Abstract
The major concern for severe acute respiratory syndrome (SARS), caused by the SARS-associated coronavirus (SARS-CoV), is the lack of diagnostic and therapeutic agents. Using a phage display technology in a chicken system, high-affinity monoclonal antibody fragments against the SARS-CoV spike protein were characterized. Ten truncated spike protein gene fragments were expressed in Escherichia coli cells. Following the immunization of chickens with these recombinant spike proteins, two single-chain variable fragment (scFv) antibody libraries were established with short or long linkers to contain 5 × 107 and 9 × 106 transformants, respectively. After four rounds of panning selection, the scFv antibodies of randomly chosen clones were demonstrated by Coomassie blue staining, and verified by western blot analysis. In a comparison of nucleotide sequences with the chicken germline gene, we found that all clones varied in the complementarity-determining regions, that two scFv antibodies reacted significantly with SARS-CoV-infected Vero cells, and that those two specific scFv antibodies recognized the same region of the spike protein spanning amino acid residues 750–1000. In conclusion, the results suggest that the chicken scFv phage display system can be a potential model for mass production of high-affinity antibodies against the SARS-CoV spike protein.
Abbreviations: SARS-CoV, severe acute respiratory syndrome-associated coronavirus; S, spike; scFv, single-chain variable fragment; E. coli, Escherichia coli; RT-PCR, reverse-transcription polymerase chain reaction; CDR, complementarity-determining region; FR, framework region; VH, heavy-chain variable region; VL, light-chain variable region
Keywords: SARS-CoV, Phage display technology, Spike protein, scFv
1. Introduction
The severe acute respiratory syndrome (SARS) is a newly emerging disease caused by a SARS-associated coronavirus (SARS-CoV) (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). The virion consists of the following four major structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) (Marra et al., 2003, Rota et al., 2003). The S protein has two functional domains (S1 and S2) based on the predicted localization of their amino acid residues: 17–680 and 681–1255, respectively (He et al., 2004). A region located between amino acids 300 and 510 on the S1 domain serves as a receptor-binding site (Dimitrov, 2003, Li et al., 2003, Wang et al., 2004). The C-terminal S2 domain has been shown to mediate membrane fusion during SARS-CoV infection (Tripet et al., 2004). Further, immunization of mice with recombinant S protein can protect them from SARS-CoV infection (Bisht et al., 2004, Yang et al., 2004). The results suggested that the S protein is a good candidate for developing vaccines and antiviral drugs, and that generating monoclonal antibodies to recognize specifically the S protein would be valuable.
Although monoclonal antibodies with high specificities have been favored for both research and clinical applications in recent years, using the traditional hybridoma approach to generate human monoclonal antibodies for therapeutic purposes is still difficult because it is a tedious and expensive process (Groves and Morris, 2000, Lillehoj and Malik, 1993). In contrast, the phage display system is a safe and effective procedure because the process involves the in vitro cloning of antibody repertoires, and subsequent isolation of monoclonal antibodies from combinatorial antibody libraries (Barbas et al., 1991, Winter et al., 1994). Of many recombinant antibody forms, the single-chain variable fragment (scFv) is a small protein entity retaining the variable regions of both heavy and light chains of an entire antibody molecule (Bird et al., 1988, Huston et al., 1988) which can be efficiently generated in a phage display system (Chi et al., 2002, Pavoni et al., 2006, Wang et al., 2006).
Antibody production in the chicken is efficient (Abouzid et al., 2006, LeClaire et al., 2002). It has been reported that constructing chicken antibody libraries using the phage display technology can generate high-affinity scFvs for diagnostic applications (Fehrsen et al., 2005, Finlay et al., 2006, Park et al., 2005). Performing reverse-transcription polymerase chain reaction (RT-PCR) to amplify the entire V region repertoire using one set of primers is simple and convenient because all avian immunoglobulin genes are derived from single light- and heavy-chain variable (V L and V H) germline sequences (Andris-Widhopf et al., 2000, McCormack et al., 1993, Yamanaka et al., 1996). Using the phage display technology, monospecific scFv and Fab antibodies neutralizing the SARS-CoV infection have been generated from non-immunized individuals and convalescent SARS patients (Kang et al., 2006, Sui et al., 2004). The current study aimed to show that monoclonal IgY scFv antibodies which bind specifically to the S protein and SARS-CoV-infected Vero cells can be isolated from chickens immunized with Escherichia coli-derived S proteins.
