Abstract
Monoclonal antibodies (Mabs) against the Urbani strain of the SARS-associated coronavirus (SARS-CoV) were developed and characterized for reactivity to SARS-CoV and SARS-CoV S, N, M, and E proteins using enzyme-linked immunoabsorbent (ELISA), radioimmunoprecipitation, immunofluorescence, Western Blot and microneutralization assays. Twenty-six mAbs were reactive to SARS-CoV by ELISA, and nine were chosen for detailed characterization. Five mAbs reacted against the S protein, two against the M protein, and one each against the N and E proteins. Two of five S protein mAbs neutralized SARS-CoV infection of Vero E6 cells and reacted to an epitope within amino acids 490–510 in the S protein. While two of the three non-neutralizing antibodies recognized at second epitope within amino acids 270–350. The mAbs characterized should prove useful for developing SARS-CoV diagnostic assays and for studying the biology of infection and pathogenesis of disease.
Keywords: SARS-coronavirus, Monoclonal antibody, Immunoassay, Epitope, Neutralizing
1. Introduction
Coronaviruses (CoVs) are large, enveloped, positive-stranded RNA viruses that cause a variety of illnesses in humans and animals (Lai, 1990, Lavi et al., 1999, Perlman, 1998, Snijder and Horzinek, 1993, Snijder et al., 1993, Zhou et al., 2004). Most human coronaviruses (HCoVs) fall into one of two serotypes, OC43-like and 229E-like; however, the global outbreak of severe acute respiratory syndrome (SARS) was quickly linked to infection with a novel CoV, SARS-CoV (Ksiazek et al., 2003, Peiris et al., 2003), and HCoV-NL63, has been recognized recently as a human pathogen (van der Hoek et al., 2004). The CoV genome encodes numerous non-structural proteins and four or five structural proteins including the spike (S), nucleocapsid (N), membrane (M), small envelope (E), and (in some) strains a hemagglutinin-esterase (HE) protein (Lai, 1990). SARS-CoV has four structural proteins, the S, N, M, and E proteins that have various functions. The S protein forms spikes on the virion surface and is crucial for viral attachment and entry into the host cell. It also induces protective immunity, and is associated with host range, tissue tropism and virulence (Sanchez et al., 1999). The N protein forms the nucleocapsid; the M protein interacts with the nucleocapsid and forms the internal viral core; and the E protein is associated with the viral envelope. SARS-CoV is genetically distinct from previously described coronaviruses, which have been placed into three antigenic groups: I, II, and III. Human coronaviruses, i.e., 229E-like and OC43-like, belong to groups I and II, respectively, and are recognized as the second most common cause of upper respiratory disease, but are associated infrequently with serious lower respiratory tract disease (El-Sahly et al., 2000, Hendley et al., 1972, Makela et al., 1998, Falsey et al., 2002). However, SARS-CoV is usually associated with serious lower respiratory tract disease, having a fatality rate as ranging between 10% and 15% that may be as high as 50% in patients >60 years of age (Drosten et al., 2003, Enserink, 2003, Holmes, 2003, Ksiazek et al., 2003, Poutanen et al., 2003, Rota et al., 2003). Although the global spread of SARS-CoV was stopped in June 2003, six instances of laboratory-acquired infections have been confirmed and a cluster of sporadic cases have been detected in Guangdong Province, China, between December 2003 and April 2004 (Liang et al., 2004), demonstrating the potential for SARS to re-emerge and possibly become pandemic.
Anticipating the need for improved immunological reagents to aid identification and characterization of SARS-CoV, monoclonal antibodies (mAbs) to SARS-CoV proteins were produced. This report describes nine such mAbs that include antibodies reactive against each of the four structural proteins, including two S protein reactive antibodies that neutralize SARS-CoV.
