Abstract
This review presents a retrospective of the studies carried out in the last 10 years (2006–2016) using spectroscopic methods as a research tool in the field of virology. Spectroscopic analyses are sensitive to variations in the biochemical composition of the sample, are non-destructive, fast and require the least sample preparation, making spectroscopic techniques tools of great interest in biological studies. Herein important chemometric algorithms that have been used in virological studies are also evidenced as a good alternative for analyzing the spectra, discrimination and classification of samples. Techniques that have not yet been used in the field of virology are also suggested. This methodology emerges as a new and promising field of research, and may be used in the near future as diagnosis tools for detecting diseases caused by viruses.
Keywords: Spectroscopy, Virus identification, Chemometrics, Classification algorithms, Multivariate analysis
Graphical abstract
Highlights
-
•
A retrospective study of 2006–2016 using spectroscopic methods as a research tool in the field of virology.
-
•
Chemometric algorithms used in virological studies were evidenced.
-
•
This review emerges as a new and promising field of research in virology.
1. Introduction
The purpose of this review is to present a great tool to the scientific community working in the field of virology for detecting the biochemical changes caused by the presence of viruses in biological samples. Blout and Mellors [1], and Woernley [2] were pioneers in using spectroscopy with biological perspectives. They investigated IR spectra of homogenized tissues to find disease indicators, but used a single beam and manually scanned instruments which resulted in low sensitivity and reproducibility. In addition, the field was abandoned [3] due to a lack of development in interpretation of observed spectra.
With the advancement of technology and consequently advanced spectroscopy, the interest of researchers in spectroscopic techniques in biological studies has grown. This field of science is known as biospectroscopy, and means the use of spectroscopy to analyze biological samples [4]. Several studies have been conducted involving identification of bacteria [5], [6], viruses [7], [8], cancer diagnosis [9], and even in the field of forensic entomotoxicology [10], demonstrating that spectroscopic techniques are capable of detecting biochemical changes in biological matrices.
Viruses are submicroscopic infectious agents and obligate intracellular parasites. They are totally dependent on a host cell because they are not able to generate energy to conduct all biological processes. There is no structural pattern among all existing viruses, but usually the nucleic acid (RNA for retrovirus, and DNA for adenovirus) is surrounded by a protein membrane arranged in either helical or icosahedral symmetry called the capsid (or nucleocapsid), which in turn may be protected by a lipid bilayer called the envelope, and which may have encrusted spicules usually formed of glycoproteins [11].
Virus infections can cause various health damage, from a simple fever to death. Viral haemorrhagic fevers (VHFs) are examples of acute infections with high death rates caused by different viruses such as the Marburg virus (MBGV), Ebola virus (EBOV), Lassa virus (LASV), Junin, Machupo, Sabia, Guanarito viruses, Crimean-Congo hemorrhagic fever virus (CCHFV), Rift Valley fever virus (RVFV), Hanta viruses, Yellow fever virus (YFV) and Dengue virus (DENV) [12]. The two methods most commonly used in clinical diagnoses of viruses are enzyme-linked immunosorbent assay, with the best known being the ELISA method and real-time polymerase chain reaction (PCR). These methods have brought benefits such as high levels of repeatability and reproducibility, ease in handling and robustness. However, they have also some negative points [13]. Table 1 summarizes some advantages and disadvantages of these methods.
Table 1.
Advantages and disadvantages of ELISA and PCR methods in virus diagnosis.
| Method | Advantages | Disadvantages |
|---|---|---|
| ELISA | Cost effective; robust; easy to use; scalable to testing large numbers of samples; high levels of repeatability and reproducibility. | Requires high quality antisera; in some situations it is not suitable for identifying specific viral species/strains; and is destructive to the samples. |
| PCR | High specificity and sensitivity; high levels of repeatability and reproducibility; ease in handling; robust. | Problems with post-PCR contamination due to high sensitivity (false positive problems, except for RT-PCR and q-PCR); and destructive to the samples. |
Thus, there is a need for techniques that are as advantageous as ELISA and PCR techniques, and which have fewer disadvantages. The potential of spectroscopic techniques in the detection and identification of virus-infected cells has been studied using statistical methods as a sensitive, rapid and reliable methodology. The ability to discriminate between contaminated and non-contaminated samples in a short time with good sensitivity and specificity is pragmatic, suggesting that biospectroscopy is a field that should be more studied in virology [7], [8], [14].
An expected difficulty in the use of biospectroscopy in virology is related to the fact that humans have a great diversity of virus circulating in their organism, and each human has a unique microbiome. With this, obtaining a fingerprint would be more difficult in view of the specificity of each organism. The solution to this problem seems to be the use of a broad and well-trained database, and a correct evaluation of biomarkers changes obtained by multivariate statistical analysis, differentiating these alterations [15].
The main spectroscopic techniques that have been used in virological studies are nuclear magnetic resonance (NMR) spectroscopy [16], Raman spectroscopy [7], infrared spectroscopy (IR) [8] and molecular fluorescence spectroscopy [17]. These techniques are known to provide rapid responses and reliable data, as well as having powerful structural elucidation capability.
For example, Shanmukh et al. [18] used surface-enhanced Raman spectroscopy (SERS) and multivariate statistical analysis techniques to identify and classify respiratory syncytial virus (RSV). The multivariate statistical methods used were principal component analysis (PCA) and hierarchical cluster analysis (HCA). The results showed that SERS combined with these multivariate methods can be used as a rapid methodology for identification and viral classification. In the study by Moor et al. [14] on non-invasive and label-free determination of virus-infected cells by Raman spectroscopy, it was also observed that Raman spectroscopy is a powerful tool for detecting virus-infected cells. In this study, 293 human embryonic kidney cells were infected by an adenovirus and were successfully detected at 12, 24 and 48 h after infection started. The PCA algorithm was able to discriminate infected cells from uninfected cells, providing a rapid, non-invasive and non-destructive method for virus detection.
Such advantages highlight the possibility of identifying and classifying different types of virus using spectroscopic techniques. In this paper, studies using biospectroscopy coupled to statistical methods of classification in virological investigations are emphasized. First, we will discuss the most commonly used spectroscopic techniques, then we will discuss the computational processes used to extract useful information from the obtained spectra (spectral preprocessing, multivariate classification algorithms, performance evaluation), and finally we will discuss some works published in the period from 2006 to 2016 using spectroscopy and multivariate analysis in studies involving viruses.
2. Spectroscopic techniques
The main spectroscopic techniques that have been investigated in virological studies are nuclear magnetic resonance (NMR) spectroscopy [16], Raman spectroscopy [7], infrared spectroscopy (IR) [8] and molecular fluorescence spectroscopy [17].
Spectroscopic methods of analysis are based on the ability of atoms or molecules to interact with electromagnetic radiation. The electromagnetic spectrum covers a large range of energy, where the wavelength, frequency and energy are characteristic of each technique. The most widely used spectroscopic techniques in virology studies are presented hereafter.
2.1. Nuclear magnetic resonance (NMR) spectroscopy
NMR is a spectroscopic technique based on the magnetic properties of some atoms. The principle behind this technique is that a specific isotope has a nuclear spin value and is electrically charged so that when exposed to an external magnetic field, transitions between fundamental and excited spin states are possible [19]. In these transitions, energy transfers occur at wavelengths that correspond to radio frequencies, and the spin returns to the fundamental state, therefore energy of the same frequency is released [19]. This transfer can be measured, yielding an NMR spectrum. From the produced spectrum, it is possible to identify information regarding the chemical structure of the samples being analyzed.
Many applications of NMR spectroscopy in biological matrices are in the field of metabolomics, which is defined as the quantitative measure of dynamic multiparametric metabolic response of living systems to pathophysiological stimulus or genetic modification [19]. This technique is recognized as a promising tool in the evaluation of global metabolic changes, and it can be very useful in the process of clinical diagnosis to distinguish diseases, determine disease severity, and evaluate therapeutic response, among others. The rationale for metabolomics is that a disease causes changes in the concentrations of metabolites in biological fluids or tissues. Key steps in this approach are patient fluid collection, sample preparation, NMR signal acquisition, data processing, and analysis [19].
Examples for virological studies include the use of NMR spectroscopy in determining of membrane topology of NS2B from dengue serotype 4 (DENV-4) [16], where very useful results were obtained for additional functional and structural analysis of NS2B. Meyer and Peters (2003) [20] in their review on the use of NMR spectroscopy techniques in screening and identifying ligand binding to protein receptors make a good discussion on the use of the transferred NOE effect for detecting and characterizing the ligand binding, the use of chemical-shift changes in identification of the ligand binding and the binding pocket of the receptor, as well as the use of relaxation times and diffusion to identify ligand binding and the conditions for NMR spectroscopy screening and binding characterization [20]. Wang et al. (2014) [21] in their study compared the plasma metabolic profiles of patients with Autoimmune Hepatitis (AIH), primary biliary cirrhosis (PBC), PBC/AIH overlap syndrome (OS) and drug-induced liver injury (DILI) with those from healthy individuals attempting to identify biomarkers for AIH. From the information obtained by NMR, it was concluded that there are 9 biomarkers with greatest discriminant significance: citrate, glutamine, acetone, pyruvate, β-hydroxyisobutyrate, acetoacetate, histidine, dimethylamine, and creatinine. Significantly higher amounts of these metabolites were found in patients with AIH compared to healthy controls or other diseases. The diagnostic capacity based on these biomarkers was evaluated, where sensitivity, specificity and accuracy were obtained above 93% in discriminating the AIH from PBC, DILI and OS [21]. In addition, Galal et al. (2016) [22] in their study on chronic viral hepatitis C in the pediatric age group used 1H magnetic resonance spectroscopy (MRS), where they concluded that 1H MRS is a non-invasive technique that has good potential as a diagnostic tool for evaluating staging and fibrosis chronic asymptomatic hepatitis C [22].
2.2. Raman spectroscopy
Raman is a non-invasive, high performance spectroscopic technique capable of obtaining spectra from chemical entities according to its polarizability changes [23]. Using Raman spectroscopy it is possible to detect the presence of a wide range of polar or non-polar chemical bonds, including cellular changes. In this technique, the photons from a monochromatic light source interact with the chemical bonds present in the sample. These photons are absorbed (increasing the vibrational energy of the molecules), and then released (causing the vibrational energy of the molecules to return to their initial state). In this process an inelastic scattering phenomenon can occur, which is when the molecule does not return to a vibratory state of initial energy; then the released photon has a frequency deviation, which maintains the equilibrium of the system. This happens with less than 1% of absorbed photons and 99% are emitted due to elastic scattering. This anomalous scattering can be measured to give what is known as a Raman signal [24]. The inelastic scattering is composed of Stokes and anti-Stokes scattering, which occurs in 1 in 10 million absorbed photons; where the first (Stoke scattering) occurs when the molecule absorbs part of the energy of the incoming wavelength, thereby emitting a wavelength of less energy than the wavelength received; and the anti-Stokes scattering occurs when the molecule releases a wavelength of greater energy than that absorbed. This happens under certain circumstances where the molecule is in a partially excited energy state before absorbing the wavelength received.
Elastic scattering is filtered so that only Raman scattering is detected and used in the production of a spectrum containing bands with information corresponding to the wavelengths at which Raman scattering occurred. An important feature of this technique is that water has insignificant Raman scattering, making this approach more feasible in biological studies.
