Abstract
Although mucosal immune responses are critical for protection of hosts from clinical illness and even mortality caused by mucosal pathogens, the molecular mechanism of mucosal immunity, which is independent of systemic immunity, remains elusive. To explore the mechanistic basis of mucosal protective immunity, gene transcriptional profiling in mucosal tissues was evaluated after the primary and secondary immunization of animals with an attenuated avian infectious bronchitis virus (IBV), a prototype of Coronavirus and a well-characterized mucosal pathogen. Results showed that a number of innate immune factors including toll-like receptors (TLRs), retinoic-acid-inducible gene-1 (RIG-1), type I interferons (IFNs), complements, and interleukin-1 beta (IL-1β) were activated locally after the primary immunization. This was accompanied or immediately followed by a potent Th1 adaptive immunity as evidenced by the activation of T-cell signaling molecules, surface markers, and effector molecules. A strong humoral immune response as supported by the significantly up-regulated immunoglobulin (Ig) gamma chain was observed in the absence of innate, Th1 adaptive immunity, or IgA up-regulation after the secondary immunization, indicating that the local memory response is dominated by IgG. Overall, the results provided the first detailed kinetics on the molecular basis underlying the development of primary and secondary mucosal immunity. The key molecular signatures identified may provide new opportunities for improved prophylactic and therapeutic strategies to combat mucosal infections.
Abbreviations: CCR, chemokine (C-C motif) receptor; CTL, cytotoxic T lymphocyte; CXCR, chemokine (C-X-C motif) receptor; DC, dendritic cell; IBV, infectious bronchitis virus; IFN, interferon; Ig, immunoglobulin; IL, interleukin; TLR, toll-like receptor
Keywords: Transcriptional regulation, Mucosal immunity, Molecular mechanism, IBV, Primary and secondary immunity
1. Introduction
Mucosal and systemic immunity are generally believed to be independently regulated and to act differently in distinct disease processes. This concept is based in large part on observations from studies of specific viral or bacterial infections in animal model systems. Increasing evidence also indicates that mucosal vaccination can induce both systemic and local mucosal immunity, while systemic immunization generally fails to elicit strong mucosal immunity (Valosky et al., 2005, Zhang et al., 2002). This suggests that the development of mucosal immunity relies on the direct interaction of local epithelial cells with pathogens (Cavanagh, 2003, Dhinakar and Jones, 1997). However, the exact molecular transcriptions that regulate the development of antigen-specific mucosal immunity are not well defined.
It has been long recognized that innate immunity is indispensable for the subsequent induction of potent adaptive immunity at both the local (mucosal surfaces) and the systemic levels. The long-recognized innate factors that are important in enhancing adaptive immunity include, but are not limited to, toll-like receptors (TLRs), complements, type I interferons (IFNs), inflammatory cytokines, and chemokines. Indeed, cytokines, complements, and molecules derived from microorganisms such as cholera toxin (CT) and CpG DNA, which are capable of activating TLRs, cytokines, and chemokines, have been employed as vaccine adjuvants to enhance local and systemic adaptive immunity (Fearon, 2000, Holmgren et al., 2003, Staats and Ennis, 1999). But it still remains elusive on the molecular basis for the regulation of mucosal immune response introduced by mucosal vaccination, including the induction of mucosal innate immunity, the communication and transition from local innate to local adaptive immunity, as well as the commitment to memory response which is rapidly recruited upon reencounter of the same mucosal pathogen.
It was the aim of this study to characterize the specific gene expression profiles of the upper respiratory tract (the trachea) at defined time points after vaccination of animals with a model virus and to uncover the molecular mechanisms underlying the development of mucosal immunity. The disease model we chose to use is chicken infectious bronchitis (IB). Chicken IB is a well-studied disease localized mainly to a mucosal site (the trachea) and is caused by infectious bronchitis virus (IBV), the prototype of the Coronavirus family (Cavanagh, 2005, Cavanagh, 2003, Dhinakar and Jones, 1997). The disease can be prevented by vaccination of chickens with attenuated live virus vaccine only when administered locally (intranasal or intra-ocular), not systemically (Cavanagh, 2005, Gillette, 1981, Seo and Collisson, 1997). Studies also indicated that local cytotoxic T cells (CTLs) play an essential role in the clearance of IBV during early IBV infection (Seo and Collisson, 1997), while mucosal antibody response plays a critical role against subsequent infections (Gillette, 1981). Additionally, the kinetics of epithelial cells and antigen specific immune cells during IBV infection or after vaccination has been well characterized in previous studies (Kotani et al., 2000a, Kotani et al., 2000b). In light of the well-characterized mucosal immunity of chicken IBV and the availability of a well-designed and validated chicken cDNA microarray for global gene expression study (Burnside et al., 2005), we believe that IBV serves as an excellent model to better understand the molecular regulation of mucosal immunity by looking at the gene expression profiles at defined times after vaccination. We are particularly interested in identifying the sequential molecular regulation of epithelial cells that may contribute to the activation and recruitment of antigen specific immune cells in the trachea. Because the mucosal surfaces are under constant exposure to a variety of pathogenic organisms, a better understanding of the molecular regulation of mucosal immunity is critical in designing novel strategies for prophylactic and therapeutic control of mucosal-related infectious diseases.
2. Materials and methods
2.1. Virus and animal immunization
Attenuated IBV-Massachusetts (IBV-Mass) grown on primary chicken embryo kidney cell culture was used for the primary and secondary immunizations (Wang et al., 2006). Three-week-old SPF chickens (SPAFAS, Storrs, CT) were administered intranasally with either 50 μl of 104.2 EID50 of IBV-Mass or the same amount of sterilized water as controls. Eight chickens from either vaccinated or control groups were euthanized at 1, 3, 5, 8, 12, and 21 days post-inoculation (d1, d3, d5, d8, d12, and d21). Trachea tissues were separated from the surrounding connecting tissue, aseptically removed, and soaked in RNAlater (Ambion, Austin, TX) to preserve the integrity of tissue RNA. A secondary immunization was performed at d21 with the same protocol. Tracheal samples were collected at 1 and 3 days later (d22 and d24). Animal protocols used in this study were approved by the institutional IACUC (03-A008).
2.2. RNA preparation and array hybridization
The mucosal lining of the trachea was carefully peeled off with forceps and samples from two chickens at the same time point were pooled for total RNA extraction with RNeasy mini kit (Qiagen, Valencia, CA) (Wang et al., 2006). To get enough RNA, total RNA thus extracted was then subject to RNA amplification (Ambion, Foster City, CA). RNA quality and quantity was assured by gel electrophoresis and NanoDrop spectrophotometer (Wilmington, DE) before further analysis. A mixed loop-design as illustrated in Fig. 1 was used for the microarray experiment, which allowed direct comparison of data among all time points with enhanced statistical precision, power, and robustness (Tempelman, 2005). Four different RNA samples were used for the four different hybridizations at each time point with the chicken 13 k cDNA microarray slides (FHCRC, Seattle, WA). RNA amplification, cDNA synthesis, probe labeling and hybridization were performed as recommended (http://www.fhcrc.org/science/shared_resources/genomics/dna_array/spotted_arrays/chicken_array/). After washing, arrays were scanned immediately and spot intensities of the hybridized images were collected by using GenePixPro6.0 (Molecular Devices, Sunnyvale, CA).