2. Materials and methods
2.1. Biosafety
All work with live SARS-CoV was done in biosafety level 3 (BSL-3) containment laboratories at the Centers for Disease Control and Prevention, Atlanta, Georgia. SARS-CoV was inactivated by 60Co gamma irradiation at 2 × 106 rad prior to its use as an immunogen or as an antigen in an ELISA. Within the limits of detecting viable virus, 2 × 106 rads gamma irradiation was sufficient to inactivate all infectivity.
2.2. Virus preparation
Vero E6 cells were maintained in Dulbecco's minimal essential media (DMEM, Invitrogen Corp., Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine sera (FBS, Hyclone, Logan, UT) and 2 mM l-glutamine (Invitrogen). The Urbani strain of SARS-CoV was plaque-purified, grown to stock titers in Vero E6 cells, purified by polyethylene glycol (PEG) precipitation as described previously (Kiley et al., 1980), and frozen at −70 °C until use. Viral antigen used for ELISA was prepared by detergent extraction of SARS-CoV-infected Vero E6 cells and subsequent gamma irradiation (Ksiazek et al., 2003).
2.3. B cell hybridoma production
Immunizations were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee. Female, 4–6-week-old, specific pathogen-free BALB/c mice (Jackson Laboratories, Bar Harbor, ME) were intraperitoneally (i.p.) administered 200 μl of PEG-purified SARS-CoV inactivated by gamma-irradiation and diluted in PBS, followed by two similar immunizations 3 and 6 weeks later. Mice were euthanized by exsanguination under Avertin (2,2,2-tribromoethanol) anesthesia 3 days after the last immunization, and their spleens were removed.
Splenocytes were fused with non-secretor SP2/0 myeloma cells at a 2:1 ratio using 50% polyethylene glycol 1000 (PEG 1000, Sigma Chemical Company, St. Louis, MO) and cultured in 24-well plates as described previously (Reimer et al., 1984). B cell hybridomas were selected using DMEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, and 1× sodium hypoxanthine, aminopterin, and thymidine (HAT; Invitrogen), and culture fluids were screened for reactivity against a SARS-CoV infected Vero E6 cell lysate and against a similarly prepared uninfected Vero E6 cell lysate by ELISA. The mean optical density (OD) of the mAbs reactive to SARS-CoV infected Vero E6 cell lysate (positive control, P) was divided by the mean OD of mAbs reactive to a similarly prepared uninfected Vero E6 cell lysate (negative control, N) and were termed P/N ratios. B cell hybridoma cells from wells that gave positive-to-negative (P/N) ratios ≥3 were cloned by limiting dilution. Hybridomas cell lines were grown in DMEM supplemented with 10% FBS.
Monoclonal immunoglobulin class and isotype were determined by use of the IsoStrip mouse mAb isotyping kit (Roche Diagnostics Corporation, Indianapolis, IN) and confirmed for isotype by ELISA (Amersham Biosciences, Piscataway, NJ) as described by the manufacturer.
2.4. Recombinant proteins
2.4.1. Recombinant SARS-CoV N protein
Escherichia coli BL21 cells (Invitrogen) were transformed with an inducible pET vector expressing 6× histidine-tagged SARS-CoV N protein (Invitrogen), and expressed 6× histidine-tagged SARS-CoV N protein was purified by metal-chelate chromatography (ProBond resin; Invitrogen) following the manufacturer's recommended protocols. The purified 6× histidine-tagged SARS-CoV N protein was eluted in lysis buffer (100 mM sodium phosphate monobasic, 10 mM Tris–HCl, 300 mM NaCl in PBS containing EDTA-free proteinase inhibitor (pH 8.0) adjusted to pH 4.5. The eluate was immediately adjusted to pH 7.5 and dialyzed against PBS. The purified N protein was tested by ELISA for reactivity to hyperimmune mouse sera and anti-SARS N mAb.
