Abstract
Bovine coronavirus (BCoV), a positive sense single-stranded RNA virus, is an important causative agent of neonatal diarrhoea in calves from beef and dairy cattle worldwide. The routine detection and diagnosis of BCoV have been mainly dependent on assays with low sensitivity. The aim of the present study was to develop and evaluate a semi-nested PCR (SN-PCR) to amplify a 251 bp fragment of BCoV N gene from fresh (n = 25) and frozen (n = 25) diarrhoeic faecal samples of naturally infected calves. To improve detection of BCoV in faecal samples by the SN-PCR an internal control was developed, and the results were compared with a conventional RT-PCR assay. The rates of positive samples by SN-PCR and RT-PCR were 24% (12/50) and 8% (4/50), respectively (K = 0.43). Only fresh samples were positive in RT-PCR while the SN-PCR detected BCoV in both fresh and frozen faecal samples. The sensitivity of SN-PCR was determined by 10-fold serial dilutions of the BCoV Kakegawa strain (HA titre: 256) that was detected until 10−7 dilution. The specificity of the amplicons was assessed by restriction fragment length polymorphism and sequence analysis. The inclusion of an internal control provides a way to detect assay inhibition in faecal samples and failure of nucleic acid extraction that allow reduction of the number of false-negative results.
Keywords: Calves, Diarrhoea, Bovine coronavirus, SN-PCR
1. Introduction
The neonatal diarrhoea complex is one of the main causes of calf morbidity and mortality causing major economic losses in many dairy and beef herds. Diarrhoea in calves is considered a multifactorial disease. Several environmental, managemental, nutritional and physiological factors may occur either alone or in synergy with the different infectious agents such as enterotoxigenic Escherichia coli, Cryptosporidium sp., bovine group A rotavirus, calicivirus, torovirus and bovine coronavirus (BCoV) (Snodgrass et al., 1986, Duckmanton et al., 1998, Naciri et al., 1999, Van Der Poel et al., 2000). This emphasizes the difficulties in investigating and diagnosing the infectious aetiology of calf diarrhoea. Diarrhoea caused by BCoV is clinically important and the virus also infects the respiratory tract of cattle and has been associated with winter dysentery in adult cattle (Clark, 1993, Saif, 1990).
BCoV is a member of the order Nidovirales, Coronaviridae family, that possesses a single-stranded, non-segmented RNA genome of positive polarity. The genome of coronaviruses is the largest among the RNA viruses, 26–30 kb long, and encodes a nested set of multiple subgenomic mRNAs during infection (Lai and Cavanagh, 1997). The virion contains five major structural proteins: the nucleocapsid protein (N), the transmembrane protein (M), the hemaglutinin esterase protein (HE), the spike protein (S) and the small membrane protein (E). The N protein is a phosphoprotein of 50–60 kd that interacts with genomic RNA to form the viral nucleocapsid and may play a role in replication of viral RNA (Lapps et al., 1987, Clark, 1993).
Electron microscopy (EM) may be used for the direct detection of BCoV in faecal samples (Collins et al., 1987, Bulgin et al., 1989). However, due to the common presence of pleomorphic membranous structures similar to the coronavirus it may be difficult to recognize BCoV by EM (Saif, 1990). Virus isolation in human rectal adenocarcinoma (HRT-18) cells is rarely used as a means of diagnosis because BCoV is difficult to isolate. Furthermore, virus isolation is a laborious method and requires a long time to obtain conclusive results (Dea et al., 1980, Tahir et al., 1995, Kapil et al., 1996). The haemagglutination (HA) test, performed directly with the faecal samples must be carefully interpreted since the faeces contain non-specific haemagglutinins providing false-positive results. The presence of colostral IgG1 and local IgA in faeces may also interfere with the HA results. The haemagglutination inhibition (HI) test is performed concurrently to improve the specificity of the HA test. Besides, another disadvantage of the HA/HI test is the need to maintain mouse colonies as sources of the fresh erythrocytes that are required for this diagnostic test (Sato et al., 1977, Kapil et al., 1999). Enzyme-linked immunosorbent assays (ELISA) are probably the most widely used diagnostic test for BCoV although this test lacks sensitivity (Sato and Akashi, 1993, Tsunemitsu and Saif, 1995, Schoenthaler and Kapil, 1999, Silva et al., 1999).
More sensitive tests are required to detect BCoV especially from faecal samples of calves at early or late stages of disease when they have low levels of viral shedding. Specificity is equally important to avoid false positive results.