Fig. 1.
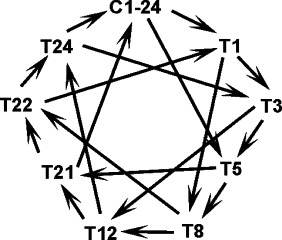
A flowchart schematically shows the microarray experiment design. C1-24, pooled samples from age-matched control chickens; T, IBV-Mass immunized chickens; the numbers indicated the day after the primary immunization. Total RNA from each group was used to compare with RNA from other four groups as indicated by the lines between the groups. The arrowhead pointed samples were labeled with Cy5 and hybridized with the samples on the other end of the lines, which were labeled with Cy3. Four different RNA samples were used for the four different hybridizations at each time point with the chicken 13 k cDNA microarray slides (FHCRC, Seattle, WA).
2.3. Microarray data analysis
R/BioConductor “LIMMA (LInear Model for MicroArray)” package was used for background correction, normalization and model fitting (Smyth, 2004). Those genes with an ANOVA adjusted p-value for false-discovery rate <0.01 and an absolute fold change ≥2 at two time points or more were considered as significantly expressed genes (Tempelman, 2005). Functional annotation of the significant genes was analyzed with KAAS (http://www.genome.jp/kegg/kaas) or TIGR (http://www.tigr.org). Key immune components were selected for further hierarchical clustering and pathway analysis with GenMAPP (Dahlquist et al., 2002). All data were deposited into GEO database (http://www.ncbi.nih.gov/geo/info/linking.html, GSE6198).
2.4. Real-time RT-PCR and array data validation
Selected genes were subjected to real-time RT-PCR to validate the microarray data (Supplementary Table 1). Viral glycoprotein gene (IBV-S) was also quantified to determine the viral RNA copies. Furthermore, transcriptional profiling of key cytokines not included in the array was assessed. Real-time PCR was performed with SYBR® Green (Applied Biosystems, Foster City, CA) on Mx3000P® (Stratagene, La Jolla, CA). Primer-dimer formation was assessed by both melting curves and gel electrophoresis. Series of diluted cDNAs were used to establish standard curves for RNA quantification (Bai et al., 2006).
3. Results
We examined the gene expression profiles in the trachea mucosal lining at 1, 3, 5, 8, 12, and 21 days (d1, d3, d5, d8, d12, and d21) after primary immunization and at 1 and 3 days (d22 and d24) after secondary immunization with IBV-Mass. Of the 13,008 usable cDNA features on the array (Burnside et al., 2005), a total of 1752 elements were changed by 2-fold or more for at least two time points (Supplementary Tables 2 and 4). The majority of changes occurred upon primary immunization, while relatively few elements changed upon the secondary immunization. The significantly altered immune-related genes, which are grouped based on their functions and their expression patterns, are described below.
3.1. Innate immune response
3.1.1. Factors identified for pathogen recognition
TLRs are one of the key molecules of the innate immune system, which detect conserved structures found in a broad range of pathogens. Type 1 TLR2 (2.9- and 4.2-fold for d1 and d3), TLR3 (4.0-fold for d1), TLR6 (3.5-fold for d5), and TLR7 (3.8-, 5.1-, 3.0-fold for d1, d3, and d5) were significantly up-regulated upon primary IBV immunization. TLR3 is induced by the intermediate dsRNAs during viral replication and its role in viral immunology is well established (Le Goffic et al., 2007, Liu et al., 2007, Wang et al., 2006). TLR7 is required for single stranded RNA virus recognition and the subsequent cytokine production by plasmacytoid dendritic cells (pDCs) and activation of B cells (Lund et al., 2004). TLR2/6 pathway is responsible for recognition of peptidoglycan and lipoprotein from Gram-positive bacteria, mycoplasmas, mycobacteria, and spirochetes (Hoebe et al., 2005). However, unlike its counterpart of type 2, type 1 TLR2 possesses less ability to act as a receptor for bacterial LPS and is likely to be a novel sentinel for viral infection (Fukui et al., 2001). Overall, it appeared that TLR3, TLR7, and type 1 TLR2/6 all played some roles in the local recognition of the RNA virus, IBV-Mass.
Viral RNA replication can also be sensed in a TLR-independent mechanism. Retinoic-acid-inducible gene-1 (RIG-1)-mediated antiviral activity, which leads to type 1 IFN production, was revealed by the induction of RIG E3 protein (increased 8.2-, 6.1-, 6.8, 3.5-fold from d1 to d8) (Gack et al., 2007), together with the repression of cytochrome p450 (−2.5-, −3.2-, −2.0-fold for d1, d3, and d5) for retinoic acid degradation. RIG-1 and other cytoplasmic RNA helicases ensure viruses are detected even when they have escaped initial recognition by TLRs. Taken together, we identified two sets of infection sensors, suggesting two independent strategies employed for the elicitation of local innate immunity in response to IBV infection.
3.1.2. Cytokine related activities
Cytokines, the master regulators of immune system, play a pivotal role in local mucosal immunity. Each cytokine binds to a specific cell-surface receptor to initiate a cascade of intracellular signaling for specific cell functions (Kruth, 1998). The activated cytokines and their receptors (Fig. 2 ) encompassed IL-1β (10.8-, 5.5-, 2.1-, and 2.5-fold for d1, d3, d5, and d21), IL-10R2 (6.9-, 2.4-, and −2.3-fold for d1, d3, and d12), and common cytokine receptor γ (increased 2.4-, 4.0-, 4.4-, and 2.9-fold from d1 to d8) to bind IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21. Since cytokines function in almost every phase of the host response to viral infections, including control of inflammation and induction of the antiviral state, their stimulation here indicates the activation of the host immune system.
Fig. 2.