2. Materials and methods
2.1. Truncated S fragment preparation
Ten sets of primers were synthesized to amplify S gene fragments using the SARS-CoV RNA genome as a template (GenBank accession no. NC_004718). The entire procedure was performed using a one-step RT-PCR kit as described by the manufacturer (Qiagen, Valencia, CA, USA). The amplified S gene products of 300–750 bp in length were individually digested with BamHI and XbaI restriction enzymes and ligated into the pET-21 expression vector (Novagen, Darmstadt, Germany). The resultant plasmids were transformed into the E. coli BL-21 (DE3) strain for protein expression. Clones were grown in 5 ml LB medium containing ampicillin (100 μg/ml) at 37 °C overnight. The bacterial culture was then diluted 10-fold in the same LB medium and further grown until the OD600 reached 0.6. For His-fused S protein expression, iso-propyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM in the culture for induction. The cell pellet was resuspended in 2 ml of binding buffer (100 mM NaCl, 10 mM Tris–Cl, and 8 M urea; pH 8.0) and lysed by three cycles of freezing (−70 °C) and thawing (37 °C). After centrifugation, the resulting cellular lysate was loaded onto a Ni2+-charged resin column for protein purification according to the manufacturer (Amersham Biosciences, Uppsala, Sweden).
2.2. Chicken immunization and phage display scFv library construction
Female white leghorn (Gallus domesticus) chickens were immunized with mixed S protein fragments (10 μg/each) in an equal volume of Freund's complete adjuvant by intramuscular injection. Three additional immunizations were carried out in intervals of 7 days. After each immunization, blood was obtained and titrated by an enzyme-linked immunosorbent assay (ELISA) to determine the presence of an antigen-specific immune response. The spleens of these chickens were harvested for RNA isolation. Phage libraries displaying scFv antibodies were constructed according to published protocols with minor modifications (Andris-Widhopf et al., 2000, Barbas et al., 2001).
After PCR amplification with chicken-specific primers, gene products of heavy-chain (V H) and light-chain variable (V L) regions were subjected to a second round of PCR with a short or long linker to form full-length scFvs, which were cloned further into the pComb3H vector. To increase the cloning efficiency, 7 μg of pComb3H and 3.5 μg of the full-length scFv product were applied in the ligation reaction. The recombinant DNAs were transformed into the E. coli XL-1 blue strain by electroporation, recombinant phage production was initiated by the addition of the helper phage, VCS-M13, and were precipitated with 4% polyethylglycol 8000 and 3% NaCl (w/v) and finally resuspended in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and stored at 4 °C.
2.3. Biopanning and expression of specific scFv binders
Microtiter plates precoated with mixed S protein fragments (0.5 μg/well) at 4 °C overnight were blocked with 3% BSA for 1 h at 37 °C. Then, 1011 plaque-forming units (pfu) of recombinant phages from the library preparation was added to each well, and the plates were incubated at 37 °C for 2 h. Unbound phages were removed, and the wells were washed vigorously with TBST (TBS with 0.05% Tween 20) buffer. Bound phages were eluted with 0.1 M HCl/glycine (pH 2.2)/0.1% BSA and neutralized with 2 M Tris–base buffer. Eluted phages were used to infect the E. coli XL-1 strain in the log phase for amplification, and they were recovered with 4% polyethylglycol 8000 and 3% NaCl for the next round of selection. The panning procedure was repeated in the wells four times. Phagemid DNA from the final enriched phage was prepared and digested with NheI and SpeI to remove the phage protein III gene. The digested DNA with compatible cohesive ends was self-ligated, and the resultant phagemid was electroporated into E. coli. Individual clones were grown in the presence of 0.5 mM IPTG for protein induction. Cells were pelleted and lysed by three cycles of freezing and thawing. After the final thawing, the supernatants were harvested by centrifugation for subsequent assays.