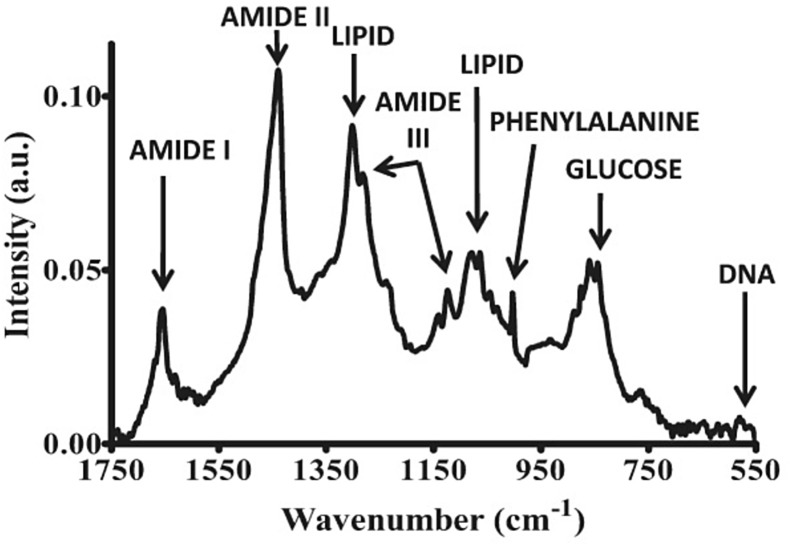
There are several technologies for many applications that make use of Raman spectroscopy, including spatially offset Raman spectroscopy (SORS) and surface-enhanced Raman spectroscopy (SERS) [23], [24]. For example, the potential of Raman spectroscopy followed by statistical methods in detecting and identifying Herpes Simplex Virus type 1 (HSV-1) infections as a sensitive, rapid and reliable method has been evaluated by Salman et al. [25]. Differentiation between a control group and infected cells was observed with sensitivity close to 100%. The main structural changes were mainly related to structures of proteins, lipids and nucleic acids (1195–1726 cm−1 range of the Raman spectrum) [25]. Fig. 1 shows the characteristic bands of biomolecules in a Raman spectrum.
Fig. 1.
Peak assignments in the fingerprint region of biochemical species for a Raman spectrum.
Taken from Kelly et al. [23].
In their paper, Butler et al. (2015) [26] demonstrated the potential of 150 nm gold nanoparticles to generate surface-enhanced Raman spectroscopy (SERS) signals to analyze biological samples, noting that high Raman regions co-localize with the presence of these nanoparticles. Compared with smaller nanoparticles (40 nm), the larger nanoparticles (150 nm) are more easily detectable. Moreover, instead of the signal spreading throughout the cell surface, it appears to be highly localized in the regions surrounding these nanoparticles, giving support to the SERS effect theory. Another advantage of larger nanoparticles is that although their aggregates, even individual particles can be optically observed. This allows areas with abundance of these nanoparticles to be manually oriented for analysis, making it possible to acquire highly enhanced spectra more easily [26]. However, in view of the nanoscale dimension of the viral particle, the use of nanoparticles is inconsistent in the reproducibility of spectral enhancement. As a solution to this inconsistency, a specific marker such as made by Fogarty et al. (2014) [27] can be used, where a two-step protocol with cationic gold particles followed by silver intensification to generate silver nanoparticles on the cell surface was used with SERS of endothelial cell membrane, facilitating the collection of enhanced spectra. This methodology generates a 100-fold increase of the SERS spectral signal [27]. Thus, one way of indirectly identifying the presence of a specific virus would be to detect the presence of antibodies induced by the virus tagged with nanoparticles through SERS signals.
Raman spectroscopy has already been used in virological studies as a structural characterization of 5′ untranslated RNA from hepatitis C virus [28], investigation of a single tobacco mosaic virus [29], study of chicken embryo cells infected with ALVAC virus [30], characterization of different virus strains [31] and identification of new emerging influenza viruses [32]. All these studies proved that Raman spectroscopy is a powerful technique in virus studies, as well as being non-destructive, fast and having a simple procedure.
2.3. Infrared (IR) spectroscopy
Chemical bonds have vibratory motions like bending, stretching, rocking or scissoring that allow the molecules to absorb infrared radiation related to its specific vibrational energy levels. This absorption is only active if the molecular bonds have an electric dipole moment changeable by atomic displacement due to its vibrations [33]. Spectral acquisition is made using a Fourier transform (FT) filter to change the time domain to frequency, thus generating the term FT-IR.
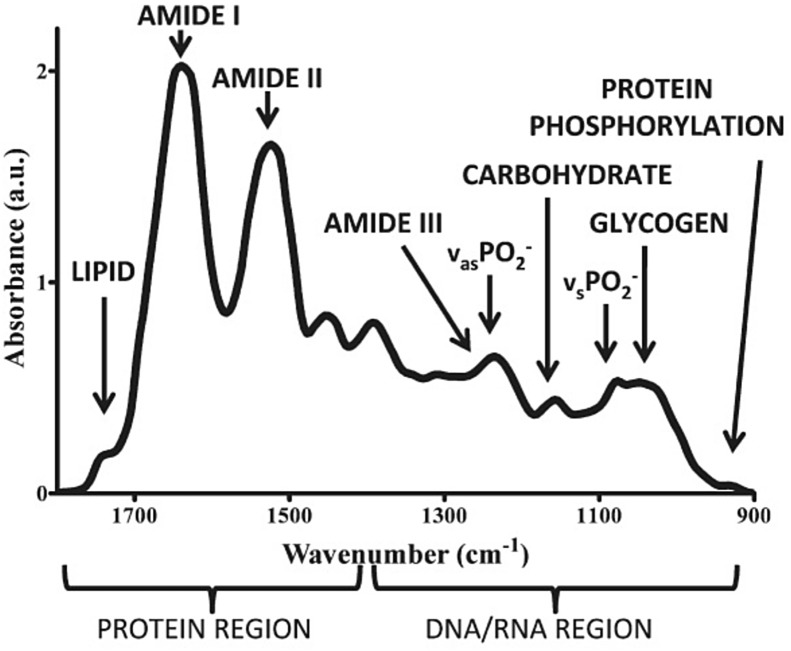
The infrared region can be subdivided into near-IR, far-IR and mid-IR. Among these regions, the mid-IR region is of particular interest in biological studies because in this range there is the fingerprint region of biological samples , also called the “biofingerprint” region; that is, the region where there are spectral bands related to biomolecules such as lipids (∼1750 cm−1), carbohydrates (∼1155 cm−1), proteins (Amide I, ∼1650 cm−1, Amide II, ∼1550 cm−1, Amide III, ∼1260 cm−1), and DNA/RNA (∼1225 cm−1, 1080 cm−1), among others [23], [33], [34]. Table 2 shows these principal absorptions and Fig. 2 shows a mid-IR spectrum at the fingerprint region with the bands corresponding to the main biomarker fragments [23], [35].
Table 2.
| Band | Assignment |
|---|---|
| 970 cm−1 | of phosphorylated proteins |
| 1030 cm−1 | Glycogen |
| 1080 cm−1 | of phosphodiester groups of nucleic acids |
| 1155 cm−1 | ν(C–O) of carbohydrates |
| 1225 cm−1 | in RNA and DNA |
| 1260 cm−1 | Amide III: ν(C–N) in proteins |
| 1550 cm−1 | Amide II: δ(N–H) coupled to ν(C–N) |
| 1650 cm−1 | Amide I: ν(C O) |
| 1750 cm−1 | ν(C C) of lipids |
νs = symmetric stretching; νas = asymmetric stretching; δ = bending.
Fig. 2.
Peak assignments in the fingerprint region of biochemical species for an infrared spectrum.
Taken from Kelly et al. [23].
Although mid-IR (MIR) spectroscopy is more widely used in studies with biological samples, near-IR (NIR, 700–2500 nm) spectroscopy is also commonly used. NIR spectroscopy is a technique based on the overtones of the fundamental vibrational modes observed in the mid-IR region, and it provides fast data acquisition combined with no reagent requirements and minimum sample preparation. These characteristics make NIR spectroscopy a reliable and non-invasive approach with potential for rapid diagnosis of viral diseases and infections. When working with NIR spectroscopy in the investigation of biological samples, the most useful region is between 650 and 1000 nm (called the “optical window”), because there is great absorption of hemoglobin and water below 650 nm and above 1000 nm, respectively, hindering other signals in these regions. Therefore, in the clinical diagnostic perspective the use of the 650–1000 nm region is more suitable for virological studies to analyze biochemical alterations [36].
2.4. Molecular fluorescence spectroscopy
Although molecular fluorescence spectroscopy has been little used in studies in the field of virology, it is also an interesting approach with great potential in this perspective. This technique analyzes the fluorescence capacity of a sample [17], where a beam of high energy light (usually in the ultraviolet region) is irradiated on the sample to be excited into a higher electronic energy level; then the fluorophore molecule will rapidly lose energy to this environment through non-radiative modes (called internal conversion) and will return to the lowest vibrational level of the lowest electronic excited state. The molecule persists at this vibronic level for a period of time known as the fluorescence lifetime, and then returns to the fundamental electronic state by emitting a photon with energy lower than the irradiated one [37]. The excitation and emission spectrum are recorded by the instrument and is generally used to build excitation-emission (EEM) fluorescence matrices.
Another commonly-employed form of fluorescence technique is fluorescence correlation spectroscopy (FCS), which is used for temporal and spatial analysis of molecular interactions of biomolecules present in solution at extremely low concentrations. This technique is based on the principle that a fluorophore molecule has a specific free diffusion rate that is directly related to its size. This basic principle, for example, can be used to study protein interactions. As with other spectroscopic techniques, molecular fluorescence spectroscopy provides rapid results with high sensitivity and specificity, and is non-destructive, making this technique a tool of interest in the field of virology [17].
Table 3 summarizes the main advantages and disadvantages of NMR, Raman, IR and molecular fluorescence spectroscopy to analyze biological materials.
Table 3.
Some advantages and disadvantages of spectroscopy techniques.
| Spectroscopy technique | Advantages | Disadvantages |
|---|---|---|
| NMR | Limit of detection normally micromolar; high reproducibility; easy identification of the metabolite using 1D or 2D spectra and database; more than 200 known identifiable metabolites; easy sample preparation; non-destructive method; requires small amount of sample; low cost of sample analysis; quick results [19]. | Only detects metabolite if there are specific isotopes in the molecule; expensive instrumentation. |
| Raman | High molecular specificity; ability to derivate label-free and non-destructive spectral information; minimal sample preparation; high penetration depth [38]. | Low sensitivity caused by the low-probability of Raman scattering event; fluorescence interference; local thermal decomposition of the sample, in particular when using ultraviolet or visible wavelengths lasers [38]. |
| Mid-infrared (MIR) | High signal-to-noise ratio; reduced scattering; high spatial resolution; analysis of large target area; nondestructive data acquisition; minimum sample preparation; relatively low-cost instrumentation; automated stages of analysis [33]. | Pressure over the sample when using ATR module can be destructive; air interfering, in particular CO2; sample thickness issues [33]. |
| Near-infrared (NIR) | Fast analysis; low-cost instrumentation; minimum sample preparation; portable instruments are highly available; small amount of sample is require; high resolution; non-destructive analysis; high reproducibility [39], [40]. | Low signal-to-noise ratio; many spectral superposition; dependence on reference methods and chemometric analysis [39], [40]. |
| Molecular fluorescence | High sensitivity and specificity; relatively low-cost instrumentation; small concentration of sample is required; high signal-to-noise ratio. | Sample preparation is relatively complex; large time of analysis; signal saturation is often observed; presence of Rayleigh scattering. |
3. Computational analysis
When spectroscopically interrogating biological samples, spectra with a lot of information are generated. To facilitate the spectral analysis, it is generally necessary to use computational tools which facilitate the information analysis and extraction. For this, pre-processing and multivariate analysis techniques are employed.