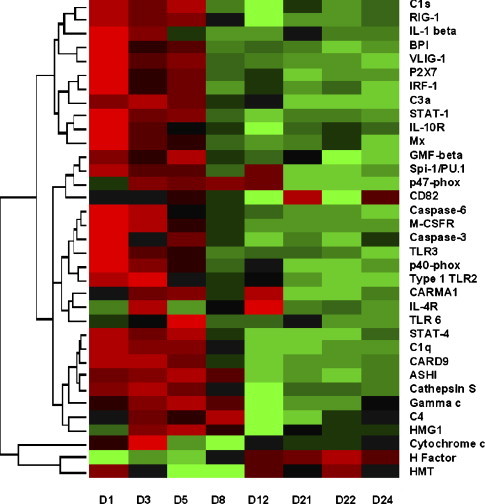
Expression profiles of local innate immunity after attenuated IBV-Mass immunizations. Rows are individual genes, and columns are the measurement taken at different time points. Colors represent the relative gene expression level. Green indicates an expression below the mean value for the gene, black indicates an expression near the mean, and red indicates an expression above the mean. log2 fold changes of genes used in clustering analysis are provided in Supplementary Table 3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
IFNα/β are synthesized by infected cells and immature DCs and establish the antiviral state. Firstly, IFN regulatory factor-1 (IRF-1, 4.3-, 2.0-, 2.4-fold for d1, d3, and d5) was responsible for sustained IFN transcription after infection. Secondly, IFNs triggered a Jak/Stat signaling cascade and induced p53-dependent antiviral protection (Shin-Ya et al., 2005). This was revealed by the activation of STAT (STAT-1, 6.4-, 3.4-, 3.7-fold for d1, d3, and d5; STAT-4, 3.4-, 2.5-, 3.0-fold for d1, d3, and d5) p53 (p53-inducible proapoptotic scotin, 9.1-, 5.5-, 6.8-, 2.4-, 4.0-, and 2.3-fold for d1, d3, d5, d8, d12, and d24; CD82/R2 leukocyte antigen, 2.0-, 2.2-, 2.4-, 3.9-, and 2.6-fold for d1, d3, d5, d12, and d24; liver LKB1-interacting protein, 2.6-, 1.9-, 2.1-, and 2.1-fold for d1, d3, d5, and d21) and caspase cysteine proteases (caspase-6, 2.7-, 2.6-fold for d1 and d3; and CARD-9, 3.1-, 3.6-, 2.4-fold for d1, d3, and d5) (Fig. 2). Thirdly, IFN antiviral state was enhanced via IFN-responsive genes. Myxovirus resistance protein Mx (increased 18.4-, 4.1-, and 2.7-fold for d1, d3, and d5) has intrinsic GTPase to sense nucleocapsid-like structures during infection and traps viral components to locations unavailable for new particle generation (Li et al., 2007). The novel very large inducible GTPase VLIG-1 (increased 20.2-, 5.3-, and 8.8-fold for d1, d3, and d5) is responsible for regulation of several hundred genes in pathogen defense at the transcriptional level (Klamp et al., 2003). Taken together, the activation of cytokines like IL-1β, MIP-1β, and IFN signaling pathways may be critical in bridging the innate and adaptive local immunity by modulating the local microenvironment.
3.1.3. Complement system
A biochemical cascade of the complement system helps virus clearance and forms the central core in innate immunity (Ogundele, 2001). Recent studies also suggested that the complement system is important in T-cell activation (Kemper and Atkinson, 2007). Complement participation in local immunity was confirmed by the induction of complement C1q (3.0-, 2.5-, 2.4-fold for d1, d3, and d5), C1s (8.6-, 6.3-, 8.5-, 2.4-, and 2.0-fold for d1, d3, d5, d21, and d24), anaphylatoxin C3a receptor (4.8-, 5.6-, 4.1-, 2.1-, and 2.3-fold from d1 to d12), and complement C4 (2.0-, and 3.0-fold for d3 and d8), as well as inhibition of its negative regulator factor H (decreased −2.1-, −1.8-, and −2.0-fold for d1, d3, and d5) (Fig. 2). C1q and C4b are involved in the opsonization process, suggesting the activation of the classical complement pathway. C3a and C4a stimulate a local inflammatory response to limit infection. This together with the stimulation of phagocytes demonstrate that the various complement components at mucosal sites are actively involved in the primary defense and communication between the innate and adaptive immune systems.
3.1.4. Chemokine and other factors for immune cell trafficking
One of the major functions of chemokines is to guide the migration of cells during immune surveillance. Chemokines identified here included CXCR4 (2.1-, 2.1-, 3.0-, 2.2-fold for d1, d3, d5, and d12), CCR6 (2.0-, 2.5-, 2.2-fold for d1, d3, and d5), chemokine-like receptor 1/CHEMR23 (3.0- and 2.9-fold for d1 and d3), stromal cell-derived factor SDF4/Cab45 (2.8-, 2.0-, 2.9-fold for d3, d5, and d58), and the functionally unknown ah221 (5.6-, 2.4-, 2.5-, 2.2-fold for d1, d3, d5, and d12) and ah294 (7.0-, 3.8-, 6.4- and 2.0-fold from d3 to d12). CXCR4 is important for the regulation of stem/progenitor cell trafficking (Tamamura et al., 2006). It has been reported that CCR6 is responsible in regulating the positioning and recruitment of DCs and T cells during inflammatory and immunological responses in small intestine (Lugering et al., 2003). CHEMR23 interacts with chemerin and plays a key role to direct macrophage and immature DC trafficking from blood to tissue sites (Zabel et al., 2005). SDF4 is a novel calcium-binding protein that localizes to the Golgi compartment, and its role in immune cell recruiting remains to be defined. Chicken has a different repertoire of chemokines from those of mammals, and little is known about the function of ah221 and ah294, which could be an orthologue of mammalian CCL5 (Hughes et al., 2007). Since chemokine activated integrins are responsible for leukocyte chemotaxis in microenvironments (Kim, 2004), the activation of integrin β2 (CD18, 2.3-, 3.4-, 2.5-fold for d1, d3, and d5) provided further evidence for the participation of chemokine and their activated molecules in directing the appropriate influx of immune cells to local tissues.
In addition to chemokines, matrix metalloproteinase (MMPs) have recently been implicated in the migration of immune cells (Faveeuw et al., 2001, Gurney et al., 2006, Li et al., 2004). For example, a recent study suggested that mice deficient in MMP3 exhibited a transient block of CD4+ T-cell migration to the lamina propria of intestines during a bacterial infection, confirming the role of MMP3 in the migration of T cells to local tissues (Li et al., 2004). We observed that MMP3 was significantly up-regulated after primary immunization (164.8-, 47.6-, 28.1-, 16.0-, 6.2-, and 2.4-fold from d1 to d21), suggesting its potential role in the migration of activated T cells.
3.1.5. Phagocytosis and inflammation
Molecules involved in monocyte/macrophage signaling were shown to be stably elevated (Fig. 2), which included Spi-1/PU.1 (2.7-, 2.1-, 2.1-, 2.3-fold for d1, d3, d5, and d12), glia maturation factor GMF-β (2.3-, 2.1-, 2.5-fold for d1, d3, and d5), and macrophage colony stimulating factor receptor M-CSFR (3.5-, 2.8-fold for d1 and d3). Abundant transcription factor Spi-1 promotes macrophage differentiation (DeKoter and Singh, 2000). GMF is an intracellular signal transduction regulator to stimulate p38, NF-κB, and GM-CSF and promote CD4−/CD8+ cell differentiation (Yamazaki et al., 2005). M-CSF is essential for macrophage linage development. Neutrophils induce respiratory burst through NADPH oxidase enzymes (p47-phox, 2.0-, 2.8-, 2.6-, 2.9-, 2.7-fold from d1 to d12; p40-phox, 4.2-, 2.9-fold for d1 and d3; NADPH dehydrogenase, 2.1-, 1.9-, 2.3-, and 3.1-fold for d1, d3, d5, and d21; NADH-ubiquinone oxidoreductase ASHI, 2.3-, 2.7-, 3.0-, 2.1-fold from d1 to d8), which produce large quantities of reactive oxygen species to kill the phagocytosed pathogens. Neutrophil function was also accomplished via degranulation, a process that release antimicrobial cytotoxic molecules, such as protease cathepsin S (2.2-, 2.4-, 2.0-fold for d1, d3, and d5) and bactericidal permeability-increasing protein BPI (25.8-, 4.0-, 4.9-fold for d1, d3, and d5). BPI is recognized as a “molecular shield” for mucosal surface protection (Canny et al., 2002). Since neutrophils are generally specific to bacterial defense, its unexpected influx locally may suggest a previously unappreciated protective mechanism against virus infections.