2.4. Western blotting and ELISA
The scFv-expressing lysates were subjected to sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred onto nitrocellulose membranes (Amersham Biosciences, Little Chalfont, UK), which were then blocked with 5% skim milk in TBST for 1 h. Polyclonal goat anti-chicken IgY antibodies (Bethyl Laboratories, Montgomery, TX, USA) were added and incubated for an additional hour. The membranes were washed with TBST three times for 5 min each. The bound antibodies were detected by adding horseradish peroxidase (HRP)-conjugated donkey anti-goat immunoglobulin (Ig) antibodies (Sigma, St. Louis, MO, USA). After three washings, the membranes were developed with diaminobenzidine (DAB) substrate until the desired intensity was achieved.
To examine the binding ability of the scFv-expressing lysate against the S protein, microtiter plates precoated with SARS-CoV-infected Vero cell lysates (Euroimmun, Lueberk, Germany) were used for the ELISA. After rinsing with PBST (PBS with 0.05% Tween 20), test lysates were distributed to wells in duplicate and incubated at 37 °C for 1 h. The bound scFvs were detected with enzyme-labeled antibodies as described above.
2.5. Sequence analysis
The nucleotide sequences of heavy- and light-chain genes from chosen clones were determined by an auto-sequencer machine (ABI PRISM 377; Perkin-Elmer, National Health Research Institute). The sequencing primer, ompseq (5′-AAGACAGCTATCGCGATTGCAGTG-3′) as described by Andris-Widhopf et al. (2000), was used for the V L and V H gene analyses.
2.6. Immunocytochemical staining analysis
Biochip slides (Euroimmun) were used to test the binding reactivities of antibodies in sera or scFv-expressing lysates to native S protein in SARS-CoV-infected Vero cells. Slides were incubated with either sera or lysates at room temperature (RT) for 1 h. After washing with TBST, the bound antibodies or scFvs were incubated with mouse anti-human (Sigma) or goat anti-chicken antibodies (Bethyl Laboratories) at RT for 1 h. After 3 washings, slides were incubated for 1 h at RT with corresponding fluorescein isothiocyanate (FITC)-labeled antibodies (Sigma). Finally, slides were coated with mounting oil and examined with an immunofluorescent microscope.
2.7. Epitopic mapping and competitive inhibition assays
Epitopes were mapped with specific scFvs by detecting their reactivities with truncated S protein fragments by western blot analysis. Ten purified His-fused S fragments were transferred onto a nitrocellulose membrane after electrophoresis. Following blocking with 5% skim milk, membranes were incubated with scFv-expressing lysates for 2 h at RT. Then, the bound scFv antibodies were detected as described above.
The binding affinities of specific scFvs were determined by competitive inhibition assays. Briefly, 50 μl of each scFv-expressing lysate were first incubated with equal volumes of a series of two-fold-diluted soluble S fragments (2.5–80 μg/ml) at RT for 2 h. The reactivities of the mixtures with each fragment individually precoated on a microtiter plate (0.5 μg/well) were measured as described above. The dissociation constant (K d) of the scFv binders was calculated according to the Klotz method (Friguet et al., 1985).
3. Results
3.1. S protein fragment preparation
Ten truncated fragments of the SARS-CoV S gene were amplified by PCR, cloned into the pET-21 vector, and expressed as His-fused recombinant proteins. Table 1 lists the locations and predicted gene lengths of these fragments. After transformation and induction, these proteins were expressed successfully in E. coli BL-21 cells and purified using Ni2+-charged resin. Fig. 5A shows that these S protein fragments, S1–S10, had the predicted molecular weights of about 12–30 kDa after being analyzed by SDS-PAGE.
Table 1.
Residual locations and predicted molecular weights of truncated fragments of S protein
| S fragment | Location (bp) | MW (kDa) |
|---|---|---|
| S1 | 451–1200 | 29 |
| S2 | 1051–1368 | 13 |
| S3 | 1201–1500 | 12 |
| S4 | 1369–1950 | 23 |
| S5 | 1801–2400 | 23 |
| S6 | 2251–3000 | 29 |
| S7 | 2629–3000 | 15 |
| S8 | 2851–3300 | 18 |
| S9 | 3136–3600 | 18 |
| S10 | 3301–3600 | 12 |
Abbreviation: MW denotes molecular weights of predicted protein.
Fig. 5.