Although BCoV has been established as an agent of diarrhoea in calves, its prevalence has been investigated to only a limited extend because of the diagnostic difficulties. However, more sensitive and specific tests for BCoV detection will permit more conclusive and larger epidemiological studies.
A reverse transcription-polymerase chain reaction (RT-PCR) assay for amplification of BCoV RNA from faecal samples has been described and its sensitivity and specificity has been reported although the detection was performed only in clinical samples from experimentally inoculated calves (Tsunemtitsu et al., 1999, Cho et al., 2001). Studies that describe the development of RT-PCR for BCoV detection from naturally infected diarrhoeic calves are still sporadic. Besides, there are no comparative studies on the ability of different primer sets to detect BCoV in clinical samples. Also, faeces remains the most difficult clinical sample for nucleic acid extraction and amplification due to the presence of PCR inhibitors that may yield false negative results (Monteiro et al., 1997). The use of an internal control in the PCR reaction has been most commonly applied to monitor and evaluate these failures, but this approach has not been used in assays for BCoV detection (Beld et al., 2004, Dingle et al., 2004, Hoorfar et al., 2004).
In this study we have developed a semi-nested PCR assay (SN-PCR) with an internal control to detect BCoV in faecal samples from naturally infected calves and have compared it with a RT-PCR assay previously described.
2. Materials and methods
2.1. Virus and cells
The HRT-18 cells were grown in Dulbecco's Modified Eagle's Medium (D-MEM, Gibco BRL, USA), supplemented with 10% fetal bovine serum (FBS, Gibco BRL, USA), 55 μg/ml gentamicine (Sigma Co., USA) and 2.5 μg/ml amphotericin B (Sigma Co., USA). The Kakegawa strain of BCoV was propagated in HRT-18 cells cultured in FBS free D-MEM and used in all standardization procedures of the RT-PCR and SN-PCR assays.
2.2. Clinical samples
Faecal samples from beef and dairy cattle herds were obtained from 50 calves up to 30 days old with signs of diarrhoea. Of the 50 faecal specimens 25 were stored at −20 °C for 2 years and the 25 remaining samples were fresh samples without previous freezing. Another 15 faecal samples from asymptomatic calves were included in this study as a control group. The samples were prepared either as 10% (w/v) suspensions of solid (control) or semi-solid faeces in 0.01 M phosphate-buffered saline (PBS) pH 7.2 (137 mM NaCl; 3 mM KCl; 8 mM Na2HPO4; 15 mM KH2PO4) or as 50% (v/v) suspensions of liquid faeces in 0.01 M PBS and centrifuged at 3000 × g for 15 min at 4 °C. The supernatants were used for RNA extraction.
2.3. RNA extraction
Aliquots of 400 μl from faecal suspensions were treated with SDS at a final concentration of 1% (v/v), homogenized by vortexing and kept at 56 °C for 30 min. For RNA extraction a combination of phenol/chloroform/isoamyl alcohol and silica/guanidinium isothiocyanate methods was performed according to Barreiros et al. (2004) with slight modifications. Briefly, 400 μl of phenol/chloroform/isoamyl alcohol (25:24:1) were added, vortexed and heated at 56 °C for 15 min (Sambrook et al., 1989). The mixture was centrifuged at 10,000 × g for 10 min and the supernatant was transferred into a new tube and processed by the silica/guanidinium isothiocyanate method (Boom et al., 1990). The RNA was eluted from the silica pellet with 50 μl of ultrapure (MilliQ®) sterile water by 15 min incubation at 56 °C and centrifugation at 10,000 × g for 10 min. The supernatant fraction was kept at −20 °C until use. Aliquots of ultrapure sterile water were included as negative controls in all the RNA extraction procedures.
2.4. RT-PCR
The RT-PCR was performed using the oligonucleotide primers upstream 5′-GCCGATCAGTCCGACCAATC-3′ (nt 79–98) and downstream 5′-AGAATGTCAGCCGGGGTAT-3′ (nt 467–485) that amplify a 407 base pair (bp) fragment of the N gene of BCoV. The technique was carried out as described by Tsunemtitsu et al. (1999).
