Abstract
High-level protein expression is an important means of obtaining large amounts of viral proteins to investigate further their biological properties. To express the membrane (M) protein of SARS–CoV at high-level in vitro, the M gene fragment was amplified and cloned it into the Pichia Pastoris expression vector pPICZαA. SDS–PAGE and Western blotting analysis of the induced products of recombinant yeast transformant indicated that successful high-level expression of M protein was achieved, and that the expression product was similar antigenically to the natural protein. Purified recombinant M protein was used subsequently as an ELISA antigen for detection of eight serum samples screened previously by whole virus ELISA and immunofluorescence assay, and consistent results were obtained. These findings suggest that the recombinant M protein may be useful as a diagnostic reagent.
Keywords: SARS coronavirus, Recombinant M protein, Antigenicity
1. Introduction
Severe acute respiratory syndrome (SARS), also called atypical pneumonia, is a human severe respiratory infectious disease that emerged recently in Asia, North America and Europe. The transmission of SARS occurs mainly by direct contact, and its incubation period is 2–7 days. Infection is usually characterized by high fever, which is followed a few days, later by a dry non-productive cough and shortness of breath, and may progress to generalized interstitial infiltrates in the lung. Death from progressive respiratory failure occurs in between 3–10% of cases (Rota et al., 2003). At present, no effective therapy or vaccine is available.
A novel coronavirus, named as SARS–CoV, has been identified as the main causative agent of SARS. The virions are 80–140 nm in diameter, with 20–40 nm complex surface projections surrounding the periphery. Hemagglutinin esterase type glycoprotein projections were not seen (Ksiazek et al., 2003). The genome of SARS–CoV is a single-stranded, positive-sense polyadenylated RNA molecule of approximately 29,727 nucleotides. The genomic organization is typical of coronaviruses, having the characteristic gene order [5′-replicase (rep), spike (S), envelope (E), membrane (M), and nucleocapsid (N)-3′] and short untranslated regions at both termini. The SARS–CoV rep gene is predicted to encode two polyproteins that undergo co-translational proteolytic processing. There are four open reading frames (ORF) downstream of rep that are predicted to encode the four structural proteins S, E, M and N, which are common to all known coronavirus. Additionally, SARS–CoV also encodes several non-structural proteins (Rota et al., 2003).
The clinical case definition of SARS is essentially one of fever and pneumonia, with or without a contact history. There are many causes of pneumonia and in the absence of a definite history of contact with other patients with SARS, therefore, it is difficult to differentiate SARS from other causes of pneumonia in clinic diagnosis. Laboratory tests that can confirm a diagnosis of SARS–CoV infection early in the course of the illness are, therefore, a critical clinical need (Riley et al., 2003). Since the outbreak of SARS in 2003, several laboratory diagnostic methods have been established, including real-time RT-PCR assay, whole-virus-based immunofluorescence assay (IFA), recombinant protein-based enzyme-linked immunosorbent assay (ELISA) and immunochromatographic tests, antigen-capturing enzyme-linked immunosorbent assay, and Western blot (WB) assay. These methods were critically important for controlling the transmission of SARS; furthermore, many new laboratory diagnostic methods are being developed (Poon et al., 2003, Poon et al., 2004, Shi et al., 2003, Guan et al., 2004, He et al., 2004).
The M protein is an important structural protein of the coronavirus. In the cases of other known coronaviruses, it is the most abundant among the structural proteins and can induce antibody-dependent complement-mediated virus neutralization (Woods et al., 1987). In addition, the M protein is also involved in the assembly and budding of virions together with the E protein. (Rottier, 1995, de Haan et al., 2000, Molekamp and Spaan, 1997, Narayanan and Makino, 2001, Corse and Machamaer, 2000, Vennema et al., 1996). The M protein contains highly conserved glycosylated sequences, and its glycosylation may be related to the interaction between virus and host (de Haan et al., 2002, de Haan et al., 1998). The important role of the M protein in the life cycle of coronaviruses and inducing immunity make it an attractive target for anti-SARS drug research, vaccine development and the establishment of a serological detection assay. Here, we report the expression of recombinant M protein in P. Pastoris and its antigenicity.
2. Materials and methods
2.1. Gene and vector
The genomic cDNA of SARS–CoV strain BJ01 was provided by Huada Gene Company. The P. Pastoris expression vector pPICZαA and yeast host strain GS115 were all purchased from Invitrogen company.
