Summary
Objectives
Many adult patients hospitalised with acute respiratory illness have viruses detected but the overall importance of viral infection compared to bacterial infection is unclear.
Methods
Patients were recruited from two acute hospital sites in Leicester (UK) over 3 successive winters. Samples were taken for viral and bacterial testing.
Results
Of the 780 patients hospitalised with acute respiratory illness 345 (44%) had a respiratory virus detected. Picornaviruses were the most commonly isolated viruses (detected in 23% of all patients). Virus detection rates exceeded 50% in patients with exacerbation of asthma (58%), acute bronchitis and Influenza-like-illness (64%), and ranged from 30 to 50% in patients with an exacerbation of COPD (38%), community acquired pneumonia (36%) and congestive cardiac failure (31%). Bacterial detection was relatively frequent in patients with exacerbation of COPD and pneumonia (25% and 33% respectively) but was uncommon in all other groups. Antibiotic use was high across all clinical groups (76% overall) and only 21% of all antibiotic use occurred in patients with detectable bacteria.
Conclusions
Respiratory viruses are the predominant detectable aetiological agents in most hospitalised adults with acute respiratory illness. Antibiotic usage in hospital remains excessive including in clinical conditions associated with low rates of bacterial detection. Efforts at reducing excess antibiotic use should focus on these groups as a priority.
Registered International Standard Controlled Trial Number: 21521552.
Keywords: Respiratory viruses, Acute respiratory illness, Hospitalisation, Adults, Antibiotics
Highlights
-
•
We tested nearly 800 inpatients with respiratory illness for viruses and bacteria.
-
•
Detection of respiratory viruses was common across all clinical groups.
-
•
Bacterial detection was uncommon except in patients with COPD and pneumonia.
-
•
Antibiotic use was high in all patient groups including asthma and bronchitis.
Introduction
Acute respiratory illness is responsible for a large proportion of acute hospital admissions in adults.1 It is accepted that respiratory viruses are responsible for the majority of acute respiratory illness in infants and children, including those admitted to secondary care.2, 3 Whilst respiratory viral infection has been increasingly recognised in the aetiology of acute respiratory illness in adults, its overall importance compared to bacteria is unclear. Respiratory viruses are detected in the majority of exacerbation of asthma in adults (41–78%) and in a smaller proportion of patients with exacerbation of COPD (22–44%) and community acquired pneumonia (15–54%).4, 5, 6, 7, 8, 9, 10 Picornaviruses (rhinoviruses and enteroviruses) are the most frequent cause of the common cold and are also consistently the most commonly detected viruses in adults with exacerbations of asthma,4 chronic obstructive pulmonary disease (COPD)5, 7 and in most studies of aetiology in community acquired pneumonia.8, 9 In addition, recent evidence suggests that respiratory viruses are associated with a significant proportion of acute respiratory disease amongst hospitalised adults and the elderly.8, 9, 10, 11, 12, 13
Antibiotic use is common in hospitals. A recent US study demonstrated that over 50% of hospital inpatients received antibiotics at some stage during their stay, with acute respiratory illness being the commonest reason for antibiotic use.14 In addition to the frequent isolation of viruses, there is also a lack of evidence of clinical benefit from antibiotic use in many acute respiratory conditions presenting to secondary care including; acute bronchitis,15 exacerbations of asthma16 and mild-moderate cases of exacerbation of chronic obstructive airways disease.17, 18 Widespread use of antibiotics in hospitals and the community has led to the emergence and proliferation of antibiotic resistant bacteria19, 20 and to increasing rates of health care associated infections (HCAIs) such as Clostridium difficile. Effective antibiotic stewardship and reduction in excess antibiotic use in hospitals has been shown to be effective in reducing antimicrobial resistance and HCAIs and has therefore become a global priority.21
In this study we sought to define the microbial aetiology in hospitalised adults with a variety of common acute respiratory syndromes and to describe their clinical characteristics and antimicrobial use.
Methods
Subjects
Subjects were participants in a prospectively recruited trial of rapid diagnostic testing for Streptococcus pneumoniae and influenza and met the following inclusion criteria; aged ≥18 years; able and willing to give written informed consent (or a relative or carer is able and willing to give informed assent); have an acute exacerbation of chronic cardio-pulmonary illness of <168 h (7 days) duration (including exacerbation of COPD, asthma and congestive cardiac failure) OR an acute pulmonary illness <7 days duration (including pneumonia, bronchitis and influenza-like illness); can be recruited to the study within a 16-h period of initial assessment on the Medical Admissions Unit; able and willing to adhere to the procedures stated in the protocol. Full details of inclusion and exclusion criteria are given in Appendix A. Patients were recruited during the winter months in three successive years (December 2005 to May 2006; October 2006 to May 2007; September 2007 to May 2008) across two large hospital sites with acute medical admission units, within the University Hospitals of Leicester NHS Trust, UK. The study was approved by the Leicestershire, Northamptonshire and Rutland Ethics Committee and all patients gave written informed consent.
Clinical data
Demographic and clinical data (presenting symptoms and signs, co-morbidity, medications use, prior antibiotic use, smoking history, and vaccination status) were collected at enrolment and recorded on a standardised case report form. Radiological and laboratory data were collated retrospectively using computerised imaging and laboratory systems. All patients had a chest radiograph performed within 24 h of admission. Individual clinical groups were identified by discharge ICD-10 (International classification of disease, tenth edition) code classification and confirmed by case note review.22 Full details of clinical groups and definitions are given in Appendix B.
Laboratory methods
Standard microbiological testing
Blood, urine, sputum and nasopharyngeal swab samples were collected at enrolment. Blood cultures were incubated aerobically and anaerobically using the BacT/ALERT 3D blood culture system (bioMerieux, Durham, North Carolina, USA). Sputum was examined by Gram stain microscopy and cultured on sheep blood agar and chocolate agar. Plates were incubated aerobically in CO2 at 37 °C, were read at 24 h and at 48 h. Urine was tested for S. pneumoniae antigens using the NOW S. pneumoniae urinary antigen tests (Binax, Portland, Maine, USA). Nasopharyngeal swabs were stored in viral transport medium and frozen at −80 °C prior to testing. Urine was tested for Legionella pneumophilia (Binax, Portland, Maine, USA) antigen at physician's discretion in patients with pneumonia.