3.1. Preprocessing
The spectrum obtained in spectroscopic analysis is composed of the analyte information plus noise. The noise can be caused by chemical interference, which can cause band superposition by additive effects; and by physical interference, which can cause baseline deviations due to light scattering. In addition, random noise from environmental effects is always present in the spectra of real samples.
To correct this and improve the signal-to-noise ratio of the spectrum, pre-processing techniques are commonly employed before data analysis. To understand the functions of some commonly used pre-processes, a few examples are shown hereafter.
3.1.1. Spectral cut
Many times only a region of interest of the spectrum being analyzed is utilized to build a chemometric model. This occurs when there is notable interference in other parts of the spectrum, such as water or solvent absorptions, or when there is irrelevant information for the analysis being performed. Depending on the spectroscopic technique used the biological fingerprinting will be different. For example, in biological samples being analyzed with FTIR spectroscopy the region of interest (referent to the biofingerprint region) is between 1800 and 900 cm−1 due to the fundamental absorption of fragments of key biochemical molecules [34]. On the other hand, for Raman spectroscopy the selected region is between 2000 and 500 cm−1 [23].
3.1.2. Baseline correction
Baseline deviation frequently occurs in first-order spectra. For each sample there are wavenumbers which are not absorbed; where their absorbance must have zero value. However, this is often not observed. Spectra are generally raised to values above zero due to a phenomenon called Mie scattering [23], [41]. This phenomenon occurs when some biochemical structures through which IR radiation passes have a size comparable to or greater than the wavelength of IR light, causing light scattering [25]. In solid materials, the baseline is also affected by non-homogenous particle sizes, which likewise cause light scattering. In addition, the baseline slope is affected by reflection, temperature, concentration or anomalies of the instrument used. These effects can be minimized through a variety of baseline correction techniques, such as multiplicative scatter correction (MSC) [42], standard normal variate (SNV) [42], and derivative [43], among others [33], [44], [45].
3.1.3. Normalization
Spectral normalization techniques are used when it is necessary to remove spectral changes responsible for the thickness or concentration of the sample, making the normalized spectra become comparable to each other [23]. Among the possible normalizations, there is the min-max normalization, which can be applied when there is a known peak that is stable and consistent between the specimens [23]; or scaling methods to equalize the importance of each variable in multivariate data [46]. In biological samples, the amide I (∼1650 cm−1) or amide II (∼1550 cm−1) peak normalization [33], [34] are typically used; or vector normalization, where each spectrum is divided by its Euclidean norm (appropriate normalization after using differentiation as pre-processing) [33], [34].
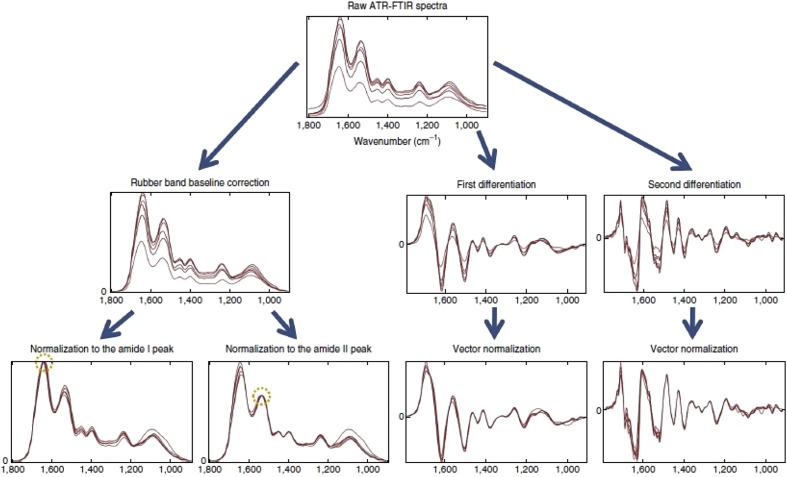
Fig. 3 shows the visual effect after using these pre-processing techniques on a set of FTIR spectra.
Fig. 3.
Visual effect of different pre-processing on a set of FTIR spectra.
Taken from Baker et al. [33].
3.2. Multivariate analysis
Multivariate analysis techniques are employed to analyze multivariate data, meaning data having two or more variables per object [46]. Examples are first-order data (such as FTIR, NIR, Raman spectrum) and second-order data (such as EEM fluorescence). Some multivariate classification algorithms that are widely used in biospectroscopy studies are discussed below, as well as some biospectroscopy studies in which these techniques were used.
3.2.1. Principal component analysis (PCA)
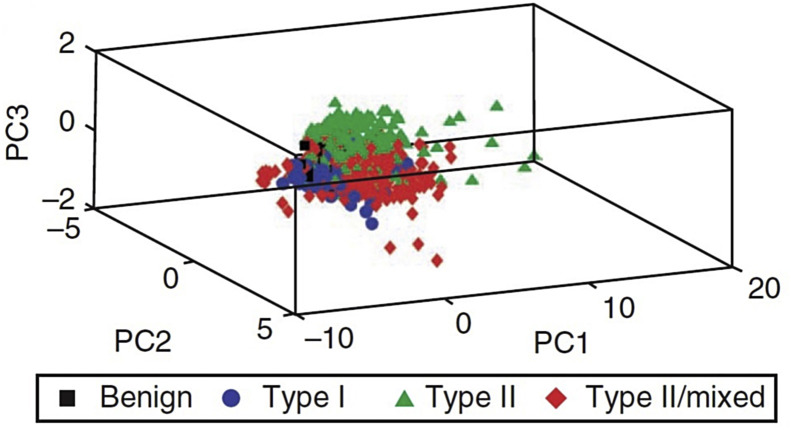
PCA is an unsupervised multivariate analysis technique widely used in biological studies. This technique is used to reduce the dimensionality of the sample's data and generate a new visualization. Dimensionality reduction occurs through a linear transformation of the original variables, generating orthogonal variables called principal components (PC). The first main component has a greater ability to explain the observed variance in the data than the second PC, which in turn has a greater explanation of the data than the third PC, and so on. In this way, it is possible to choose the smallest number of principal components with the largest explained variance, so that the dimensionality can be reduced with the certainty that important information of the samples is not lost [23]. Generally, few PC's (10–20) provide more than 99% of the observed variance of the original data [47]. Each PC is composed by a score (projections of the samples on the PC direction) and a loading (angle cosines of the variables projected on the PC direction) [48], [49]. PCA allows visualization of the data set in reduced size where segregation between classes can be revealed (Fig. 4 ) [47].
Fig. 4.
Example of PCA score plot for different types of endometrial tissues. The classes for this example are: benign (black squares); carcinoma type I (blue circles); carcinoma type II (green triangles); and type II/mixed carcinoma (red diamonds).
Taken from Martin et al. [47].
When the biological samples are divided into classes (e.g., category of samples or patients identity), PCA can be applied to identify spectra clusters and the main contributory information for this distribution [49], [50]. Cluster vector approaches have been proposed in combination with PCA to analyze this type of samples [50]. For this, a median score is calculated for each of the 3 PCs that represent the best samples' clustering in a tridimensional space; thereafter, the three loadings vectors for these PCs weighted by the median scores are summed. As a result, a new loading vector is generated as an effective loadings plot that represents the cluster.
However, PCA has some disadvantages, such as the risk for artificial discrimination depending on the number of PCs selected during model construction, wherein selecting an optimum number of PCs is a critical and difficult process; and the ambiguity to obtain an optimum grouping into a specific cluster, since many PCs combinations may reveal different clustering [50].
Saade et al. (2008) [7] used PCA in their study on spectral differentiation between healthy and contaminated samples with hepatitis C in vitro based on human serum. In this study, twenty-nine samples were examined by near-infrared Raman spectroscopy, being 17 healthy and 12 contaminated; and PCA was used in extracting the main spectral characteristics for classification. Table 4 summarizes the obtained results. As can be seen, a total of 15 out of 17 healthy samples were classified as healthy (88% specificity), and a total of 11 out of 12 contaminated samples were classified as actually being contaminated with hepatitis C (92% sensitivity) [7].
Table 4.
Sensitivity and specificity results obtained by PCA for the diagnosis of hepatitis C based on Raman spectroscopy: HS – health serum; CS – hepatitis C serum.
| Traditional diagnosis | PCA analysis |
||||
|---|---|---|---|---|---|
| HS | CS | Total | Sensitivity (%) | Specificity (%) | |
| Healthy | 15 | 2 | 17 | – | 88 |
| Hepatitis C | 1 | 11 | 12 | 92 | – |
Obtained from Saade et al. [7].
Shanmukh et al. (2008) [18] in their study to identify and classify respiratory syncytial virus (RSV) also used PCA technique combined with Raman spectroscopy. In this study, Raman spectra of A/Long, B1, A2 strains and the recombinant A2 G gene deletion mutant (ΔG) strain of the respiratory syncytial virus (RSV) were recorded. Based on the intrinsic spectra for each sample, the 3 virus strains were detected and identified. Chemometric results showed that PCA was able to segregate the 3 virus strains (A/Long, B1and A2), as can be seen in the scores plot of PC1 versus PC2 (Fig. 5 ), demonstrating the potential of this technique [18].
Fig. 5.
PCA scores on PC1 versus PC2 calculated from the Raman spectra for • RSV strains A/Long,  B1,
B1,  A2, and the A2 strain-related G gene mutant virus (ΔG).
A2, and the A2 strain-related G gene mutant virus (ΔG).
Taken from Shanmukh et al. [18].