2.5. SN-PCR
For the SN-PCR assay we designed another primer set to detect BCoV from faecal samples. Nucleotide sequence data of BCoV strains were obtained from Entrez database (http://www.ncbi.nlm.nih.gov/Entrez/). For the design, sequence alignment and the preliminary evaluation of the primers used in this assay and for the restriction endonucleases selection used in the evaluation of the amplicon specificity the following softwares were used: Gene Runner Version 3.05 (Hastings Software Inc., Hastings, NY) (http://www.generunner.com/), CLUSTAL W Multiple Sequence Alignment Program (http://www.ebi.ac.uk/clustalw/) (Thompson et al., 1997), and BLAST (http://www.ncbi.nlm.nih.gov/BLAST). The oligonucleotide primers were designed from the highly conserved region of the N gene of the Mebus strain (GenBank accession number U00735). The sequence of primers (positions calculated from the start codon of the N gene) were: BCoV1 sense: 5′-CGATGAGGCTATTCCGAC-3′ (nt 504–521) and BCoV2 antisense: 5′-TGTGGGTGCGAGTTCTGC-3′ (nt 940–957) for the PCR reaction; BCoV3 sense: 5′-TTGCTAGTCTTGTTCTGGC-3′ (nt 707–725) and BCoV2 antisense for the SN-PCR reaction. The predicted PCR and SN-PCR products were 454 and 251 bp, respectively.
To check the efficiency of the nucleic acid extraction and amplification an internal control (BOV1 and BOV2 primers) was included for each reaction. BOV1: 5′-ATACGCCTTCATTACCAG-3′ (nt 12,231–12,248) and BOV2: 5′-TTGAATGGAGTAGTGCTG-3′ (nt 12,839–12,856) primers amplified a 626 bp fragment of the ND5 gene present in the bovine mitochondrial DNA (GenBank accession number NC_001567) (Wosiacki et al., 2005). The internal control was used in an individual reaction or in Multiplex-PCR together with BCoV1 sense and BCoV2 antisense primers in the first round of the reaction (PCR).
The reverse transcription (RT) reaction was performed with 5 μl of extracted RNA and 1 μl of the primer BCoV2 antisense (20 pmol) that was incubated at 97 °C for 4 min. Subsequently, it was placed on ice for 5 min and 10 μl of RT mix containing 1× RT buffer (50 mM Tris–HCl, pH 8.3, 3 mM MgCl2, 75 mM KCl), 0.1 mM of each dNTP (Invitrogen™ Life Technologies, USA), 10 mM DTT, 100 units of M-MLV Reverse Transcriptase (Invitrogen™ Life Technologies, USA) and ultrapure sterile water to a final volume of 20 μl were added and incubated at 42 °C for 30 min and followed by inactivation at 95 °C for 5 min.
For the first round of amplification, 8 μl of the RT reaction were added to 42 μl of the PCR mix consisting of 1.5× PCR buffer (30 mM Tris–HCl pH 8.4 and 75 mM KCl), 2 mM MgCl2, 0.2 mM of each dNTP, 1 μl (20 pmol) of each primer (BCoV1 sense/BCoV2 antisense and BOV1/BOV2 in single or multiplex reaction), 2.5 units Platinum Taq DNA polymerase (Invitrogen™ Life Technologies, USA) and ultrapure sterile water to a final volume of 50 μl. The reaction was performed in a thermocycler (PTC-200, MJ Research Co. Water Town, MA, USA) with the following time and temperature conditions: one step of 4 min/94 °C; followed by 40 cycles of 1 min/94 °C, 1 min/55 °C, 1 min/72 °C and a final step of 7 min/72 °C.
For the second round of amplification (SN-PCR) 3 μl of the first amplification product (PCR assay) were added to 47 μl of the SN-PCR mix containing 1× PCR buffer (20 mM Tris–HCl pH 8.4 and 50 mM KCl), 2 mM MgCl2, 0.2 mM of each dNTP, 1 μl (20 pmol) of each SN-PCR primer (BCoV3 sense and BCoV2 antisense), 2.5 units of Platinum Taq DNA polymerase and ultrapure sterile water to a final volume of 50 μl. The reaction was performed with the following time and temperature conditions: one step of 4 min/94 °C; followed by 30 cycles of 1 min/94 °C, 1 min/55 °C, 1 min/72 °C and a final step of 7 min/72 °C.
2.6. Analysis of RT-PCR and SN-PCR
The products were analyzed by electrophoresis in a 2% agarose gel in TBE buffer pH 8.4 (89 mM Tris; 89 mM boric acid; 2 mM EDTA), ethidium bromide (0.5 μg/ml) stained and visualized under UV light.
2.7. Restriction analysis
The specificity of the SN-PCR amplicons from the first and second amplification rounds were firstly confirmed by restriction fragment length polymorphism (RFLP) with Hae III enzyme (Invitrogen™ Life Technologies, USA). The reaction was performed following the instructions of the manufacturer.