Multiplex PCR for respiratory viruses
The quadriplex influenza A/B/H5 real-time assay which includes an internal control (bacteriophage MS2) has been detailed elsewhere.23 Three additional multiplex real-time (TaqMan®) PCR assays were employed to detect the remaining respiratory viruses including respiratory syncytial virus (RSV) A and B, parainfluenza (PIV) types 1–4, adenovirus, enterovirus, rhinovirus, human metapneumovirus (hMPV), group 1 coronaviruses (HCoV-229E and HCoV-NL63), group 2 coronaviruses (HCoV-OC43 and HCoV-HKU1), and SARS associated coronavirus. Full details of the PCR methods including primer and probe sequences are given in Appendix C.
Statistical analysis
Statistical analysis was performed using PRISM Version 6 (Graphpad Software, La Jolla, CA) and SPSS version 18 (SPSS Inc, Chicago, IL). Non-parametric data were described as median and inter-quartile range and the Mann–Whitney U test was used to compare groups. Proportions were compared using χ2 and Fisher's exact test. The Clopper-Pearson method was used to calculate 95% confidence intervals (CI) for proportions. A p value of <0.05 was taken as the threshold for statistical significance. A CRP of <5 mg/L was considered to be equal to 3 mg/L for statistical purposes.
Results
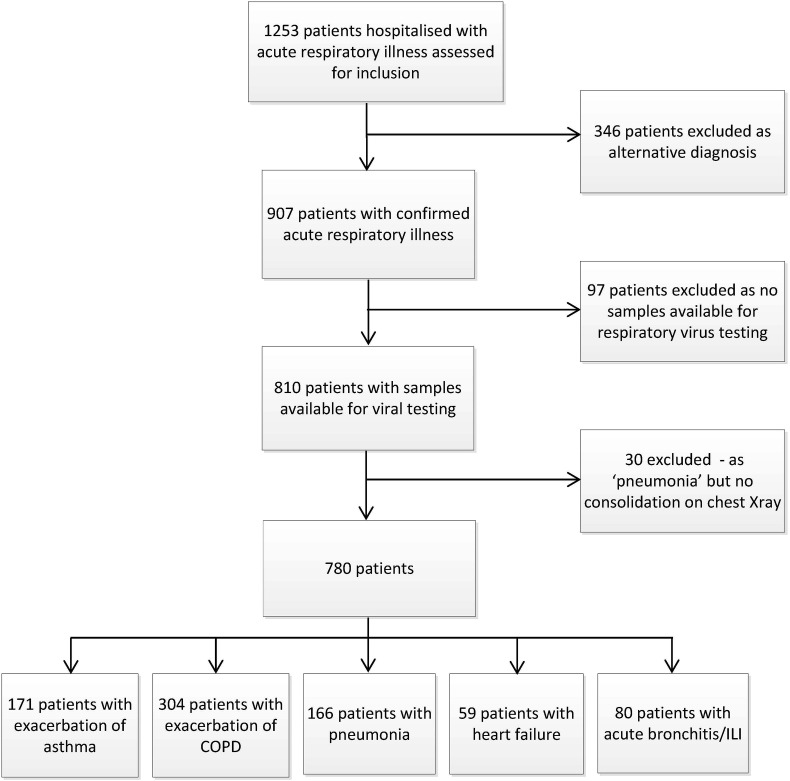
A total of 780 patients were studied. A trial profile is shown in Fig. 1 . Demographic, clinical, laboratory and outcome data by clinical group are shown in Table 1 .
Figure 1.
Trial profile. ILI, influenza-like illness.
Table 1.
Demographic, clinical and outcome data for all patients combined and by clinical group. Data are presented as number (percentage) and median [inter-quartile range].
| Missing data | All patients | Asthma | AECOPD | Pneumonia | Heart failure | Bronchitis/ILI | |
|---|---|---|---|---|---|---|---|
| Number | 780 | 171 | 304 | 166 | 59 | 80 | |
| Age, years | 0 | 64 [48–76] | 39 [31–52] | 70 [62–77] | 67 [51–84] | 74 [69–85] | 52 [38–72] |
| Aged over 65 | 0 | 376 (48) | 16 (9) | 197 (65) | 86 (52) | 47 (80) | 24 (30) |
| Male | 0 | 386 (49) | 85 (50) | 141 (46) | 87 (52) | 39 (66) | 35 (44) |
| White ethnicity | 0 | 706 (90) | 135 (79) | 293 (96) | 158 (95) | 51 (87) | 69 (87) |
| Current smoker | 2 | 225 (29) | 50 (29) | 107 (35) | 40 (24) | 7 (12) | 21 (26) |
| Influenza vaccinea | 10 | 449 (58) | 77 (45) | 220 (72) | 86 (53) | 41 (69) | 25 (31) |
| Co-Morbidity | |||||||
| Cardiac disease | 10 | 362 (47) | 41 (24) | 157 (53) | 78 (47) | 56 (95) | 30 (38) |
| Chronic Respiratory Disease | 9 | 574 (74) | 166 (98) | 291 (97) | 90 (54) | 18 (31) | 8 (10) |
| Diabetes mellitus | 9 | 101 (13) | 19 (11) | 34 (11) | 26 (16) | 14 (25) | 8 (10) |
| Liver disease | 9 | 12 (2) | 1 (1) | 5 (2) | 4 (2) | 1 (2) | 1 (1) |
| Renal disease | 9 | 19 (2) | 1 (1) | 8 (3) | 7 (4) | 3 (5) | 0 (0) |
| Immune compromise | 9 | 27 (4) | 6 (4) | 5 (2) | 10 (6) | 4 (7) | 2 (3) |
| Duration of symptoms, daysb | 3 | 4 [2–6] | 4 [2–7] | 4 [2–7] | 4 [2–7] | 5 [3–7] | 4 [2–7] |
| Antimicrobial use | |||||||
| Antibiotics prior to admission | 0 | 187 (24) | 43 (25) | 84 (28) | 38 (23) | 8 (14) | 14 (18) |
| Antibiotics during admission | 19 | 582 (76) | 98 (59) | 235 (79) | 156 (98) | 33 (58) | 60 (75) |
