Highlights
► The diagnostic sensitivity and specificity of the HCoV-OC43 truncated N immunoassay were 90.9% (10/11) and 82.9% (39/47), respectively. ► The assay demonstrated 100% diagnosis agreement with the previous testing methods. ► This assay offers may be a useful tool in coronavirus seroepidemiological studies.
Keywords: Coronavirus, Immunoassay, Nucleocapsid, Serology, HCoV-OC43
Abstract
Human coronaviruses are one of the main causes of upper respiratory tract infections in humans. While more often responsible for mild illness, they have been associated with illnesses that require hospitalization. In this study, an assay for one of the human coronaviruses, OC43, was developed using a truncated recombinant nucleocapsid (N) protein antigen in an enzyme immunosorbent assay (ELISA) and evaluated using serum collected from HCoV-OC43-infected patients, healthy adults, and patients with other respiratory virus infections. Results showed that the diagnostic sensitivity and specificity of the assay were 90.9% (10/11) and 82.9% (39/47), respectively. To evaluate the clinical utility of the ELISA, serum samples collected from patients during an outbreak of HCoV-OC43 infection and previously identified as positive by HCoV-OC43 whole N ELISA were screened resulting in 100% diagnosis agreement between the testing methods. These results suggest that this assay offers a reliable method to detect HCoV-OC43 infection and may be a useful tool in coronavirus seroepidemiological studies.
1. Introduction
Human coronaviruses (HCoVs) were first identified in the 1960s (Hamre et al., 1967), but with the worldwide outbreak of severe acute respiratory syndrome (SARS) in 2003 and the subsequent isolation of new HCoV family members (van der Hoek et al., 2004, Woo et al., 2005), considerable attention has recently been focused on understanding their molecular diversity, prevalence, and clinical impact. Sequence analyses have classified the four identified HCoVs into two groups with group 1b containing HCoV-229E and HCoV-NL63 and group2a containing HCoV-OC43 and HCoV-HKU1 (Woo et al., 2009). Seroprevalence estimates for HCoVs range from 5 to 30% of all respiratory infections (Woo et al., 2005). Though infections are primarily associated with mild respiratory symptoms, HCoV-OC43 and other coronaviruses have been linked to severe infections, neuroinvasive disease, and even fatalities in vulnerable subpopulations such as the elderly, prompting further interest in HCoV-OC43 epidemiology (Arbour et al., 2000, Patrick et al., 2006, Rota et al., 2003). However, epidemiologic surveys and large scale surveillance studies have been limited by the sensitivity and specificity of current diagnostic approaches (Lehmann et al., 2008). HCoV infection can induce antibodies that cross react among the HCoVs making it difficult to estimate prevalence of infection for the different HCoV viruses without doing neutralization assays. Development of more specific enzyme immunoassays (EIAs) for the HCoV strains would help to clarify strain-specific patterns of infection. A highly specific, rapid diagnostic assay would significantly improve surveillance and the investigation of HCoV-OC43 distribution.
Coronaviruses are made up of a single positive-strand RNA genome encoding four major structural proteins, spike (S), nucleocapsid (N), membrane (M), and envelope (E). The nucleocapsid protein has been recognized as an important target in the development of HCoV diagnostics because it reliably induces a good antibody response (Mourez et al., 2007). However, the N protein has sequences that are conserved among HCoVs and, therefore, antibodies detected may have been induced by other HCoVs resulting in a false positive results for the HCoV strains of interest (Chan et al., 2009, Mourez et al., 2007, Severance et al., 2008). Analysis of the aa sequence of HCoV N proteins suggests that parts of the protein could provide a more specific antigen for EIAs. To investigate this possibility, serologic assays using truncated recombinant HCoV-OC43 N proteins were developed and evaluated in indirect enzyme-linked immunosorbant assay (ELISA) against a serum samples from HCoV-OC43 (group 2a), HCoV-229E (group 1b), and SARS-CoV (group 2b) infected patients. Each antigen was assessed for its diagnostic accuracy, optimal cut-off, and sensitivity and specificity. One of the truncated OC43 N proteins gave minimal cross-reactivity and allowed for serological differentiation of past HCoV-OC43 infection from other CoV infections and to estimate seroprevalence of HCoV-OC43 in 162 healthy adults.
