Abstract
Human coronavirus NL63 was identified in 2004 in the Netherlands. Due to the high prevalence and world-wide distribution of this pathogen, it is essential to develop a sensitive and specific detection assay suitable for use in a routine diagnostic laboratory. Techniques based on PCR or real-time PCR are laborious and expensive. Detailed analysis of the HCoV-NL63 genome permitted the identification of a conserved nucleic acid sequential motif, which was sufficient for the design of a loop-mediated isothermal amplification (LAMP) assay. Evaluation of the method showed that the test is specific to HCoV-NL63 and that it does not cross-react with other respiratory viruses. The detection limit was found to be 1 copy of RNA template per reaction in cell culture supernatants and clinical specimens.
Keywords: Coronavirus, HCoV-NL63, LAMP, Loop-mediated isothermal amplification
Human coronavirus NL63 (HCoV-NL63) is a human respiratory pathogen with high worldwide prevalence (Pyrc et al., 2004, Pyrc et al., 2006, Pyrc et al., 2007a, Pyrc et al., 2007b, van der Hoek et al., 2004, van der Hoek et al., 2005). The virus belongs to the Coronaviridae family, has a positive single-stranded RNA genome of >27 kb and a typical corona-like appearance, and employs a characteristic discontinuous replication strategy. Infection with HCoV-NL63 is associated with upper and lower respiratory tract illnesses, which occur commonly during the winter season with more severe symptoms in children, elderly, and immunocompromised individuals (Arden et al., 2005, Bastien et al., 2005, Choi et al., 2006, Ebihara et al., 2005, Esper et al., 2010, Gerna et al., 2006, Kaiser et al., 2005, Moes et al., 2005, Pyrc et al., 2007b, Vabret et al., 2005, van der Hoek et al., 2005). The virus is believed to be the most important pathogen for the development of croup in young children and one of the most relevant human coronaviruses (Dijkman et al., 2008, van der Hoek et al., 2004, van der Hoek et al., 2005, Fielding, 2011). Diagnosis of HCoV-NL63 is performed routinely with RT-PCR or real-time PCR techniques, which, although relatively laborious and cost-ineffective are specific and sensitive (Arden et al., 2005, Bastien et al., 2005, Choi et al., 2006, Ebihara et al., 2005, Esper et al., 2010, Gerna et al., 2006, Kaiser et al., 2005, Moes et al., 2005, Pyrc et al., 2007b, Vabret et al., 2005, van der Hoek et al., 2005).
The technique of loop-mediated isothermal amplification (LAMP) is based on the principle of the strand displacement reaction, which occurs under isothermal conditions with the generation of cauliflower-like DNA structures. As the target is recognized by six distinct primers, amplification of a target sequence by the LAMP method is highly specific (Notomi et al., 2000, Nagamine et al., 2002). When the LAMP reaction is combined with reverse transcription, RNA-to-gel electrophoresis takes 60 min. The goal of this study was to develop and evaluate one-step LAMP assays specific for HCoV-NL63.
LAMP primers specific for HCoV-NL63 were designed with Primer Explorer V4 software (http://primerexplorer.jp/e/) based on a conserved fragment of the nucleocapsid gene. The primers, including outer primers (F3 and B3), inner primers (FIP and BIP) and loop primers (LF and LB), are shown in Table 1 . In the design process, the complete genome sequences of fifteen different HCoV-NL63 isolates were used as templates (the GenBank accession numbers: NC_005831, AY567487, DQ445912, DQ445911, DQ462769, DQ462763, DQ462768, DQ462764, DQ462765, DQ462766, DQ462767, EF081296, DQ846901, AY563108 and AY563107), and homology was analyzed using ClustalX 2.0 (http://www.clustal.org/) and Bioedit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html).
Table 1.