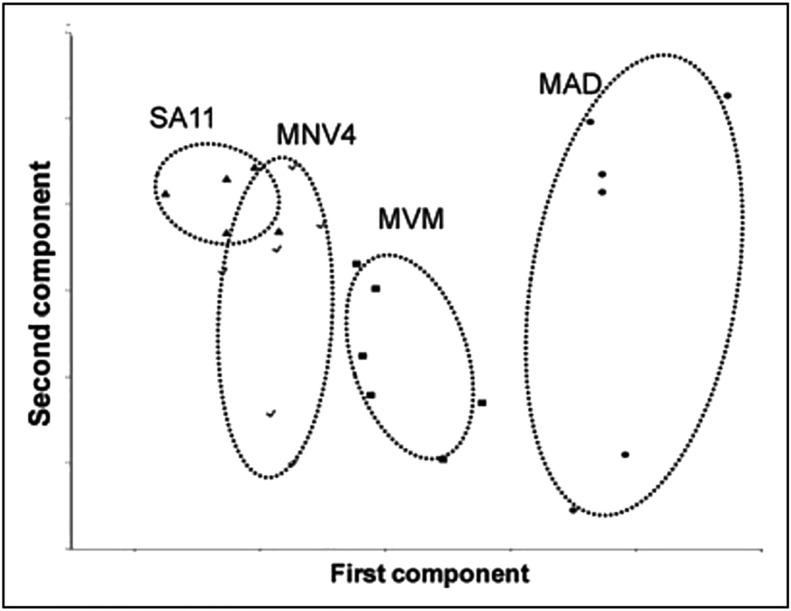
Fan et al. (2010) [51] used Raman spectroscopy to detect and discriminate 7 food and water viruses (including norovirus, adenovirus, parvovirus, rotavirus, coronavirus, paramyxovirus and herpes virus). PCA was conducted based on the Raman spectra of 4 non-enveloped virus samples (including norovirus strain – MNV4, simian rotavirus strain – SA11, adenovirus strain – MAD and parvovirus strain – MVM). As can be observed in Fig. 6 , the two-dimensional plot of the scores on the two first PCs (PC1 versus PC2) showed good class segregation ability; although some SA11 and MNV4 samples were partially overlapping. Similar results were obtained for enveloped virus samples [51].
Fig. 6.
PCA scores based on Raman spectra, using only the spectral range of 200–1800 cm−1, acquired from 4 undeveloped virus strains: MAD, MNV4, SA11 and MVM.
Taken from Fan et al. [51].
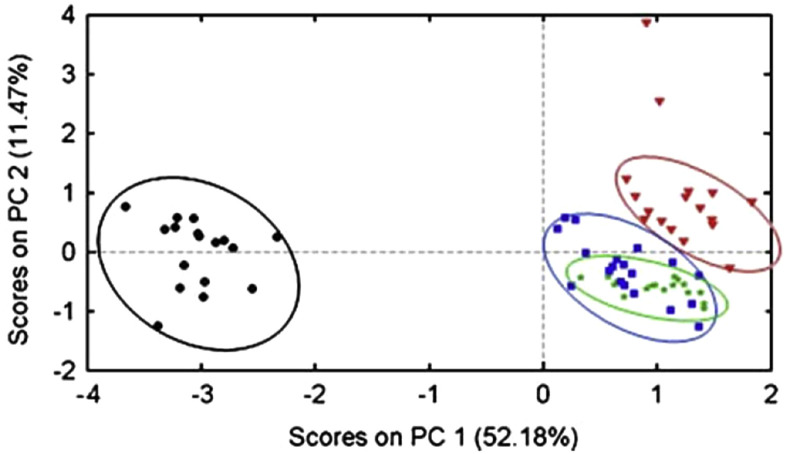
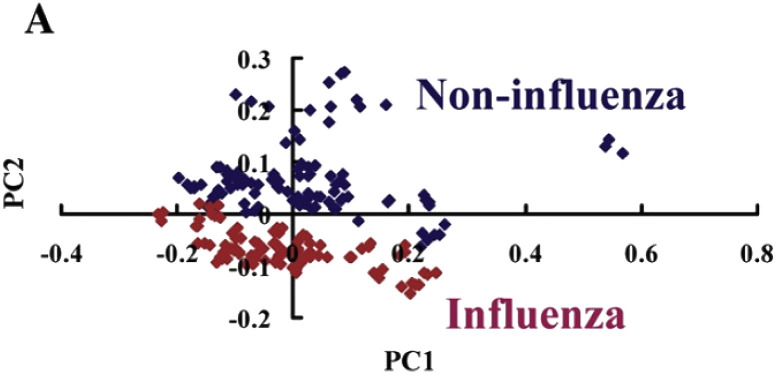
Sakudo et al. (2012) [8] analyzed near-infrared spectra using molecular clones of various HIV-1 subtypes. The spectra obtained were subjected to PCA to extract information and exploratory analysis, where it was observed that the presence of HIV-1 in the medium altered wavelength absorption at around 950 and 1030 nm, suggesting that HIV-1 changes the vibrations of OH in water [8]. In addition, absorption variations related to different subtypes have been observed, suggesting that different subtypes directly interact with the medium [8]. Still in 2012, Sakudo et al. [52] performed a study with the objective of discriminating nasal fluid samples from patients infected with influenza virus through Vis-NIR spectroscopy. Samples from 33 healthy and 34 influenza patients were used. The results of the PCA scores (Fig. 7 ) using the two-dimensional plot of the scores on PC1 versus PC2 shown good segregation between the two classes [52].
Fig. 7.
Result of PCA scores employed in the differentiation of patients with  Influenza and
Influenza and  Non-Influenza.
Non-Influenza.
Taken from Sakudo et al. [52].
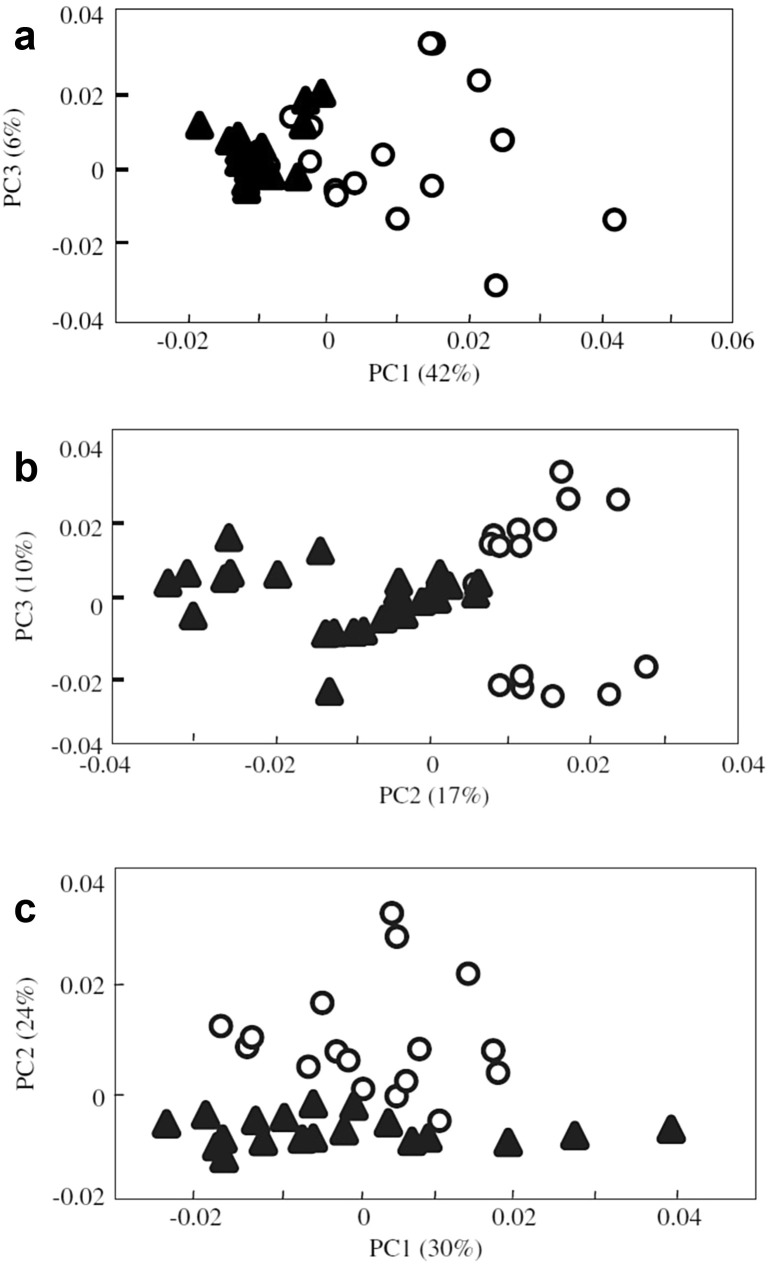
Moor et al. (2013) [53] analyzed control (uninfected) cell samples, cells after 24 h of adenovirus infection, and cells after 7 days of adenovirus infection by Raman spectroscopy. When submitted to PCA, the Raman spectra of the three-class samples showed results suggesting that the infection induces rapid (24 h) production of a specific protein in the cells, which can differentiate infected samples from uninfected ones [53]. In 2014, Moor et al. [14] continued their work on the power of Raman spectroscopy as a tool for detecting virus-infected cells, and developed a study where they successfully detected embryonic kidney cells of 293 adenovirus-infected humans at 12, 24 and 48 h after infection onset. The PCA score plot was effective in discriminating the spectra and segregating the uninfected cell classes of the infected cells after 12 h (Fig. 8 a), 24 h (Fig. 8b) and 48 h (Fig. 8c) [14].
Fig. 8.
PCA scores for ○ control cells and ▴ cells with virus: (a) after 12 h of infection; (b) after 24 h of infection; (c) after 24 h of infection.
Taken from Moor et al. [14].
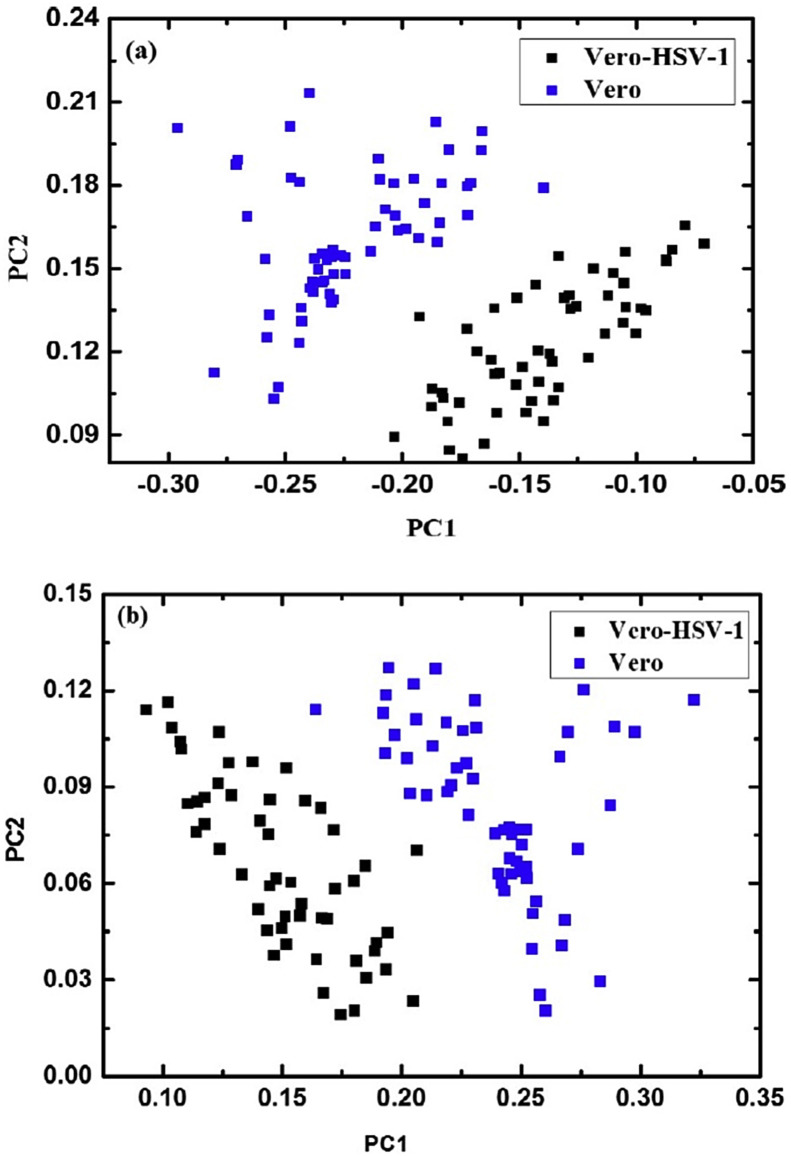
Wood et al. (2014) [54] performed a study of the diagnosis of malaria-infected cells based on synchrotron Fourier transform infrared (FTIR) imaging. The images were extracted and preprocessed by second derivative and normalization. The application of PCA to spectral data showed the potential of the technique to identify and discriminate uncontaminated from contaminated samples [54]. Salman et al. (2014) [25] evaluated the potential of Raman spectroscopy as a sensitive, reliable and rapid method for detecting and identifying viral infection by Herpes simplex virus type 1 (HSV-1) in cell culture. The spectral data were submitted to PCA, where a good segregation tendency was observed between the infected and non-infected classes (Vero-HSV-1), mainly when the region of 600–1726 cm−1 (Fig. 9 a) and 1195–1726 cm−1 (Fig. 9b) are analyzed [25].