2. Materials and methods
2.1. Serum specimens
Acute and convalescent phase sera from HCoV-229E (n = 17 acute; n = 10 convalescent) and HCoV-OC43 (n = 15 acute; n = 11 convalescent) patients were kindly provided by Dr. Ann Falsey (University of Rochester School of Medicine). These sera exhibited at least fourfold rise in HCoV-229E and HCoV-OC43 antibody titers when tested by whole virus EIA (A. Falsey, personal communication). Serum samples were collected from 20 patients with laboratory-confirmed SARS-CoV infection and 162 healthy adult donors with no exposure to SARS-CoV. An additional 19 acute (n = 8) and convalescent (n = 11) specimens collected from 12 patients (n = 10 HCoV-OC43 positive; n = 2 HCoV-OC43 negative) during a respiratory outbreak in a Wisconsin nursing home in the Fall of 2008 with HCoV-OC43 infection status determined by real-time PCR (Wisconsin State Laboratory of Hygiene and CDC) and/or serology testing (CDC). Specimens used in this study were exempt from CDC Institutional Review Board review under 45 CFR 46.101 (b) (4). Polyclonal mouse sera against HCoV-HKU1 and HCoV-NL63 was collected from 10 to 12 week old mice subcutaneously (s.c.) immunized with 50 ug/ml of recombinant HCoV-HKU1 N or HCoV-NL63 N proteins emulsified 1:1 with TiterMax Gold (Sigma) the hind limb. Mice were boosted 2–3 weeks post-immunization using the same dose of antigen in Dulbecco's phosphate buffered saline by intraperitoneal administration. Two weeks after the final boost, animals were euthanized and blood was collected. Antibody titers were determined by screening serum samples against the immunizing antigen. All studies were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee. All serum samples were kept in aliquots at <−20 °C until use.
2.2. Nucleocapsid protein sequence homology among CoVs
The protein sequence of HCoV-OC43 N protein (GenBank ID: AY585228) was compared by National Center for Biotechnical Information (NCBI) pair wise BLAST to the N protein sequence for group 1 HCoVs 229E (GenBank ID: DQ243939) and NL63 (GenBank ID: AY567487), and group 2 HCoV HKU1 (GenBank ID: DQ778921) as well as SARS-CoV (GenBank ID: AY310120). Sequences corresponding to the truncated HCoV-OC43 N proteins were also aligned with the whole N protein sequences of other CoVs.
2.3. Expression and purification of truncated recombinant nucleocapsid proteins
HCoV-OC43 RNA was extracted from the lysate of infected HRT18 cells (gift from D. Erdman, CDC) using QIAMP™ Viral RNA Kit (Qiagen, Valencia, CA). Truncated HCoV-OC43 N genes were amplified by reverse transcriptase (RT)-polymerase chain reaction (PCR) with primers designed from previously reported nucleotide sequence of HCoV-OC43 N gene (GenBank ID: NC_005147). The oligonucleotide primers for the truncated HCoV-OC43 N1 (aa1-119), HCoV-OC43 N2 (aa120-332), and HCoV-OC43 N3 (aa333-448) were OC43 N1 forward primer 5′-GGATCCCATATGTCTTTTACTCCTGGTAAGC-3′, and reverse primer 5′-CTCGAATTCTTATTATTGACGCTGGTTGCCATCGGC-3′, HCoV-OC43 N2 forward primer 5′-GGATCCCATATGCTGCTGCCACGATGGTATTTTTAC-3′ and reverse primer 5′-CTCGAATTCTTATTATAATCT TGATCCAAAGAAAAACGC-3′, HCoV-OC43 N3 forward primer 5′-GGATCCCATATGGAGTTGGCCAAAGTGCAGAAT-3′, and reverse primer 5′-CTCGAATTCTTATTATAT TTCTGAGGTGTCTTCAGT-3′. The amplified genes were cloned into a pET-28 vector encoding a C-terminal polyhistidine (His)6 tag (gift from Dr. Xin Zhang, Emory University). Sequence identity of expression constructs was confirmed by sequencing. The pET-28 constructs were transformed into Escherichia coli strain BL21 (DE3) cells (Invitrogen, Carlsbad, CA), and the expression of recombinant proteins was induced by addition of isopropyl-l-thio-d-galactopyranoside (IPTG). The recombinant proteins were purified by metal affinity chromatography (QIAGEN, Valencia, CA), separated (2 μg/well)on a 4% stacking, 20% resolving Criterion® gradient gel (BioRad Laboratories, Hercules, CA) and analyzed by Coomassie Blue stain. The control antigen, purified protein from E. coli containing pET-28 vector without the N gene, was expressed as described above. The whole HCoV-OC43 N protein was expressed and purified as described above. Purified protein was analyzed by SDS-PAGE and Western blot and the expected protein size (∼55 kD) and reactivity was confirmed (data not shown).
