated down-regulation of survivin inhibits bladder cancer cell growth. Int J Oncol. 2004;25(4):1065–1071. [PubMed] [Google Scholar]
- Omi K., Tokunaga K., Hohjoh H. Long-lasting RNAi activity in mammalian neurons. FEBS Lett. 2004;558(1–3):89–95. doi: 10.1016/S0014-5793(04)00017-1. [DOI] [PubMed] [Google Scholar]
- Osta M.A., Christophides G.K., Kafatos F.C. Effects of mosquito genes on plasmodium development. Science. 2004;303(5666):2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- Osta M.A., Christophides G.K., Vlachou D., Kafatos F.C. Innate immunity in the malaria vector Anopheles gambiae: comparative and functional genomics. J Exp Biol. 2004;207(Pt 15):2551–2563. doi: 10.1242/jeb.01066. [DOI] [PubMed] [Google Scholar]
- Osta W.A., Chen Y., Mikhitarian K., Mitas M., Salem M., Hannun Y.A. EpCAM Is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64(16):5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428(6981):427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Park C.W., Chen Z., Kren B.T., Steer C.J. Double-stranded SiRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem Biophys Res Commun. 2004;323(1):275–280. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- Paroo Z., Corey D.R. Challenges for RNAi in Vivo. Trends Biotechnol. 2004;22(8):390–394. doi: 10.1016/j.tibtech.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Persengiev S.P., Zhu X., Green M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (SiRNAs) RNA. 2004;10(1):12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri W.A., Jr. Pathogenesis of amebiasis. Curr Opin Microbiol. 2002;5(4):443–447. doi: 10.1016/s1369-5274(02)00335-1. [DOI] [PubMed] [Google Scholar]
- Pille J.Y., Denoyelle C., Varet J., Bertrand J.R., Soria J., Opolon P. Anti-RhoA and Anti-RhoC SiRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther. 2005;11(2):267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Pistelli R., Lange P., Miller D.L. Determinants of prognosis of COPD in the elderly: mucus hypersecretion, infections, cardiovascular comorbidity. Eur Respir J. 2003;Suppl 40:10s–14s. doi: 10.1183/09031936.03.00403403. [DOI] [PubMed] [Google Scholar]
- Poulin G., Nandakumar R., Ahringer J. Genome-wide RNAi screens in Caenorhabditis elegans: impact on cancer research. Oncogene. 2004;23(51):8340–8345. doi: 10.1038/sj.onc.1208010. [DOI] [PubMed] [Google Scholar]
- Pusch O., Boden D., Silbermann R., Lee F., Tucker L., Ramratnam B. Nucleotide sequence homology requirements of HIV-1-specific short hairpin RNA. Nucleic Acids Res. 2003;31(22):6444–6449. doi: 10.1093/nar/gkg876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.F., An D.S., Chen I.S., Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100(1):183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G., Grakoui A., Rice C.M. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100(1):235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G., Rice C.M. Interfering With hepatitis C virus RNA replication. Virus Res. 2004;102(1):19–25. doi: 10.1016/j.virusres.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rennard S.I. Inflammation and repair processes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160(5 Pt 2):S12–S16. doi: 10.1164/ajrccm.160.supplement_1.5. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational SiRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rosahl T.W. Validation of GABA(A) receptor subtypes as potential drug targets by using genetically modified mice. Curr Drug Targets CNS Neurol Disord. 2003;2(4):207–212. doi: 10.2174/1568007033482823. [DOI] [PubMed] [Google Scholar]
- Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33(3):401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Rudd B.D., Burstein E., Duckett C.S., Li X., Lukacs N.W. Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J Virol. 2005;79(6):3350–3357. doi: 10.1128/JVI.79.6.3350-3357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco O., Silvestri M., Sabatini F., Sale R., Defilippi A.C., Rossi G.A. Epithelial cells and fibroblasts: structural repair and remodelling in the airways. Paediatr Respir Rev. 2004;5(Suppl A):S35–S40. doi: 10.1016/s1526-0542(04)90008-5. [DOI] [PubMed] [Google Scholar]