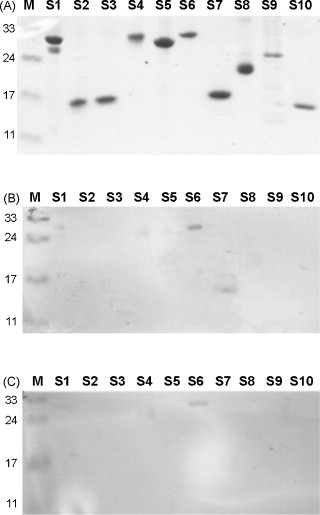
Epitopes mapped with specific single-chain variable fragment (scFv) antibodies using western blot analysis. Panel (A) shows 10 truncated S fragments on a Coomassie blue-stained polyacrylamide gel. Both Ssc35 (B) and Lsc18 (C) scFv antibodies but not cellular lysate (not shown) recognized a truncated S6 fragment spanning amino acid residues 750–1000.
3.2. scFv library construction
To construct the scFv libraries, two female white leghorn chickens were chosen for immunization. After the final immunization step, the chickens were killed, and the RNA was extracted from their spleen cells. One set of specific primers were designed to amplify the V L and V H regions of the scFv fragment genes. After a second and consecutive PCR step, the V L and V H genes were joined with a short (GGSSRSS) or long linker (GGSSRSSSSGGGGSGGGG) to form a full-length scFv gene conformation (data not shown). Two libraries, Ssc (with the short linker) and Lsc (with the long linker), containing 5 × 107 and 9 × 106 transformants, respectively, were constructed in this study.
3.3. Panning and scFv antibody expression
Fig. 1A shows that scFv antibodies were well amplified and expressed. As predicted, the molecular weights of these expressed scFv antibodies with short or long linkers were 24 and 28 kDa, respectively. Their identities were verified further using specific anti-chicken IgY antibodies in the western blot analysis (Fig. 1B).
Fig. 1.
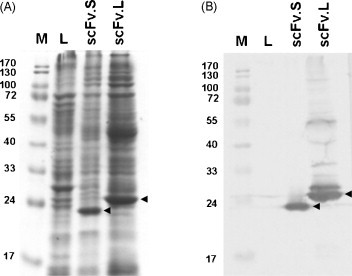
Total cellular lysates seen using Coomassie blue dye (A), while single-chain variable fragment (scFv) antibodies were shown by the western blotting technique (B). The recombinant scFv.S (with short linker) and scFv.L (with long linker) antibodies were identified as 24 and 28 kDa bands, respectively. Total cellular lysates from XL-1 bacteria were loaded into lane L. Lane M denotes prestained protein markers.
3.4. ELISA and gene sequencing
The binding abilities of 10 clones against the S protein from each library were analyzed using ELISA. Fig. 2A shows five clones obtained from the Ssc library to be strongly positive (with ODs of 1.1–1.8) for binding activity, while others showed weak or no reactivity. Fig. 2B illustrates four clones selected from the Lsc library to have higher activity (with ODs of 0.7–1.0). The nucleotide sequences of the V L and V H regions of 14 clones (seven each from the Ssc and Lsc libraries) with different binding abilities were analyzed. The CDRs of the V L region of five of the 14 clones showed more than 50% variation. From among those 14 clones, Ssc20 showed the lowest and Ssc37 the highest mutation rates, of 23% and 57%, respectively. On the other hand, the V H region analysis showed that ten of the 14 clones showed more than 50% CDR variations. Among those 14 clones, Ssc29 showed the lowest and Ssc33 the highest mutation rates, of 44% and 63%, respectively (Fig. 3 ).
Fig. 2.
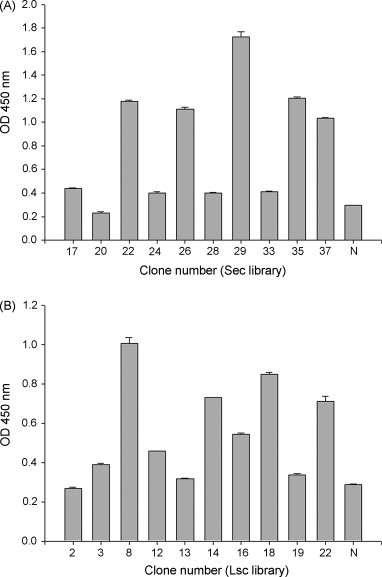
Binding activity of randomly selected clones analyzed by ELISA. Cellular lysates containing single-chain variable fragment (scFv) antibodies from various Ssc (A) and Lsc (B) library clones were examined for their binding to SARS-CoV-infected cell lysates using a commercially available kit. Negative control (N) is a cellular lysate lacking scFv expression. Bound scFv was detected using anti-chicken light-chain antibodies and was measured at 450 nm. The ELISA data were represented as mean of the duplicated wells ± S.E.M. using SigmaPlot Statistical Analysis software.