2.4. Western blot analysis
Purified His-tagged recombinant nucleocapsid proteins (0.25 μg/well) were separated by SDS-PAGE (4–20%) (BioRad Laboratories, Hercules, CA) and transferred onto PVDF membranes (Amersham Biosciences). PageRuler™ Plus (Fermentas, Glen Burnie, Maryland) was used as a molecular weight standard. After transfer, membranes were incubated for 1 h at room temperature in blocking buffer containing 20 mM Tris 137 mM NaCl, 0.1% Tween-20 (TBS-T) and 3% bovine serum albumin (BSA). The membranes were washed twice with 20 mM TBS-T and then incubated for 1 h at room temperature with 1:3000 dilution of Tetra-His antibody (Qiagen, Valencia, CA) in blocking buffer. Membranes were washed twice in 20 mM Tris and incubated at room temperature for 1 hr. with 1:3000 dilution of horseradish perioxidase (HRP)-conjugated goat anti-mouse IgA + IgM + IgG antibody (KPL, Gaithersburg, MD) in blocking buffer. The membranes were washed and reacted with enhanced chemiluminescence fluorescence system (GE Healthcare, Piscataway, NJ) for 5 min. Chemiluminiscent signal was acquired with a Typhoon 9400 imager (GE Healthcare, Piscataway, NJ) and analyzed using ImageQuant software. After analysis with pooled convalescent HCoV-OC43 sera, the faint bands seen at N1 and N2 fragments were determined to be non-specific.
2.5. ELISA optimization for recombinant N protein fragments and whole N protein antigens
Recombinant HCoV-OC43 N protein indirect ELISA was developed using a modified version of the recombinant SARS-CoV N protein ELISA as previously described (Haynes et al., 2007). Briefly, flat bottom Immulon ELISA plates (Thermo Scientific, Rochester, NY) were coated with purified recombinant HCoV-OC43 N protein (6.75 ng/well), HCoV-OC43 N1 protein (6.75 ng/well), HCoV-OC43 N2 protein (12.5 ng/well), or HCoV-OC43 N3 protein (25 ng/well) and control antigen (6.75–25 ng/well depending on positive antigen) in sterile phosphate buffered saline (PBS) pH7.4 and incubated overnight at 4 °C. Optimal concentration for each antigen was determined by checkerboard titration (data not shown). Plates were washed three times with PBS containing 0.05% Tween-20 (PBS-T), and incubated with 1:200 dilution of serum in PBS containing 5% skim milk and 0.05% Tween-20 (PBS-T-M) for 1 hr at 37 °C. After incubations, the plates were washed three times in PBS-T and incubated with 1:3000 dilution of HRP-conjugated goat anti-human IgG (H+L, KPL, Gaithersburg, MD) in PBS-T-M for 1 h at 37 °C. After subsequent washing, 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonate) ABTS® peroxidase substrate (KPL, Gaithersburg, MD) was added to each well and incubated at 37 °C for 30 min. The reaction was stopped using ABTS® peroxidase stop solution and read at an absorbance of 405 nm with a 490 nm reference filter. The OD values of the negative control wells were subtracted from the OD values of antigen-coated wells and the average OD values of the antigen-coated wells were calculated. Mean OD values above 0.2 from non-HCoV-OC43 serum samples were considered cross-reactive. ELISA results were confirmed by evaluating a subset of samples by Western blot.
2.6. Statistical analyses
Receiver-operating characteristic (ROC) analysis, area under the curve (AUC), and optimal test cut-offs were determined using Microsoft Excel as described previously (Greiner, 1995, Greiner et al., 1995). Briefly, diagnostic sensitivity (Se) and specificity (Sp) were estimated by comparing ELISA test results with “Gold Standard” results (as specified for each virus above) for HCoV-OC43 positive and negative samples. Diagnostic accuracy was demonstrated with ROC analysis of the ELISA test by plotting Se against 1-Sp, and was summarizes in the AUC value. Optimal test cut-off values were ascertained by plotting the percent sensitivity and specificity against OD405 values, and determining the intersection of the two curves.
3. Results
3.1. Expression and purification of recombinant truncated HCoV-OC43 nucleocapsid proteins
Truncated HCoV-OC43 N proteins were amplified, cloned, and expressed in a pET-28 vector with a C-terminal (His)6-tag, as described in Section 2. After purification, the proteins were analyzed by SDS-PAGE (Fig. 1A) and Western blot for reactivity to anti-His antibody (Fig. 1B). As expected, bands were observed at approximately 55 kDa for HCoV-OC43 N (data not shown), 17 kDa for HCoV-OC43 N1 (aa1–119), 25 kDa for HCoV-OC43 N2 (aa120–332), and 17.5 kDa for HCoV-OC43 N3 (aa333–448). The resulting proteins were used as antigens in subsequent immunoassays.
Fig. 1.