| IV antibiotics | 19 | 229 (30) | 22 (13) | 67 (22) | 104 (65) | 16 (28) | 20 (25) |
| NI use during admission | 19 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Temperature,°C | 16 | 36.8 [36.3–37.4] | 36.8 [36.3–37.3] | 36.6 [36.2–37.2] | 37.0 [36.6–37.8] | 36.5 [36.0–37.0] | 37.1 [36.7–37.5] |
| WCC, ×109/L | 11 | 10.8[8.3–14.5] | 10.7 [8.2–13.0] | 10.6 [8.3–13.7] | 14.1 [11.1–18.7] | 8.4[6.6–11.1] | 8.9 [5.8–12.3] |
| CRP, mg/L | 60 | 33 [14–93] | 10 [3–26] | 17 [6–46] | 174 [77–279] | 20 [9–46] | 21 [8–51] |
| Length of stay, days | 0 | 3 [1–7] | 1 [1–3] | 3 [1–6] | 4 [2–9] | 8 [3–13] | 1 [1–4] |
| 30 day mortality | 0 | 23 (3) | 0 (0) | 6 (2) | 11 (7) | 4 (7) | 2 (2) |
IQR, inter-quartile range; AECOPD, Acute exacerbation of COPD. ILI, influenza-like illness; WCC, white cell count. NI, Neuraminidase inhibitor (Oseltamivir, Zanamivir).
For current influenza season.
Duration of symptoms prior to admission.
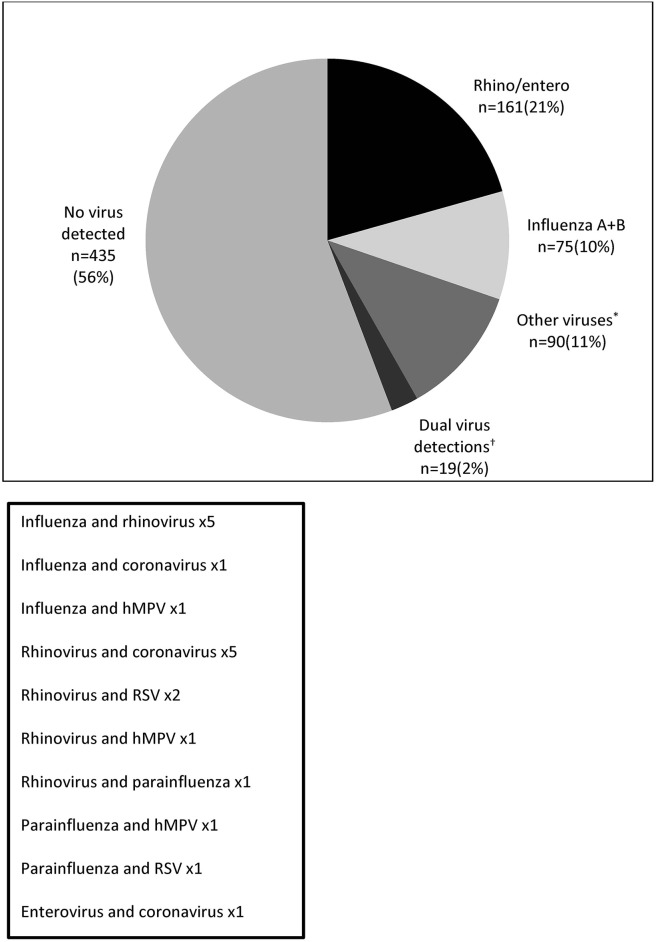
Overall 44% (345/780) of patients hospitalised with acute respiratory illness had respiratory viruses detected. Picrornaviruses (91% rhinovirus, 9% enterovirus) were the most frequently identified virus and were detected in 23% (176/780) of patients, either alone (21%) or with another respiratory virus (2%). Influenza A and B viruses were detected in 11% (82/780) of patients. Altogether human metapneumovirus, coronaviruses (group 1 and 2) and parainfluenza virus (types 1–4) were detected in 11% (85/780) patients. Few admissions were associated with RSV or adenovirus detection; 3% (22/780) of patients combined. Co-detection of two different respiratory viruses occurred in 2% (19/780) of patients. Table 2 shows the rate of respiratory virus detection for all patients and by clinical group. Fig. 2 details single and dual viral detections.
Table 2.
Respiratory virus detection rate for all patients combined and by clinical diagnosis.
| All patients | Asthma | AECOPD | Pneumonia | Heart failure | Bronchitis/ILI | p valuea | |
|---|---|---|---|---|---|---|---|
| 780 | 171 | 304 | 166 | 59 | 80 | ||
| Patients with viral detection, n (%) | 345 (44) | 99 (58) | 117 (38) | 60 (36) | 18 (31) | 51 (64) | <0.001 |
| [95% CI] | [41–48] | [48–63] | [33–44] | [29–44] | [19–44] | [52–74] | |
| Total number of virus detections | 364 | 103 | 125 | 64 | 19 | 53 | |
| Single virus detection (%) | 326 (42) | 95 (56) | 109 (35) | 56 (34) | 17 (29) | 49 (61) | |
| Picornavirus [Rhino/enterovirus] (%) | 161 (21) | 58 (34) | 48 (16) | 30 (18) | 9 (15) | 16 (20) | <0.001 |
| Influenza A + B (%) | 75 (10) | 19 (11) | 20 (7) | 13 (8) | 2 (3) | 21 (26) | <0.001 |
| hMPV (%) | 28 (4) | 6 (4) | 11 (4) | 2 (1) | 3 (5) | 6 (8) | 0.17 |
| HCoV Groups 1 + 2 (%) | 24 (3) | 5 (3) | 13 (4) | 3 (2) | 0 (0) | 3 (4) | 0.37 |
| Parainfluenza 1–4 (%) | 19 (2) | 3 (2) | 8 (3) | 4 (2) | 2 (3) | 2 (3) | 0.96 |
| RSV (%) | 12 (2) | 2 (1) | 8 (3) | 2 (1) | 0 (0) | 0 (0) | 0.31 |
| Adenovirus (%) | 7 (1) | 2 (1) | 1 (0) | 2 (1) | 1 (2) | 1 (1) | 0.74 |
| Dual virus detections (%) | 19 (2) | 4 (2) | 8 (3) | 4 (2) | 1 (2) | 2 (3) | 0.99 |
AECOPD, Acute exacerbation of COPD. ILI, influenza like illness. CI, confidence interval. hMPV, human metapneumovirus.