2.8. DNA sequencing
Amplicons from the first and second amplification round were purified by QIAquick PCR Purification Kit (Qiagen, USA) and sequenced in Mega Bace 1000/Automated 96 Capillary DNA Sequencer (Amersham Biosciences, UK). The quality of each sequence obtained was analyzed with Phred/Phrap/Consed (http://www.phrap.org/) software and the sequence similarity was checked against sequences deposited in the GenBank using the BLAST software.
2.9. Sensitivity
To assess the lowest detection limit of the SN-PCR the cell culture adapted BCoV Kakegawa strain (HA titre: 256) was 10-fold serially diluted in maintenance medium. RNA was extracted using the nucleic acid extraction method as mentioned earlier.
2.10. Specificity
For evaluation of the SN-PCR assay specificity faecal specimens were included containing enterotoxigenic E. coli (n = 5), bovine group A rotavirus (n = 10) and Cryptosporidium sp. (n = 5), detected respectively, by bacteriological routine tests, polyacrylamide gel electrophoresis technique and modified Ziehl-Nielsen method.
2.11. Statistical method
The kappa statistic (K) was calculated to evaluate the agreement between RT-PCR and SN-PCR systems. When K = 0 there is no agreement, K < 0.3 the agreement is poor, K between 0.3 and 0.5 is acceptable, K between 0.5 and 0.7 is good and K > 0.7 is excellent (Martin and Bonnet, 1987).
3. Results
Among the 50 diarrhoeic faecal samples included in this study the N gene of BCoV was detected in 4 (8%) samples by RT-PCR and in 12 (24%) samples by SN-PCR (K = 0.43). In fresh faecal samples (n = 25) the BCoV was detected in four samples by both, RT-PCR and SN-PCR in the first amplification round. No frozen faecal sample was BCoV positive by RT-PCR. However, in the SN-PCR developed in this study eight frozen faecal samples (n = 25) were BCoV positive in the second amplification round (Fig. 1 ). No positive results were obtained in normal faecal samples from asymptomatic calves by either of the PCR assays.
Fig. 1.
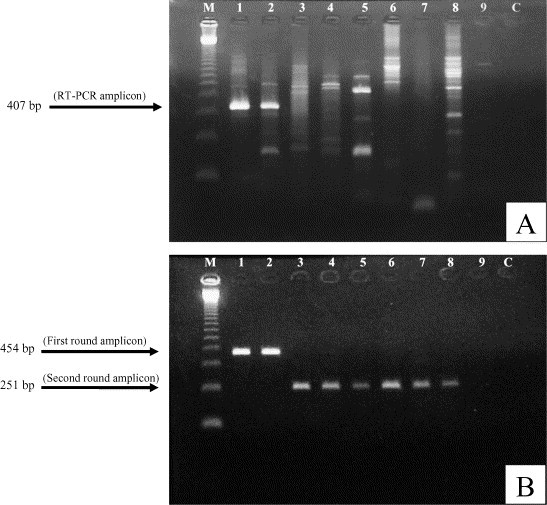
Amplification of bovine coronavirus (BCoV) N gene in faecal samples from naturally infected calves by RT-PCR (panel A) and by SN-PCR (panel B). The BCoV specific amplicons are indicated by arrows in the left margin. Lane M: 123 bp ladder (Invitrogen™ Life Technologies, USA); lane 1: positive control (BCoV Kakegawa strain); lanes 2–9: faecal samples from diarrhoeic calves; lane C: negative control.
The specificity of SN-PCR amplicons from the first and second amplification rounds of the BCoV Kakegawa strain and the clinical samples were confirmed by RFLP with the Hae III enzyme. The amplicons of 454 bp (first round) and 251 bp (second round) from SN-PCR yielded fragments of, respectively, 366 and 88 bp, and 163 and 88 bp (Fig. 2 ). In addition, these amplicons were sequenced and submitted to BLAST analysis. Both techniques confirm that the 454 and 251 bp amplicons were BCoV specific.
Fig. 2.