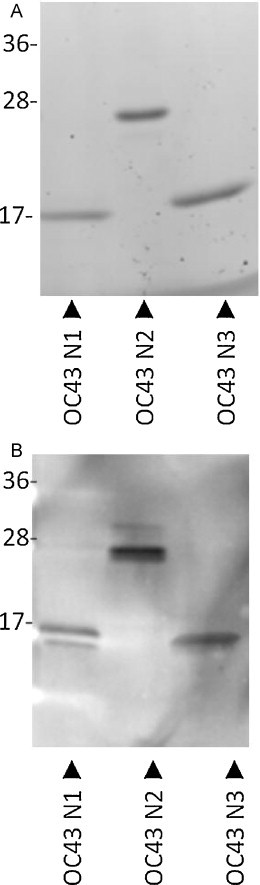
Analysis of the purified recombinant truncated HCoV-OC43 nucleocapsid (N) proteins by SDS-PAGE (A) and Western blot (B). Proteins were separated by SDS-PAGE and analyzed by immunoblotting using anti-His antibody. Lane 1: HCoV-OC43 N1 truncated protein (aa1–119); lane 2: HCoV-OC43 N2 protein (aa120–332); lane 3: HCoV-OC43 N3 protein (aa333–448). Expected band sizes: HCoV-OC43 N1 (aa1–119) 17 kDa, HCoV-OC43 N2 (aa120–332) 25 kDa, and HCoV-OC43 N3 (aa333–448) 17.5 kDa.
The full length N protein of HCoV-OC43 showed between 6.5% and 64.9% homology with other CoV N proteins. HCoV-OC43 showed the highest sequence homology with HCoV-HKU1, another group 2a HCoV. The truncated HCoV-OC43 N proteins showed less sequence homology with other CoVs, with HCoV-OC43 N3 showing the least sequence homology overall. Sequence homology (% amino acid identities) of the truncated HCoV-OC43 proteins with other CoVs is shown in Table 1 .
Table 1.
Amino acid sequence homology between coronavirus nucleocapsid proteins.
| CoVb | HCoV-OC43a |
|||
|---|---|---|---|---|
| N | N1 | N2 | N3 | |
| SARS-CoV | 29.9%c | 39.5% | 45.1% | 19.8% |
| HCov-229E | 24.7% | 18.5% | 38.0% | 7.8% |
| HCov-HKU1 | 64.9% | 81.5% | 73.7% | 48.3% |
| HCoV-NL63 | 6.5% | – | 17.3% | 18.1% |
Human Coronavirus OC43 (GenBank ID: AY585228) full-length Nucleocapsid protein (N), and truncated proteins N1 (aa1–119), N2 (aa120–332), and N3 (aa 333–448).
Sequences for the whole nucleocapsid protein for each coronavirus was compared to sequences for whole or truncated HCoV-OC43 N protein. Nucleocapsid protein accession numbers: SARS-CoV (GenBank ID: AY310120), Human coronavirus 229E (GenBank ID: DQ243939), Human coronavirus HKU1 (GenBank ID: DQ778921), Human coronavirus NL63 (GenBank ID: AY567487).
Values represent percent amino acid identities as determined using National Center for Biotechnical Information (NCBI) Blast.
3.2. Screening HCoV positive sera for immunoreactivity to HCoV-OC43 N protein fragments
To evaluate the antibody response to truncated HCoV-OC43 N proteins, the antigen specific IgG titers of twenty-six acute (n = 15) and convalescent sera (n = 11) from HCoV-OC43 patients were screened for reactivity as shown in Fig. 2 . All three HCoV-OC43 N proteins were detected by antibodies in convalescent-phase serum specimens with an average OD greater than 0.3; however, detection by acute-phase sera was limited (Fig. 2A). The predominant HCoV-OC43 antibody response was against HCoV-OC43 N3 protein (11 out of 11) followed by responses to HCoV-OC43 N2 (4 out of 11) and HCoV-OC43 N1 (3 out of 11). An additional 47 serum samples from convalescent SARS-CoV (n = 20) and acute and convalescent (n = 27) HCoV-229E–infected patients were also screened by ELISA to determine the reactivity to the HCoV-OC43 antigens (Fig. 2A). Two serum samples, one acute and one convalescent, from different HCoV-229E patients showed reactivity to HCoV-OC43 N3 antigen (7.4%); while the convalescent sera from six SARS patients (30%) showed reactivity to the HCoV-OC43 N3 protein fragment. In contrast to HCoV-OC43 N3 fragment, 20 out of 27 (74%) serum samples from HCoV-229E patients and all SARS-CoV serum samples (100%) showed reactivity to the whole HCoV-OC43 N protein (Fig. 2B). The level of absorbance in convalescent-phase sera from HCoV-229E and SARS–CoV infected persons against HCoV-OC43 N3 antigen was 88–90% lower than that against the HCoV-OC43 whole N antigen suggesting less cross-reactivity of the non-HCoV-OC43 sera to the truncated HCoV-OC43 N3 ELISA (Fig. 2B). Due to limited availability, serum samples from confirmed HCoV-HKU1 (group 2a) or HCoV-NL63 (group 1b) – infected patients were not evaluated. However, hyperimmune sera from mice immunized against HCoV-HKU1 N protein or HCoV-NL63 N protein were tested and showed no reactivity to HCoV-OC43 N3 protein and minimal reactivity to whole HCoV-OC43 N protein (data not shown). Based on the strong reactivity of HCoV-OC43 sera to the HCoV-OC43 N3 protein, with minimal cross-reactivity to other CoV sera, this antigen was chosen for further evaluation.