Inflammation is mediated by granulocytes, complements, and histamines, as well as its own regulatory factors. We observed a down-regulation of histamine methyltransferase HMT (−2.0, −5.3-, −5.2, −2.2, −2.2-fold for d3, d5, d8, d21, and, d24), which is dominant in histamine biotransformation in bronchial epithelium. Moreover, specific regulatory factors like purinergic receptor P2X7 (5.3-, 2.3-, 3.1-fold for d1, d3, and d5) and high mobility group box HMGB1 (2.4-, 3.1-, −4.0-fold for d3, d5, and d12) (Fig. 2) showed dramatic alteration, indicating a well-controlled inflammation. P2X7 functions as a major regulator in IL-1 β-mediated inflammation via its cation channel (Chen and Brosnan, 2006). HMGB1 is derived from activated macrophages and functions as a late mediator of systemic inflammation (Yan et al., 2000). Thus, the process of inflammation is orchestrated by an array of mediators including IFNs and SOCS3 as described later, which may be important in fine-tuning the type of potent local adaptive immunity to be induced.
3.2. Th1 adaptive response
3.2.1. Antigen presentation and T-cell activation
Activation of the adaptive immune system is triggered by processes of antigen presentation and T-cell activation (Viret and Janeway, 1999). IBV infected cells were recognized by the engagement of TCR on CD8+ T cells (TCRβ, increased 4.0-, 5.6-, 8.1-, 6.7-, 6.2-, 1.8-, 2.0-, and 3.4-fold from d1 to d24; TCRζ, increased 2.5-, 3.8-, 4.3-, 3.0-, 2.7-fold from d1 to d12) with peptides presented by MHC class I (increased 2.3-, 1.8-, and 1.9-fold from d3 to d8) on macrophages or DCs. Exogenous viral antigen was processed and presented by MHC class II (β chain, 3.1-, 2.6-, 2.0-fold for d1, d3, and d5; invariant chain, increased 2.2-, 2.4-, 2.5-, 2.0-fold from d1 to d8) on mature DCs (mDC marker CD83, 4.4-, 1.9-, 2.0-fold for d1, d3, and d5). Interaction of TCR on CD4+ T cells with the MHC class II-peptide complex resulted in activation and differentiation of T lymphocytes via a series of biochemical events. Full activation required the interaction of other surface proteins and costimulatory molecules, e.g., CD8α (2.3-, 3.1-fold for d5 and d8) is activated by lymphoid-restricted zinc finger transcription factor Ikaro (2.5-, 3.9-fold for d3 and d5) (Urban and Winandy, 2004), and CD3 complex (CD3δ/γ homolog, increased 3.6-, 4.8-, 8.1-, 6.9-, 2.3-, 1.9-, 2.1-, 3.3-fold from d1 to d24; CD3ɛ, 3.1-, 3.6-, 7.0-, 3.4-, and 2.5-fold for d1, d3, d5, d8, and d24). Upon activation, the tyrosine kinase (2.8-, 2.1-, 2.6-fold for d1, d3, and d5) and its associated GRAP (GRB2-related adaptor protein, 2.4-, 2.9-, 2.3-fold for d3, d5, and d8) (Parry et al., 2006) phosphorylated the intracellular portions of the CD3 complex, which in turn, interacted with ζ chain associated protein ZAP-70 (increased 2.4-, 2.5-, 4.1-, 2.2-fold from d1 to d8) for transcription of genes in T-cell differentiation and proliferation. In addition, T-cell activation was further facilitated by lymphocyte cytosolic protein LCP-1/l-plastin (11.4-, 9.7-, 9.5-, 3.7-, and 2.2-fold for d1, d3, d5, d8, and d24), which binds to actin to promote surface transport of the T-cell activation molecules CD69 and CD25 (Wabnitz et al., 2007). Overall, the results clearly revealed the molecular kinetics of local T-cell activation.
3.2.2. Elevated Th1 molecular makers
T-cell polarization plays an important role in immune responses against pathogens. Molecules exclusively linked to Th1 cells were activated immediately upon IBV immunization. The elevated Th1-specific factors comprised of Ras-related C3 botulinum toxin substrate 2 (Rac2, 3.6-, 3.3-, 4.4-, 3.5-, and 2.3-fold for d1, d3, d5, d8, and d24) (Li et al., 2000), bovine antigen WC1.1 (5.3-, 4.4-, 2.3-fold for d1, d3, and d5) on γ/δ T cells (Rogers et al., 2005), and Tim3 (T-cell Ig and mucin domains-containing protein 3, increased 2.5-, 2.4-, 2.5, 2.3 and 2.4-fold from d1 to d12) (Chae et al., 2004) (Fig. 3 ). All of them are selectively expressed on Th1 lymphocytes and are associated with IFN-γ production. In addition, the activation of IFN-γ, the Th1 hallmark cytokine, could be inferred from the activation of IFN-γ inducible Mg11 (7.6-, 3.2-, 4.2-fold for d1, d3, and d5), an immunity-related GTPase (IRG) for host resistance to intracellular pathogens (Taylor, 2007). In summary, these molecular markers are strong indicators that proliferating T helper cells differentiate into Th1 subtypes.
Fig. 3.
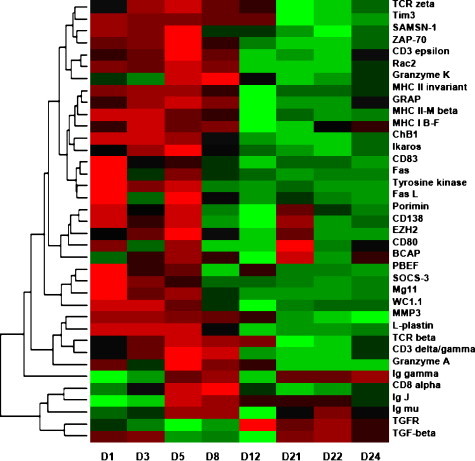
Expression profiles of local adaptive immunity after attenuated IBV-Mass immunizations. Rows are individual genes, and columns are measurement taken at different time points. Colors represent the relative gene expression level. Green indicates an expression below the mean value for the gene, black indicates an expression near the mean, and red indicates an expression above the mean. log2 fold changes of genes used in clustering analysis are provided in Supplementary Table 3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2.3. Elevated Th1 effector factors
CTLs, one of the major sources of IFN-γ, are superbly equipped Th1 lethal effector cells capable of selectively eliminating virus-infected cells. Markers for CTLs such as granzyme A (4.6-, 2.0-, 18.1-, 6.4- and 4.1-fold from d1 to d12), membrane mucin porimin (5.6-, 2.2-, 5.5-, and 3.4-fold for d1, d3, d5, and d21), and the Fas/FasL system (Fas ligand, 2.4- and 2.3-fold for d1 and d5; Fas, 4.5- and 2.1-fold for d1 and d5) were activated after primary immunization (Fig. 3). Granzyme A is an important effector of CTLs and NK for target cell lysis by reactive oxygen species and mitochondrial transmembrane potential loss (Pardo et al., 2004). Granzyme K triggers DNA fragmentation and is involved in apoptosis. Unlike cytolysis or apoptosis induced by granzymes and Fas/Fas ligand system, porimin does lethal damage to cell membrane and induces cell death with features similar to oncosis (Ma et al., 2001). Activated CTLs start to express Fas ligand (2.4- and 2.3-fold for d1 and d5), which binds to Fas (4.5- and 2.1-fold for d1 and d5) expressed on infected cells to induce apoptotic cell death (Cianga et al., 2004). Overall, the early induction of CTL responses may account for the overt purging of virus from local infection sites. Consistent with this, local T lymphocytes resident in the oral mucosa provide essential protection against oral candidiasis following active immunization by the oral route (Farah and Ashman, 2005).