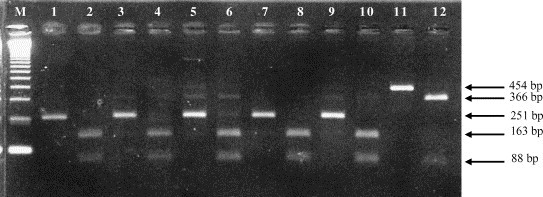
Ethidium bromide stained agarose gel electrophoresis of restriction fragment length polymorphism (RFLP) with Hae III enzyme of BCoV N gene amplicons. Lane M: 123 bp ladder (Invitrogen™ Life Technologies, USA); lanes 1, 3, 5, 7 and 9: BCoV N gene SN-PCR amplicons; lanes 2, 4, 6, 8 and 10: RFLP of BCoV N gene SN-PCR amplicons digested with Hae III; lane 11: BCoV N gene first round amplicon; lane 12: RFLP of BCoV N gene first round amplicon digested with Hae III. Lanes 1–8, 11 and 12: diarrhoeic faeces from naturally infected calves; lanes 9 and 10: BCoV Kakegawa strain.
The extracted RNA of the BCoV Kakegawa strain (HA titre: 256) was amplified by SN-PCR until 10−7 dilution showing the sensitivity of the assay. No cross-reactivity was found when the SN-PCR was applied to faecal samples collected from calves with neonatal diarrhoea caused by enterotoxigenic E. coli, bovine group A rotavirus and Cryptosporidium sp. infections. No false positive or false negative results were observed in SN-PCR reactions with ultrapure sterile water or BCoV Kakegawa strain that were both included in all experiments.
The internal control primers (BOV1 and BOV2) used in a multiplex reaction in the first amplification round of the SN-PCR, or separately in two reactions, yielded a specific 626 bp amplicon in all diarrhoeic faecal samples analyzed in this study (Fig. 3 ).
Fig. 3.
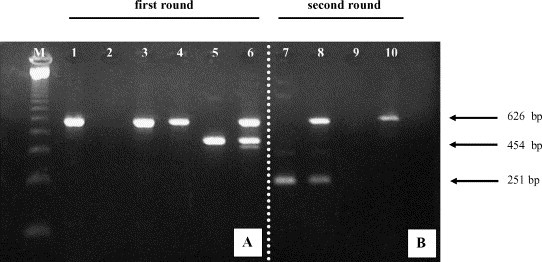
Amplicon analysis by agarose gel electrophoresis of bovine coronavirus N gene (454 bp for first round and 251 bp for second round) and bovine mitochondrial ND5 gene (internal control, 626 bp) from bovine faecal samples analyzed by SN-PCR. Lane M: 123 bp ladder (Invitrogen™, Life Technologies, USA). Panel A: Faecal sample from asymptomatic (lanes 1–3, calf 55) and diarrhoeic (lanes 4–6, calf 17) calves. First round amplification with internal control primers (lanes 1 and 4) and BCoV specific primers (lanes 2 and 5) in single reaction, and in multiplex reaction (lanes 3 and 6). Panel B: Faecal sample from diarrhoeic (lanes 7 and 8, calf 15) and asymptomatic (lanes 9 and 10, calf 58) calves. Second round amplification in single reaction with BCoV specific primers (lanes 7 and 9) and in multiplex reaction with BCoV and internal control primers (lanes 8 and 10).
4. Discussion
To improve the detection rate of BCoV in diarrhoeic fecal samples from naturally infected calves we developed a SN-PCR assay with an internal control and compared with a RT-PCR already described. The agreement between the two assays was only acceptable (K = 0.43). We worked with previously diluted samples subjected to long-term storage in freezer and with fresh faecal samples to verify the sensitivity assay under of the storage conditions. There was agreement between the two PCR methods in the detection rate of BCoV in fresh samples. However, the RT-PCR assay failed to amplify BCoV in frozen faeces. Of the 25 frozen samples the BCoV N gene could be detected in eight of them by the SN-PCR only. The employment of the second amplification round allowed a 200% increase in BCoV positive results in the samples included in this study.
We can exclude the possibility that our positive results in the frozen faecal samples were the result of intersample contamination. Careful measures were adopted to avoid the carry-over risk. Negative and positive controls accompanied the clinical samples in all the steps (nucleic acid extraction, RT, PCR and SN-PCR) where each step was carried-out in separate rooms. Besides, all the positive results were reanalysed (nucleic acid extraction until SN-PCR procedures) including the controls and the normal faeces in the same run.
A study by Van der Hoek et al. (1995), demonstrated that no appreciable loss in PCR signal was seen with long-term storage of faecal samples. They detected HIV RNA in faeces which had been stored for 9 years at −70 °C by RT-PCR. Therefore, in a retrospective study, SN-PCR developed in this study might be useful to detect BCoV in stored samples.