HCoV, human coronavirus. RSV, respiratory syncytial virus.
χ2 across all clinical groups except ‘all patients’ group.
Figure 2.
Number and proportion of all patients with respiratory viruses detected (n = 780). *RSV, parainfluenza (types 1–4), human metapneumovirus (hMPV), coronavirus (group 1 and 2) and adenovirus. †Dual viral detections are detailed in the box below.
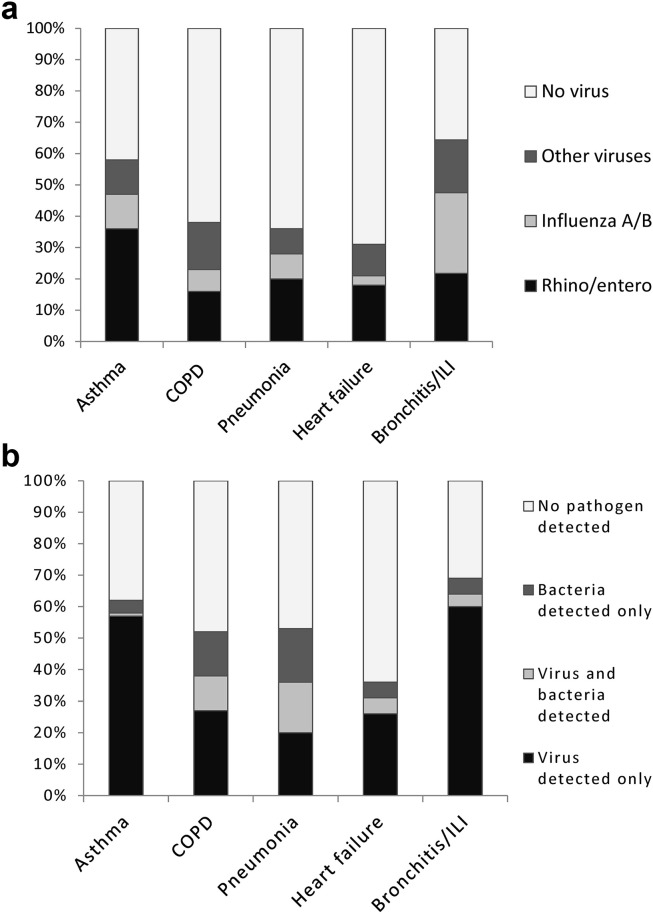
Respiratory virus detection exceeded 50% in patients with exacerbation of asthma (58% [99/171] of patients) and acute bronchitis/ILI combined (64% [51/80] of patients); and ranged from 30 to 50% in patients with exacerbation of COPD (38% [117/304] of patients), pneumonia (34% [60/166] of patients) and heart failure (31% [18/59] of patients). Picornaviruses were more commonly detected in patients with asthma compared to other clinical groups (p < 0.001) whereas influenza virus detection was more commonly associated with acute bronchitis/ILI (p < 0.001). Fig. 3 a shows respiratory virus detection rate by clinical condition.
Figure 3.
a. Proportions of respiratory viruses detected by clinical group (n = 780). ILI, influenza-like illness. Other viruses includes; RSV, parainfluenza (virus type 1–4), hMPV, coronaviruses (group 1 and 2), and adenovirus. b. Proportion of patients with viruses only detected, bacteria only detected, viruses and bacteria detected together and no pathogen detected, by clinical group (n = 780). ILI, influenza-like illness. Data represented are for all patients including those with incomplete sampling for bacteria.
Bacteria were detected in 19% (146/780) patients with acute respiratory illness overall. Table 3 shows bacterial detection by clinical group. Bacterial detection was more common in patients with pneumonia (33% [55/166] of patients) and exacerbation of COPD (25% [75/304] of patients) compared to other clinical groups (p < 0.001). S. pneumoniae was the most commonly identified bacterial pathogen and was detected in 53% (78/146) patients with positive tests for bacteria. Mixed viral and bacterial detection was also more common in patients with pneumonia (16% [26/166] patients), and exacerbation of COPD (11% [34/304] patients) compared to the other clinical groups (p < 0.001). Detection of bacteria was uncommon in patients hospitalised with exacerbation of asthma (5% [9/171] of patients), heart failure (8% [6/59] of patients) and acute bronchitis/ILI (6% [5/80] of patients). Fig. 3b shows the proportions of patients with viruses detected alone, bacteria detected alone, bacteria and viruses detected together, and cases where no pathogens were isolated.
Table 3.