2.4.2. Synthetic SARS S protein fragments
The full-length soluble portion of the SARS-CoV S protein (amino acids 1–1190; S1190) and S protein fragments containing amino acids 1–269, 1–350, 1–490, 1–510 and 270–510 (S269, S350, S490, S510, S270–510) were synthesized, expressed, and purified as described previously (Babcock et al., 2004). Protein fragments were analyzed by Coomassie staining and by Western Blot using human SARS-convalescent serum or mouse anti-synthetic S protein for detection. The convalescent serum recognized all the S protein fragments with the exception of S269, while the anti-synthetic S protein recognized all the S protein fragments.
2.5. Construction of replicons expressing SARS-CoV S, M, N, and E proteins
RNA was extracted from a preparation of the Urbani strain of SARS-CoV in Trizol LS reagent (Invitrogen) following the manufacturer's protocol. Reverse transcription reactions for generation of SARS-CoV cDNA were performed with a Super Script III kit (Invitrogen) using random hexamers. The SARS-CoV cDNA was PCR-amplified with the gene-specific primer pairs using Platinum Taq Hifi (Invitrogen). Following 18 cycles of amplification, a sample of each reaction was analyzed by gel electrophoresis, and fragments of the appropriate size were noted for each of the four genes (data not shown). The gene PCR products were sequenced and the expected sequences were confirmed. The SARS-CoV M, N, and E gene PCR products were cloned directly into the venezuelan equine encephalitis (VEE) replicon vector using EcoRV and AscI restriction sites. The SARS-CoV S gene PCR product was directly cloned into the VEE replicon vector using AscI and PmeI restriction sites. RNA was in vitro transcribed from each SARS gene replicon DNA using a RiboMax T7 RNA transcription kit (Promega) following the manufacturer's protocol.
2.6. Venezuelan equine encephalitis (VEE) replicons expressing SARS-CoV S, M, N, and E proteins
Vero cells were electroporated with replicon RNA expressing either SARS M, N, S or E genes. After electroporation, the cells were incubated in media containing OptiPRO media (Invitrogen) for 16 h at 37 °C. Electroporated cells were mixed at a 2.3:1 ratio with non-electroporated cells and 50 μl of the cell mixture was placed on each spot of a 12-well spot slide (VWR International, Buffalo Grove, IL). The slides were incubated at 37 °C for 3.5 h, washed with PBS, and then fixed with acetone and methanol (1:1) for 5 min at room temperature. The fixed monolayers were analyzed in an indirect immunofluorescence assay (IFA), using SARS convalescent human plasma as the primary antibody and Alexa Fluor 488 chicken anti-human IgG (H + L) (Molecular Probes, Eugene, OR) as the secondary antibody. Cells electroporated with the VEE replicon RNAs containing the SARS-CoV S, M, N, and E genes were intensely fluorescent in these assays (data not shown).
2.7. IFA
IFA was performed on 2% paraformaldehyde-fixed Vero E6 cells infected with 1000 TCID50 SARS-CoV for 27 h, mock-infected Vero E6 or Vero E6 cell lines expressing SARS-CoV S, N, M or E as described previously (Harcourt et al., 2004).
2.8. ELISA
An indirect ELISA using detergent-extracted SARS-CoV inactivated by gamma-irradiation or similarly prepared Vero E6 cell lysate was used to determine the mAb reactivity as described previously (Ksiazek et al., 2003). A similar ELISA protocol was used to detect mAb reactivity to recombinant SARS-CoV N protein, untransformed E. coli BL21 cell lysate control, S1190 protein, or angiotensin-converting enzyme 2 antigen control.