The three primers used in SN-PCR were designed from the published sequence of the N gene of the BCoV Mebus strain. Careful primer design can aid PCR optimization and improve greatly the assay (Butler et al., 2001). The SN-PCR primers possess very similar melting temperatures, with only two degrees of difference between them. Furthermore, only one primer-dimer formation was noted. Also of importance is the target region of BCoV RNA to be amplified. We chose the N gene because it is highly conserved among BCoV strains. It was concluded that the N protein is the most abundant antigen in coronavirus-infected cells because its RNA template is the smallest and it has the most abundant sgRNA (subgenomic RNA) during transcription (Hiscox et al., 2001). This indicates that there is more available RNA for the N gene than for the other BCoV protein genes. Consequently, detection of the N gene RNA might be advantageous due to its high abundance in cells, facilitating a high sensitivity of the diagnostic technique.
Tsunemtitsu et al. (1999) described the detection of BCoV RNA in faecal samples by RT-PCR and they also chose the N gene as target region for amplification. However, these authors worked with samples recently collected from experimentally inoculated adult cows while our BCoV positive results were obtained in faecal samples from calves with neonatal diarrhoea by natural infection. We tested the primers designed by Tsunemtitsu et al. (1999) (RT-PCR) to compare the efficiency of the amplification in frozen and fresh samples from naturally infected calves. Although it was efficient for amplification of the positive control (Kakegawa strain), the same was not noted for clinical samples. The presence of unspecific amplicons and smears or failure to amplify the expected product disabled the diagnostic use. No absolute rules could be established to predict the ability of a primer pair to faithfully amplify a specific gene. However, the difference in detection rate may be the result of primer design. Analysis of the RT-PCR primers disclosed dimers and internal loop formation, palindrome sequences and different melting temperatures between the primers that can be related to the bad quality of the amplified products.
An adequate viral RNA extraction method is very important for molecular diagnosis. Four RNA extraction methods (TRIzol™, phenol/chloroform/isoamyl alcohol, guanidinium isothiocyanate, and a combination of phenol/chloroform/isoamyl alcohol and silica/guanidinium isothiocyanate) were tested and compared for BCoV detection in faecal samples. SN-PCR detection rates after RNA extraction using the combination of phenol/chloroform/isoamyl alcohol and silica/guanidinium isothiocyanate methods were better than after other RNA extraction protocols (data not shown). The combination of phenol/chloroform/isoamyl alcohol and silica/guanidinium isothiocyanate methods was performed according to Barreiros et al. (2004) with slight modifications. Faecal samples remain the most difficult specimens for nucleic acid extraction and amplification due the presence of inhibitors (Monteiro et al., 1997). Guanidinium isothiocyanate alone or in combination with phenol–chloroform inactivates ribonucleases and therefore improves the stability of the extracted RNA genome (Romero, 1999). Van der Hoek et al. (1995), Hale et al. (1996), Rassol et al. (2002), Paula et al. (2003) also demonstrated that the guanidinium isothiocyanate method successfully removed inhibitors from faecal samples.
An internal control was included in the SN-PCR reactions to avoid false-negative results due to the presence of PCR inhibitors. This is particularly critical for faecal samples, in which the presence of inhibitors might affect the amplification process.
Epidemiological data on the frequency of BCoV occurring in South American livestock farms is very limited. In studies conducted in France, Spain, Costa Rica and Britain, BCoV was detect by antigen-capture ELISA in 6.8%, 7.3%, 9% and 14%, respectively (Reynolds et al., 1986, De la Fuente et al., 1998, Pérez et al., 1998, Naciri et al., 1999). However, the real frequency of the BCoV in neonatal calf diarrhoea outbreaks depends of the diagnostic technique. Cho et al. (2001) showed that nested PCR was 5000 times more sensitive than the antigen-capture ELISA. Therefore, the applicability of a sensitive technique such as PCR is fundamental to not underestimating the real prevalence of BCoV. Cho et al. (2001) also described a second round of amplification with the same primers described by Tsunemtitsu et al. (1999). However, this primer pair was tested only in fresh samples from experimentally infected calves and without an internal control.
We conclude that the SN-PCR described here is a useful tool for the diagnosis of BCoV in faeces from naturally infected calves. Differences in the ability to amplify and the characteristics of the PCR systems available for BCoV detection in clinical samples should be considered before use a test in future epidemiological studies. The rate of 24% positive diarrhoeic faecal samples confirms that the BCoV is an important etiological agent of neonatal diarrhoea in dairy and beef Brazilian cattle herds. The availability of a sensitive and specific diagnostic technique such as the SN-PCR described in this work will make it possible to undertake larger epidemiological studies of BCoV involved in the neonatal calf diarrhoea aetiology.