Bacterial detection for all patients and by clinical group.
| All patients | Asthma | AECOPD | Pneumonia | Heart failure | Bronchitis/ILI | p valuea | |
|---|---|---|---|---|---|---|---|
| All patients (n = 780) | 780 | 171 | 304 | 166 | 59 | 80 | |
| Positive pneumococcal ag (%) | 63/733 (9) | 5/159 (3) | 20/285 (7) | 29/156 (19) | 5/55 (9) | 4/78 (5) | |
| Positive blood culture (%) | 13/636 (2) | 0/133 (0) | 1/245 (0) | 12/143 (8)b | 0/46 (0) | 0/69 (0) | |
| Positive sputum culture (%) | 94/324 (29) | 4/52 (8) | 60/152 (39) | 28/78 (36) | 1/15 (7) | 1/27 (4) | |
| All patients (n = 780) | |||||||
| Bacterial detection, total (%) | 146 (19) | 9 (5) | 75 (25) | 55 (33) | 6 (10) | 5 (6) | <0.001 |
| Pneumococcal detection, total (%) | 78 (10) | 5 (3) | 29 (10) | 35 (21) | 5 (8) | 4 (5) | <0.001 |
| Mixed viral/bacterial detection (%) | 69 (9) | 2 (1) | 34 (11) | 26 (16) | 3 (5) | 4 (5) | <0.001 |
| Patients with complete sampling (n = 269)c | |||||||
| Bacterial detection, total (%) | 100 (37) | 5 (11) | 52 (43) | 38 (55) | 1 (9) | 3 (13) | <0.001 |
| Pneumococcal detection, total (%) | 39 (14) | 2 (4) | 15 (12) | 19 (28) | 0 (0) | 2 (9) | 0.003 |
| Mixed viral/bacterial detection (%) | 48 (18) | 1 (2) | 24 (20) | 19 (27) | 1 (9) | 3 (13) | 0.01 |
AECOPD, Acute exacerbation of COPD. ILI, influenza like illness. CI, confidence interval.
χ2 across all clinical groups except 'all patients' group.
Streptococcus pneumoniae x9, Streptococcus milleri group x1, Staphylococcus aureus x1, Klebsiella pneumoniae x1.
Complete sampling is defined as; blood culture, sputum culture and urine antigen testing for Streptococcus pneumoniae.
Although all patients had nasopharyngeal swabs tested for viruses and the vast majority of patients had blood cultures and pneumococcal antigen testing performed, only 342/780 (42%) were able to produce sputum samples for culture. We therefore performed a separate analysis of only those patients with complete sampling for bacteria (sputum culture, blood culture, pneumococcal antigen testing). Among this subgroup the overall rate of bacterial detection was higher than in the main cohort, for patients with pneumonia (55% versus 33%) and exacerbation of COPD (43% versus 25%), but remained low in asthma (9%), heart failure (13%) and acute bronchitis/ILI (11%), shown in Table 3.
Overall, antibiotics were given in 76% (582/761) of patients hospitalised with acute respiratory illness and at least one dose of intravenous (IV) antibiotic was administered in 30% (229/761). Antibiotic use was high (>50%) across all clinical groups. Antibiotics were given in 98% (156/160) of patients with pneumonia and 65% (104/160) received at least 1 dose of IV antibiotic. 79% (235/299) of patients with exacerbation of COPD received antibiotics and 22% (67/299) received IV antibiotics. Antibiotics were given in 59% (98/165) of patients with exacerbation of asthma and 13% (22/165) received IV antibiotics. 58% (33/57) of patients with heart failure and 75% (60/80) of patients with acute bronchitis/ILI received antibiotics. No patients were treated with neuraminidase inhibitors or other antiviral agents whilst hospitalised. Antimicrobial treatment by clinical group is shown in Table 1.
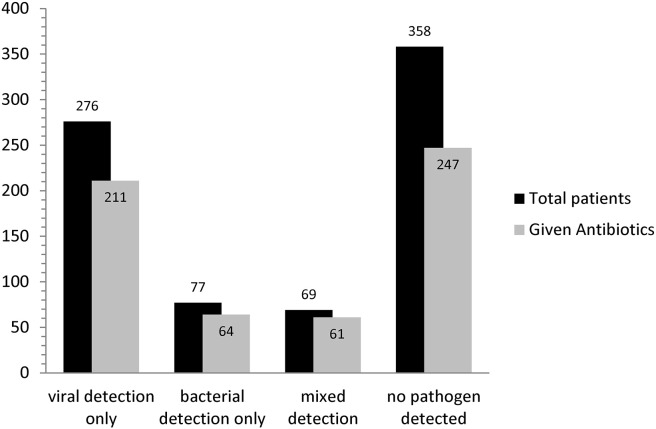
Examining antibiotic use by aetiology showed that overall 78% (211/270) of patients with viruses detected alone were given antibiotics, representing 36% (211/582) of all antibiotic use in acute respiratory illness. Only 21% (125 of 582) of all antibiotic use occurred in patients with bacteria detected – either alone (11%) or concurrently with a virus (10%). 70% (247/351) of patients with no pathogen detected received antibiotics representing 42% (247/582) of all antibiotic use. Antibiotic use by aetiology is shown in Fig. 4 .
Figure 4.
Antibiotic use in patients by aetiology (n = 758). Mixed detection refers to the concurrent detection of viruses and bacteria in the same patient.
Discussion
This is one of the largest studies examining microbial aetiology in hospitalised adults with acute respiratory illness and contains within it the largest cohort of hospitalised patients with COPD systematically tested for respiratory viruses. In agreement with other recent studies it demonstrates the importance of respiratory viruses8, 9, 10, 11, 12, 13 and the large burden of picornavirus infection. In addition to triggering exacerbations of asthma and COPD4, 5 and causing lower respiratory tract illness in community-dwelling elderly24 this work suggests that picornaviruses are important in a large proportion of adult hospitalisations with acute respiratory illness. This study shows that bacterial detection is relatively frequent in patients with exacerbation of COPD and pneumonia (where mixed detection of bacteria and viruses is also common) compared to all other clinical groups, where detection of bacteria is rare. In agreement with other similar studies13 antibiotic use was high across all clinical groups and was not related to aetiology. The vast majority of antibiotic use took place in patients without a detectable bacterial infection and this included a significant amount of intravenous antibiotic use.