3.3. Humoral response
A number of genes important in triggering B cell activation were up-regulated after primary immunization. The signaling molecules responsible for B cell activation and differentiation included CD80-like co-stimulator (2.7-, 2.8-, 3.9-fold for d1, d5, and d21), B cell maturation promoting cytokine PBEF (pre-B cell enhancing factor, 5.6-, 2.3-, 2.5-, 2.0-fold for d1, d3, d5, and d12), Th2 promotion factor SOCS3 (suppressor of cytokine signaling 3, 8.3-, 2.8-, 2.1-fold for d1, d3, and d5) (Seki et al., 2003), B cell adaptor protein (BCAP, 2.8-, 5.0-, 2.6-, 6.4-, 3.0-fold for d3, d5, d8, d21, and d24), adaptor and scaffold protein of SAM-domain protein SAMSN-1 (3.6-, 3.1-, 5.5-fold for d1, d3, and d5) (Zhu et al., 2004), avian B cell differentiation antigen chB1 (3.3-, 3.9-, 3.1-fold for d1, d3, and d5) (Goitsuka et al., 2001), zeste homolog enhancer EZH2 (2.1-, 2.9-, 6.5-, 2.7-fold for d1, d3, d5 and d21) for IgH rearrangement regulation, immunoglobulin heavy chain binding protein BIP (3.5-, 3.3-, 6.0-, 6.2-, and 2.8-fold for d1, d3, d5, d21, and d24), and syndecan-1/CD138 (4.4-, 2.3-, 4.2-, 3.4-fold for d1, d3, d5, and d21) on Ig secreting plasma cells (Fig. 3). These are accompanied or followed by the transient activation of Ig μ chain (IgM) (1.9-, 1.7-fold for d3 and d5) and its linkage component of Ig J chain (−2.8-, −2.0-, 2.5-fold for d1, d3, and d5) (Fig. 3). Ig γ chain was activated after primary immunization and surged immediately after the secondary immunization (5.1-, 6.6-, 1.0-, 5.1-, 4.9-, 7.8-fold from d5 to d24). Surprisingly, no activation of Ig α chain (IgA) was observed. Instead, data obtained from real-time RT-PCR revealed that the transcription of Ig α chain was suppressed (−27.33-, −17.86-, −4.51-fold for d8, d12, and d22, respectively). This is further supported by the suppression of TGF-β (−2.4-, −2.2-, −3.6-fold for d5, d8, and d12, respectively), a regulator for IgA. Taken together, the results suggested that IgA might not be an important arm of local primary and secondary immunity against IBV-Mass and the dominant local IgG immunity after secondary immunization provided protection from virus entry by neutralizing viruses.
4. Discussion
Since transcriptional control is one of the most important aspects in regulating any biological process, transcriptional profiling has been widely used to uncover the underlying mechanisms of a number of complex biological processes including development and diseases. Here, we have intended to identify the molecular mechanism of mucosal immunity by examining the local gene transcription profile at defined time points after primary and secondary immunizations. To our knowledge, this is the first attempt to characterize the molecular control of the protective mechanisms in mucosal immunity. We recognized that a diversity of cellular and molecular factors are involved in the induction and regulation of mucosal immunity. A number of innate factors such as TLRs and RIG-1 worked synergistically in the initial sensing of the danger signals, which is followed by the induction of type I IFNs, IL-1β, MIP-1β, SOCS 3, and other soluble mediators which not only triggered the antiviral state of adjacent cells, but also facilitated the activation and migration of T cells to local tissues by modifying the local microenvironment and dictating the type of adaptive immunity. More recent studies have suggested that TLR3 and RIG-1 are also involved in the local recognition of other respiratory RNA viruses including influenza A virus and respiratory syncytial virus (Le Goffic et al., 2007, Liu et al., 2007), suggesting that multiple universal innate factors are activated in the local respiratory tract upon virus entry and they may play a critical role in triggering a potent local adaptive immunity by working synergistically. We observed that potent innate and Th1 adaptive local immunity were induced by IBV-Mass primary immunization and they were mainly responsible for the viral clearance from the local infection sites. A strong humoral response dominated by mucosal IgG, the specific antibody already primed earlier, was activated after the secondary immunization and provided protection against virus entry by neutralizing virus particles. These observations are consistent with the previous studies showing the importance of local CTL response in primary infection and local neutralizing antibody response against secondary exposure to the same virus (Gillette, 1981, Seo and Collisson, 1997). The molecular transcription profile further provides proof for the hypothesis that strong innate and adaptive local immunity are efficiently induced locally by mucosal vaccination (Cavanagh, 2003, Dhinakar and Jones, 1997), and it is this regional immune response that plays a critical role in providing effective protection against mucosal pathogens.
Despite the long-recognized type I IFNs induced genes such as Mx activated by IBV (Enjuanes et al., 2006), other well-characterized and important signature molecules for mucosal immunity such as IL-1β and MIP-1β were also significantly up-regulated immediately after primary immunization (Mattapallil et al., 1998, Staats and Ennis, 1999). Surprisingly, T-cell activation was detected as early as day 1 after primary immunization, suggesting the efficient trafficking of T cell to local tissues. Several chemokines including CXCR4, CCR6, and cell adhesion molecules such as syndecan-4 were activated locally and may be partly responsible for the migration of activated T cells. New evidence clearly suggests that cytokines such as IL-1β might be critical for the induction MMPs and the subsequent migration of immune cells such as neutrophils and CD4+ T cells to local tissues (Flannery et al., 1999, Gurney et al., 2006, Koshy et al., 2002, Li et al., 2004, Wertheimer et al., 1995), a process that has long been recognized to be mediated mainly by chemokines. A transient block of CD4+ T-cell migration to the lamina propria of the intestinal tract was observed in MMP3-deficient mice (Li et al., 2004). We reason that MMP3, possibly activated by IL-1β as described previously (Flannery et al., 1999, Wertheimer et al., 1995), may facilitate the migration of T cells to local tissues, leading to development of a Th1 dominant local immunity and the eradication of virus from local tissues. This is supported by the observation that a significant increase in MMP3 from d1 to d8 after primary immunization occurred concurrently with the up-regulation of T-cell receptors locally, suggesting that MMP3 may be important in the migration of T cell to local respiratory mucosal tissues, similar to what was found in the gut and intestine tract (Li et al., 2004). Further investigations are needed to confirm the potential role of MMP3 in facilitating the migration of activated T cells to the respiratory tract.