Acknowledgements
The authors would like to thank Dr. J.A. Jerez (Universidade de São Paulo, São Paulo, Brazil) for providing the HRT-18 cells. We would also like to thank the Brazilian Institutes CNPq, CAPES and Fundação Araucária (FAP/PR) for financial support. Alfieri, A.A. is recipient of CNPq fellowship.
References
- Barreiros M.A., Alfieri A.F., Medici K.C., Leite J.P., Alfieri A.A. G and P genotypes of group A rotavirus from diarrhoeic calves born to cows vaccinated against the NCDV (P[1] G6) rotavirus strain. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2004;51:104–109. doi: 10.1111/j.1439-0450.2004.00737.x. [DOI] [PubMed] [Google Scholar]
- Beld M., Minnar R., Weel J., Sol C., Damen M., van der Avoort H., Wertheim-van Dillen P., van Breda A., Boom R. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 2004;42:3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R., Sol C.J.A., Salimans M.M.M., Jansen C.L., Wertheim-van Dillen P.M.E., Van Der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgin M.S., Ward A.C.S., Barret D.P., Lane V.M. Detection of rotavirus and coronavirus shedding in two beef cow herds in Idaho. Can. Vet. J. 1989;30:235–239. [PMC free article] [PubMed] [Google Scholar]
- Butler J.M., Ruitberg C.M., Vallone P.M. Capillary electrophoresis as a tool for optimization of multiplex PCR reactions. Fresenius J. Anal. Chem. 2001;369:200–205. doi: 10.1007/s002160000641. [DOI] [PubMed] [Google Scholar]
- Cho K.O., Hasoksuz M., Nielsen P.R., Chang K.O., Lathrop S., Saif L.J. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.A. Bovine coronavirus. Br. Vet. J. 1993;149:51–70. doi: 10.1016/S0007-1935(05)80210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.K., Riegel C.A., Olson J.D., Fountain A. Shedding of enteric coronavirus in adult cattle. Am. J. Vet. Res. 1987;48:361–365. [PubMed] [Google Scholar]
- Dea S., Roy R.S., Begin M.E. Bovine coronavirus isolation in continuous cell lines. Am. J. Vet. Res. 1980;41:30–38. [PubMed] [Google Scholar]
- De la Fuente R., Garcia A., Ruiz-Santa-Quiteria J.A., Luzón M., Cid D., García S., Orden J.A., Gómez-Bautista M. Proportional morbidity rates of enteropathogens among diarrheic dairy calves in central Spain. Prev. Vet. Med. 1998;36:145–152. doi: 10.1016/S0167-5877(98)00077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K.E., Crook D., Jeffery K. Stable and noncompetitive RNA internal control for routine clinical diagnostic reverse transcription-PCR. J. Clin. Microbiol. 2004;42:1003–1011. doi: 10.1128/JCM.42.3.1003-1011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36:1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale A.D., Green J., Brown D.W. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J. Virol. Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoorfar J., Malorny B., Abdulmawjood A., Cook N., Wagner M., Fach P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 2004;42:1863–1868. doi: 10.1128/JCM.42.5.1863-1868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapil S., Richardson K.L., Radi C., Chard-Bergstrom C. Factors affecting isolation and propagation of bovine coronavirus in human rectal tumor-18 cell line. J. Vet. Diagn. Invest. 1996;8:96–99. doi: 10.1177/104063879600800115. [DOI] [PubMed] [Google Scholar]
- Kapil S., Richardson K.L., Maag T.R., Goyal S.M. Characterization of bovine coronavirus isolates from eight different states in the USA. Vet. Microbiol. 1999;67:221–230. doi: 10.1016/S0378-1135(99)00042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.W., Bonnet B. Clinical epidemiology. Can. Vet. J. 1987;28:318–325. [PMC free article] [PubMed] [Google Scholar]
- Monteiro L., Bonnemaison D., Vekris A., Petry K.G., Bonnet J., Vidal R., Cabrita J., Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naciri M., Lefay M.P., Mancassola R., Poirier P., Chermette R. Role of Cryptosporidium parvum as a pathogen in neonatal diarrhea complex in suckling and dairy calves in France. Vet. Parasitol. 1999;85:245–257. doi: 10.1016/S0304-4017(99)00111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula V.S., Villar L.M., Gaspar A.M.C. Comparison of four extraction methods to detect hepatitis A virus RNA in serum and stool samples. Braz. J. Infect. Dis. 2003;7:135–141. doi: 10.1590/s1413-86702003000200007. [DOI] [PubMed] [Google Scholar]