Fig. 9.
Scores on PC1 versus PC2 for  Vero-control and ■ Vero-HSV-1 cells, measured from spectra in the region of: (a) 600–1726 cm−1 (entire range); (b) 1195–1726 cm−1 (high range).
Vero-control and ■ Vero-HSV-1 cells, measured from spectra in the region of: (a) 600–1726 cm−1 (entire range); (b) 1195–1726 cm−1 (high range).
Taken from Salman et al. [25].
3.2.2. Cluster analysis (CA)
Cluster analysis (CA) techniques are unsupervised methods of pattern recognition that aim to group the spectra into groups when there is no information about the classes [23]. These techniques are exploratory, therefore they group the samples based on their similarity between spectra. CA techniques include k-means clustering (KMC), fuzzy c-means cluster analysis (FCA) and hierarchical cluster analysis (HCA) [34], [55]. HCA (using Ward method [56]) is considered the most capable of correlating spectral data with histopathology, but is more time-consuming [34]. Another difference between these techniques is that HCA does not require making assumptions about the number of data classes, whereas KMC and FCA require this information. HCA starts with separate clusters, and merges the clusters that have the closest distance from each other (usually based on Euclidian distance) in each step, so there is a reduced number of clusters in each step until only one remains [34], [55].
In the study by Shanmukh et al. (2008) [18], HCA was used to identify and classify respiratory syncytial virus (RSV) based on Raman spectroscopy. The results showed that HCA was able to easily distinguish an A2 strain-related G gene mutant virus (ΔG) from an A2 strain. HCA showed good classification results for the four strain classes [A/Long, B1, A2 and the ΔG] of the respiratory syncytial virus (RSV), as can be seen in Table 5 [18].
Table 5.
Classification of 4 RSV virus strains based on hierarchical cluster analysis (HCA).
| Viral strain | Correctly classified | Falsely classified | Also classified as | Sensitivitya | Specificityb |
|---|---|---|---|---|---|
| RSV A/Long | 17 | 0 | – | 1.0 | 1.0 |
| RSV B1 | 17 | 0 | – | 1.0 | 0.92 |
| RSV ΔG | 15 | 2 | A2(2) | 0.88 | 0.94 |
| RSV A2 | 12 | 7 | ΔG(3), B1(4) | 0.63 | 0.96 |
Probability of assigning a class as positive when it really is positive.
Probability of assigning a class as negative when it really is negative.
Obtained from Shanmukh et al. [18].
3.2.3. Partial least squares (PLS)
PLS is a multivariate calibration technique that finds factors (latent variables, LVs) in the spectra set that explain the maximum variance in the reference variables set, using the simultaneous decomposition of the two [46]. For this, PLS finds a set of new maximally correlated variables orthogonal to each other, similar to PCA. However, it makes use of the respective object labels for this decomposition, thus being a supervised technique [23]. For discriminatory purpose, partial least squares discriminant analysis (PLS-DA) is employed. PLS-DA is a linear classification technique for which the classification criterion is obtained by PLS analysis [46]. Therefore, it makes use of PLS to find a straight line that divides the data space into two-regions, where each region is related to the space of each class [57].
Lee-Montiel et al. (2011) [58] used ATR-FTIR spectroscopy and cell culture for detecting and quantifying poliovirus infection in buffalo green monkey kidney (BGMK) cells. The cells were infected with different virus concentrations, and after 1–12 h of post-infection (h.p.i), PLS was used to analyze spectra from different infection trites. The results of this study showed that the detection and quantification of poliovirus through ATR-FTIR spectroscopy, PLS and cell culture is a methodology that could be adapted for use in areas such as water safety monitoring and clinical diagnosis [58].
Petisco et al. (2011) [59] also implemented PLS in the field of virology. They developed a rapid method based on near infrared spectroscopy to detect viral RNA present in Epichloë festucae strains isolated from Festuca rubra plants. Forty two samples were used as the data set, where a correct classification of 75% of the uninfected isolates and 86% of the infected isolates were obtained using PLS-DA, demonstrating that this technique is also promising for detecting viral infections in fungus samples, being a faster and a more cost-saving alternative than the conventional analyzes of reference [59]. Wang et al. (2014) [21] used NMR spectroscopy in conjunction with multivariate analysis algorithms such as PLS to characterize the metabolic profile of Autoimmune Hepatitis (AIH) and to identify biomarkers with diagnostic potential for AIH. The results were encouraging and also demonstrate the potential of the technique [21].
3.2.4. Linear discriminant analysis (LDA)
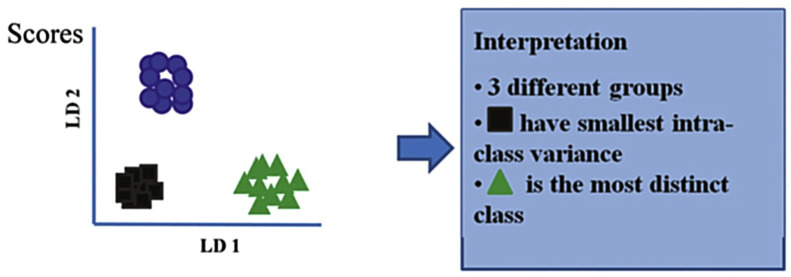
Linear discriminant analysis (LDA) is a supervised technique widely used for class discrimination. It maximizes the between-class variance over the within-class variance [23] in order to create a linear decision boundary between them [60]. Additionally, linear discriminant analysis has its operation enhanced when associated with other dimensionality reduction algorithms. For example, LDA is often combined with PCA or PLS in many virology studies [21], [25], [58], [59], [61]. An illustrative result obtained when performing LDA is the scores shown in Fig. 10 . It is possible to do some interpretations on the studied data, such as the number of classes (groups), which class has the lowest intra-class variance, and which class has the largest intra-class variance [22].
Fig. 10.
Example of some interpretations that can be taken from the scores produced by LDA.
Taken from Kelly et al. [23].
Examples using PCA-LDA and stepwise-LDA have been also demonstrated for cluster vector analysis [50]. This can solve the ambiguity problems generated by PCA providing a more reliable class differentiation. For PCA-LDA the cluster vector is obtained by calculating the mean cluster scores matrix and the loading matrix for each cluster, and then the cluster loading matrix is obtained by summing the loadings weighted by the mean cluster scores [50]. On the other hand, for stepwise-LDA the cluster vector is obtained by firstly selecting a single prominent spectral peak and then LDA is applied to this wavenumber across all data, generating a Gaussian posterior density fitted to each group. Thereafter, one data point is left out and the Gaussian is used to estimate the predicted posterior class probability for that spectrum across all class categories (at that one specific wavenumber). This is reaped for all spectra and after for all wavenumbers. At the end, the predicted class probability for each data point is compared with the known true class value (e.g., 0 or 1); and these absolute deviations for each wavenumber are summed to give a sum of absolute posterior misclassification, where a plot of this error against wavenumber are then analogous to peaks on a PCA loadings plot, in which they indicate wavenumbers that best distinguish one particular class from the rest [50].
There are other algorithms that have shown excellent results in several biological studies, but have not yet been used in virological studies [5], [6], [9], [10], [62]. At this point, we would like to suggest two algorithms that can be used as variable selection techniques in problems of identification, classification and diagnostics for viruses, but which have not been used in this perspective; they are the successive projections algorithm (SPA) and genetic algorithm (GA), which are discussed below.
3.2.5. Successive projections algorithm (SPA)
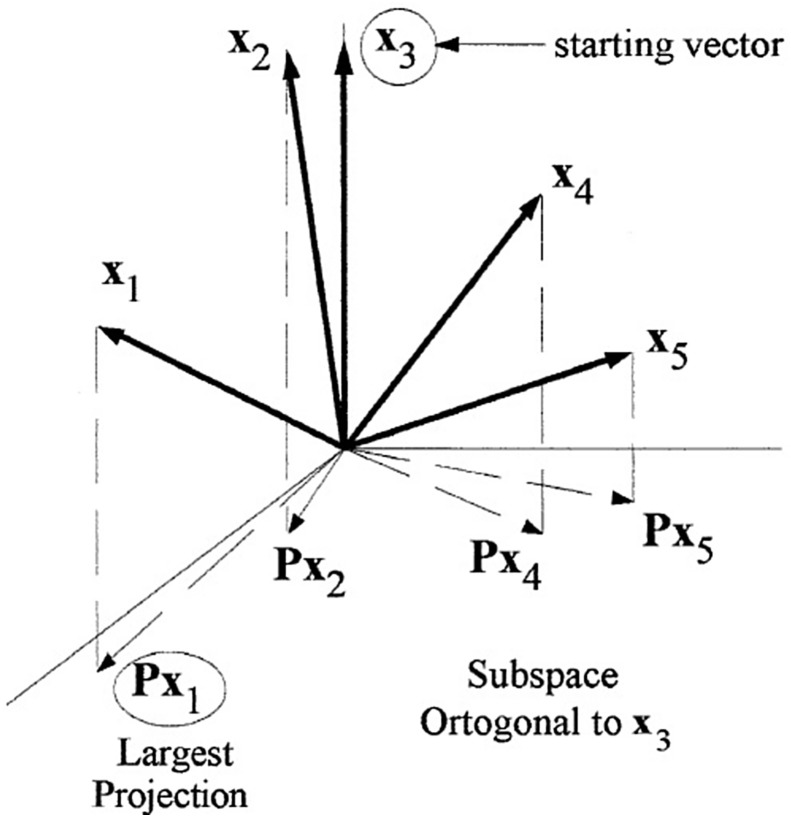
SPA is a progressive variable selection technique. This means that it starts with a variable (wavelength or wavenumber, for example) and adds new variables in each interaction until an optimal number is selected. This technique uses multicollinearity minimization as a criterion for variable selection. For this, each variable representing a vector during SPA has a projection in an orthogonal subspace. The variables with the largest projections (minimum multicollinearity) are selected (as shown in Fig. 11 ), where the result of the interaction is the selected vector x 1 [63], [64], [65].