Fig. 3.
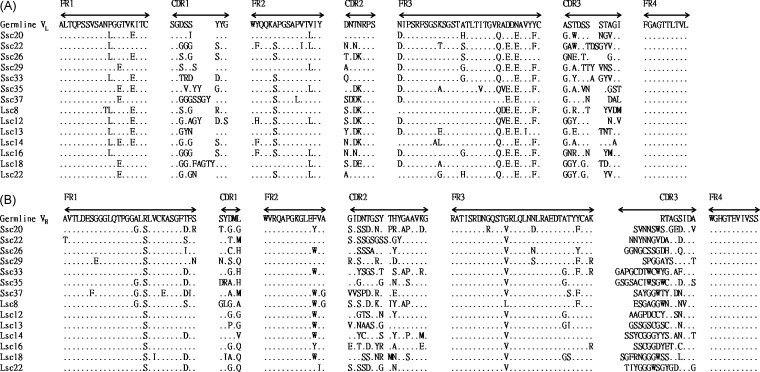
Sequence analysis of selected single-chain variable fragment (scFv) genes. Light-chain (VL) (A) and heavy-chain variable (VH) (B) region gene sequences were compared with that of the chicken germline gene showing the similarity. Sequence gaps were introduced to maximize alignment as indicated by blank spaces. Framework region (FR) and complementarity-determining region (CDR) boundaries are indicated above each germline sequence.
3.5. Immunocytochemical staining analysis
The binding activities against the S protein expressed by SARS-CoV-infected cells of 10 clones with OD values greater than 0.5 from the two constructed libraries (Ssc22, 26, 29, 35, and 37 from the Ssc library and Lsc8, 14, 16, 18, and 22 from the Lsc library) were assessed (Table 2 ). As represented in Fig. 4 , the immunocytochemical staining results showed that clones Ssc22 (C), Ssc35 (D), and Lsc18 (E) had better or similar reactivities compared with that of convalescent serum (A), while clone Lsc22 (F) exhibited no significant binding signal. Two negative controls including normal human serum (B) and bacterial cell lysate without scFv expression (G) exhibited no reactivity at all. Panels H–N were used to demonstrate the total cell numbers, morphology, and distribution under light microscopy.
Table 2.
Immunocytochemical detection of SARS-infected Vero cells using monoclonal scFv binder
| scFv clone | Linker type | Flurorescence intensity |
|---|---|---|
| Ssc22 | Short | ++ |
| Ssc26 | Short | + |
| Ssc29 | Short | + |
| Ssc35 | Short | ++++ |
| Ssc37 | Short | +/− |
| Lsc8 | Long | + |
| LscU | Long | + |
| Lsc16 | Long | + |
| Lsc18 | Long | ++++ |
| Lsc22 | Long | − |
| PC | ++ | |
| NC | − | |
Abbreviations: PC denotes convalescent patient serum; NC denotes normal human serum.
Fig. 4.
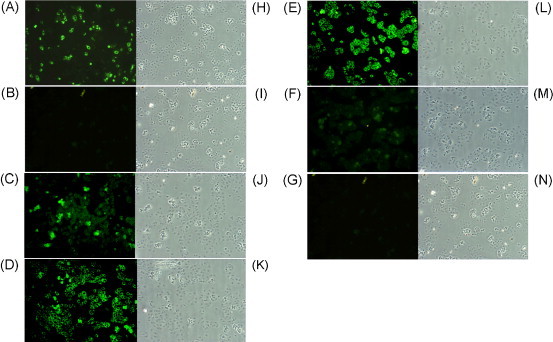
Specific binding of recombinant single-chain variable fragment (scFv) antibodies to SARS-CoV-infected Vero E6 cells. The method of detection was described in Section 2. Biochips precoated with SARS-CoV-infected cells were immunostained with convalescent serum (A), normal serum (B), and Ssc22 (C), Ssc35 (D), Lsc18 (E), and Lsc22 (F) scFv antibodies. Infected cells probed with bacterial cell lysate lacking scFv (G). Panels (H–N) show the morphology and distribution of SARS-CoV-infected cells under a light microscope before treatment.