- Pérez E., Kummeling A., Janssen M.M., Jimenez C., Alvarado R., Cabal M., Donaldo P., Dwinger R.H. Infectious agents associated with diarrhoea of calves in the canton of Tilarán, Costa Rica. Prev. Vet. Med. 1998;33:195–205. doi: 10.1016/S0167-5877(97)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassol N.B.G., Monroe S.S., Glass R.I. Determination of a universal nucleic acid extraction procedure for PCR detection of gastroenteritis viruses in faecal specimens. J. Virol. Methods. 2002;100:1–16. doi: 10.1016/s0166-0934(01)00379-2. [DOI] [PubMed] [Google Scholar]
- Reynolds D.J., Morgan J.H., Chanter N., Jones P.W., Bridger J.C., Debney T.G., Bunch K.J. Vet. Rec. 1986;119:34–39. doi: 10.1136/vr.119.2.34. [DOI] [PubMed] [Google Scholar]
- Romero J.R. Reverse-trancription polymerase chain reaction detection of the enteroviruses. Arch. Pathol. Lab. Med. 1999;123:1161–1169. doi: 10.5858/1999-123-1161-RTPCRD. [DOI] [PubMed] [Google Scholar]
- Saif L.J. A review of evidence implicating bovine coronavirus in the etiology of winter dysentery cows: an enigma resolved? Cornell Vet. 1990;80:303–311. [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. second ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sato K., Inaba Y., Kurogi H., Takahashi E., Satoda K., Omori T., Matsumoto M. Hemagglutination by calf diarrhea coronavirus. Vet. Microbiol. 1977;2:83–87. [Google Scholar]
- Sato M., Akashi H. Detection of bovine coronavirus by enzyme-linked immunosorbent assay using monoclonal antibodies. J. Vet. Med. Sci. 1993;55:771–774. doi: 10.1292/jvms.55.771. [DOI] [PubMed] [Google Scholar]
- Schoenthaler S.L., Kapil S. Development and applications of a bovine coronavirus antigen detection enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 1999;6:130–132. doi: 10.1128/cdli.6.1.130-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.R., O’Reilly K.L., Lin X., Stine L., Storz J. Sensitivity comparison for detection of respiratory bovine coronaviruses in nasal samples from feedlot cattle by ELISA and isolation with the G clone of HRT-18 cells. J. Vet. Diagn. Invest. 1999;11:15–19. doi: 10.1177/104063879901100102. [DOI] [PubMed] [Google Scholar]
- Snodgrass D.R., Terzolo H.R., Sherwood D., Campbell I., Menzies J.D., Synge B.A. Aetiology of diarrhea in young calves. Vet. Rec. 1986;119:31–34. doi: 10.1136/vr.119.2.31. [DOI] [PubMed] [Google Scholar]
- Tahir R.A., Pomeroy K.A., Goyal S.M. Evaluation of shell via cell culture technique for the detection of bovine coronavirus. J. Vet. Diagn. Invest. 1995;7:301–304. doi: 10.1177/104063879500700301. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu H., Saif L.J. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch. Virol. 1995;140:1303–1311. doi: 10.1007/BF01322757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemtitsu H., Smith D.R., Saif L.J. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 1999;144:167–175. doi: 10.1007/s007050050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoek L., Boom R., Goudsmit J., Snijders F., Sol C.J.A. Isolation of human immunodeficiency virus type 1 (HIV-1) RNA from feces by a simple method and difference between HIV-1 subpopulations in feces and serum. J. Clin. Microbiol. 1995;33:581–588. doi: 10.1128/jcm.33.3.581-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Poel W.H.M., Vinjé J., Van Der Heide R., Herrera M.I., Vivo A., Koopmans M.P.G. Norwalk-like calicivirus genes in farm animals. Emerging Infect. Dis. 2000;6:36–41. doi: 10.3201/eid0601.000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosiacki S.R., Barreiro M.A.B., Alfieri A.F., Alfieri A.A. Semi-nested PCR for detection and typing of bovine papillomavirus type 2 in urinary bladder and whole blood from cattle with enzootic haematuria. J. Virol. Methods. 2005;126:215–219. doi: 10.1016/j.jviromet.2005.01.021. [DOI] [PubMed] [Google Scholar]