- Saxena S., Jonsson Z.O., Dutta A. Small RNAs with imperfect match to endogenous MRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278(45):44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(7):1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers R.M., Ansari A., Xu J., Zhou Q., Tang Q., Storm G. Cancer SiRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Semizarov D., Kroeger P., Fesik S. SiRNA-mediated gene silencing: a global genome view. Nucleic Acids Res. 2004;32(13):3836–3845. doi: 10.1093/nar/gkh714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M.X., Nakanaga T., Nadel J.A. Cigarette smoke induces MUC5AC mucin overproduction via tumor necrosis factor-alpha-converting enzyme in human airway epithelial (NCI-H292) cells. Am J Physiol Lung Cell Mol Physiol. 2004;287(2):L420–L427. doi: 10.1152/ajplung.00019.2004. [DOI] [PubMed] [Google Scholar]
- Shu X., Wu W., Mosteller R.D., Broek D. Sphingosine kinase mediates vascular endothelial growth factor-induced activation of ras and mitogen-activated protein kinases. Mol Cell Biol. 2002;22(22):7758–7768. doi: 10.1128/MCB.22.22.7758-7768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Chang K., Hannon G.J., Rivas F.V. RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene. 2004;23(51):8401–8409. doi: 10.1038/sj.onc.1208176. [DOI] [PubMed] [Google Scholar]
- Siolas D., Lerner C., Burchard J., Ge W., Linsley P.S., Paddison P.J. Synthetic ShRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23(2):227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sioud M., Sorensen D.R. Systemic delivery of synthetic SiRNAs. Methods Mol Biol. 2004;252:515–522. doi: 10.1385/1-59259-746-7:515. [DOI] [PubMed] [Google Scholar]
- Siripurapu V., Meth J., Kobayashi N., Hamaguchi M. DBC2 significantly influences cell-cycle, apoptosis cytoskeleton and membrane-trafficking pathways. J Mol Biol. 2005;346(1):83–89. doi: 10.1016/j.jmb.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Snove O., Jr., Holen T. Many commonly used SiRNAs risk off-target activity. Biochem Biophys Res Commun. 2004;319(1):256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J. RNA interference targeting fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Sorensen D.R., Leirdal M., Sioud M. Gene silencing by systemic delivery of synthetic SiRNAs in adult mice. J Mol Biol. 2003;327(4):761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M. Therapeutic silencing of an endogenous gene by systemic administration of modified SiRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Stege A., Priebsch A., Nieth C., Lage H. Stable and complete overcoming of MDR1/P-glycoprotein-mediated multidrug resistance in human gastric carcinoma cells by RNA interference. Cancer Gene Ther. 2004;11(11):699–706. doi: 10.1038/sj.cgt.7700751. [DOI] [PubMed] [Google Scholar]
- Sugimoto A. Throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004;72(2–3):81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- Sun L., Deng L., Ea C.K., Xia Z.P., Chen Z.J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14(3):289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- Takabatake, Y., Isaka, Y., Mizui, M., Kawachi, H., Shimizu, F., & Ito, T., et al. (in press). Exploring RNA interference as a therapeutic strategy for renal disease.Gene Ther. [DOI] [PubMed]
- Taniyama Y., Tachibana K., Hiraoka K., Aoki M., Yamamoto S., Matsumoto K. Development of safe and efficient novel nonviral gene transfer using ultrasound: enhancement of transfection efficiency of naked plasmid DNA in skeletal muscle. Gene Ther. 2002;9(6):372–380. doi: 10.1038/sj.gt.3301678. [DOI] [PubMed] [Google Scholar]
- Thakker D.R., Natt F., Husken D., Maier R., Muller M., van der P.H. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci U S A. 2004;101(49):17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., Ketting R.F., Plasterk R.H. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. Epub@2002 Jun 11. [DOI] [PubMed] [Google Scholar]