3.6. Antigenic epitope mapping and competitive ELISA
Clones Ssc35 and Lsc18 were used to identify possible antigenic sites on the S protein with western blot analysis. The purity of the 10 S protein fragments (S1 to S10) was examined and are shown in Fig. 5 A. Fig. 5B and C shows that Ssc35 and Lsc18 scFv antibodies recognized mainly the S6 protein, suggesting that a potential antigenic epitope is located in the region of amino acid residues 750–1000 of the intact S protein. Fig. 6 illustrates the results of the competitive inhibition assays of two representative scFv antibodies against the S6 fragment. Eighy-three percent and 68% inhibitory effects were found on the binding reactivity of Ssc35 and Lsc18 antibodies, respectively, against the S6 fragment in the presence of a concentration of 80 μg/ml of the free S6 fragment. The K d values calculated using the Klotz plot for scFvs Ssc35 and Lsc18 were 1.17 × 10−6 and 1.26 × 10−6 M, respectively.
Fig. 6.
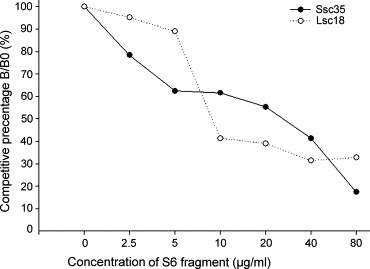
Competitive inhibition assay of two representative single-chain variable fragment (scFv) antibodies against the S6 fragment. The amount of bound Ssc35 and Ssc18 scFv in the presence of free S6 inhibitor was measured and expressed as a percentage of the binding of scFv in the absence of an inhibitor. B and B0 are the amounts of bound scFv in the presence and absence of the inhibitor, respectively.
4. Discussion
To face a possible SARS outbreak in the future, it is necessary to have better diagnostic and therapeutic agents. To overcome the problems which occur frequently during traditional antibody production, such as obtaining low quantities of monoclonal antibodies (mAbs) and the occasional loss of their efficiency due to persistent culturing, a phage display system was used to develop avian anti-SARS-CoV antibodies. The highly conserved 5′ and 3′ regions of FR1 and FR4 facilitate the use of a set of oligonucleotide primers to amplify the V region repertoire for library construction (Davies et al., 1995, McCormack and Thompson, 1990). Two scFv antibody libraries were established which contained 5 × 107 and 9 × 106 clones showing that construction of the chicken scFv antibody library was easier compared to the construction of antibody libraries for humans or other mammals. Thus, the clinical and scientific aspects of chicken scFv generation have received recently much attention (Fehrsen et al., 2005, Finlay et al., 2006, Park et al., 2005).
As shown in Fig. 1A and B, the expression and presence of scFv antibodies were examined by Coomassie blue staining and western blot analysis. Using ELISA, it was observed that more than half of the selected clones in both the Ssc and Lsc libraries could react significantly with SARS-CoV-infected cell lysates (Fig. 2). Interestingly, the Ssc29 and Lsc8 scFv antibodies showed strong reactivity in the ELISA, but weak binding signals in the subsequent immunocytochemical staining analyses. This discrepant result may have been due to the conformational modification of the S protein in the different preparation processes. Some chosen enriched binders against target antigens immobilized on ELISA wells have been found to react poorly against the same antigens located on the surface of cells (Parren et al., 1996). Despite this potential problem in our study, three clones (Ssc22, Ssc35, and Lsc18) were found to bind equally or more efficiently to the S protein compared with the patients’ sera by immunocytochemical staining (Fig. 4).
As shown in Fig. 3, sequence comparison of the 14 clones (seven each from the Ssc and Lsc libraries) with the chicken germline revealed that all of the CDRs were dissimilar. Somatic hypermutations to increase antibody affinity have been found to occur more frequently in CDRs than FRs of the rearranged V gene (Gearhart and Bogenhagen, 1983). In fact, the Ig genes of the selected clones from hyperimmunized chickens had more mutations in the CDRs as a result of the affinity selection of B cells. Therefore, this conclusion is in agreement with that obtained by other researches (Finlay et al., 2005, Finlay et al., 2006, Sapats et al., 2006) and suggests that high mutation rates in these variable genes indicate an antigen-driven response in the chicken induced by the S proteins.