Compared to a recent similar study of hospitalised adults with acute respiratory illness we found lower rates of bacterial co-detection among patients with viruses detected.13 This is likely to be explained by the authors use of procalcitonin(PCT) levels >0.25 ng/ml to define bacterial infection, in addition to the actual detection of bacteria. Whilst PCT levels in patients with acute respiratory illness tend to be higher in patients with bacteria detected compared to those with viruses detected,25 viral infection has been frequently associated with PCT levels of >0.25 ng/ml independently of bacteria.26
Strategies to reduce excess antibiotic use are urgently needed but must be balanced against the risks of undertreating genuine and severe bacterial infection. In patients with pneumonia where complications and mortality rates are high27 and evidence exists for the benefit of timely antibiotic treatment28 such a high rate of antibiotic use seems justifiable and appropriate. Although viruses are frequently detected in patients with pneumonia, co-detection with bacteria is also common6, 7, 8, 9 and secondary bacterial pneumonia following respiratory viral infection is known to occur.29, 30 Primary viral pneumonia occurs in non-immunocompromised adults with influenza virus infection and may also occur with other respiratory viruses.30 However, definitely excluding a concurrent bacterial infection in these patients who are often severely unwell is difficult to achieve in practice and so strategies to reduce antibiotic use in hospitalised patients with pneumonia are unlikely to be appropriate or successful.
The detection rate of respiratory viruses in patients with exacerbation of COPD was around 40%, which is consistent with the findings of a recent systematic review.5 Although antibiotic use was very common, occurring in around 80% of patients, bacterial detection and co-detection of viruses and bacteria together were also relatively common suggesting that viral identification in patients with exacerbation of COPD cannot be used in isolation to withhold antibiotic therapy. Although bacteria are frequently detected from patients with exacerbation of COPD their exact role and importance compared to viruses is the subject of ongoing debate. Antibiotics are currently recommended in COPD exacerbation according to clinical criteria18 however the evidence for clinical benefit in mild to moderate exacerbations is limited17, 18 and must be balanced against the risks of adverse events. Studies using the serum biomarker Procalcitonin(PCT) have demonstrated that antibiotics can be safely withheld in patients with exacerbation of COPD and low levels of PCT. Biomarker directed antibiotic use probably represent the most promising strategy currently available for reducing excess antibiotic use in this group31 and studies further evaluating this are ongoing.
In keeping with previous studies4 we identified a respiratory virus in around 60% of patients hospitalised with an exacerbation of asthma, the majority of which were picornaviruses. Influenza virus was detected in over 10% of all asthma exacerbations in our study and it is notable that asthma was a risk factor for hospital admission with laboratory confirmed influenza during the recent 2009 (H1N1) pandemic.32 In our cohort bacterial detection in patients with exacerbation of asthma was unusual. Although we did not test for the atypical bacteria Mycoplasma pneumoniae or Chlamydia pneumoniae their role in triggering exacerbations of asthma is not established and trials of antibiotics active against these organisms have not shown benefit in asthmatic subjects.4, 33 Despite the high prevalence of viral infection, the lack of evidence of clinical benefit for antibiotics16 and national guidelines discouraging their use,34 antibiotics were still used in the majority of hospitalised asthmatic patents.
Whilst it is often cited as reason for cardiac decompensation, a causal relationship between acute decompensated congestive cardiac failure and respiratory virus infection is not well established. A large study from the US demonstrated that RSV and influenza viruses were both associated with a significant proportion of hospitalisations with congestive heart failure.11 In our study around one third of patients admitted with decompensated heart failure had a respiratory virus detected with bacteria being detected infrequently and antibiotics being used in the majority.
We detected respiratory viruses in around two thirds of patients with acute bronchitis and ILI combined, which is comparable to findings from previous studies.35, 36, 37 In contrast bacteria were rarely detected although antibiotic use was again very common. A recent randomised placebo controlled trial of antibiotics for acute bronchitis (in patients without chronic lung disease) demonstrated no benefit from antibiotics and a trend towards harm in those over aged 60 years.38 A Cochrane review has concluded that there is limited if any evidence for the use of antibiotics in acute bronchitis and has discouraged their use in this group.15 In our study a greater proportion of patients with acute bronchitis/ILI had influenza virus detected compared to other clinical groups although other virus types were also frequently detected.
In total influenza virus was detected in more than 10% of all patients with acute respiratory illness. Influenza is the only respiratory virus for which specific antiviral agents exist, however no patients in our study were prescribed neuraminidase inhibitors (NIs) either before or during their hospital admission. In contrast to the US CDC guidelines, UK NICE guidance advocates the use of NIs only when started within 48 h of symptom onset39 irrespective of hospitalisation status or severity. Using even these restrictive criteria, nineteen of the 82 (23%) patients with laboratory confirmed influenza in our study presented within 48 h of the onset of symptoms and should therefore have been considered for treatment. In addition evidence from recent large observational studies suggests that NIs may reduce mortality from pandemic H1N1 influenza among hospitalised adults even when started after 48 h of symptom duration.40
There are several limitations to our study. Our results are only applicable to hospitalised adults and cannot be extrapolated to children or to patients presenting to primary care. The seasonal nature of our recruitment means that the results are only applicable to patients admitted outside of the summer months and may overestimate the overall annual incidence of respiratory viral detection in these patient groups. In addition, because the exact dates of patient recruitment were different year by year there is a potential for underestimating the prevalence of respiratory viruses with seasonal periodicity such as RSV and influenza. This may explain our relatively low rate of RSV detection compared to some other studies. Our definition of bacterial detection includes the isolation of potentially pathogenic bacteria in sputum which may overestimate the true numbers of bacterial infections as the presence of bacteria in sputum may represent colonisation rather than infection. As we did not test for M. pneumoniae or C. pneumoniae in our study we may have underestimated the overall proportion of bacterial detection. However the role of these organisms in clinical groups other than pneumonia and bronchitis is not established and similar studies in hospitalised adults have shown low rates of detection even when using molecular methods.13 In our cohort a large number of patients were unable to provide adequate sputum samples for analysis and culture which could have underestimated the overall rate of bacterial detection. However in our subgroup analysis of patients with complete sampling the overall rate of bacterial detection was higher in patients with COPD and pneumonia but for other groups was not significantly different. Finally, around a quarter of our patients were already taking antibiotics at the time of hospitalisation which could have led to a reduction in the detection of bacteria and so an underestimation of bacterial detection rate. However it is notable that the detection rates of bacteria in our study are consistent with other similar studies including those where patients already on antibiotics were excluded.13
In summary this study demonstrates the predominance of respiratory viruses in adults hospitalised with acute respiratory illness and shows that excess antibiotic use occurs in clinical groups where viral detection is common, bacterial detection is rare and there is a lack of evidence for clinical benefit from antibiotics. Efforts at reducing excess antibiotic use in hospitals should focus in these groups as a priority.