Primer sets used for detection of HCoV-NL63 by the LAMP method. Nucleocapsid was chosen as the target gene.
| Primer type | Sequence | |
|---|---|---|
| 1 | F3 | TTTGGCTTTAAAGAACTTAGGT |
| 2 | B3 | ACCATTCTGAACAAGATCTGA |
| 3 | FIP | GGTTGAGAAAGAGGCTTATTAGGTTTTTGATAACCAGTCGAAGTCA |
| 4 | BIP | TCGTTGGAAGCGTGTTCCTATGTGATTAAAATCACGAGGAC |
| 5 | LF | TCTTAGGAGTGGAAGTACCAGAAG |
| 6 | LB | CAGAGAGGAAAATGTTATTCAGTGC |
An HCoV-NL63 (Amsterdam I strain) cell culture supernatant generated from infected LLC-MK2 cells was used as input material. LLC-MK2 cells were maintained in minimal essential medium (MEM), containing 2 parts Hank's MEM and 1 part Earle's MEM (PAA Laboratories, Pasching, Austria) supplemented with 3% heat-inactivated fetal bovine serum (PAA Laboratories, Pasching, Austria), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were cultured in 6-well plates (Sarstedt, Nümbrecht, Germany) at 37 °C with 5% CO2 and were infected with HCoV-NL63 at TCID50 of 400. Following infection, cells were incubated for 6 subsequent days at 32 °C with 5% CO2. The cells were then lysed by two freeze–thawing cycles and the virus-containing fluid was aliquoted and stored at −80 °C. A control LLC-MK2 cell lysate from mock infected cells was prepared in the same manner as the virus stocks. Total nucleic acid was isolated using RNA mini kit (A&A Biotechnology, Gdynia, Poland) and was quantified by real-time PCR as described previously (Pyrc et al., 2010).
LAMP assay conditions were optimized, and the following conditions were tested: temperature (54–64 °C), magnesium concentration (2–12 mM), betaine concentration (0.6–1.6 M), outer/inner primers ratio (1/2–1:10), outer/loop primers ratio (1/2–1:10) and Bsm polymerase concentration (1–4 U/reaction). Eventually, the LAMP reaction was carried out in a volume of 10 μL containing 1× Bsm buffer (Fermentas, Vilnius, Lithuania), 4.0 mM MgSO4, 0.8 M betaine (Sigma–Aldrich, St. Louis, USA), 1.2 mM dNTPs, 0.2 μM each of outer primers, 1.6 μM each of inner primers, 0.4 μM each of loop primers, 1 U maxima reverse transcriptase (Fermentas, Vilnius, Lithuania) and 1 U of Bsm polymerase (Fermentas, Vilnius, Lithuania) with 1 μL total RNA as template. After quantitation, viral RNA was serially diluted to obtain 105–100 copies/reaction. The amplification was performed at 60 °C in a laboratory water bath or thermocycler for 1 h. Analysis was conducted by agarose gel electrophoresis. A ladder-like pattern was detected in positive samples while no signal was detected in negative controls. As shown in Fig. 1 , careful optimization of the reaction allowed the detection of as little as 1 copy per reaction. The identity of the amplified product was confirmed by sequencing.
Fig. 1.
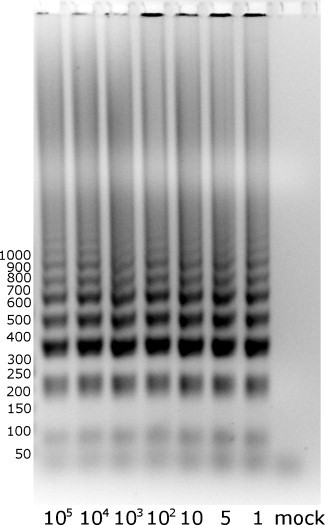
Sensitivity of the HCoV-NL63 LAMP assay. The assay was performed with serial dilutions of viral RNA (105, 104, 103, 102, 10, 5, and 1 copies per reaction). Mock: negative control from mock-infected LLC-MK2 cells.
The time required to complete the reaction was assessed using the SYBR Green reagent (A&A Biotechnology, Gdynia, Poland) and appeared to be dependent on initial virus yield. The time varied from 20 to 40 min. Changes in the fluorescence signal were monitored using the ABI 7500 fast real-time PCR apparatus. RNA quantification was possible with qLAMP, although only within a limited range of concentrations, and was inferior to that described previously for real-time PCR (Pyrc et al., 2010).