2.2. Human sera
Sera from healthy people and SARS patients were collected from Peking Union Medical College Hospital between April and June 2003. Sample # 1–4 were from healthy people, sample # 5–8 were from four SARS patients (age 28–40; two males, two females) during the acute phase of infection, which were collected at day 31, 26, 22, 19 post SARS onset, respectively. All four SARS patients had a history of contact with other patients infected with SARS, and exhibited symptoms including persistent fever (>38.0 °C), cough and shortness of breath for several days before they were hospitalized. Patients were subsequently confirmed to be infected with SARS by clinical diagnosis combined with laboratory diagnostic methods.
2.3. PCR amplification and sequence analysis of M gene fragment
According to the published genomic sequence of SARS–CoV strain TOR (Poutanen et al., 2003), a pair of primers were designed and synthesized. The sequence of the primers were 5′-GAGCCGCGGCCGCTCAATGTGGTCATTC-3′(forward primer) and 5′-CGTTCTAGATGTACTAGCAAAGCAATA-3′ (reverse primer), which carried a SacII and XbaI restriction site, respectively.
The primers were used to amplify the M gene fragment without its transmembrane domain from the genomic cDNA of SARS–CoV strain BJ-01. The PCR reaction contained 5 μl of cDNA, 25 pmol of each of two primers, 5 μl of buffer concentrate, 4 μl of dNTP and 0.5 μl of Ex Taq enzyme (TaKaRa, Japan), and sterile distilled water up to 50 μl. PCR was performed with the following settings: 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 50 °C for 45 s and 72 °C for 1 min, and ending with 72 °C for 10 min.
The amplified products were then sent to the TaKaRa Biotechnology (Dalian) Co. Ltd. for sequencing. The determined sequence was analyzed further by DNAMAN biological software.
2.4. Construction of the expression plasmid
The M gene fragment and expression vector pPICZαA were digested by SacII and XbaI, respectively. The larger fragments were purified by QIAquick Gel Extraction kit (Qiagen, Germany). The concentrations of purified products were measured by Beckman DV-600 (Beckman, U.S.A). After mixing in a 3:1 molecular ratio, the purified products were ligated by T4 DNA ligase at 16 °C overnight and then transformed into Escherichia coli Top10 competent cells. Transformants selected on low-salt LB plates containing Zeocin (25 μg/ml) were screened by direct colony PCR, and by restriction digestion of purified plasmids. The sequence of the inserts were verified. The sequence data obtained were compared with the sequence of the M gene of SARS–CoV strain BJ01 (accession number AY278488).
2.5. Expression and purification of recombinant protein
The transformation of yeast cells and screening of transformants, as well as expression in P. Pastoris, were performed as described previously (David and James, 1998). The recombinant plasmid was linearized by the restriction enzyme SacI, the purified products were transformed into yeast cells GS115 and transformants were selected on low-salt MD plates containing Zeocin (25 μg/ml). The colonies of recombinant yeast cells with high copies of the M gene were screened on low-salt MD plates containing different concentrations of Zeocin (500 μg/ml, 1 and 2 mg/ml). A screened colony of yeast cells was selected and inoculated into 100 ml of BMMY in the presence of Zeocin (25 μg/ml). The culture was grown at 28–30 °C until the optical density at 600 nm (OD600) reached 4.0. The culture was then centrifuged at 4 °C at 3000 × g for 5 min, the cells were re-suspended in 1/10 prime volume of MMY, and continued to grow at 28–30 °C until the optical density at 600 nm (OD600) reached 2–6. Methanol was added every 24 h to a final concentration of 0.5% to induce the expression of recombinant M protein, and the incubation was continued for further 3–4 days. Control cultures were processed in parallel. The recombinant M protein was secreted into the culture supernatant as soluble form. The supernatant was harvested by centrifugation and used for analysing the expression level of recombinant M protein by SDS–PAGE.