Fig. 2.
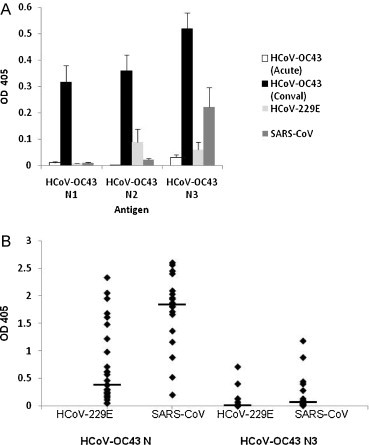
Specificity and cross-reactivity of patient sera samples for truncated human coronavirus OC43 (HCoV-OC43) N protein antigen candidates. (A) Sera samples acute HCoV-OC43 (n = 15), convalescent OC43 (n = 11), HCoV-229E (n = 27), and SARS-CoV serum samples (n = 20) were screened for reactivity to HCoV-OC43 N1, N2 and N3 truncated proteins. Error bars represent the standard error of the mean. (B) Sera from HCoV-229E patients (n = 27), and SARS-CoV patients (n = 18), were screened for cross-reactivity against HCoV-OC43 whole nucleocapsid and HCoV-OC43 N3 proteins by ELISA. Specimens were run in duplicates and the optical densities at 405 nm (OD405) with a 490 nm reference filter for each samples are shown. The line represents the median OD values for each sample set.
3.3. Determination of assay cut-off values for positive test results
A panel of known HCoV-OC43 positive and negative samples was screened and Receiver Operating Characteristic (ROC) analysis was utilized to determine the assay cut-off. ROC analysis allows for data that is not normally distributed and for evaluation of sensitivity and specificity to be determined separately. The null hypothesis of ROC analysis is that the testing method is no better than flipping a coin, as would be demonstrated by a diagonal line on the ROC curve and an area under the curve (AUC) equal to 0.5 while a perfect test would have an AUC of 1. Given the limited detection of the HCoV-OC43 acute sera by HCoV-OC43 N3 protein (Fig. 2A), ROC analysis was completed based on the results of screening 11 HCoV-OC43 positive convalescent and 47 negative serum specimens (Fig. 3 ). AUC value for HCoV-OC43 N3 was 0.894 (data not shown). Optimal cut-off values for positive test results were determined by plotting percent sensitivity and specificity against OD cut-off values (Fig. 3A). The cut-off value was set at 0.161 (OD405 value at intersectional point in Fig. 3A) with a corresponding sensitivity of 90.1% and specificity of 82.9%. With this cut-off value, 10 out of 11 HCoV-OC43 convalescent samples had positive test results with the remaining sample producing a borderline result. The sensitivity of the HCoV-OC43 N3 protein ELISA was comparable to that of the whole HCoV-OC43 N protein ELISA (Fig. 3B). While the overall mean OD values for both acute and convalescent specimens were higher using the whole HCoV-OC43 N protein ELISA, the HCoV OC43 N3 ELISA had a better ability to detect rise in antibody titer between acute and convalescent specimens than the whole HCoV-OC43 N protein ELISA. Using the established cut-off value, the 47 HCoV-229E (n = 27) and SARS (n = 20) serum samples were re-screened by HCoV-OC43 N3 ELISA. One acute and one convalescent serum sample from two HCoV-229E patients and convalescent sera from six SARS patients showed reactivity to HCoV-OC43 N3 protein for 82.9% specificity (Fig. 4A). The overall assay performance is summarized in Table 2 .
Fig. 3.