A comparison of sensitivity of the LAMP assay with that of previously described PCR and real-time PCR based methods showed that the new assay provides similar to better sensitivity, although the procedure is less laborious and more cost-effective. For instance, the cost of the complete reaction is about 7 times lower than for the standard real-time PCR assay.
To demonstrate the specificity of the method, and to exclude the cross-reaction of primers with other respiratory tract pathogens, samples containing high levels of seven different human respiratory viruses were tested, including two other members of the Coronaviridae family (HCoV-229E and HCoV-HKU1), and five RNA and DNA viruses that belong to other families, including respiratory syncytial virus, echovirus 9, human metapneumovirus, influenza A virus and adenovirus type 1. Stock samples containing respiratory syncytial virus, influenza A virus, human echovirus 9, human rhinovirus, human parainfluenza 3 virus and human adenovirus were kindly provided by Marcel Muller; human coronavirus 229E and OC43 were a kind gift from Lia van der Hoek; human metapneumovirus was kindly provided by Oliver Schildgen. The results clearly indicate that the reaction is highly specific for HCoV-NL63 with no cross-reactivity being observed for other viruses (Fig. 2 ).
Fig. 2.
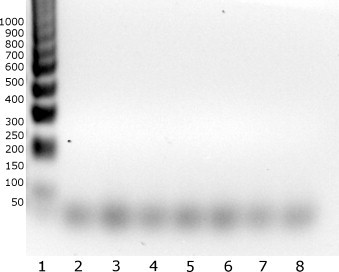
Cross-reactivity of the HCoV-NL63 LAMP assay. 1: HCoV-NL63; 2: HCoV-229E; 3: HCoV-HKU1; 4: respiratory syncytial virus; 5: echovirus 9; 6: human metapneumovirus; 7: influenza A virus; 8: adenovirus type 1.
As the method is intended for use on clinical material, its performance with various types of clinical materials was evaluated, including nose wash, bronchioalveolar lavage, sputum, and human sera. Briefly, 0.5 μL of culture supernatant from infected LLC-MK2 cells was inoculated into 10 μL of clinical sample that was negative for coronaviruses. Reactions were performed according to the protocol described above and the resulting products were analyzed on 0.8% agarose gel. No inhibition of the reaction was observed (Fig. 3 ).
Fig. 3.
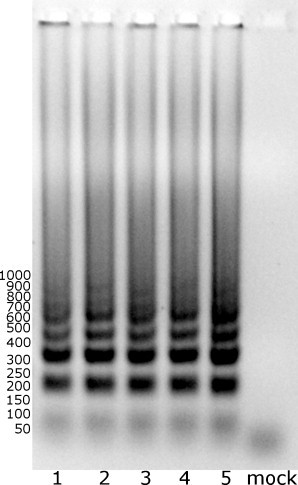
Detection of HCoV-NL63 in clinical specimens with the LAMP assay. 1: Cell culture supernatant; 2: bronchoalveolar lavage; 3: nose wash; 4: sputum; 5: human sera; Mock: negative control from mock-infected LLC-MK2 cells.
The LAMP method has been used to detect a number of pathogens, including RNA and DNA viruses, bacteria, and fungi and has the potential to be used as a simple test for the rapid laboratory confirmation of the occurrence of infectious diseases in resource-limited settings (Cardoso et al., 2010, Chen et al., 2008, Chen et al., 2010a, Chen et al., 2010b, Hong et al., 2004, Poon et al., 2005). Human coronavirus NL63 has been reported to be present worldwide, affecting 1–10% of patients with respiratory diseases. In healthy individuals, HCoV-NL63 infections present relatively mild symptoms, including moderate fever, cough, sore throat, and rhinitis (Bastien et al., 2005). On the other hand, several reports have shown that this virus can cause severe disease in children, elderly and immunocompromised individuals with sometimes a fatal outcome (Bastien et al., 2005, Fouchier et al., 2004, Oosterhof et al., 2010, van der Hoek et al., 2004). HCoV-NL63 infections have been linked to the development of croup in children of less than 3 years of age and HCoV-NL63 appears to be a major player in the development of this condition (Sung et al., 2010, van der Hoek et al., 2005, van der Hoek et al., 2006). HCoV-NL63 has also been associated with Kawasaki disease, which is one of the most common forms of childhood vasculitis, although the results of other multiple studies contradict this link (Baker et al., 2006, Chang et al., 2006, Dominguez et al., 2006, Esper et al., 2005, Lehmann et al., 2009, Shimizu et al., 2005).