It is generally believed that mucosal IgA is the predominant immune effector molecule induced after mucosal immunization to prevent the entry of invading bacterial and viral pathogens (Harriman et al., 1999). Surprisingly, we did not detect any expression of Ig α chain after primary and secondary immunizations. Instead, local IgG was induced after primary immunization and surged immediately after secondary immunization, suggesting a dominant local IgG in the stimulation of memory response against viral re-infection. Our data is consistent with the recent study showing that chlamydial re-infection is successfully prevented by heightened IgG2a and IgG2b responses in secretions in IgA deficient (IgA−/−) mice (Morrison and Morrison, 2005). More recent studies have also suggested an important, protective role of IgG against influenza virus infection in mice, where IgA is not indispensable to confer protection, and IgG1, IgG2a, and IgM may be effective in preventing influenza virus infection in IgA knockout mice (Mbawuike et al., 1999). Similarly, complete protection of both the upper and lower airways was correlated with the presence of respiratory syncytial virus (RSV)-specific IgG in mucosal secretions at the time of challenge (Valosky et al., 2005). Others have elucidated that IgA might be only important in non-pathogen induced antibody responses (Harriman et al., 1999, Mbawuike et al., 1999). Taken together, local IgG response is one of the dominant responses in secondary immunity against pathogens and may provide an in situ defense of the mucosa against invasion. Results from our and others’ studies challenged the role of IgA in secondary immunity in virus infection. It will be of interest to further examine the secondary immunity against other bacterial and viral infections to see if this is a universal mechanism.
Although potent innate and local CTL responses were induced after the primary immunization and were responsible for the clearance of virus from local tissues, we did not detect any activation of these molecules after the secondary immunization. Together with the rapid increase of IgG after secondary immunization, we suspect that the virus was immediately neutralized by local IgG after administration to avoid recognition locally by innate factors and the subsequent activation of CTLs. Therefore, it is reasonable to speculate that complete protection by local IgG is the reason why no local innate and CTL responses were induced. The role of memory CTLs in preventing local virus entry appeared to be minimal, if any. The result is contradictory to the previous study showing that memory CTLs mainly reside in local nonlymphoid tissues and is ready to react after second pathogen exposure (Masopust et al., 2001). Since the authors did not examine the activation of the memory CTLs following a second exposure to the same pathogen, the role of memory CTLs in preventing pathogen entry remains unclear. We prefer the notion that memory CTLs may only be activated if the virus breaches the innate barriers and enters the host and that they are important in combating local infection and eradicating infected cells. Further investigations are necessary to evaluate the role of memory CTLs in local protection.
Overall, we have provided data on the molecular control of mucosal primary and secondary immunity using avian IBV as a model system. As schematically shown in Fig. 4 , multiple innate factors were activated, worked synergistically, and orchestrated the activation of T cells as supported by the up-regulation of Fas, granzyme, porimin, and other weapons of CTLs used to kill virus-infected cells, which is followed by the activation of B cells and increased expression of Ig gamma chain locally. The kinetics of molecular signatures identified here by transcriptional profiling not only provides a proof for the mechanistic basis of mucosal immunity to respiratory viral pathogens but also confirms the previous observations on mucosal immunity defined using traditional immunology tools (Gillette, 1981, Seo and Collisson, 1997). It should be noted that the molecular signatures defined here may not be complete due to the lack of certain genes on the 13 k cDNA array and the inevitably missed time points. Nevertheless, expressional kinetics reveals the molecular mechanisms of primary and secondary mucosal immunity and provides guidance on how vaccines could be best designed to confer efficient protection against mucosal infectious diseases in respiratory tracts.
Fig. 4.
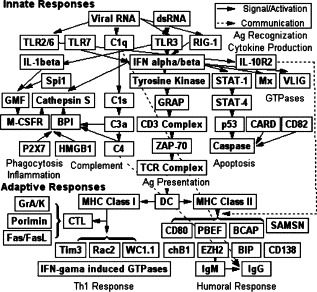
A proposed model on the molecular kinetics of mucosal immunity following local immunizations of IBV-Mass. IBV antigen is firstly recognized by two independent innate mechanisms including TLR and RIG-1. Cytokines such as IL-1β and IFNs activated by the innate pathways regulated the activation of T cells and induced cells to an anti-viral state. In addition, a number of other innate responses are also induced, including phagocytosis, complement, inflammation, cell death, and antigen presentation, which created an ideal microenvironment for T-cell activation and served as a bridging factor for connecting the local innate and adaptive immune systems. Once the T cells are activated, two branches of adaptive immune responses are initiated with CTLs being activated initially, followed by humoral immunity. CTLs, together with parts of the innate immunity, are responsible for the virus clearance from the local infection sites. Local IgG is the only and dominant arm of local secondary immunity against subsequent virus exposure.
Acknowledgements
The authors thank Mrs. Diane Baker, Mrs. Jessica Freeling, and Mr. Ping Lu for help with the animals and RNA extraction. This study was supported by grants from NSF/EPSCoR (EPS-0091948) and by the State of South Dakota.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vetimm.2007.09.016.