2.9. Radioimmunoprecipitation
SARS-CoV-infected Vero E6 cells or mock-infected Vero E6 cells (2.5 × 105) were radiolabeled with [35S]-methionine/cysteine (0.1 mCi/ml; ICN, Irvine, California) and reacted with individual mAbs to determine antigen specificity as described previously (Benaroch et al., 1995). Briefly, radiolabelled cells were collected by centrifugation and lysed in lysis buffer (0.1% SDS; 1.0% TritionX-100; 1% NaDOC, 150 mM NaCl, 10 mM Tris–HCl (pH 7.2) and 5 mM EDTA) containing 1 mM PMSF and 100 mM Pefablock (Boehringer Mannheim, Mannheim, Germany). Antigen-antibody complexes were precipitated using Protein G sepharose beads (Amersham Biosciences, Uppsala, Sweden), and the bound radiolabeled immunoprecipitate was analyzed by 12.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
2.10. MAb epitope mapping
Immunoprecipitation and Western Blot analysis were employed to determine the epitopes recognized by the anti-S protein mAbs. HEK-293 cells were transiently transfected with DNA encoding various S protein fragments (S269, S510, S490, S350, S270–510, S1190), and culture supernatants were harvested 48 h post-transfection. Equal amounts of S protein-containing supernatants were incubated with respective anti-S protein mAbs and protein G-Sepharose for 2 h at room temperature while shaking. The beads were washed in PBS, and protein was eluted in Laemmli sample buffer by boiling the sample for 5 min. Samples were resolved by 10% SDS-PAGE and transferred to an Immobilon P membrane (Millipore, Billerica, MA). Membranes were washed, using 1% non-fat dried milk in PBS–Tween, and incubated with anti-His6 mAb (Invitrogen). Blots were washed twice in 1% non-fat dried milk in PBS–0.05% Tween 20, incubated with horseradish peroxidase-conjugated (HRP) anti-mouse IgG (Jackson Laboratories, Bar Harbor, ME) for 45 min at room temperature, and washed, and bands were detected by use of an enhanced chemiluminescence reagent (Amersham) and exposed to X-Omat-AR film for various times. For Western blotting, 1 ml of each S protein fragment supernatant was incubated with Ni-NTA agarose (Invitrogen) for 2 h at room temperature. Resin was washed with PBS, mixed with reducing Laemmli buffer, and separated by 10% SDS-PAGE. The proteins were transferred to Immobilon P (Millipore); the membranes were blocked using 5% non-fat dried milk in PBS–Tween overnight, and incubated for 1 h with respective anti-S protein mAbs. The blots were washed and incubated with HRP-conjugated anti-mouse IgG. After being washed, the bands were detected as described above (Babcock et al., 2004).
2.11. Virus microneutralization assay
The mAbs were tested for their ability to neutralize SARS-CoV infection of Vero E6 cells by microneutralization assay as described (Sui et al., 2004). The microneutralization titer of test antibody was the reciprocal of the highest dilution of test antibody that showed inhibition in all triplicate wells. Controls were included for each microneutralization assay performed and included back titration, inclusion of positive control antibody (i.e., serum from a convalescent SARS patient) and human serum negative for SARS-CoV-specific antibody.
3. Results
3.1. Characterization of mAbs to SARS-CoV
Twenty-six B cell hybridoma cell lines were made that produced mAbs reactive to SARS-CoV by ELISA. On the basis of ELISA reactivity of the mAbs reactive for SARS-CoV-infected Vero E6 cell lysate (positive control, P) and a similarly prepared uninfected Vero E6 cell lysate (negative control, N), nine B cell hybridomas with P/N ratios ≥3 were subcloned and expanded, and the mAbs were characterized further. Four of the nine mAbs were of the subclass IgM, four were of subclass IgG2a and one was of subclass IgG1 (Table 1 ).
Table 1.