Due to the high prevalence of HCoV-NL63 and its association with disease in humans, it is essential to include the virus in the diagnostic panel for respiratory pathogens. In this study, a method for HCoV-NL63 detection in cell cultures and clinical specimens was developed. Furthermore, optimization of the temperature range has shown that the method is equally effective in the temperature range of 56–62 °C (data not shown), indicating that it can be performed using a low quality water bath. In summary, a specific, sensitive, cost- and time-efficient method for HCoV-NL63 detection that allows high-throughput screening was developed.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
This work was in part supported by Foundation for Polish Science within the Homing Programme (KP), grant from the Ministry of Scientific Research, Poland (0095/B/P01/2009/37) (KP), Iuventus Plus grant from the Ministry of Science and Higher Education, Poland (IP 2010 033870) (KP), the Jagiellonian University statutory funds DS/9/WBBiB (JP and KP), and grants from the Department of Scientific Research, Polish Ministry of Science and Education (1642/B/P01/2008/35) (JP) and the Foundation for Polish Science (TEAM project DPS/424-329/10) (JP). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of the structural funds from the European Union (grant no.: POIG.02.01.00-12-064/08 – “Molecular Biotechnology for Health”). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Arden K.E., Nissen M.D., Sloots T.P., Mackay I.M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 2005;75:455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.C., Shimizu C., Shike H., Garcia F., van der Hoek L., Kuijper T.W., Reed S.L., Rowley A.H., Shulman S.T., Talbot H.K., Williams J.V., Burns J.C. Human coronavirus-NL63 infection is not associated with acute Kawasaki disease. Adv. Exp. Med. Biol. 2006;581:523–526. doi: 10.1007/978-0-387-33012-9_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N., Anderson K., Hart L., Van Caeseele P., Brandt K., Milley D., Hatchette T., Weiss E.C., Li Y. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 2005;191:503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso T.C., Ferrari H.F., Bregano L.C., Silva-Frade C., Rosa A.C., Andrade A.L. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Mol. Cell. Probes. 2010 doi: 10.1016/j.mcp.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.Y., Chiang B.L., Kao C.L., Wu M.H., Chen P.J., Berkhout B., Yang H.C., Huang L.M. Lack of association between infection with a novel human coronavirus (HCoV), HCoV-NH, and Kawasaki disease in Taiwan. J. Infect. Dis. 2006;193:283–286. doi: 10.1086/498875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.T., Zhang J., Ma Y.P., Ma L.N., Ding Y.Z., Liu X.T., Cai X.P., Ma L.Q., Zhang Y.G., Liu Y.S. Reverse transcription loop-mediated isothermal amplification for the rapid detection of infectious bronchitis virus in infected chicken tissues. Mol. Cell. Probes. 2010;24:104–106. doi: 10.1016/j.mcp.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.T., Zhang J., Sun D.H., Ma L.N., Liu X.T., Quan K., Liu Y.S. Reverse transcription loop-mediated isothermal amplification for the detection of highly pathogenic porcine reproductive and respiratory syndrome virus. J. Virol. Methods. 2008;153:266–268. doi: 10.1016/j.jviromet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li J., Fang X.E., Xiong W. Detection of swine transmissible gastroenteritis coronavirus using loop-mediated isothermal amplification. Virol. J. 2010;7:206. doi: 10.1186/1743-422X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.H., Lee H.J., Kim S.J., Eun B.W., Kim N.H., Lee J.A., Lee J.H., Song E.K., Kim S.H., Park J.Y., Sung J.Y. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin. Infect. Dis. 2006;43:585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., El Idrissi N.B., Pyrc K., Muller M.A., Kuijpers T.W., Zaaijer H.L., van der Hoek L. Human coronavirus NL63 and 229E seroconversion in children. J. Clin. Microbiol. 2008;46:2368–2373. doi: 10.1128/JCM.