Appendix A. Supplementary data
References
- Bai L., Deng Y.M., Dodds A.J., Milliken S., Moore J., Ma D.D. A SYBR green-based real-time PCR method for detection of haemopoietic chimerism in allogeneic haemopoietic stem cell transplant recipients. Eur. J. Haematol. 2006;77:425–431. doi: 10.1111/j.1600-0609.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- Burnside J., Neiman P., Tang J., Basom R., Talbot R., Aronszajn M., Burt D., Delrow J. Development of a cDNA array for chicken gene expression analysis. BMC Genomics. 2005;6:13. doi: 10.1186/1471-2164-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny G., Levy O., Furuta G.T., Narravula-Alipati S., Sisson R.B., Serhan C.N., Colgan S.P. Lipid mediator-induced expression of bactericidal/permeability-increasing protein (BPI) in human mucosal epithelia. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3902–3907. doi: 10.1073/pnas.052533799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Chae S.C., Song J.H., Pounsambath P., Yuan H.Y., Lee J.H., Kim J.J., Lee Y.C., Chung H.T. Molecular variations in Th1-specific cell surface gene Tim-3. Exp. Mol. Med. 2004;36:274–278. doi: 10.1038/emm.2004.37. [DOI] [PubMed] [Google Scholar]
- Chen L., Brosnan C.F. Regulation of immune response by P2X7 receptor. Crit. Rev. Immunol. 2006;26:499–513. doi: 10.1615/critrevimmunol.v26.i6.30. [DOI] [PubMed] [Google Scholar]
- Cianga C., Cianga P., Cozma L., Diaconu C., Carasevici E. The significance of Fas/FasL molecular system in breast cancer. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2004;108:440–444. [PubMed] [Google Scholar]
- Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Dhinakar R.G., Jones R.C. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol. 1997;26:677–706. doi: 10.1080/03079459708419246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Almazan F., Sola I., Zuniga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- Farah C.S., Ashman R.B. Active and passive immunization against oral Candida albicans infection in a murine model. Oral. Microbiol. Immunol. 2005;20:376–381. doi: 10.1111/j.1399-302X.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- Faveeuw C., Preece G., Ager A. Transendothelial migration of lymphocytes across high endothelial venules into lymph nodes is affected by metalloproteinases. Blood. 2001;98:688–695. doi: 10.1182/blood.v98.3.688. [DOI] [PubMed] [Google Scholar]
- Fearon D.T. Innate immunity—beginning to fulfill its promise? Nat. Immunol. 2000;1:102–103. doi: 10.1038/77773. [DOI] [PubMed] [Google Scholar]
- Flannery C.R., Little C.B., Caterson B., Hughes C.E. Effects of culture conditions and exposure to catabolic stimulators (IL-1 and retinoic acid) on the expression of matrix metalloproteinases (MMPs) and disintegrin metalloproteinases (ADAMs) by articular cartilage chondrocytes. Matrix Biol. 1999;18:225–237. doi: 10.1016/s0945-053x(99)00024-4. [DOI] [PubMed] [Google Scholar]
- Fukui A., Inoue N., Matsumoto M., Nomura M., Yamada K., Matsuda Y., Toyoshima K., Seya T. Molecular cloning and functional characterization of chicken toll-like receptors. A single chicken toll covers multiple molecular patterns. J. Biol. Chem. 2001;276:47143–47149. doi: 10.1074/jbc.M103902200. [DOI] [PubMed] [Google Scholar]
- Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gillette K.G. Local antibody response in avian infectious bronchitis: virus-neutralizing antibody in tracheobronchial secretions. Avian Dis. 1981;25:431–443. [PubMed] [Google Scholar]
- Goitsuka R., Mamada H., Kitamura D., Cooper M.D., Chen C.L. Genomic structure and transcriptional regulation of the early B cell gene chB1. J. Immunol. 2001;167:1454–1460. doi: 10.4049/jimmunol.167.3.1454. [DOI] [PubMed] [Google Scholar]
- Gurney K.J., Estrada E.Y., Rosenberg G.A. Blood-brain barrier disruption by stromelysin-1 facilitates neutrophil infiltration in neuroinflammation. Neurobiol. Dis. 2006;23:87–96. doi: 10.1016/j.nbd.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Harriman G.R., Bogue M., Rogers P., Finegold M., Pacheco S., Bradley A., Zhang Y., Mbawuike I.N. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J. Immunol. 1999;162:2521–2529. [PubMed] [Google Scholar]
- Hoebe K., Georgel P., Rutschmann S., Du X., Mudd S., Crozat K., Sovath S., Shamel L., Hartung T., Zahringer U., Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- Holmgren J., Czerkinsky C., Eriksson K., Mharandi A. Mucosal immunisation and adjuvants: a brief overview of recent advances and challenges. Vaccine. 2003;21(Suppl. 2):S89–S95. doi: 10.1016/s0264-410x(03)00206-8. [DOI] [PubMed] [Google Scholar]
- Hughes S., Poh T.Y., Bumstead N., Kaiser P. Re-evaluation of the chicken MIP family of chemokines and their receptors suggests that CCL5 is the prototypic MIP family chemokine, and that different species have developed different repertoires of both the CC chemokines and their receptors. Dev. Comp. Immunol. 2007;31:72–86. doi: 10.1016/j.dci.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Kemper C., Atkinson J.P. T-cell regulation: with complements from innate immunity. Nat. Rev. Immunol. 2007;7:9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- Kim C.H. Chemokine-chemokine receptor network in immune cell trafficking. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2004;4:343–361. doi: 10.2174/1568008043339712. [DOI] [PubMed] [Google Scholar]
- Klamp T., Boehm U., Schenk D., Pfeffer K., Howard J.C. A giant GTPase, very large inducible GTPase-1, is inducible by IFNs. J. Immunol. 2003;171:1255–1265. doi: 10.4049/jimmunol.171.3.1255. [DOI] [PubMed] [Google Scholar]
- Koshy P.J., Lundy C.J., Rowan A.D., Porter S., Edwards D.R., Hogan A., Clark I.M., Cawston T.E. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: a time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 2002;46:961–967. doi: 10.1002/art.10212. [DOI] [PubMed] [Google Scholar]
- Kotani T., Shiraishi Y., Tsukamoto Y., Kuwamura M., Yamate J., Sakuma S., Gohda M. Epithelial cell kinetics in the inflammatory process of chicken trachea infected with infectious bronchitis virus. J. Vet. Med. Sci. 2000;62:129–134. doi: 10.1292/jvms.62.129. [DOI] [PubMed] [Google Scholar]
- Kotani T., Wada S., Tsukamoto Y., Kuwamura M., Yamate J., Sakuma S. Kinetics of lymphocytic subsets in chicken tracheal lesions infected with infectious bronchitis virus. J. Vet. Med. Sci. 2000;62:397–401. doi: 10.1292/jvms.62.397. [DOI] [PubMed] [Google Scholar]
- Kruth S.A. Biological response modifiers: interferons, interleukins, recombinant products, liposomal products. Vet. Clin. North Am. Small Anim. Pract. 1998;28:269–295. doi: 10.1016/s0195-5616(98)82005-6. [DOI] [PubMed] [Google Scholar]
- Le Goffic R., Pothlichet J., Vitour D., Fujita T., Meurs E., Chignard M., Si-Tahar M. Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- Li B., Yu H., Zheng W., Voll R., Na S., Roberts A.W., Williams D.A., Davis R.J., Ghosh S., Flavell R.A. Role of the guanosine triphosphatase Rac2 in T helper 1 cell differentiation. Science. 2000;288:2219–2222. doi: 10.1126/science.288.5474.2219. [DOI] [PubMed] [Google Scholar]
- Li C.K., Pender S.L., Pickard K.M., Chance V., Holloway J.A., Huett A., Goncalves N.S., Mudgett J.S., Dougan G., Frankel G., MacDonald T.T. Impaired immunity to intestinal bacterial infection in stromelysin-1 (matrix metalloproteinase-3)-deficient mice. J. Immunol. 2004;173:5171–5179. doi: 10.4049/jimmunol.173.8.5171. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Qu L.J., Hou Z.C., Yao J.F., Xu G.Y., Yang N. Genomic structure and diversity of the chicken Mx gene. Poult. Sci. 2007;86:786–789. doi: 10.1093/ps/86.4.786. [DOI] [PubMed] [Google Scholar]