Fig. 11.
Visual illustration of the projections involved in SPA. The variables in this technique are seen as vectors (x) with their orthogonal projections (Px), and then selected to eliminate multicollinearity problems. In this example, the interaction resulted in selecting the variable related to vector x1.
Taken from Araújo et al. [64].
3.2.6. Genetic algorithm (GA)
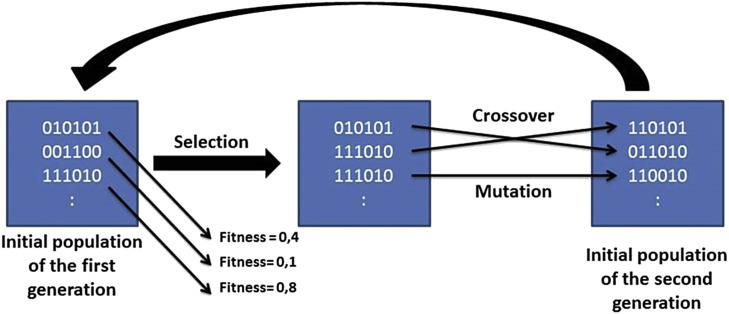
The genetic algorithm is a technique that mimics Darwin's theory of evolution, where evolution occurs by natural selection in which the more adapted organisms have a greater chance of survival. In the case of GA, the variable selection process begins with a randomly formed population of variables. This initial population consists of subsets of variables called chromosomes, where each variable is assigned a value of 0 or 1, with variables of 0 being the ones initially not selected by the model, and 1 being those initially selected to participate in the model. Each chromosome is assigned an aptitude through a mathematical function called fitness function, where chromosomes with the highest fitness value are copied and the chromosomes with the lowest fitness value are eliminated in a step called the selection step. After the selection step, genetic operators are probabilistically applied. The mutation operator makes a variable that is selected to be unselected or vice-versa, and the crossover operator crosses the chromosomes. This process is involved in a generation, and can be repeated for as many generations as are requested. In the end, the population with the better variables is selected based on a cost function [66] (see Fig. 12 ).
Fig. 12.
Operational scheme of the genetic algorithm (GA). In this scheme an initial population with 3 chromosomes is shown. A fitness value is assigned for each chromosome through the fitness function (F). Note that the chromosome with less fitness is discarded in the selection stage, while the larger one is doubly copied and the second largest fitness receives a copy. It is observed that the chromosome is mutated through the mutation operator in the second moment, and the other two chromosomes are crossed through the crossover operator. This process is repeated for a defined number of generations.
As mentioned earlier, LDA has its operation enhanced when used in conjunction with other techniques that reduce the size of the data. Therefore, a good alternative to using SPA and GA is to join them with LDA (SPA-LDA and GA-LDA). In this, SPA and GA will reduce the data to a smaller number of selected variables, and the LDA only based on these more discriminant variables will work on maximizing the differences between the classes.
4. Performance evaluation techniques
In most of the studies presented in this review, the authors evaluated their technique based mainly on sensitivity and specificity. However, other quality measures may be used in order to assess whether the technique is effective or not. Here we present seven figures of merit that can be used in the evaluation stage of the classification technique aiming for its validation. They are the sensitivity, specificity, positive predictive values, negative predictive values, Youden's index, positive likelihood ratio and negative likelihood ratio.
Sensitivity (SENS) can be defined as the confidence that a positive result for a sample of the labeled class is obtained; specificity (SPEC) is the confidence that a negative result for a sample of non-labeled classes is obtained; positive predictive value (PPV) measures the proportion of positives that are correctly assigned; negative predictive value (NPV) measures the proportion of negatives that are correctly assigned; Youden's index (YOU) evaluates the classifier's ability to avoid failure; positive likelihood ratio (LR+) represents the ratio between the probability of predicting a sample as positive when it truly is positive and the probability of predicting a sample as positive when it is actually not positive; and the negative likelihood ratio (LR−) represents the ratio between the probability of predicting a sample as negative when it is actually positive and the probability of predicting a sample as negative when it is truly negative [67]. The mathematical formulas for each of these figures of merit are shown as follows [67]:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where TP is defined as true positive, TN as true negative, FP as false positive, and FN as false negative.
5. Diagnostic virology
Diagnostic virology continues to evolve rapidly. Viral testing is now essential for the care of a number of patient groups, including hospitalized patients with acute respiratory infections; transplant recipients and other immunocompromised patients; patients infected with human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV); and infants with possible congenital infection. Multiple test methods continue to be used, but molecular tests are emerging as the dominant technology. A variety of commercial molecular assays have been or are in the process of being approved or cleared as in vitro diagnostic tests by the Food and Drug Administration (FDA). This is an important development because it makes viral diagnostic testing available to more laboratories, and it improves the standardization of diagnostic testing. The scope of diagnostic virology has broadened [68]. General categories of viral diagnosis/research testing and the viruses included in those categories are shown in Table 6 .
Table 6.
Categories of testing performed in diagnostic and research virology.
| Category of testing | Specific viruses | Methodology |
|---|---|---|
| Respiratory viruses | Influenza A and B, RSV, PIV 1–4, hMPV, rhinoviruses, enteroviruses, coronaviruses, adenoviruses | Rapid antigen tests (influenza A and B, RSV), fluorescent antibody staining (influenza A and B, RSV, PIV 1–3, adenoviruses, hMPV), culture, multiplex NAAT, RS [14], SERS [18] |
| Gastrointestinal viruses | Rotavirus, norovirus, adenovirus, astrovirus | Rapid antigen tests (rotavirus, norovirus, adenovirus), NAAT |
| Mucocutaneous viruses | HSV, VZV, HPV | Fluorescent antibody staining (HSV and ZVZ), culture (HSV and VZV), NAAT |
| Central nervous system viruses | HSV, VZV, CMV, EBV, HHV-6, JCV, enteroviruses, parechoviruses, West Nile virus, other arboviruses | NAAT, serology (West Nile and other arboviruses) |
| Opportunistic agents | CMV, EBV, BKV, HHV-6, adenoviruses | NAAT, antigen detection (CMV pp65 assay), cytology (BKV) |
| Mononucleosis syndrome in non-immunocompromised individuals | EBV, CMV, HIV | Serology, NAAT, IR [8] |
| HIV, HCV, HBV viral loads | HIV, HCV, HBV | NAAT, RS [7] |
| Viral Genotyping | HCV, HBV, HPV | Nucleotide sequencing, reverse hybridization, NAAT (Cleavase reaction for HPV) |
| HIV, HCV, HBV diagnosis | HIV, HCV, HBV | Serology, NAAT |
| Systemic infections of childhood | Parvovirus B19, measles virus, rubella virus, mumps virus | Serology, NAAT |
| Tropical and emerging infections | Dengue virus, Zika virus, Yellow Fever virus and other flaviviruses; Chikungunya and other alphaviruses; hemorrhagic fever viruses including arenaviruses, bunyaviruses, and filoviruses; Hendra and Nipah viruses | Serology, culture, NAAT (hemorrhagic fever testing is done in BSL-4 laboratories), NMR spectroscopy [16] |
| Unknown virus | Any | Culture, microarray, nucleotide sequencing, NGS, MSF [17] |
BKV, BK virus; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HHV-6, human herpes virus 6; HIV, human immunodeficiency virus; hMPV, human metapneumovirus; HPV, human papillomavirus; HSV, herpes simplex virus; IR, Infrared spectroscopy; JCV, JC virus; MFS, molecular fluorescence spectroscopy; NAAT, nucleic acid amplification testing; NGS, next-generation sequencing; NMR, nuclear magnetic resonance spectroscopy; PIV, parainfluenza virus; RS, Raman spectroscopy; RSV, respiratory syncytial virus; SERS, surface-enhanced Raman spectroscopy; VZV, varicella-zoster virus.
Updated from Storch and Wang [68].
Modern diagnostic virology dates to the first growth of human viruses in tissue culture reported by Weller and Enders in 1948 [69]. This and other landmarks in the history of diagnostic virology are shown in Table 7 . Table 8 highlights 13 relevant studies carried out since 2006, making use of biospectroscopy for virological purposes.
Table 7.
Landmarks in the history of diagnostic virology.
| Year | Landmark |
|---|---|
| 1892 | Intranuclear and intracytoplasmic inclusions noted at the base of smallpox lesions [70] |
| 1898 | Discovery by loeffler and Frosch that foot-and-mouth disease of cattle is caused by a filterable agent, referred to as a virus |
| 1929 | Complement fixation method described for detection of antibodies to smallpox, vaccinia, and varicella-zoster viruses [71] |
| 1948 | First growth of pathogenic human viruses in tissue culture [69] |
| 1949 | Use of spectroscopy with biological perspectives [1] |
| 1956 | Detection of influenza virus in respiratory secretions using fluorescent antibody staining [72], [73] |
| 1975 | Development of monoclonal antibodies as diagnostic reagents [74] |
| 1985 | Discovery of polymerase chain reaction [75] |
| 1992 | Development of real-time PCR [76] |
| 2002 | Beginning of systematic approaches to virus discovery [76], [77], [78] |
Updated from Storch and Wang [68].
Table 8.
Some relevant biospectroscopy studies carried out in the field of virology since 2006.
| Year | Spectroscopic technique | Virus/class of virus studied | Objective |
|---|---|---|---|
| 2006 | Raman and FTIR | Hepatitis C virus | To characterize the structure of the region 5′ untranslated (5′ UTR, 342-mer RNA) of the HCV genome [24]. |
| 2008 | Near-infrared Raman | Hepatitis C virus | To differentiate between healthy human blood serum and human serum with hepatitis C contamination in vitro[5]. |
| Surface-enhanced Raman | RSV | Identification and classification of respiratory syncytial virus (RSV) strains [11]. | |
| Tip-enhanced Raman scattering | Tobacco mosaic virus | To provide spectroscopic vibration information with a spatial resolution of less than 50 nm to characterize unique viruses at the molecular level [25]. | |
| 2010 | Surface-enhanced Raman | Food and Waterborne viruses | To detect and discriminate 7 food and aquatic viruses, including norovirus, adenovirus, parvovirus, rotavirus, coronavirus, paramyxovirus and herpes virus [41]. |
| NMR | Hepatitis C virus | To apply metabonomics to identify patients infected with the hepatitis C virus (HCV) through an analysis of 1H NMR spectra of urine samples [48]. | |
| 2011 | FTIR | Poliovirus | To detect and quantify poliovirus infection in cell culture [46]. |
| 2012 | Near-infrared (NIR) | HIV-1 | To analyze spectroscopic changes caused by the presence of HIV-1 [6]. |
| Near-infrared (NIR) | Influenza virus | To identify nasal fluids contaminated with influenza virus [42]. | |
| 2014 | Raman | Adenovirus | To detect adenovirus-infected cells [12]. |
| 2015 | NMR | Dengue virus | Reveal NS2B membrane topology of the dengue virus [9]. |
| 2016 | NMR | Hepatitis C virus | To assess viral activity and hepatic fibrosis [21]. |
| 2017 | ATR-FTIR | Dengue virus | Identification of Dengue-3 viral load in serum and blood [79]. |
6. Conclusions
Diseases caused by viral infections are one of the biggest problems for global health; and as methods involved in diagnostics are getting faster and more efficient, treatment is the fastest. This retrospective study aimed to explore biospectroscopy applications in studies in the field of virology in order to provide a theoretical support for the techniques used, and to suggest the applying tools that have not been used.