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez S.R., Anderson M.S., Glode M.P., Robinson C.C., Holmes K.V. Blinded case–control study of the relationship between human coronavirus NL63 and Kawasaki syndrome. J. Infect. Dis. 2006;194:1697–1701. doi: 10.1086/509509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T., Endo R., Ma X., Ishiguro N., Kikuta H. Detection of human coronavirus NL63 in young children with bronchiolitis. J. Med. Virol. 2005;75:463–465. doi: 10.1002/jmv.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Ou Z., Huang Y.T. Human coronaviruses are uncommon in patients with gastrointestinal illness. J. Clin. Virol. 2010;48:131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Shapiro E.D., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Association between a novel human coronavirus and Kawasaki disease. J. Infect. Dis. 2005;191:499–502. doi: 10.1086/428291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C. Human coronavirus NL63: a clinically important virus? Future Microbiol. 2011;6:153–159. doi: 10.2217/fmb.10.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A., Baldanti F. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J. Med. Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser L., Regamey N., Roiha H., Deffernez C., Frey U. Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr. Infect. Dis. J. 2005;24:1015–1017. doi: 10.1097/01.inf.0000183773.80217.12. [DOI] [PubMed] [Google Scholar]
- Lehmann C., Klar R., Lindner J., Lindner P., Wolf H., Gerling S. Kawasaki disease lacks association with human coronavirus NL63 and human bocavirus. Pediatr. Infect. Dis. J. 2009;28:553–554. doi: 10.1097/inf.0b013e31819f41b6. [DOI] [PubMed] [Google Scholar]
- Moes E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P., Pyrc K., Berkhout B., van der Hoek L., Van Ranst M. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof L., Christensen C.B., Sengelov H. Fatal lower respiratory tract disease with human corona virus NL63 in an adult haematopoietic cell transplant recipient. Bone Marrow Transplant. 2010;45:1115–1116. doi: 10.1038/bmt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Leung C.S., Chan K.H., Lee J.H., Yuen K.Y., Guan Y., Peiris J.S. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Berkhout B., van der Hoek L. Identification of new human coronaviruses. Expert. Rev. Anti Infect. Ther. 2007;5:245–253. doi: 10.1586/14787210.5.2.245. [DOI] [PubMed] [Google Scholar]
- Pyrc K., Berkhout B., van der Hoek L. The novel human coronaviruses NL63 and HKU1. J. Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Dijkman R., Deng L., Jebbink M.F., Ross H.A., Berkhout B., van der Hoek L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Sims A.C., Dijkman R., Jebbink M., Long C., Deming D., Donaldson E., Vabret A., Baric R., van der Hoek L., Pickles R. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010;84:11255–11263. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C., Shike H., Baker S.C., Garcia F., van der Hoek L., Kuijpers T.W., Reed S.L., Rowley A.H., Shulman S.T., Talbot H.K., Williams J.V., Burns J.C. Human coronavirus NL63 is not detected in the respiratory tracts of children with acute Kawasaki disease. J. Infect. Dis. 2005;192:1767–1771. doi: 10.1086/497170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J.Y., Lee H.J., Eun B.W., Kim S.H., Lee S.Y., Lee J.Y., Park K.U., Choi E.H. Role of human coronavirus NL63 in hospitalized children with croup. Pediatr. Infect. Dis. J. 2010;29:822–826. doi: 10.1097/INF.0b013e3181e7c18d. [DOI] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Dina J., van der Hoek L., Gouarin S., Petitjean J., Brouard J., Freymuth F. Human coronavirus NL63, France. Emerg. Infect. Dis. 2005;11:1225–1229. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Sure K., Ihorst G., Stang A., Pyrc K., Jebbink M.F., Petersen G., Forster J., Berkhout B., Uberla K. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Sure K., Ihorst G., Stang A., Pyrc K., Jebbink M.F., Petersen G., Forster J., Berkhout B., Uberla K. Human coronavirus NL63 infection is associated with croup. Adv. Exp. Med. Biol. 2006;581:485–491. doi: 10.1007/978-0-387-33012-9_86. [DOI] [PMC free article] [PubMed] [Google Scholar]