The highly expressing Pichia colony was selected for inoculation into 500 ml of BMMY, and grown at 28–30 °C until the culture reached an OD600 = 4.0. The culture was centrifuged at 4 °C at 3000 × g for 5 min, the cells were re-suspended in 1/10 prime volume of MMY, and continued to grow at 28–30 °C until the optical density at 600 nm (OD600) reached 2–6. Glycerol and methanol were added every 24 h to final concentrations of 0.5 and 0.8%, respectively, to induce the expression of recombinant M protein, and the incubation was continued for a further 3–4 days. The supernatant of culture was harvested by centrifugation at 8000 × g for 30 min at 4 °C, then ammonium sulphate was added to a final saturation of 70% to precipitate proteins. The pellet was collected by centrifugation at 8000 × g for 1 h at 4 °C and dialyzed to 2000 ml of PBS overnight. The dialysis buffer was replaced every 2 h until it was confirmed that the ammonium sulphate was completely removed. The prepared sample was loaded onto an NTA column equilibrated with NiSO4. This process was repeated three times. The loaded column was first washed with a 10-fold column volume of MCAC0 (20 mM Tris, pH 8.0; 500 mM NaCl) to remove the contaminant proteins, and then eluted with 10 ml of 200 mM imidazole in MCAC0. The different elution parts were analyzed by SDS–PAGE, and the part containing recombinant M protein was concentrated by superfiltration.
2.6. Western blotting
Two microgram of the purified recombinant M protein was separated by SDS–PAGE and transferred to polyvinylidene difluoride membrane (PVDF) by standard methods. After blocking with PBST (14 mmol/l NaCl, 2.7 mmol/l KCl, 10 mmol/l NaHPO4, 1.8 mmol/l KH2PO4 and 0.02% Tween-20) containing 5% non-fat milk for 1 h at 4 °C, the membrane was incubated for 3 h at room temperature with SARS–CoV-positive human serum (1:3000 dilution). After washing three times in an appropriate volume of PBST, the membrane was incubated for 1 h with peroxidase labeled goat anti-human IgG (1:1000, No: A-0170, Sigma). The membrane was washed three times with PBS-T and once with distilled water, for 10 min each time. Finally, the washed membrane was transferred into an appropriate volume of ECL solution (Amersham) and reacted at room temperature for about 1 min, then put onto a sensitization film for 1–3 min to develop and fix.
2.7. ELISA
The purified recombinant M protein was diluted with 50 mmol/l carbonate buffer (15 mmol/l NaCO3, 35 mmol/l NaHCO3, [pH 9.6]), and then used to coat 96-well plates (Costar Inc.) (1 μg/well) overnight at 4 °C. After washing with PBS-T (PBS containing 0.5% Tween-20, pH 7.4) for three times, each well of the plate was incubated with blocking solution (3% BSA in carbonate buffer) for 2 h at 37 °C. The wells were washed three times with phosphate-buffered saline (PBS) containing 0.05% Tween-20 (PBS-T). Hundred microlitres of the primary antibody (SARS patient sera), diluted 1:800 in PBS-T, was added in duplicate and the plates were incubated for 1 h at 37 °C. After four washes with PBS-T, plates were then incubated with goat anti-human IgG antibody labeled with peroxidase (1:50,000, No.: A-0170, Sigma) for 1 h at 37 °C. TMB [3,3′, 5,5′-tetramethylbenzidine] substrate (100 μl/well) was added after seven washes with PBS-T and the wells were incubated for 10 min at 37 °C. Fifty microlitres of stop buffer (2 mol/l H2SO4) was added to each well and the optical density (OD) was read at 450 nm in Bio-RAD550. The cut-off value was defined as the mean OD plus three standard deviations calculated from the four negative samples used as control.
3. Results
3.1. Amplification and sequence analysis of M gene
The M gene fragment without transmembrane domain was amplified from the genomic cDNA of SARS–CoV strain BJ01. Sequence analysis indicated that the nucleotide sequence of that amplified fragment is completely identical to that of SARS–CoV strain BJ01 (accession number AY278488). The deduced amino acid sequence of the M protein of SARS–CoV strain BJ01 has identities of 41.0% or less with the M proteins of other human or animal coronaviruses (Fig. 1 ).
Fig. 1.