- Liu P., Jamaluddin M., Li K., Garofalo R.P., Casola A., Brasier A.R. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugering A., Kucharzik T., Soler D., Picarella D., Hudson J.T., III, Williams I.R. Lymphoid precursors in intestinal cryptopatches express CCR6 and undergo dysregulated development in the absence of CCR6. J. Immunol. 2003;171:2208–2215. doi: 10.4049/jimmunol.171.5.2208. [DOI] [PubMed] [Google Scholar]
- Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Zhang C., Prasad K.V., Freeman G.J., Schlossman S.F. Molecular cloning of Porimin, a novel cell surface receptor mediating oncotic cell death. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9778–9783. doi: 10.1073/pnas.171322898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Vezys V., Marzo A.L., Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- Mattapallil J.J., Smit-McBride Z., McChesney M., Dandekar S. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1beta expression and display antiviral cytotoxic activity despite severe CD4(+) T-cell depletion in primary simian immunodeficiency virus infection. J. Virol. 1998;72:6421–6429. doi: 10.1128/jvi.72.8.6421-6429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbawuike I.N., Pacheco S., Acuna C.L., Switzer K.C., Zhang Y., Harriman G.R. Mucosal immunity to influenza without IgA: an IgA knockout mouse model. J. Immunol. 1999;162:2530–2537. [PubMed] [Google Scholar]
- Morrison S.G., Morrison R.P. The protective effect of antibody in immunity to murine chlamydial genital tract reinfection is independent of immunoglobulin A. Infect. Immun. 2005;73:6183–6186. doi: 10.1128/IAI.73.9.6183-6186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogundele M. Role and significance of the complement system in mucosal immunity: particular reference to the human breast milk complement. Immunol. Cell Biol. 2001;79:1–10. doi: 10.1046/j.1440-1711.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- Pardo J., Bosque A., Brehm R., Wallich R., Naval J., Mullbacher A., Anel A., Simon M.M. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J. Cell Biol. 2004;167:457–468. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry R.V., Whittaker G.C., Sims M., Edmead C.E., Welham M.J., Ward S.G. Ligation of CD28 stimulates the formation of a multimeric signaling complex involving grb-2-associated binder 2 (gab2), SRC homology phosphatase-2, and phosphatidylinositol 3-kinase: evidence that negative regulation of CD28 signaling requires the gab2 pleckstrin homology domain. J. Immunol. 2006;176:594–602. doi: 10.4049/jimmunol.176.1.594. [DOI] [PubMed] [Google Scholar]
- Rogers A.N., Vanburen D.G., Hedblom E.E., Tilahun M.E., Telfer J.C., Baldwin C.L. Gammadelta T cell function varies with the expressed WC1 coreceptor. J. Immunol. 2005;174:3386–3393. doi: 10.4049/jimmunol.174.6.3386. [DOI] [PubMed] [Google Scholar]
- Seki Y., Inoue H., Nagata N., Hayashi K., Fukuyama S., Matsumoto K., Komine O., Hamano S., Himeno K., Inagaki-Ohara K., Cacalano N., O’Garra A., Oshida T., Saito H., Johnston J.A., Yoshimura A., Kubo M. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat. Med. 2003;9:1047–1054. doi: 10.1038/nm896. [DOI] [PubMed] [Google Scholar]
- Seo S.H., Collisson E.W. Specific cytotoxic T lymphocytes are involved in in vivo clearance of infectious bronchitis virus. J. Virol. 1997;71:5173–5177. doi: 10.1128/jvi.71.7.5173-5177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin-Ya M., Hirai H., Satoh E., Kishida T., Asada H., Aoki F., Tsukamoto M., Imanishi J., Mazda O. Intracellular interferon triggers Jak/Stat signaling cascade and induces p53-dependent antiviral protection. Biochem. Biophys. Res. Commun. 2005;329:1139–1146. doi: 10.1016/j.bbrc.2005.02.088. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article 3) [DOI] [PubMed] [Google Scholar]
- Staats H.F., Ennis F.A., Jr. IL-1 is an effective adjuvant for mucosal and systemic immune responses when coadministered with protein immunogens. J. Immunol. 1999;162:6141–6147. [PubMed] [Google Scholar]
- Tamamura H., Tsutsumi H., Fujii N. The chemokine receptor CXCR4 as a therapeutic target for several diseases. Mini Rev. Med. Chem. 2006;6:989–995. doi: 10.2174/138955706778195135. [DOI] [PubMed] [Google Scholar]
- Taylor G.A. IRG proteins: key mediators of interferon-regulated host resistance to intracellular pathogens. Cell Microbiol. 2007;9:1099–1107. doi: 10.1111/j.1462-5822.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- Tempelman R.J. Assessing statistical precision, power, and robustness of alternative experimental designs for two color microarray platforms based on mixed effects models. Vet. Immunol. Immunopathol. 2005;105:175–186. doi: 10.1016/j.vetimm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Urban J.A., Winandy S. Ikaros null mice display defects in T cell selection and CD4 versus CD8 lineage decisions. J. Immunol. 2004;173:4470–4478. doi: 10.4049/jimmunol.173.7.4470. [DOI] [PubMed] [Google Scholar]
- Valosky J., Hishiki H., Zaoutis T.E., Coffin S.E. Induction of mucosal B-cell memory by intranasal immunization of mice with respiratory syncytial virus. Clin. Diagn. Lab. Immunol. 2005;12:171–179. doi: 10.1128/CDLI.12.1.171-179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret C., Janeway C.A., Jr. MHC and T cell development. Rev. Immunogenet. 1999;1:91–104. [PubMed] [Google Scholar]
- Wabnitz G.H., Kocher T., Lohneis P., Stober C., Konstandin M.H., Funk B., Sester U., Wilm M., Klemke M., Samstag Y. Costimulation induced phosphorylation of l-plastin facilitates surface transport of the T cell activation molecules CD69 and CD25. Eur. J. Immunol. 2007;37:649–662. doi: 10.1002/eji.200636320. [DOI] [PubMed] [Google Scholar]
- Wang X., Rosa A.J., Oliverira H.N., Rosa G.J., Guo X., Travnicek M., Girshick T. Transcriptome of local innate and adaptive immunity during early phase of infectious bronchitis viral infection. Viral Immunol. 2006;19:768–774. doi: 10.1089/vim.2006.19.768. [DOI] [PubMed] [Google Scholar]
- Wertheimer S., Katz S., Rowan K., Lugo A., Levin W., Hanglow A.C. Stromelysin expression in IL-1 beta stimulated bovine articular cartilage explants. Inflamm. Res. 1995;44(Suppl. 2):S119–S120. doi: 10.1007/BF01778291. [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Tateyama H., Asai K., Fukai I., Fujii Y., Tada T., Eimoto T. Glia maturation factor-beta is produced by thymoma and may promote intratumoral T-cell differentiation. Histopathology. 2005;47:292–302. doi: 10.1111/j.1365-2559.2005.02224.x. [DOI] [PubMed] [Google Scholar]
- Yan L., Galinsky R.E., Bernstein J.A., Liggett S.B., Weinshilboum R.M. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000;10:261–266. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- Zabel B.A., Silverio A.M., Butcher E.C. Chemokine-like receptor 1 expression and chemerin-directed chemotaxis distinguish plasmacytoid from myeloid dendritic cells in human blood. J. Immunol. 2005;174:244–251. doi: 10.4049/jimmunol.174.1.244. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pacheco S., Acuna C.L., Switzer K.C., Wang Y., Gilmore X., Harriman G.R., Mbawuike I.N. Immunoglobulin A-deficient mice exhibit altered T helper 1-type immune responses but retain mucosal immunity to influenza virus. Immunology. 2002;105:286–294. doi: 10.1046/j.0019-2805.2001.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.X., Benn S., Li Z.H., Wei E., Masih-Khan E., Trieu Y., Bali M., McGlade C.J., Claudio J.O., Stewart A.K. The SH3-SAM adaptor HACS1 is up-regulated in B cell activation signaling cascades. J. Exp. Med. 2004;200:737–747. doi: 10.1084/jem.20031816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