The result of the present study showed that three clones (Ssc22, Ssc35, and Lsc18) had significant binding signals to intact SARS-CoV-infected Vero cells (Fig. 4). To map the potential antigenic epitopes with western blotting, it was found that clones Ssc35 and Lsc18 recognized the purified S6 fragment (Fig. 5B and C), but that clone Ssc22 showed no reactivity (data not shown). Conformational folding of the S protein present on the surface of SARS-CoV-infected cells differs from that of the truncated S proteins immobilized on nitrocellulose membranes, thus leading to differential recognition of clone Ssc22's scFv antibody. The western blotting results in the study indicated further an antigenic epitope that spanned amino acids 750–1000. Previous studies have shown an antigenic domain that spanned amino acid residues 797–1192 can produce antibodies against SARS-CoV infection (Chou et al., 2005, Wang et al., 2005). Based on these results, it is concluded that the Ssc35 and Lsc18 scFv antibodies have specific binding abilities against the S6 protein and may be able to neutralize the infectivity of SARS-CoV in susceptible host cells. In conclusion, the identified epitope is a potential candidate for vaccine development, and these specific monoclonal scFv antibodies can help in developing valuable reagents for both scientific and clinical applications.
Acknowledgments
This study was supported by a grant (NSC 92-2751-B-038-001-Y) from the National Science Council (NSC) of Taiwan. Prof. Winston W. Shen made editing comments on a previous version of this manuscript.
References
- Abouzid K., Ndeboko B., Durantel S., Jamard C., Zoulim F., Buronfosse T., Cova L. Genetic vaccination for production of DNA-designed antibodies specific to Hepadnavirus envelope proteins. Vaccine. 2006;24:4615–4617. doi: 10.1016/j.vaccine.2005.08.085. [DOI] [PubMed] [Google Scholar]
- Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F., III Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods. 2000;242:159–181. doi: 10.1016/s0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Barbas C.F., III, Kang A.S., Lerner R.A., Benkovic S.J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C.F., III, Burton D.R., Scott J.K., Silverman G.J. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Phage Display: A Laboratory Manual. [Google Scholar]
- Bird R.E., Hardman K.D., Jacobson J.W., Johnson S., Kaufman B.M., Lee S.M., Lee T., Pope S.H., Riordan G.S., Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.S., Landt Y., Crimmins D.L., Dieckgraefe B.K., Ladenson J.H. Isolation and characterization of rabbit single chain antibodies to human RegI alpha protein. J. Immunol. Methods. 2002;266:197–207. doi: 10.1016/s0022-1759(02)00117-5. [DOI] [PubMed] [Google Scholar]
- Chou T.H., Wang S., Sakhatskyy P.V., Mboudjeka I., Lawrence J.M., Huang S., Coley S., Yang B., Li J., Zhu Q., Lu S. Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2005;334:134–143. doi: 10.1016/j.virol.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E.L., Smith J.S., Birkett C.R., Manser J.M., Anderson-Dear D.V., Young J.R. Selection of specific phage-display antibodies using libraries derived from chicken immunoglobulin genes. J. Immunol. Methods. 1995;186:125–135. doi: 10.1016/0022-1759(95)00143-x. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fehrsen J., van Wyngaardt W., Mashau C., Potgieter A.C., Chaudhary V.K., Gupta A., Jordaan F.A., du Plessis D.H. Serogroup-reactive and type-specific detection of bluetongue virus antibodies using chicken scFvs in inhibition ELISAs. J. Virol. Methods. 2005;129:31–39. doi: 10.1016/j.jviromet.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Finlay W.J., deVore N.C., Dobrovolskaia E.N., Gam A., Goodyear C.S., Slater J.E. Exploiting the avian immunoglobulin system to simplify the generation of recombinant antibodies to allergenic proteins. Clin. Exp. Allergy. 2005;35:1040–1048. doi: 10.1111/j.1365-2222.2005.02307.x. [DOI] [PubMed] [Google Scholar]