The views and opinions expressed in this manuscript are those of the authors and do not necessarily reflect those of the HTA programme, NIHR, NHS or the Department of Health.
Author contributions
TWC was involved in the design of the study, performed the molecular laboratory work, data collection and statistical analysis and wrote the manuscript. MM was involved in the design of the study, performed the molecular laboratory work and was involved in writing the manuscript. SB was involved in the design of the study and the writing of the manuscript. MC was involved in the design of the study (the molecular methods) and the writing of the manuscript. SP was involved in the design of the study (the molecular methods) and the writing of the manuscript. KGN was involved in the design of the study, data analysis and the writing of the manuscript. TWC acts as guarantor of the paper, taking responsibility for the integrity of the work as a whole, from inception to published article.
Funding source
This work was supported by the National Institute of Health Research through the Health Technology Assessment programme HTA grant number [03/39/18].
Competing interests
None declared for TWC, MM, IS, SB, MC and SP. KGN was a founding member of the European Scientific Working Group on Influenza and resigned in 2001; within the past 5 years he received H5 vaccines from Novartis for MRC-funded research and H1N1 pandemic vaccines from GlaxoSmithKline and Baxter for NIHR-funded research.
Ethics approval
The study was approved by the Leicestershire, Northamptonshire and Rutland Ethics Committee and all patients gave informed written consent.
Acknowledgements
We would like to thank all the hospital staff at UHL involved in the care of the patients in this study and the laboratory staff involved in the processing of their samples. The assays used were developed as part of an ongoing collaboration between HPA East of England and the Respiratory Virus reference laboratory at the Centre for Infections, London. We thank Alison Bermingham for her assistance in the validation of some of the PCR assays used in this study.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jinf.2014.07.023.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Department of Health hospital episode statistics 1998–2008 . 2009. NHS HES online.http://www.hesonline.nhs.uk Available at: [Google Scholar]
- 2.Mansbach J.M., McAdam A.J., Clark S., Hain P.D., Flood R.G., Acholonu U. Prospective multicenter study of the viral etiology of bronchiolitis in the emergency department. Acad Emerg Med. 2008;15(2):111–118. doi: 10.1111/j.1553-2712.2007.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juvén T., Mertsola J., Waris M., Leinonen M., Meurman O., Roivainen M. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19(4):293–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Papadopoulos N.G., Christodoulou I., Rohde G., Agache I., Almqvist C., Bruno A. Viruses and bacteria in acute asthma exacerbations – A GA2LEN-DARE* systematic review. Allergy. 2011;66:458–468. doi: 10.1111/j.1398-9995.2010.02505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohan A., Chandra S., Agarwal D., Guleria R., Broor S., Gaur B. Prevalence of viral infection detected by PCR and RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology. 2010;15(3):536–542. doi: 10.1111/j.1440-1843.2010.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jennings L.C., Anderson T.P., Beynon K.A., Chua A., Laing R.T., Werno A.M. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63(1):42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 7.Johansson N., Kalin M., Tiveljung-Lindell A., Giske C.G., Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50(2):202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Templeton K.E., Scheltinga S.A., van den Eeden W.C., Graffelman A.W., van den Broek P.J., Claas E.C. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41(3):345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnstone J., Majumdar S.R., Fox J.D., Marrie T.J. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest. 2008;134(6):1141–1148. doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman D., Shimoni A., Shemer-Avni Y., Keren-Naos A., Shtainberg R., Lieberman D. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138(4):811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352(17):1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 12.Widmer K., Zhu Y., Williams J.V., Griffin M.R., Edwards K.M., Talbot H.K. Rates of hospitalizations for respiratory syncytial virus, human metapneumovirus, and influenza virus in older adults. J Infect Dis. 2012;206(1):56–62. doi: 10.1093/infdis/jis309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falsey A.R., Becker K.L., Swinburne A.J., Nylen E.S., Formica M.A., Hennessey P.A. Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis. 2013;208(3):432–441. doi: 10.1093/infdis/jit190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridkin S., Baggs J., Fagan R., Magill S., Pollack L.A., Malpiedi P. Centers for Disease Control and Prevention (CDC). Vital signs: improving antibiotic use among hospitalized patients. MMWR Morb Mortal Wkly Rep. 2014;63(9):194–200. [PMC free article] [PubMed] [Google Scholar]
- 15.Smith S.M., Fahey T., Smucny J., Becker L.A. Antibiotics for acute bronchitis. Cochrane Database Syst Rev. 2014;3:CD000245. doi: 10.1002/14651858.CD000245.pub3. [DOI] [PubMed] [Google Scholar]
- 16.Graham V.A., Milton A.F., Knowles G.K., Davies R.J. Routine antibiotics in hospital management of acute asthma. Lancet. 1982;1(8269):418–420. doi: 10.1016/s0140-6736(82)91619-1. [DOI] [PubMed] [Google Scholar]
- 17.Ram F.S., Rodriguez-Roisin R., Granados-Navarrete A., Garcia-Aymerich J., Barnes N.C. Antibiotics for exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2006 doi: 10.1002/14651858.CD004403.pub2. (2)(2):CD004403. [DOI] [PubMed] [Google Scholar]