Spectroscopic methods have the characteristic of providing fast results and reliable information related to the composition of the samples. The studies presented herein have shown promising results in a field of science that needs to be better explored. It has been shown that multivariate analysis techniques are of great importance to analyze spectroscopic data, providing the potential to identify and classify biological samples. We hope that with advancement in this field of study, portable spectroscopic devices could be used in clinics and hospitals in the near future, so that the samples could be analyzed in loco for screening or diagnosis strategies.
Acknowledgments
Marfran C.D. Santos and Camilo L.M. Morais would like to thank PPGQ/UFRN and CAPES grant (070/2012) for financial support. Kássio M.G. Lima would like to acknowledge the CNPq grant (305962/2014-0) for financial support.
References
- 1.Blout E.R., Mellors R.C. Infrared spectra of tissues. Science. 1949;110:137–138. doi: 10.1126/science.110.2849.137. [DOI] [PubMed] [Google Scholar]
- 2.Woernley D.L. IR absorption curves for normal and neoplastic tissues and related biological substances. Cancer Res. 1952;12:516–523. [PubMed] [Google Scholar]
- 3.Diem M., Romeo M., Boydston-White S., Miljkovic M., Matthäus C. A decade of vibrational micro-spectroscopy of human cells and tissue (1994–2004) Analyst. 2004;129:880–885. doi: 10.1039/b408952a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell A.L., Gajjar K.B., Theophilou G., Martin F.L., Martin-Hirsch P.L. Vibrational spectroscopy of biofluids for disease screening or diagnosis: translation from the laboratory to a clinical setting. J. Biophotonics. 2014;7:153–165. doi: 10.1002/jbio.201400018. [DOI] [PubMed] [Google Scholar]
- 5.Marques A.S., de Melo M.C.N., Cidral T.A., de Lima K.M.G. Feature selection strategies for identification of Staphylococcus aureus recovered in blood cultures using FT-IR spectroscopy successive projections algorithm for variable selection: a case study. J. Microbiol. Methods. 2014;98:26–30. doi: 10.1016/j.mimet.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Marques A.S., Moraes E.P., Júnior M.A.A., Moura A.D., Neto V.F.A., Neto R.M., Lima K.M.G. Rapid discrimination of Klebsiella pneumonia carbapenemase 2-producing and non-producing Klebsiella pneumoniae strains using near-infrared spectroscopy (NIRS) and multivariate analysis. Talanta. 2015;134:126–131. doi: 10.1016/j.talanta.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Saade J., Pacheco M.T.T., Rodrigues M.R., Silveira L., Jr. Identification of hepatitis C in human blood serum by near-infrared Raman spectroscopy. Spectroscopy. 2008;22:387–395. [Google Scholar]
- 8.Sakudo A., Suganuma Y., Sakima R., Ikuta K. Diagnosis of HIV-1 infection by near-infrared spectroscopy: analysis using molecular clones of various HIV-1 subtypes. Clin. Chim. Acta. 2012;413:467–472. doi: 10.1016/j.cca.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Theophilou G., Lima K.M.G., Martin-Hirsch P.L., Stringfellow H.F., Martin F.L. ATR-FTIR spectroscopy coupled with chemometric analysis discriminates normal, borderline and malignant ovarian tissue: classifying subtypes of human cancer. Analyst. 2016;141:585–594. doi: 10.1039/c5an00939a. [DOI] [PubMed] [Google Scholar]
- 10.Baia T.C., Gama R.A., de Lima L.A.S., Lima K.M.G. FTIR microspectroscopy coupled with variable selection methods for the identification of flunitrazepam in necrophagous flies. Anal. Methods. 2016;8:968–972. [Google Scholar]
- 11.Cann A.J. fourth ed. Elsevier Academic Press; Burlington: 2005. Principles of Molecular Virology. [Google Scholar]
- 12.Drosten C., Göttig S., Schilling S., Asper M., Panning M., Schmitz H., Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and Yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boonham N., Kreuze J., Winter S., van der Vlugt R., Bergervoet J., Tomlinson J., Mumford R. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Moor K., Ohtani K., Myrzakozha D., Zhanserkenova O., Andriana B.B., Sato H. Noninvasive and label-free determination of virus infected cells by Raman spectroscopy. J. Biomed. Opt. 2014;19:067003. doi: 10.1117/1.JBO.19.6.067003. [DOI] [PubMed] [Google Scholar]
- 15.Jin N., Zhang D., Martin F.L. Fingerprinting microbiomes towards screening for microbial antibiotic resistance. Integr. Biol. (Camb.) 2017;9:406–417. doi: 10.1039/c7ib00009j. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Li Q., Wong Y.L., Liew L.S.Y., Kang C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta. 2015;1848:2244–2252. doi: 10.1016/j.bbamem.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Shahzad A., Köhler G., Knapp M., Gaubitzer E., Puchinger M., Edetsberger M. Emerging applications of fluorescence spectroscopy in medical microbiology field. J. Transl. Med. 2009;7:99. doi: 10.1186/1479-5876-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanmukh S., Jones L., Zhao Y.-P., Driskell J.D., Tripp R.A., Dluhy R.A. Identification and classification of respiratory syncytial virus (RSV) strains by surface-enhanced Raman spectroscopy and multivariate statistical techniques. Anal. Bioanal. Chem. 2008;390:1551–1555. doi: 10.1007/s00216-008-1851-0. [DOI] [PubMed] [Google Scholar]
- 19.Amathieu R., Triba M.N., Goossens C., Bouchemal N., Nahon P., Savarin P., Le Moyec L. Nuclear magnetic resonance based metabolomics and liver diseases: recent advances and future clinical applications. World J. Gastroenterol. 2016;22:417–426. doi: 10.3748/wjg.v22.i1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer B., Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Pu S., Sun Y., Li Z., Niu M., Yan X., Zhao Y., Wang L., Qin X., Ma Z., Zhang Y., Li B., Luo S., Gong M., Sun Y., Zou Z., Xiao X. Metabolomic profiling of autoimmune hepatitis: the diagnostic utility of nuclear magnetic resonance spectroscopy. J. Proteome Res. 2014;13:3792–3801. doi: 10.1021/pr500462f. [DOI] [PubMed] [Google Scholar]
- 22.Galal S.M., Aal F.H.A., Mohammed A.E., Mohamed M.Z., El-Rahman Y.G.A. Chronic viral hepatitis C in pediatric age group; assessment of viral activity and hepatic fibrosis by 1H magnetic resonance spectroscopy and diffusion weighted imaging in asymptomatic patient. Egypt. J. Radiol. Nucl. Med. 2016;47:739–748. [Google Scholar]
- 23.Kelly J.G., Trevisan J., Scott A.D., Carmichael P.L., Pollock H.M., Martin-Hirsch P.L., Martin F.L. Biospectroscopy to metabolically profile biomolecular structure: a multistage approach linking computational analysis with biomarkers. J. Proteome Res. 2011;10:1437–1448. doi: 10.1021/pr101067u. [DOI] [PubMed] [Google Scholar]
- 24.Lambert P.J., Whitman A.G., Dyson O.F., Akula S.M. Raman spectroscopy: the gateway into tomorrow's virology. Virol. J. 2006;3:51. doi: 10.1186/1743-422X-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salman A., Shufan E., Zeiri L., Huleihel M. Characterization and detection of Vero cells infected with Herpes Simplex Virus type 1 using Raman spectroscopy and advanced statistical methods. Methods. 2014;68:364–370. doi: 10.1016/j.ymeth.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Butler H.J., Fogarty S.W., Kerns J.G., Martin-Hirsch P.L., Fullwood N.J., Martin Francis L. Gold nanoparticles as a substrate in bio-analytical near-infrared surface-enhanced Raman spectroscopy. Analyst. 2015;140:3090–3097. doi: 10.1039/c4an01899k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogarty S.W., Patel I.I., Martin F.L., Fullwood N.J. Surface-enhanced Raman spectroscopy of the endothelial cell membrane. PLoS One. 2014;9(9):e106283. doi: 10.1371/journal.pone.0106283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Casado A., Bartolomé J., Carreño V., Molina M., Carmona P. Structural characterization of the 5′ untranslated RNA of hepatitis C virus by vibrational spectroscopy. Biophys. Chem. 2006;124:73–79. doi: 10.1016/j.bpc.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Cialla D., Deckert-Gaudig T., Budich C., Laue M., Möller R., Naumann D., Deckert V., Popp J. Raman to the limit: tip-enhanced Raman spectroscopic investigations of a single tobacco mosaic virus. J. Raman Spectrosc. 2009;40:240–243. [Google Scholar]
- 30.Misra A.K., Kamemoto L.E., Hu N., Dykes A.C., Yu Q., Zinin P.V., Sharma S.K. Proc. SPIE 8025, Smart Biomedical and Physiological Sensor Technology VIII. 2011. Micro-Raman spectroscopy study of ALVAC virus infected chicken embryo cells; p. 80250C. [Google Scholar]
- 31.Hermann P., Hermelink A., Lausch V., Holland G., Möller L., Bannert N., Naumann D. Evaluation of tip-enhanced Raman spectroscopy for characterizing different virus strains. Analyst. 2011;136:1148–1152. doi: 10.1039/c0an00531b. [DOI] [PubMed] [Google Scholar]
- 32.Lim J., Nam J., Yang S., Shin H., Jang Y., Bae G.-U., Kang T., Lim K., Choi Y. Identification of newly emerging influenza viruses by surface-enhanced Raman spectroscopy. Anal. Chem. 2015;87:11652–11659. doi: 10.1021/acs.analchem.5b02661. [DOI] [PubMed] [Google Scholar]
- 33.Baker M.J., Trevisan J., Bassan P., Bhargava R., Butler H.J., Dorling K.M., Fielden P.R., Fogarty S.W., Fullwood N.J., Heys K.A., Hughes C., Lasch P., Martin-Hirsch P.L., Obinaju B., Sockalingum G.D., Sulé-Suso J., Strong R.J., Walsh M.J., Wood B.R., Gardner P., Martin F.L. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014;9:1771–1791. doi: 10.1038/nprot.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trevisan J., Angelov P.P., Carmichael P.L., Scott A.D., Martin F.L. Extracting biological information with computational analysis of Fourier-transform infrared (FTIR) biospectroscopy datasets: current practices to future perspectives. Analyst. 2012;137:3202–3215. doi: 10.1039/c2an16300d. [DOI] [PubMed] [Google Scholar]
- 35.Movasaghi Z., Rehman S., ur Rehman I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008;43:134–179. [Google Scholar]