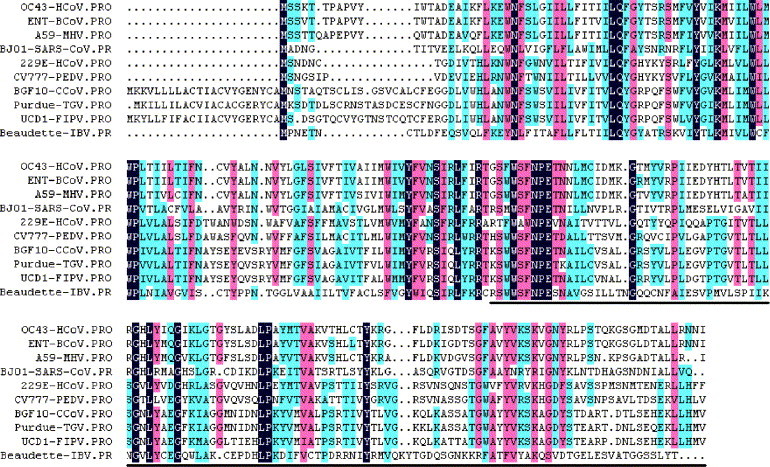
Deduced amino acid sequence alignment for the M proteins of selected coronavirus isolates. The sequence alignment was generated by DNAMAN sequence analysis software, and the region of amplified M gene fragment was indicated on the bottom line. GenBank accession numbers for M protein sequences of selected isolated are as follow: BJ01-SARS–CoV (SARS coronavirus, strain BJ01), AAP30034; 229E-HcoV (human coronavirus, strain 229E), NP_073555; OC43-HcoV (human coronavirus, strain OC43), NP_937953; Beaudette-IBV (avian infectious bronchitis virus, strain Beaudette), NP_040835; A59-MHV (murine hepatitis virus, strain MHV-A59), NP_045301; BGF10-CcoV (canine coronavirus, strain BGF10), AAQ17224; CV777-PEDV (porcine epidemic diarrhea virus, strain CV777), NP_598313; ENT-BcoV (bovine coronavirus, isolate BCoV-ENT), NP_150082; Purdue-TGV (transmissible gastroenteritis virus, strain Purdue), NP_058427; UCD1-FIPV (feline infectious peritonitis virus, strain UCD1), BAC01156.
3.2. Construction of the expression plasmid
The amplified product of 348 bp was cloned in the pPICZαA expression vector, and the sequences of inserts were verified by sequencing (data not shown). The nucleotide sequence alignment performed by DNAMAN software revealed 100% nucleotide identity between the inserted sequence and that of SARS–CoV BJ01.
3.3. Expression and purification of recombinant protein
The M gene fragment was cloned in pPICZαA expression vector and transformed into yeast host strain GS115. The colonies of recombinant yeast cells with high copies of M gene were screened. After inducing with methanol, the supernatant of culture was harvested by centrifugation. The recombinant protein with a polyhistidine tag at the C-terminus was produced in soluble form in the culture supernatant (Fig. 2 ).
Fig. 2.
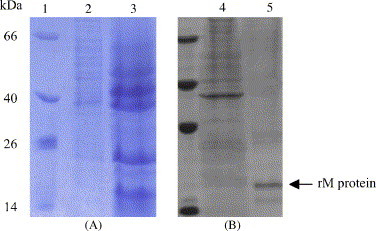
SDS–PAGE analysis of expression of M protein in yeast (A) and purity of recombinant M protein after purified by affinity chromatography (B). Lane 1: molecular weight marker. Lane 2: supernatant of the induced culture of recombinant yeast without insert of M gene fragment. Lane 3: supernatant of the induced culture of recombinant yeast containing insert of M gene fragment. Lane 4: flow-through from nickel affinity column. Lane 5: recombinant M protein eluted from the column.
Twenty microlitres of the supernatant (20 μg of protein) was subjected to SDS–PAGE and Coomassie blue staining, and a protein band corresponding to the expected molecular mass of 14 kDa was revealed in supernatant, which was not present either in the culture before the induction or in the control culture before and after the induction.
The recombinant M protein was purified by nickel affinity chromatography and the purity was confirmed by single banding in SDS–PAGE (Fig. 2). Upon elution from the NTA column, the protein was >80% pure as estimated from SDS–PAGE. Typically, 6 mg of pure recombinant M protein is obtained per liter of P. pastoris culture.
3.4. Western blotting
In order to test the reactivity of the recombinant M protein, Western blotting was carried out using SARS–CoV-positive human serum. The protein band of 14 kDa showed a strong and specific reaction with the SARS–CoV-positive serum, whereas the supernatant of the induced culture of recombinant yeast without the insert of the M gene fragment did not react with the SARS–CoV-positive serum (Fig. 3 ). These results confirmed the authenticity of the recombinant M protein.
Fig. 3.
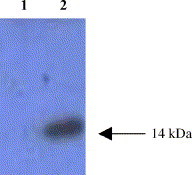
Western blotting of the purified recombinant M protein. Lane 1: supernatant of the induced culture of recombinant yeast without insert of M gene fragment. Lane 2: purified recombinant M protein.