MAbs to SARS-CoV
| Subclone | Isotype | IFA-specificitya | ELISA-SARS-CoV reactiveb | ELISA-recombinant S protein reactivec | ELISA-recombinant N protein reactived | Neutralizing titere |
|---|---|---|---|---|---|---|
| 19C | IgM | M | 8.2 | ≤1.3 | ≤1 | <1:10 |
| 42C | IgM | N | 9.2 | ≤1.3 | 2 | <1:10 |
| 154C | IgM | S | 9.5 | 7.0 | ≤1 | <1:10 |
| 240C | IgG2a | S | 13.2 | 34.9 | ≤1 | <1:10 |
| 283C | IgG1 | M | 3.7 | ≤1.3 | ≤1 | <1:10 |
| 341C | IgG2a | S | 13.8 | 24.2 | ≤1 | 1:1280 |
| 472C | IgM | E | 4.3 | ≤1.3 | ≤1 | <1:10 |
| 534C | IgG2a | S | 6.9 | 22.5 | ≤1 | 1:80 |
| 560C | IgG2a | S | 18.7 | 17.5 | ≤1 | <1:10 |
MAb specificity shown here was determined by IFA reactivity using Vero cells expressing SARS-CoV S, N, M, or E proteins.
Hybridoma supernatants were tested at a 1:80 dilutions for reactivity against SARS-CoV infected cell lysate and uninfected Vero E6 cell lysate by ELISA. The positive-to-negative (P/N) ratios are indicated.
Hybridoma supernatants were tested at a 1:80 dilution for reactivity against the full-length, soluble S protein (aa 1–1190), and a similarly prepared ACE-2 protein control by ELISA. The positive-to-negative (P/N) ratios are indicated.
Hybridoma supernatants were tested at a 1:40 dilution for reactivity against recombinant N protein, and a similarly prepared E. coli BL21 control antigen by ELISA. The positive-to-negative (P/N) ratios are indicated.
Neutralizing antibody titers were measured as the reciprocal of the highest antibody dilution that completely inhibited Vero E6 cell lysis in triplicate wells according to Sui et al. (2004).
3.2. Specificity of the mAbs
The specificities of the nine mAbs were determined by immunofluorescence antibody staining of transfected Vero cells expressing SARS-CoV E, M, N or S proteins. The results showed that two mAbs reacted against the M protein, five reacted against the S protein, one reacted weakly against the E protein, and one reacted weakly against N protein. A representative IFA using an S protein-specific mAb (mAb 341) and M protein-specific mAb (mAb 292) reactive to SARS-CoV infected Vero E6 cells is shown in Fig. 1 . As expected, all the mAbs examined reacted strongly with SARS-CoV-infected Vero E6 cells but not mock-infected cells.
Fig. 1.
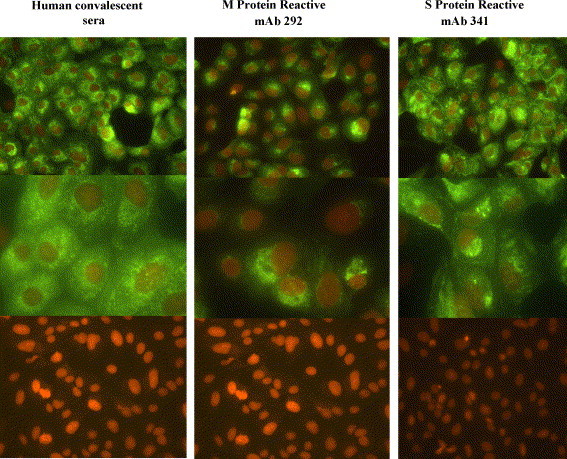
Immunofluorescence staining (IFA) of SARS-CoV infected Vero E6 cells. A representative IFA is shown for an S protein-specific mAb (mAb 341) and M protein-specific mAb (mAb 292). Cell staining was visualized at 40× and 100× using a Zeiss Axioskop microscope with an Axiovert BlueH 485 nm filter and an RT Color Spot digital camera.
Radioimmunoprecipitation analysis confirmed mAbs reactive to the S (mAb 341, 534, 560, 240), M (mAb 283), and N (mAb 42) proteins (Fig. 2 ). In addition, anti-N protein mAb specificity was confirmed by ELISA using recombinant N protein, and by Western Blot, using PEG-purified SARS-CoV-inactivated by gamma-irradiation (data not shown), and anti-S protein mAb specificity was confirmed by reactivity against recombinant S protein by ELISA and by Western Blot analysis, using inactivated SARS-CoV, full-length soluble S protein, and S protein fragments.