- 36.Sakudo A., Suganuma Y., Kobayashi T., Onodera T., Ikuta K. Near-infrared spectroscopy: promising diagnostic tool for viral infections. Biochem. Biophys. Res. Commun. 2006;341:279–284. doi: 10.1016/j.bbrc.2005.12.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bachmann L., Zezell D.M., Ribeiro A.C., Gomes L., Ito A.S. Fluorescence spectroscopy of biological tissues – a review. Appl. Spectrosc. Rev. 2006;41:575–590. [Google Scholar]
- 38.Butler H.J., Ashton L., Bird B., Cinque G., Curtis K., Dorney J., Esmonde-White K., Fullwood N.J., Gardner B., Martin-Hirsch P.L., Walsh M.J., McAinsh M.R., Stone N., Martin F.L. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016;11:664–687. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 39.Pasquini C. Near infrared spectroscopy: fundamentals, practical aspects and analytical applications. J. Braz. Chem. Soc. 2003;14:198–219. [Google Scholar]
- 40.Manley M. Near-infrared spectroscopy and hyperspectral imaging: non-destructive analysis of biological materials. Chem. Soc. Rev. 2014;43:8200–8214. doi: 10.1039/c4cs00062e. [DOI] [PubMed] [Google Scholar]
- 41.Bassan P., Byrne H.J., Bonnier F., Lee J., Dumas P., Gardner P. Analyst. 2009;134:1586–1593. doi: 10.1039/b904808a. [DOI] [PubMed] [Google Scholar]
- 42.Fearn T., Riccioli C., Garrido-Varo A., Guerrero-Ginel J.E. On the geometry of SNV and MSC. Chemometr. Intell. Lab. Syst. 2009;96:22–26. [Google Scholar]
- 43.de Oliveira L.H., Trindade M.A.G. Baseline-corrected second-order derivative electroanalysis combined with ultrasound-assisted liquid–liquid microextraction: simultaneous quantification of fluoroquinolones at low levels. Anal. Chem. 2016;88:6554–6562. doi: 10.1021/acs.analchem.6b01379. [DOI] [PubMed] [Google Scholar]
- 44.Bassan P., Kohler A., Martens H., Lee J., Jackson E., Lockyer N., Dumas P., Brown M., Clarke N., Gardner P. RMieS-EMSC correction for infrared spectra of biological cells: extension using full Mie theory and GPU computing. J. Biophotonics. 2010;3:609–620. doi: 10.1002/jbio.201000036. [DOI] [PubMed] [Google Scholar]
- 45.Li N., Li X.-Y., Zou Z.-X., Lin L.-R., Li Y.-Q. A novel baseline-correction method for standard addition based derivative spectra and its application to quantitative analysis of benzo(a)pyrene in vegetable oil samples. Analyst. 2011;136:2802–2810. doi: 10.1039/c0an00751j. [DOI] [PubMed] [Google Scholar]
- 46.Hibbert D.B. Vocabulary of concepts and terms in chemometrics (IUPAC Recommendations 2016) Pure Appl. Chem. 2016;88:407–443. [Google Scholar]
- 47.Martin F.L., Kelly J.G., Llabjani V., Martin-Hirsch P.L., Patel I.I., Trevisan J., Fullwood N.J., Walsh M.J. Distinguishing cell types or populations based on the computational analysis of their infrared spectra. Nat. Protoc. 2010;5:1748–1760. doi: 10.1038/nprot.2010.133. [DOI] [PubMed] [Google Scholar]
- 48.Geladi P., Kowalski B.R. Partial least-squares regression: a tutorial. Anal. Chim. Acta. 1986;185:1–17. [Google Scholar]
- 49.Bro R., Smilde A.K. Principal component analysis. Anal. Methods. 2014;6:2812–2831. [Google Scholar]
- 50.Martin F.L., German M.J., Wit E., Fearn T., Ragavan N., Pollock H.M. Identifying variables responsible for clustering in discriminant analysis of data from infrared microspectroscopy of a biological sample. J. Comput. Biol. 2007;14:1176–1184. doi: 10.1089/cmb.2007.0057. [DOI] [PubMed] [Google Scholar]
- 51.Fan C., Hu Z., Riley L.K., Purdy G.A., Mustapha A., Lin M. Detecting food- and waterborne viruses by surface-enhanced Raman spectroscopy. J. Food Sci. 2010;75:M302–M307. doi: 10.1111/j.1750-3841.2010.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakudo A., Baba K., Ikuta K. Discrimination of influenza virus-infected nasal fluids by Vis-NIR spectroscopy. Clin. Chim. Acta. 2012;414:130–134. doi: 10.1016/j.cca.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 53.Moor K., Kitamura H., Hashimoto K., Sawa M., Andriana B.B., Ohtani K., Yagura T., Sato H. Proc. SPIE 8587, Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XI. 2013. Study of virus by Raman spectroscopy; p. 85871X-1. [Google Scholar]
- 54.Wood B.R., Bambery K.R., Dixon M.W.A., Tilley L., Nasse M.J., Mattson E., Hirschmugl C.J. Diagnosing malaria infected cells at the single cell level using focal plane array Fourier transform infrared imaging spectroscopy. Analyst. 2014;139:4769–4774. doi: 10.1039/c4an00989d. [DOI] [PubMed] [Google Scholar]
- 55.Wang L., Mizaikoff B. Application of multivariate data-analysis techniques to biomedical diagnostics based on mid-infrared spectroscopy. Anal. Bioanal. Chem. 2008;391:1641–1654. doi: 10.1007/s00216-008-1989-9. [DOI] [PubMed] [Google Scholar]
- 56.Ward J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. [Google Scholar]
- 57.Brereton R.G., Lloyd G.R. Partial least squares discriminant analysis: taking the magic away. J. Chemometrics. 2014;28:213–225. [Google Scholar]
- 58.Lee-Montiel F.T., Reynolds K.A., Riley M.R. Detection and quantification of poliovirus infection using FTIR spectroscopy and cell culture. J. Biol. Eng. 2011;5:16. doi: 10.1186/1754-1611-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petisco C., Garcia-Criado B., Zabalgogeazcoa I., Vázquez-de-Aldana B.R., Garcia-Ciudad A. A spectroscopy approach to the study of virus infection in the endophytic fungus Epichloë festucae. Virol. J. 2011;8:286. doi: 10.1186/1743-422X-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixon S.J., Brereton R.G. Comparison of performance of five common classifiers represented as boundary methods: Euclidean distance to centroids, linear discriminant analysis, quadratic discriminant analysis, learning vector quantization and support vector machines, as dependent on data structure. Chemometr. Intell. Lab. Syst. 2009;95:1–17. [Google Scholar]
- 61.Godoy M.M.G., Lopes E.P.A., Silva R.O., Hallwass F., Koury L.C.A., Moura I.M., Gonçalvez S.M.C., Simas A.M. Hepatitis C virus infection diagnosis using metabonomics. J. Viral Hepat. 2010;17:854–858. doi: 10.1111/j.1365-2893.2009.01252.x. [DOI] [PubMed] [Google Scholar]
- 62.Gajjar K., Trevisan J., Owens G., Keating P.J., Wood N.J., Stringfellow H.F., Martin-Hirsch P.L., Martin F.L. Fourier-transform infrared spectroscopy coupled with a classification machine for the analysis of blood plasma or serum: a novel diagnostic approach for ovarian cancer. Analyst. 2013;138:3917–3926. doi: 10.1039/c3an36654e. [DOI] [PubMed] [Google Scholar]
- 63.Galvão R.K.H., Pimentel M.F., Araujo M.C.U., Yoneyama T., Visani V. Aspects of the successive projections algorithm for variable selection in multivariate calibration applied to plasma emission spectrometry. Anal. Chim. Acta. 2001;443:107–115. [Google Scholar]
- 64.Araújo M.C.U., Saldanha T.C.B., Galvão R.K.H., Yoneyama T., Chame H.C., Visani V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemometr. Intell. Lab. Syst. 2001;57:65–73. [Google Scholar]
- 65.Pontes M.J.C., Galvão R.K.H., Araújo M.C.U., Moreira P.N.T., Neto O.D.P., José G.E., Saldanha T.C.B. The successive projections algorithm for spectral variable selection in classification problems. Chemometr. Intell. Lab. Syst. 2005;78:11–18. [Google Scholar]
- 66.Broadhurst D., Goodacre R., Jones A., Rowland J.J., Kell D.B. Genetic algorithms as a method for variable selection in multiple linear regression and partial least squares regression, with applications to pyrolysis mass spectrometry. Anal. Chim. Acta. 1997;348:71–86. [Google Scholar]
- 67.Siqueira L.F.S., Lima K.M.G. MIR-biospectroscopy coupled with chemometrics in cancer studies. Analyst. 2016;141:4833–4847. doi: 10.1039/c6an01247g. [DOI] [PubMed] [Google Scholar]
- 68.Storch G.A., Wang D. In: Fields Virology. sixth ed. Knipe D.M., Howley P.M., editors. 2013. Diagnostic virology. [Google Scholar]
- 69.Weller R.H., Enders J.F. Production of hemagglutinin by mumps and influenza A viruses in suspended cell tissue cultures. Proc. Soc. Exp. Biol. Med. 1948;69:124–128. doi: 10.3181/00379727-69-16638. [DOI] [PubMed] [Google Scholar]
- 70.Guarnieri G. Recherche sulla Pathogenesi ed Etiologia dell'infezione vaccinica e variolosa. Arch. Sci. Med. 1892;16:403–423. [Google Scholar]
- 71.Bedson S., Bland J. Complement-fixation with filterable viruses and their antisera. Br. J. Exp. Pathol. 1929;10:393–404. [Google Scholar]
- 72.Gardner P.S., McQuillin J. Application of Immunofluorescence. second ed. Butterworths; London: 1980. Rapid virus diagnosis. [Google Scholar]
- 73.Liu C. Rapid diagnosis of human influenza. Proc. Soc. Exp. Biol. Med. 1956;92:883–887. doi: 10.3181/00379727-92-22642. [DOI] [PubMed] [Google Scholar]
- 74.Kohler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 75.Saiki R.K., Scharf S., Faloona F., Mullis K.B., Horn G.T., Erlich H.A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 76.Higuchi R., Dollinger G., Walsh P.S., Griffith R. Simultaneous amplification and detection of specific DNA sequence. Biotechnology (NY) 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 77.Nagel M.A., Cohrs R.J., Mahalingam R., Wellish M.C., Forghani B., Schiller A., Safdieh J.E., Kamenkovich E., Ostrow L.W., Levy M., Greenberg B., Russman A.N., Katzan I., Gardner C.J., Häusler M., Nau R., Saraya T., Wada H., Goto H., de Martino M., Ueno M., Brown W.D., Terborg C., Gilden D.H. The varicella zoster virus vasculopathies: clinical, CSF, imaging, and virologic features. Neurology. 2008;70:853–860. doi: 10.1212/01.wnl.0000304747.38502.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber G., Shendure J., Tanenbaum D.M., Church G.M., Meyerson M. Identification of foreign gene sequences by transcript filtering against the human genome. Nat. Genet. 2002;30:141–142. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- 79.Santos M.C.D., Nascimento Y.M., Araújo J.M.G., Lima K.M.G. ATR-FTIR spectroscopy coupled with multivariate analysis techniques for the identification of DENV-3 in different concentrations in blood and serum: a new approach. RSC Adv. 2017;7:25640–25649. [Google Scholar]