- 18.National Clinical Guideline Centre . National Clinical Guideline Centre; London: 2010. Chronic obstructive pulmonary disease: management of chronic obstructive pulmonary disease in adults in primary and secondary care.http://guidance.nice.org.uk/CG101/Guidance/pdf/English Available from: [Google Scholar]
- 19.Hawkey P.M., Jones A.M. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64(Suppl 1):i3–10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 20.Steinman M.A., Gonzales R., Linder J.A., Landefeld C.S. Changing use of antibiotics in community-based outpatient practice, 1991–1999. Ann Intern Med. 2003;138(7):525–533. doi: 10.7326/0003-4819-138-7-200304010-00008. [DOI] [PubMed] [Google Scholar]
- 21.Dellit T.H., Owens R.C., McGowan J.E., Jr., Gerding D.N., Weinstein R.A., Burke J.P. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;44(2):159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]
- 22.Bramer G.R. International statistical classification of diseases and related health problems. Tenth revision. World Health Stat Q. 1988;41(1):32–36. [PubMed] [Google Scholar]
- 23.Ellis J.S., Curran M.D. Simultaneous molecular detection and confirmation of influenza A H5, with internal control. Diagnostic virology protocols (Methods in Molecular Biology series) In: Stephenson J.R., Warnes A., editors. 2nd ed. Vol. 665. Humana Press; 2011. pp. 161–181. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson K.G., Kent J., Hammersley V., Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 1997;315(7115):1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krüger S., Ewig S., Papassotiriou J., Kunde J., Marre R., von Baum H., CAPNETZ Study Group Inflammatory parameters predict etiologic patterns but do not allow for individual prediction of etiology in patients with CAP: results from the German competence network CAPNETZ. Respir Res. 2009;10:65. doi: 10.1186/1465-9921-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingram P.R., Inglis T., Moxon D., Speers D. Procalcitonin and C-reactive protein in severe 2009 H1N1 influenza infection. Intensive Care Med. 2010 Mar;36(3):528–532. doi: 10.1007/s00134-009-1746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim W.S., Baudouin S.V., George R.C., Hill A.T., Jamieson C., Le Jeune I. Pneumonia guidelines committee of the BTS standards of care committee. BTS guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax. 2009;64(Suppl. 3) doi: 10.1136/thx.2009.121434. iii1–55. [DOI] [PubMed] [Google Scholar]
- 28.Houck P.M., Bratzler D.W., Nsa W., Ma A., Bartlett J.G. Timing of antibiotic administration and outcomes for medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med. 2004;164(6):637–644. doi: 10.1001/archinte.164.6.637. [DOI] [PubMed] [Google Scholar]
- 29.Cesario T.C. Viruses associated with pneumonia in adults. Clin Infect Dis. 2012;55(1):107–113. doi: 10.1093/cid/cis297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brundage J.F. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 2006;6:303–312. doi: 10.1016/S1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolz D., Christ-Crain M., Bingisser R., Leuppi J., Miedinger D., Müller C. Antibiotic treatment of exacerbations of COPD: a randomized, controlled trial comparing procalcitonin-guidance with standard therapy. Chest. 2007;131(1):9–19. doi: 10.1378/chest.06-1500. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen-Van-Tam J.S., Openshaw P.J., Hashim A., Gadd E.M., Lim W.S., Semple M.G. Influenza Clinical Information Network (FLU-CIN). Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave. Thorax. May-September 2009;2010(65):645–651. doi: 10.1136/thx.2010.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland E.R., King T.S., Icitovic N., Ameredes B.T., Bleecker E., Boushey H.A. National Heart, Lung and Blood Institute's Asthma Clinical Research Network. A trial of clarithromycin for the treatment of suboptimally controlled asthma. J Allergy Clin Immunol. 2010;126(4):747–753. doi: 10.1016/j.jaci.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.British Thoracic Society Scottish Intercollegiate Guidelines Network BTS guidelines: British guideline on the management of Asthma. Thorax. 2008;63:iv1–iv121. doi: 10.1136/thx.2008.097741. [DOI] [PubMed] [Google Scholar]
- 35.Gonzales R., Sande M.A. Uncomplicated acute bronchitis. Ann Intern Med. 2000;133(12):981–991. doi: 10.7326/0003-4819-133-12-200012190-00014. [DOI] [PubMed] [Google Scholar]
- 36.Hombrouck A., Sabbe M., Van Casteren V., Wuillaume F., Hue D., Reynders M. Viral aetiology of influenza-like illness in Belgium during the influenza A(H1N1)2009 pandemic. Eur J Clin Microbiol Infect Dis. 2012;31(6):999–1007. doi: 10.1007/s10096-011-1398-4. [DOI] [PubMed] [Google Scholar]
- 37.Thiberville S.D., Ninove L., Vu Hai V., Botelho-Nevers E., Gazin C., Thirion L. The viral etiology of an influenza-like illness during the 2009 pandemic. J Med Virol. 2012;84(7):1071–1079. doi: 10.1002/jmv.23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little P., Stuart B., Moore M., Coenen S., Butler C.C., Godycki-Cwirko M. Amoxicillin for acute lower-respiratory-tract infection in primary care when pneumonia is not suspected: a 12-country, randomised, placebo-controlled trial. Lancet Infect Dis. 2013;13(2):123–129. doi: 10.1016/S1473-3099(12)70300-6. [DOI] [PubMed] [Google Scholar]
- 39.National Institute for Health and Clinical Excellence . 2009. Amantadine, oseltamivir, and zanamivir for the treatment of influenza London: NICE. [Google Scholar]
- 40.Muthuri S.G., Myles P.R., Venkatesan S., Leonardi-Bee J., Nguyen-Van-Tam J.S. Impact of neuraminidase inhibitor treatment on outcomes of public health importance during the 2009–2010 influenza A(H1N1) pandemic: a systematic review and meta-analysis in hospitalized patients. J Infect Dis. 2013 Feb15;207(4):553–563. doi: 10.1093/infdis/jis726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.