- Tiscornia G., Singer O., Ikawa M., Verma I.M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci U S A. 2003;100(4):1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G., Tergaonkar V., Galimi F., Verma I.M. CRE recombinase-inducible RNA interference mediated by lentiviral vectors. Proc Natl Acad Sci U S A. 2004;101(19):7347–7351. doi: 10.1073/pnas.0402107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T. Expanding small RNA interference. Nat Biotechnol. 2002;20(5):446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- Ulanova M., Puttagunta L., Marcet-Palacios M., Duszyk M., Steinhoff U., Duta F. Syk tyrosine kinase participates in beta1-integrin signaling and inflammatory responses in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288(3):L497–L507. doi: 10.1152/ajplung.00246.2004. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B., Werth S., Abuharbeid S., Czubayko F., Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed SiRNA in vivo. Gene Ther. 2005;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- van Haaften G., Plasterk R.H., Tijsterman M. Genomic instability and cancer: scanning the Caenorhabditis elegans genome for tumor suppressors. Oncogene. 2004;23(51):8366–8375. doi: 10.1038/sj.onc.1208011. [DOI] [PubMed] [Google Scholar]
- Vanhecke D., Janitz M. High-throughput gene silencing using cell arrays. Oncogene. 2004;23(51):8353–8358. doi: 10.1038/sj.onc.1208027. [DOI] [PubMed] [Google Scholar]
- Vanhecke D., Janitz M. High-throughput gene silencing using cell arrays. Oncogene. 2004;23(51):8353–8358. doi: 10.1038/sj.onc.1208027. [DOI] [PubMed] [Google Scholar]
- Vanhecke D., Janitz M. Functional genomics using high-throughput RNA interference. Drug Discov Today. 2005;10(3):205–212. doi: 10.1016/S1359-6446(04)03352-5. [DOI] [PubMed] [Google Scholar]
- Vayssie L., Vargas M., Weber C., Guillen N. Double-stranded RNA mediates homology-dependant gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol. 2004;138(1):21–28. doi: 10.1016/j.molbiopara.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Verdrager J. Localized permanent epidemics: the genesis of chloroquine resistance in Plasmodium falciparum. Southeast Asian J Trop Med Public Health. 1995;26(1):23–28. [PubMed] [Google Scholar]
- Verma U.N., Surabhi R.M., Schmaltieg A., Becerra C., Gaynor R.B. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9(4):1291–1300. [PubMed] [Google Scholar]
- Vernick K.D., Waters A.P. Genomics and malaria control. N Engl J Med. 2004;351(18):1901–1904. doi: 10.1056/NEJMcibr042899. [DOI] [PubMed] [Google Scholar]
- Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278(9):7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- Wall N.R., Shi Y. Small RNA: can RNA interference be exploited for therapy? Lancet. 2003;362(9393):1401–1403. doi: 10.1016/S0140-6736(03)14637-5. [DOI] [PubMed] [Google Scholar]
- Wang J., Barr M.M. RNA interference in Caenorhabditis elegans. Methods Enzymol. 2005;392:36–55. doi: 10.1016/S0076-6879(04)92003-4. [DOI] [PubMed] [Google Scholar]
- Wang Q.C., Nie Q.H., Feng Z.H. RNA interference: antiviral weapon and beyond. World J Gastroenterol. 2003;9(8):1657–1661. doi: 10.3748/wjg.v9.i8.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J Virol. 2004;78(14):7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C., Karasarides M., Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5(11):875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Wen F.Q., Liu X., Kobayashi T., Abe S., Fang Q., Kohyama T. Interferon-gamma inhibits transforming growth factor-beta production in human airway epithelial cells by targeting smads. Am J Respir Cell Mol Biol. 2004;30(6):816–822. doi: 10.1165/rcmb.2002-0249OC. [DOI] [PubMed] [Google Scholar]
- Whittaker P.A. Genes for asthma: much ado about nothing? Curr Opin Pharmacol. 2003;3(3):212–219. doi: 10.1016/s1471-4892(03)00035-3. [DOI] [PubMed] [Google Scholar]