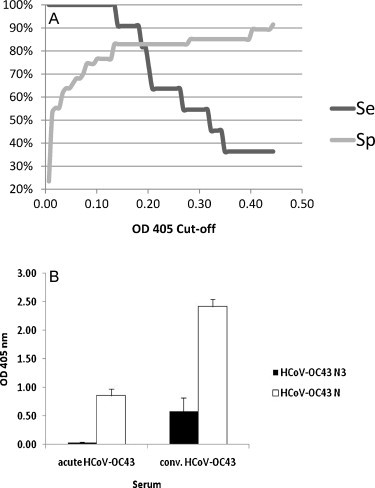
Cut-off analysis for Human coronavirus OC43 (HCoV-OC43) N3 based ELISA and comparison with HCoV-OC43 whole N based ELISA. (A) The association between sensitivity (Se) and specificity (Sp) and OD value cut-offs for the HCoV-OC43 N3 based ELISA was determined (B) Comparison of average optical densities (OD405) for acute (n = 15) and convalescent (n = 11) HCoV-OC43 samples against HCoV-OC43 N3 and whole HCoV-OC43 N based ELISAs.
Fig. 4.
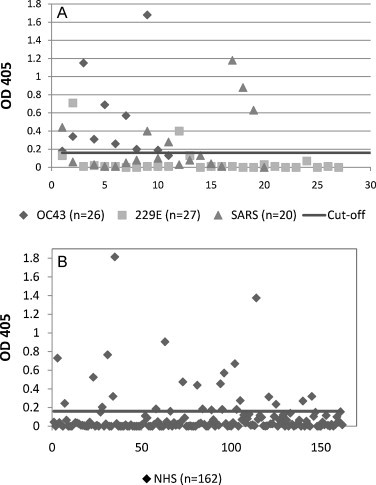
Scatter chart of absorbance values with representative sera from (A) HCoV-OC43 (OC43), HCoV-229E (229E), SARS patients and (B) healthy U.S. blood donors (NHS) for IgG antibodies tested by HCoV-OC43 N3 protein based ELISA. Results are plotted as optical density values at 405 nm with a 490-nm reference filter. The black line indicates the cutoff value of 0.161 as determined by ROC analysis.
Table 2.
Summary of human coronavirus OC43 N3 based ELISA test results.
| Sample type | % Sensitivity (pos/total) | % Specificity (neg/total) | % Seropositive |
|---|---|---|---|
| HCoV-OC43a | 90.1% (10/11) | – | – |
| Non HCoV-OC43 respiratory virusesb | – | 82.9% (39/47) | – |
| Healthy donorsc | – | – | 18.5% (30/162) |
Human coronavirus OC43 (HCoV-OC43) positive convalescent human patient samples (n = 11). Data is based on the cut-off determined in the ROC analysis.
Non-HCoV-OC43 respiratory samples: human samples positive for human coronavirus 229E (n = 27) and SARS coronavirus (n = 20).
Healthy adult donor samples (n = 162).
3.4. Estimation of HCov-OC43 seroprevalence from a panel of healthy adults
A panel of sera collected from 162 healthy adult donors was evaluated to determine HCoV-OC43 seroprevalance (Fig. 4B). Sera from 25 adult donors were positive for the presence HCoV-OC43 N3 antibodies. Results that fell within the 95% confidence interval of the cut-off (0.13 ≤ OD < 0.16) value were considered borderline results (n = 5). Borderline positive samples were confirmed positive by Western blot analysis (data not shown). The resulting seroprevalence for HCoV-OC43 infection was 18.5% (30/162).
3.5. Assay utility in surveillance outbreak setting
To evaluate the surveillance performance of the assay, nineteen acute and convalescent serum specimens collected from 12 patients during an outbreak of HCoV-OC43 infection in a long-term care facility located in Wisconsin were screened. The infection status of the patients was previously determined by RT-PCR and/or serological diagnostic analysis. The N3 assay detected 6 out of 8 (75%) acute and 8 out of 9 (88.9%) convalescent specimens as positive (Fig. 5 ). Two additional convalescent serum specimens from two patients were negative (data not shown). Consistent with previous diagnostic testing, 10 out of the 12 patients were considered seropositive for HCoV-OC43 and the two remaining patients were seronegative as determined by this ELISA.
Fig. 5.
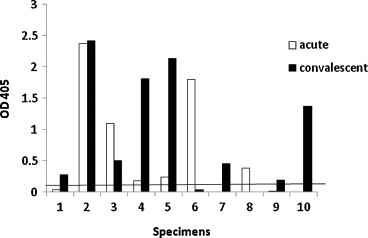
Graph of absorbance values with acute (n = 8) and convalescent (n = 9) specimens from 10 patients collected during an HCoV-OC43 outbreak for IgG antibodies to recombinant HCoV-OC43 N3 protein. Results are plotted as optical density values at 405 nm with a 490-nm reference filter. The black line indicates the cut-off value of 0.161.