Fig. 2.
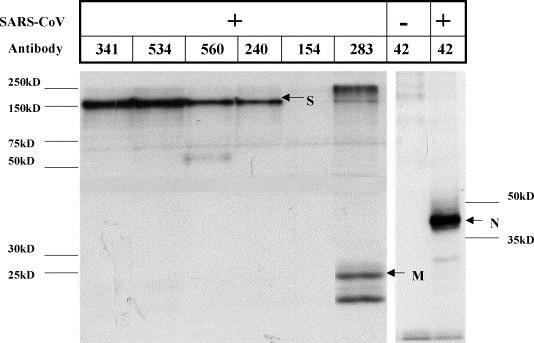
Radioimmunoprecipitation of mAbs with SARS-CoV lysate. SARS-CoV-infected Vero E6 cells (+) or mock-infected Vero E6 cells (−) were radiolabeled with [35S]-methionine/cysteine and reacted with individual mAbs to determine antigen specificity as described previously in Section 2. The anti-SARS-CoV S (S) mAbs (341, 534, 560, and 240) detected a protein with an apparent molecular mass of approximately 150 kDa, while the anti-matrix (M, 283) and anti-nucleocapsid (N, 42) mAbs detected proteins with an apparent molecular mass of approximately 25 kDa and 45 kDa, respectively. No bands were detected in the uninfected Vero E6 lysate control. The lack of signal with mAb 154 is likely due to the differential binding properties of protein G.
The S protein fragments contained amino acids (aa) 1–269, 1–350, 1–490, 1–510, 1–1190 and 270–510 (Fig. 3 ). The development and characterization of these fragments has been described previously (Babcock et al., 2004). The reactivity patterns against the S protein fragments indicated the region recognized by these antibodies. S protein mAbs 154C and 560C reacted to fragments S350, S490, S510, S1190, and S270–510 indicating these antibodies recognize an epitope within amino acids 270–350. Monoclonal antibodies 240C, 341C, 534C reacted to S protein fragments S510, S1190, and S270–510, suggesting that these antibodies recognized a second epitope within amino acids 490–510 (Table 2 ). A representative Western Blot for a neutralizing anti-S protein monoclonal, 341C (Fig. 4a), and a non-neutralizing anti-S protein monoclonal, 560C (Fig. 4b), are shown.
Fig. 3.
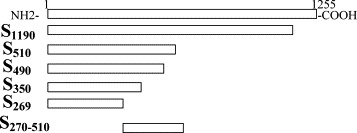
Schematic diagram of the various S glycoprotein fragments synthesized. The top box represents the whole length of SARS-CoV S glycoprotein (aa 1–1255) and the relative sizes of the selected S protein fragments are shown.
Table 2.
Epitopes recognized by anti-SARS-CoV S MAbs
| Subclone | Isotype | Neutralizing titer | S1190a | S510a | S490a | S350a | S270–510a | S269a | Epitopeb |
|---|---|---|---|---|---|---|---|---|---|
| 154C | IgM | <1:10 | + | + | + | + | + | − | 270–350 |
| 240C | IgG2a | <1:10 | + | + | − | − | + | − | 490–510 |
| 341C | IgG2a | 1:1280 | + | + | − | − | + | − | 490–510 |
| 534C | IgG2a | 1:80 | + | + | − | − | + | − | 490–510 |
| 560C | IgG2a | <1:10 | + | + | + | + | + | − | 270–350 |
The epitopes recognized by the anti-S mAbs were determined by Western Blot with C-terminally truncated S proteins covering the region between amino acids (aa) 269 and 1190 of the S glycoprotein. S1190, aa 1–1190; S510, aa 1–510; S490, aa 1–490; S350, aa 1–350; S270–510, aa 270–510; S269, aa 1–269; (+): positive reaction; (−): negative reaction. The Western Blot results were confirmed by immunoprecipitation.