- Whittaker P.A. What is the relevance of bioinformatics to pharmacology? Trends Pharmacol Sci. 2003;24(8):434–439. doi: 10.1016/S0165-6147(03)00197-4. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I.G., Arya S. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A. 2003;100(5):2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler K.E. Killing the messenger. Nat Rev Drug Discov. 2004;3(10):823–824. doi: 10.1038/nrd1527. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M., Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77(16):8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Mao Q., Paulson H.L., Davidson B.L. SiRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20(10):1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10(8):816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Xu J., Li L., Qian Z., Hong J., Shen S., Huang W. Reduction of PTP1B by RNAi upregulates the activity of insulin controlled fatty acid synthase promoter. Biochem Biophys Res Commun. 2005;329(2):538–543. doi: 10.1016/j.bbrc.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Yang G., Thompson J.A., Fang B., Liu J. Silencing of H-ras gene expression by retrovirus-mediated SiRNA decreases transformation efficiency and tumor growth in a model of human ovarian cancer. Oncogene. 2003;22(36):5694–5701. doi: 10.1038/sj.onc.1206858. [DOI] [PubMed] [Google Scholar]
- Yang Z.G., Chen Z., Ni Q., Xu N., Shao J.B., Yao H.P. Inhibition of hepatitis B virus surface antigen expression by small hairpin RNA in vitro. World J Gastroenterol. 2005;11(4):498–502. doi: 10.3748/wjg.v11.i4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z.H., Ren C.P., Li F., Yang X.Y., Li H., Zhao M. Suppression of Bcl-2 gene by RNA interference increases chemosensitivity to cisplatin in nasopharyngeal carcinoma cell line CNE1. Acta Biochim Biophys Sin (Shanghai) 2004;36(11):749–753. doi: 10.1093/abbs/36.11.749. [DOI] [PubMed] [Google Scholar]
- Yokota T., Miyagishi M., Hino T., Matsumura R., Tasinato A., Urushitani M. SiRNA-based inhibition specific for mutant SOD1 with single nucleotide alternation in familial ALS, compared with ribozyme and DNA enzyme. Biochem Biophys Res Commun. 2004;314(1):283–291. doi: 10.1016/j.bbrc.2003.12.098. [DOI] [PubMed] [Google Scholar]
- Yu J.Y., DeRuiter S.L., Turner D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99(9):6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaas D.W., Duncan M.J., Li G., Wright J.R., Abraham S.N. Pseudomonas invasion of type i pneumocytes is dependent on the expression and phosphorylation of caveolin-2. J Biol Chem. 2005;280(6):4864–4872. doi: 10.1074/jbc.M411702200. [DOI] [PubMed] [Google Scholar]
- Zambrowicz B.P., Turner C.A., Sands A.T. Predicting drug efficacy: knockouts model pipeline drugs of the pharmaceutical industry. Curr Opin Pharmacol. 2003;3(5):563–570. doi: 10.1016/j.coph.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Zender L., Hutker S., Liedtke C., Tillmann H.L., Zender S., Mundt B. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A. 2003;100(13):7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nolan E., Kreitschitz S., Rabussay D.P. Enhanced delivery of naked DNA to the skin by non-invasive in vivo electroporation. Biochim Biophys Acta. 2002;1572(1):1–9. doi: 10.1016/s0304-4165(02)00270-2. [DOI] [PubMed] [Google Scholar]
- Zhang R., Guo Z., Lu J., Meng J., Zhou C., Zhan X. Inhibiting severe acute respiratory syndrome-associated coronavirus by small interfering RNA. Chin Med J (Engl) 2003;116(8):1262–1264. [PubMed] [Google Scholar]
- Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X. Silencing SARS-CoV spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560(1–3):141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Y., Zhang Y.F., Bryant J., Charles A., Boado R.J., Pardridge W.M. Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin Cancer Res. 2004;10(11):3667–3677. doi: 10.1158/1078-0432.CCR-03-0740. [DOI] [PubMed] [Google Scholar]
- Zhang W., Yang H., Kong X., Mohapatra S., Juan-Vergara H., Hellermann G. Inhibition of respiratory syncytial virus infection with intranasal SiRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11(1):56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(1):135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.S., Liu D.P., Liang C.C. Challenges and strategies: the immune responses in gene therapy. Med Res Rev. 2004;24(6):748–761. doi: 10.1002/med.20009. [DOI] [PubMed] [Google Scholar]