3.5. ELISA
To test whether the recombinant M protein is effective as an ELISA antigen for detecting SARS–CoV patient serum, the sera from four healthy people and four SARS patients were used. The mean and the standard deviation obtained using four SARS–CoV-negative sera were 0.106 and 0.003, respectively. Therefore, the cut-off value of ODs was determined as 0.206. Sera from four SARS patients were determined to be positive using only the M protein as an ELISA antigen. The mean and standard deviation were 0.610 and 0.004, respectively (see Fig. 4 for detailed data).
Fig. 4.
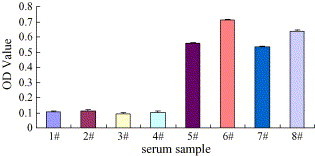
Detection results of eight human sera by ELISA using purified recombinant M protein as antigen.# 1–4: sera from four healthy people, respectively, # 5–8: sera from four SARS patients, respectively.
The same sera from four SARS patients were also positive by whole SARS–CoV ELISA in parallel, using a diagnostic kit for antibodies to coronavirus (ELISA) obtained from Beijing BGI-GBI Biotech Co., Ltd., China (unpublished results).
4. Discussion
Coronaviruses are important pathogens in human and animals, where they cause mostly respiratory and enteric diseases. SARS–CoV is a newly identified human coronavirus that differs from all previously known human coronaviruses (for example, HCOV-229E and HCOV-OC43) in both the speed of transmission and the severity of disease. Both the lack of understanding and effective methods to control the virus means that the threat of SARS still exists, and therefore, there is an urgent need to develop effective vaccines and anti-viral drugs. The key role of the M protein in coronavirus assembly, budding and inducing virus neutralization make it an attractive target for the development of vaccines and drugs, as well as for effective diagnostic reagents. SARS–CoV differs from other coronaviruses in nucleotide sequence, and the exact function of its M protein is unknown. Since SARS–CoV is a pathogen with high infectivity and mortality, special facilities are required when the live virus is used to prepare viral proteins given the high risk of infection. Therefore, it is important to obtain large amounts of M protein by expression in vitro.
The M protein is a structural glycoprotein, and the glycosylation modification is important for its activity. A previous report indicated that the recombinant M protein, when expressed in inclusion body form in E. coli, reacted weakly with human SARS sera (Yi et al., 2003). P. Pastoris is a eukaryotic expression system with high efficiency, and proteins expressed in this system can get appropriate post-translational modifications, including glycosylation. Therefore, the P. Pastoris system is more suitable for expression of the M protein in vitro than E. coli (Grinner and Tschopp, 1989, Kukuruzinska et al., 1987, Romanos et al., 1992). However, we cannot rule out the possibility that the glycosylation sites of the SARS–CoV M gene in P. Pastoris, while suitable for detection of antibodies, may be quite different from those present in the virus.
We expressed successfully the M protein of SARS–CoV strain BJ01 in P. Pastoris, and the expressed M protein was similar to the native protein in its immune response. This point can be demonstrated by the reaction of recombinant M protein with SARS-positive sera in Western blotting and ELISA. The availability of large amounts of recombinant M protein offers a better chance to study its biological characteristics and its role in SARS immunity during infection.
Due to the lack of an effective vaccine or special therapeutic drugs, the control of SARS infection relies mainly on early detection of the virus or its antibodies, and rigid quarantine measures. At present, well-established laboratory diagnostic methods for SARS include real-time PCR, whole-virus-based indirect immunofluorescence assay and ELISA. In these assays, however, the viral nucleic acid and antigens needed were commonly prepared from live pathogen, so there is a strong associated risk of individual exposure to this pathogen. Compared with these methods, preparing viral antigens by expression in vitro is both easier and safer. Among the proteins encoded by the SARS coronavirus, the M protein can induce an immunological response and thus may be used for detection of SARS–CoV antibodies. ELISA with the expressed recombinant M protein of canine coronavirus has been reported, and was demonstrated to be a valid and effective detection method (Elia et al., 2003). In order to evaluate whether our expressed recombinant M protein could be applied as a diagnostic antigen for SARS, we used it as an ELISA antigen to detect eight collected SARS-positive and SARS-negative human sera. The results were in complete accordance with those of other assays, thus indicating that the recombinant M protein may be useful as an ELISA antigen for detecting specific antibodies to SARS–CoV in human sera. Furthermore, the recombinant M protein offers several advantages over the cell-prepared SARS–CoV antigen.