- Finlay W.J., Shaw I., Reilly J.P., Kane M. Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl. Environ. Microbiol. 2006;72:3343–3349. doi: 10.1128/AEM.72.5.3343-3349.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet B., Chaffotte A.F., Djavadi-Ohaniance L., Goldberg M.E. Measurements of the true affinity constant in solution of antigen–antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- Gearhart P.J., Bogenhagen D.F. Clusters of point mutations are found exclusively around rearranged antibody variable genes. Proc. Natl. Acad. Sci. U.S.A. 1983;80:3439–3443. doi: 10.1073/pnas.80.11.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves D.J., Morris B.A. Veterinary sources of nonrodent monoclonal antibodies: interspecific and intraspecific hybridomas. Hybridoma. 2000;19:201–214. doi: 10.1089/02724570050109602. [DOI] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Huston J.S., Levinson D., Mudgett-Hunter M., Tai M.S., Novotny J., Margolies M.N., Ridge R.J., Bruccoleri R.E., Haber E., Crea R. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Yang B.A., Hu Y., Zhao H., Xiong W., Yang Y., Si B., Zhu Q. Human neutralizing Fab molecules against severe acute respiratory syndrome coronavirus generated by phage display. Clin. Vaccine Immunol. 2006;13:953–957. doi: 10.1128/CVI.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- LeClaire R.D., Hunt R.E., Bavari S. Protection against bacterial superantigen staphylococcal enterotoxin B by passive vaccination. Infect. Immun. 2002;70:2278–2281. doi: 10.1128/IAI.70.5.2278-2281.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj E.P., Malik V.S. The new antibody technologies. Adv. Appl. Microbiol. 1993;38:149–209. doi: 10.1016/s0065-2164(08)70216-8. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- McCormack W.T., Thompson C.B. Chicken IgL variable region gene conversions display pseudogene donor preference and 5′–3′ polarity. Genes Dev. 1990;4:548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- McCormack W.T., Tjoelker L.W., Thompson C.B. Immunoglobulin gene diversification by gene conversion. Prog. Nucleic Acid Res. Mol. Biol. 1993;45:27–45. doi: 10.1016/s0079-6603(08)60865-x. [DOI] [PubMed] [Google Scholar]
- Park K.J., Park D.W., Kim C.H., Han B.K., Park T.S., Han J.Y., Lillehoj H.S., Kim J.K. Development and characterization of a recombinant chicken single-chain Fv antibody detecting Eimeria acervulina sporozoite antigen. Biotechnol. Lett. 2005;27:289–295. doi: 10.1007/s10529-005-0682-8. [DOI] [PubMed] [Google Scholar]
- Parren P.W., Fisicaro P., Labrijn A.F., Binley J.M., Yang W.P., Ditzel H.J., Barbas C.F., III, Burton D.R. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type 1 envelope. J. Virol. 1996;70:9046–9050. doi: 10.1128/jvi.70.12.9046-9050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavoni E., Flego M., Dupuis M.L., Barca S., Petronzelli F., Anastasi A.M., D’Alessio V., Pelliccia A., Vaccaro P., Monteriu G., Ascione A., De Santis R., Felici F., Cianfriglia M., Minenkova O. Selection, affinity maturation, and characterization of a human scFv antibody against CEA protein. BMC Cancer. 2006;6:41. doi: 10.1186/1471-2407-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sapats S.I., Trinidad L., Gould G., Heine H.G., van den Berg T.P., Eterradossi N., Jackwood D., Parede L., Toquin D., Ignjatovic J. Chicken recombinant antibodies specific for very virulent infectious bursal disease virus. Arch. Virol. 2006;151:1551–1566. doi: 10.1007/s00705-006-0729-8. [DOI] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chou T.H., Sakhatskyy P.V., Huang S., Lawrence J.M., Cao H., Huang X., Lu S. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 2005;79:1906–1910. doi: 10.1128/JVI.79.3.1906-1910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.H., Zhang J.B., Zhang Z.P., Zhou Y.F., Yang R.F., Chen J., Guo Y.C., You F., Zhang X.E. Construction of single chain variable fragment (ScFv) and BiscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 2006;78:997–1004. doi: 10.1021/ac0512352. [DOI] [PubMed] [Google Scholar]
- Winter G., Griffiths A.D., Hawkins R.E., Hoogenboom H.R. Making antibodies by phage display technology. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- Yamanaka H.I., Inoue T., Ikeda-Tanaka O. Chicken monoclonal antibody isolated by a phage display system. J. Immunol. 1996;157:1156–1162. [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]