4. Discussion
The CoV nucleocapsid protein has been used as antigen for the serological diagnosis of infection, but conservation in sequence homology among various CoV N proteins has led to the need for more specific antigens to detect antibodies against a specific HCoV. Included in the CoV N protein, is a highly conserved region (FYYLGTGP; aa125–132 of HCoV-OC43) that occurs in the N-terminal portion (Rota et al., 2003) which, if immunogenic, would induce cross-reactivity antibodies against other coronaviruses (Chan et al., 2009, Sun and Meng, 2004, Woo et al., 2004, Patrick et al., 2006). One approach to reduce antigenic cross-reactivity was to divide the HCoV-OC43 N protein into three regions and evaluate the resultant truncated proteins with serum specimens from HCoV-OC43-infected, HCoV-229E-infected, or SARS CoV-infected persons in these antibody assays. As expected, both HCoV-229E and SARS-CoV positive sera showed reactivity to the N2 protein but no reactivity to the N1 protein. The HCoV-OC43 positive sera showed comparable reactivity to both N1 and N2 fragments. Serum samples positive for other CoV infections (i.e. SARS and HCoV-229E) showed low to moderate reactivity and convalescent-phase HCoV-OC43 sera showed high reactivity to the N3 protein. Acute phase HCoV-OC43 sera showed minimal reactivity to all truncated N proteins consistent with their origin from individuals susceptible to infection. The lack of reactivity against these acute-phase specimens also demonstrates that assays based on these proteins are more specific than those based on full length N (Fig. 4B). Of the three truncated proteins, the HCoV-OC43 N3 fragment had the lowest sequence homology with other coronavirus nucleocapsid proteins and was associated with the highest signal in convalescent HCoV-OC43 sera. It was chosen as the antigen for the additional evaluation.
The ROC analysis provided a means to determine assay cut-off value in this population in whom prior infection status and expected antibody positivity was unknown. With this cutoff value, the HCoV-OC43 N3 based ELISA exhibited a high diagnostic accuracy. Although the sample size of HCoV-OC43 specimens limited the power of the cut-off calculations, the HCoV-OC43 N3 based assay demonstrated a strong ability to confirm positive results with 10 out of 11 convalescent HCoV-OC43 specimens confirmed, and the remaining specimen having a borderline positive result. Expansion of the HCoV serum panel to include other group 2a samples and increasing the overall sample number could improve the diagnostic accuracy and refine the cut-off values for positive and borderline test results.
Using the truncated HCoV-OC43 N3 ELISA, a seroprevalence of 18.5% in 162 healthy adults for HCoV-OC43 infection was demonstrated and is consistent with early epidemiologic surveys showing that HCoV-OC43 is responsible for 15–30% of upper respiratory tract infections (McIntosh et al., 1974, Wege et al., 1982, Larson et al., 1980). While consistent with early seroepidemiologic studies, our seroprevalence rates are lower than recently reported rates for HCoV-OC43 of 88–100% using recombinant whole nucleocapsid-based IgG ELISA, IFA and Western blot assays (Mourez et al., 2007, Severance et al., 2008). In those studies the use of whole HCoV-OC43 N protein may have given too high a rate of past infection because it detected antibodies induced by other HCoVs that cross-react with the N protein. In this study, the HCoV-OC43 N3 ELISA detected antibodies in two serum samples from HCoV-229E patients and sera from six SARS patients. These antibodies could represent prior HCoV-OC43 infection or antibodies that induced by these viruses that cross react with HCoV-OC43. Due to the limited availability of human sera collected after confirmation of HCoV-HKU1 and HCoV-NL63 infection, polyclonal sera from mice immunized with HCoV-HKU1 or HCoV-NL63 N proteins were screened and did not show any reactivity to HCoV-OC43 N3 protein further demonstrating the specificity of the assay.
Respiratory outbreaks associated with HCoV-OC43 have been reported and highlight the potential virulence of HCoV-OC43 infections in vulnerable populations, such as the elderly in care facilities and hospitalized individuals with underlying chronic conditions (Patrick et al., 2006, Birch et al., 2005, Falsey et al., 2002, El-Sahly et al., 2000, Glezen et al., 2000). This study suggests that the HCoV-OC43 N3 assay provides a serologic tool to assist in investigations of HCoV outbreaks and surveillance studies.
Acknowledgements
This research was also supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy and CDC. The authors wish to thank Dr. Ann Falsey (Univ. of Rochester School of Medicine, Rochester, NY) for providing sera from patients with HCoV-229E and HCoV-OC43 infections. Thanks also to Dr. Suxiang Tong (CDC) for technical assistance with HCoV fragments and Dr. Dean Erdman (CDC) for the HCoV-OC43 virus stock.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Contributor Information
Elisabeth G. Blanchard, Email: HZQ0@cdc.gov.
Congrong Miao, Email: bdu7@cdc.gov.