In summary, the P. Pichia system is a viable tool to express the M protein at high-levels in vitro. The availability of large amounts of SARS–CoV recombinant M protein offers an opportunity to better investigate the biological properties of the M protein during SARS–CoV infection as well as its immunological role, and thus provide a basis for development of SARS–CoV vaccine and therapeutic drugs. The recombinant M protein expressed in P. Pastoris may have better antigenicity than that expressed in E. coli, and is more suitable for use as a diagnostic antigen.
Acknowledgements
We are grateful to Hai Pang from Tsinghua University for technical assistance, and to Gewei Lian and Hongkui Deng from Peking University for providing us with sera and for help with ELISA experiments. We also thank Hui Wang from Oxford University for comments and critical reading. This work was supported by Project 973 & 863 of the Ministry of Science and Technology of China (Grant no. GZ236(202/9): “Structural proteomics of SARS coronavirus”).
References
- Corse E., Machamaer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R.H., James M.C. Humana Press Inc.; New Jersey: 1998. Pichia Protocols. [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., de Wit M., Kuo L., Montalto C., Masters P.S., Weiss S.R., Rottier P.J. O-glycosylation of the mouse hepatitis coronavirus membrane protein. Virus Res. 2002;82:77–81. doi: 10.1016/S0168-1702(01)00390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia G., Fiermonte G., Pratelli A., Martella V., Camero M., Cirone F., Buonavoglia C. Recombinant M protein-based ELISA test for detection of antibodies to canine coronavirus. J. Virol. Methods. 2003;109:139–142. doi: 10.1016/S0166-0934(03)00064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinner L.S., Tschopp J.F. Size distribution and general structural features of N-linked oligosaccharides from the methyltrophic yeast Picha pastoris. Yeast. 1989;5:107–115. doi: 10.1002/yea.320050206. [DOI] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Foo S.Y., Tan Y.J., Goh P.Y., Wee S.H. Recombinant protein-based enzyme-linked imunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diag. Lab. Immunol. 2004;11:287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Chong K.H., Chng H.H., Leung B., Ling A.E., Wei T., Chan S.W., Ooi E., Kwang J. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin. Diag. Lab. Immunol. 2004;11:417–422. doi: 10.1128/CDLI.11.2.417-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska M.A., Bergh M.L., Jackson B.J. Protein glycosylation in yeast. Annu. Rev. Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Molekamp R., Spaan W.J. Identification of a specific interaction between the mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75:9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chan K.H., Wong O.K., Yam W.C., Yuen K.Y., Guan Y., Lo Y.M., Peiris J.S. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J. Clin. Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chan K.H., Wong O.K., Cheung T.K., Ng I., Zheng B., Seto W.H., Yuen K.U., Guan Y., Peiris J.S. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.M., Lam T.H., Thach T.Q., Chau P., Chan K.P., Lo S.V., Leung P.Y., Tsang T., Ho W., Lee K.H., Lau E.M., Ferguson N.M., Anderson R.M. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Romanos M.A., Scorer C.A., Clare J.J. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- Rota, P.A., Oberste, M.S., Monroe, S.S., Nix, W.A., Campagnoli, R., Icenogle, J.P., Peneranda, S., Bankamp, B., Maher, K., Chen, M.H., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J.L., Chen, Q., Wang, D., Erdman, D.D., Peret, T.C., Burns, C., Kziazek, T.G., Rollin, P.E., Sanchez, A., Liffick, S., Holloway, B., Limor, J., McCaustland, K., Olsen-Rasmussen, M., Fouchier, R., Gunther, S., Osterhaus, A.D., Drosten, C., Pallansch, M.A., Anderson, L.J. and Bellini, W.J., 2003. Characterization of a novel coronavirus associated with severe acute respiratory science. Science 300, 1394–1399. [DOI] [PubMed]
- Rottier P.J.M. The coronavirus membrane protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 115–139. [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godecke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R.D., Wesley R., Kapke P.A. Complement-dependent neutralization of transmissible gastroenteritis virus by monoclonal antibodies. Adv. Exp. Med. Biol. 1987;218:493–500. doi: 10.1007/978-1-4684-1280-2_64. [DOI] [PubMed] [Google Scholar]
- Yi Y., Li C., Shi Y., Li L., Li P., Huang W., Wang S., Ma Q., Cao C. Over-expression in Escherichia coli and purification of nucleocapsid and membrane protein of SARS coronavirus. Chin. J. Biotechnol. 2003;19:392–396. [PubMed] [Google Scholar]