Based on monoclonal reactivity, the target linear epitopes within the S protein were predicted.
Fig. 4.
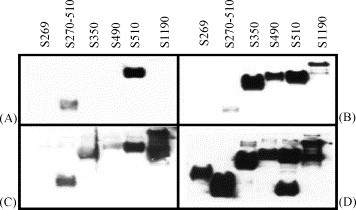
Epitope mapping of neutralizing and non-neutralizing anti-S mAbs. The epitopes recognized by the anti-S protein mAbs were determined by Western Blot with C-terminally truncated S proteins covering the region between amino acids 269 and 1190 of the S protein. S1190, aa 1–1190; S510, aa 1–510; S490, aa 1–490; S350, aa 1–350; S270–510, aa 270–510; S269, aa 1–269. A representative Western Blot for a neutralizing anti-S protein monoclonal, 341C (A), a non-neutralizing anti-S protein monoclonal, 560C (B), SARS convalescent-phase sera (C) and anti-(His)6 antibody (D) are shown.
3.3. Neutralization of SARS-CoV
Five of the nine mAbs were reactive to the S protein, and mAbs 341C and 534C neutralized SARS-CoV infection of Vero E6 cells. Of note, both neutralizing antibodies were reactive to the same epitope (i.e., aa 490–510), while non-neutralizing mAbs, (with the exception of antibody 240C), recognized a second epitope, i.e., aa 270–350 (Table 2). The reactivity patterns combined with two functional assays, virus neutralization and receptor blocking results (data not shown), were consistent with described previously sites on the S protein involved in binding to ACE-2 and in virus neutralization.
4. Discussion
The mAbs described in this study will facilitate detection of the structural proteins of SARS-CoV, provide a means to diagnose infection, and aid in the study of the biology of SARS-CoV infection and disease pathogenesis. The ability to detect specific proteins has proven useful for assessing protein-specific antibody responses and for developing diagnostic assays. On the basis of results from studies of other coronavirus infections in animals (Daniel and Talbot, 1990, De Diego et al., 1992, Ignjatovic and Galli, 1993, Kusters et al., 1989, Saif, 1993) and on the serum antibody response of patients with SARS (Che et al., 2003), the antibody response to SARS-CoV appears to be dominated by anti-N protein and anti-S protein antibodies. Thus, mAbs reactive to N or S proteins can be used in antibody capture assays and provide a key reagent for high-quality IgM and IgA antibody assays. In addition, the anti-S protein mAbs can be used to study virus neutralization, and to better understand the function of the two epitopes against which they react.
There is a well-recognized need for mAbs reactive to SARS-CoV proteins. A recent study described the first neutralizing murine mAbs reactive to SARS-CoV (Berry et al., 2004). In that study, two of five mAbs reactive to S protein neutralized SARS-CoV infection of Vero cells, one mAb reacted to N protein, and eleven mAbs had undetermined specificity (Berry et al., 2004). In addition, three groups have described neutralizing human mAbs reactive against SARS-CoV (Sui et al., 2004, Traggiai et al., 2004, Zhou et al., 2004). In this study, the authors show the availability of murine mAbs reactive to the four structural proteins of SARS-CoV and two S protein epitopes that fill an important gap in immunological tools needed to further explore the biology of SARS-CoV infection and pathogenesis of disease. The mAbs should help the development of diagnostic assays and studies examining the biology of infection and pathogenesis of disease.