Thomas E. Haupt, Email: Thomas.Haupt@dhs.wisconsin.gov.
Larry J. Anderson, Email: larry.anderson@emory.edu.
Lia M. Haynes, Email: loh5@cdc.gov.
References
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch C.J., Clothier H.J., Seccull A., Tran T., Catton M.C., Lambert S.B., Druce J.D. Human coronavirus OC43 causes influenza-like illness in residents and staff of aged-card facilities in Melbourne, Australia. Epidemiol. Infect. 2005;133:273–277. doi: 10.1017/s0950268804003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tse H., Wong S.S.Y., Woo P.C.Y., Lau S.K.P., Chen L., Zheng B.J., Huang J.D., Yuen K.Y. Examination of seroprevalence of coronavirus HKU1 infection with S protein-based ELISA and neutralization assay against viral spike pseudotyped virus. J. Clin. Virol. 2009;45:54–60. doi: 10.1016/j.jcv.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B. Spectrum of clinical illness in hospitalized patients with ‘common cold’ virus infections. Clin. Infect. Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Walsh E.E., Hayden F.G. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J. Infect. Dis. 2002;185:1338–1341. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezen W.P., Greenberg S.B., Atmar R.L., Piedra P.A., Couch R.B. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA. 2000;283:499–505. doi: 10.1001/jama.283.4.499. [DOI] [PubMed] [Google Scholar]
- Greiner M. Two-graph receiver operating characteristic (TG-ROC): a Microsoft-EXCEL template for the selection of cut-off values in diagnostic tests. J. Immunol. Methods. 1995;185:145–146. doi: 10.1016/0022-1759(95)00078-o. [DOI] [PubMed] [Google Scholar]
- Greiner M., Sohr D., Göbel P. A modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J. Immunol. Methods. 1995;185:123–132. doi: 10.1016/0022-1759(95)00121-p. [DOI] [PubMed] [Google Scholar]
- Hamre D., Kindig D.A., Mann J. Growth and intracellular development of a new respiratory virus. J. Virol. 1967;1:810–816. doi: 10.1128/jvi.1.4.810-816.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes L.M., Miao C., Harcourt J.L., Montgomery J.M., Le M.Q., Dryga S.A., Kamrud K.I., Rivers B., Babcock G.J., Oliver J.B., Comer J.A., Reynolds M., Uyeki T.M., Bausch D., Ksiazek T., Thomas W., Alterson H., Smith J., Ambrosino D.M., Anderson L.J. Recombinant protein-based assays for detection of antibodies to severe acute respiratory syndrome coronavirus spike and nucleocapsid proteins. Clin. Vaccine Immunol. 2007;14:331–333. doi: 10.1128/CVI.00351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson H.E., Reed S.E., Tyrell D.A. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J. Med. Virol. 1980;5:221–229. doi: 10.1002/jmv.1890050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann C., Wolf H., Xu J., Zhao Q., Shao Y., Motz M., Lindner P. A line immunoassay utilizing recombinant nucleocapsid proteins for detection of antibodies to human coronaviruses. Diagn. Microbiol. Infect. Dis. 2008;61:40–48. doi: 10.1016/j.diagmicrobio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Chao R.K., Krause H.E., Wasil R., Mocega H.E., Mufson M.A. Coronavirus infection in acute lower respiratory tract disease of infants. J. Infect. Dis. 1974;130:502–507. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourez T., Vabret A., Han Y., Dina J., Legrand L., Corbet S., Freymuth F. Baculovirus expression of HCoV-OC43 nucleocapsid protein and development of a Western blot assay for detection of human antibodies against HCoV-OC43. J. Virol. Methods. 2007;139:175–180. doi: 10.1016/j.jviromet.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J. Infect. Dis. Med. Microbiol. 2006;17:330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance E.G., Bossis I., Dickerson F.B., Stallings C.R., Origoni A.E., Sullens A., Yolken R.H., Viscidi R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. 2008;15:1805–1810. doi: 10.1128/CVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.F., Meng X.J. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group 1 animal coronaviruses:implication for SARS diagnosis. J. Clin. Microbiol. 2004;42:2351–2352. doi: 10.1128/JCM.42.5.2351-2352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Peñaranda S., Bankamp B., Maher K., Chen M.-h., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Siddell S., ter Meulen V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.-m., Chan K.-h., Tsoi H.-w., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.-k., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K.-y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with Pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Chan K.H., Hui W.T., Kwan G.S., Peiris J.S., Couch R.B., Yuen K.Y. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E recitified by Western blotting with recombinant SARS-CoV spike polypeptide. J. Clin. Microbiol. 2004;42:5885–5888. doi: 10.1128/JCM.42.12.5885-5888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