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroch P., Yilla M., Raposo G., Ito K., Miwa K., Geuze H.J., Ploegh H.L. MHC class II molecules reach the endocytic pathway. EMBO J. 1995;14:37–49. doi: 10.1002/j.1460-2075.1995.tb06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J., Nicolas B., Andonova M., Ranada F., Gubbins M.J., Blake Ball T., Kitching P., Li Y., Kabani A., Plummer F. Development and characterization of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120:87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Hao W., Qiu L.W., Pan Y.X., Liao Z.Y., Xu H., Chen J.J., Hou J.L., Woo P.C., Lau S.K., Kwok Y.Y., Huang Z. Antibody response of patients with severe acute respiratory syndrome (SARS) to nucleocapsid antigen of SARS-associated coronavirus. Di Yi Jun Yi Da Xue Xue Bao. 2003;23:637–639. [PubMed] [Google Scholar]
- Daniel C., Talbot P.J. Protection from lethal coronavirus infection by affinity-purified spike glycoprotein of murine hepatitis virus, strain A59. Virology. 1990;174:87–94. doi: 10.1016/0042-6822(90)90057-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego M., Laviada M.D., Enjuanes L., Escribano J.M. Epitope specificity of protective lactogenic immunity against swine transmissible gastroenteritis virus. J. Virol. 1992;66:6502–6508. doi: 10.1128/jvi.66.11.6502-6508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink M. Infectious diseases. Calling all coronavirologists. Science. 2003;300:413–414. doi: 10.1126/science.300.5618.413. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Walsh E.E., Hayden F.G. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J. Infect. Dis. 2002;185:1338–1341. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Smith C., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendley J.O., Fishburne H.B., Gwaltney J.M., Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- Holmes K.V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Galli L. Structural proteins of avian infectious bronchitis virus: role in immunity and protection. Adv. Exp. Med. Biol. 1993;342:449–453. doi: 10.1007/978-1-4615-2996-5_71. [DOI] [PubMed] [Google Scholar]
- Kiley M.P., Regnery R.L., Johnson K.M. Ebola virus: identification of virion structural proteins. J. Gen. Virol. 1980;49:333–341. doi: 10.1099/0022-1317-49-2-333. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kusters J.G., Jager E.J., Lenstra J.A., Koch G., Posthumus W.P., Meloen R.H., van der Zeijst B.A. Analysis of an immunodominant region of infectious bronchitis virus. J. Immunol. 1989;143:2692–2698. [PubMed] [Google Scholar]
- Lai M.M. Coronavirus: organization, replication and expression of genome. Ann. Rev. Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lavi E., Schwartz T., Jin Y.P., Fu L. Nidovirus infections: experimental model systems of human neurologic diseases. J. Neuropathol. Exp. Neurol. 1999;58:1197–1206. doi: 10.1097/00005072-199912000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Chen Q., Xu J., Liu Y., Lim W., Peiris J.S., Anderson L.J., Ruan L., Li H., Kan B., Di B., Cheng P., Chan K.H., Erdman D.D., Gu S., Yan X., Liang W., Zhou D., Haynes L., Duan S., Zhang X., Zheng H., Gao Y., Tong S., Li D., Fang L., Qin P., Xu W., SARS Diagnosis Working Group Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province. Chin. Emerg. Infect. Dis. 2004;10:1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimaki M., Blomqvist S., Hyypia T., Arstila P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. Pathogenesis of coronavirus-induced infections. Review of pathological and immunological aspects. Adv. Exp. Med. Biol. 1998;440:503–513. [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Reimer C.B., Phillips D.J., Aloisio C.H., Moore D.D., Galland G.G., Wells T.W., Black C.M., McDougal J.S. Evaluation of thirty-one mouse mAbs to human IgG epitopes. Hybridoma. 1984;3:263–275. doi: 10.1089/hyb.1984.3.263. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Coronavirus immunogens. Vet. Microbiol. 1993;37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C., Spaan W.J. The coronaviruslike superfamily. Adv. Exp. Med. Biol. 1993;342:235–244. doi: 10.1007/978-1-4615-2996-5_37. [DOI] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthew L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;24:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human mAbs from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyre K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wothers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang H., Luo D., Rowe T., Wang Z., Hogan R.J., Qiu S., Bunzel R.J., Huang G., Mishra V., Voss T.G., Kimberly, Luo R.M. An exposed domain in severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 2004;78:7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


